Abstract
Recent studies have revealed that normal microbiota interacts with the host through four mechanisms: the normal microbiome acts as a barrier against pathogens; second, as modulators of the permeability of host mucosa; third, as modulators of energy extraction from, and metabolic utilization of ingested food; and lastly, as modulators of the immune system. An alteration of the normal microbiota increases predisposition of the host to diseases through these four mechanisms.
Introduction
The discovery of the human microbiota has broadened our understanding of germs. Infections by microbes, collectively termed germs (i.e. bacteria, fungi, viruses and helminthes), have been a persistent cause of morbidity and mortality in humans for thousands of years. Pasteur and others developed the Germ Theory over 150 years ago, which posits that eliminating germs eliminates disease. Germ Theory has contributed immensely to human health. For a thousand years prior to the construction of Germ Theory the average life expectancy had been 20 years or less. As a consequence of improved sanitation, hygiene and public health efforts to minimize the transmission of germs, life span doubled.1 New culturing techniques were established to grow and identify pathogens. Development of methods to culture pathogens led to the development of new antibiotics and tests for sensitivity to antibiotics. In addition to culturing, development of staining and structural techniques identified a number of virulence factors, products made by a microbe that determines the degree of pathogenicity to the host.
More recently, organisms could be identified without culturing because of the advent of polymerase chain reaction (PCR) amplification and decreased cost of DNA sequencing. Culture independent identification has revealed communities of microbes colonize many anatomical surfaces, such as skin and mucosa (oral, respiratory, urogenital and gastrointestinal) of healthy individuals.2 These results show that very few germs are harmful (i.e. pathogens), while the majority of microbes are neutral (commensals) and some are beneficial to the host (symbionts).
One shortcoming of Germ Theory is the inability to explain the variance of host response to infections. Typically, the risk of, and ability to survive, an infection has been attributed to host genetics. Surprisingly, recent research shows that the configuration and constituents of a host’s normal germ flora influences the risk for disease. An abnormal flora, called dysbiosis, can increase the risk of obesity and inflammatory diseases. Additionally, recent studies make a compelling argument for dysbiosis increasing risk of autoimmune and cognitive diseases. Thus, the unexpected finding that the normal flora plays a crucial role in maintaining health (homeostasis) has emerged. In this review, we analyze recent studies where an abnormal microbial flora promotes disease by four different mechanisms. We argue that an abnormal microbial flora appears to be a risk factor for disease. Moreover, the spectrum between a normal and abnormal microbial flora may contribute to disease risk as much as the genetics (allelic differences) in the population.
Definitions, Nomenclature, and Methods for Measuring the Microbiota
Microbial communities exist within any given environment. The collection of microbes is referred to as the microbiota. The collective genomes of the microbiota are referred to as the microbiome. Such communities could not be identified previously because less than 1% (but 40 to 50% of pathogens) bacteria can be currently cultured. Shotgun sequencing of the microbiome is termed metagenomics. In some instances, it is desirable to simply identify the bacterial and fungal species (i.e. “who is present?”), rather than sequencing entire genomes. This is accomplished by amplification of the 16S ribosomal RNA (16S rRNA) gene. The 16S rRNA is an essential component of the small subunit of the ribosome that performs protein translation in bacteria. The 16S rRNA gene can be readily amplified because there are invariant blocks (See green segments in Figure 1) of sequences surrounding variable regions. Primers for PCR amplification designed for the invariant regions can amplify (nearly) all species in the kingdom Bacteria. The variable regions (See gray segments in Figure 1) vary between species but are conserved within a species, and thus the sequence can be used to identify the organism. Massively parallel sequencing (also called next generation sequencing) of the PCR amplicons is used to identify bacterial species and the number of times a sequence is observed is used to quantitate the amount of bacteria present, in any collected sample requiring no culturing.
Figure 1.
Structure of 16S ribosomal RNA gene in kingdom Bacteria. Primers in the conserved (green) regions are used to amplify segments of the gene and are subject to DNA sequencing. The sequence of the variable regions (V1 through 9) varies between species, but is nearly identical within species (and similar between related species, e.g. Staphylococcus aureus and S. epidermis). The sequence of the variable region is thus used to identify the organism present in the sample. Although, any variable region can be sequenced for identification, sequencing of V4 segment provides robust results.
Using these culture-independent techniques, investigators have shown that the skin, upper airway mucosal surfaces, the mouth, gums, tongue, the entire digestive and urogenital tract have a unique community of microbes at each anatomical site. The communities are dynamic and respond to non-physiological (e.g. infections, trauma and antibiotics) and physiological (e.g. pregnancy) changes in the host. For instance, the gut microbes change with age, fiber intake,3 and the vaginal microbiome changes during pregnancy and with the gestational age of the fetus.4–6 There are now several examples where changes in the microbiota lead to disease states. We review a few of these studies because they document that the microbiota plays an important role in nutrient uptake and utilization (conversion of food to usable calories for the host), in altering permeability of barriers that exist within the host and in altering the immune system.
The Microbiota Influences Energy Extraction and Utilization Leading to Obesity
The relationship between the gut microbiota and diet has been an area of interest for many years. For instance, it was noted in the 1960s that germ-free mice (referred to as Gnotobiotic mice, these mice are born by Cesarean section, then maintained under a sterile environment, including being fed autoclaved sterile food and water) require 30% more calories to maintain the same weight as their conventionally raised siblings.7 Theses studies with mice suggested that efficient energy extraction from food requires a gut microflora. In the last decade, the role of gut microbiota in modulation of energy extraction in humans has been elucidated in the context of obesity and is described next.
Obesity is a multifactorial process that includes, but is not limited to, host genetics, environment, socioeconomic and dietary variables.8,9 It is associated with a number of comorbidities including fatty liver disease, cardiovascular disease, insulin resistance and diabetes. Within this complex set of interactions lies the microbiota. A number of recent reviews have discussed the role of the microbiota in obesity. 10–12 The hypothesis that the microbiota and obesity are correlated is supported by observations by Turnbaugh et al. which demonstrated that the microbiomes of lean and obese monozygotic twins were different.13,14 Further, the obesity–associated microbiota could be transferred from human host to individual germ-free mice as well. Observations such as these have lead to studies that have uncovered potential pathways for the microbiota to support obesity. Such pathways include the breakdown of food to usable nutrients (e.g. plant starch to sugars), the release of metabolic products by bacteria that aid absorption of nutrients and the production of cofactors (e.g. vitamins B and K) that are needed by the host.
In addition to the bacterial metabolic products directly affecting the host metabolism, increased immune activity has been recognized in obesity and associated with the microbiota. Lipopolysaccharides (LPS) from gram-negative bacteria in the microbiota are found to increase in obesity. LPS activates host’s immune system through Toll-like receptors on monocytes. Lean individuals have lower concentrations of LPS in the blood compared to obese individuals15,16. Furthermore, there is an association between elevated LPS and decreased insulin sensitivity.15,16 These studies establish a fundamental link between obesity and insulin sensitivity through inflammation.
Endocrine functions have also been identified as a mechanism by which the gut microbiota can lead to obesity. Bacterial fermentation products can induce the secretion of hormone peptide YY and leptin, by signaling through G protein–coupled receptors (GPCR) which are present in the small intestine, colon and adipocytes. Changes in these hormones can affect caloric intake. As an example, genetically obese mice have distinct differences in the microbiota resulting in different leptin levels.17 Together, these studies have established a number of mechanisms by which dysbiosis (directly or indirectly) leads to risk of obesity. These mechanisms include short chain fatty acid (SCFA) byproducts, inflammation, endocrine functions, endo-cannabinoid tone and bile acid production.
The Host Microbiota Acts as a Barrier or Deterrent for Pathogens
Clostridium difficile infection (CDI) is an example of how the intestinal microbiota acts as a barrier. Loss of a healthy microbiota leads to conditions that allow C. difficile to infect and to cause disease. In 1977, it was noted that CDI required previous antibiotic exposure of the patient.18,19 Multiple studies in humans and mice have subsequently shown that antibiotics can have a profound and sometimes long standing effect on the gastrointestinal (GI) microbiota. These changes are produced by alterations in the metabolites from both GI enterocytes and the microbiota,20,21 as well as by the overall structure of the microbiota.22 The mechanisms by which antibiotics cause dysbiosis (abnormal microbiota) and subsequent CDI have been shown to include toll-like receptor (TLR) signaling, changes in T-helper 17 T-cells (TH17), and changes in epithelial permeability.23 Supporting the idea that changes in the microbiota lead to CDI is the compelling evidence that restoring the diversity of the microbiota by fecal transplantation can lead to resolution and resistance to CDI. Patients with CDI can eliminate C. difficile after transplantation of healthy microbiota24. Studies are currently evolving to define how healthy microbiota from different individuals with considerable variability can have the same result of resolution of CDI. It is possible that fecal transplant may support the growth of suppressed indigenous microbiota leading to CDI resolution and resistance22.
The Gut Microbiota Can Influence Mucosal Barrier Permeability
All biological systems are organized into smaller subsystems. For example, the human body anatomically consists of five critical subsystems or organs, viz. heart, lungs, liver, kidneys and brain. Indeed, there are over 60 organs. Each of these anatomical subsystems are separated or compartmentalized by a barrier, connected by lymphatic and circulatory subsystem. Flow of materials across the barrier is highly regulated. For instance, ingested food is broken down and digested in the stomach and some of the resulting nutrients are absorbed in the stomach and intestines to be used by the rest of the body. It has become evident that species within the gut microbiota not only play an important role in metabolizing the ingested food but some of the resident bacteria, directly or through the immune system, regulate the permeability of the gut mucosa to determine the absorption of the nutrients. Additionally, species within the microbiota encode for a number of enzymes, whose products alter gut permeability. For example, production of histamine via microbial encoded histidine decarboxylase increases mucosal permeability.25 Altering the permeability not only affects that transfer of nutrients and bacterial metabolites, but also affects the immune system. A rather spectacular example of how gut dysbiosis can affect the central nervous system, across several barriers, is in autism spectrum disorders (ASD).
ASD is a disease of unknown origin but it has been linked to several triggers including pre- or post natal exposure to chemicals and drugs, air pollutants, maternal infection, stress and dietary factors.26 Several comorbidities have also been associated with ASD including gastrointestinal distress.27 Studies in both murine and human systems have defined potential pathways that connect ASD to changes in the microbiota of the gut. Mouse strains that have symptoms of ASD behavior have been shown to have gut barrier functional defects and dysbiosis.28 In one study the authors showed that providing Bacteriodes fragillis to the microbiota of these same mice early in development reduced autism behavior. This same study implicated microbiota produced metabolic products such as 4-ethylphenylsulfate were directly linked to symptoms of ASD in mice.28 Pregnant mice treated with valproic acid, a compound shown to change the murine gut microbiota, resulted in ASD phenotypes in offspring.29,30 Gnotobiotic mice have reduced sociability, social cognition deficits and repetitive grooming, similar to behaviors observed in ASD. These symptoms can be at least partially reversed by colonization of the germ free mice.31
In human studies, children with ASD have a distinct and less diverse gut microbiota and gut community structure.32 In one study, children with ASD were found to have lower levels of the species Bifidobacterium and higher levels of Lactobacillus leading to change in the metabolites acetate, propionate and valerate.33 Another set of studies have shown changes in metabolites, LPS and potential neuromodulating compounds such as IL-6 in ASD patients.34 IL-6 has been shown to increase permeability of the blood brain barrier and LPS has been linked to an increase in GI permeability. Interestingly, children with ASD treated with antibiotics directed against bacteria that produce LPS resulted in improvement of some of their cognitive skills.35 Taken together the above examples indicate that the gut microbiota contributes to the development of ASD by affecting the barrier permeability of the gut and possibly the blood-brain barrier.
Direct Link Between Microbiota and the Host Immune System
The host immune system regulates the constituents and the abundance of the microbiota through a number of mechanisms. First, epithelial cells produce a number of anti-microbial proteins (AMP). These peptides belong in three families: the defensins, cathelicidins, and histamins30. Second, IgA secreted by B-cells or plasma cells shapes the abundance of the microbes.31, 32 T-cells, like B-cells are part of the adaptive immune system, are educated during early development to recognize self-antigens. Similarly, exposure to bacterial and viral antigens during early development leads to education of the adaptive immune system. As a consequence, the immune system does not respond to these organisms. Moreover, these bacterial antigens are needed for the development of a normal immune system as is evident in germ-free mice.36 The commensal gut microbes produce low-level activation of both the innate and adaptive immune system.37 This low-level activation sets the threshold that must be exceeded to produce an appropriate immune response to pathogens. Perhaps simplistically, in the absence of a normal microbiome the activation of the immune system is set to a too low threshold, and consequently the immune system is hyper-responsive. Correspondingly, epidemiological evidence over the last three decades indicates that changes in the microbiota can alter the risk factors for allergies, autoimmune and other inflammatory diseases.38 These epidemiological studies parallel the so-called hygiene hypothesis39–41 further supports the important role of the normal microbiome in maintaining homeostasis of the immune system.
Interactions Between These “Host-Factors”
As we have documented above, the normal microbiome affects the host through a number of mechanisms. The main four mechanisms are: first, by modulating calorie extraction from the diet and nutrient utilization; second, by acting as a deterrent to pathogen colonization; third, by modulating the immune response; and finally, and by modulating barrier permeability directly through secretion of chemical mediators and indirectly through the immune system. Moreover, there is considerable crosstalk between the immune system and barriers and metabolism, which are in turn influenced by genetics and the environment (See Figure 3). The crosstalk increases the complexity of the system in the sense that we must understand each subsystem (e.g. how an immune response is regulated; See Figure 2), but also in the sense that risk or benefit cannot be assigned to any one factor or component (e.g. how effective the immune response is). This crosstalk evolved to increase the overall robustness of the system because transient loss or decrease in function in one subsystem can be compensated for by the other subsystems.
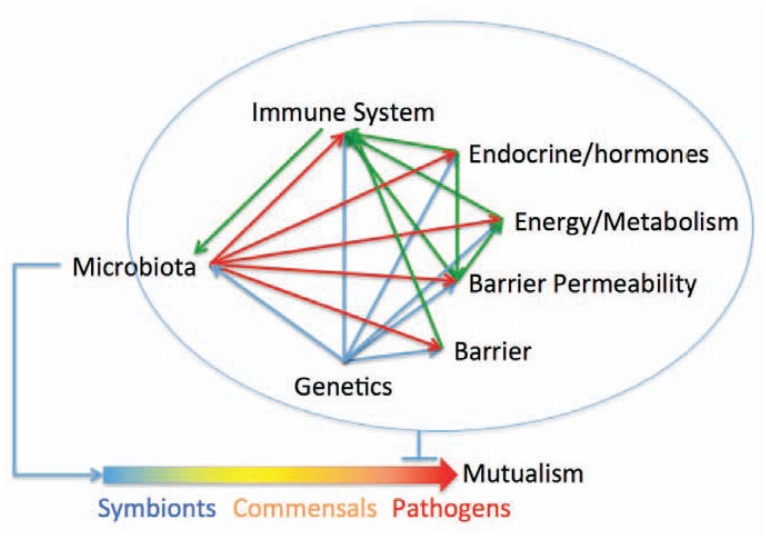
Figure 3.
Interactions between the microbiome and amongst the host subsystems that may account for variability in response to pathogens. The response of different individuals to pathogens can vary from being resistant, to asymptomatic carriers, to completely sensitive. Host genetics only partially account for this variability. Additional non-heritable influences such as prior exposure to related pathogens and nutritional status of the host could also factor into the response. We argue that the microbiota, because it influences the host health status, also influences the health of the host. Thus, dysbioses is a host risk factor that determines susceptibility to pathogens such as C. difficile infections, and to obesity and autism. The crosstalk between the subsystems provides robustness and maintains health. The crosstalk provides robustness because other subsystems can compensate for the transient loss of one subsystem. The contribution of genetics is shown by blue lines, physiological interactions are shown in green and the influence of microbiota is shown in red.
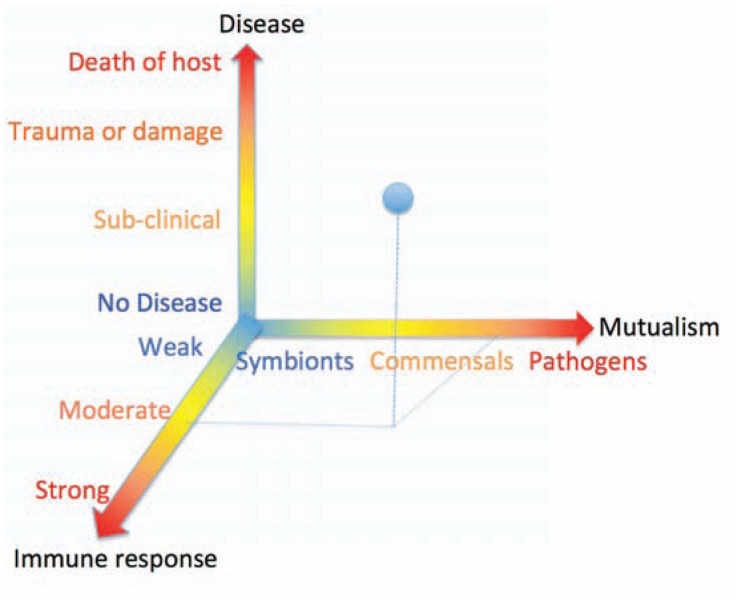
Figure 2.
The relationship between germs, immune response and disease: Germs (viruses, bacteria, fungi, helminthes) are shown as a spectrum of mutualism: on one end of the spectrum are the symbionts (in blue) and one the other extreme, the pathogens (red). Pathogens can be more or less pathogenic. Between the two extremes (in yellow) of the spectrum are commensals, which are neither beneficial nor harmful. The other axis depicted at the bottom is the host immune response, which maintains the normal flora and responds to pathogens. The vertical axis depicts the disease states, which can vary from healthy (no disease) to death of host. A healthy immune response to a moderate pathogen (shown by dashed lines) can eliminate the pathogen or lead to subclinical disease (shown as blue dot). This representation also shows that an aberrant immune response to commensals (or self antigens) can also lead to chronic inflammation and disease.
Conclusion
We began this review by pointing out the strengths and shortcomings of Germ Theory. The strength of Germ Theory is that it produced an increased awareness of pathogens that led to some relatively simple solutions like sewers, clean water sources, and improved hygiene to prevent the spread of these pathogens. The primary shortcoming of Germ Theory has been that it cannot readily explain the variation in host response (e.g. why some individuals are asymptomatic carriers and others succumb to the infections). Conventionally, it has been argued that host genetics (heritable alleles) are in large part responsible for this variation. However, emerging data indicates that epigenetics and environmental factors that are non-heritable contribute significantly. The variation occurs through both heritable and non-heritable factors. We propose that the normal (healthy) microbiota composition, although influenced by genetic factors is a non-heritable component that contributes critically to preventing disease in three significant ways: first, it contributes to setting thresholds of the immune response. As we note above, a weak or overly strong response is harmful to the host because it causes damage. Second, the microbiota forms a barrier by competing for nutrients and space in the host, and further by altering the permeability of mucosa, which prevents pathogens from colonizing, crossing the mucosa and dissemination. Third, the microbiota modulates the extraction and utilization of energy and nutrients from food consumed. As undernourished and overnourished hosts have hypo- or hyper-responsive immune activation respectively, the healthy microbiota contributes to response to a pathogen. Together, these factors contribute to the ability of the host to fight pathogens and to recover from pathogens. Thus, the microbiota is a non-heritable host factor that contributes to maintaining and restoring health by maintaining homeostasis (balance). Given that different populations of patients could have different and unique microbiota, it is interesting to consider how the measurements of microbiota will be incorporated into the practice of precision medicine in the future.
Acknowledgments
We thank Eileen Moran for her aid in improving the clarity of this manuscript. We also thank our students, fellows and colleagues for invaluable discussions.
Biography
Rajeev Aurora, PhD, (left), is in the Molecular Microbiology and Immunology Department, and Thomas Sanford, MD, (right) is in Department of Otolaryngology; both are at the Saint Louis University School of Medicine.
Contact: aurorar@slu.edu


Footnotes
Disclosure
None reported.
References
- 1.Casanova JL, Abel L. The genetic theory of infectious diseases: a brief history and selected illustrations. Annu Rev Genomics Hum Genet. 2013;14:215–243. doi: 10.1146/annurev-genom-091212-153448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero R, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aagaard K, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008599. 237ra265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero R, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2:4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coates ME. Gnotobiotic animals in research: their uses and limitations. Lab Anim. 1975;9:275–282. doi: 10.1258/002367775780957296. [DOI] [PubMed] [Google Scholar]
- 8.Serra-Majem L, Bautista-Castano I. Etiology of obesity: two “key issues” and other emerging factors. Nutr Hosp. 2013;28(Suppl 5):32–43. doi: 10.3305/nh.2013.28.sup5.6916. [DOI] [PubMed] [Google Scholar]
- 9.Gupta D, Krueger CB, Lastra G. Over-nutrition, obesity and insulin resistance in the development of beta-cell dysfunction. Current diabetes reviews. 2012;8:76–83. doi: 10.2174/157339912799424564. [DOI] [PubMed] [Google Scholar]
- 10.Moran CP, Shanahan F. Gut microbiota and obesity: role in aetiology and potential therapeutic target. Best Pract Res Clin Gastroenterol. 2014;28:585–597. doi: 10.1016/j.bpg.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Caricilli AM, Saad MJ. Gut microbiota composition and its effects on obesity and insulin resistance. Curr Opin Clin Nutr Metab Care. 2014;17:312–318. doi: 10.1097/MCO.0000000000000067. [DOI] [PubMed] [Google Scholar]
- 12.Cani PD. Metabolism in 2013: The gut microbiota manages host metabolism. Nat Rev Endocrinol. 2014;10:74–76. doi: 10.1038/nrendo.2013.240. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2008 doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 15.Wakayama S, et al. Lipopolysaccharide impairs insulin sensitivity via activation of phosphoinositide 3-kinase in adipocytes. Immunopharmacol Immunotoxicol. 2014;36:145–149. doi: 10.3109/08923973.2014.887096. [DOI] [PubMed] [Google Scholar]
- 16.Piya MK, McTernan PG, Kumar S. Adipokine inflammation and insulin resistance: the role of glucose, lipids and endotoxin. J Endocrinol. 2013;216:T1–T15. doi: 10.1530/JOE-12-0498. [DOI] [PubMed] [Google Scholar]
- 17.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis. 1977;136:701–705. doi: 10.1093/infdis/136.5.701. [DOI] [PubMed] [Google Scholar]
- 19.Gorbach SL, Thadepalli H. Isolation of Clostridium in human infections: evaluation of 114 cases. J Infect Dis. 1975;131(Suppl):S81–85. doi: 10.1093/infdis/131.supplement.s81. [DOI] [PubMed] [Google Scholar]
- 20.Theriot CM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature communications. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theriot CM, Young VB. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut microbes. 2014;5:86–95. doi: 10.4161/gmic.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology. 2014;146:1547–1553. doi: 10.1053/j.gastro.2014.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borody TJ, Brandt LJ, Paramsothy S, Agrawal G. Fecal microbiota transplantation: a new standard treatment option for Clostridium difficile infection. Expert Rev Anti Infect Ther. 2013;11:447–449. doi: 10.1586/eri.13.26. [DOI] [PubMed] [Google Scholar]
- 25.Landete JM, De las Rivas B, Marcobal A, Munoz R. Updated molecular knowledge about histamine biosynthesis by bacteria. Critical reviews in food science and nutrition. 2008;48:697–714. doi: 10.1080/10408390701639041. [DOI] [PubMed] [Google Scholar]
- 26.Dietert RR, Dietert JM, Dewitt JC. Environmental risk factors for autism. Emerging health threats journal. 2011;4:7111. doi: 10.3402/ehtj.v4i0.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buie T, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125(Suppl 1):S1–18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Theije CG, et al. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 2014;37:197–206. doi: 10.1016/j.bbi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 30.de Theije CG, et al. Intestinal inflammation in a murine model of autism spectrum disorders. Brain Behav Immun. 2014;37:240–247. doi: 10.1016/j.bbi.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Borre YE, Moloney RD, Clarke G, Dinan TG, Cryan JF. The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. Adv Exp Med Biol. 2014;817:373–403. doi: 10.1007/978-1-4939-0897-4_17. [DOI] [PubMed] [Google Scholar]
- 32.Kang DW, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068322. e68322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finegold SM, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Emanuele E, et al. Low-grade endotoxemia in patients with severe autism. Neurosci Lett. 2010;471:162–165. doi: 10.1016/j.neulet.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn M, Grave S, Bransfield R, Harris S. Long term antibiotic therapy may be an effective treatment for children co-morbid with Lyme disease and autism spectrum disorder. Med Hypotheses. 2012;78:606–615. doi: 10.1016/j.mehy.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 36.Kamada N, Nunez G. Role of the gut microbiota in the development and function of lymphoid cells. J Immunol. 2013;190:1389–1395. doi: 10.4049/jimmunol.1203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly D, Mulder IE. Microbiome and immunological interactions. Nutr Rev. 2012;70(Suppl 1):S18–30. doi: 10.1111/j.1753-4887.2012.00498.x. [DOI] [PubMed] [Google Scholar]
- 38.Kosiewicz MM, Zirnheld AL, Alard P. Gut microbiota, immunity, and disease: a complex relationship. Frontiers in microbiology. 2011;2:180. doi: 10.3389/fmicb.2011.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bendiks M, Kopp MV. The relationship between advances in understanding the microbiome and the maturing hygiene hypothesis. Curr Allergy Asthma Rep. 2013;13:487–494. doi: 10.1007/s11882-013-0382-8. [DOI] [PubMed] [Google Scholar]
- 40.Romagnani S. The increased prevalence of allergy and the hygiene hypothesis: missing immune deviation, reduced immune suppression, or both? Immunology. 2004;112:352–363. doi: 10.1111/j.1365-2567.2004.01925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]