Abstract
Planarian flatworms have an indefinite capacity to regenerate missing or damaged body parts owing to a population of pluripotent adult stems cells called neoblasts (NBs). Currently, little is known about the importance of the epigenetic status of NBs and how histone modifications regulate homeostasis and cellular differentiation. We have developed an improved and optimized ChIP-seq protocol for NBs in Schmidtea mediterranea and have generated genome-wide profiles for the active marks H3K4me3 and H3K36me3, and suppressive marks H3K4me1 and H3K27me3. The genome-wide profiles of these marks were found to correlate well with NB gene expression profiles. We found that genes with little transcriptional activity in the NB compartment but which switch on in post-mitotic progeny during differentiation are bivalent, being marked by both H3K4me3 and H3K27me3 at promoter regions. In further support of this hypothesis, bivalent genes also have a high level of paused RNA Polymerase II at the promoter-proximal region. Overall, this study confirms that epigenetic control is important for the maintenance of a NB transcriptional program and makes a case for bivalent promoters as a conserved feature of animal stem cells and not a vertebrate-specific innovation. By establishing a robust ChIP-seq protocol and analysis methodology, we further promote planarians as a promising model system to investigate histone modification–mediated regulation of stem cell function and differentiation.
The promoters of developmental genes in mammalian embryonic stem cells (ESCs) are frequently marked with both the silencing H3K27me3 mark and active H3K4me3 marks. It has been proposed that this “bivalent” state precedes resolution into full transcriptional activation or repression, depending on ultimate cell type commitment (Bernstein et al. 2006; Voigt et al. 2013; Harikumar and Meshorer 2015). The advantage is that bivalency represents a poised or transcription-ready state, whereby a developmental gene is silenced in ESCs, but can be readily rendered active during differentiation to a defined lineage. Evidence for this comes from the finding that 51% of bivalent promoters in ESCs are bound by paused polymerase (RNAPII-Ser5P), compared with 8% of nonbivalent promoters (Brookes et al. 2012; Lesch and Page 2014), demonstrating a strong but not complete association. Bivalency may also protect promoters against less reversible suppressive mechanisms, such as DNA methylation (Lesch and Page 2014). Bivalent chromatin has also been discovered in male and female germ cells at many of the gene promoters that regulate somatic development and may underpin the gametes’ ability to generate a zygote capable of producing all cellular lineages (Lesch et al. 2013; Sachs et al. 2013; Lesch and Page 2014).
It remains unclear whether the poised bivalent promoters of developmental genes are an epigenetic signature of vertebrates or arose earlier in the ancestor of all animals. Recently, the orthologs of bivalent genes that sit at the top of transcriptional hierarchies in mammalian development were also found to be poised in chicken male germ cells (Lesch et al. 2016). Sequential ChIP has also established H3K4me3/H3K27me3 co-occupancy of promoters in zebrafish blastomeres (Vastenhouw et al. 2010). Conversely, comparatively few bivalent domains were identified in Xenopus embryos undergoing the midblastula transition (Akkers et al. 2009). Xenopus genes which appear to have signals for both H3K4me3 and H3K27me3 originate from cells in distinct areas of the embryo, and as such, the observed bivalency can be explained by cellular heterogeneity (Akkers et al. 2009).
As ESC pluripotency requires bivalent chromatin, planarian adult stem cells or neoblasts represent another possible scenario in which poised promoters could have an important role in invertebrates, if this regulatory feature is conserved. Planarian NBs are a population of adult dividing cells that collectively produce all differentiated cells during homeostatic turnover and regeneration (Aboobaker 2011; Rink 2013). Several RNA-binding proteins, such as piwi and vasa, typically associated with nuage of germ cells are also expressed in planarian NBs where they function in the maintenance of pluripotency (Reddien et al. 2005; Palakodeti et al. 2008; Solana 2013; Shibata et al. 2016; Lai and Aboobaker 2018). Moreover, the ability of NBs to differentiate upon demand must also require well-regulated transcriptional and epigenetic processes, and poised, bivalent promoters may constitute an effective way of coordinating the differentiation of these stem cells.
Here, we develop an optimized ChIP-seq methodology for planarian NBs and combine this with informatics approaches to establish robust approaches for studying histone modifications at transcriptional starts sites (TSSs). This enabled us to identify genes with inactive/low expression in the NB population, but with greatly increased expression in post-mitotic NB progeny that are actively differentiating, with bivalent promoters in planarian NBs by combining transcriptomic and epigenetic analyses. Our findings indicate that bivalent promoters in pluripotent stem cells are not just a facet of vertebrates, but may have a role in regulating pluripotency in embryonic and adult stem cells across animals.
Results
Genome-wide annotation of transcribed loci in asexual Schmidtea mediterranea genome and categorization by proportional expression in FACS populations
We sought to produce an annotation of all transcribed loci on the asexual Schmidtea mediterranea genome (SmedAsxl v1.1) utilizing both de novo assembled transcriptomes and 164 independent RNA-seq data sets covering RNAi knockdown-, regenerating-, whole worm-, and cell compartment-specific data sets (Fig. 1A; Supplemental File 1). The inclusion of these diverse data sets was to improve the overall representation of the genome and is useful for discovering potential noncoding RNAs and protein-coding genes expressed at low levels, both of which may not have been fully covered by individual studies limited by read number or reliant on homology-based annotation processes such as MAKER (Cantarel et al. 2008).
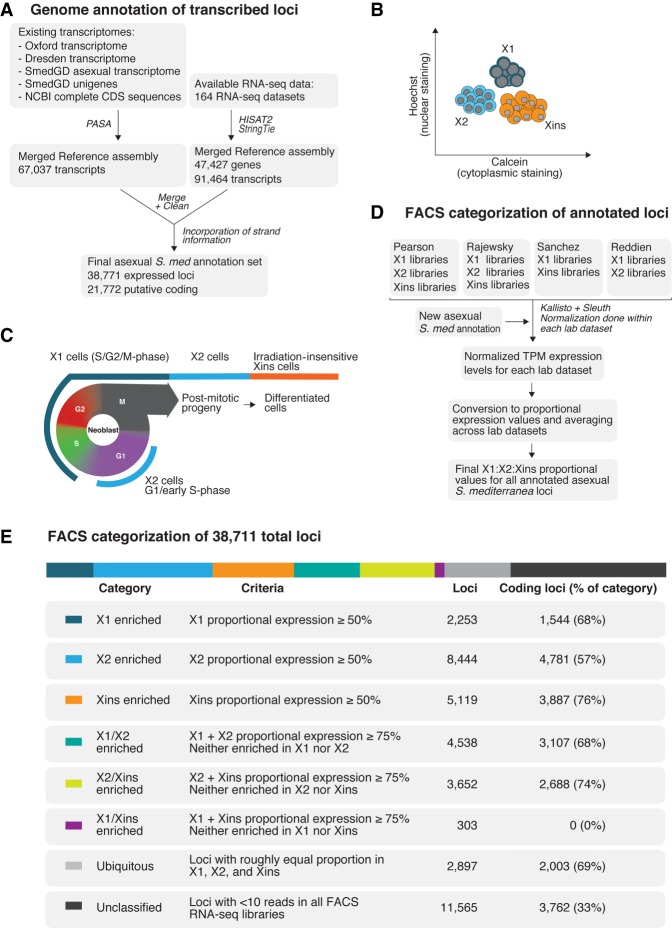
Figure 1.
(A) Overview of methodology for annotating the Schmidtea mediterranea asexual genome based on expression. One hundred sixty-four RNA-seq data sets, four de novo transcriptome assemblies, NCBI complete CDS sequences, and Smed Unigenes were mapped to the SmedGD Asxl v1.1 genome. Reference assemblies were merged, cleaned to remove potential splice variant redundancies, and the best representative transcript for each genomic locus was chosen. Strand information was obtained by BLAST to UniProt, prediction of longest ORF, and data from strand-specific libraries. This process yielded a total set of 38,771 loci. (B) Methodology for gating X1, X2, Xins cell populations based on nuclear to cytoplasmic ratio during Fluorescent Activated Cell Sorting (FACS). (C) Diagram depicting how X1, X2, Xins FACS population gates relate to cell cycle and differentiation stage. (D) Overview of methodology for categorization of annotated loci based on FACS RNA-seq data sets documented in Supplemental Figure 2. FACS RNA-seq data sets were mapped to our annotated genome using Kallisto and normalized with Sleuth. Normalization was done individually for each of the laboratories’ data sets. Normalized TPMs were converted to proportions between available FACS categories of each laboratory, and a final consensus X1:X2:Xins proportion was calculated. (E) A table presenting number of loci in different FACS classification groups, as well as number of protein-coding genes in each group based on Transdecoder evidence.
Our new expression-based annotation identified 38,711 expressed loci, 21,772 of which are predicted to be coding (Fig. 1A). Moreover, compared to the current available annotation of the Schmidtea mediterranea asexual genome (Smed GD 2.0) (Robb et al. 2008), our annotation discovered 10,210 new potential protein-coding loci that are expressed at similar overall levels to previously annotated protein-coding genes. A total of 6300 genes from the existing MAKER homology-based annotation were not present in our expression-driven annotation. Further analysis of these MAKER-specific genes shows that they generally have no or very little potential expression within the 164 RNA-seq libraries utilized for our annotation (Supplemental Fig. 1).
In the absence of transgenic approaches and antibodies for confirmed cell lineage markers, Fluorescence Activated Cell Sorting (FACS) gating cell populations stained with Hoechst and calcein is the best available tool for isolating NBs, progeny, and differentiated cells (Hayashi et al. 2006; Romero et al. 2012). FACS allows for two irradiation sensitive compartments to be discerned: the ‘X1’ gate representative of S/G2/M-phase NBs with more than 2C DNA content; and the ‘X2’ gate representative of G1 phase NBs and post-mitotic progeny with 2C DNA content. The third FACS population, ‘Xins,’ represents an irradiation-insensitive population with a higher cytoplasmic to nuclear ratio (Fig. 1B,C). These cell compartments are heterogeneous, with subpopulations of NBs expressing epidermal, gut, and other lineage-specific markers present within the X1 population (Scimone et al. 2014; Van Wolfswinkel et al. 2014; Wurtzel et al. 2015), and the X2 compartment consisting of an amalgam of G1 NBs and lineage-committed post-mitotic progeny (Baguñá and Romero 1981; Hayashi et al. 2006; Zhu et al. 2015; Molinaro and Pearson 2016).
We used the publicly available RNA-seq data sets for these three different FACS populations in order to compare the expression of our annotated loci in these three distinct compartments (Labbé et al. 2012; Önal et al. 2012; Van Wolfswinkel et al. 2014; Duncan et al. 2015; Zhu et al. 2015). We first looked at the normalized TPM expression levels for annotated loci in our annotated genome in the FACS population data sets originating from four different planarian laboratories (Supplemental Fig. 2). This revealed a rough congruence between different FACS populations from different laboratories (Supplemental Fig. 3A).
We transformed absolute TPM expression values into proportional values for each FACS compartment in each of the data sets (Fig. 1D; Supplemental Fig. 3B). These proportional values were then averaged across data sets, to produce a final set of X1:X2:Xins proportions for 27,206 loci (18,010 of which are predicted to be protein coding) that had at least 10 reads mapped in at least one FACS RNA-seq library.
We were now able to sort all annotated genes according to whether their predominant expression (i.e., ≥50% expression) is in X1 (S/G2/M-phase NBs), X2 (NBs and stem cell progeny), or Xins (differentiated cells) (Fig. 1E). We confirmed this analysis by Gene Ontology (GO) analyses and verification of the proportional expression profiles for known genes previously described as being enriched in X1, X2, or Xins (Supplemental Fig. 4; Labbé et al. 2012; Önal et al. 2012; Solana et al. 2012). We also reanalyzed FACS single-cell RNA-seq data sets in the context of our genome annotation, and visualization of the data by breakdown into our defined FACS expression categories was entirely consistent with this data (Supplemental Fig. 5). In particular we note that those genes with the highest proportion of expression in the X2 compartment are indicative of genes expressed in post-mitotic undifferentiated NB progeny, with only very little expression in NBs themselves (Supplemental Fig. 5).
Together these analyses provide a set of annotations and expression values that are directly related to the genome assembly, allowing integration of ChIP-seq data to investigate correlations between epigenetic marks and gene expression in the different planarians cell FACS compartments.
An optimized ChIP-seq protocol reveals that H3K4me3 and H3K36me3 levels correlate with gene expression in planarian NBs
Research into the epigenetic mechanisms governing stem cell pluripotency in planarian NBs is still in its infancy (Dattani et al. 2018). Previous work uncovered a lack of endogenous DNA methylation in the Schmidtea mediterranea genome and characterized loss of function phenotypes for members of the NURD complex (Scimone et al. 2010; Jaber-Hijazi et al. 2013; Vásquez-Doorman and Petersen 2016), COMPASS, and COMPASS-like families (Hubert et al. 2013; Duncan et al. 2015; Mihaylova et al. 2017). The first study to utilize ChIP-seq in planarians documented the effects of mll1/2 and set1 RNAi with respect to the activation mark H3K4me3 (Duncan et al. 2015). However, we revisited this data and noted that the total number of ChIP-seq reads from −1 million X1 sorted NBs was comparatively low in comparison to those from Drosophila melanogaster S2 “carrier” cells.
We developed an optimized ChIP-seq protocol for FACS-sorted X1 NBs without the addition of excess “carrier” cells. Instead, a 3% Drosophila S2 spike-in was added to our chromatin before immunoprecipitations (IP) simply as a method to normalize any technical differences in IPs across our replicate libraries (Orlando et al. 2014). We were able to generate high-quality uniquely mapped reads to our annotated Schmidtea mediterranea genome using only 150,000–200,000 X1 cells per IP—five to seven times less material than the previously established planarian protocol (Duncan et al. 2015). With our protocol, Drosophila spike-in reads accounted for an average of 27% of X1 H3K4me3 libraries compared to an average of 87% in the previous study's X1 H3K4me3 libraries. Moreover, Drosophila spike-in reads accounted for 9% of our X1 H3K36me3 libraries compared with 99% of the single X1 H3K36me3 replicate included in a previous study (Supplemental Fig. 6; Duncan et al. 2015).
We tested the robustness of our ChIP-seq protocol with reference to both H3K4me3 and H3K36me3—epigenetic marks that are known to positively correlate with gene expression in other model systems. H3K4me3 is laid down by the trithorax group (trxG) complexes containing SET or MLL enzymes at both active and bivalent promoters near TSSs (Hu et al. 2013; Bledau et al. 2014; Denissov et al. 2014). H3K36me3 is a mark of transcriptional elongation and is deposited on histones as they are displaced by RNA Polymerase II and as such this modification is enriched toward the 3′ end of genes (Li et al. 2002; Wagner and Carpenter 2012). H3K36me3 is hypothesized to prevent spurious transcriptional initiation at cryptic promoter-like sequences within exons and, in yeast, this is achieved by the recruitment of histone deacetylase complexes (HDAC) that erases elongation-associated acetylation (Carrozza et al. 2005; Joshi and Struhl 2005).
As predicted, ChIP-seq of H3K4me3 in X1 NBs revealed a high average peak around the TSSs of genes characterized as being X1 enriched (Fig. 2A). Conversely, we observed comparatively lower H3K4me3 deposition at the TSSs of Xins-enriched genes not expressed or expressed only at very low levels in X1 cells. Intermediate levels of H3K4me3 in the X2 compartment are consistent with this FACS population being a mixture of NBs and post-mitotic progeny. Indeed, genes with the highest proportion of X2 expression (i.e., high-ranking X2 genes) indicative of expression in post-mitotic progeny but not NBs had lower levels of H3K4me3 in X1 cells compared with low ranking X2 genes that retain expression in cycling NBs (Fig. 2B). A base-by-base Spearman's rank correlation of ChIP-seq signal to FACS proportional expression values of annotated loci across a 2.5-kb region on either side of the TSS shows a positive correlation between genes defined by high X1 proportional expression and the levels of H3K4me3 deposition close to the TSS (Supplemental Fig. 7A). On the other hand, there is a negative correlation between H3K4me3 deposition and genes with high Xins proportional expression across the same region. Thus, a high H3K4me3 ChIP-seq signal reflects higher expression of a locus in X1 NBs, whereas lower H3K4me3 signal reflects lower X1 NB gene expression but higher expression in the differentiated Xins compartment.
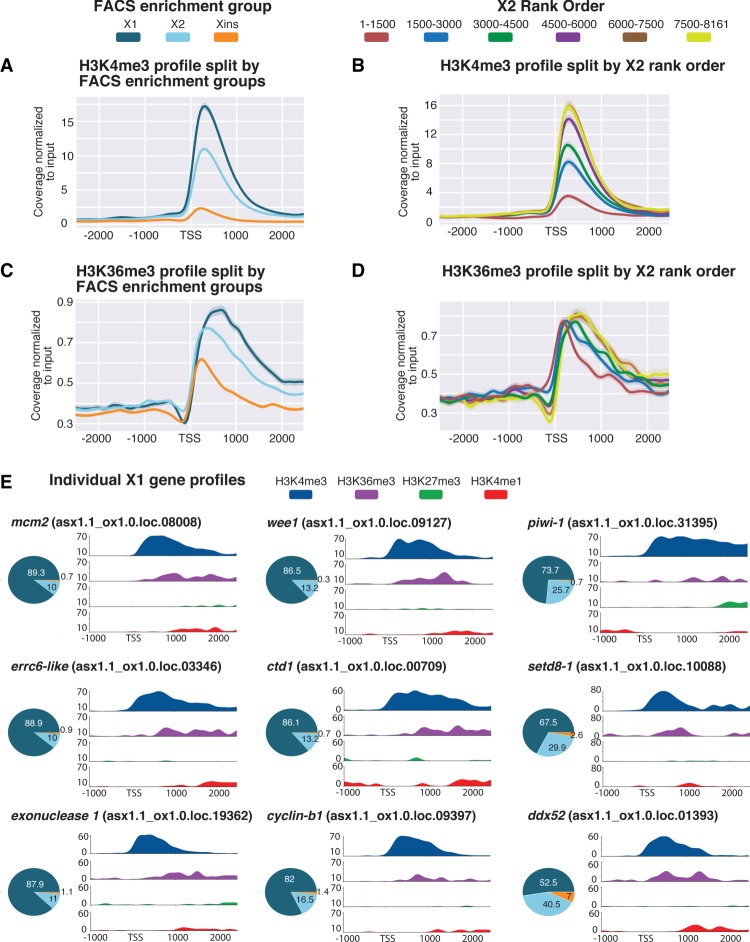
Figure 2.
Histone marks for actively transcribed genes in X1 NBs. (A) Average H3K4me3 ChIP-seq coverage profiles across X1-, X2-, and Xins-enriched loci in X1 NBs across biological replicates following outlier removal. The y-axis represents the difference in coverage between sample and input, and the x-axis represents 2.5 kb upstream of and downstream from the TSS. H3K4me3 signal is highest around the promoter-proximal region close to the TSS for X1-enriched loci in NBs consistent with the role of H3K4me3 in active transcription. (B) H3K4me3 ChIP-seq profiles following outlier removal for X2 genes ranked from high to low X2 proportional expression. H3K4me3 signal in NBs decreases with an increase in proportion of X2 expression, indicative of high-ranking X2 genes having a predominant role in post-mitotic progeny as opposed to NBs. (C) Average H3K36me3 ChIP-seq profile across X1-, X2-, and Xins-enriched loci in X1 NBs across biological replicates following outlier removal. The y-axis represents the difference in coverage between sample and input, and the x-axis represents 2.5 kb upstream of and downstream from the TSS. Signal for H3K36me3 is promoter-proximal for Xins genes, whereas the magnitude of signal is greater and shifted 3′ for X1 genes. (D) H3K36me3 ChIP-seq profiles following outlier removal for X2 genes from high to low X2 proportional ranking. H3K36me3 signal in NBs shifts to the 3′ end with a decrease in X2 proportion, consistent with these lowly ranked genes having transcriptional activity in NBs. (E) H3K4me3 and H3K36me3 (active marks) and H3K4me1 and H3K27me3 (suppressive marks) ChIP-seq profiles for highly expressed X1 genes in NBs. The y-axis represents percentage coverage for each mark and allows for the four epigenetic marks to be directly compared. The x-axis represents 1.0 kb upstream of and 2.5 kb downstream from the TSS. Pie charts represent proportional expression for each gene in X1 (dark blue), X2 (light blue), and Xins (orange).
ChIP-seq plots of H3K36me3 split by FACS gene expression revealed, as predicted, a higher average peak around X1-enriched genes when compared with the X2 and Xins FACS enrichment categories (Fig. 2C). Importantly, the average peak for X1 genes is located toward the 3′ end of genes, whereas the smaller Xins peak is promoter-proximal in comparison. This can be explained by a higher level of transcriptional elongation of X1 transcripts in NBs compared with Xins genes that have a predominant expression in the differentiated compartment. When splitting X2-enriched genes by rank order, we observed that genes with highest expression in the X2 compartment and, as a consequence lowest transcript abundance in NBs, have an enrichment for H3K36me3 at the promoter-proximal end of the gene (Fig. 2D). Conversely, with decreasing X2 proportional expression and a concomitant increase in transcriptional activity in the NB compartment, the average peak of H3K36me3 is shifted downstream from the TSS toward the 3′ ends of genes.
We also looked at the individual H3K4me3 and H3K36me3 profiles of genes known to be highly expressed in NBs and compared this to the signal for the suppressive marks H3K4me1 and H3K27me3 (discussed later). We confirmed that known metazoan genes associated with stem cell maintenance, such as cell cycle and replication related genes (i.e., mcm2, cyclin-B1, wee1, ctd1), RNA-binding proteins (piwi-1, ddx52), DNA-damage response (DDR) genes (errc6-like, exonuclease 1), and epigenetic-related genes (setd8-1), all have high levels of H3K4me3 at the promoter-proximal end and H3K36me3 in the gene body, but a comparatively low signal for the suppressive marks H3K4me1 and H3K27me3 (Fig. 2E).
Levels of repressive histone marks H3K27me3 and H3K4me1 at TSSs in NBs correlate with gene expression
Utilizing our optimized ChIP-seq protocol, we investigated the occurrence of two additional histone modifications: H3K27me3, a repressive promoter mark catalyzed by the PRC2 complex, and H3K4me1, a mark mediated by the MLL3/4 family of histone methyltransferases that correlates both with active enhancers and inactive promoter regions (Calo and Wysocka 2013; Cheng et al. 2014).
Genes that are categorized as being X1 enriched have low levels of H3K27me3 deposition at the TSS, compared with Xins-enriched genes, which are silenced in NBs (Fig. 3A). A positive correlation is observed between the level of H3K27me3 and expression in the Xins compartment in a window from the TSS to 1 kb downstream. This fairly broad domain of H3K27me3 deposition is consistent with previous studies in mammals (Supplemental Fig. 7B; Pauler et al. 2009; Hawkins et al. 2011). Conversely, a negative correlation at the TSS is observed between H3K27me3 signal and genes with high X1 expression (Supplemental Fig. 7B). Consequently, the genome-wide pattern for H3K27me3 is the opposite to that observed for H3K4me3. When splitting X2 genes by rank, we note that genes with higher transcriptional enrichment in the post-mitotic compartment have a higher overall level of H3K27me3 at the promoter-proximal region compared to genes that have NB expression (Fig. 3B).
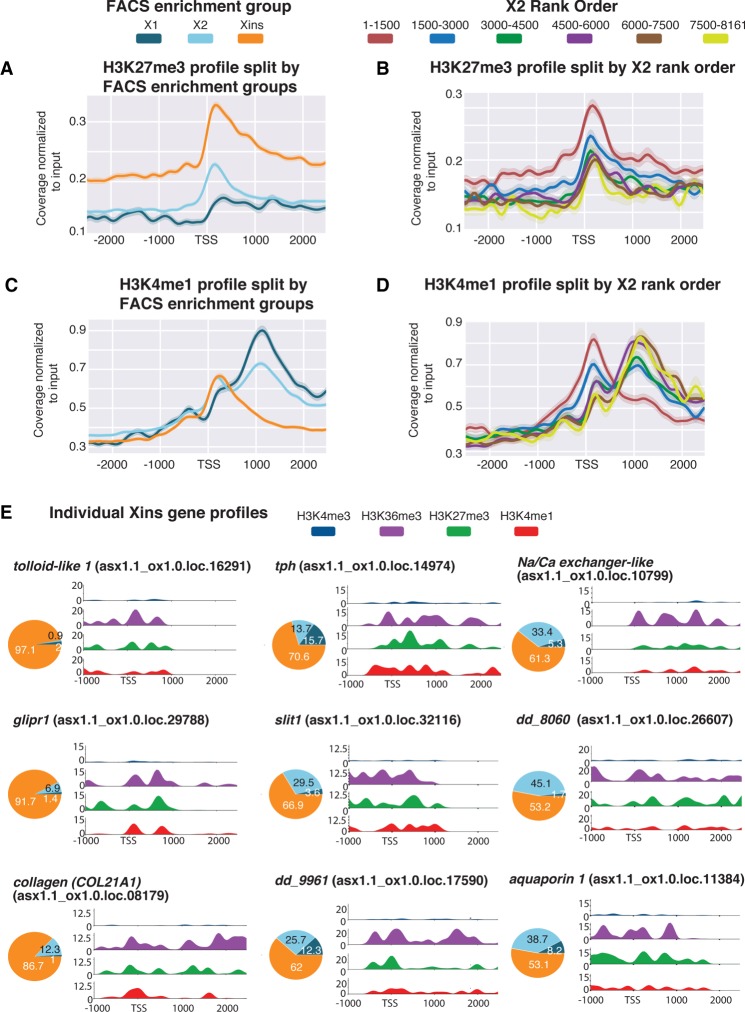
Figure 3.
Histone marks for inactive genes in X1 NBs. (A) Average H3K27me3 ChIP-seq profile across X1-, X2-, and Xins-enriched loci in X1 NBs across three biological replicates following outlier removal. The y-axis represents the difference in coverage between sample and input, and the x-axis represents signal 2.5 kb upstream of and downstream from the TSS. (B) H3K27me3 ChIP-seq profiles following outlier removal for X2 genes from high to low X2 proportional ranking. H3K27me3 signal increases with an increase in proportion of X2 gene expression, indicative of these high-ranking X2 genes being transcriptionally silenced or lowly expressed in NBs. (C) Average H3K4me1 ChIP-seq profiles following outlier removal across X1-, X2-, and Xins-enriched loci in X1 NBs. The y-axis represents the absolute difference in coverage between sample and input, and the x-axis represents signal 2.5 kb upstream of and downstream from the TSS. (D) H3K4me1 ChIP-seq profiles following outlier removal for X2 genes from high to low X2 proportional ranking. Highly ranked X2 genes have a H3K4me1 signal at the promoter-proximal region, and a decrease in X2 ranking coincides with a peak shift −1 kb downstream from the TSS. (E) H3K4me3, H3K36me3, H3K4me1, and H3K27me3 NB ChIP-seq profiles for highly expressed Xins genes. The y-axis scale represents percentage coverage for each mark, and the x-axis represents 1.0 kb upstream of and 2.5 kb downstream from the TSS. Pie charts represent proportional expression for each gene in X1 (dark blue), X2 (light blue), and Xins (orange).
The distribution of the H3K4me1 mark is noticeably different compared to that observed for either H3K27me3 or H3K4me3. Specifically, Xins loci have high levels of H3K4me1 at the TSS in X1 NBs, consistent with these genes being expressed at low levels in NBs, whereas X1 loci have H3K4me1 peaks that are on average −1 kb downstream from the TSS (Fig. 3C). This data suggests that the H3K4me1 signal shifts away from the TSS for genes that are actively expressed in NBs, in agreement with previous observations in mammals (Cheng et al. 2014). Further evidence of this peak shifting comes from analysis of X2-enriched genes sorted by rank order of expression (Fig. 3D). Highly ranked X2 genes are marked with H3K4me1 at the promoter-proximal region. As the proportion of X2 enrichment decreases, indicative of increasing expression the G1 NB compartment, the average H3K4me1 profile becomes bimodal, eventually shifting downstream from the TSS (Fig. 3D).
We plotted the epigenetic profiles of individual genes known to have high Xins proportional expressions and that have validated expression patterns both by single-cell RNA sequencing data and in situ hybridizations (Fig. 3E; Fincher et al. 2018; Plass et al. 2018). For example, these genes are expressed almost exclusively in the muscle (COL21A1, slit1), parenchyma (glipr1, tolloid-like 1), cathepsin-positive cells (dd961, aquaporin 1), nonciliated neurons (tph, dd8060), and protonephridia (Na/Ca exchanger-like), all have high H3K27me3 signal at the TSS consistent with these genes being silenced in NBs. Moreover, these Xins-enriched genes all have a high H3K4me1 signal at the TSS that anti-correlates with H3K4me3 deposition, in support of an earlier hypothesis that H3K4me1 limits the role of H3K4me3 interacting proteins (Cheng et al. 2014). We also observed an atypical placement of H3K36me3 at the TSS of individual Xins genes, which supports the previous suggestion that H3K36me3 may silence genic loci when placed at a promoter-proximal region of a gene (Wu et al. 2011).
Correlations of H3K27me3 and H3K4me3 profiles against FACS proportions provide evidence for promoter bivalency in NBs
Having demonstrated that known active and suppressive marks correlate with gene expression in planarian NBs, we investigated whether promoter bivalency could act to keep genes in a poised state prior to the onset of differentiation. Bivalent promoters are characterized by the presence of both the activating mark H3K4me3 and repressive mark H3K27me3. The simultaneous presence of both these marks keeps the gene in a poised transcriptional state, with low or no expression, and upon differentiation resolves such that only one of the two marks is dominant. We reasoned that loci that are off or have relatively low proportional expression in X1 NBs, but which are up-regulated during the differentiation process in post-mitotic progeny (high X2 expression), would be good candidates for potential regulation by bivalent promoters in NBs. In addition, in the absence of sequential or co-ChIP-seq technologies for planarians, using genes with no or very low expression in NBs greatly reduces the likelihood that any bivalent signals are due to cell heterogeneity. This is because these genes would not be expected to have high levels of H3K4me3 in any (or at least very few cells) in the X1 NB compartment.
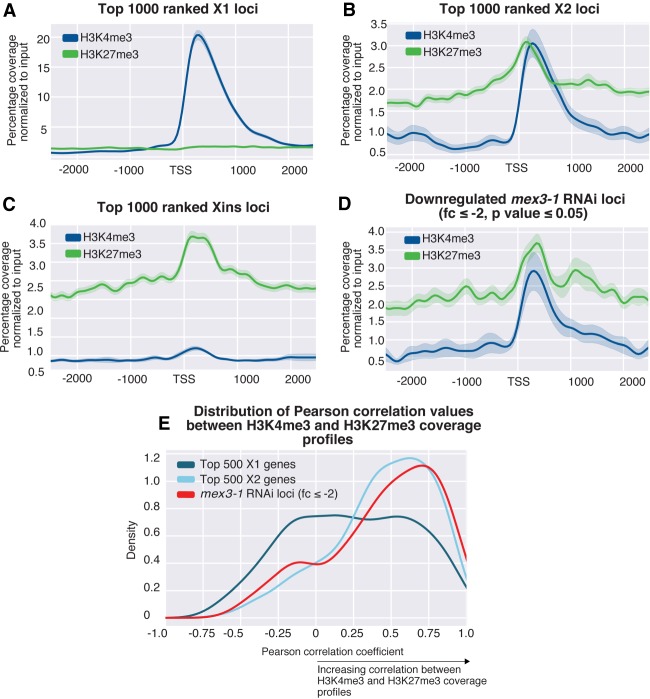
We plotted the percentage of maximum coverage for both H3K4me3 and H3K27me3 for the top 1000 genes for each of the three FACS enrichment categories (Fig. 4A–C). A plot for the top 1000 X1 genes shows that these genes have a higher level of H3K4me3 compared to H3K27me3 (Fig. 4A), whereas the top 1000 Xins genes have on average a much higher H3K27me3 signal compared to H3K4me3 (Fig. 4C). Consistent with our hypothesis, the top 1000 X2 genes, have peaks that are of similar magnitude for both of these functionally opposing epigenetic marks (Fig. 4B).
Figure 4.
(A–E) Average H3K4me3 and H3K27me3 ChIP-seq profiles in X1 NBs across three biological replicates. The y-axis is percentage coverage after normalization to input to allow both ChIP-seq profiles to be directly compared. Plots are shown for the following: (A) top 1000 ranked X1 genes by expression; (B) top 1000 ranked X2 genes; (C) top 1000 ranked Xins genes; and (D) 285 Smed-mex3-1 down-regulated loci with greater than twofold change (P < 0.05). (E) A distribution of Pearson correlation values for the top 500 X1 expressed loci, the top 500 X2 expressed loci, and 285 loci ≥ twofold down-regulated after mex3-1(RNAi). The Pearson correlation coefficient was calculated between the H3K4me3 and H3K27me3 values at each 50-bp window −1000 bp and +1500 bp around the TSS.
We also plotted the epigenetic profiles of genes that are down-regulated following RNAi of the planarian homolog of the RNA-binding protein MEX3 (Zhu et al. 2015). Previously, mex3-1 was shown to be necessary for generating the differentiated cells of multiple lineages and, consistent with a role in the differentiation process, we found that the down-regulated genes (down-regulated twofold; P-value ≤0.05) had a higher average X2 proportional expression value (62.4%) compared with that of X1 (12.5%) (Supplemental File 2). As expected, we note a paired H3K4me3 and H3K27me3 ChIP-seq signal for these mex3-1 down-regulated genes (Fig. 4D).
One possibility is that our observations are as a result of some highly ranked X2 genes having only the H3K4me3 mark, whereas other genes exist in a H3K27me3-only state in NBs. This would produce an average profile that appears bivalent when many genes are looked at simultaneously. To rule out this possibility, we plotted the distribution of Pearson correlation coefficients between H3K4me3 and H3K27me3 for the top 500 ranked X1, X2, and 285 mex3-1(RNAi) down-regulated loci. This showed a strong positive correlation between H3K4me3 and H3K27me3 for top 500 X2 loci and mex3-1 down-regulated loci compared to a weak or no average correlation for X1 loci (Fig. 4E). This is consistent with the interpretation that bivalency is present at promoters of genes that are highly enriched for expression in the X2 compartment.
Planarian orthologs of mammalian bivalent genes are marked by H3K4me3, H3K27me3, and paused RNA Pol II at the promoter-proximal region
RNA Polymerase II (RNAPII) pausing at genes that are highly inducible has been hypothesized to play a pivotal role in preparing genes for rapid induction in response to environmental or developmental stimuli. In a number of mammalian cellular contexts, bivalent genes have been shown to have a high density of paused RNA Pol II at the promoter-proximal region compared to genes that are actively transcribed, therefore allowing genes to be maintained in a transcriptionally poised state (Stock et al. 2007; Ferrai et al. 2017; Liu et al. 2017). Paused RNA Pol II can be distinguished from other forms by a phosphorylation at Ser5 (Ser5P) of the YSPTSPS heptad repeat at the C terminus of the largest subunit of the Pol II complex. This heptad repeat is conserved across metazoans and is found in S. mediterranea (Corden 2013; Yang and Stiller 2014).
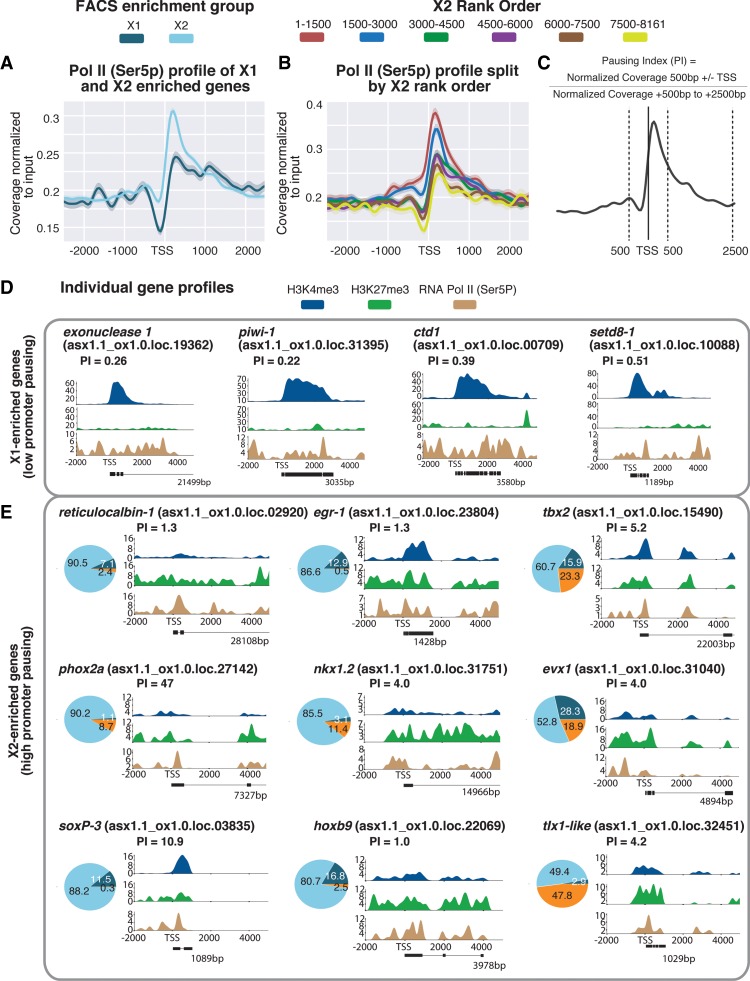
ChIP-seq for RNAPII-Ser5P in NBs revealed that X2-enriched genes have a higher level of paused RNA Pol II at the promoter-proximal region compared to X1 genes (Fig. 5A). More significantly, highly ranked X2 genes with high expression in post-mitotic progeny and little expression in NBs have the highest amount of paused RNA Poll II close to the TSS, and with increasing expression in NBs, the enrichment for this mark decreases (Fig. 5B).
Figure 5.
(A) Average paused RNAPII-Ser5P ChIP-seq profile across X1- and X2-enriched loci in X1 NBs across biological replicates following outlier removal. The y-axis represents the difference in coverage between sample and input, and the x-axis represents signal 2.5 kb upstream of and downstream from the TSS. (B) RNAPII-Ser5P ChIP-seq profiles following outlier removal for X2 genes from high to low X2 proportional ranking. RNAPII-Ser5P signal increases with an increase in proportion of X2 gene expression, indicative of these high-ranking X2 genes being transcriptionally silenced but maintained in a permissive state for rapid induction. (C) Calculation for pausing index (PI) of genes ≥1 kb. We divided normalized coverage between ±500 bp TSS by normalized coverage +500 bp to +2.5 kb. For genes <2.5 kb, we inspected RNAPolI-Ser5P profiles visually to confirm whether Pol II pausing was enriched at the promoter-proximal region. (D) Individual profiles for H3K4me3 and H3K27me3 of highly enriched NB X1 genes. X1 genes have a high level of H3K4me3, and levels of H3K27me3 correspond to intron regions and are not enriched at the promoter-proximal region. RNAPII-Ser5P signal is not enriched at the promoter-proximal region compared with the gene body; as a result, PI <1. (E) We selected highly enriched X2 genes with a PI ≥1 that have both H3K4me3 and H3K27me3 enriched at the promoter-proximal region, together with an enrichment of RNAPII-Ser5P close to the TSS. Pie charts represent proportional expression for each gene in X1 (dark blue), X2 (light blue), and Xins (orange).
We calculated the pausing index (PI) for all annotated genes in our genome that have a total annotated length ≥1 kb. For our particular genome annotation, we calculated the PI as the read coverage (normalized to input) ±500 bp either side of the annotated TSS divided by the normalized read coverage from +500 to +2500 bp from the TSS (Fig. 5C). We applied a conservative definition of a gene as being significantly stalled for transcription if the PI ≥1. As expected, individual genes highly expressed in the NB compartment had both a low PI and were not enriched for RNAPII-Ser5P at the promoter-proximal region, thereby confirming our methodology was accurate at the gene level (Fig. 5D). We also found that X2 genes with high PI scores had, on average, higher Pearson correlation coefficients between H3K4me3 and H3K27me3 (indicative of a bivalent state) compared with both X1 and X2 genes that have lower PI scores (Supplemental Fig. 8). Given this correlation, we chose individual X2-enriched genes with high PI values and plotted the ChIP-seq profiles for H3K4me3, H3K27me3, and RNAPII-Ser5P as a percentage of maximum coverage for each mark.
Among genes enriched for these three signatures of bivalent promoters were those that have orthology to transcription factor (TF) families and include the Hox (hoxb9), Nkx (nkx1.2), Even-skipped (evx-1), Paired-like (phox2A), and T-box (tbx2) and Tlx (tlx1-like) gene classes (Fig. 5E). Indeed, previous studies in both mouse ESCs (Bernstein et al. 2006) and quiescent muscle stem cells (Liu et al. 2013) have shown that members of these gene families are typically marked by both H3K4me3 and H3K27me3. A paired level of these marks at the TSS for these individual genes suggests the existence of bivalent chromatin states at these conserved developmental genes and confirms our correlational analysis of X2 loci (Fig. 4E). Moreover, single-cell sequencing data and pseudotime analyses plots made from single-cell data show that these genes are expressed at detectable levels in very few, if any, piwi-1-positive cells (the archetypal NB marker) and are instead enriched in post-mitotic cells of specific lineages (Supplemental Fig. 9; Fincher et al. 2018; Plass et al. 2018).
One caveat of our analyses is that the bivalent profiles of X2-enriched differentiation related genes may, for some individual genes that appear bivalent, reflect admixture of transcriptionally active and repressed states within the X1 NB compartment. For example, previous work has shown that the X1 compartment is highly heterogeneous with subsets of piwi-1-positive NBs expressing lineage-specific TFs (Van Wolfswinkel et al. 2014). These genes, such as SoxP-3 and egr-1, which are in fact X2-enriched according to our data set and others (Labbé et al. 2012), appear to have a paired H3K4me3 and H3K27me3 signal (Fig. 5E). Because they are known to be expressed in a subset of cells in the X1 compartment and are definitive markers of lineage-primed NB subsets that will go through one more cell division (as validated by in situ hybridization, condensin knockdown studies) (Van Wolfswinkel et al. 2014; Lai et al. 2018) and single-cell RNA-seq data (Wurtzel et al. 2015; Fincher et al. 2018; Plass et al. 2018), no definitive conclusions concerning bivalency of these particular genes can be reached.
Discussion
In this study, we produced a Schmidtea mediterranea asexual genome annotation based on gene expression and integrated FACS RNA-seq data sets from different laboratories to calculate consensus proportional expression values for each annotated locus in the X1, X2, and Xins cellular compartments. We developed an optimized ChIP-seq protocol and used this to generate robust genome-wide profiles of the active H3K4me3 and H3K36me3 marks and repressive H3K4me1 and H3K27me3 marks in planarian NBs.
We found that the active marks H3K4me3 and H3K36me3, and suppressive H3K4me1 and H3K27me3 marks in X1 NBs correlate with the proportion of total transcript expression of these loci in X1 cells, validating our NB ChIP-seq methodology. These analyses showed that genes associated with stem cell differentiation, and which are expressed at low levels in X1 population but activated at high levels in the X2 population, are marked with both H3K4me3 and H3K27me3 marks at comparable levels at the TSS. Moreover, these genes were also highly marked with paused RNA polymerase (RNAPII-Ser5P) at the promoter region consistent with the definition of transcriptionally poised bivalent genes. Although we cannot entirely rule out cell heterogeneity within the X1 NB population as a factor contributing to our observation of promoter bivalency, our focus on both genes with high X2 expression (post-mitotic NB progeny) and orthology to vertebrate transcription factors known to have bivalent profiles, provide strong evidence that bivalent histone marks may be involved in poising of genes for activation upon NB commitment and differentiation in planarians.
The existence of promoter bivalency in invertebrates, prior to our work here, has been contentious. For example, the mammalian orthologs of bivalent genes in Drosophila germ cells were found to have only repressive H3K27me3 deposited at their promoters (Schuettengruber et al. 2009; Gan et al. 2010; Lesch et al. 2016). However, in a more recent study using fly embryos, the Pc-repressive complex 1 (PRC1) that binds to H3K27me3 was shown to copurify with both the Fsh1 (ortholog of mammalian BRD4) that binds to acetylated histone marks and to Enok/Br140 (orthologs to subunits of mammalian MAZ/MORF histone acetyltransferase complex). ChIP-seq identified two groups of PRC1/Br140 genomic binding sites that were either defined by strong H3K27me3 signal or strong H3K27ac signal (i.e., actively transcribed genes). Both groups were also marked with narrow peaks of H3K4me3 at the TSS (Kang et al. 2017). These recent findings also argue for the existence of bivalent-like promoters outside of vertebrates, at least with respect to the binding of chromatin regulatory complexes, and extends the model to suggest that acetylation may be important in the resolution of bivalent protein complexes during development.
One key role of bivalence may be to allow the maintenance of pluripotency in ESCs, by having genes involved in differentiation and commitment both silent but competent to switch on if the right signals are received. Our data suggest that this mechanism is likely to be important for pluripotency in planarian pluripotent NBs, as genes that can switch on rapidly upon differentiation appear to be the bivalent. Indeed, these genes also included planarian orthologs to mammalian TFs that have been documented to be bivalent in ESCs. Consequently, we are able to present a case for promoter bivalency in planarian NBs and in doing so demonstrate that this process is not necessarily vertebrate-specific. This novel finding adds to the growing body of evidence that suggests a deep conservation of regulatory mechanisms involved in stem cell function (Juliano et al. 2010; Solana 2013; Alié et al. 2015; Solana et al. 2016; Lai and Aboobaker 2018) as well as combinatorial patterns of post-translational modifications (Schwaiger et al. 2014; Sebé-Pedrós et al. 2016; Gaiti et al. 2017). Epigenetic studies in the unicellular relative of metazoans, Capsaspora owczarzaki (Sebé-Pedrós et al. 2016), could not find any evidence of bivalency given the absence of H3K27me3, and epigenetic studies in the sponge Amphimedon queenslandica (Gaiti et al. 2017) and the cnidarian Nematostella vectensis (Schwaiger et al. 2014) have also not revealed any evidence for this approach to gene regulation. Further work will be required to establish when bivalent chromatin evolved in animals.
Overall our development of a robust ChIP-seq protocol for use with planarian sorted NBs, together with good coverage for four definitive and essential epigenetic marks, establishes a resource for the future planarian studies investigating the epigenetic regulation of stem cell function.
Methods
Reference assembly and annotations
Previous transcriptome assemblies—Oxford (ox_Smed_v2), Dresden (dd_smed_v4), SmedGD Asexual, Smed GD Unigenes—were downloaded from PlanMine (Brandl et al. 2016) and Smed GD 2.0 (Robb et al. 2008). NCBI complete CDS sequences for Schmidtea mediterranea were also downloaded. Sequences were aligned to the SmedGD Asexual 1.1 genome with GMAP (Wu and Watanabe 2005) and consolidated with PASA (Haas et al. 2003). An independent reference assembly was also performed by mapping 164 available RNA-seq data sets with HISAT2, and assembly was performed using StringTie (Pertea et al. 2015, 2016). The PASA consolidated and StringTie annotations were merged with StringTie.
An intron Jaccard score (intersection of introns/union of introns) was calculated for all overlapping transcripts. Pairwise Jaccard similarity scores of 0.9 or greater were used to create a graph of similar annotations. From the resultant cliques of transcripts, one was chosen to be the representative transcript for the locus, by prioritizing transcript length, ORF length, and BLAST homology.
Strand information for annotations was assigned by utilizing in-house strand-specific RNA-seq libraries, BLAST homology, and longest ORF length. Transdecoder was run utilizing PFAM and UniProt evidence to identify protein-coding transcripts in the annotations. Detailed methods are recorded in an IPython notebook (Supplemental File 3).
FACS proportional expression value generation for annotated loci
Kallisto (Bray et al. 2016) was used to pseudoalign RNA-seq libraries originating from four laboratories (Supplemental Fig. 2; Labbé et al. 2012; Önal et al. 2012; Van Wolfswinkel et al. 2014; Duncan et al. 2015; Zhu et al. 2015) to our expression-based annotation of the asexual S. mediterranea genome. This generated TPM values for each annotated locus. Sleuth was used to calculate a normalization factor for each library (Pimentel et al. 2017). For each locus, the TPM values of member transcripts (potential isoforms) were summed to generate a consensus TPM value and then normalized accordingly. Replicates within each laboratory data set were then averaged.
Normalized TPM values for each laboratory data set were converted to a proportional value as a representation of expression in FACS categories. We next calculated three sets of pairwise ratios (X1:X2, X1:Xins, X2:Xins) using these proportional values. Given two of the three ratios, a third ratio can be “predicted.” Consequently, we calculated three “observed” ratios and three “predicted ratios.” A good Spearman's rank correlation was observed for the X2:Xins ratio and as such we kept these observed proportions and calculated an inferred X1 proportion. Detailed methodology is documented in Supplemental File 4, and full list of X1, X2, and Xins proportional values is available in Supplemental File 5.
ChIP-seq protocol
For each experimental replicate, 600,000–700,000 planarian X1 cells were isolated (sufficient for ChIP-seq of three histone marks and an input control) by utilization of a published FACS protocol (Romero et al. 2012). We dissociated cells from an equal number of head, pharyngeal, or tail pieces from 3-d regenerating planarians. For whole worm ChIP-seq, wild-type worms were starved for 2 wk prior to dissociation.
Following FACS, cells were pelleted. The pellet was resuspended in Nuclei Extraction Buffer (0.5% NP-40, 0.25% Triton X-100, 10 mM Tris-Cl at pH 7.5, 3 mM CaCl2, 0.25 mM Sucrose, 1 mM DTT, phosphatase cocktail inhibitor 2, phosphatase cocktail inhibitor 3). A 3% Drosophila S2 cell spike-in was added at this point. This was followed by 1% formaldehyde fixation for 7 min, which was quenched with the addition of glycine to a final 125 mM concentration. The nuclei pellet was resuspended in SDS lysis buffer (1% SDS, 50 mM Tris-Cl at pH 8.0, 10 mM EDTA) and incubated on ice, followed by the addition of ChIP dilution buffer. Samples were sonicated and one-tenth volume of Triton X-100 was added to dilute SDS in the solution. Samples were pelleted, and supernatant was collected that contained the sonicated chromatin. Test de-crosslinking was performed on one-eighth of the sonicated chromatin and analyzed using a 4200 TapeStation (Agilent) DNA HS tape to verify the DNA fragment range was between 100 and 500 bp. Commercial Drosophila S2 chromatin (Active Motif 53083) spike-in was added at this point (at 3% of the amount of amount of S. mediterranea prepared chromatin) if S2 cells had not been added earlier before chromatin preparation.
Protein A-covered Dynabeads were used for immunoprecipitation (IP). Fifty microliters of Dynabeads were incubated overnight at 4°C with 7 µg of antibody (H3K4me3 Abcam ab8580; H3K36me3 Abcam ab9050; H3K4me1 Abcam ab8895; H3K27me3 Abcam ab6002; RNAPII-Ser5P Abcam ab5131) diluted in 0.5% BSA/PBS. Following incubation, Dynabeads were washed with 0.5% BSA/PBS, and one-fourth of the total isolated chromatin was added per IP. Following overnight incubation, washes were performed six times with RIPA buffer (50 mM HEPES-KOH at pH 8.0, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% DOC, protease inhibitors). Dynabeads were washed with TE buffer and resuspended in Elution Buffer (50 mM Tris-Cl at pH 8.0, 10 mM EDTA, 1% SDS). Protein was separated from Dynabeads by incubating for 15 min at 65°C on a shaking heat block at 1400 rpm. Eluates were de-crosslinked at 65°C overnight. Input chromatin (one-eighth of the total chromatin amount) was also de-crosslinked at this point. Following incubation, RNase A (0.2 µg) and Proteinase K (0.2 µg) was added to each sample and incubated for 1 h. DNA was purified with phenol:chloroform extraction followed by ethanol precipitation. DNA is resuspended in TE and quantified with Qubit dsDNA HS kit. NEB Ultra II kit was used for library preparation, and clean-up was performed with Agencourt Ampure XP beads. Samples were paired-end sequenced at a length of 75 nt on the Illumina NextSeq.
ChIP-seq analysis
Reads were trimmed with Trimmomatic (Bolger et al. 2014) and aligned to a concatenated file containing both our annotated Schmidtea mediterranea genome as well as the Drosophila melanogaster release 6 reference genome (Hoskins et al. 2015) using BWA-MEM 0.7.12 (Li and Durbin 2009). Only uniquely mapping reads were considered further. Paired reads that map to both species were also removed. Picard tools-1.115 (https://broadinstitute.github.io/picard/) was used to remove duplicate reads. Reads were separated into sets that mapped to Drosophila or S. mediterranea using custom Python scripts (documented in the IPython notebook in Supplemental File 6). The number of reads aligning to the Drosophila genome were calculated for use in normalization calculations. For each paired or single map read, coordinates representing 100 bp at the center of the sequence were parsed and written to a BED file.
The genomecov function was used in BEDTools 2.27.0 (Quinlan and Hall 2010) to generate coverage tracks in bedgraph format. The resultant bedgraph file was converted to bigwig format using UCSC's bedgraphtoBigWig tool (Kent et al. 2010). deepTools2's computeMatrix was used to extract coverage around 2.5 kb or 5 kb either side of the annotated TSS for each annotated locus in 50-bp windows for each sample and corresponding input (Ramírez et al. 2016). A normalization factor was calculated using the number of mapped reads corresponding to the Drosophila spike-in to control for between IP technical variation (Orlando et al. 2014). A scaling factor for input ChIP-seq libraries was calculated using the deepTools2 Python API that uses the SES method (Diaz et al. 2012). The mean normalized coverage was calculated for each sample and input. The normalized input coverage was subtracted from the normalized sample coverage to generate a final coverage track for downstream visualization and analyses. Individual gene profiles for given ChIP-seq tracks could then be visualized, and sequences for those genes plotted in this paper are given in Supplemental File 7.
To calculate the correlation of ChIP-seq signal coverage to proportional FACS expression, two vector values were calculated. The first vector was proportional FACS expression for all genomic loci. The second vector was ChIP-seq coverage at each 50-bp position 2.5 kb either side of the TSS. A Spearman's rank correlation was performed on both vectors yielding a correlation value for the assayed position. The correlation value for each nonoverlapping 50-bp window was then plotted on a graph.
For comparison of profiles between different epigenetic marks, a percentage coverage was calculated for each mark. The maximum coverage was found across all 5- or 10-kb regions for all loci. Absolute normalized coverage for each 50-bp window was then divided by the maximum coverage observed for that mark in the genome, resulting in a percentage coverage in each 50-bp window for each mark.
For calculation of a pausing index for individual genes, we divided normalized coverage to input 500 bp either side of the annotated TSS by the coverage between 500 bp and 2.5 kb downstream from the TSS.
Detailed methods for ChIP-seq analysis are documented in Supplemental File 6.
Data access
ChIP short read data from this study have been submitted to the NCBI BioProject (BioProject; https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA471851 and PRJNA338116. Annotations made on the S. mediterranea genome and used in this study are available as a compressed GFF file (Supplemental File 8).
Supplementary Material
Acknowledgments
This work was funded by grants from the Medical Research Council (MRC) (MR/M000133/1) and the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/K007564/1) awarded to A.A.A. A.D. is funded by a BBSRC studentship (BB/J014427/1).
Author contributions: A.A.A. originally conceived and designed the study, upon which A.D. innovated. A.D., Y.M., and P.A. performed ChIP-seq experiments. S.H., S.S., and A.L. assisted with optimization of ChIP-seq and RNA-seq protocols. A.D. and D.K. performed bioinformatic analyses. A.D., D.K., and A.A.A. wrote, reviewed, and revised the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.239848.118.
References
- Aboobaker AA. 2011. Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol 21: 304–311. [DOI] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs KJ, Stunnenberg HG, Veenstra GJC. 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell 17: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alié A, Hayashi T, Sugimura I, Manuel M, Sugano W, Mano A, Satoh N, Agata K, Funayama N. 2015. The ancestral gene repertoire of animal stem cells. Proc Natl Acad Sci 112: E7093–E7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñá J, Romero R. 1981. Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia 84: 181–194. [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bledau AS, Schmidt K, Neumann K, Hill U, Ciotta G, Gupta A, Torres DC, Fu J, Kranz A, Stewart AF, et al. 2014. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 141: 1022–1035. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl H, Moon HK, Vila-Farré M, Liu SY, Henry I, Rink JC. 2016. PlanMine—a mineable resource of planarian biology and biodiversity. Nucleic Acids Res 44: D764–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Brookes E, De Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. 2012. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Alvarado AS, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. 2014. A role for H3K4 mono-methylation in gene repression and partitioning of chromatin readers. Mol Cell 53: 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL. 2013. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem Rev 113: 8423–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattani A, Sridhar D, Aziz Aboobaker A. 2018. Planarian flatworms as a new model system for understanding the epigenetic regulation of stem cell pluripotency and differentiation. Semin Cell Dev Biol. 10.1016/j.semcdb.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. 2014. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141: 526–537. [DOI] [PubMed] [Google Scholar]
- Diaz A, Park K, Lim DA, Song JS. 2012. Normalization, bias correction, and peak calling for ChIP-seq. Stat Appl Genet Mol Biol 11 10.1515/1544-6115.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EM, Chitsazan AD, Seidel CW, Sánchez Alvarado A. 2015. Set1 and MLL1/2 target distinct sets of functionally different genomic loci in vivo. Cell Rep 13: 2741–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, Torlai Triglia E, Risner-Janiczek JR, Rito T, Rackham OJ, de Santiago I, Kukalev A, Nicodemi M, Akalin A, Li M, et al. 2017. RNA polymerase II primes Polycomb-repressed developmental genes throughout terminal neuronal differentiation. Mol Syst Biol 13: 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW. 2018. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360: eaaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiti F, Jindrich K, Fernandez-Valverde SL, Roper KE, Degnan BM, Tanurdžić M. 2017. Landscape of histone modifications in a sponge reveals the origin of animal cis-regulatory complexity. eLife 6: e22194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, Chen X. 2010. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol 11: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A, Meshorer E. 2015. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep 16: 1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, et al. 2011. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res 21: 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. 2006. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ 48: 371–380. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al. 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A. 2013. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol 20: 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert A, Henderson JM, Ross KG, Cowles MW, Torres J, Zayas RM. 2013. Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases. Epigenetics 8: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber-Hijazi F, Lo PJ, Mihaylova Y, Foster JM, Benner JS, Tejada Romero B, Chen C, Malla S, Solana J, Ruzov A, et al. 2013. Planarian MBD2/3 is required for adult stem cell pluripotency independently of DNA methylation. Dev Biol 384: 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Swartz SZ, Wessel GM. 2010. A conserved germline multipotency program. Development 137: 4113–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Jung YL, McElroy KA, Zee BM, Wallace HA, Woolnough JL, Park PJ, Kuroda MI. 2017. Bivalent complexes of PRC1 with orthologs of BRD4 and MOZ/MORF target developmental genes in Drosophila. Genes Dev 31: 1988–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé RM, Irimia M, Currie KW, Lin A, Zhu SJ, Brown DD, Ross EJ, Voisin V, Bader GD, Blencowe BJ, et al. 2012. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30: 1734–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AG, Aboobaker AA. 2018. EvoRegen in animals: time to uncover deep conservation or convergence of adult stem cell evolution and regenerative processes. Dev Biol 433: 118–131. [DOI] [PubMed] [Google Scholar]
- Lai AG, Kosaka N, Abnave P, Sahu S, Aboobaker AA. 2018. The abrogation of condensin function provides independent evidence for defining the self-renewing population of pluripotent stem cells. Dev Biol 433: 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Page DC. 2014. Poised chromatin in the mammalian germ line. Development 141: 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. 2013. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci 110: 16061–16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Silber SJ, McCarrey JR, Page DC. 2016. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet 48: 888–894. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277: 49383–49388. [DOI] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. 2013. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 4: 189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu X, Zhang H, Pfeifer GP, Lu Q. 2017. Dynamics of RNA polymerase II pausing and bivalent histone H3 methylation during neuronal differentiation in brain development. Cell Rep 20: 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova Y, Abnave P, Kao D, Hughes S, Lai A, Jaber-Hijazi F, Kosaka N, Aboobaker A. 2017. Conservation of epigenetic regulation by the MLL3/4 tumour suppressor in planarian pluripotent stem cells. 10.1101/126540. [DOI] [PMC free article] [PubMed]
- Molinaro AM, Pearson BJ. 2016. In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol 17: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önal P, Grün D, Adamidi C, Rybak A, Solana J, Mastrobuoni G, Wang Y, Rahn HP, Chen W, Kempa S, et al. 2012. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J 31: 2755–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. 2014. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9: 1163–1170. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. 2008. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14: 1174–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. 2009. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. [DOI] [PubMed] [Google Scholar]
- Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N. 2018. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360: eaaq1723. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. 2005. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1327–1330. [DOI] [PubMed] [Google Scholar]
- Rink JC. 2013. Stem cell systems and regeneration in planaria. Dev Genes Evol 223: 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb SM, Ross E, Sánchez Alvarado A. 2008. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res 36: D599–D606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero BT, Evans DJ, Aboobaker AA. 2012. FACS analysis of the planarian stem cell compartment as a tool to understand regenerative mechanisms. Methods Mol Biol 916: 167–179. [DOI] [PubMed] [Google Scholar]
- Sachs M, Onodera C, Blaschke K, Ebata K, Song J, Ramalho-Santos M. 2013. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep 3: 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, Van Lohuizen M, Tanay A, Cavalli G. 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Schönauer A, Rendeiro AF, Pribitzer C, Schauer A, Gilles AF, Schinko JB, Renfer E, Fredman D, Technau U. 2014. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res 24: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Meisel J, Reddien PW. 2010. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development 137: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, Reddien PW. 2014. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports 3: 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Ballaré C, Parra-Acero H, Chiva C, Tena JJ, Sabidó E, Gómez-Skarmeta JL, Di Croce L, Ruiz-Trillo I. 2016. The dynamic regulatory genome of Capsaspora and the origin of animal multicellularity. Cell 165: 1224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Kashima M, Ishiko T, Nishimura O, Rouhana L, Misaki K, Yonemura S, Saito K, Siomi H, Siomi MC, et al. 2016. Inheritance of a nuclear PIWI from pluripotent stem cells by somatic descendants ensures differentiation by silencing transposons in planarian. Dev Cell 37: 226–237. [DOI] [PubMed] [Google Scholar]
- Solana J. 2013. Closing the circle of germline and stem cells: the Primordial Stem Cell hypothesis. Evodevo 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Kao D, Mihaylova Y, Jaber-Hijazi F, Malla S, Wilson R, Aboobaker A. 2012. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol 13: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Irimia M, Ayoub S, Orejuela MR, Zywitza V, Jens M, Tapial J, Ray D, Morris Q, Hughes TR, et al. 2016. Conserved functional antagonism of CELF and MBNL proteins controls stem cell-specific alternative splicing in planarians. eLife 5: e16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435. [DOI] [PubMed] [Google Scholar]
- Van Wolfswinkel JC, Wagner DE, Reddien PW. 2014. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15: 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez-Doorman C, Petersen CP. 2016. The NuRD complex component p66 suppresses photoreceptor neuron regeneration in planarians. Regeneration (Oxf.) 3: 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. 2013. A double take on bivalent promoters. Genes Dev 27: 1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. [DOI] [PubMed] [Google Scholar]
- Wu SF, Zhang H, Cairns BR. 2011. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res 21: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW. 2015. A generic and cell-type-specific wound response precedes regeneration in planarians. Dev Cell 35: 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Stiller JW. 2014. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci 111: 5920–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Hallows SE, Currie KW, Xu C, Pearson BJ. 2015. A mex3 homolog is required for differentiation during planarian stem cell lineage development. eLife 4: e07025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.