Abstract
SPI1 (also known as PU.1) is a dominant but transient regulator in early T-cell precursors and a potent transcriptional controller of developmentally important pro-T-cell genes. Before T-lineage commitment, open chromatin is frequently occupied by PU.1, and many PU.1 sites lose accessibility when PU.1 is later down-regulated. Pioneering activity of PU.1 was tested in this developmentally dynamic context by quantitating the relationships between PU.1 occupancy and site quality and accessibility as PU.1 levels naturally declined in pro-T-cell development and by using stage-specific gain- and loss-of-function perturbations to relate binding to effects on target genes. PU.1 could bind closed genomic sites, but rapidly opened many of them, despite the absence of its frequent collaborator, CEBPA. RUNX motifs and RUNX1 binding were often linked to PU.1 at open sites, but highly expressed PU.1 could bind its sites without RUNX1. The dynamic properties of PU.1 engagements implied that PU.1 binding affinity and concentration determine its occupancy choices, but with quantitative trade-offs for occupancy between site sequence quality and stage-dependent site accessibility in chromatin. At nonpromoter sites, PU.1 binding criteria were more stringent than at promoters, and PU.1 was also much more effective as a transcriptional regulator at nonpromoter sites where local chromatin accessibility depended on the presence of PU.1. Notably, closed chromatin presented a qualitative barrier to occupancy by the PU.1 DNA-binding domain alone. Thus, effective pioneering at closed chromatin sites also depends on requirements beyond site recognition, served by non-DNA-binding domains of PU.1.
In development, a cell's regulatory history determines different preexisting chromatin accessibility landscapes, which in turn may bias a transcription factor (TF) to engage different sites in different contexts (Treiber et al. 2010). However, questions remain about how much of a barrier chromatin state may pose, or whether there is a clear distinction between transcription factors that are highly chromatin-limited and other “pioneer” factors that might engage their sites impervious to chromatin state. SPI1 (also known as PU.1) works both to mediate developmental choices of blood progenitor cells and to serve the alternative developmental fates, associated with different epigenetic landscapes that emerge from these choices. Its role in T-cell development is confined to the early stages leading up to T-cell lineage commitment, where it occupies tens of thousands of genomic sites in uncommitted cells but disappears in a programmed way during commitment. This dynamic transition offers a rare window into the quantitative contributions of affinity, chromatin context, factor concentration, and developmental history as predictors of PU.1 function.
The best studied roles for PU.1 involve its sustained actions as a lineage determining transcription factor in myeloid, dendritic-cell, and B-cell development, where it can open closed chromatin and recruit other transcription factors including CEBPA, NF-κB complexes, and IRF4 or IRF8 (Escalante et al. 2002; Carotta et al. 2010; Heinz et al. 2010; Natoli et al. 2011; Ostuni et al. 2013; McAndrew et al. 2016; for review, see Gosselin and Glass 2014). In the macrophage context, PU.1 can act as a pioneer factor, capable of evicting nucleosomes (Barozzi et al. 2014), but collaboration with known partners also affects PU.1 site choices (Heinz et al. 2013). PU.1 is also crucial for multiple other hematopoietic lineages (Back et al. 2005; Iwasaki et al. 2005; Nutt et al. 2005) including the earliest stages of T-cell development (Champhekar et al. 2015). Although most mature T cells express no PU.1, early-stage pro-T cells maintain substantial PU.1 through multiple cell divisions before down-regulating it during T-cell lineage commitment (Yui et al. 2010; Zhang et al. 2012). CEBP or IRF family factors that PU.1 cooperates with in myeloid and B cells are expressed little if at all in early T-lineage cells, which should affect PU.1 genome-wide binding profiles (Heinz et al. 2013). Yet its potential for pioneering activity in other cell types, enhanced by its unusual protein stability (Kueh et al. 2013), could make it a strong contributor to “epigenetic memory” in early pro-T cells. Here, we have investigated how PU.1 sets and responds to the chromatin context in which the T-cell program emerges.
PU.1 expression is well established in multilineage lymphoid precursors before they enter the thymus. Within the thymus, pro-T cells proliferate and differentiate through stages called DN1, DN2a, DN2b, and DN3a before they express T-cell receptor proteins. They become committed to the T-cell fate only during the DN2a to DN2b transition (Fig. 1A), which follows multiple cell divisions under the influence of thymic Notch pathway signaling (for review, see Yui and Rothenberg 2014). Commitment encompasses a global shift in 3D chromatin associations (Hu et al. 2018) and in regulatory gene expression (Rothenberg et al. 2016), and this is when PU.1 expression is finally shut down. Until commitment, PU.1 binds to more than 30,000 sites in DN1 and DN2a pro-T cells, distinct from sites occupied in B and myeloid lineage cells (Zhang et al. 2012). It supports proliferation and restrains specific alternative lineage genes, but slows progression toward commitment, thus enforcing correct timing of access to T-lineage genes (Champhekar et al. 2015). Previously, we identified impacts of PU.1 on specific, developmentally relevant genes by acute gain- and loss-of-function perturbations (Anderson et al. 2002; Dionne et al. 2005; Franco et al. 2006; Del Real and Rothenberg 2013; Champhekar et al. 2015). PU.1 repressed as well as activated genes (Dionne et al. 2005; Franco et al. 2006; Del Real and Rothenberg 2013), antagonizing Notch signaling in early T-cell development and delaying activation of many T-cell genes until commitment. However, PU.1 binding sites globally appeared to be associated with active chromatin and actively expressed genes (Zhang et al. 2012), leaving the repression unexplained. Furthermore, most genes linked to PU.1 occupancy in vivo do not change expression as PU.1 levels decrease in T-cell development (Zhang et al. 2012), implying that most PU.1 binding events do not control transcription. Here, therefore, we have used gain- and loss-of-function perturbations coupled with genome-wide analyses of the changes they induce in binding, chromatin state, and transcription of developmentally significant genes to show how PU.1 selects the sites that it will occupy in changing chromatin contexts, respecting or reshaping local chromatin states, and how its binding at certain classes of sites does control early T-cell gene expression.
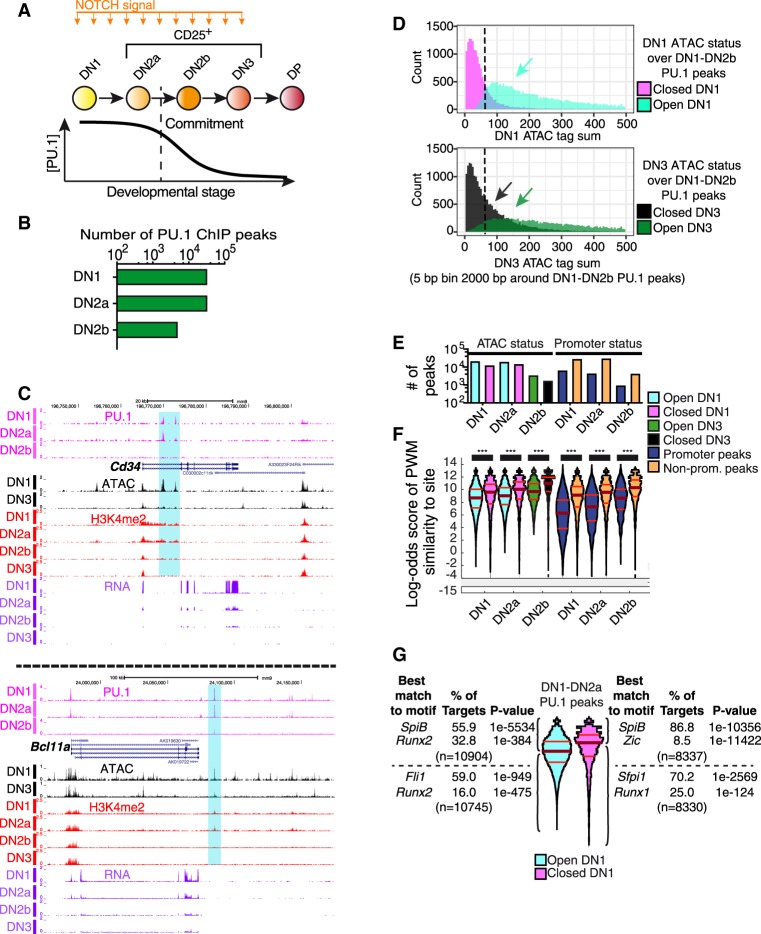
Figure 1.
Endogenous PU.1 binding in pro-T cells: distinct affinity thresholds for binding at open and closed sites and promoter and nonpromoter sites. (A) Outline of early T-cell development with schematic depicting PU.1 expression levels. (DP) A later stage. (B) Number of endogenous PU.1-occupancy peaks detected in DN1, DN2a, and DN2b pro-T cells. (C) Cd34 and Bcl11a UCSC Genome Browser tracks (http://genome.ucsc.edu) showing endogenous PU.1 ChIP, ATAC-seq, H3K4me2 ChIP, and RNA-seq in the DN1, DN2a, DN2b, and/or DN3 stages. Samples from in vitro differentiation from fetal liver precursors or from thymus (ATAC, DN1, DN3). Data from GSE31235; GSE93755. (D) Histogram of ATAC tag counts at DN1 (top) or DN3 (bottom) stages across all pro-T-cell PU.1 binding sites. Sites in regions defined as ATAC “open” or “closed” (Methods) are plotted separately to aid visualization. (Y-axes) Number of sites at indicated ATAC signal level. (E) Number of PU.1 peaks in indicated stages in ATAC-open and ATAC-closed as well as promoter and nonpromoter regions. The same color key is used in E and F. (F) Distribution of PU.1 motif log-odds scores at binding sites in open (cyan: open in DN1; green: open in DN3) and closed regions (magenta: closed in DN1; black: closed in DN3), and in promoters (dark blue) and nonpromoter (orange) elements. Scores from a DN1-DN2b-derived PU.1 PWM-matrix (Supplemental Table S1). Kruskal–Wallis statistical test: (***) P ≤ 0.0001. (G) Motif analyses of PU.1 sites in DN1-DN2a cells, classified by PWM scoring and ATAC accessibility in DN1.
Results
Dynamic PU.1 occupancy at open and closed chromatin sites in precommitment pro-T cells
In early, precommitment T-cell development (Fig. 1A), PU.1 occupancy starts at approximately 30,000 genomic sites in DN1 and DN2a stages, globally distinct from the PU.1 sites in non-T-lineage cells, then decreases to a subset of about 5000 scorable sites in newly committed DN2b cells (Fig. 1B), before PU.1 disappears at later stages (Zhang et al. 2012). To evaluate how PU.1 might select these sites, we examined whether chromatin status at the sites and the binding specificity of the protein itself could determine occupancy preferences. We used a sequence-agnostic criterion of regulatory site activity during the natural decrease of PU.1 during commitment (from DN1 and DN2a to DN2b and DN3). Activity states of chromatin were mapped at high resolution as changes in DNA accessibility (“openness”) using Analysis of Transposase-Accessible Chromatin (ATAC)-seq (Buenrostro et al. 2013). Figure 1C shows examples of PU.1 occupancy patterns at the Cd34 and Bcl11a target genes, which are most highly expressed in precommitment DN1 cells, correlated with ATAC status of the relevant sites before (DN1) and just after commitment (DN3). Although “permissive” H3K4me2 chromatin modifications change little across these stages in general (Zhang et al. 2012), these loci are linked with specific sites where developmental loss of PU.1 binding during commitment is accompanied by losses of ATAC accessibility and H3K4me2 (Fig. 1C, highlighted regions). Considering all sites genome-wide where PU.1 bound during any DN1-DN2b stage, ATAC criteria showed that more of these sites were likely to be “open” in DN1 stage (Fig. 1D, cyan) than in DN3 stage, when PU.1 levels were low (Fig. 1D, dark green). This suggests that PU.1 either causes these sites to open or is preferentially recruited to them because of their accessibility.
Changes in ATAC status of PU.1 sites between DN1 and DN3 stage were strongly correlated with changes in expression of the closest linked gene. As shown by an empirical cumulative distribution frequency (ECDF) plot (Supplemental Fig. S1A), genes linked to PU.1 sites that lost accessibility during commitment (“closing sites”) were more likely to be down-regulated (DN3/DN2a ratio <1, adjusted P-value [P.adj] <0.1, blue arrow) than genes without PU.1 sites. Accordingly, genes linked to sites that increased accessibility (“opening sites”) were more likely to be up-regulated (DN3/DN2a ratio >1, P.adj <0.1, red arrow). Taken together with the enrichment of open regions among all PU.1-occupied sites, this suggests that PU.1 could positively regulate hundreds of genes specific to precommitment cells. However, PU.1 binding across the genome was not confined to open sites (Fig. 1D,E). In DN1 and DN2a cells, PU.1 occupied similar numbers of closed and open sites (Fig. 1E). Therefore, PU.1 does not simply engage sites epigenetically primed by other factors, nor does its binding always cause chromatin opening as scored by ATAC-seq.
Distinct PU.1 affinity thresholds in open and closed chromatin, and at promoter and nonpromoter sites
In macrophages, PU.1 is a pioneer factor capable of displacing nucleosomes from its binding sites (Barozzi et al. 2014). In pro-T cells, where PU.1 expression is inherited from prethymic precursors but its best-characterized partner factors are absent, it was not clear whether PU.1 led or followed other factors to select its sites. The definition of pioneering (Zaret and Carroll 2011) implies that the DNA binding specificity of the factor itself guides its binding to sites in initially closed chromatin. We tested whether PU.1 intrinsic specificity guides its binding to “open” and “closed” sites by scoring PU.1 occupancy sites genome-wide for log-odds match to an optimal position weight matrix (PWM), calculated de novo (Supplemental Table S1; Supplemental Fig. S1B) from its pro-T-cell binding sites (Zhang et al. 2012). The scoring pattern of sites across the genome (Fig. 1E,F) agreed well with the pattern scored using a more permissive PWM, calculated from PU.1 sites occupied in macrophages, which was reported to reflect PU.1 binding preferences on naked DNA (Supplemental Fig. S1B,C; Pham et al. 2013). The canonical PU.1 motif was the most enriched at all classes of sites (Supplemental Fig. S1D). However, at each stage, PU.1-occupied sites in open regions had a broader range of quality scores, with lower median and quartile quality scores, than the PU.1-bound sites in closed regions, implying that higher affinity was needed to establish binding at closed sites. The “closed” PU.1 binding sites accordingly showed a greater enrichment of a PU.1 subfamily (Sfpi1, SpiB) ETS motif than the “open” sites (Fig. 1G; Supplemental Fig. S1D). On the other hand, open sites and the lower-quality half of the closed sites were distinguished from higher-quality closed sites by their high coenrichment of RUNX family motifs (Fig. 1G; Supplemental Fig. S1D).
The changes in PU.1 occupancy across the genome as PU.1 levels naturally decrease in development implied that PU.1 binding site selection is governed by its own mass action. As PU.1 tag counts at individual sites and the number of scorable binding regions genome-wide both dropped during commitment (Fig. 1B), the sites still occupied were those with higher motif quality (Fig. 1F; Supplemental Fig. S1C,E). However, distinct binding criteria applied in open and closed chromatin regions. Both open and closed regions lost PU.1 occupancy at DN2b stage (Fig. 1E, left), but the site quality difference between them persisted where PU.1 remained bound (Fig. 1F, left; Supplemental Fig. S1E), consistent with a continuing affinity penalty in the closed regions. Matching site quality distributions at “closed” DN2a and “open” DN2b sites suggested that closure reduced PU.1 sensitivity by 4× to 5×. In the same cell nuclei, declining PU.1 levels also differentially affected binding at promoter and nonpromoter (distal) sites. Promoter sites were occupied even when they had poor-quality target motifs (Fig. 1F, right), and when PU.1 fell to lower levels, the promoter sites still occupied were poorer quality than nonpromoter sites. This global difference in binding criteria at promoter and nonpromoter sites was seen at all stages, and even when open and closed promoter and nonpromoter sites were compared separately (Supplemental Fig. S1E). Thus, PU.1 is recruited differently to promoters and enhancer elements.
Nonpromoter PU.1 sites control chromatin configuration and function
Genome-wide PU.1 binding dynamics were correlated with changes in local chromatin status. Figure 2, A and B, shows the ∼40K sites of PU.1 occupancy in the pro-T genome, clustered based on PU.1 binding, ATAC accessibility, and H3K4me2 modification status from DN1 to DN2b or DN3 stages. The majority of sites fell into either of two major groups (Group 1 and Group 2; k-means) (Fig. 2A). One large group of sites comprised open sites that stayed highly accessible, independent of developmental stage or level of PU.1 binding (site Group 1, “static”) (Fig. 2A,B). In contrast, ∼60% of PU.1 occupancy sites in DN1 and DN2a cells lost accessibility, and sometimes even the “permissive” histone mark H3K4me2, as PU.1 expression declined in DN2b/DN3 stages (site Group 2, “closing”) (Fig. 2A,B). A few sites also “opened” as PU.1 levels declined (see below), but did not cluster efficiently here. The static sites (Group 1) were highly enriched for annotated promoters (39.8% in promoter regions; χ2 test P < 0.0001), whereas the closing sites (Group 2) were overwhelmingly in nonpromoter elements (only 4.6% peaks in promoter regions). Thus, the dominant division of PU.1 occupancies was into sites with constitutive accessibility and sites whose accessibility decreased in parallel with PU.1 expression itself.
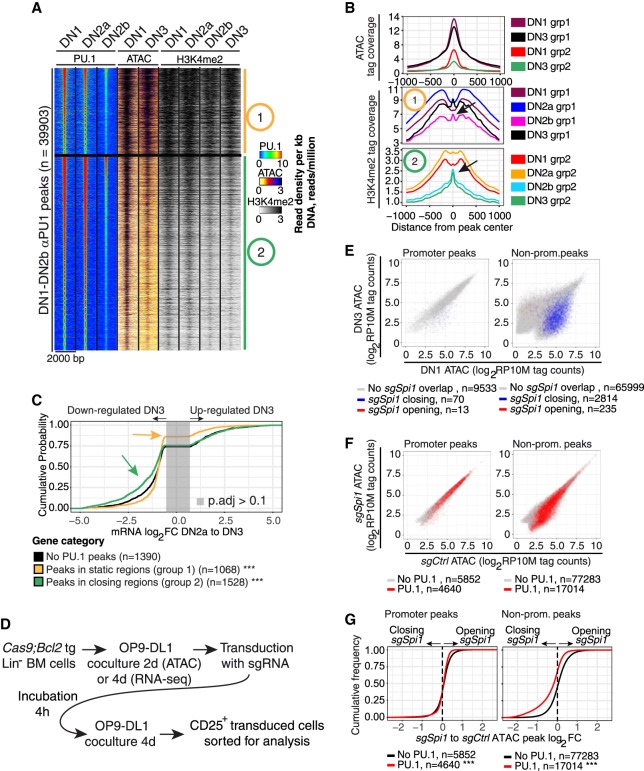
Figure 2.
PU.1 loss reduces chromatin accessibility at nonpromoter sites. (A) Heatmap of PU.1 and ATAC tag count distribution in early DN-pro-T development. Regions 2-kb wide were k-means clustered (k = 2) based on PU.1 binding, ATAC, and H3K4me2 patterns. Group 1: constitutively open sites. Group 2: sites losing accessibility from DN1 to DN3. (B) ATAC and H3K4me2 distributions at the indicated stages at sites in Groups 1 and 2. Arrows in the H3K4me2 plot: nucleosome reconstitution after PU.1 loss. (C) Natural changes in gene expression across commitment (DN2a to DN3, genes changing with P.adj < 0.1) linked to Group 1 (static) and 2 (closing) sites: (***) Kolmogorov–Smirnov P-value ≤0.0001 relative to genes without PU.1 peaks. (D) Schematic of protocol for CAS9-mediated disruption of PU.1 using transduction with sgSpi1 or control guide RNAs. (E) Effects of disruption of PU.1 in DN2 cells on accessibility of promoter and nonpromoter sites: (x-axis) ATAC accessibility of individual sites in DN1 stage; (y-axis) accessibility in DN3 stage; (blue) sites that lose accessibility upon PU.1 deletion in DN2 cells; (red) sites that gain accessibility (rare). (F) Enrichment of PU.1-binding sites among genomic sites that lose ATAC accessibility upon PU.1 deletion. ATAC accessibility of genomic sites after PU.1 deletion (y-axis) is plotted against their accessibility in controls (x-axis): (red) sites overlapping with PU.1 binding in DN1, DN2a, and/or DN2b cells. (G) Effects of PU.1 knockout on accessibility of promoter and nonpromoter PU.1 binding sites: (***) Kolmogorov–Smirnov P-value ≤0.0001.
PU.1 binding at nonpromoter sites, but not at promoters, was correlated with gene expression specifically before commitment. Genes linked only to Group 1 binding sites changed little in expression between DN2a (high PU.1) and DN3 (low PU.1) stages (Fig. 2C), even less (yellow arrow, P < 0.0001) than genes without any PU.1 binding peaks at all (Fig. 2C, black curve). In contrast, genes linked to Group 2 PU.1 sites (Fig. 2C, green) changed expression much more than genes without peaks, most of them losing expression in this developmental transition (green arrow, P < 0.0001, skew to left), despite similar kinetics of retention of PU.1 between Group 1 and Group 2 sites (Supplemental Fig. S1F). These results suggest that PU.1 binding in pro-T cells is more stringent, more important for chromatin site accessibility, and better correlated with transcription when it binds at nonpromoter regions than at promoters.
To test whether PU.1 itself was responsible for the dynamics of ATAC accessibility at these nonpromoter sites, we exploited the Cas9 transgenic mouse strain (Platt et al. 2014) to provide T-cell precursors in which PU.1 could be acutely deleted at a specific developmental stage (Fig. 2D; Hosokawa et al. 2018). Hematopoietic precursors from these mice were isolated and induced to begin T-lineage differentiation in vitro by coculture with OP9-DL1 stromal cells. When PU.1 was at its height during DN1 to DN2a stages, specific guide RNAs were introduced by retroviral transduction (sgSpi1 or sgControl), and effects on chromatin accessibility were assayed by ATAC-seq after four more days of culture. This deletes targeted exons biallelically (Supplemental Fig. S1G) and removes endogenous PU.1 expression (Hosokawa et al. 2018). Although promoter sites were unaffected when PU.1 was deleted, many nonpromoter sites specifically lost accessibility, far more than gained accessibility, and these affected sites were normally open in DN1 but not in DN3 stage (Fig. 2E). The sites with PU.1-dependent accessibility were indeed direct targets, for they were also significantly enriched among distal sites that normally bound PU.1 in control DN1 cells (P ≤ 0.0001) (Fig. 2F,G). The whole accessibility distribution of PU.1 binding sites in nonpromoter regions shifted toward closure when Spi1 was disrupted (Fig. 2G, right). In contrast, PU.1 deletion had little if any effect on promoter sites, with or without PU.1 binding (Fig. 2E–G). Thus, sustained PU.1 activity is needed to maintain ATAC accessibility at many nonpromoter PU.1 binding sites.
PU.1 can bind its target sites independently of RUNX1
The motifs most highly coenriched with PU.1 motifs were for RUNX factors. In a related study, we showed that PU.1 physically interacts with RUNX1 in pro-T cells and recruits it to PU.1 binding sites (Hosokawa et al. 2018). Supplemental Figure S2A shows that a large fraction of endogenous PU.1 occupancy sites that were open in primary DN1 cells were in fact co-occupied by RUNX1 (data from Hosokawa et al. 2018), with highest RUNX1 binding enrichment among those sites with lower-quality PU.1 motifs (below median PWM score). RUNX1 co-occupancy was much lower at PU.1 sites in closed regions, consistent with motif enrichments (Fig. 1G; Supplemental Fig. S1D). These results raised the question of how much RUNX1 was contributing to the ability of PU.1 to establish occupancy at different classes of sites.
Whereas primary cell recovery is severely inhibited by combined deletion of Runx1 and forced expression of PU.1, we could test the effect of RUNX1 loss on binding of exogenous PU.1 in the Scid.adh.2C2 model cell system. These are immortal, clonal DN3-like cells that do not express endogenous PU.1, but respond sensitively to introduction of exogenous PU.1 with responses similar to those of primary pro-T cells (Fig. 3A; Dionne et al. 2005; Del Real and Rothenberg 2013). Scid.adh.2C2 cells were transduced with Cas9 (Cas9-GFP) followed by either control sgRNA (sgCtrl) or sgRNA against Runx1 (sgRunx1), and then transduced with exogenous PU.1 (Fig. 3B). This treatment biallelically disrupted the region around the first Met codon shared by all RUNX1 isoforms (Supplemental Fig. S2B) and eliminated RUNX1 protein expression (Hosokawa et al. 2018). Figure 3C shows how this affected PU.1 binding to nonpromoter sites. The sites bound by exogenous PU.1 were more likely to be initially open in Scid.adh.2C2 cells if near sites where RUNX1 bound previously (Hosokawa et al. 2018) (tallied below plots in Fig. 3C). However, high-quality PU.1 binding to these nonpromoter sites was essentially unchanged by Runx1 disruption, whether they had been previous sites of RUNX1 occupancy (Fig. 3C, right panel) or not (left panel). Thus, although RUNX1 can be important for PU.1 function in pro-T cells (Hosokawa et al. 2018), PU.1 does not depend on RUNX1 to establish its binding at open or closed sites.
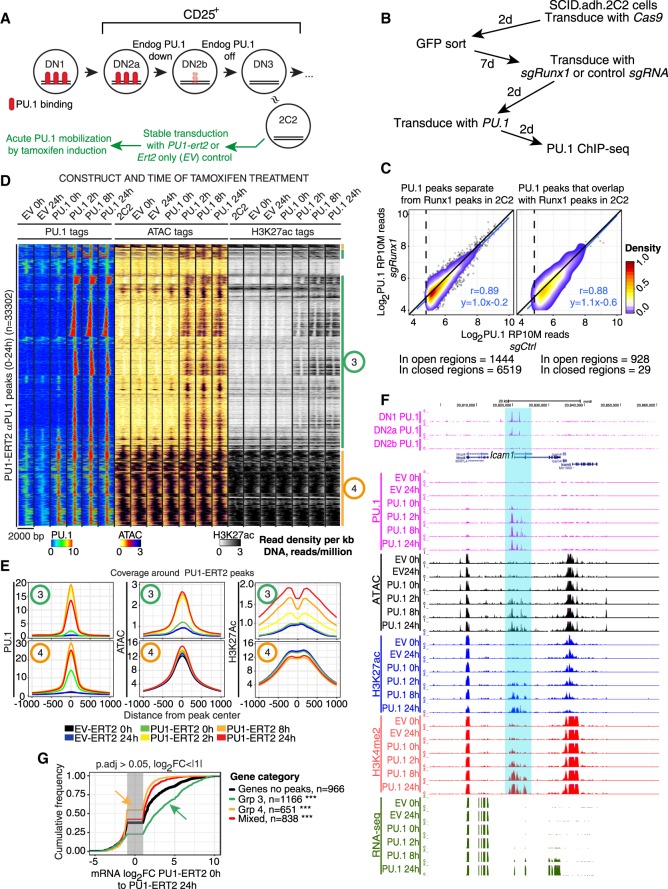
Figure 3.
Induction of PU.1 activity leads to chromatin opening, altered histone modifications, and cis-regulatory element-dependent gene activation. (A) Schematic showing approximate relationship of Scid.adh.2C2 cells to normal program of T-cell development, with experimental plan for panels D–G. (B) Schematic of test in Scid.adh.2C2 cells to determine whether exogenous PU.1 site binding is RUNX1-dependent. (C) Results of protocol shown in B. PU.1 occupancy scores for PU.1 binding in cells after Runx1 disruption (y-axis) are plotted against scores for PU.1 binding in controls (detected with αPU.1). Sites are stratified according to their ATAC accessibility and RUNX1 occupancy in unmanipulated Scid.adh.2C2 cells (Hosokawa et al. 2018). Pearson's r and linear trend lines are shown for peaks with a peak score (Homer findPeaks) greater than 30 in controls. (D) Time courses of PU.1 binding, induced changes in chromatin accessibility, and histone H3K27 acetylation after 0–24 h of mobilization by 4-hydroxytamoxifen (4-OHT), in Scid.adh.2C2 stably transduced to express PU1-ERT2 or EV-ERT2. Sites in heatmap were hierarchically clustered on Pearson correlation with average linkage. Shown are manually derived groups of sites that open and gain H3K27ac upon PU1-ERT2 mobilization (Group 3) or that are already open and associated with H3K27ac at 0 h (Group 4). (E) Quantitative distribution plots of PU.1, ATAC, and H3K27ac signals within 1 kb of Groups 3 and 4 PU.1 bound sites. (F) PU.1 binding to the positively regulated Icam1 gene in DN1-DN2b cells (top) and samples from the time course shown in D. Highlight indicates chromatin regions opened by PU.1 binding prior to RNA expression. (G) Association of PU1-ERT2 binding site Groups 3 and 4 with changes in linked gene expression between 0 and 24 h of 4-OHT induction: (***) Kolmogorov–Smirnov P-value ≤0.0001.
Pioneering by PU.1 in DN3-like cells: site choice and chromatin opening in a closed epigenetic landscape
PU.1 not only maintained the open status of its normal binding sites but could also cause sites to open in closed chromatin, as shown using the Scid.adh.2C2 cell line to provide a defined, low-background pro-T-cell context. To place PU.1 activity under tight temporal control, Scid.adh.2C2 cells were transduced stably with a vector encoding full-length PU.1 linked to the ligand-binding domain of a tamoxifen-dependent estrogen receptor (PU1-ERT2) (Fig. 3A). Most of the PU.1 expressed in these cells is tethered in the cytoplasm, but the PU.1 can translocate quickly to the nucleus in response to 4-hydroxytamoxifen (4-OHT). This down-regulates the pro-T-cell surface marker IL2RA (also known as CD25) beginning by 24 h (Supplemental Fig. S2C), and up-regulates the myeloid-cell surface marker ITGAM (also known as CD11b) later (Dionne et al. 2005; Del Real and Rothenberg 2013). From 0 to 24 h of stimulation with 0.1 µM 4-OHT, we compared the kinetics of PU.1 binding to the genome with changes in H3K27ac and ATAC accessibility at these sites (Fig. 3D,E; Supplemental Fig. S2D). Baselines were defined by control cells transduced with an “empty” control vector encoding ERT2 only (“EV”, “EV-ERT2”) (Fig. 3B,C; for gene expression effects, see Supplemental Table S2). Two site groups were seen (Groups 3 and 4). There was some background signal at 0 h: In Scid.adh.2C2 cells stably expressing PU1-ERT2, low levels of PU1-ERT2 reached the nucleus even without 4-OHT, unlike EV-ERT2 samples (Fig. 3D; Supplemental Fig. S2D,E). Sites occupied in the presence of these low levels of nuclear PU.1 were restricted to already-active chromatin sites, based on ATAC accessibility and H3K27ac in nontransduced or EV-transduced cells (Fig. 3D, Group 4; Supplemental Fig. S2D,E). However, 4-OHT treatment induced nearly 5× more PU.1 binding sites within 2 h (Supplemental Fig. S2D,E), and most of this new binding was to Group 3 sites (Fig. 3D) that had no PU1-ERT2 binding at 0 h and were closed in unperturbed Scid.adh.2C2 cells (Fig. 3E; Supplemental Fig. S2D,E). Examples are the highlighted sites in the Icam1 gene, which were also bound in DN1-DN2a cells by endogenous PU.1 (Fig. 3F). Like Group 1 sites of endogenous PU.1 in primary cells (Fig. 2A), 51% of Group 4 sites were at promoters, contrasting with only 3.1% of Group 3 sites (χ2 P-value <0.0001), and the PWM match quality for Group 3 sites was much higher than for Group 4 sites (Supplemental Fig. S2F). Binding to Group 3-type sites was confirmed with another exogenous PU.1 construct, PU1WTHA (see below), which preferentially occupied “closed” sites in these cells unlike “open” sites bound by endogenous ETS1 (Supplemental Fig. S2G).
In this cell context, PU1-ERT2 exerted its main positive impact at closed, nonpromoter Group 3 sites, rapidly making them ATAC accessible. This response was maximal already within 2 h (Fig. 3D,E; Supplemental Fig. S2D). Slightly later, ∼50% of the Group 3 sites also acquired the active histone mark H3K27ac de novo, beginning at 2 h but with most signal observed by 8–24 h of PU.1 binding (Fig. 3D,E; Supplemental Fig. S2D). Genome-wide, genes responding to PU1-ERT2 binding at linked Group 3 sites also usually increased expression over the 24 h period (Fig. 3G, green). In contrast, those linked only to binding at Group 4 sites showed even less increase than genes without closely linked PU.1 sites at all (Fig. 3G, yellow).
Many functionally important PU.1 sites were bound immediately: The sites occupied at 2 h were as likely to predict expression of linked genes at 24 h as sites that were occupied only at 8 or 24 h (Supplemental Fig. S3A). However, the kinetics of transcriptional response implied a stepwise process. The pattern of expression of DEGs closely linked to new PU1-ERT2 sites began to change from 2 h onward (Supplemental Fig. S3B), but only about one-third of the genes that were eventually activated showed up-regulation by 8 h, despite maximal ATAC accessibility already; most were only activated by 24 h (all P < 10−9). Thus, PU.1 binding rapidly initiated local ATAC accessibility in previously closed chromatin sites, enabling further steps resulting in local target gene activation, consistent with pioneering activity.
Whereas binding of PU.1 to nonpromoter sites most often exerted positive effects, if any, on linked genes, PU.1 introduction also repressed many genes in these cells (Dionne et al. 2005; Del Real and Rothenberg 2013). Supplemental Figure S3B shows that such repression could start early, often before most positive target genes were up-regulated (2 h versus 8 h) (Supplemental Table S2). Although much of this repression was likely indirect (Del Real and Rothenberg 2013; Champhekar et al. 2015; Hosokawa et al. 2018), some was accompanied by direct binding. For example, Il7r was substantially repressed as PU.1 bound to an open Group 4 site at its promoter (Supplemental Fig. S3C).
Opening of closed chromatin by PU.1 mediates its developmental gene regulatory function
Data in Figures 1 and 2 have shown that PU.1 binding in steady state is correlated both with chromatin openness and with local gene expression, but correlation alone could not prove that PU.1 normally regulates these genes. Data in Figure 3 show that PU.1 could induce chromatin opening and gene activation when introduced at high levels in a PU.1-negative cell-line context. To confirm whether PU.1 serves its normal function in pro-T cells through opening chromatin around its nonpromoter binding sites, acute perturbations of PU.1 had to be carried out in a primary cell developmental context with dosage changes close to the normal range and with changes in its local binding tested for direct linkage to genes showing changes in expression. The evidence is detailed in Supplemental Figures S4 and S5, Supplemental Tables S3–S7, and summarized in Figure 4.
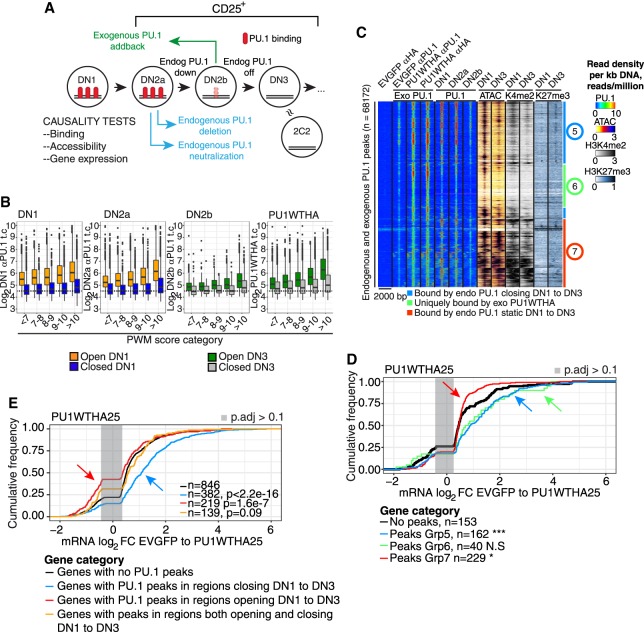
Figure 4.
Causality test: sites bound by exogenous PU.1 that mediate function in pro-T cells. (A) Schematic of experiments shown in this figure and Supplemental Figures S4 and S5: acute gain and loss of function of PU.1 in developing primary pro-T cells. CD25+ cells (DN2 to DN3) are cells within the T-cell pathway. (B) Comparison of site occupancies by endogenous PU.1 (DN1-DN2b, detected by αPU.1) and by exogenous PU.1 (PU1WTHA, detected by αHA) as a function of PWM score, at “open” and “closed” sites. (C) Heatmap of binding of exogenous PU1WTHA in CD25+ DN2b/DN3 primary cells (Exo PU.1) compared with endogenous PU.1 in DN1, DN2a, and DN2b, ATAC-seq in DN1 and DN3, and H3K4me2 and H3K27me3 in DN1 and DN3 cells. Color scales: tag count densities. (D) Association of Groups 5, 6, and 7 sites with changes in linked gene expression induced by exogenous PU1WTHA in CD25+ cells. (***) Kolmogorov–Smirnov P-value ≤0.0001; (*) P-value ≤0.05. (E) Changes in gene expression induced by acute PU1WTHA introduction in genes linked to sites that would normally close (blue) or open (red) from DN1 to DN3 as compared to genes with no PU1WTHA peaks. Genes with a mixture of opening and closing peaks are also shown. Genes are those with significant differential expression induced in cells remaining CD25+.
Briefly, we determined the RNAs changing expression after acute gains of PU.1 function at the DN2b/DN3 stages, when endogenous activity was diminishing, and reciprocally, after losses of PU.1 function (Figs. 2D–F, 4A). We introduced epitope-tagged exogenous PU.1 (PU1WTHA) so that changes in transcription could be directly attributed to local binding of the exogenous PU.1 constructs. Supplemental Figure S4, A and B, shows the cellular features of this system. Exogenous PU1WTHA established occupancy at sites with the same site quality spectrum that the endogenous PU.1 had occupied earlier (Fig. 4B). Supplemental Figure S4, C–F, shows the patterns of gene expression change that resulted and their relationship both to normal pro-T development (Supplemental Tables S3, S4) and to likely physiological target genes affected by acute Spi1 deletion (Supplemental Tables S5, S6). Supplemental Figure S4, E–G, shows the criteria used to focus on cells affected by PU.1 within the normal pro-T developmental trajectory (“PU1WTHA25”), minimizing second-order effects. The exogenous Spi1 in these experiments was expressed at about 3× to 4× its endogenous DN1-cell level (Supplemental Table S3). Exogenous PU.1 in these cells altered gene expression in rough agreement with normal developmental trends of the affected genes (Supplemental Fig. S4H), and especially well in a curated “developmental index” gene set (Supplemental Fig. S4I; Supplemental Table S4). Most genes affected in PU1WTHA25 cells were up-regulated, especially those with direct PU.1 binding (Supplemental Fig. S4J).
PU1WTHA established occupancy at most sites that endogenous PU.1 had occupied before (“Exo PU.1” versus “PU.1”) (Fig. 4C) at both nonpromoter (Group 5) and promoter-enriched (Group 7) sites, as well as binding a group of sites that were normally not occupied by endogenous PU.1 (Group 6). The Group 5 and Group 7 sites respectively resembled Group 2 and Group 1 sites of endogenous PU.1 binding in chromatin dynamics and site qualities (Fig. 4C; Supplemental Fig. S5A,B). Group 6 sites had high-quality motifs but were normally closed and not bound by endogenous PU.1 (Supplemental Fig. S5A,B; Supplemental Table S7). However, the genes up-regulated in PU1WTHA25 cells were enriched not only for Group 5 sites but also for Group 6 sites (Fig. 4D; Supplemental Fig. S5C), i.e., where default chromatin states without PU.1WTHA would have been closing or closed. PU1WTHA binding to promoter-enriched Group 7 sites again resulted in minimal change.
Globally, the dynamic status of site accessibility in normal development predicted the direction of response of a linked gene (Supplemental Fig. S5D–F): PU1WTHA tended to activate genes linked to sites that normally closed as endogenous PU.1 declines (“closing sites”, Kolmogorov–Smirnov P < 2.2 × 10−16) (Fig. 4E, blue arrow), whereas it had little or negative effect on genes linked to sites that would normally open as endogenous PU.1 declines (“regions opening” ) (Fig. 4E, red arrow). Thus, acute transcriptional changes caused by PU.1 in pro-T cells were tightly linked with its engagement at nonpromoter sites where chromatin accessibility normally follows PU.1 expression. This supports a normal mechanism of PU.1 action via chromatin accessibility.
PU.1 requires its non-DNA-binding domains to occupy sites in closed chromatin
To assess the domains needed for pioneering activity within the PU.1 protein, we mapped the genomic sites occupied by the isolated PU.1 DNA-binding domain alone (PU1ETSHA) as compared to full-length PU.1 (PU1WTHA). To relate this binding to function comparable to PU1WTHA, we also tested a dominant negative PU1ETSHA derivative, PU1ENGHA (constructs diagrammed in Supplemental Fig. S6A), which primarily repressed targets rather than activating them (Supplemental Fig. S6B). Both truncated forms bound to DNA with site quality criteria similar to those of PU1WTHA (Supplemental Fig. S6C), and in primary pro-T cells, PU1ENGHA exerted its effects preferentially on sites that would normally close from DN1 to DN3 (Supplemental Fig. S6D), like sites preferred for function by PU1WTHA (cf. Fig. 4E). However, when PU1WTHA, PU1ETSHA, and PU1ENGHA were introduced into Scid.adh.2C2 cells, both truncated constructs bound quite differently from full-length PU.1 (Supplemental Fig. S6E,F), with much greater biases to promoter sites and reduced binding at nonpromoter sites. Thus, whereas PU1WTHA bound both to normal sites of PU.1 occupancy in pro-T cells and some additional ones, the truncated constructs in these cells bound well only to a subset of natural PU.1 sites (Supplemental Fig. S6G, Group B).
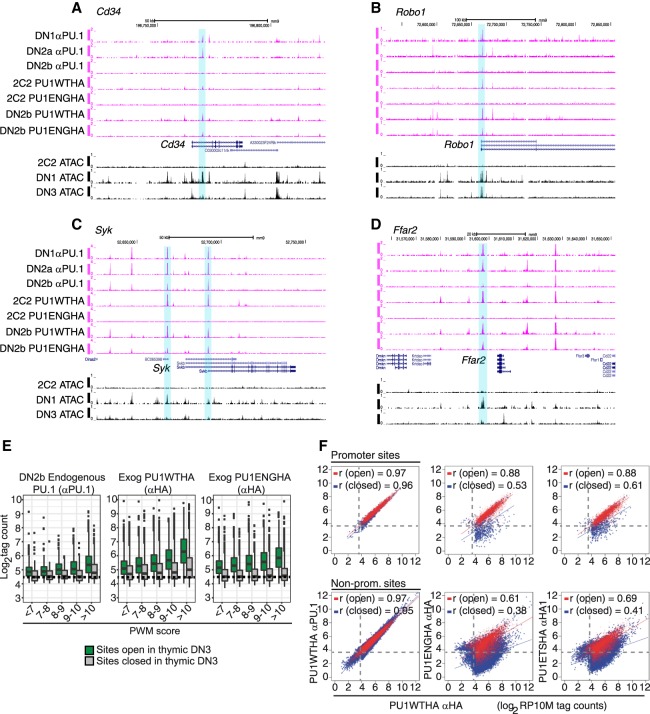
The sites that were poorly bound by PU1ETSHA and PU1ENGHA in Scid.adh.2C2 cells were not poor quality in sequence, for in primary pro-T cells these sites (Supplemental Fig. S6G,H, Group A sites) could be bound by PU1ENGHA as well as the sites that were accessible in the Scid.adh.2C2 cells (Group B sites). In primary cells, also, the Group A sites were even better at mediating the repressive effect of PU1ENGHA (Supplemental Fig. S6I) than the Group B sites, which were constitutively open and promoter-enriched. The difference, however, was that Group A sites were in much less accessible chromatin in Scid.adh.2C2 cells (Supplemental Fig. S6G). Figure 5, A–D, shows Cd34, Robo1, Syk, and Ffar2, all positively regulated by full-length PU.1 and repressed by PU1ENGHA in primary cells (Supplemental Table S3), which exemplify this context-dependent differential binding (highlighted) at Group A-type sites. Supplementary Figure S7 shows additional examples: Cd44, Vav1, Elovl5, Flt3, Notch2, and Myd88.
Figure 5.
Full-length PU.1 is required for access to closed chromatin. (A–D) UCSC Genome Browser tracks comparing binding of endogenous PU.1, full-length exogenous PU1WTHA, and PU1ENGHA to open and closed regions in primary DN2b/3 CD25+ cells and Scid.adh.2C2 cells. Shown are the genes: (A) Cd34; (B) Robo1; (C) Syk; (D) Ffar2. (E) Comparison of PU1ENGHA binding with that of PU1WTHA and baseline binding of endogenous PU.1 in primary DN2b cells. Analysis as in Figure 4B. (Green bars) Open sites in primary DN2b-DN3 cells; (gray bars) closed sites; (dashed line) background. (F) Comparison of exogenous PU1WTHA with PU1ENGHA or PU1ETSHA binding in transduced Scid.adh.2C2 cells at open and closed promoter and nonpromoter sites. PU1WTHA αHA counts in open (red) and closed (blue) regions are plotted against PU1WTHA αPU.1 (left), PU1ENGHA αHA (middle), or PU1ETSHA αHA (right) tag counts in promoters (top three panels) or nonpromoter genomic sites (bottom three panels). Peaks at open promoters: n = 4498; peaks at closed promoters: n = 810; peaks at open nonpromoter elements: n = 8719; peaks at closed nonpromoter elements: n = 33158. Pearson's r shown. Dashed lines represent the tag threshold for peaks considered bound.
The failure of the isolated PU.1 DNA-binding domain (or its derivative) to interact with these sites in the context of the DN3-like cell line suggested that PU.1 without its native trans-activation and protein-interaction domains could be more sensitive to chromatin state for access to its binding sites than full-length PU.1. We tested whether the truncated constructs really have a qualitative requirement for open chromatin or whether they simply have lower affinity or reach lower effective protein concentrations. First, we compared binding success of PU1WTHA and PU1ENGHA in primary CD25+ cells at sites of different PWM quality scores with open versus closed ATAC status. Here, PU1ENGHA bound comparably to PU1WTHA at open sites (Fig. 5E). However, although PU1WTHA showed the expected affinity cost for entry into closed chromatin, PU1ENGHA showed much less binding at closed sites, even those with very high-quality motifs (Fig. 5E). Wild-type and truncated constructs also showed sharply different binding to target sites in SCID.adh.2C2 cells depending on whether they were ATAC-open or ATAC-closed. Figure 5F shows the occupancies of the same open and closed sites by PU1WTHA, PU1ENGHA, or PU1ETSHA, all compared to PU1WTHA (detected by a different antibody). In these graphs, identical binding falls on the diagonal, as seen when the same PU1WTHA was detected with αPU.1 or with αHA antibodies. Global DNA binding efficiency differences between a truncated construct and the full-length form should shift the whole trend line downward on the log/log plot or change its slope. Instead, Figure 5F shows that the truncated constructs gave a split pattern. Both truncated constructs closely matched PU1WTHA on open sites (red), but were specifically penalized, within the same cells, with lower binding slopes and high dispersions at closed sites (blue). Thus, the non-DNA-binding domains of PU.1 strongly and selectively affected the ability of the PU.1 DNA-binding domain to establish occupancy in closed chromatin.
Discussion
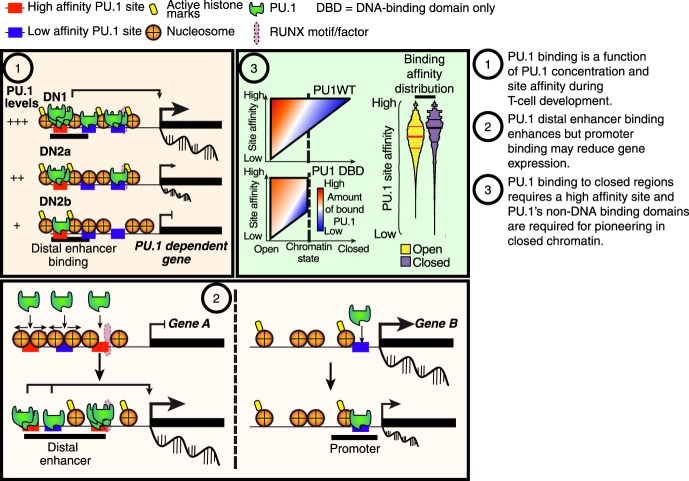
PU.1 is known to act as a pioneer in the role of a lineage-determining factor that initiates and sustains identity of myeloid cells (Natoli et al. 2011; Barozzi et al. 2014). In this study, we asked whether its inherited high activity in the earliest pro-T cells also shapes the epigenetic landscape in this context, although its activity in this lineage is hit and run and many of its known partners are not expressed. Criteria for pioneering involve the ability to open closed chromatin and to initiate occupancy of a cis-regulatory site by a complex of transcription factors (Zaret and Carroll 2011). Logically, this implies that pioneer factors can (1) find and bind to sites in closed chromatin; (2) bind to target sequences based on their own specificity, even before other factors are available to collaborate; and (3) initiate the opening of chromatin as new partners are recruited. In the myeloid system, PU.1 often plays these roles as part of a partnership with C/EBP family factors, in which either PU.1 or the C/EBP factor can help nucleate binding by the other (Laiosa et al. 2006; Feng et al. 2008; Heinz et al. 2010, 2013). As we show here, PU.1 also fulfills most of the criteria for pioneering in pro-T cells. It can bind closed chromatin as well as open. Its binding is primarily guided by its own well-defined specificity, although its binding to closed chromatin sites in these cells requires a consistently higher affinity of site recognition than binding to open chromatin. Its binding preferences as its concentrations drop during T-cell development are consistent with mass action in which the dissociation constant is dominated simply by PU.1's own specificity. Finally, it can trigger chromatin opening and is required continuously to maintain accessibility at many of its target sites. These functions are exerted especially at a major subset of its nonpromoter sites, where PU.1 can rapidly induce transposase accessibility followed by recruitment of histone acetyltransferases. It may cooperate with RUNX1 in causing chromatin to open and to activate its positive target genes (Hosokawa et al. 2018), but does not require RUNX1 for its own binding. This is summarized in Figure 6.
Figure 6.
Schematic of PU.1 site choice criteria and functional impacts in pro-T cells. Summary of mechanisms regulating PU.1 binding and function (Discussion).
ChIP-seq analysis, short-term forced expression, and acute synchronized CAS9-mediated deletion tests have robustly defined the positive regulatory targets of direct PU.1 binding in early pro-T cells. A core group of about 250 genes is positively regulated by PU.1 based on gain and loss of function in pro-T cells (Supplemental Table S6), and these genes are linked to nonpromoter sites of PU.1 binding that depend on PU.1 for ATAC accessibility and lose accessibility when PU.1 levels decline. We also identified a minority of negative regulation targets that are also likely to be direct. These high-confidence target genes of PU.1, both positive and negative, show direct binding, chromatin features at PU.1 sites, and developmental expression, all implying that normal endogenous PU.1 activity is a major controller of their site accessibilities during normal pro-T-cell development.
The high developmental resolution possible in this system sheds light on features of PU.1 action that have much broader implications. The first is that motif quality of sites, magnitudes of occupancy, promoter-association, and immediate degrees of ATAC accessibility are useful but not sufficient criteria to identify loci of actual PU.1 regulatory action. Promoters are by far the most accessible sites for PU.1 binding: Not only are they constitutively open as measured by ATAC-seq, but also they offer an exceptionally permissive affinity threshold for PU.1 engagement, and they do not require the PU.1 transactivation domains for efficient PU.1 binding. In myeloid cells, PU.1 is commonly bound at promoter-proximal regulatory elements of functional target genes (Gonzalez et al. 2015). However, although PU.1 is frequently bound to promoters of active genes in pro-T cells as well, in this cell context the actual functional impact of its activity at these promoters is often weak or null, unless supplemented by local binding to nonpromoter sites as well. Instead, it is those dynamic distal elements, even those with less-pronounced PU.1 binding, that are best associated with function.
PU.1 binding is excluded by H3K27me3-marked repressive chromatin (Zhang et al. 2012), but modestly elevated PU.1 can also enter other regions that appear “inaccessible” by criteria of ATAC-seq, and upon binding can mediate rapid chromatin opening and local gene activation. However, we have shown that the ability to establish binding in such closed sites depends on a function of PU.1 structurally distinct from its site recognition. As in the case of EBF1 (Boller et al. 2016), although the PU.1 DNA-binding domains define its nucleotide sequence specificity, the non-DNA-binding domains of PU.1 are required for its access to closed sites. Thus, the DNA-binding domain alone is not only deprived of discrete transactivation or protein interaction functions, but also of access to a large subset of potential genomic targets. This effect explains some of the functional specificity of the obligate-repressor PU1ENGHA form as previously reported (Champhekar et al. 2015), which can compete with endogenous PU.1 at open binding sites but cannot gain access to those sites once they have developmentally closed. The non-DNA-binding domains of PU.1 are important for its interactions with protein partners, at least including RUNX1 and SATB1 in early pro-T cells (Hosokawa et al. 2018), IRFs (Escalante et al. 2002), and factors important in erythroid development (Zhang et al. 2000; Stopka et al. 2005). Although RUNX1 itself may not explain this specific binding requirement, other interacting proteins (Hosokawa et al. 2018) remain as candidates to assist PU.1 binding in closed chromatin of pro-T cells. Thus, the full site-selectivity of PU.1 as a pioneer depends not only on its ability to find high-affinity sites, but also on the ability of its protein-interaction domains to convert recognition of sites in closed chromatin into stable, functional occupancy.
Thus, despite PU.1's fulfillment of simple criteria of pioneer function, its impact on early T-lineage cells is determined by the intersection of its own powerful binding activity and its dynamic interaction with other determinants of epigenetic state. The effect of closed chromatin on full-length PU.1 appears to be quantitatively compensated by the higher binding affinities at nearer-optimal sites, but the transcriptional effect of this PU.1 binding on neighboring genes depends on other factors that influence whether the site bound is constitutively open, constitutively closed, or may be dynamically opening or closing in development. This reconciles the biochemical pioneering-like activities of PU.1 with its distinctive lineage-specific occupancy patterns and functional impacts in the earliest stages of T-cell development.
Methods
Detailed methods and a full reagent list are provided in Supplemental Material.
Mice and cells
As a source of primary cells, C57BL/6 mice and mice with Cas9 and Bcl2 transgenes on a C57BL/6 background were used as described previously (Hosokawa et al. 2018). Fetal liver precursors (Zhang et al. 2012; Del Real and Rothenberg 2013; Champhekar et al. 2015) and bone marrow precursors for the CAS9-mediated deletion experiments (Hosokawa et al. 2018) were prepared as described. All mouse husbandry and procedures were reviewed and approved by the Caltech Institutional Animal Care and Use Committee. Scid.adh.2C2 cells (Dionne et al. 2005; Del Real and Rothenberg 2013) were cultured as in Hosokawa et al. (2018). In vitro T-lineage development using OP9-DL1 cocultures and analysis by flow cytometry were described previously (Champhekar et al. 2015; Hosokawa et al. 2018). Details of flow cytometric staining, marker terminology, antibodies, and culture conditions in each experiment are detailed in Supplemental Material.
Retroviral constructs
Exogenous PU.1 constructs in LZRS retroviral vectors and PU.1 in a pMXs derivative were previously described (Champhekar et al. 2015; Hosokawa et al. 2018). Cas9 was cloned into a GFP (mNeonGreen) derivative of pQCXIN and guide RNAs into an mTurquoise derivative of pQCXIN described in Supplemental Material.
ChIP-seq, ATAC-seq, and RNA-seq
Standard RNA-seq and ChIP-seq methods were used as in our related report (Hosokawa et al. 2018), and ATAC-seq followed standard procedures (Buenrostro et al. 2013). Data for these studies have been deposited as reported previously (GSE93755, GSE11020). Details of sample processing, antibodies, sequence processing, and data analysis are presented in full in Supplemental Material. Analyses of binding patterns and differential gene expression used HOMER (Heinz et al. 2010) and other publicly available packages. Alignments shown are to mm9 (NCBI37) for comparability with previous results (Zhang et al. 2012). All sample types were generated in 2–4 separate biological replicates, with inter-sample correlations as shown in Supplemental Figures S4 and S8. Detailed methods of statistics and bioinformatics are presented in Supplemental Material.
Supplementary Material
Acknowledgments
We thank Jeffrey Longmate (Beckman Research Institute of the City of Hope), Hao Yuan Kueh (Caltech and University of Washington), and Barbara Wold (Caltech) for generous advice; Rothenberg group members and previous group members, Marissa Del Real (City of Hope) and Ameya Champhekar (UCLA), for constructs and helpful discussion; Diana Perez, Jaime Tijerina, and Rochelle Diamond for cell sorting and advice; Ingrid Soto for mouse colony care; Vijaya Kumar for library preparation and sequencing; Henry Amrhein and Diane Trout for computational system assistance; and Igor Antoshechkin for sequencing facility management. This research was supported by fellowships from the Swedish Research Council (to J.U.), the Manpei Suzuki Diabetes Foundation (to H.H.), grants from the US Public Health Service (to E.V.R.) (R01HD076915 [Eunice Kennedy Shriver National Institute of Child Health and Human Development]; R01AI95943 [National Institute of Allergy and Infectious Diseases]), by the L.A. Garfinkle Memorial Laboratory Fund and the Al Sherman Foundation, Facility and Resource Center support from the Beckman Institute at Caltech, and the Albert Billings Ruddock Professorship (to E.V.R.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231423.117.
References
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. 2002. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16: 285–296. [DOI] [PubMed] [Google Scholar]
- Back J, Allman D, Chan S, Kastner P. 2005. Visualizing PU.1 activity during hematopoiesis. Exp Hematol 33: 395–402. [DOI] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54: 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, Schübeler D, Grosschedl R. 2016. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity 44: 527–541. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S, Dakic A, D'Amico A, Pang SH, Greig KT, Nutt SL, Wu L. 2010. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 32: 628–641. [DOI] [PubMed] [Google Scholar]
- Champhekar A, Damle SS, Freedman G, Carotta S, Nutt SL, Rothenberg EV. 2015. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev 29: 832–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Real MM, Rothenberg EV. 2013. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development 140: 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. 2005. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol 280: 448–466. [DOI] [PubMed] [Google Scholar]
- Escalante CR, Brass AL, Pongubala JM, Shatova E, Shen L, Singh H, Aggarwal AK. 2002. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol Cell 10: 1097–1105. [DOI] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. 2008. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci 105: 6057–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. 2006. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci 103: 11993–11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez AJ, Setty M, Leslie CS. 2015. Early enhancer establishment and regulatory locus complexity shape transcriptional programs in hematopoietic differentiation. Nat Genet 47: 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Glass CK. 2014. Epigenomics of macrophages. Immunol Rev 262: 96–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. 2013. Effect of natural genetic variation on enhancer selection and function. Nature 503: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa H, Ungerbäck J, Wang X, Matsumoto M, Nakayama KI, Cohen SM, Tanaka T, Rothenberg EV. 2018. Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity 48: 1119–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Fang D, Hirose S, Wang X, Wangsa D, Jin W, Ried T, Liu P, Zhu J, et al. 2018. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity 48: 227–242.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106: 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. 2013. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 341: 670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. 2006. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25: 731–744. [DOI] [PubMed] [Google Scholar]
- McAndrew MJ, Gjidoda A, Tagore M, Miksanek T, Floer M. 2016. Chromatin remodeler recruitment during macrophage differentiation facilitates transcription factor binding to enhancers in mature cells. J Biol Chem 291: 18058–18071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Ghisletti S, Barozzi I. 2011. The genomic landscapes of inflammation. Genes Dev 25: 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. 2005. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med 201: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171. [DOI] [PubMed] [Google Scholar]
- Pham TH, Minderjahn J, Schmidl C, Hoffmeister H, Schmidhofer S, Chen W, Längst G, Benner C, Rehli M. 2013. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res 41: 6391–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M et al. 2014. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159: 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Ungerbäck J, Champhekar A. 2016. Forging T-lymphocyte identity: intersecting networks of transcriptional control. Adv Immunol 129: 109–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. 2005. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J 24: 3712–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET, Grosschedl R. 2010. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity 32: 714–725. [DOI] [PubMed] [Google Scholar]
- Yui MA, Rothenberg EV. 2014. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 14: 529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Feng N, Rothenberg EV. 2010. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol 185: 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96: 2641–2648. [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.