Abstract
Naive pluripotency exists in epiblast cells of mouse pre-implantation embryos. However, whether the naive pluripotency is transient or nonexistent in primate embryos remains unclear. Using RNA-seq in single blastomeres from 16-cell embryos through to hatched blastocysts of rhesus monkey, we constructed the lineage segregation roadmap in which the specification of trophectoderm, epiblast, and primitive endoderm is initiated simultaneously at the early blastocyst stage. Importantly, we uncovered the existence of distinct pluripotent states in monkey pre-implantation embryos. At the early- and middle-blastocyst stages, the epiblast cells have the transcriptome features of naive pluripotency, whereas they display a continuum of primed pluripotency characteristics at the late and hatched blastocyst stages. Moreover, we identified potential regulators that might play roles in the transition from naive to primed pluripotency. Thus, our study suggests the transient existence of naive pluripotency in primates and proposes an ideal time window for derivation of primate embryonic stem cells with naive pluripotency.
The development of an organism begins with a fertilized one-cell embryo. At early cleavage stage, the blastomere undergoes mitotic division without cell fate segregation. In mouse, blastomeres acquire apical-basal polarity and are located inside or outside of the embryo following the eight-cell stage. The different location and polarity properties of the cells provide them with cues toward the first cell lineage segregation, in which the inside cells become the inner cell mass (ICM) while the outside cells develop into extra-embryonic trophectoderm (TE) (Stephenson et al. 2012). Following the first cell lineage determination, the inner cell mass continues to segregate into extra-embryonic primitive endoderm (PrE) and pluripotent epiblast (EPI), and the latter develops into the embryo proper (Schrode et al. 2013). While the regulation of the two cell fate determination events has been extensively explored in mouse, rudimentary knowledge has been obtained in human or nonhuman primates. Several recent studies examined the lineage specification of human pre-implantation embryos by large-scale single-cell RNA-sequencing analysis and reported the overall similarities as well as differences of lineage regulation between human and mouse (Xue et al. 2013; Nakamura et al. 2016; Petropoulos et al. 2016). Despite these advances, large gaps remain in understanding the regulation of cell fate determination in early embryogenesis of human and nonhuman primates.
Epiblasts at differential developmental stages exhibit distinct pluripotent states, namely the naive and primed pluripotent states. The two pluripotent states differ in many cellular and molecular aspects (Theunissen et al. 2016; Weinberger et al. 2016), including the chimeric and differentiation potentials, specific markers, transposon element expression profiles, X Chromosome activation in female cells, the core pluripotency regulatory circuitry, and the epigenetic and metabolic states. In mouse, the in vivo naive and primed pluripotent states exist in epiblast cells of pre-implantation and early post-implantation embryos, respectively. The naive pluripotent state can be stably captured in embryonic stem cells (ESCs) derived from pre-implantation blastocysts, whereas the primed pluripotent state is captured in epiblast stem cells (EpiSCs) derived from post-implantation embryos (embryonic day 5.5) (Brons et al. 2007; Tesar et al. 2007). In contrast, the human and monkey ESCs derived from pre-implantation embryos closely resemble mouse EpiSCs and display the characteristics of primed pluripotency (Rossant and Tam 2017). Although there are limited studies reporting the varying degree of success in generating human and monkey naive pluripotent stem cells (PSCs) (Fang et al. 2014; Takashima et al. 2014; Theunissen et al. 2014; Ware et al. 2014; Chen et al. 2015; Guo et al. 2016b; Pastor et al. 2016), the experiences of stem cell derivation and differentiation in human and monkey suggested that the pluripotency dynamics in primates may be different from that in mice (Rossant and Tam 2017). Thus, it is essential to understand the pluripotency dynamics in primates.
Rhesus monkey is an ideal nonhuman primate animal model to study various human diseases and physiology. Our recent study reported a high degree of similarity in regulation of pre-implantation embryogenesis between human and rhesus monkey using single embryo and pooled embryos (Wang et al. 2017). Moreover, genome editing by CRISPR/Cas9 or TALEN has achieved success in monkeys (Liu et al. 2014; Niu et al. 2014). These works highlight the importance of studying cell lineage segregation as well as pluripotency dynamics in monkeys. Herein, we collected single cells from rhesus monkey early embryos encompassing the stages of 16-cell, early morula, late morula, early blastocyst, middle blastocyst, late blastocyst, and hatched blastocyst. By utilizing single-cell RNA-sequencing analysis, we investigated the regulation of cell lineage segregation and pluripotency dynamics.
Results
Single-cell RNA-sequencing of blastomeres from pre-implantation rhesus monkey embryos
Rhesus monkey 16-cell embryos, early morulae (EM), late morulae (LM), early blastocysts (EB), middle blastocysts (MB), late blastocysts (LB), and hatched blastocysts (HB) were collected to isolate single blastomeres (Fig. 1A). The definition of developmental stages and the criteria of embryo selection are described in Methods. To tell the spatial position (inner or outer) of an isolated single cell within an embryo, we incubated the embryos with PKH26 dye, which labels the live outer cells with red fluorescence. Cells marked with red fluorescence were considered as outer cells, whereas those without fluorescence were inner cells. Both outer and inner cells were collected at developmental stages from 16-cell to HB. Single-cell RNA-sequencing was performed using the Illumina sequencing platform (Supplemental Table S1). The read qualities were examined by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the filtered reads were aligned to rhesus monkey genome MacaM_v7 (Zimin et al. 2014). The total reads, mapped reads, and the numbers of detectable genes (defined as fragment per kilobase per million [FPKM] > 1) were calculated (Supplemental Fig. S1A; Supplemental Table S2). The saturation analysis revealed a minimum of 0.5 million reads required for robust single-cell gene expression quantification (Supplemental Fig. S1B). We removed the potential batch effects by sva (Leek et al. 2012) and assessed the effectiveness by Principal component analysis (PCA) (Supplemental Fig. S1C). To test whether these samples were sufficient to capture all cell fates at each developmental stage, we compared the single-cell RNA-sequencing average profile to that of single-embryo RNA-sequencing at each matching stage (Wang et al. 2017) using our iCpSc package (Sun et al. 2017). The detection of major cell types at stages from EM through to HB apparently reached saturation level at our current coverage depth (Supplemental Fig. S1D).
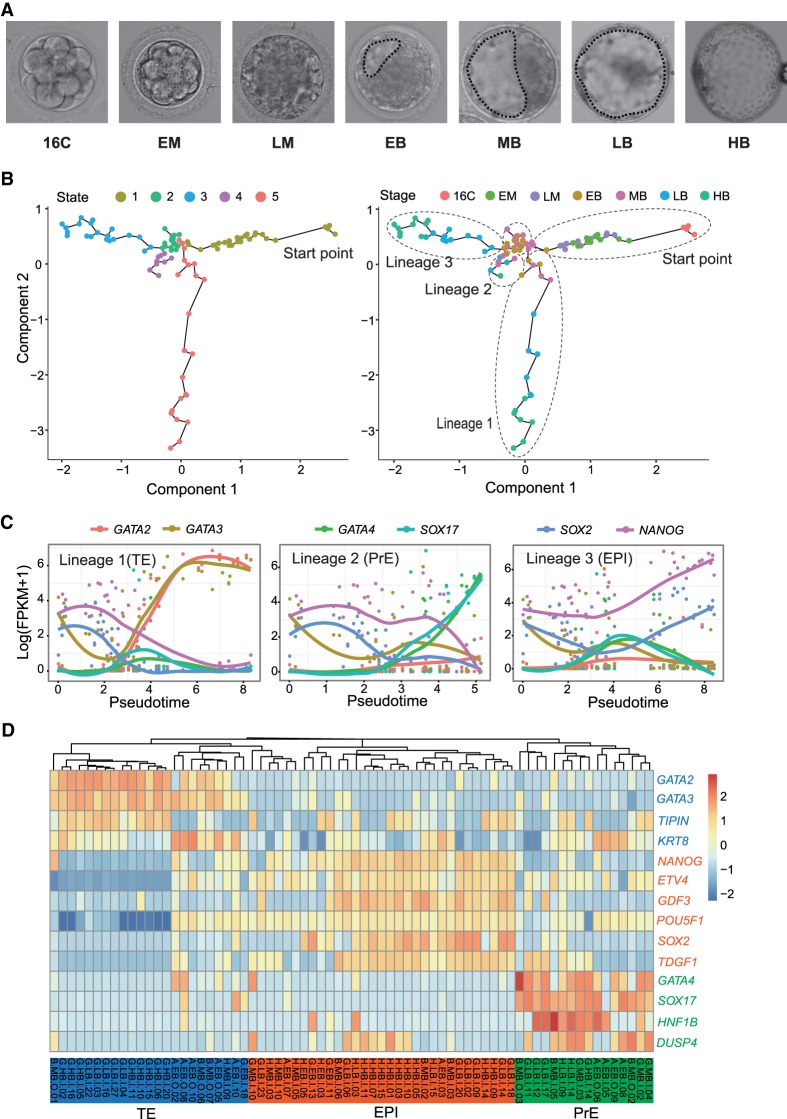
Figure 1.
Lineage segregation in rhesus monkey early embryos. (A) Representative photos of rhesus monkey embryos at the stages of 16-cell (16C), early morula (EM), late morula (LM), early blastocyst (EB), middle blastocyst (MB), late blastocyst (LB), and hatched blastocyst (HB). Dashed lines depict the blastocyst cavities. (B) Lineage segregation path constructed by Monocle based on most variable genes. Five cell states and three lineages were identified. (C) Expression pattern of known marker genes of TE (GATA2, GATA3), PrE (GATA4, SOX17), and EPI (SOX2, NANOG) across the pseudotime assigned by Monocle. (D) Hierarchical clustering based on conserved lineage marker genes identified three cell lineages and fate-undetermined cells.
Trajectories of cell lineage segregation in rhesus monkey early embryo development
To investigate the cell lineage segregation, we first evaluated the relative extent of heterogeneity of single cells at different stages by calculating the sum of standard deviations (SSDs) based on the expression level of all genes. The SSDs increased at late morula stage and thereafter, suggesting an increasing level of cell diversification (Supplemental Fig. S1E). We then explored the cell fate commitment and lineage formation in an unsupervised manner. To this end, we first identified the top 600 most variable genes across all stages of samples, balancing the mean and variation in single-cell RNA-seq (Supplemental Fig. S1F; Guo et al. 2016a). The most variable genes included several key cell lineage markers of TE (KRT18, GATA2, GATA3), EPI (NANOG), and PrE (SOX17, GATA4) (Mitsui et al. 2003; Adjaye et al. 2005; Shimoda et al. 2007; Niakan et al. 2010; Artus et al. 2011; Bai et al. 2011). Using the top 600 most variable genes, we constructed the lineage segregation trajectories by Monocle (Trapnell et al. 2014). The blastomere of the 16-cell stage was set as the starting point, and five developmental states and three branches representing three cell lineages were identified (Fig. 1B; Supplemental Table S3). Examination of the expression patterns of several conserved lineage markers (GATA2 and GATA3 for TE; GATA4 and SOX17 for PrE; and NANOG and SOX2 for EPI) across the pseudotime revealed each lineage identity (Fig. 1C). To corroborate the lineage segregation obtained through Monocle, we performed hierarchical clustering analysis of all cells from the EB through to the HB stage, based on the gene expression of three well-known lineage markers. Consistently, three cell clusters were identified corresponding to TE lineage (23 cells), EPI lineage (31 cells), and PrE lineage (16 cells), respectively (Fig. 1D). Each lineage contained blastomeres collected from the EB stage (Fig. 1D), and lineage-uncommitted cells (state 2 cells) also included blastomeres from EB and MB stages (Fig. 1B). These observations suggested that the three lineages specification occurred at as early as the EB stage and in an asynchronized manner. Concordantly, the PCA analysis on samples from EB through to HB stages, using the most variable genes within these stages, showed three directions of separation (Supplemental Fig. S1G), with the PCA loading genes corresponding to the known cell lineages of EPI, PrE, and TE (Supplemental Fig. S1H).
To further investigate the lineage segregation in detail, we separately conducted the PCA analysis on samples from EB to MB and from LB to HB. Consistently, cells at each stage of blastocyst were segregated (Supplemental Fig. S2A,B). Unsupervised clustering analysis also revealed that some blastomeres of EB and MB already showed the preferential expression of certain lineage markers (Supplemental Fig. S2C), but some blastomeres still expressed two lineage marker genes at a similar level, indicative of their noncommitment and bipotential state (Supplemental Fig. S2C). However, this bipotential state no longer existed in cells from LB and HB stages (Supplemental Fig. S2D), suggesting the completion of cell fate commitment. In addition, no cell lineage segregation was detected in embryos from the 16-cell stage through to LM by PCA and clustering analyses. Although the inner and outer cells were separated apart and many differentially expressed genes (DEGs) were identified at each stage (Supplemental Fig. S2E–G; Supplemental Table S4), the DEGs neither contained known lineage markers (Supplemental Table S4), nor were enriched for terms related to the TE or ICM properties (Supplemental Table S5). Taken together, these data support the notion that the segregation of three cell lineages is initiated at the EB stage and completed at the LB stage in monkey, which is similar to human (Petropoulos et al. 2016) but different from mouse embryogenesis (Cockburn and Rossant 2010).
Cell lineage markers and the regulation of lineage specification
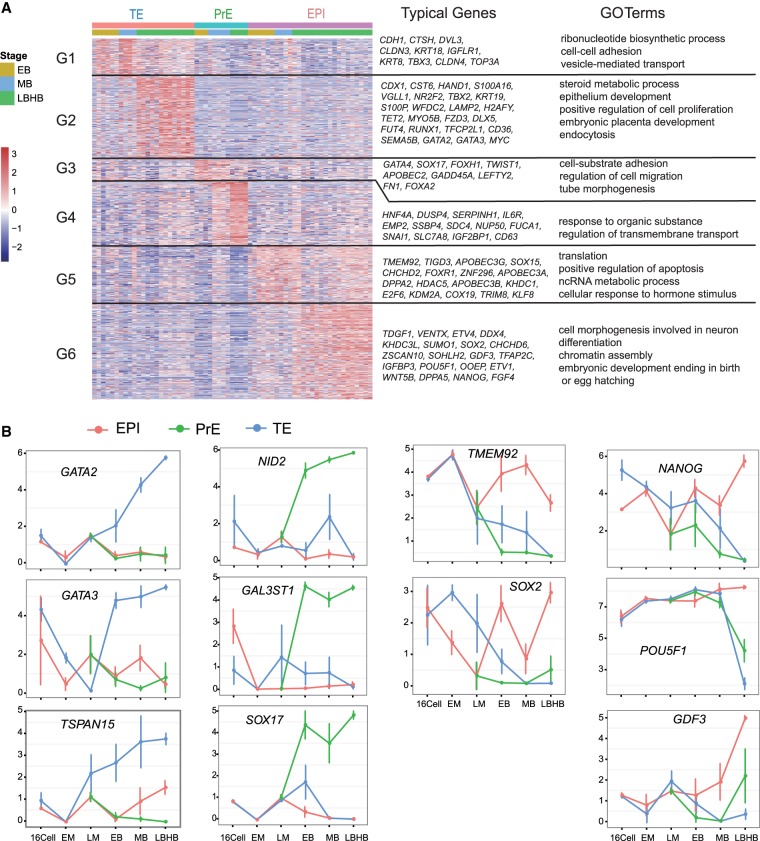
Having defined the lineage identity of each cell collected from blastocysts, we went on to identify new cell lineage markers by comparing one lineage against the other two at each blastocyst stage (Petropoulos et al. 2016). Genes with a false discovery rate (FDR) < 0.01 were considered as lineage specific. Based on this criterion, we identified a list of lineage markers for TE, PrE, and EPI, respectively, at each stage (Fig. 2A; Supplemental Table S6). The enriched GO terms of these lineage markers reflected the lineage properties (Fig. 2A). While many markers displayed a stage-specific expression pattern and could only mark a lineage at certain developmental stages, a limited number of genes persistently labeled a lineage throughout all the blastocyst stages. These genes include: TE markers GATA2, GATA3, TSPAN15, ATP6V1B1, SLC12A3, and TMEM54; PrE markers NID2, GAL3ST1, SOX17, SERPINH1, GATA4, and HMOX2; and EPI markers TMEM92 and SOX2. Pluripotency markers including NANOG, POU5F1, GDF3, UTF1, and PRDM14 displayed restrictive expression at middle and/or late blastocyst stages (Fig. 2B; Supplemental Fig. S3A).
Figure 2.
Lineage-specific genes in rhesus monkey early embryos. (A) Clustering of specific genes of three cell lineages and their GO term enrichment (FDR < 0.01). (B) Expression pattern of some marker genes for TE, EPI, and PrE lineages. Data were obtained from batch 1 samples including 23 TE cells, 16 PrE cells, and 31 EPI cells.
To understand the upstream regulators involved in the segregation of TE from ICM, we performed DEG analysis between TE cells and ICM/EPI cells at the EB stage. A total of 183 and 476 genes were significantly highly expressed in TE and ICM (FDR < 0.05), respectively (Supplemental Fig. S3B; Supplemental Table S7). We noticed that several DNA damage repair pathways were the most enriched GO terms of ICM (Supplemental Table S8). DNA repair pathways, such as base excision repair, are integral parts of DNA de-methylation machinery (Hajkova et al. 2010; He et al. 2011; Kohli and Zhang 2013). These suggest that the DNA repair proteins might be actively involved in the establishment of ICM lineage, probably via regulating DNA demethylation.
To investigate the segregation of PrE from EPI, we analyzed the transcriptomes of individual ICM cells from EB, MB, and LB-HB stages. Most DEGs displayed stage-specific expression patterns (Supplemental Fig. S3C; Supplemental Tables S9, S10), suggesting that the PrE and EPI lineage formation was dynamically regulated by a different set of genes at different stages. For instance, SIN3A and DNMT3B were highly expressed in PrE lineage at MB, coincident with the establishment of de novo methylation and repressive epigenetic features in PrE. On the other hand, GATA4 and SOX17 play critical roles at all stages of PrE formation and maturation. NANOG and SOX2 displayed mutually exclusive expression patterns with GATA4 and SOX17, whereas POU5F1 was expressed at a similar level in two cell lineages throughout the blastocyst stages. Of note, at the MB stage, EPI-specific genes are highly enriched for terms of caspase activity regulation and apoptosis induction, while PrE-specific genes were enriched in terms of macromolecular catabolic processes, membrane organization, cell junction assembly, and focal adhesion (Supplemental Fig. S3C; Supplemental Tables S9, S10). These terms reflected the event of cell sorting in middle blastocyst, which is accompanied by cell migration, apoptosis of uncommitted cells, and the morphological change of PrE to epithelium-like cells (Plusa et al. 2008; Meilhac et al. 2009; Xenopoulos et al. 2015). At the LB stage, the EPI-specific genes are enriched in DNA and RNA metabolic processes, translation, cell cycle, telomere maintenance, DNA damage response and repair, and the TP53 signaling pathway. These terms and pathways suggested active anabolism, catabolism, and genome stability surveillance in highly proliferative epiblast cells.
Existence of distinct pluripotent states in rhesus monkey pre-implantation embryos
In primates, the in vivo dynamics of the pluripotent state remain unclear. To understand it, we examined the pluripotent state of monkey EPI cells from early, mid, late, and hatched blastocysts. Due to batch effect, only 25 out of 31 EPI cells were selected for further study (five cells from EB, four cells from MB, seven cells from LB, and nine cells from HB) (Supplemental Fig. S4A). PCA and unsupervised clustering analyses revealed that EPI cells from EB and MB tended to cluster together, whereas those from LB and HB aligned together (Supplemental Fig. S4B,C). We also identified the DEGs among the EPI cells of four stages and grouped them into five classes by BICSKmeans (Supplemental Fig. S4D; Supplemental Table S11; Zhang et al. 2013). Consistently, the largest number of DEGs was detected between the earlier two and latter two stages (DEGs of group 2 and group 4). These results suggested the possible existence of two discrete cellular states of EPI cells in blastocysts.
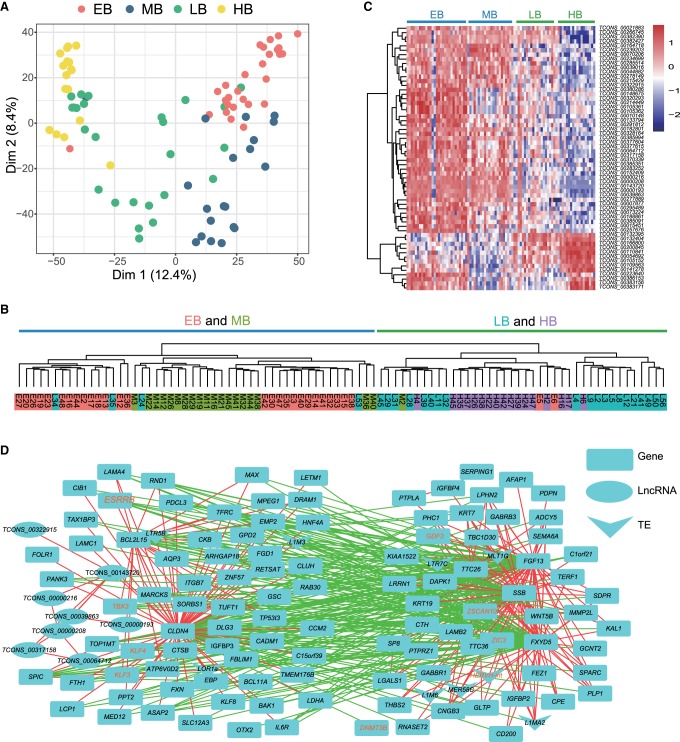
To validate this observation, we collected an additional 161 individual blastomeres from EB through to HB (Supplemental Table S1, batch 2). The cDNAs were amplified using Smart-seq2 protocol (Picelli et al. 2013, 2014) and were sequenced as described above. All of the single cells passed the sequencing quality control (Supplemental Fig. S5A). Among these 161 cells, we identified 90 EPI cells based on their expression of typical lineage markers (29 cells from EB, 20 cells from MB, 24 cells from LB, and 17 cells from HB) (Supplemental Fig. S5B; Supplemental Table S12). We then estimated the consistence of the two batches of EPI samples (batch 1 and 2) by PCA analysis. Although there exist potential batch effects on PC2, the two batches of EPI cells displayed a similar segregation pattern on PC1, in which the earlier two stages (EB and MB) were clearly separated from the latter two stages (LB and HB) (Supplemental Fig. S5C). Further PCA and unsupervised clustering analyses on the 90 batch 2 EPI cells also revealed the alignment of EB with MB, and of LB with HB (Supplemental Fig. S5D,E). Thus, we conclude that the EPI cells from EB and MB are distinct from those from LB and HB. In the following studies, we will present the data from 90 batch 2 EPI cells in the main text, as they contain a bigger sample size and, whenever relevant, show the results from the 25 batch 1 EPI samples in the Supplemental Information as further support.
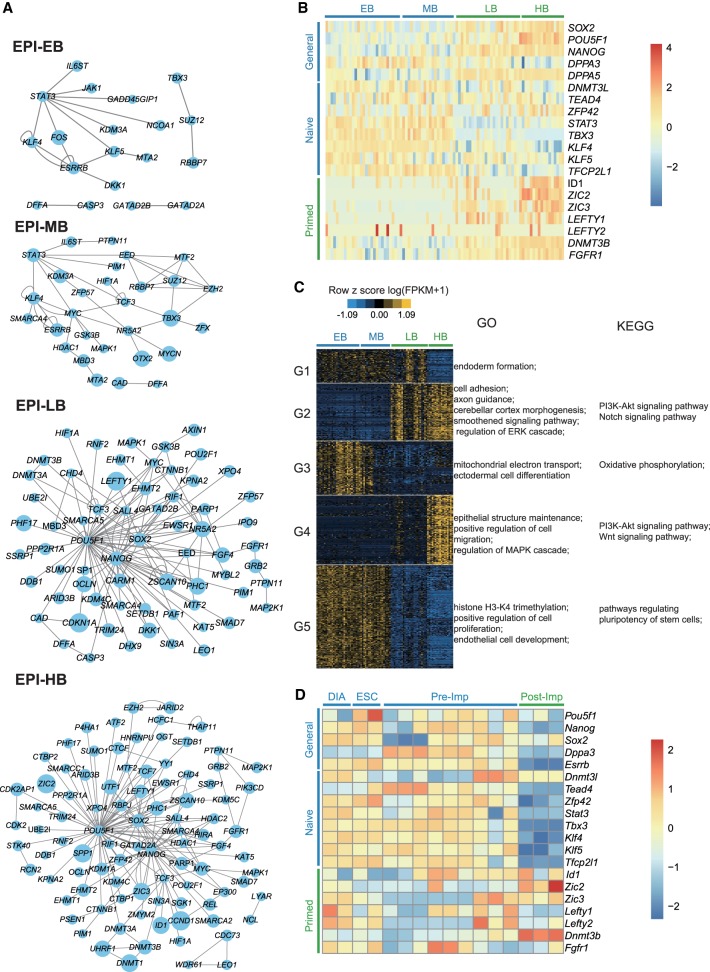
Pluripotency is one of the key features of EPI cells, and its dynamics remains elusive in primates. We wondered whether the EPI cells from different developmental stages have distinct pluripotent states. To this end, we performed PluriNetWork analysis to construct the pluripotency regulatory networks (Boroviak et al. 2015), in which the expression levels of the genes were normalized and the preferentially active regulators were displayed as network nodes. Analysis of the batch 2 (Fig. 3A) and batch 1 (Supplemental Fig. S6A) EPI cells consistently revealed the following drastic changes of the network topology at the transition from the mid- to late-blastocyst stage: (1) At the EB and MB stages, the key naive pluripotency regulators (e.g., STAT3, KLF4, ESRRB, and TBX3) dominated the networks (van Oosten et al. 2012; Dunn et al. 2014; Adachi et al. 2018), whereas at the LB and HB stages, the general core pluripotency regulators (NANOG, POU5F1, SALL4, and SOX2) took over the pluripotency regulation; (2) the network nodes of the latter two stages included MEK/ERK cascade components FGF4, FGFR1, and GRB2, which trigger the ESC lineage commitment (Kunath et al. 2007; Kang et al. 2013) and are required for the in vitro propagation of primed human ESCs (Xu et al. 2005); (3) the enzymes DNMT3A and DNMT3B responsible for de novo DNA methylation were detected at the latter two stages, corresponding to the overall increased methylation levels in the primed pluripotent state. Thus, these network topology changes suggested the existence of two different pluripotent states in monkey pre-implantation blastocysts and their shift in between the middle- and late-blastocyst stages.
Figure 3.
Distinct pluripotent states exist in rhesus monkey pre-implantation embryos. (A) PluriNetWork analysis of EPI cells from four blastocyst stages revealed a transition of network topology from mid- to late-blastocyst stage. (B) Expression pattern of marker genes for naive pluripotency, primed pluripotency, and general pluripotency in EPI cells of EB, MB, LB, and HB in rhesus monkey. (C) BICSKmeans clustering of differentially expressed genes (DEGs) (FDR < 0.01) in EPI cells from EB through to HB stages. The GO term and KEGG pathway enrichment of each group of DEGs are shown. In A–C, data were from 90 batch 2 EPI cells (EB, 29 cells; MB, 20 cells; LB, 24 cells; and HB, 17 cells). (D) Expression pattern of marker genes for naive pluripotency, primed pluripotency, and general pluripotency in EPI cells of pre-implantation and post-implantation embryos in mouse. Analysis was performed on the archival data from Boroviak et al. (2015).
To further corroborate this hypothesis, we compared the expression level of marker genes for naive pluripotency, primed pluripotency, and general pluripotency among the EPI cells of the four blastocyst stages (Stuart et al. 2014; Guo et al. 2016b; Sahakyan et al. 2016). As shown in Figure 3B (batch 2 EPI cells) and Supplemental Figure S6B (batch 1 EPI cells), the naive pluripotent markers were overall highly expressed in EPI cells from EB and MB. Conversely, the primed pluripotent markers were preferentially expressed in EPI cells of LB and HB. Of note, the general pluripotent markers SOX2, NANOG, and POU5F1 displayed increased expression at LB and HB stages. This was in line with the previous observation on cynomolgus monkeys pre- and post-implantation embryo development (Nakamura et al. 2016). As a control, the housekeeping genes showed similar expression levels across the four blastocyst stages (Supplemental Fig. S6C,D).
To further understand the differences of EPI cells from the four blastocyst stages, we identified the DEGs among the 90 batch 2 EPI cells and grouped them into five clusters by BICSKmeans (Fig. 3C; Supplemental Table S13; Zhang et al. 2013). The enriched biological processes and pathways in each group were presented (Fig. 3C; Supplemental Table S14). Specifically, DEGs highly expressed in EPI cells of EB and MB were enriched for the processes of oxidative phosphorylation, histone H3-K4 trimethylation, and regulation of pluripotency of stem cells (Fig. 3C; Supplemental Fig. S4C; Supplemental Table S14). In contrast, DEGs highly expressed in EPI cells of LB and HB were overrepresented in the processes of cell adhesion, axon guidance, cerebellar cortex morphogenesis, epithelial structure maintenance, and smoothened signaling, which may reflect the initiation of morphogenetic events of EPI cells heading for gastrulation at the LB and HB stages. The highly expressed genes in LB and HB were also enriched for the signaling pathways including MAPK/ERK and Notch pathways (Fig. 3C; Supplemental Fig. S4C; Supplemental Table S14), which drive the transition out of the naive state to the primed state and promote stem cell differentiation (Schroeder et al. 2006; Hackett and Surani 2014). Blockage of MAPK/ERK pathways facilitates the derivation of naive pluripotent ESCs (Ying et al. 2008; Fang et al. 2014; Hassani et al. 2014; Guo et al. 2016b). We also assessed the transcriptome similarity among monkey EPI cells, human naive ESCs, and primed ESCs, using two sets of human ESC transcriptome data from different studies (Takashima et al. 2014; Theunissen et al. 2014). PCA analysis consistently revealed that monkey EPI cells from EB and MB aligned closer to human naive ESCs, whereas EPI cells from LB and HB aligned with human primed ESCs (Supplemental Fig. S6E,F).
Taken together, these lines of evidence support that naive pluripotency transiently appears at EB and MB stages but starts transition to the primed pluripotency thereafter. At the LB stage, EPI cells display high heterogeneity, with some close to HB cells, some close to EB/MB cells, and some in the intermediate (Supplemental Figs. S5C,D, S6E,F). This observation may suggest that EPI cells in LB are in a continuum state, acquiring primed pluripotency. In sharp contrast, naive pluripotency persists in mouse pre-implantation embryos and progresses to primed pluripotency after implantation, as evidenced by the expression pattern of various pluripotent markers (Fig. 3D; Boroviak et al. 2015).
Expression profiles of transposable elements and lncRNAs in rhesus monkey early embryos
Transposable elements are mobile genetic entities and occupy nearly half of the human genome. They play essential roles in early embryonic development via rewiring the gene expression networks (Gifford et al. 2013; Gerdes et al. 2016) and serve as reliable markers of pluripotent states (Theunissen et al. 2016). To examine the profile of transposable elements in rhesus monkey early embryos, we quantified their expression by the unit of subfamily (FPKM > 1) and identified the expressed subfamilies in embryos at stages from 16-cell through to HB in batch 1 samples (Supplemental Fig. S7A; Supplemental Table S15). We then used their expression profiles to re-evaluate the cell fate segregation. PCA analysis showed that the blastomeres at differential developmental stages were clearly separated (Supplemental Fig. S7B). Likewise, the blastomeres of three lineages defined by protein-coding genes were also segregated by transposable elements (Supplemental Fig. S7C,D). Thus, the transcription from transposable elements is also dynamically regulated during monkey early embryogenesis and lineage segregation.
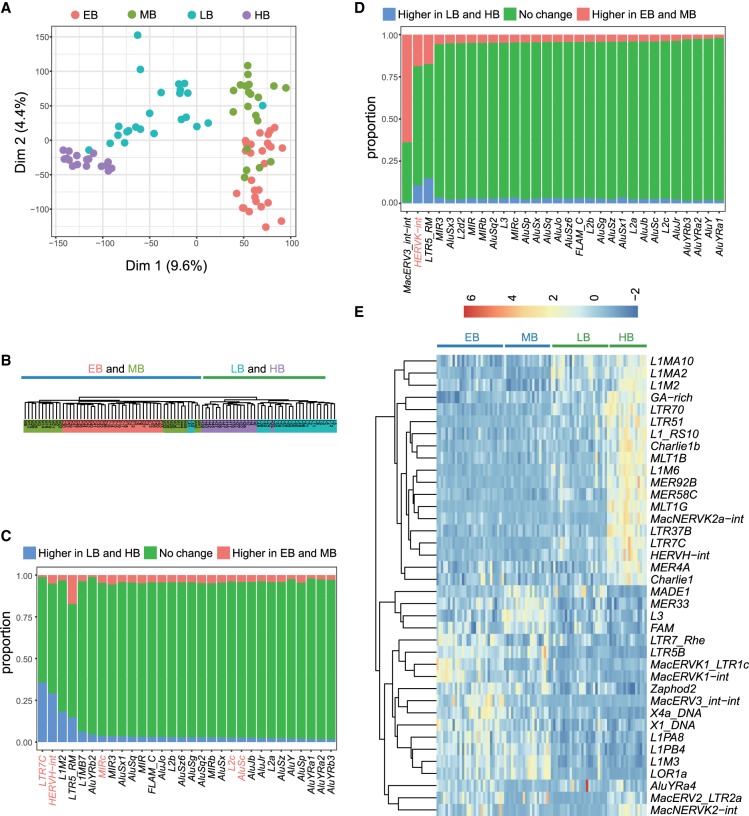
Because transposable elements are reliable markers to define the pluripotent states (Theunissen et al. 2016), we used the expression profiles of transposable elements in batch 1 and 2 EPI cells (Supplemental Tables S16, S17) from EB through to HB to further validate the existence of distinct pluripotent states in pre-implantation monkey embryos. The numbers of detected transposable element subfamilies in EPI cells were comparable among four blastocyst stages (Supplemental Fig. S8A,B). PCA (Fig. 4A; Supplemental Fig. S8C) and unsupervised clustering (Fig. 4B; Supplemental Fig. S8D) analyses revealed that EPI cells of EB and MB were clustered together and separated from those of LB and, especially, HB. This pattern was similar to the one obtained by analyzing protein-coding genes (Supplemental Fig. S5C,D). To find out the potential transposable elements that may play roles in regulating the cellular state changes of EPI cells between the earlier two and latter two blastocyst stages, we identified the differentially expressed loci of transposable elements in the batch 2 EPI cells and summarized them by subfamily (Fig. 4C,D). Some LB-HB highly expressed transposable elements, such as HERVH-int, LTR7C, AluJb, AluSc, L2c, MIRc, and L2d2 (Fig. 4C), were previously shown to be molecular signatures of the primed pluripotency (Theunissen et al. 2016). Likewise, the known naive pluripotency marker HERVK-int (Theunissen et al. 2016) was highly expressed in EB and MB (Fig. 4D). Similar results were obtained when the differentially expressed transposable elements (FDR < 0.01) were calculated by the unit of subfamily (Fig. 4E; Supplemental Table S18). Analyses of batch 1 EPI cells also showed consistent results (Supplemental Fig. S8E,F). Taken together, the transposable element expression supports the existence of distinct pluripotent states in pre-implantation monkey embryos.
Figure 4.
Transposable elements define the distinct cellular states of EPI cells from EB through to HB. PCA (A) and hierarchical clustering (B) analyses using the expression profiles of transposable elements segregated the EPI cells of EB and MB from those of LB and HB. (C) Transposable element subfamilies that showed biased expression in LB and HB stages. (D) Transposable element subfamilies that showed biased expression in EB and MB stages. In C and D, columns are split into segments, representing loci with higher expression in LB and HB (P-values < 0.05) (Higher in LB and HB), loci with higher expression in EB and MB (Higher in EB and MB), and loci with no expression difference in all stages (No change). (E) Differentially expressed transposable element subfamilies (FDR < 0.05) also identified the naive-primed transition at blastocyst stages. All data were obtained from 90 batch 2 EPI cells (EB, 29 cells; MB, 20 cells; LB, 24 cells; and HB, 17 cells).
Long noncoding RNAs (lncRNAs) play a wide range of regulatory roles in embryonic development and are highly tissue-specific (Deveson et al. 2017). To explore the dynamic expression pattern of lncRNAs and their possible involvement in monkey early embryogenesis, we assembled 909 expressed lncRNAs (FPKM > 1) from the 16-cell through to the HB stage in batch 1 samples (Supplemental Table S19). Their expressions at each stage (Supplemental Fig. S9A) and cell lineage (Supplemental Fig. S9B) were summarized. Like the protein-coding genes and transposable elements, lncRNA expression profiles could also define the developmental stages (Supplemental Fig. S9C) as well as the cell lineages (Supplemental Fig. S9D) of the single blastomeres. We next examined the lncRNA expression profiles in both batch 1 and 2 EPI cells (Supplemental Tables S20, S21). Likewise, the EPI cells of the four blastocyst stages were segregated into two discrete clusters (EB-MB clusters versus LB-HB clusters) by PCA (Fig. 5A; Supplemental Fig. S9E) and unsupervised clustering analyses (Fig. 5B; Supplemental Fig. S9F), based on their lncRNA expression profiles. To find out the lncRNA species which may regulate the naive-primed pluripotency transition, we identified a list of stage-specific lncRNAs among the batch 2 EPI cells of the four blastocyst stages (Fig. 5C; Supplemental Table S22). The majority of these lncRNAs were grouped into two classes, representing EB-MB-specific and LB-HB-specific. These stage-specific lncRNAs may play potential roles in the regulation of different pluripotent states and their transition.
Figure 5.
Two differential cellular states of EPI cells defined by the lncRNAs and co-expression networks. PCA (A) and hierarchical clustering (B) analyses based on lncRNA expression profiles separated the EPI cells of EB and MB stages from those of LB and HB stages. (C) Differentially expressed lncRNAs in EPI cells of four blastocyst stages (FDR < 0.05). (D) Co-expression networks constructed with the protein-coding genes, transposable elements, and lncRNAs contained two anti-correlated modules. The two modules may regulate the two cellular states of EPI cells in EB-MB stages and LB-HB stages, respectively. Red and green edges indicated that the two nodes were transcriptionally correlated and anti-correlated, respectively. All data were obtained from 90 batch 2 EPI cells (EB, 29 cells; MB, 20 cells; LB, 24 cells; and HB, 17 cells).
To better understand the pluripotency regulation as a whole, using the batch 2 EPI cells we conducted co-expression network analysis of three layers of regulators comprised of protein-coding genes, transposable elements, and lncRNAs. Stage-specific protein-coding genes, transposable elements, and lncRNAs in EPI cells from EB through to HB formed a highly contacted co-expression network, constructed using WGCNA (Fig. 5D; Langfelder and Horvath 2008). In the network, the transcriptional correlation and anti-correlation between two nodes were annotated as red and green edges, respectively. The network was composed of two anti-correlated modules, of which one contained naive pluripotency-related factors (e.g., TBX3, KLF3, KLF4), whereas the other was enriched with primed pluripotency relevant factors (e.g., GDF3, ZIC3, DNMT3B, HERVH-int, LTR7C, MLT1G, L1M6, MER58C, L1MA2). The dense connections between the two modules suggest that they might collaborate to regulate the two alternative cellular states of EPI cells and their transition.
Discussion
By utilizing single-cell RNA-sequencing analysis, we mapped the developmental trajectory of the earliest cell lineage segregation in rhesus monkey embryogenesis at the transcriptome level and identified a list of markers specific for the three cell lineages of TE, EPI, and PrE. Compared to the reports on human and mouse (Guo et al. 2010; Xue et al. 2013; Petropoulos et al. 2016), we found that monkey and human are very similar in lineage segregation regulation but are distinct from mouse. Notably, we uncovered the existence of distinct pluripotent states in EPI cells of monkey pre-implantation blastocysts. EPI cells at the EB and MB stages display the properties of naive pluripotency. Thereafter, they lose the naive pluripotency and progressively acquire the primed pluripotency. This pluripotency dynamic is distinct from that of mouse, in which naive pluripotency persists for the whole pre-implantation stage and shifts to primed pluripotency after implantation (Wu and Izpisua Belmonte 2015). Thus, this study proposed an appropriate time-window to derive primate ESCs with naive pluripotency from pre-implantation embryos. It should be mentioned that the primate pre-implantation embryo studies are all exclusively conducted on the in vitro fertilized and cultured embryos. However, based on the high efficiency of in vitro embryo culture system in primates, we proposed that the results obtained from the in vitro embryos might reflect the bona fide in vivo situations.
We analyzed the EPI cells at more developmental stages than the other related studies on monkey or human embryos (Nakamura et al. 2016; Petropoulos et al. 2016). This detailed time-series analyses allowed us to capture the dynamics of pluripotency in monkey embryos. We presented several lines of evidence supporting that naive pluripotency transiently exists at early- and middle-blastocyst stages and gradually advances to primed pluripotency thereafter. The EPI cells collected from four developmental stages (early-, middle-, late-, and hatched-blastocyst stages) consistently formed two distinct clusters (early-middle stage cluster and late-hatched stage cluster), whether the analyses were based on the differentially expressed protein-coding genes, transposable elements, or lncRNAs. This implicated that EPI cells of the earlier two and latter two developmental stages were in distinct cellular states. Specifically, EPI cells of the two clusters showed differences in pluripotency regulation and the expression patterns of naive and primed pluripotency markers including genes and transposable elements. In addition, the potential regulators of EPI cells identified by co-expression network analysis also formed two anti-correlated modules, of which one contained the central regulatory factors for naive pluripotency and the other contained primed pluripotency-relevant factors. In line with our observations, Theunissen et al. (2016) also reported that human naive pluripotent stem cells have stronger convergence with morulae than with EPI cells of late blastocysts (Theunissen et al. 2016). This transient existence of naive pluripotency in monkey pre-implantation embryos provides an explanation for the frequent failure in deriving and generating naive pluripotent primate ESCs. Due to the high similarity of the regulation of early embryo development between rhesus monkey and human, we envision that similar pluripotency dynamics might also exist in human.
Methods
Animals, treatment, oocyte recovery, in vitro fertilization, and embryo culture
All experimental procedures and animal care were approved by the Ethics Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences. Adult rhesus monkeys with a successful reproductive history were used in this study. Cycling female monkeys were ovarian-stimulated by injection of rhFSH (recombinant human follitropin alfa, GONAL-F, Merck Serono), 18 U twice daily for 8 d from the onset of the menstrual cycle. On day 9, 1000 U of rhCG (recombinant human chorionic gonadotropin alfa, OVIDREL, Merck Serono) were injected. After 32–35 h of rhCG administration, mature oocytes were collected by laparoscopic follicular aspiration (Niu et al. 2010). Oocytes were removed of cumulus cells and those with extrusion of the first polar body (metaphase II stage) were collected for downstream experiments. Semen collection, sperm capacitation, and in vitro fertilization were performed as described previously (Zheng et al. 2001). Successful fertilization was assessed by the presence of two pronuclei. Embryos were then cultured at 37°C, 5% CO2 in 50-μL drops of equilibrated HECM-9 containing 10% FBS (HyClone Laboratories Inc.). Culture medium was changed every other day. Morphologically normal embryos with few fragments were used in the study. Sixteen-cell embryos with good morphology were collected at about embryonic day 3 (E3). Early morulae with compacted blastomeres were collected at about E4. Late morulae with more blastomeres were collected at about E4.5. Early blastocysts having a small cavity were collected at about E5. Middle blastocysts with a cavity occupying around half the size of the embryos were collected at about E5.5. Late blastocysts with a fully expanded cavity were collected at about E6. Hatched blastocysts from the zona pellucida were collected at about E7.
Single blastomere dissociation
To dissociate embryos into single blastomeres, batch 1 embryos with good quality were transferred into 0.5% Pronase in TL-HEPES to remove the zona pellucida, followed by three washes in PVP-BSA. To distinguish the outer and inner cells, embryos were incubated with PKH26 dye (Sigma) which labels the live outer cells with red fluorescence. After labeling, embryos were rinsed with Ca2+/Mg2+-free PVP-PBS and treated with 0.125% trypsin-EDTA for at least 5 min at 37°C until the cell surface looked rough. Disaggregating was done by a mouth pipette aided by several finely pulled glass capillaries of different internal diameter. After most cells were separated apart, single cells were individually transferred into small droplets of PVP-PBS and the inner/outer cell identity was identified by the fluorescent microscopy. Cells showing no fluorescence were inner cells, whereas those with red fluorescence were outer cells.
To dissociate batch 2 cells, blastocysts with good quality were transferred into 0.1% Pronase (Merck) in PBS to remove the zona pellucida, followed by three washes in 0.05% PVP-PBS. Blastocysts were then incubated with PKH26 dye (Sigma) and washed in PVP-PBS. The blastocysts were cut off with a 1-mL syringe and were dissociated as described above.
Single-cell transcriptome generation
For batch 1 samples, single-cell cDNA amplification was prepared according to the protocol from Tang et al. (2009). Briefly, single cells were washed three times in PVP-PBS, and those with a clear round shape were added into cell lysis buffer, followed by cDNA synthesis, primer removal, poly(A) tailing, second-strand cDNA synthesis, and PCR amplification. PCR products were purified by Ampure XP beads and used for library construction. Libraries were deep-sequenced on an Illumina HiSeq 2500 platform, using a 100-bp single-end strategy.
For batch 2 cells, single-cell cDNAs were prepared using Smart-seq2 (Picelli et al. 2013, 2014). Briefly, single cells were washed three times in 0.1% BSA-PBS and were manually picked into lysis buffer containing ERCC spike-in (Baker et al. 2005). Full-length poly(A)-tailed RNA was reverse-transcribed and amplified with 21 PCR cycles to increase the cDNA amount. Sequencing was performed using the Illumina X Ten platform with paired-end 150-bp reads.
Read mapping and quantification
Clean reads were mapped with TopHat v2.1.0 (Kim et al. 2013) to rhesus genome MacaM_v7 (Zimin et al. 2014), with the parameters “-g 1 -N 5 --read-edit-dist 5 --microexon-search”. Only uniquely mapped reads were used to calculate the counts of each gene by HTSeq (Anders et al. 2015). Reads counts were normalized to library size by DESeq2 (Love et al. 2014) with default parameters, and normalized reads were then used to calculate the FPKM (Fragments Per Kilobase of transcript per Million fragments mapped). A gene was defined as expressed if the FPKM value is above 1. The potential batch effect was removed by the sva (Leek et al. 2012) package in R (R Core Team 2008).
Saturation curve analysis
To evaluate how many reads are sufficient to quantify expressed genes, we first randomly and separately selected 0.00025, 0.0005, 0.0025, 0.005, 0.025, 0.05, 0.25, 0.5, 2.5, and 5 million reads from each sample as a pseudolibrary and then mapped these reads to the reference genome and quantified the expressed genes.
To evaluate if the numbers of single cells we collected at each stage are sufficient to capture all cell fates at each stage, we calculated the Pearson correlation coefficient (PCC) of expression level of expressed genes between single-embryo RNA-sequencing data (Wang et al. 2017) and the average profile of single-cell RNA-sequencing data.
Lowess coefficient of dispersion (LCOD) analysis for identifying the most variable genes
To identify the most variable genes, we first fitted a Lowess curve to the log2 of the mean versus the log2 of the standard deviation of all these cells by loess function in R (R Core Team 2008). We then calculated the distance of each gene to the curve and selected the top 600 genes for Monocle analysis.
Monocle and PCA analyses
Principal component analysis (PCA) was performed by FactoMineR package in R (R Core Team 2008). Monocle analysis was performed by the Monocle package in R with default parameters, and the 16-cell stage was set as the starting point.
Differentially expressed gene identification and functional enrichment analysis
Differentially expressed genes were identified by DESeq2 (Love et al. 2014) for pairwise stages. Genes were identified as differentially expressed between two developmental stages if the FDR < 0.00001. Functional enrichment analysis was conducted by Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al. 2008; Sherman and Lempicki 2009).
Lineage-specific gene identification
To find lineage-specific genes, we combined the Z scores of each gene obtained from the differential expression analysis of one lineage against the other two (Stouffer's method) (Petropoulos et al. 2016). Genes that have an FDR < 0.01 were considered as lineage-specific genes.
Construction of PluriNetWork
The network of pluripotency (PluriNetWork) was constructed as previous described (Som et al. 2010). First, we obtained all the pluripotency-related genes and regulatory relationships from a published database (Som et al. 2010). For each stage, we only visualized genes that preferentially expressed at this stage, and the network edge was required to have active source and target nodes from the database (Som et al. 2010). Last, we visualized the network by Cytoscape (Smoot et al. 2010).
Transposable element analysis
Transposable element counts were generated by iteres (https://github.com/lidaof/iteres) based on the repeat annotations from UCSC (https://genome.ucsc.edu/), with the version Mmul_8.0.1. When using unique mapping for transposon sequences, there might be a bias toward older transposons. Transposable elements that do not have at least three samples with 10 reads were discarded. Differentially expressed transposable elements were identified by DESeq2 packages in R (R Core Team 2008) by requiring that P-values < 0.05.
LncRNA analysis
Because the annotation of lncRNAs in monkey was insufficient, we assembled the transcripts of lncRNAs according to the previous report (Hezroni et al. 2015). The analysis pipeline of lncRNAs was similar to coding genes.
Cross-species transcriptome comparison
We performed a human-monkey cross-species comparison as previous described (Wang et al. 2014). We first scaled the expression matrix within each species by Z-score and then combined them into a matrix based on the homology genes annotation downloaded from Ensembl (https://asia.ensembl.org). This matrix was further subjected to PCA analysis to measure the transcriptome similarity.
Data access
Single-cell RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE117219.
Supplementary Material
Acknowledgments
We thank Prof. Naihe Jing and Dr. Guangdun Peng of the Shanghai Institute of Biochemistry and Cell Biology, CAS for help in setting up the single-cell RNA-seq techniques and Dr. Chaohui Li at Yunnan University and Dr. Lei Ao for assistance in blastomere dissociation. This work was supported by the “Strategic Priority Research Program” of the CAS, Grant No. XDB13010600 and XDA01010203 to P.Z., and grants from China Ministry of Science and Technology 2015CB964803 and 2016YFE0108700, National Natural Science Foundation of China 91749205 and 31210103916, and Chinese Academy of Sciences XDA01010303 to J-D.J.H.
Author contributions: X.W., D.H., and C.S. collected the monkey embryos and prepared single-cell cDNA samples for RNA-sequencing. X.H. and L.Y. performed the egg collection, in vitro fertilization, and embryo culture. D.L. and X.W. conducted the data analysis. Y.L. performed the cross-species transcriptome comparison. P.Z. designed the experiments. P.Z. and J-D.J.H. supervised the project and wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.233437.117.
References
- Adachi K, Kopp W, Wu G, Heising S, Greber B, Stehling M, Arauzo-Bravo MJ, Boerno ST, Timmermann B, Vingron M, et al. 2018. Esrrb unlocks silenced enhancers for reprogramming to naive pluripotency. Cell Stem Cell 23: 266–275.e6. [DOI] [PubMed] [Google Scholar]
- Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, et al. 2005. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells 23: 1514–1525. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Piliszek A, Hadjantonakis A-K. 2011. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol 350: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Sakurai T, Someya Y, Konno T, Ideta A, Aoyagi Y, Imakawa K. 2011. Regulation of trophoblast-specific factors by GATA2 and GATA3 in bovine trophoblast CT-1 cells. J Reprod Dev 57: 518–525. [DOI] [PubMed] [Google Scholar]
- Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M. 2005. The External RNA Controls Consortium: a progress report. Nat Methods 2: 731–734. [DOI] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, Nichols J, Smith A, Bertone P. 2015. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell 35: 366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. [DOI] [PubMed] [Google Scholar]
- Chen Y, Niu Y, Li Y, Ai Z, Kang Y, Shi H, Xiang Z, Yang Z, Tan T, Si W, et al. 2015. Generation of cynomolgus monkey chimeric fetuses using embryonic stem cells. Cell Stem Cell 17: 116–124. [DOI] [PubMed] [Google Scholar]
- Cockburn K, Rossant J. 2010. Making the blastocyst: lessons from the mouse. J Clin Invest 120: 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveson IW, Hardwick SA, Mercer TR, Mattick JS. 2017. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet 33: 464–478. [DOI] [PubMed] [Google Scholar]
- Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. 2014. Defining an essential transcription factor program for naive pluripotency. Science 344: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Liu K, Zhao Y, Li H, Zhu D, Du Y, Xiang C, Li X, Liu H, Miao Z, et al. 2014. Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell 15: 488–496. [DOI] [PubMed] [Google Scholar]
- Gerdes P, Richardson SR, Mager DL, Faulkner GJ. 2016. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol 17: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford WD, Pfaff SL, Macfarlan TS. 2013. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol 23: 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18: 675–685. [DOI] [PubMed] [Google Scholar]
- Guo G, Pinello L, Han X, Lai S, Shen L, Lin TW, Zou K, Yuan GC, Orkin SH. 2016a. Serum-based culture conditions provoke gene expression variability in mouse embryonic stem cells as revealed by single-cell analysis. Cell Rep 14: 956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A, Nichols J. 2016b. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports 6: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. 2014. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 15: 416–430. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani SN, Totonchi M, Sharifi-Zarchi A, Mollamohammadi S, Pakzad M, Moradi S, Samadian A, Masoudi N, Mirshahvaladi S, Farrokhi A, et al. 2014. Inhibition of TGFβ signaling promotes ground state pluripotency. Stem Cell Rev 10: 16–30. [DOI] [PubMed] [Google Scholar]
- He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. 2015. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep 11: 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2008. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, Hadjantonakis AK. 2013. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development 140: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. 2013. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502: 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134: 2895–2902. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, et al. 2014. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 14: 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac SM, Adams RJ, Morris SA, Danckaert A, Le Garrec J-F, Zernicka-Goetz M. 2009. Active cell movements coupled to positional induction are involved in lineage segregation in the mouse blastocyst. Dev Biol 331: 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M. 2016. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537: 57–62. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, et al. 2010. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev 24: 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Yu Y, Bernat A, Yang S, He X, Guo X, Chen D, Chen Y, Ji S, Si W. 2010. Transgenic rhesus monkeys produced by gene transfer into early-cleavage–stage embryos using a simian immunodeficiency virus-based vector. Proc Natl Acad Sci 107: 17663–17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156: 836–843. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE, Clark AT. 2016. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 18: 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Reyes AP, Linnarsson S, Sandberg R, Lanner F. 2016. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165: 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10: 1096–1098. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9: 171–181. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis A-K. 2008. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135: 3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Rossant J, Tam PP. 2017. New insights into early human development: lessons for stem cell derivation and differentiation. Cell Stem Cell 20: 18–28. [DOI] [PubMed] [Google Scholar]
- Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, Kuoy E, Langerman J, Clark AT, Jaenisch R, et al. 2016. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20: 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N, Xenopoulos P, Piliszek A, Frankenberg S, Plusa B, Hadjantonakis AK. 2013. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis 51: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T, Meier-Stiegen F, Schwanbeck R, Eilken H, Nishikawa S, Hasler R, Schreiber S, Bornkamm GW, Nishikawa S, Just U. 2006. Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech Dev 123: 570–579. [DOI] [PubMed] [Google Scholar]
- Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Shimoda M, Kanai-Azuma M, Hara K, Miyazaki S, Kanai Y, Monden M, Miyazaki J-I. 2007. Sox17 plays a substantial role in late-stage differentiation of the extraembryonic endoderm in vitro. J Cell Sci 120: 3859–3869. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. 2010. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som A, Harder C, Greber B, Siatkowski M, Paudel Y, Warsow G, Cap C, Schöler H, Fuellen G. 2010. The PluriNetWork: an electronic representation of the network underlying pluripotency in mouse, and its applications. PLoS One 5: e15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson RO, Rossant J, Tam PP. 2012. Intercellular interactions, position, and polarity in establishing blastocyst cell lineages and embryonic axes. Cold Spring Harb Perspect Biol 4: a008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart HT, van Oosten AL, Radzisheuskaya A, Martello G, Miller A, Dietmann S, Nichols J, Silva JC. 2014. NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr Biol 24: 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yu X, Li F, Liu D, Suo S, Chen W, Chen S, Song L, Green CD, McDermott J, et al. 2017. Inference of differentiation time for single cell transcriptomes using cell population reference data. Nat Commun 8: 1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158: 1254–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6: 377–382. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. 2014. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15: 471–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Friedli M, He Y, Planet E, O'Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, et al. 2016. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten AL, Costa Y, Smith A, Silva JC. 2012. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat Commun 3: 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. 2014. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516: 405–409. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu D, He D, Suo S, Xia X, He X, Han JJ, Zheng P. 2017. Transcriptome analyses of rhesus monkey preimplantation embryos reveal a reduced capacity for DNA double-strand break repair in primate oocytes and early embryos. Genome Res 27: 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, et al. 2014. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci 111: 4484–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH. 2016. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 17: 155–169. [DOI] [PubMed] [Google Scholar]
- Wu J, Izpisua Belmonte JC. 2015. Dynamic pluripotent stem cell states and their applications. Cell Stem Cell 17: 509–525. [DOI] [PubMed] [Google Scholar]
- Xenopoulos P, Kang M, Puliafito A, Di Talia S, Hadjantonakis A-K. 2015. Heterogeneities in Nanog expression drive stable commitment to pluripotency in the mouse blastocyst. Cell Rep 10: 1508–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Rosler E, Jiang J, Lebkowski JS, Gold JD, O'Sullivan C, Delavan-Boorsma K, Mok M, Bronstein A, Carpenter MK. 2005. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cells 23: 315–323. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. 2013. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu Y, Sun N, Wang D, Boyd-Kirkup J, Dou X, Han J-Dong J. 2013. Integrating genomic, epigenomic, and transcriptomic features reveals modular signatures underlying poor prognosis in ovarian cancer. Cell Rep 4: 542–553. [DOI] [PubMed] [Google Scholar]
- Zheng P, Bavister BD, Ji W. 2001. Energy substrate requirement for in vitro maturation of oocytes from unstimulated adult rhesus monkeys. Mol Reprod Dev 58: 348–355. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, et al. 2014. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.