Spontaneous CLIP dissociation from an autoimmunity-associated MHC II protein enhances presentation of peptides released by insulin-producing β cells. Presentation of such extracellular peptides does not require endosomal antigen processing and augments islet infiltration by CD4 T cells.
Abstract
A number of autoimmunity-associated MHC class II proteins interact only weakly with the invariant chain–derived class II–associated invariant chain peptide (CLIP). CLIP dissociates rapidly from I-Ag7 even in the absence of DM, and this property is related to the type 1 diabetes–associated β57 polymorphism. We generated knock-in non-obese diabetic (NOD) mice with a single amino acid change in the CLIP segment of the invariant chain in order to moderately slow CLIP dissociation from I-Ag7. These knock-in mice had a significantly reduced incidence of spontaneous type 1 diabetes and diminished islet infiltration by CD4 T cells, in particular T cells specific for fusion peptides generated by covalent linkage of proteolytic fragments within β cell secretory granules. Rapid CLIP dissociation enhanced the presentation of such extracellular peptides, thus bypassing the conventional MHC class II antigen-processing pathway. Autoimmunity-associated MHC class II polymorphisms therefore not only modify binding of self-peptides, but also alter the biochemistry of peptide acquisition.
Introduction
The MHC locus plays a central role in susceptibility to many human autoimmune diseases, including type 1 diabetes (T1D; Davies et al., 1994; Hu et al., 2015). In many of these diseases, the strongest association is observed with particular alleles of MHC class II (MHC II) genes, providing strong evidence for a critical role of antigen presentation to CD4 T cells. T1D is an excellent example for this general principle: susceptibility is most closely associated with certain alleles of the DQB1 gene, in particular those encoding HLA-DQ8 (DQB1*03:02) and HLA-DQ2 (DQB1*02:01; Todd et al., 1987; Concannon et al., 2009). The DQB1*03:02 and DQB1*02:01 alleles share an important polymorphism: position 57 of the β chain encodes a nonaspartic residue in T1D-associated alleles, but aspartic acid in most other DQB1 alleles. This polymorphism is also relevant for the spontaneous mouse model of T1D in non-obese diabetic (NOD) mice because β57 of I-Ag7 is also a nonaspartic acid residue (serine; Acha-Orbea and McDevitt, 1987). Crystal structures of DQ8, DQ2, and I-Ag7 have demonstrated that this polymorphic position has a major impact on the charge of the P9 pocket of the peptide binding groove (Corper et al., 2000; Lee et al., 2001; Kim et al., 2004). An aspartic acid at β57 forms a salt bridge with arginine 76 of the DQ or I-A α chains, allowing binding of hydrophobic amino acids in the P9 pocket (Brown et al., 1993; Scott et al., 1998). In contrast, the absence of a negative charge at β57 of DQ8, DQ2, and I-Ag7 results in a P9 pocket with a positive charge that has a strong preference for negatively charged peptide side chains (Corper et al., 2000; Lee et al., 2001; Kim et al., 2004). The major hypothesis for the role of the β57 polymorphism in the pathogenesis of T1D has been that it enables binding of pathogenic peptides (Todd et al., 1987; Quartey-Papafio et al., 1995). As we will discuss here, the β57 polymorphism also has an impact on the affinity of the invariant chain–derived class II–associated invariant chain peptide (CLIP), and may therefore also modulate the biochemistry of peptide binding.
MHC II proteins associate with the invariant chain in the ER, and this complex is targeted to the endosomal compartment, where the invariant chain is degraded, leaving CLIP in the binding groove (Avva and Cresswell, 1994; Denzin and Cresswell, 1995). Textbooks state that H2-DM (abbreviated as DM) induces CLIP dissociation and thereby enables binding of peptides generated by proteolysis of exogenous antigens. However, the affinity of CLIP differs by four orders of magnitude among MHC II proteins because many polymorphic residues shape the specificity of the peptide binding groove (Sette et al., 1995). We previously demonstrated that the diabetes-associated I-Ag7 protein binds CLIP with very low affinity, allowing CLIP to rapidly dissociate in a DM-independent manner at an acidic pH characteristic for the endosomal peptide loading compartment (Hausmann et al., 1999). The low affinity of CLIP for I-Ag7 is related to the β57 polymorphism: the hydrophobic P9 anchor of CLIP (methionine) is a poor fit for the positively charged P9 pocket, and substitution of the P9 anchor of CLIP to alanine or aspartic acid increases the affinity of CLIP for I-Ag7. A number of MHC II proteins associated with human autoimmune diseases have been shown to have a low affinity for CLIP (Reed et al., 1997; Patil et al., 2001). CLIP was also shown to bind with rather low affinity to HLA-DQ8, and peptide elution studies showed that HLA-DQ2 binds CLIP in an unusual alternative register with relatively low affinity (Fallang et al., 2008; Wiesner et al., 2008). Both HLA-DQ2 and HLA-DQ8 also interact only weakly with DM (Fallang et al., 2008; Zhou et al., 2016). Rapid CLIP dissociation may enable binding of peptides in compartments that lack DM and may also favor presentation of low-affinity peptides that are not edited by DM. This hypothesis is consistent with data demonstrating that the bound repertoire of peptides has a lower average half-life for I-Ag7 compared with other I-A proteins (Carrasco-Marin et al., 1996; Kropshofer et al., 1996).
These alterations in the biochemistry of peptide binding by I-Ag7 may have a significant impact on the pathogenesis of T1D in NOD mice. There is increasing evidence for the importance of unique types of antigens that are released as proteolytic fragments by pancreatic β cells. One of the major identified antigens is the insulin B9-23 peptide, which binds in at least two registers to I-Ag7 (Daniel et al., 1995; Mohan et al., 2011). Proinsulin undergoes proteolytic processing in secretory granules of β cells, and proteolytic fragments containing the insulin B9-23 epitope are present in these secretory granules (Mohan et al., 2010). Indeed, CD4 T cells specific for the insulin B12-20 peptide (a lower-affinity register) are only activated when APCs are incubated with the peptide, but not the intact protein, suggesting that this peptide binds to I-Ag7 on the cell surface or early endosomal compartments of APCs (Mohan et al., 2011). Also, hybrid insulin peptides (HIPs) were recently reported that result from covalent linkage of proinsulin peptides to other proteolytic fragments in β cells (Delong et al., 2016). Thus, both major types of antigens (insulin, HIP) are released as peptides/proteolytic fragments by β cells and do not require (extensive) processing by APCs. Spontaneous dissociation of CLIP from I-Ag7 may facilitate presentation of such antigens by creating empty molecules in compartments that lack DM, thus sidestepping DM-mediated editing.
We generated knock-in NOD mice with a single amino acid substitution of CLIP that moderately increased its affinity for I-Ag7. These knock-in mice allowed us to examine the impact of CLIP affinity on the spontaneous development of T1D without changing the binding specificity of I-Ag7. We found that these knock-in mice developed T1D with a lower incidence than control NOD mice, and that T cell infiltration into islets was significantly diminished. MHC II tetramer labeling experiments demonstrated reduced infiltration of islets by HIP-specific CD4 T cells, which was due to reduced presentation of HIP by APCs.
Results
Knock-in mice with a single amino acid substitution of CLIP have a reduced incidence of T1D
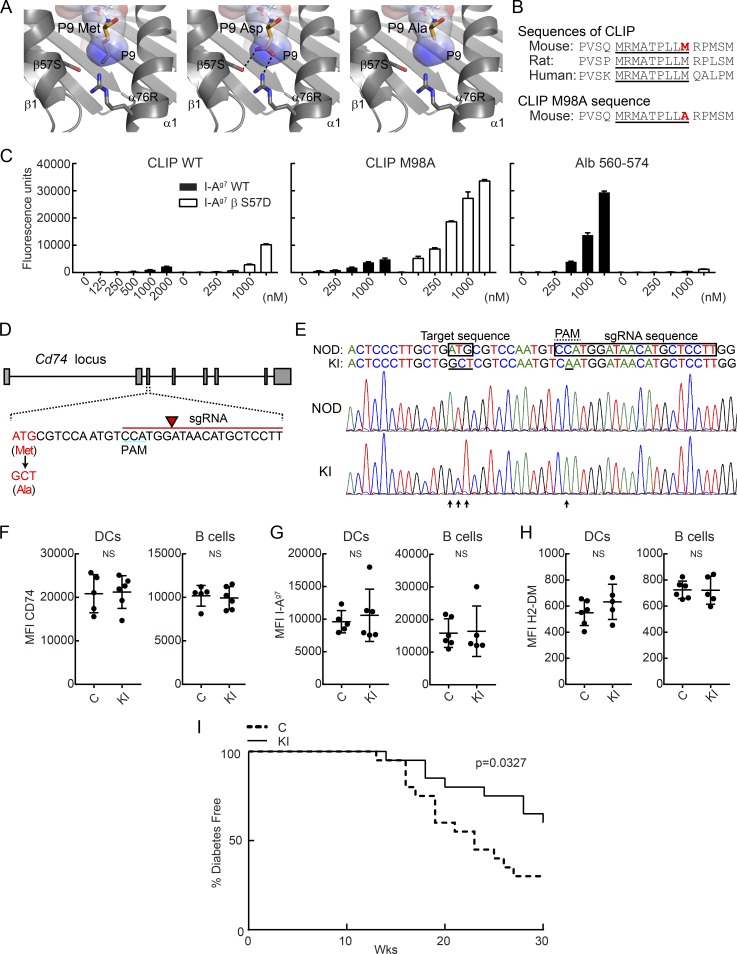
We generated knock-in mice with a single amino acid substitution of the P9 anchor of CLIP. The methionine to alanine (ATG to GCT) substitution was designed to result in a moderate increase in CLIP affinity for I-Ag7 (Fig. 1, A and B; Hausmann et al., 1999). Peptide binding studies demonstrated very little to no binding of WT CLIP to I-Ag7 at pH 5.2, which corresponds to the pH of the endosomal peptide loading compartment. In contrast, binding was clearly detected for I-Ag7 with a serine to aspartic acid mutation at position β57 (β S57D), indicating that the disease-associated β57 polymorphism impacts CLIP binding. A single amino acid substitution at the P9 position of CLIP from methionine to alanine (M98A) resulted in a moderate increase of CLIP binding to I-Ag7, while a high-affinity positive control peptide derived from albumin (Alb 560–574) showed strong binding to I-Ag7, as expected (Fig. 1 C). These results were consistent with previous peptide binding studies with I-Ag7 that demonstrated an IC50 of >125 µM for WT CLIP, 15 µM for CLIP M98A, and 0.5 µM for CLIP M98D at an acidic pH. Previous work had also demonstrated that CLIP binds to I-Ag7 at neutral pH, which is relevant for assembly of I-Ag7 with invariant chain in the ER (Hausmann et al., 1999).
Figure 1.
Knock-in mutation in CLIP segment of invariant chain reduces incidence of T1D in NOD mice. (A) P9 pocket of I-Ag7–peptide complex. P9 Met residue of I-Ag7–GAD65 complex (left, PDB accession no. 1ES0) was mutated to Asp (middle) or Ala (right). P9 pocket is shown as a surface colored by electrostatic potential (blue = positive, red = negative). (B) Amino acid sequence of CLIP and CLIP M98A, highlighting the core sequence required for MHC II binding (underlined) and the P9 anchor residue (red). (C) Binding of peptides to soluble I-Ag7 WT or βS57D mutant protein. Indicated biotinylated peptides (125–2,000 nM) were incubated with soluble I-Ag7 proteins, and bound peptide was quantified by ELISA for triplicate samples. Mean + SD are shown. (D) sgRNA design used to introduce CLIP M98A mutation in the Cd74 gene. The sgRNA sequence is indicated in red, with arrow highlighting the site of DNA cleavage. (E) Nucleotide sequence of WT and M98A knock-in allele. The introduced mutation (ATG→GCT) is indicated as well as the silent mutation in the PAM (C→A). Chromatograms are shown for WT NOD and M98A/M98A (KI) mice. (F–H) Protein expression level of CD74 (F), I-Ag7 (G), and H2-DM (H) by splenic CD45+ I-Ag7+ CD11c+ CD11b− DCs and CD19+ I-Ag7+ B cells. Results are shown as mean ± SD with five to six mice/group. MFI, mean fluorescence intensity. (I) Diabetes incidence in control mice (C; n = 20) and cohoused M98A/M98A mice (KI; n = 20). Control mice were bred from same founders as M98A/M98A mice. Data are representative of two (C and F–H) or three independent experiments (I). Statistical analysis was performed with Mann-Whitney (F–H) or Gehan-Breslow-Wilcoxon test (I).
The CLIP M98A mutation was introduced directly into the genome of WT NOD mice by micro-injection of fertilized oocytes with Cas9 mRNA, a guide RNA (gRNA) targeting the invariant chain gene (Cd74) and a single-stranded DNA oligonucleotide for homology-directed repair of the double-strand break introduced by Cas9. A small molecule inhibitor of nonhomologous end joining was also coinjected, which was previously shown to reduce off-target lesions and enhance the efficiency of homology-directed repair (Maruyama et al., 2015). Furthermore, we used a gRNA with a minimal length of 17 bases, which was shown to greatly diminish the frequency of off-target lesions (Fu et al., 2014). One founder mouse was obtained with the desired mutation, which was confirmed by DNA sequencing and TaqMan SNP genotyping assay (Fig. 1, D and E). The heterozygous founder (M98A/WT) was backcrossed three times to WT NOD mice to dilute potential off-target lesions (twofold for each cross except for those located on the same chromosome as Cd74). The three times backcrossed heterozygous mice (M98A/WT) were intercrossed to generate a homozygous M98A/M98A knock-in line (abbreviated as KI in figures) and a homozygous WT/WT (M98A) control line (referred to as control, abbreviated as C in figures); offspring of these two lines were used for all comparisons. We performed a computational analysis of potential off-target sites (Table S1) using the Benchling tool. One intergenic region on chromosome 10 with a single basepair mismatch gave a high off-target score, but sequencing of five M98A/M98A mice showed that this mutation was absent. All other predicted off-target sites had two or three mismatches; three of these off-target sites were located in genes (Tpp2, Hist1h2be, and Mug1), but off-target mutations were not detected at these sites (Fig. S1).
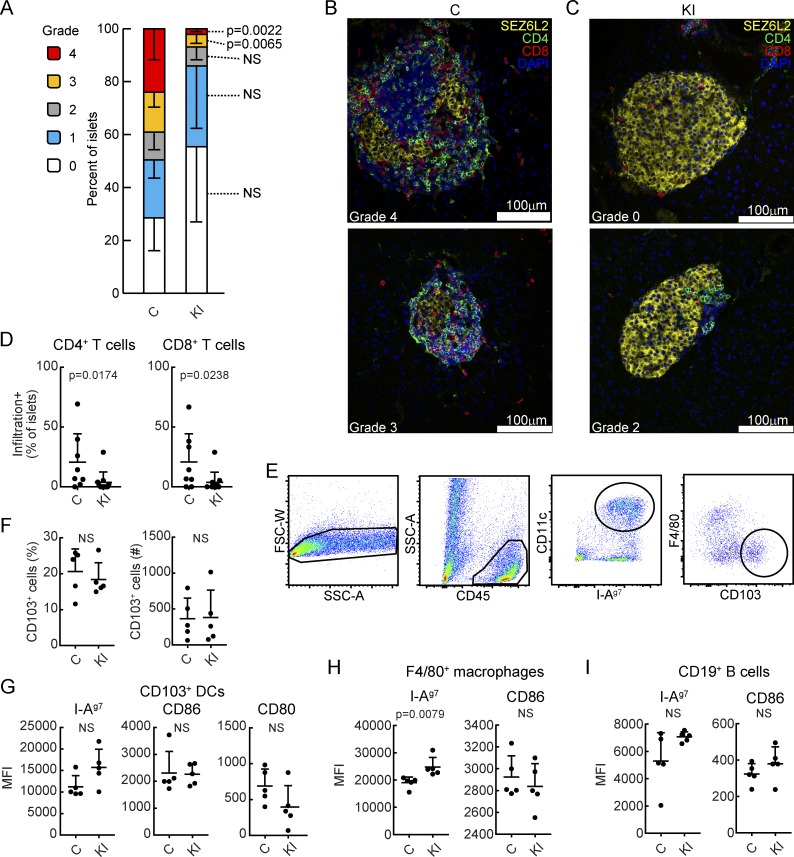
Homozygous knock-in mice were normal in terms of development, appearance, and fertility, and expressed normal levels of CD74, I-Ag7, and DM proteins in splenic APCs (Fig. 1, F–H). Female M98A/M98A mice developed T1D with a significantly lower incidence compared with control mice, and there was also a significant delay in disease development (Fig. 1 I). The introduced mutation did not represent a classical loss-of-function mutation, but rather a fairly subtle single amino acid substitution that modestly increased the affinity of CLIP for I-Ag7. Immunofluorescence analysis of tissue sections from the pancreas of 12-wk-old mice showed that a significantly reduced fraction of islets was highly infiltrated by T cells in M98A/M98A compared with control mice (Fig. 2, A–C; and Fig. S2). We further assessed infiltration of islets at an earlier disease stage and found that a lower percentage of islets from 9-wk-old M98A/M98A mice were infiltrated by CD4+ or CD8+ T cells compared with control mice (Fig. 2 D). Batf3-dependent CD103+ dendritic cells (DCs) have been shown to play an essential role in the development of T1D in NOD mice (Ferris et al., 2014). The knock-in mutation did not affect the number of islet-infiltrating CD103+ DCs (Fig. 2, E and F) and also did not change the expression level of I-Ag7, CD80, and CD86 by CD103+ DCs (Fig. 2 G). Although there was a modest increase in the I-Ag7 surface protein level on macrophages in M98A/M98A mice compared with control mice, B cells had similar I-Ag7 and CD86 cell surface protein levels in both strains (Fig. 2, H and I). These results demonstrate that the M98A mutation significantly reduced the fraction of islets that was severely infiltrated by T cells, but did not impair recruitment or the activation state of CD103+ DCs.
Figure 2.
Attenuated infiltration of T cells into islets of M98A/M98A NOD mice. (A) Histological assessment of T cell infiltration into pancreatic islets in M98A/M98A mice (KI) or cohoused control mice (C) at 12 wk of age (n = 5 to 6 mice/group). Grade 0: no infiltration; grade 1: T cells at islet perimeter; and grades 2–4: infiltration of T cells into ≤25%, ≤75%, and >75% of islet area, respectively. 27–107 islets/mouse were analyzed. Error bars indicate SD. Data are combined from two independent experiments. (B and C) Representative confocal microscopy images of islets from control (C) (B) and M98A/M98A (KI) (C) mice from A labeled with antibodies specific for SEZ6L2 (islet surface marker, yellow), CD4 (green) or CD8 (red); nuclear stain with DAPI (blue). Scale bars, 100 µm. (D) Quantification of percentage of islets infiltrated by CD4 or CD8 T cells at 9 wk of age in control (C) or M98A/M98A (KI) mice (n = 8 to 11 mice/group). Data are combined from four independent experiments. (E–I) Analysis of CD103+ DCs, CD11c+ F4/80+ macrophages, and CD19+ B cells in islets of 12-wk-old control mice (C) or M98A/M98A mice (KI; n = 5 mice/group). (E) Gating strategy for CD103+ DCs. (F) Percentage and number of F4/80− CD103+ DCs (of CD45+ CD11c+ I-Ag7+ cells) in individual mice. (G) Protein expression level of I-Ag7, CD86, and CD80 by CD103+ DCs. MFI, mean fluorescence intensity. (H and I) Expression level of I-Ag7 and CD86 by F4/80+ macrophages (H) and CD19+ B cells (I). Data are representative of three independent experiments. Statistical significance was calculated using the Mann-Whitney test for A, D, and F–I; results represent mean and SD.
Reduced infiltration of islets by T cells specific for HIPs
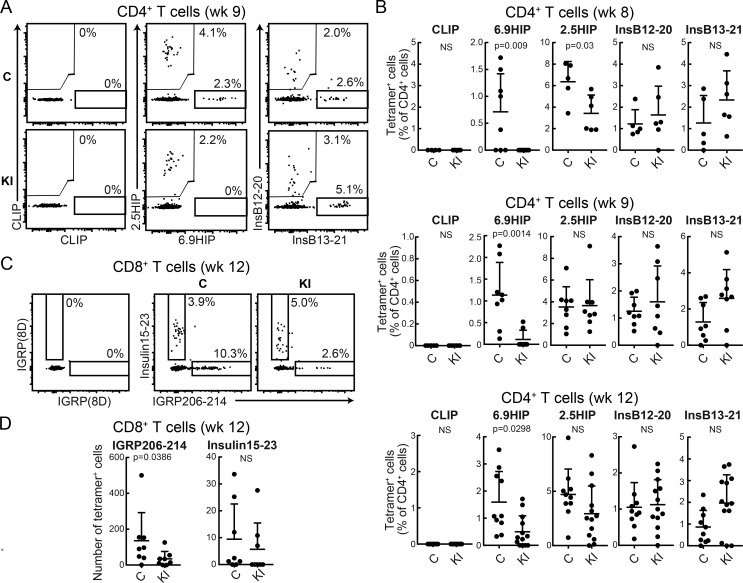
The very low affinity of CLIP for I-Ag7 at an endosomal pH may affect presentation of particular pancreatic β cell antigens to CD4 T cells. We therefore performed a comprehensive analysis of islet-infiltrating CD4 T cell specificities implicated in the disease. A substantial body of work implicated the insulin B9-23 peptide in the pathogenesis of T1D in NOD mice and identified at least two binding registers (InsB12-20 and InsB13-21 peptides; Mohan et al., 2011). Also, a recent study identified two unusual HIPs formed by covalent cross-linking of proinsulin fragments to other peptides present in β cell secretory granules (Delong et al., 2016; Wiles et al., 2017). These peptides contain C-terminal fragments of chromogranin A (2.5HIP, recognized by BDC-2.5 TCR) and islet amyloid polypeptide pro-peptide 2 (6.9HIP, recognized by BDC-6.9 TCR). We performed two-color tetramer labeling to enable analysis of all tetramer-positive cells in islets from individual mice and ensure specificity of binding (Fig. 3 A). I-Ag7 tetramers loaded with these peptides identified distinct CD4 T cell populations in islets of WT NOD mice, while labeling with the CLIP control tetramer resulted in little background staining. Interestingly, the frequency of CD4 T cells labeled with the 6.9HIP tetramer was greatly diminished in islets from M98A/M98A compared with control mice. 6.9HIP tetramer–positive CD4 T cells were almost undetectable at both 8 and 9 wk in M98A/M98A mice, while they were readily detectable in control mice (Fig. 3, A and B). At 12 wk, 6.9HIP tetramer–positive CD4 T cells were detected in islets from a fraction of M98A/M98A mice. Nevertheless, the percentage of 6.9HIP tetramer–positive CD4 T cells remained smaller in M98A/M98A compared with control mice (Fig. 3 B). The frequency of 2.5HIP–specific CD4 T cells was also reduced in M98A/M98A mice at week 8, but not at weeks 9 or 12, compared with control mice. In contrast, insulin-specific T cells identified with InsB12-20 or InsB13-21 tetramers were present at similar frequencies in both knock-in and control mice (Fig. 3, A and B).
Figure 3.
Impact of CLIP mutation on islet-infiltrating CD4 T cell populations specific for fusion peptides. (A) Representative examples of tetramer labeling of islet-infiltrating CD4 T cells from 9-wk-old control (C) or M98A/M98A (KI) mice. Two-color tetramer stains (PE and allophycocyanin) are shown for negative control CLIP, 6.9HIP, and 2.5HIP peptides as well as InsB13-21 and InsB12-20 peptides. (B) Summary of tetramer labeling data for islet-infiltrating CD4 T cells (% tetramer+ cells of CD4+ cells) from 8-wk-, 9-wk-, and 12-wk-old mice (n = 5–13 mice/group). (C and D) Flow-cytometric analysis of islet-infiltrating CD8+ T cells from 12-wk-old control (C) or M98A/M98A (KI) mice. Two-color tetramer stains (PE and allophycocyanin) with IGRP (8D) control peptide as well as IGRP206-214 and Ins15-23 peptides (n = 8 mice/group). All data represent two independent experiments. Statistical analyses were performed with the Mann-Whitney test; mean + SD are shown.
We also assessed whether changes in CD4 T cell populations could impact CD8 T cell responses to β cell antigens. The numbers of IGRP (islet-specific glucose-6-phosphatase catalytic subunit-related protein)-specific CD8 T cells, but not of insulin B15-23 (InsB15-23)–specific CD8 T cells, were significantly lower in M98A/M98A compared with control mice at 12 wk (Fig. 3, C and D; Wong et al., 1999; Lieberman et al., 2003). These data demonstrate that the CLIP M98A mutation reduces the accumulation of CD4 and CD8 T cells with particular antigen specificities (2.5HIP, 6.9HIP, and IGRP) in islets, indicating that the affinity of CLIP for I-Ag7 has an impact on spontaneous priming of T cell responses to particular β cell antigens. These data are also consistent with the hypothesis that CD4 T cells provide help for the priming/expansion of β cell–specific CD8 T cells.
A subpopulation of insulin B9-23–specific T cells expresses forkhead box P3, while islet-infiltrating HIP tetramer–positive T cells exhibit an inflammatory T helper type 1 cell phenotype
We next examined the potential functional properties of tetramer-labeled CD4 T cell populations. A large proportion of CD4 T cells labeled with InsB12-20 or InsB13-21 tetramers expressed forkhead box P3 (FoxP3), a T regulatory cell (T reg cell)–specific transcription factor (Fig. 4, A and B). In contrast, the majority of CD4 T cells identified with 2.5HIP and 6.9HIP tetramers were negative for FoxP3 staining. In particular, FoxP3 labeling was almost undetectable in CD4 T cells labeled with the 6.9HIP tetramer at week 9 (Fig. 4, A and B). The proportion of FoxP3-expressing T cells was dependent on epitope specificity (highest for insulin peptides), but not related to the genotype of mice (WT/WT versus M98A/M98A) with the exception of 6.9HIP tetramer+ CD4 T cells, for which the percentage of FoxP3-expressing T cells was larger in M98A/M98A compared with control mice at week 12 (Fig. 4 C). InsB12-20 tetramer+ FoxP3-expressing T cells did not produce IFN-γ upon ex vivo stimulation with PMA and ionomycin, and expressed the T reg cell markers GITR, Helios, and CTLA-4 (Fig. 4, D–F). In islets of 9-wk-old NOD mice, the proportion of Ki67+ cells was higher among FoxP3+ than FoxP3− CD4 T cells, indicating that a higher fraction of T reg cells were proliferating compared with effector CD4 T cells (Fig. S3, A and B). Moreover, all FoxP3-expressing InsB12-20 and InsB13-21 tetramer+ CD4 T cells were Ki67+, indicating that these β cell–specific T reg cells had a high proliferative capacity in islets at least at this early stage of the disease process (Fig. S3, A and C).
Figure 4.
Insulin-specific CD4 T cells show FoxP3+ T reg cell phenotype in islets of NOD mice. (A) Representative flow-cytometric analysis of tetramer+ CD4+ T cells from pancreatic islets of 9-wk-old NOD mice labeled with FoxP3-specific or isotype control antibodies. (B) Percentage of FoxP3+ cells among islet-infiltrating CD4+ T cells from 9-wk-old NOD mice labeled with four different I-Ag7 tetramers (% FoxP3+ of tetramer+ CD4+ T cells; n = 8 mice/group). (C) Analysis of FoxP3 labeling for tetramer+ CD4 T cells from islets of control mice (C) or M98A/M98A mice (KI) at 9 wk or 12 wk of age (n = 7 to 11 mice/group). (D) Percentage of IFN-γ+ cells among tetramer+ FoxP3+ CD4 T cells from islets of 9-wk-old NOD mice. Cells were stimulated with PMA and ionomycin and stained with IFN-γ antibody (n = 4–5 mice/group). (E) Representative flow-cytometric analysis of GITR, Helios, and CTLA-4 expression in InsB12-20–labeled FoxP3+ CD4+ T cells from pancreatic islets of 9-wk-old NOD mice. Cell surface GITR and intracellular Helios as well as CTLA-4 were examined. (F) Summary plots of GITR+, Helios+, or CTLA-4+ cells for InsB12-20–labeled FoxP3+ CD4+ T cells (n = 6 to 12 mice/group). Statistical analyses were performed with the one-way ANOVA test (B) and the Mann-Whitney test (C), mean + SD (B–D), or mean – SD (F) are shown (*, 0.01 ≤ P < 0.05; **, 0.001 ≤ P < 0.01). Data represent two independent experiments.
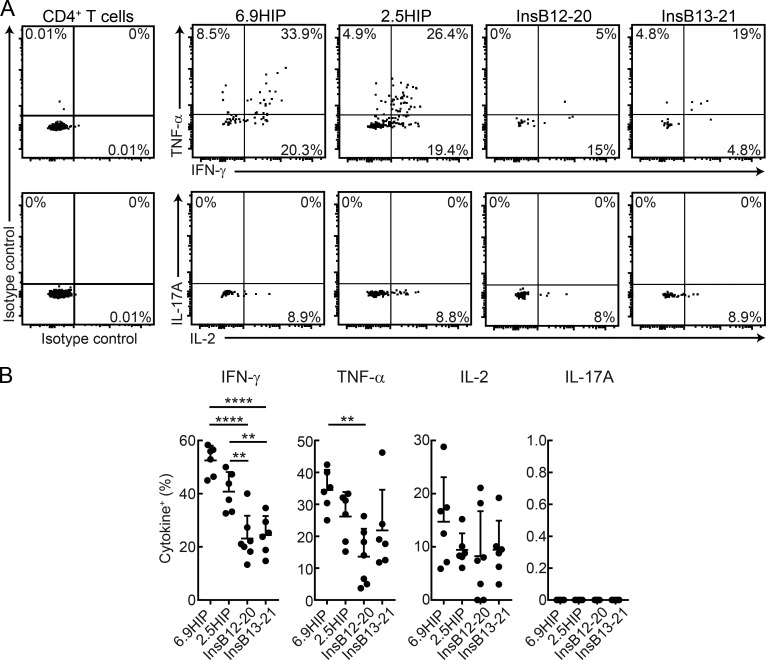
We next examined which of these β cell antigen-specific CD4 T cells had the capacity to secrete inflammatory cytokines. T cells were isolated from islets of WT NOD mice, and intracellular cytokine staining was performed following ex vivo stimulation with PMA and ionomycin. Larger proportions of 2.5HIP- and 6.9HIP-specific CD4 T cells produced IFN-γ compared with InsB12-20– or InsB13-21–specific CD4 T cell populations (Fig. 5, A and B). In addition, the percentage of TNF-α–producing cells was also larger for 6.9HIP- than InsB12-20–specific CD4 T cells (Fig. 5, A and B). The production of these pro-inflammatory cytokines was particularly pronounced for CD4 T cells labeled with the 6.9HIP tetramer. The frequency of IL-2–producing T cells was similar for all peptide-specific populations, and IL-17A–producing cells were not detected for these CD4 T cell specificities (Fig. 5, A and B). These results demonstrate that 6.9HIP- and 2.5HIP-specific CD4 T cells exhibit an inflammatory T helper type 1 cell (Th1 cell) phenotype in islets of NOD mice.
Figure 5.
Th1 cell phenotype of fusion peptide-specific CD4 T cells in islets of NOD mice. (A) Representative flow-cytometric analysis of indicated tetramer+ CD4+ T cells from islets of 9-wk-old NOD mice. Cells were stimulated with PMA and ionomycin and stained with anti-cytokine antibodies (TNF-α, IFN-γ, IL-17A, and IL-2). (B) Summary plots of cytokine+ cells among tetramer+ CD4 T cell populations (% of tetramer+ CD4+ T cells; (n = 6 mice/group). Statistical analyses were performed with one-way ANOVA. Mean and SD are shown (**, 0.001 ≤ P < 0.01; and ****, P < 0.0001). Data represent two independent experiments.
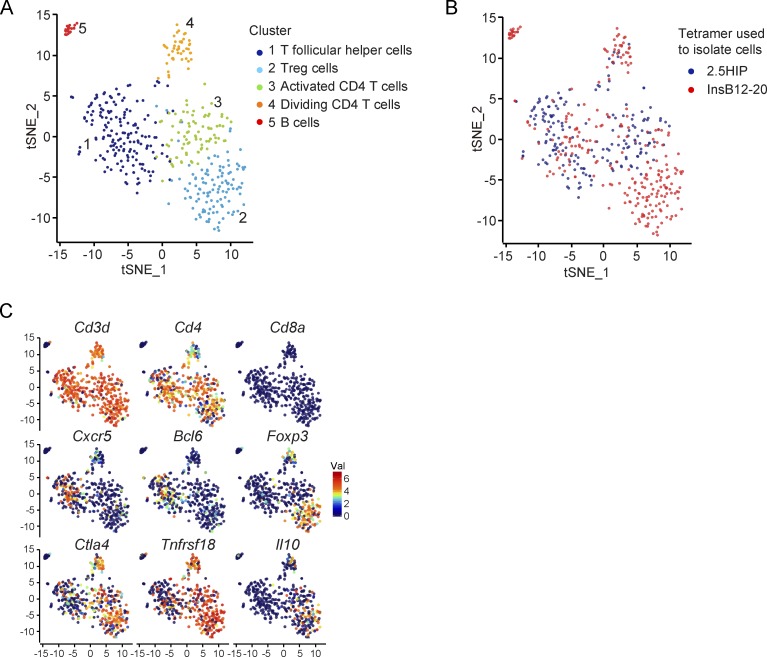
We sought to further characterize these CD4 T cell populations by single-cell RNA sequencing (RNA-seq) of islet-infiltrating CD4 T cells labeled with InsB12-20 or 6.9HIP tetramers. Unsupervised clustering of the combined samples partitioned the cells into five clusters (Fig. 6, A and B). Clusters 1, 2, 3, and 4 expressed Cd3d and Cd4 but not Cd8a genes, confirming that they were CD4 T cells (Fig. 6 C). High-level expression of B cell–related genes in cluster 5 indicated that they represented contaminating B cells (Fig. S3 D).
Figure 6.
Single-cell RNA-seq analysis of islet-specific CD4 T cell populations. (A–C) t-distributed stochastic neighborhood embedding (tSNE) plots of single-cell RNA-seq profiles of CD4 T cells isolated with InsB12-20 and 6.9HIP tetramers. T cells were labeled post hoc based on cluster assignment (A), tetramers used to isolate cells (B), or expression of indicated genes (C). One experiment.
Cluster 2 was enriched for cells with high level of expression of T reg cell signature genes, including Foxp3, Ctla4, Tnfrsf18 (encoding GITR), and Il10. Interestingly, all cells in this T reg cell cluster had been isolated with the InsB12-20 tetramer. In striking contract, CD4 T cells isolated with the 6.9HIP tetramer were absent in this T reg cell cluster, consistent with our functional data (Fig. 6, A–C; and Fig. S3 D). High-level expression of Bcl6 and Cxcr5 genes by cells in cluster 1 indicated that these CD4 T cells represented T follicular helper cells. CD4 T cells in clusters 3 and 4 represented activated cells and proliferating cells, respectively (Fig. 6, A and B; and Fig. S3 D). These data confirmed our prior findings, which had indicated that 6.9HIP- and InsB12-20–specific CD4 T cells had distinct functional properties and that a significant fraction of InsB12-20 tetramer+ CD4 T cells were FoxP3+ T reg cells.
Normal thymic differentiation of β cell–specific CD4 T cells in M98A/M98A mice
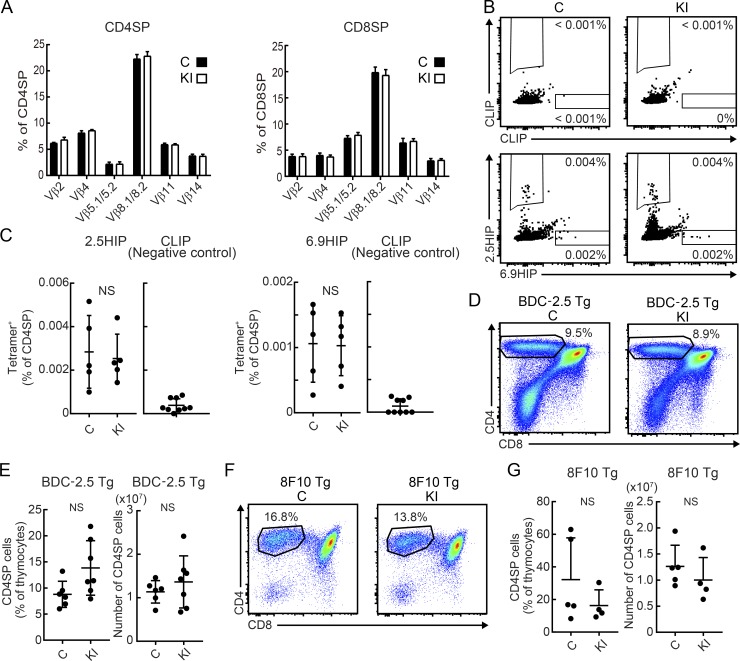
CD4 T cells are selected in the thymus by weak recognition of self-peptides bound to MHC II proteins. Given that the self-peptides responsible for positive selection of any given TCR are not known, it is possible that the altered spectrum of peptides presented on I-Ag7 in M98A/M98A mice might affect thymic development of pathogenic CD4 T cells. We therefore examined whether thymic selection of T cells could be altered in M98A/M98A mice. The fraction of CD4SP or CD8SP thymocytes expressing particular TCR Vβ chains were similar between M98A/M98A mice and control mice (Fig. 7 A). Previous studies showed that CD4 T cells recognizing BDC-2.5 mimotope peptides can be detected by tetramer labeling in the thymus of NOD mice (Jang et al., 2003; Stratmann et al., 2003). We assessed the frequency of 2.5HIP and 6.9HIP tetramer–positive CD4 single-positive (SP) T cells in the thymus by two-color tetramer labeling. CD4SP T cells labeled with the 2.5HIP tetramer could be clearly detected in the thymus, and such T cells were present at similar frequencies in control and M98A/M98A mice (Fig. 7, B and C). Also, smaller numbers of CD4SP T cells were identified with the 6.9HIP tetramer, again with similar frequencies between knock-in and control mice. In contrast, labeling with the CLIP control tetramer was low (Fig. 7, B and C).
Figure 7.
Normal thymic differentiation of β cell antigen–reactive CD4+ T cells in M98A/M98A mice. (A) Percentage of thymocytes expressing particular Vβ TCR chains in CD4SP or CD8SP cells from 12-wk-old control (C) or M98A/M98A (KI) mice (n = 4 mice/group). (B and C) Flow-cytometric analysis of CD4SP T cells from the thymus of 9-wk-old control (C) or M98A/M98A (KI) mice by two-color tetramer labeling (PE and allophycocyanin) for either negative control CLIP as well as 6.9HIP and 2.5HIP peptides (n = 5 mice/group). (D and E) Representative flow-cytometric analysis of CD4SP T cells in the thymus from 3-wk-old BDC-2.5 Tg M98A/M98A (KI) or BDC-2.5 Tg control (C) mice (littermate control; (n = 6–7 mice/group). (F and G) Flow-cytometric analysis of CD4SP T cells in the thymus from 3-wk-old 8F10 Tg M98A/M98A mice (KI) or 8F10 Tg control mice (C) (littermate control; n = 4–5 mice/group). Data represent two independent experiments. Statistical analyses were performed with the Mann-Whitney test, mean ± SD (C and E) or mean + SD (A and G) are shown.
We also used two TCR transgenic (Tg) strains to examine the potential impact of the CLIP M98A mutation on thymic selection. BDC-2.5 and 8F10 TCRs recognize 2.5HIP and InsB12-20 peptides, respectively (Mohan et al., 2013; Delong et al., 2016). The frequency and number of CD4SP T cells in the thymus were similar for BDC-2.5 Tg mice on the M98A/M98A or control background (Fig. 7, D and E). Similarly, the frequency of CD4SP cells in the thymus was similar for 8F10 Tg mice on the M98A/M98A and control background (Fig. 7, F and G). We also showed that the M98A/M98A mutation did not change the fraction of TCR Tg T cells that expressed endogenous TCR-α chains (due to lack of allelic exclusion at the TCR-α locus) or differentiated into FoxP3-expressing T reg cells (Fig. S4). Taken together, these results suggest that thymic selection of 2.5HIP- and InsB12-20–specific CD4 T cells was not altered in M98A/M98A compared with control mice.
Reduced presentation of free peptides but not full-length antigens by DCs from M98A/M98A mice
Given that 2.5HIP- and 6.9HIP-specific CD4 T cells were present in similar numbers in the thymus of M98A/M98A compared with control mice, we hypothesized that the CLIP M98A mutation may impact presentation of these peptides to CD4 T cells in pancreatic lymph nodes. The 2.5HIP peptide is recognized by the BDC-2.5 TCR, which provided an opportunity to interrogate antigen presentation to T cells with this specificity in pancreatic lymph nodes. Proliferation of BDC-2.5 CD4 T cells was assessed by adoptive transfer of CFSE-labeled BDC-2.5 TCR Tg CD4 T cells to M98A/M98A or control mice. We observed reduced levels of CFSE dilution by BDC-2.5 T cells in pancreatic lymph nodes of M98A/M98A compared with control mice. As expected, there was little proliferation of these T cells in nondraining lymph nodes (Fig. 8, A and B).
Figure 8.
Point mutation in CLIP diminishes presentation of β cell–derived peptides to CD4 T cells. (A and B) CFSE-labeled CD4+ T cells from BDC-2.5 Tg mice were transferred to control or M98A/M98A mice. On day 3, cells from nondraining lymph nodes (ndLNs) or pancreatic lymph nodes (pLNs) were analyzed for CFSE dilution. (A) Representative flow-cytometric analysis of CD4+ Vβ4+ CFSE+ cells. (B) Summary plots of percentages of CFSE-low cells in A (n = 7 samples/group). (C) CFSE-labeled CD4+ T cells from BDC-2.5 Tg mice were cultured with BM-DCs from control mice (C) or M98A/M98A mice (KI). Indicated numbers of islet cells were added to the culture. On day 5, cells were analyzed for CFSE dilution. Summary plots of CFSE dilution by CD4+ Vβ4+ cells are shown. (D and E) CFSE-labeled CD4+ T cells from BDC-2.5 Tg mice were cultured with splenic DCs from control mice (C) or M98A/M98A mice (KI). Indicated concentrations of supernatant from islet cell culture were added to the culture. On day 3, cells were analyzed for CFSE dilution. (D) Representative flow-cytometric analysis of CD4+ Vβ4+ CFSE+ cells in the presence of 30% of islet cell culture supernatant. (E) Summary plots of CFSE-low cells in D. (F–H) Presentation of soluble 2.5HIP peptide to BDC-2.5 T cells. CFSE-labeled CD4+ T cells from BDC-2.5 Tg mice were cultured with DCs from control (C) or M98A/M98A (KI) mice. Soluble 2.5HIP peptide was added at the indicated concentrations; 6.9HIP peptide (7 nM) served as a negative control. On day 3, cells were analyzed for CFSE dilution (F and G). Supernatants were analyzed by ELISA for IFN-γ (H). (I) DCs from control (C) or M98A/M98A (KI) mice were cultured with CD4+ T cells harvested from draining lymph nodes (dLNs) of WT NOD mice immunized with 6.9HIP peptide. Indicated concentrations of 6.9HIP peptide were added to the co-culture. IFN-γ production was examined by ELISPOT assay. (J and K) Hybridomas specific for InsB12-20 peptide (8F10; J) or InsB13-21 peptide (11T3; K) were cultured with splenic DCs from control (C) or M98A/M98A (KI) mice. Indicated concentrations of InsB12-20 peptide (J) or InsB13-21 peptide (K) were added to the culture. On day 3, supernatants were analyzed by ELISA for secreted IL-2. Data representative of two independent experiments. n = 4 samples/group in C, E, and G–K. Statistical analyses were performed with the Mann-Whitney test. Mean and SD (B and H–K), mean − SD (C), or mean ± SD (E and G) are shown (*, P = 0.01–0.05).
We next assessed antigen presentation to BDC-2.5 CD4 T cells by co-culture with bone marrow–derived DCs (BM-DCs), using graded numbers of freshly isolated islet cells as the antigen source. Proliferation of BDC-2.5 T cells was reduced in co-cultures with DCs from M98A/M98A compared with control mice; such a difference was not observed when large numbers of islet cells were added to the co-cultures (Fig. 8 C). Similar results were obtained when supernatants of islet cell cultures, rather than islet cells, were used as the source of antigen (Fig. 8, D and E). Diminished proliferation of BDC-2.5 T cells was also induced by DCs from M98A/M98A compared with control mice in the presence of 2.5HIP peptide, indicating that presentation of soluble extracellular peptides was reduced by the CLIP M98A mutation (Fig. 8, F–H). Likewise, DCs from M98A/M98A mice presented 6.9HIP peptide less efficiently than DCs from control mice to CD4 T cells from 6.9HIP-immunized mice (Fig. 8 I). The 2.5HIP peptide and 6.9HIP peptide are generated by fusion of two proteolytic fragments in β cells, and it is likely that both 2.5HIP and 6.9HIP fusion peptides are released by β cells at low concentrations. The in vivo and in vitro studies described above indicated that presentation of such peptides is reduced by the CLIP M98A mutation, presumably because the M98A mutation diminished spontaneous dissociation of CLIP from I-Ag7. We also observed that stimulation of T cell hybridomas by InsB12-20 or InsB13-21 peptides was diminished in co-cultures with splenic DCs from M98A/M98A compared with control mice (Fig. 8, J and K).
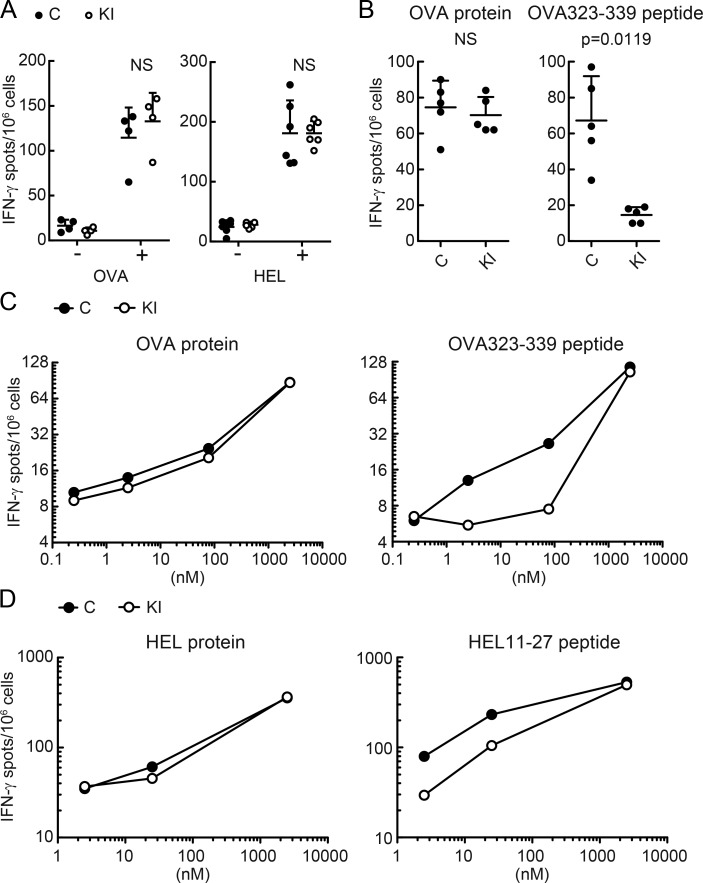
Nevertheless, M98A/M98A mice were able to mount normal CD4+ T cell responses when immunized with exogenous full-length proteins, OVA, and hen egg lysozyme (HEL; Fig. 9 A). DCs from M98A/M98A mice or control mice were co-cultured with CD4+ T cells in the presence of soluble peptides (OVA323-339 peptide or HEL11-27) or full-length proteins (OVA protein or HEL protein). This comparison demonstrated that full-length antigens were presented with similar efficiency by DCs from control and M98A/M98A mice. In striking contrast, CD4 T cell responses to both synthetic peptides were substantially diminished when these were presented by DCs from M98A/M98A compared with control mice (Fig. 9, B–D). These in vitro and in vivo studies support our conclusion that APCs from M98A/M98A mice have a normal capacity to present full-length antigens. However, they present soluble peptides at a reduced level to CD4 T cells compared with APCs from WT NOD mice.
Figure 9.
Point mutation in CLIP diminishes presentation of free peptides but does not impact presentation of full length antigens. (A) Control (C) or M98A/M98A (KI) mice were immunized with OVA or HEL protein. On day 8, CD4+ T cells from dLNs were cultured with DCs from WT NOD mice with or without the immunization protein. Each dot represents an individual mouse (n = 4 mice/group). (B and C) DCs from control (C) or M98A/M98A (KI) mice were cultured with CD4+ T cells harvested from dLNs of WT NOD mice immunized with OVA323-339 peptides. OVA protein or OVA323-339 peptide were added to the culture at a concentration of 234 nM (B; n = 5 samples/group) or at the indicated concentrations (C), respectively. (D) DCs from control mice (C) or M98A/M98A (KI) mice were cultured with CD4+ T cells harvested from dLNs of WT NOD mice immunized with HEL11-27 peptide. Indicated concentrations of HEL protein or HEL11-27 peptide were added to the culture. Each dot represents mean of duplicate samples (C and D). IFN-γ production was examined by ELISPOT. Data representative of two independent experiments. Statistical analyses were performed with the Mann-Whitney test. Mean + SD are shown (A and B).
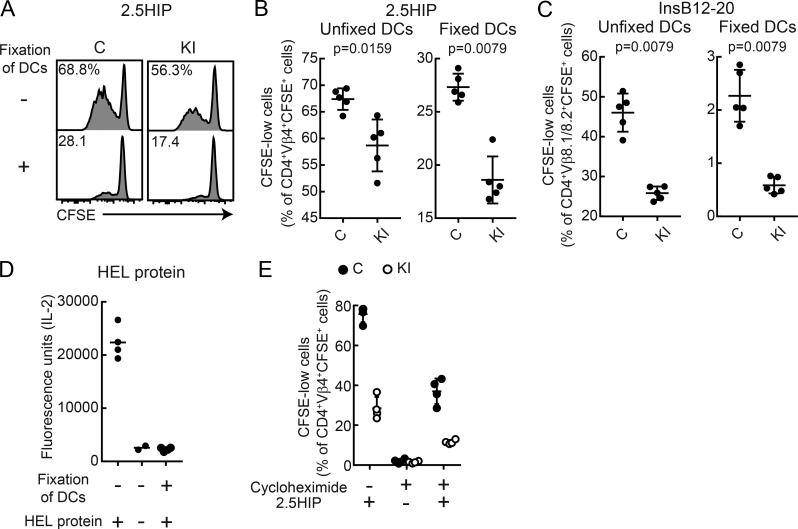
DCs fixed with a low concentration of paraformaldehyde presented soluble 2.5HIP or InsB12-20 peptide, but not full-length HEL protein, to CD4 T cells (Fig. 10, A–D). Furthermore, CD4 T cell responses to both synthetic peptides were reduced when presented by fixed DCs from M98A/M98A compared with control mice (Fig. 10, A–C). In addition, DCs in which protein synthesis was inhibited with cycloheximide presented 2.5HIP peptide to CD4 T cells, suggesting that newly synthesized I-Ag7 was not essential for 2.5HIP peptide presentation (Fig. 10 E). Also, the 2.5HIP peptide was presented at a lower level by DCs from M98A/M98A compared with control mice when DCs were been treated with cycloheximide. These results demonstrate that rapid CLIP dissociation from I-Ag7 favors an aberrant antigen presentation pathway that does not require intracellular antigen processing.
Figure 10.
Reduced presentation of extracellular peptides by DCs from knock-in mice. (A–C) Presentation of soluble 2.5HIP peptide (A and B) or InsB12-20 peptide (C) by DCs fixed with paraformaldehyde. CFSE-labeled CD4 T cells from BDC-2.5 Tg (A and B) or 8F10 Tg mice (C) were co-cultured with fixed or unfixed DCs from M98A/M98A (KI) or control (C) mice. On day 3, CD4 cells were analyzed for CFSE dilution. (A) Representative flow-cytometric analysis of CFSE dilution by T cells. (B) Summary plots of data from A. n = 5 samples/group in B and C. (D) Presentation of HEL protein by DCs fixed with paraformaldehyde. The 21.30 hybridoma was cultured with fixed or unfixed DCs from control mice. On day 1, IL-2 released into the supernatant was measured by ELISA (n = 2–4 samples/group). (E) Presentation of 2.5HIP peptide by DCs treated with cycloheximide. CFSE-labeled CD4 T cells from BDC-2.5 Tg mice were co-cultured with cycloheximide-treated or untreated DCs from M98A/M98A (KI) or control (C) mice. On day 3, CD4 cells were analyzed for CFSE dilution (n = 4 samples/group). Data representative of two independent experiments. Statistical analyses were performed with the Mann-Whitney test (B and C); mean (D) or mean ± SD (B, C, and E) are shown.
Discussion
The autoimmunity field has long pursued the hypothesis that disease-associated MHC II polymorphisms increase the risk of autoimmunity by enabling presentation of pathogenic peptides to CD4 T cells. However, such MHC II polymorphisms may also affect the interaction with the invariant chain or DM. The invariant chain–derived CLIP dissociates rapidly from I-Ag7 even in the absence of DM, due to a poor fit of the hydrophobic P9 side chain of CLIP (M98) for the positively charged P9 pocket of I-Ag7 (Hausmann et al., 1999). We introduced a fairly subtle mutation (methionine to alanine) at this position of the invariant chain gene—without mutating the peptide binding groove of I-Ag7—and found that it provided significant protection from spontaneous T1D in NOD mice. Comprehensive analysis of islet-infiltrating CD4 T cell populations with MHC II tetramers demonstrated reduced infiltration by HIP tetramer–positive CD4 T cells that recognize hybrid fusion peptides (2.5HIP and 6.9HIP). There is significant evidence for an aberrant antigen presentation mechanism in T1D: insulin and HIP peptides are generated in secretory granules of β cells and released into the extracellular space, enabling binding to MHC II proteins by tissue-resident APCs without further proteolytic processing (Mohan et al., 2011; Delong et al., 2016; Wiles et al., 2017). We hypothesize that spontaneous release of CLIP from I-Ag7 creates empty molecules that bind such extracellular peptides on the cell surface or in early endosomes, rather than through the classical MHC II presentation pathway in which DM edits the MHC II–bound peptide repertoire.
As part of this study, we performed a comprehensive analysis of islet-infiltrating CD4 T cells with MHC II tetramers. This was enabled by recent advances in the field, including the identification of insulin B9-23 binding registers for I-Ag7 and the recent discovery of hybrid fusion peptides recognized by BDC-2.5 and BDC-6.9 TCRs (Mohan et al., 2011; Delong et al., 2016). We found that the CLIP M98A mutation reduced the frequency of CD4 T cells specific for HIPs, in particular 6.9HIP. In contrast, we observed no difference in the frequency of islet-infiltrating insulin B9-23–specific T cells. These results are most likely explained by large differences in protein and peptide abundance. Insulin is by far the most abundant protein synthesized by β cells, and proteolytic fragments containing the insulin B9-23 epitope are generated in β cell secretory granules as previously shown by labeling with an insulin B9-23–specific antibody (Mohan et al., 2010). In contrast, hybrid fusion peptides are created when two proteolytic fragments are covalently linked by a protease (Berkers et al., 2009). Such hybrid peptides have been identified as CD8 T cell epitopes, but mass spectrometry analyses demonstrated that they were present in very small quantities (Vigneron et al., 2004). The generation of such hybrid fusion peptides is energetically unfavorable and also requires precise joining of two proteolytic fragments in the presence of competing peptide fragments. The C-terminal fragment of 6.9HIP originates from islet amyloid polypeptide, which is present at a level of ∼1% compared with insulin (Krizhanovskii et al., 2017). It therefore appears likely that hybrid fusion peptides are produced by β cells in small quantities. The M98A mutation may have a quantitative impact on the presentation of peptides present at low quantities in the extracellular space, but not for peptides released in larger quantities.
MHC II tetramer labeling also enabled detailed characterization of the functional properties of these T cell populations. Genetic studies have implicated both insulin and chromogranin A (encoding the C-terminal component of 2.5HIP) as important antigens for CD4 T cells, and it is quite possible that more than one antigen contributes to the disease, in particular at later stages (Nakayama et al., 2005; Baker et al., 2016). Intracellular cytokine staining demonstrated that a large fraction of 2.5HIP- and 6.9HIP-specific T cells had a pro-inflammatory phenotype characterized by IFN-γ and TNF-α production. In contrast, a smaller fraction of CD4 T cells labeled by InsB12-20 or InsB13-21 tetramers produced IFN-γ. Surprisingly, a substantial fraction of insulin-specific T cells was positive for FoxP3. The T reg cell phenotype of insulin-specific T cells was confirmed by flow cytometry using classical T reg cell markers. Also, single-cell RNA-seq analysis demonstrated a distinct population of insulin-specific CD4 T cells with a T reg cell gene expression signature. These results demonstrate that islet-infiltrating CD4 T cells with different antigen specificities have distinct functional properties. The two HIPs represent “neoantigens” resulting from peptide fusion within islets, and T cells specific for these peptides are biased toward a pro-inflammatory phenotype. It is important to keep in mind that these studies were performed in mice at a relatively early time point in the disease process (8–12 wk of age), and that the functional composition of each of these tetramer-positive populations may change during disease progression.
Previous studies demonstrated that CD4 T cells specific for the InsB12-20 register escape negative selection because the peptide is bound by I-Ag7 with low affinity. In contrast, it was proposed that InsB13-21–specific T cells are removed by negative selection due to a higher binding affinity of this peptide register for I-Ag7. This conclusion was based on immunization with synthetic peptides in complete Freund’s adjuvant: T cell responses were detected by ELISPOT following immunization with InsB12-20 but not InsB13-21 peptides (Mohan et al., 2011). However, this issue had not been addressed by tetramer labeling due to technical challenges involved in the generation of these reagents. We performed two-color tetramer labeling experiments of islet-infiltrating CD4 T cells and identified distinct CD4 T cell populations that were labeled by either the InsB12-20 or the InsB13-21 tetramer, thus confirming that there are at least two relevant registers of the insulin B9-23 peptide. It is possible that a small subpopulation of InsB13-21 CD4 T cells escaped negative selection, for example due to a lower TCR affinity. Within islets, insulin-specific CD4 T cells were highly proliferative, as shown by Ki67 staining, which could explain how small numbers of T cells that escaped negative selection accumulated within islets. Another previously reported register of the insulin B9-23 peptide was not investigated as part of this study (Crawford et al., 2011).
Finally, autoimmune diseases are caused by polymorphisms that can have rather subtle effects on immune function. However, genetic studies in mouse models are frequently based on mutations that eliminate entire cell populations or critical immune molecules/pathways. An increasingly sophisticated understanding of autoimmunity will require a more in-depth understanding of how subtle changes in immune function impact the risk for autoimmunity.
Materials and methods
Mice
NOD/ShiLtJ (NOD) and NOD.Cg-Tg (TcraBDC2.5,TcrbBDC2.5)1Doi/DoiJ (BDC-2.5) mice were obtained from The Jackson Laboratory. 8F10 Tg mice were provided from E.R. Unanue (Washington University, St. Louis, MO; Mohan et al., 2013). All mouse experiments were approved by the Institutional Care and Use Committee of the Dana-Farber Cancer Institute (protocol no. 04–103).
Generation of invariant chain knock-in mice
Zygote injections with nonhomologous end joining inhibitor were performed as previously described with minor modifications (Maruyama et al., 2015). Super-ovulated female NOD mice, 5–6 wk of age, were mated to NOD stud males. Fertilized zygotes were collected from oviducts and cultured briefly in KSOM with amino acids at 37°C and 6% CO2. Cas9 mRNA (100 ng/ml), single guide RNA (sgRNA): 5'-AAGGAGCATGTTATCCATGG-3' (50 ng/ml; Life Technologies), repair template DNA: 5'-CCTCCCTGTCCCACAGCTGCCAAACCTGTGAGCCAGATGCGGATGGCTACTCCCTTGCTGGCTCGTCCAATGTCAATGGATAACATGCTCCTTGGGGTAAGGAAGGCGGCAGGAAGGAAATGGTAGCTGGTGTTTG-3' (100 ng/ml; Integrated DNA Technologies), and Scr7 pyrazine (1 mM; Sigma-Aldrich) were mixed and microinjected into the cytoplasm of zygotes at the pronuclear stage (24 h after human chorionic gonadotropin). The injected zygotes were surgically transferred into pseudo-pregnant female ICR mice without further culturing. 2 out of 13 pups were correctly targeted on one allele (8% homologous recombination efficiency). Only one out of the two mice had the knock-in allele in the germline. The generated heterozygote founder mouse was backcrossed to NOD WT mice three times and then intercrossed to generate M98A/M98A and control breeding lines. M98A/M98A mice were cohoused with control WT/WT mice after weaning until used in experiments. BDC-2.5 Tg M98A/M98A and 8F10 Tg M98A/M98A mice were generated by intercrossing M98A/M98A mice with BDC-2.5 Tg mice and 8F10 Tg mice, respectively. Female mice were used for experiments unless otherwise stated. Genotyping was performed with Custom TaqMan SNP Genotyping Assays (Applied Biosystems; primer pair spanning mutation site in the Cd74 gene: 5'-TCCCTTGGACCTGGACTCAT-3' and 5'-GGCACCGTTCGATTGATGTG-3'; WT- and M98A-specific probes: 5'-TTGCTGATGCGTCCA-3' [WT reporter dye: VIC] and 5'-TTGCTGGCTCGTCCA-3' [M98A reporter dye: FAM]). Briefly, 100 ng/well of genomic DNA was used as a template and amplified with the TaqMan SNP genotyping assay and the AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific). PCR was performed using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with the following conditions: 95°C 10 min, followed by 40 cycles of 98°C 10 s, 64°C 30 s, and 72°C 30 s (plate reading), followed by a final 5-min extension at 72°C.
Expression of I-Ag7 and I-Ag7 S57D mutant proteins
Soluble I-Ag7 proteins were produced using a baculovirus expression system as previously described (Jang et al., 2003). Proteins were purified by affinity chromatography using mAb 10–2.16 and gel filtration chromatography. Codon β57S was mutated by site-directed mutagenesis to produce the I-Ag7 β S57D mutant.
Peptide binding assay
The linker connecting CLIP to the N-terminus of the I-Aβ chain of purified I-Ag7 or I-Ag7 β S57D was digested with thrombin to enable exchange of CLIP with test peptides. I-Ag7 or I-Ag7 βS57D (0.15 mg/ml) were incubated in Tris-buffered saline (TBS) with 20 U of thrombin per milligram of I-Ag7. Thrombin-digested I-Ag7 or I-Ag7 β S57D (150 nM) was incubated with biotinylated test peptide (0, 125, 250, 500, 1,000, and 2,000 nM) at 37°C for 18 h in a final volume of 160 µl. The buffer was 50 mM sodium citrate/50 mM sodium phosphate/100 mM NaCl, pH 5.2, containing protease inhibitor cocktail (Sigma-Aldrich). In parallel, a DELFIA yellow 96-well plate (PerkinElmer) was coated overnight at 4°C with 100 µl of 0.4 µg/well OX-6 mAb in 0.1 M sodium bicarbonate buffer, pH 9.6. Nonspecific binding sites were blocked with DELFIA Assay Buffer (PerkinElmer) for 2 h at room temperature. Wells were washed three times with TBS-Tween, and I-Ag7–peptide samples were added to the wells. After a 1-h incubation, wells were washed four times with TBS-Tween, and europium-labeled streptavidin (PerkinElmer) was added (1:2,000 in DELFIA Assay Buffer, 100 µl/well) for detection of I-Ag7–bound biotinylated peptide. After 1 h incubation at room temperature, wells were washed six times with TBS-Tween, and DELFIA enhancement solution (PerkinElmer) was added. Fluorescence was quantitated after 30 min in an EnVision Multimode Plate Reader (PerkinElmer). Samples were analyzed in triplicate. Sequences of test peptides (with C-terminal biotin) were as follows: CLIP WT, VSQMRMATPLLMRPM(K-biotin); CLIP M98A, VSQMRMATPLLARPM(K-biotin); and MSA, KATAEQLKTVMDDFA(K-biotin). Peptides were synthesized by 21st Century Biochemicals.
Analysis of potential off-target regions
Potential off-target regions were identified using the Benchling program (benchling.com). Guide parameters were set as follows: guide length, 17 bp; genome, NODSHILTJ_B37 or NODSHILTJ_B38; PAM, NGG; and query sequence, AAGGAGCATGTTATCCATGG. Four potential off-target sites were analyzed by extracting genomic DNAs from one control NOD mouse and five M98A/M98A knock-in mice. The knock-in target region of Cd74 gene and the selected potential off-target regions were amplified by PCR using primers: 5'-CACATGTCCCACCATGTGTT-3' (position 30016614 forward), 5'-TGGCCAATTCATACCAGAAA-3' (position 30016614 reverse), 5'-TGTTTCCATGCTTTCTGTGC-3' (Tpp2 forward), 5'-TGCTTAGCCGTGTATCACCA-3' (Tpp2 reverse), 5'-TGGTGAAAGCGAAGTGACAA-3' (Hist1h2be forward), 5'-ATCATTCGTGCAAATGCGTA-3' (Hist1h2be reverse), 5'-TGAAGCTCCACTGTGGGTAA-3' (Mug1 forward), 5'-GGCACAAAGAAGCAGAATCC-3' (Mug1 reverse), 5'-TCCCTTGGACCTGGACTCAT-3' (Cd74 forward), and 5'-GGCACCGTTCGATTGATGTG-3' (Cd74 reverse). The PCR reactions for the Cd74 region were performed using the Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs) with the following conditions: 98°C 30 s, followed by 35 cycles of 98°C 5 s, 68°C 10 s, and 72°C 20 s, followed by a final 2-min extension at 72°C. The PCR reactions for potential off-target regions were performed using the Phusion High-Fidelity DNA Polymerase (New England Biolabs) with the following conditions: 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 64°C for 30 s, and 72°C for 30 s, followed by a final 5-min extension at 72°C. PCR products were purified from agarose gels using the Zymoclean Gel DNA Recovery Kit (Zymo Research) followed by Sanger sequencing using primers used for PCR amplification of the gene segments.
Diabetes monitoring
Blood glucose levels were monitored weekly. Blood was collected from the tail vein and analyzed with Contour nextEZ (Bayer). Mice were considered diabetic after two consecutive glucose readings of >250 mg/dl.
Immunofluorescence analysis of frozen sections
Pancreas tissue samples were embedded in optimal cutting temperature compound and frozen in isopentane cooled with liquid nitrogen. Frozen tissue was stored at −80°C until sectioning. Sections (8 µm) were generated using a Cryotome cryostat (Thermo Fisher Scientific) cooled to −20°C. Sections were placed on Superfrost glass slides, fixed with methanol at −20°C for 10 min, air dried, rinsed twice with PBS, and blocked with 2% rat serum in PBS for 5 min. Sections were stained for 10 min at 4°C with anti-SEZ6L2/BSRP-A (11 µg/ml, a marker for pancreatic islet cells), Alexa Fluor 488–conjugated anti-CD4 (1 µg/ml), Alexa Fluor 647–conjugated anti-CD8 (1 µg/ml), and DAPI (25 µg/ml). Sections were washed twice with PBS/2% rat serum and stained with Cy3-conjugated goat anti-sheep IgG antibody (5 µg/ml) for 10 min at 4°C to detect bound SEZ6L2/BSRP-A antibodies. Images were obtained using a Leica SP5X laser scanning confocal microscope (objective lens magnification: 40×) at room temperature. Images were acquired and analyzed with Leica Application Suite software.
Grading of T cell infiltration into pancreatic islets
All islets on a section were evaluated and graded for the extent of T cell infiltration as follows: 0, no infiltration of CD4+ T cells or CD8+ T cells; 1, CD4+ T cells or CD8+ T cells contacting islet perimeter, but not penetrating into islets; 2, penetration of CD4+ T cells or CD8+ T cells into ≤25% of islet mass; 3, penetration of CD4+ T cells or CD8+ T cells into ≤75% of islet mass; and 4, penetration of CD4+ T cells or CD8+ T cells into >75% of islet mass. 27–107 islets per mouse were analyzed.
Islet isolation and dissociation
The pancreatic duct was injected under a stereomicroscope with 2.5 ml of collagenase P (0.8 mg/ml in Hanks’ balanced salt solution). The pancreatic tissue was then dissected and incubated at 37°C for 10 min for enzymatic tissue dissociation. Tubes were then shaken for 10 s, and the tissue was washed once with PBS plus 2% FBS and passed through a stainless mesh to remove undigested tissue fragments. The cell suspension containing the islets was then layered onto Histopaque-1077, followed by centrifugation for 13 min at 2,400 rpm (1,346 g) without braking at the end of the spin. The interface containing islets was aspirated, and collected islets were washed and handpicked under a stereomicroscope. Handpicking of islets was repeated to ensure purity of the preparation. Islets were dissociated into single cells by incubation with Cell Dissociation Buffer (enzyme free, EDTA based; Thermo Fisher Scientific) for 5 min on ice and intermittent pipetting. Islets from each individual mouse were pooled and analyzed by flow cytometry. For in vitro T cell proliferation assays, islets were dissociated into single cells by incubation with prewarmed trypsin-EDTA (0.25%; Thermo Fisher Scientific) for 5 min at 37°C and intermittent pipetting.
Generation of tetramers
I-Ag7/CLIP complexes with a C-terminal BirA site on the I-Aα chain were biotinylated with BirA enzyme. The linker connecting CLIP to the N-terminus of the I-Aβ chain was digested with thrombin to enable exchange of CLIP with peptides of interest. 2.5HIP and 6.9HIP were synthesized with a C-terminal epitope tag (dansyl) to enable affinity purification of the final I-Ag7–peptide complex. Peptide sequences were 2.5HIP DLQTLALWSRMDQLA(K-dansyl)-amide, and 6.9HIP DLQTLALNAARDPNR(K-dansyl)-amide. Peptide exchange reactions were performed using 0.2 mg/ml of I-Ag7 and 50 µM peptide at pH 6.0 and 25°C for 18 h. Free peptide was then removed using a Superose 12 HPLC gel filtration column, followed by anti-dansyl affinity chromatography to isolate stoichiometric complexes of I-Ag7 and the relevant peptide. I-Ag7–CLIP complexes were used as controls without cleavage of the linker (to prevent CLIP dissociation and protein aggregation).
Tetramer staining
Peptide-MHC tetramers were generated by incubating PE-conjugated or allophycocyanin-conjugated streptavidin with biotinylated-peptide I-Ag7–peptide monomers at a ratio of 4:1 for 1 h at 4°C. Islet-infiltrating CD4 T cells from individual mice were labeled with 10 µg/ml of PE-conjugated InsB12-20 tetramer plus allophycocyanin-conjugated InsB13-21 tetramer, or PE-conjugated 2.5HIP tetramer plus allophycocyanin-conjugated 6.9HIP tetramer for 1 h at room temperature. Cells were then stained with Brilliant Violet 650 anti-CD4 (RM4-5), Brilliant Violet 510 anti-CD8α (53–6.7), PerCP/Cy5.5-conjgated markers for dump channel (anti-CD11b [M1/70], anti-CD11c [N418], anti-F4/80 [BM8], anti-NKp46 [29A1.4], anti–Gr-1 [Ly6C/G], and B220) and Zombie UV (dead cell marker; Biolegend). Cells were then fixed, permeabilized, and stained with Brilliant Violet 421 anti-Ki67 (16A8), and Alexa Fluor 488 anti-FoxP3 (MF-14). PerCP/Cy5.5-conjugated dump marker–negative, Zombie UV–negative, CD8-negative, CD4-positive cells were gated and examined for tetramer labeling.
Islet-infiltrating CD8 T cells from individual mice were labeled with 20 µg/ml of PE-conjugated Ins15-23 tetramer plus allophycocyanin-conjugated IGRP206-214 tetramer for 1.5 h at room temperature. Cells were stained with Brilliant Violet 650 anti-CD8α (53–6.7), PerCP/Cy5.5-conjgated dump channel markers (anti-CD11b [M1/70], anti-CD11c [N418], anti-F4/80 [BM8], anti-NKp46 [29A1.4], anti–Gr-1 [Ly6C/G], anti-CD4 [GK1.5], and B220) and Zombie UV (dead cell marker; Biolegend). PerCP/Cy5.5-conjugated dump marker–negative, Zombie UV–negative, CD8-positive cells were gated and examined for tetramer labeling. H2-Kb tetramers were generated by the National Institutes of Health tetramer facility using the following peptides: IGRP VYLKTNVFL, IGRP(8D) VYLKTNVDL, and Ins15-23 LYLVCGERL (leucine at P9 position).
Intracellular staining
After tetramer staining, cells were stimulated with Cell Stimulation Cocktail (plus protein transport inhibitors; eBioscience) for 2.5 h, and then labeled with Brilliant Violet 650 anti-CD4, Brilliant Violet 510 anti-CD8, PerCP/Cy5.5 dump channel markers (anti-CD11b, PerCP/Cy5.5 anti-CD11c, PerCP/Cy5.5 anti-F4/80, PerCP/Cy5.5 anti-NKp46, PerCP/Cy5.5 anti–Gr-1, and PerCP/Cy5.5 anti-B220), and Zombie UV and fixed and permeabilized using BD Cytofix/Cytoperm (BD Bioscience), followed by Alexa Fluor 700 anti–IL-17A, Brilliant Violet 421 anti–IFN-γ, PE/Cy7 anti–TNF-α, and FITC anti–IL-2 staining. When FoxP3 (FITC-conjugated) was analyzed, cells were fixed and permeabilized with transcription factor staining buffer set (eBiosciences). When Helios and CTLA-4 expression in FoxP3+ T cells was examined, cells were fixed and permeabilized with a FoxP3/transcription factor staining buffer set (eBiosciences).
Antibodies
The following mAbs were used for flow cytometry (purchased from BioLegend unless indicated otherwise). Brilliant Violet 650 anti-CD4 (RM4-5), Brilliant Violet 510 anti-CD8α (53–6.7), Brilliant Violet 421 anti-Ki67 (16A8), and Alexa Fluor 488 anti-FoxP3 (MF-14) were used for experiments of tetramer-labeled cells. Brilliant Violet 421 anti-CD4 (RM4-5), Brilliant Violet 510 anti-CD8α (53–6.7), Alexa Fluor 700 anti–IL-17A (TC11-18H10.1), Brilliant Violet 421 anti–IFN-γ (XMG1.2), PE/Cy7 anti–TNF-α (MP6-XT22), and FITC anti–IL-2 (JES6-5H4) were used for experiments to analyze cytokine-producing cells. In these two experiments, PerCP/Cy5.5-conjgated anti-CD11b (M1/70), anti-CD11c (N418), anti-F4/80 (BM8), anti-NKp46 (29A1.4), anti–Gr-1 (Ly6C/G), and B220 were used for the dump channel. Brilliant Violet 510 anti-CD45 (30-F11), PE/Cy7 anti-CD11c (N418), FITC anti-rat RT1B (OX-6), allophycocyanin/Cy7 anti-F4/80 (BM8), Brilliant Violet 421 anti-CD103 (2E7), Brilliant Violet 650 anti-CD80 (16-10A1), Alexa Fluor 700 anti-CD86 (GL-1), Brilliant Violet 785 anti-CD11b (M1/70), and PE anti-CD19 (1D3) were used for analyses of APCs in islets. Anti–H2-M (2E5A), FITC anti-rat IgG1 antibody (RG11/39.4), PE anti-rat RT1B (OX-6), allophycocyanin anti-CD19 (1D3), Alexa Fluor 700 anti-CD11b (M1/70), allophycocyanin/Cy7 anti-F4/80 (BM8), Brilliant Violet 510 anti-CD45 (30-F11), Brilliant Violet 650 anti-CD74 (In-1), Brilliant Violet 421 anti-CD103 (2E7), and Brilliant Violet 785 anti-CD11c (N418) were used for analyses of splenic APCs. Brilliant Violet 421 anti-CD4 (RM4-5), Brilliant Violet 510 anti-CD8α (53–6.7), FITC anti-Vα2 TCR (B20.1), FITC anti-Vα3.2 TCR (RR3-16), FITC anti-Vα8.3 TCR (B21.14), allophycocyanin anti-FoxP3 (FJK-16s), and PE anti-Vβ4TCR (KT4) or PE anti-Vβ8.1/8.2 TCR (MR5-2) were used for staining of thymocytes. Brilliant Violet 650 anti-CD4 (RM4-5), Brilliant Violet 510 anti-CD8α (53–6.7), FITC anti-Vβ2 TCR (B20.6), FITC anti-Vβ14 TCR (14–2; BD Biosciences), allophycocyanin anti-Vβ10 TCR (8.48), Alexa Fluor 647 anti-Vβ11 TCR (KT11), PE anti-Vβ4 TCR (KT4), PE anti-Vβ8.1/8.2 TCR (KJ16-133.18), or PE/Cy7 anti-Vβ5.1/5.2 TCR (MR9-4) were used for staining of thymocytes for Vβ repertoire analysis. PE/Cy7 anti-CD4 (GK1.5), FITC-conjugated anti-CD11b (M1/70), anti-CD11c (N418), anti-F4/80 (BM8), anti-NKp46 (29A1.4), anti–Gr-1 (Ly6C/G), and B220 were used for single-cell RNA-seq experiments. PE/Cy7anti-GITR (DTA-1), Pacific Blue anti-Helios (22F6), and PE/Dazzle 594 anti-CD152 were used for staining InsB12-20–labeled FoxP3+ CD4+ T cells. Isotype control IgG was from Biolegend.
For immunofluorescence analyses, the following antibodies were used: Alexa Fluor 488 anti-CD4 (RM4-5) and Alexa Fluor 647 anti-CD8α (53–6.7) were purchased from Biolegend. Anti-SEZ6L2/BSRP-A polyclonal antibodies were purchased from R&D Systems. Cy3 goat anti-sheep IgG polyclonal antibodies were purchased from Jackson ImmunoResearch.
Flow cytometry
Flow cytometry was performed with a LSR Fortessa (BD Biosciences), and data were analyzed using FlowJo software (Tree Star).
Analysis of CD74 expression and H2-DM expression in splenic APCs
Spleens from 7-wk-old male control mice and M98A/M98A mice were treated with the Spleen Dissociation Kit (Miltenyi). Red blood cells were lysed with ACK Lysis Buffer (Life Technologies). Splenocytes were stained with mAbs for surface molecules and then with Zombie UV Viability Dye. Intracellular CD74 and H2-DM were stained using the FoxP3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific). After fixation, cells were stained with anti-CD74 antibody and anti–H2-DM antibody, washed with permeabilization buffer, and stained with FITC-conjugated anti-rat IgG1 secondary antibody.
In vivo T cell proliferation assay
Spleens and lymph nodes (cervical, brachial, axillary, inguinal, pancreatic, and mesenteric) were dissected from 5-wk-old male BDC-2.5 TCR Tg mice and dispersed into single-cell suspensions. CD4 T cells were isolated using an EasySep Mouse CD4+ T cell Isolation Kit (STEMCELL Technologies). CD4 T cells were labeled with CFSE (1 µM) for 3 min at 37°C, and the reaction was then quenched by the addition of RPMI 1640 with 10% fetal calf serum. Cells (3–4 × 106) were transferred via intravenous injection into 10-wk-old male recipient mice. On day 3, draining pancreatic lymph nodes and mesenteric lymph nodes were dissected, and CFSE dilution by CD4 T cells was analyzed by flow cytometry.
In vitro T cell proliferation assays
BM-DCs were generated and used as APCs. Bone marrow cells were collected from femur, tibia, and humerus. After lysis of erythrocytes (ACK lysis buffer from Life Technologies), 107 bone marrow cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 100 U/ml of penicillin, 100 µg/ml of streptomycin, and 55 µM 2-mercaptoethanol (2-ME) plus 20 ng/ml of GM-CSF. On day 3, half of the supernatant was replaced using the same media composition. On day 6, bone marrow cells were stimulated with 100 ng/ml of LPS. On day 7, bone marrow cells were harvested and washed twice with PBS. CD11c+ I-Ag7+ cells were sorted twice using a BD AriaIII cell sorter (final purity >85%) and used in T cell proliferation assays. T cells were collected and stained with CFSE as described above. 5 × 104 T cells and 104 BM-DCs were cultured in V-bottom 96-well plates in 200 µl of RPMI 1640 supplemented with 10% fetal calf serum, 100 U/ml of penicillin, 100 µg/ml of streptomycin, and 55 µM 2-ME plus titrated numbers of freshly isolated islet cells (272–34,000 cells/well) that were dissociated as described above. On day 3, half of the supernatants were collected for cytokine assays and replaced with the same amount of fresh media. On day 5, cells were analyzed with flow cytometry. In some experiments, 5.7 × 105 freshly isolated islet cells were cultured in 300 µl of RPMI 1640 supplemented with 10% fetal calf serum for 24 h, and cells were removed by centrifugation. Supernatant was collected and added to co-cultures of T cells and DCs.
8F10 and 11T3 T cell hybridomas were provided from E.R. Unanue (Mohan et al., 2013). When T cells or T cell hybridomas were stimulated with 2.5HIP, InsB12-20, and InsB13-21 peptides, T cells or T cell hybridomas were cultured with splenic DCs. Spleens from male control or M98A/M98A mice were treated with the Spleen Dissociation Kit (Miltenyi), and splenic CD11c+ cells were enriched using mouse CD11c MicroBeads UltraPure (Miltenyi). CD11c+ I-Ag7+ cells were sorted by Aria III (purity >93%). T cells or T cell hybridomas and sorted CD11c+ I-Ag7+ DCs were cultured with titrated amounts of peptides. On day 3, supernatants were collected, and cells were analyzed with flow cytometry. IFN-γ concentrations in culture supernatants were analyzed by mouse IFN-γ ELISA (Biolegend) according to the manufacturer’s instructions. Sequences of peptides were as follows: 2.5HIP DLQTLALWSRMDQLA(K-dansyl)-amide, 6.9HIP DLQTLALNAARDPNR(K-dansyl)-amide, InsB12-20 TEGVEALYLVCGGGS-amide, and InsB13-21 TEGEALYLVCGEGGS-amide. Peptides were generated by 21st Century Biochemicals.
For experiments with fixed APCs (Fig. 10, A–D), DCs were fixed with 0.1% paraformaldehyde for 10 min at room temperature and washed three times. CFSE-labeled T cells (2 × 105) from 8F10 mice or BDC-2.5 TCR Tg mice, or 21.30 T cell hybridomas (specific for HEL) were cultured with 4 × 104 fixed DCs with or without 10 µM InsB12-20 peptide, 10 nM 2.5HIP peptide, or 10 µM HEL protein. On day 3, CFSE dilution by T cells was analyzed with flow cytometry. Activation of 21.30 hybridoma was examined by quantifying IL-2 secretion after 24 h (ELISA; Biolegend).
For experiments with cycloheximide-treated APCs (Fig. 10 E), DCs were incubated with 5 µg/ml of cycloheximide (Sigma-Aldrich) for 3 h. 10 nM 2.5HIP peptide was added to the culture, and DCs were incubated for an additional 3 h with rocking for the first 1 h. Cells were washed twice with PBS and fixed with 0.005% glutaraldehyde (Sigma-Aldrich) for 10 s. DCs (105) were washed three times with RPMI 1640 supplemented with 10% fetal calf serum and cultured with CFSE-labeled CD4 T cells (2 × 105) from BDC-2.5 TCR Tg mice. On day 3, CFSE dilution by T cells was analyzed by flow cytometry.
Immunization and ELISPOT
Age-matched male control and M98A/M98A mice (8–12 wk old) were immunized in the hock with 100 µg of HEL protein (Sigma-Aldrich) or 100 µg of OVA protein (Sigma-Aldrich). After 8 d, the popliteal lymph nodes were removed. CD4+ T cells were enriched with the EasySep Mouse CD4+ T cell Isolation Kit (STEMCELL Technologies) according to the manufacturer’s instructions. DCs were isolated from spleens by treatment with the Spleen Dissociation Kit (Miltenyi), red blood lysis was done with ACK Lysis Buffer (Life Technologies), and CD11c+ cells were enriched with mouse CD11c MicroBeads UltraPure (Miltenyi). CD11c+ I-Ag7+ cells were then sorted by AriaIII (purity >93%). 106 CD4+ T cells were cultured with 8 × 104 CD11c+ I-Ag7+ cells with or without 2.5 µM HEL protein or OVA protein for 18–20 h at 37°C in 5% CO2. IFN-γ–producing cells were detected with the Mouse IFN-γ ELISPOT Kit (R&D Systems).
For analysis of T cell responses to defined OVA and HEL epitopes, 9-wk-old NOD mice were immunized with 8 µg of OVA323-339 peptide (Sigma-Aldrich), 9.5 µg of HEL11-27 peptide (AMKRHGLDNYRGYSLGN-amide; 21st Century Biochemicals), or 2.5 nmol 6.9HIP peptide. On day 7, CD4+ T cells were harvested as described above. CD11c+ I-Ag7+ cells were sorted from control and M98A/M98A mice as described above. 106 CD4+ T cells were co-cultured with 9 × 104 CD11c+ I-Ag7+ cells in the presence of titrated concentrations of full-length antigens (OVA and HEL) or synthetic peptides (OVA323-339, HEL11-27, or 6.9HIP) for 18 h at 37°C in 5% CO2. IFN-γ–producing cells were detected using the Mouse IFN-γ ELISPOT Kit (R&D Systems).
Single-cell RNA-seq
InsB12-20 and 6.9HIP tetramer–positive CD4 T cells were sorted into 96-well plates for single-cell RNA-seq analysis. Islets were collected from a total of 30 11-wk-old female NOD mice and divided equally into three groups. Islet cells from 10 mice per group were pooled before T cell isolation. Groups 1 and 2 were used to isolate both InsB12-20 and 6.9HIP tetramer+ T cells, while group 3 was used to isolate Ins12-20 tetramer+ T cells. Islet cells were labeled with PE–InsB12-20 tetramer and allophycocyanin-6.9HIP tetramer. After tetramer labeling, cells were stained with PE/Cy7 anti-CD4 (GK1.5), FITC-conjugated anti-CD11b (M1/70), anti-CD11c (N418), anti-F4/80 (BM8), anti-NKp46 (29A1.4), anti–Gr-1 (Ly6C/G), and B220 in the presence of 1 U/µl of RNase Inhibitor (Takara), and treated with Zombie Aqua (Biolegend) and Calcein Violet 450 a.m. Viability Dye (eBioscience). FITC-negative, Zombie Aqua–negative, Calcein Violet–positive, CD4+ tetramer-labeled cells were single-cell sorted using a BD Aria III instrument into 96-well plates, with each well containing 5 µl of lysis buffer composed of TCL buffer (QIAGEN) plus 1% 2-ME (Sigma-Aldrich) and 2 U/µl of RNase Inhibitor (Takara). After sorting, plates were spun for 1 min at 3,000 g and frozen at −80°C.
To prepare sequencing libraries, plates were thawed on ice for 1 min and spun down at 2,000 rpm for 1 min. Immediately following this step, RNA lysate was purified using a 2.2× RNAClean SPRI bead ratio (Beckman Coulter Genomics) without a final elution step (Shalek et al., 2013). RNA captured on beads was processed using a modified SMART-Seq2 protocol (Picelli et al., 2013) entailing RNA secondary structure denaturation (72°C for 3 min), reverse transcription with Maxima Reverse transcription (Life Technologies), and whole-transcription amplification (WTA) with KAPA HiFi HotStart ReadyMix 2X (Kapa Biosystems) for 22 cycles. WTA products were purified with Ampure XP beads (Beckman Coulter), quantified with a Qubit double-stranded DNA HS Assay Kit (Thermo Fisher Scientific), and quality assessed with a high-sensitivity DNA chip (Agilent). 0.2 ng of purified WTA product was used as input for the Nextera XT DNA Library Preparation Kit (Illumina). Uniquely barcoded libraries were pooled and sequenced with a NextSeq 500 high-output V2 75 cycle kit (Illumina) using 50 and 25 paired end reads.
Sequencing data were processed from raw reads to gene expression matrices as previously described (Haber et al., 2017). Briefly, sequencing reads were aligned to the UCSC mm10 mouse transcriptome using Bowtie (Langmead et al., 2009), and gene expression levels (transcripts per million [TPM]) were quantified from these alignments using RSEM (RNA-seq by expectation maximization; Li and Dewey, 2011). We excluded all cells with fewer than 500 unique genes detected. After this filtering step, 441 cells remained, with a total of 13,561 genes detected across all cells. Gene expression values were then log transformed [log(TPM/10+1)].
To identify cell types and their corresponding gene signatures, we used tools from the Seurat v2.3.1 R package (https://github.com/satijalab/seurat; Butler et al., 2018). 1,492 highly variable genes were chosen for clustering analyses using FindVariableGenes, which controls for the relationship between the mean and variance of gene expression. We centered each variable gene’s expression using ScaleData and used those values as inputs to principal components analysis with RunPCA. We used the top 20 principal components to carry out graph-based clustering using FindClusters, and for each cluster of cells, we identified differentially expressed genes using the Wilcoxon rank-sum test implemented in FindMarkers. These differentially expressed genes were combined with known cell marker genes to annotate the cell type represented by each cluster. Finally, we visualized the clustering results in two dimensions by building a t-distributed stochastic neighborhood embedding (tSNE) plot constructed from the top 20 principal components with RunTSNE.
The sequencing data shown in this paper are available in GEO under data repository accession no. GSE118322.
Statistical analysis
Statistical calculations were performed using the Prism graphing software (GraphPad Software). For comparison of kinetics of diabetes development, the Gehan-Breslow-Wilcoxon test was used because the diabetes does not develop at the same time point in all mice. Other data were analyzed using the Mann-Whitney test. A P value of <0.05 was considered to be statistically significant. Statistical analysis was performed on all data points; only technical failures were excluded. No data points were excluded on the basis of being outliers.
Online supplemental material
Table S1 shows a summary of potential off-target regions of CRISPR/Cas9 targeting with the Cd74 gRNA. Fig. S1 shows the absence of off-target mutations for chromosome 10 position 30016614, Tpp2 gene, Hist1h2be gene, and Mug1 gene. Fig. S2 shows confocal microscopy images of islets with different degrees of T cell infiltration. Fig. S3 shows Ki67 labeling of β cell–specific T reg cells in islets from NOD mice and data from single-cell RNA-seq analysis of tetramer-labeled T cells. Fig. S4 shows normal thymic differentiation of β cell antigen–reactive CD4+ T cells in M98A/M98A mice.
Supplementary Material
Acknowledgments
We would like to thank Dr. E.R. Unanue for providing 8F10 mice, insulin-specific hybridomas, and feedback on the manuscript. We would like to thank Sabrina Haag for help with the design of gRNAs for editing of the Cd74 gene and advice on the analysis of potential off-target lesions.
This study was supported by National Institutes of Health grant P01AI045757. Y. Ito was supported by a research fellowship from the Uehara Memorial Foundation. L.F. de Andrade was supported by a Friends for Life Neuroblastoma Fellowship. R.E. Tay was supported by an A*STAR Graduate Fellowship. A.M. Luoma was supported by a Cancer Immunology Training Grant (T32 CA207021). S.K. Dougan was funded by the Bill and Melinda Gates Foundation.
The authors declare no competing financial interests.
Author contributions: Y. Ito and K.W. Wucherpfennig designed the study, interpreted data, and wrote the manuscript; Y. Ito performed in vitro and in vivo experiments; S.K. Dougan generated knock-in mice; J. Pyrdol performed binding assays and generated HIP tetramers; O. Ashenberg, O. Rozenblatt-Rosen, M. Hofree, E. Christian, and A. Regev performed and analyzed single-cell RNA-seq experiments; A.M. Luoma analyzed the structure of I-Ag7; L. Teyton generated insulin tetramers; L.F. de Andrade analyzed blood glucose levels; and R.E. Tay sorted DCs.
References
- Acha-Orbea H., and McDevitt H.O.. 1987. The first external domain of the nonobese diabetic mouse class II I-A beta chain is unique. Proc. Natl. Acad. Sci. USA. 84:2435–2439. 10.1073/pnas.84.8.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avva R.R., and Cresswell P.. 1994. In vivo and in vitro formation and dissociation of HLA-DR complexes with invariant chain-derived peptides. Immunity. 1:763–774. 10.1016/S1074-7613(94)80018-9 [DOI] [PubMed] [Google Scholar]
- Baker R.L., Bradley B., Wiles T.A., Lindsay R.S., Barbour G., Delong T., Friedman R.S., and Haskins K.. 2016. Cutting Edge: Nonobese Diabetic Mice Deficient in Chromogranin A Are Protected from Autoimmune Diabetes. J. Immunol. 196:39–43. 10.4049/jimmunol.1501190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers C.R., de Jong A., Ovaa H., and Rodenko B.. 2009. Transpeptidation and reverse proteolysis and their consequences for immunity. Int. J. Biochem. Cell Biol. 41:66–71. 10.1016/j.biocel.2008.08.036 [DOI] [PubMed] [Google Scholar]
- Brown J.H., Jardetzky T.S., Gorga J.C., Stern L.J., Urban R.G., Strominger J.L., and Wiley D.C.. 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 364:33–39. 10.1038/364033a0 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Marin E., Shimizu J., Kanagawa O., and Unanue E.R.. 1996. The class II MHC I-Ag7 molecules from non-obese diabetic mice are poor peptide binders. J. Immunol. 156:450–458. [PubMed] [Google Scholar]
- Concannon P., Rich S.S., and Nepom G.T.. 2009. Genetics of type 1A diabetes. N. Engl. J. Med. 360:1646–1654. 10.1056/NEJMra0808284 [DOI] [PubMed] [Google Scholar]
- Corper A.L., Stratmann T., Apostolopoulos V., Scott C.A., Garcia K.C., Kang A.S., Wilson I.A., and Teyton L.. 2000. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science. 288:505–511. 10.1126/science.288.5465.505 [DOI] [PubMed] [Google Scholar]
- Crawford F., Stadinski B., Jin N., Michels A., Nakayama M., Pratt P., Marrack P., Eisenbarth G., and Kappler J.W.. 2011. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc. Natl. Acad. Sci. USA. 108:16729–16734. 10.1073/pnas.1113954108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel D., Gill R.G., Schloot N., and Wegmann D.. 1995. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. Eur. J. Immunol. 25:1056–1062. 10.1002/eji.1830250430 [DOI] [PubMed] [Google Scholar]
- Davies J.L., Kawaguchi Y., Bennett S.T., Copeman J.B., Cordell H.J., Pritchard L.E., Reed P.W., Gough S.C., Jenkins S.C., Palmer S.M., et al. . 1994. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 371:130–136. 10.1038/371130a0 [DOI] [PubMed] [Google Scholar]
- Delong T., Wiles T.A., Baker R.L., Bradley B., Barbour G., Reisdorph R., Armstrong M., Powell R.L., Reisdorph N., Kumar N., et al. . 2016. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 351:711–714. 10.1126/science.aad2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzin L.K., and Cresswell P.. 1995. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 82:155–165. 10.1016/0092-8674(95)90061-6 [DOI] [PubMed] [Google Scholar]
- Fallang L.E., Roh S., Holm A., Bergseng E., Yoon T., Fleckenstein B., Bandyopadhyay A., Mellins E.D., and Sollid L.M.. 2008. Complexes of two cohorts of CLIP peptides and HLA-DQ2 of the autoimmune DR3-DQ2 haplotype are poor substrates for HLA-DM. J. Immunol. 181:5451–5461. 10.4049/jimmunol.181.8.5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris S.T., Carrero J.A., Mohan J.F., Calderon B., Murphy K.M., and Unanue E.R.. 2014. A minor subset of Batf3-dependent antigen-presenting cells in islets of Langerhans is essential for the development of autoimmune diabetes. Immunity. 41:657–669. 10.1016/j.immuni.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J.D., Reyon D., Cascio V.M., and Joung J.K.. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32:279–284. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. . 2017. A single-cell survey of the small intestinal epithelium. Nature. 551:333–339. 10.1038/nature24489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann D.H., Yu B., Hausmann S., and Wucherpfennig K.W.. 1999. pH-dependent peptide binding properties of the type I diabetes-associated I-Ag7 molecule: rapid release of CLIP at an endosomal pH. J. Exp. Med. 189:1723–1734. 10.1084/jem.189.11.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Deutsch A.J., Lenz T.L., Onengut-Gumuscu S., Han B., Chen W.M., Howson J.M., Todd J.A., de Bakker P.I., Rich S.S., and Raychaudhuri S.. 2015. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47:898–905. 10.1038/ng.3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.H., Seth N.P., and Wucherpfennig K.W.. 2003. Ex vivo analysis of thymic CD4 T cells in nonobese diabetic mice with tetramers generated from I-A(g7)/class II-associated invariant chain peptide precursors. J. Immunol. 171:4175–4186. 10.4049/jimmunol.171.8.4175 [DOI] [PubMed] [Google Scholar]
- Kim C.Y., Quarsten H., Bergseng E., Khosla C., and Sollid L.M.. 2004. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. USA. 101:4175–4179. 10.1073/pnas.0306885101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovskii C., Fred R.G., Oskarsson M.E., Westermark G.T., and Welsh N.. 2017. Addition of exogenous sodium palmitate increases the IAPP/insulin mRNA ratio via GPR40 in human EndoC-βH1 cells. Ups. J. Med. Sci. 122:149–159. 10.1080/03009734.2017.1368745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropshofer H., Vogt A.B., Moldenhauer G., Hammer J., Blum J.S., and Hämmerling G.J.. 1996. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 15:6144–6154. [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Wucherpfennig K.W., and Wiley D.C.. 2001. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat. Immunol. 2:501–507. 10.1038/88694 [DOI] [PubMed] [Google Scholar]
- Li B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman S.M., Evans A.M., Han B., Takaki T., Vinnitskaya Y., Caldwell J.A., Serreze D.V., Shabanowitz J., Hunt D.F., Nathenson S.G., et al. . 2003. Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc. Natl. Acad. Sci. USA. 100:8384–8388. 10.1073/pnas.0932778100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., and Ploegh H.L.. 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33:538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan J.F., Levisetti M.G., Calderon B., Herzog J.W., Petzold S.J., and Unanue E.R.. 2010. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat. Immunol. 11:350–354. 10.1038/ni.1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan J.F., Petzold S.J., and Unanue E.R.. 2011. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. J. Exp. Med. 208:2375–2383. 10.1084/jem.20111502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan J.F., Calderon B., Anderson M.S., and Unanue E.R.. 2013. Pathogenic CD4+ T cells recognizing an unstable peptide of insulin are directly recruited into islets bypassing local lymph nodes. J. Exp. Med. 210:2403–2414. 10.1084/jem.20130582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Abiru N., Moriyama H., Babaya N., Liu E., Miao D., Yu L., Wegmann D.R., Hutton J.C., Elliott J.F., and Eisenbarth G.S.. 2005. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 435:220–223. 10.1038/nature03523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil N.S., Pashine A., Belmares M.P., Liu W., Kaneshiro B., Rabinowitz J., McConnell H., and Mellins E.D.. 2001. Rheumatoid arthritis (RA)-associated HLA-DR alleles form less stable complexes with class II-associated invariant chain peptide than non-RA-associated HLA-DR alleles. J. Immunol. 167:7157–7168. 10.4049/jimmunol.167.12.7157 [DOI] [PubMed] [Google Scholar]
- Picelli S., Björklund A.K., Faridani O.R., Sagasser S., Winberg G., and Sandberg R.. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 10:1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- Quartey-Papafio R., Lund T., Chandler P., Picard J., Ozegbe P., Day S., Hutchings P.R., O’Reilly L., Kioussis D., Simpson E., et al. . 1995. Aspartate at position 57 of nonobese diabetic I-Ag7 beta-chain diminishes the spontaneous incidence of insulin-dependent diabetes mellitus. J. Immunol. 154:5567–5575. [PubMed] [Google Scholar]
- Reed A.M., Collins E.J., Shock L.P., Klapper D.G., and Frelinger J.A.. 1997. Diminished class II-associated Ii peptide binding to the juvenile dermatomyositis HLA-DQ alpha 1*0501/DQ beta 1*0301 molecule. J. Immunol. 159:6260–6265. [PubMed] [Google Scholar]
- Scott C.A., Peterson P.A., Teyton L., and Wilson I.A.. 1998. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity. 8:319–329. 10.1016/S1074-7613(00)80537-3 [DOI] [PubMed] [Google Scholar]
- Sette A., Southwood S., Miller J., and Appella E.. 1995. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J. Exp. Med. 181:677–683. 10.1084/jem.181.2.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek A.K., Satija R., Adiconis X., Gertner R.S., Gaublomme J.T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D., et al. . 2013. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 498:236–240. 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann T., Martin-Orozco N., Mallet-Designe V., Poirot L., McGavern D., Losyev G., Dobbs C.M., Oldstone M.B., Yoshida K., Kikutani H., et al. . 2003. Susceptible MHC alleles, not background genes, select an autoimmune T cell reactivity. J. Clin. Invest. 112:902–914. 10.1172/JCI18337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J.A., Bell J.I., and McDevitt H.O.. 1987. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 329:599–604. 10.1038/329599a0 [DOI] [PubMed] [Google Scholar]
- Vigneron N., Stroobant V., Chapiro J., Ooms A., Degiovanni G., Morel S., van der Bruggen P., Boon T., and Van den Eynde B.J.. 2004. An antigenic peptide produced by peptide splicing in the proteasome. Science. 304:587–590. 10.1126/science.1095522 [DOI] [PubMed] [Google Scholar]
- Wiesner M., Stepniak D., de Ru A.H., Moustakis A.K., Drijfhout J.W., Papadopoulos G.K., van Veelen P.A., and Koning F.. 2008. Dominance of an alternative CLIP sequence in the celiac disease associated HLA-DQ2 molecule. Immunogenetics. 60:551–555. 10.1007/s00251-008-0310-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles T.A., Delong T., Baker R.L., Bradley B., Barbour G., Powell R.L., Reisdorph N., and Haskins K.. 2017. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J. Autoimmun. 78:11–18. 10.1016/j.jaut.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F.S., Karttunen J., Dumont C., Wen L., Visintin I., Pilip I.M., Shastri N., Pamer E.G., and Janeway C.A. Jr. 1999. Identification of an MHC class I-restricted autoantigen in type 1 diabetes by screening an organ-specific cDNA library. Nat. Med. 5:1026–1031. 10.1038/12465 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Reyes-Vargas E., Escobar H., Rudd B., Rockwood A.L., Delgado J.C., He X., and Jensen P.E.. 2016. Type 1 diabetes associated HLA-DQ2 and DQ8 molecules are relatively resistant to HLA-DM mediated release of invariant chain-derived CLIP peptides. Eur. J. Immunol. 46:834–845. 10.1002/eji.201545942 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.