We report a child with inherited, complete IRF9 deficiency who suffered from life-threatening influenza pneumonitis. IRF9 deficiency disrupts the activation of ISGF3 and impairs but does not abolish cellular responses to type I IFNs, as some ISGs are induced.
Abstract
Life-threatening pulmonary influenza can be caused by inborn errors of type I and III IFN immunity. We report a 5-yr-old child with severe pulmonary influenza at 2 yr. She is homozygous for a loss-of-function IRF9 allele. Her cells activate gamma-activated factor (GAF) STAT1 homodimers but not IFN-stimulated gene factor 3 (ISGF3) trimers (STAT1/STAT2/IRF9) in response to IFN-α2b. The transcriptome induced by IFN-α2b in the patient’s cells is much narrower than that of control cells; however, induction of a subset of IFN-stimulated gene transcripts remains detectable. In vitro, the patient’s cells do not control three respiratory viruses, influenza A virus (IAV), parainfluenza virus (PIV), and respiratory syncytial virus (RSV). These phenotypes are rescued by wild-type IRF9, whereas silencing IRF9 expression in control cells increases viral replication. However, the child has controlled various common viruses in vivo, including respiratory viruses other than IAV. Our findings show that human IRF9- and ISGF3-dependent type I and III IFN responsive pathways are essential for controlling IAV.
Introduction
Influenza virus is a common pathogen in children. It typically causes self-limiting respiratory disease, but can also cause severe illness with pneumonia and acute respiratory distress syndrome (ARDS; Short et al., 2014). Virus strain differences can influence the frequency and severity of infections, as illustrated by seasonal epidemics and pandemics caused by new recombinant influenza viruses, but do not explain inter-individual variability in a given population infected by the same virus (Taubenberger and Morens, 2006, 2008; Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza et al., 2010). Few human risk factors have been identified and consist mostly of acquired comorbidities, such as chronic pulmonary or cardiovascular disease and old age (Palese and Shaw, 2007; Shieh et al., 2010; Dawood et al., 2011). Thus, the pathogenesis of life-threatening pulmonary influenza in individuals without preexisting cardiac or pulmonary disease remains largely unexplained, especially in children (Centers for Disease Control and Prevention, 2013). Recently, we described autosomal recessive (AR), complete IRF7 deficiency as the first genetic etiology for life-threatening influenza ARDS in otherwise healthy humans (Ciancanelli et al., 2015). The lack of IRF7-dependent type I and type III IFN amplification by plasmacytoid dendritic cells (pDCs) or pulmonary epithelial cells, or both, probably underlies this patient’s susceptibility to influenza. GATA2 haploinsufficiency is the only other known inborn error of immunity underlying severe influenza, albeit in patients with multiple infections. Three patients had severe influenza at ages 18, 31, and 54 yr, associated with the abnormal development of multiple myeloid and lymphoid subsets, including pDCs (Sologuren et al., 2018). Influenza was the first severe infection in only one of these patients (Pasquet et al., 2013; Sole-Violan et al., 2013). The mechanism underlying severe influenza in GATA2-deficient patients is unknown, but might involve their lack of pDCs, which are among the most potent type I and type III IFN-producing cells due to their high basal expression of IRF7 (Coccia et al., 2004; Asselin-Paturel et al., 2005; Osterlund et al., 2007; Lauterbach et al., 2010; Ning et al., 2011). Intriguingly, inborn errors of TLR3- or MDA5-dependent production of IFNs have not been shown to underlie severe influenza (Zhang et al., 2007; Lamborn et al., 2017). Interestingly, STAT1-, STAT2-, JAK1-, TYK2-, IFNAR2-, and IL10RB-mutated patients, with defective type I and/or III IFN responses, were not reported to suffer from severe influenza (Glocker et al., 2009; Begue et al., 2011; Boisson-Dupuis et al., 2012; Hambleton et al., 2013; Duncan et al., 2015; Kreins et al., 2015; Shahni et al., 2015; Eletto et al., 2016; Moens et al., 2017). Moreover, patients with any of the >350 other known inborn errors of immunity, including patients lacking T cells, or B cells, or both, have not been reported to suffer from severe influenza (Bousfiha et al., 2018; Picard et al., 2018). Collectively, these data suggest that the human GATA2- and IRF7-dependent production of type I and/or type III IFNs is essential for protective immunity against influenza virus.
Mice either homozygous for an Irf7-null allele or heterozygous for a Gata2-null allele have also been characterized. Although Gata2+/− mice were found to possess significantly reduced pDC counts, they have not been studied for infectious diseases, as research has instead focused on hematopoietic and vascular integrity (Johnson et al., 2012; Collin et al., 2015; Onodera et al., 2016). Similarly, the integrity and function of the type III IFN signaling pathway remains largely unexplored in Gata2+/− mice. Irf7−/− mice display reduced induction of IFN-α/β following either viral infection or stimulation with synthetic TLR agonists, as well as increased vulnerability to infection with HSV-1 and encephalomyocarditis virus (Sato et al., 1998; Honda et al., 2005). More recently, Irf7 was identified as one of 25 differentially expressed genes between C57BL/6J and DBA/2J mice infected with influenza A virus (IAV) that overlapped with differentially expressed genes identified in other siRNA screens to be important for IAV replication (Wilk et al., 2015). Furthermore, Irf7−/− mice that were derived on a C57BL/6 background that does not contain a functional Mx1 allele, when infected by influenza virus, experienced greater morbidity and mortality than control mice (Wilk et al., 2015). Most other studies analyzing the genetics and immunology of influenza infection have also been performed in Mx1-deficient mice and thus should be reexamined in mice expressing Mx1, due to the crucial role of this IFN-stimulated gene (ISG) product in mouse immunity to influenza virus (Staeheli et al., 1988; Iwasaki and Pillai, 2014; Ciancanelli et al., 2016). Significantly, however, Ifnb1−/− mice are substantially more sensitive to IAV, even in an Mx1-positive background (Koerner et al., 2007). Similar studies in an Mx1-positive background have demonstrated crucial roles for IFNAR1, TLR7, and MyD88 signaling in defense against influenza infection (Koerner et al., 2007; Kaminski et al., 2012). Mice with impaired type III IFN immunity have been shown to display marginally elevated influenza virus replication relative to WT mice, and to be hypersensitive to influenza virus when type I IFN signaling was also ablated (Mordstein et al., 2008). Overall, type I and/or type III IFN immunity seem to be crucial for protective immunity against influenza virus in both humans and mice. Not all inborn errors of human type I and III IFN immunity, however, underlie severe influenza, at least among the few human patients reported. We therefore hypothesized that defects of type I and III IFNs other than GATA2 and IRF7 deficiency may underlie severe influenza in other individuals. Thus, we set out to analyze the whole-exome sequencing (WES) data of 20 children with severe influenza by first focusing on type I and III IFN-related genes.
Results
Homozygosity for a splice site mutation in IRF9
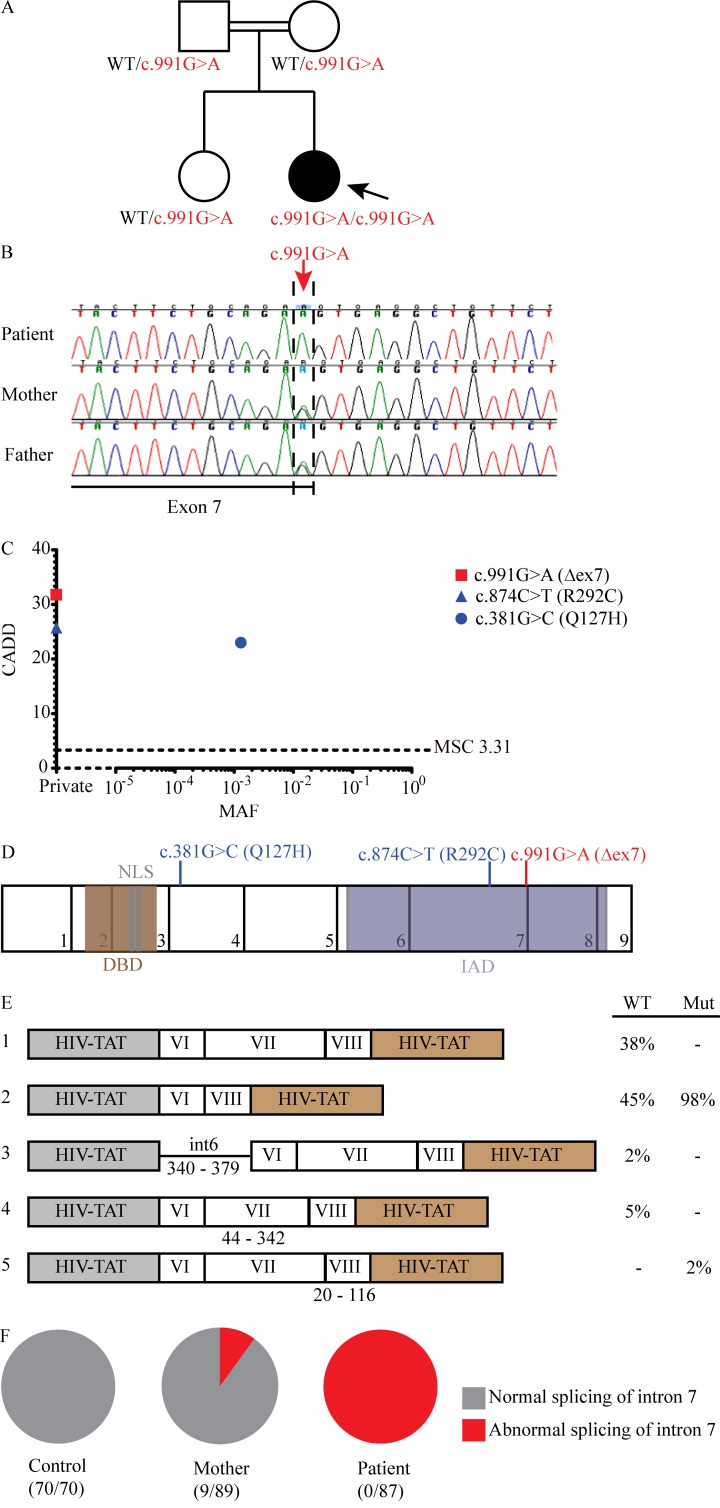
We studied a 5-yr-old girl (patient [P]) born to first-cousin Algerian parents and living in France who was hospitalized for severe infection with IAV requiring mechanical ventilation and Tamiflu treatment, and who had a history of recurrent benign bronchiolitis, biliary perforation following measles-mumps-rubella (MMR) vaccination at 1 yr of age, and recurrent fevers without a causative pathogen identified (Fig. 1 A). P also was infected with respiratory syncytial virus (RSV) within her first year of life, for which she was hospitalized but not admitted to the intensive care unit (ICU). We characterized P’s history of viral infections at age 2 yr, 7 wk before IgG i.v. substitution and after maternal antibodies disappeared, by VirScan (Xu et al., 2015). We detected significant specific antibodies in P’s serum against many other viruses, including HSV-1, human cytomegalovirus, human rhinovirus (HRV), and enterovirus, in addition to confirming infections with RSV and IAV (Fig. S1). After her recovery from IAV infection, she experienced adenovirus and parainfluenza virus (PIV) infections, both of which were also confirmed by VirScan. She was admitted to the hospital for these infections, but neither required admission to the ICU. We performed WES on this kindred (trio design) and identified a c.991G>A mutation in IRF9, encoding a critical element of the type I and III IFN signaling pathways (IRF9), which was confirmed by Sanger sequencing (Fig. 1 B; Fu et al., 1990; Veals et al., 1992; Kimura et al., 1996). P was homozygous for this mutation while both parents and her healthy sister were heterozygous, a familial segregation consistent with an AR pattern of inheritance with complete penetrance (Casanova et al., 2014). This variation was absent from public databases, including 1,000 genomes, Bravo, and Genome Aggregation Database (GnomAD)—a database that encompasses the exome aggregation consortium (ExAC) database. This mutation occurs in the final nucleotide of exon 7 and is predicted to disrupt the essential splice site at the boundary of exon 7 and intron 7, as well as to produce an aspartic acid to asparagine mutation at amino acid 331 (p.D331N). Because of the different predictions for these mutations on IRF9 protein structure, we will presently refer to this allele by its effect on the cDNA, c.991G>A. The combined annotation-dependent depletion (CADD) score of this variant was high, above the mutation significance cutoff (Fig. 1 C), further suggesting that it is deleterious (Kircher et al., 2014; Itan et al., 2016). Finally, P’s exome showed 6.8% homozygosity, consistent with her parents being first cousins (Belkadi et al., 2016). Consistently, homozygous nonsynonymous rare variations (minor allele frequency <0.01) were also found in 19 other genes, none of which is a known disease-causing gene. Genes encoding topoisomerase 2A (TOP2A) and 5′-3′ exoribonuclease 2 (XRN2), the only two connected to known primary immunodeficiency (PID) genes in a connectome analysis, carried variants that were missense (Itan et al., 2013). The only three genes that carried homozygous variations predicted to be loss-of-function were unrelated to PIDs: disheveled binding antagonist of β catenin 2 (DACT2), ENTH domain containing 1 (ENTHD1), and Forssman glycolipid synthase-like protein (GBGT1). Overall, these data suggested that P had AR IRF9 deficiency and that homozygosity for c.991G>A was the most plausible genotype responsible for severe influenza.
Figure 1.
A private IRF9 variant alters mRNA splicing in a child with severe influenza pneumonitis. (A) Pedigrees of the IRF9-deficient family. The double lines connecting the parents indicate consanguinity. The proband is indicated by an arrow. Filled shapes indicate affected individuals while open shapes identify unaffected individuals. (B) Chromatograms demonstrating c.991G>A mutation in patient PBMC-derived DNA (red arrow). (C) Population genetics of homozygous coding missense and predicted loss-of-function IRF9 mutations taken from GnomAD and in-house cohorts. The patient’s variant is private and shown in red, while two other variants, shown in blue, were also identified in our cohort. (D) Schematic illustration of the IRF9 gene. The exons are numbered 1–9, and regions corresponding to functionally significant domains are colored brown (for the DNA-binding domain, DBD), gray (nuclear localization sequence, NLS), or purple (IAD). Patient mutation indicated in red; other mutations indicated in blue. (E) IRF9 transcripts (left panel) and relative frequencies (right panel) produced during exon trapping in U2A cells. The results are representative of two independent experiments. (F) cDNA sequencing to detect the splicing of IRF9 mRNA from F-SV40 cells. Numbers of total and abnormal clones sequenced are indicated. Results representative of two experiments.
Population genetics of human IRF9
IRF9 encompasses 9 exons, including eight protein-coding exons (exons 2 through 9; Fig. 1 D). Although the gene damage index of IRF9 is moderate (1.16), its McDonald-Kreitman neutrality index of 0.088 indicates that it is under negative selection (Itan et al., 2015). Consistently, only one nonsynonymous exonic and two intronic variants were found in homozygosity in GnomAD, including whole-exome and whole-genome sequences: c.381G>C:p.Q127H, c.991+119T>G, and c.991+189C>A, with minor allele frequencies of 0.001, 0.01, and 0.001, respectively. All three variants were present in our in-house WES cohort of 4,892 patients with severe infectious diseases, with the two intronic variants found in the heterozygous state in multiple individuals and the Q127H variant found in the homozygous state in one individual. Only one of the three variants was predicted to alter protein sequence and may therefore impact IRF9 function. The latter two mutations occur in an intron, are not predicted to create a splice site, and actually do not create a splice site (data not shown). The Q127H allele exists in our cohort in the homozygous state in an individual who suffered from a nonviral infection of the brain. Furthermore, analysis of our in-house WES cohort revealed a third homozygous nonsynonymous variation in IRF9, c.874C>T:p.R292C, which was not found in public databases (Fig. 1 C). The individual harboring this variant also possessed a homozygous mutation in HOIL1 that underlied chronic autoinflammation, invasive bacterial infections, and muscular amylopectinosis, and he also experienced profound human cytomegalovirus viremia (Boisson et al., 2012). Homozygosity for any of the three exonic nonsynonymous variants (Q127H, R292C, and c.991G>A) is predicted to affect <1/10,000 individuals, which is not more common than severe influenza. The CADD scores of these three variants were all above the mutation significance cutoff, indicating that they may impact the function of IRF9 (Fig. 1 C; Itan et al., 2016). We therefore set out to assess the consequences of P’s variant on IRF9 function (c.991G>A), as well as the two other IRF9 missense alleles found in homozygosity in other, rare individuals from the general population (Q127H) or our in-house cohort (R292C).
Expression of a truncated mRNA and protein by the mutant IRF9 allele
Because the c.991G>A mutation occurs at an essential splice site at the terminal nucleotide of exon 7, we hypothesized that it would produce an aberrantly spliced transcript. We thus transiently transfected a segment of P’s allele encompassing introns 5 through 8, as well as a segment from an identical region of a WT allele, into IRF9-deficient U2A fibrosarcoma cells, a gift from S. Pellegrini (Institut Pasteur, Cytokine Signaling Unit, INSERM, Paris, France; Fig. S2; John et al., 1991). Sequencing of the resulting cDNAs revealed two transcripts produced from P’s IRF9 allele (Fig. 1 E). The dominant transcript exhibited absence of exon 7 with normal exons 6 and 8. The second, minor transcript also lacked exon 7 but used an alternative splice acceptor site in exon 8. The segment from a WT allele, however, produced four different transcripts. One transcript demonstrating correct splicing at exons 6/7 and 7/8 junctions and a second transcript lacking exon 7 but correctly using the splice sites in exons 6 and 8 were the dominant products. Two rare transcripts were also observed: one was a product of an alternative splice acceptor site in intron 5, whereas another resulted from an alternative splice acceptor site in exon 7 (Fig. 1 E). Only correctly spliced transcripts were observed in healthy control B-LCLs (EBV-transformed B lymphoblastoid cell lines) and SV40 large T antigen immortalized fibroblasts (F-SV40 cells), suggesting that the two alternative transcripts observed in the overexpression setting are not present endogenously. Moreover, cDNA sequencing of P’s B-LCLs and F-SV40 cells did not reveal any transcript that would result in a D331N variant protein, as all transcripts lacked exon 7 (Fig. 1 F). This mutant transcript (and the corresponding allele) will thus hereafter be referred to as IRF9-Δex7, as the deletion is in-frame and does not introduce a premature stop codon. Next, we evaluated IRF9 expression in P’s peripheral blood mononuclear cells (PBMCs) and F-SV40 cells. Consistent with the results of our cDNA sequencing, only IRF9 transcripts lacking exon 7 were detected in P’s PBMCs, and a reduced number of full-length IRF9 transcripts were detected in both individuals harboring the c.991G>A mutation in proportion to the dosage of this allele (Fig. 2 A). By Western blotting (WB), a polyclonal antibody detected a truncated IRF9 species in both P’s cells and those of her mother, whereas full-length IRF9 was not detected in P’s cells (Fig. 2 B). The molecular weight of this truncated protein is roughly 12 kD lower than WT IRF9, consistent with a lack of exon 7 (142 amino acids), which forms a substantial portion of the IRF association domain (IAD) where STAT proteins interact with IRF9 (Fig. 1 D). Overall, these findings showed that the patient had AR IRF9 deficiency.
Figure 2.
Impact of IRF9 Δex7 on IFN receptor-proximal signaling. (A) qRT-PCR measuring of IRF9 mRNA levels in PBMCs from the patient, her mother, and a healthy control with two probes—one probe spanning intron 7, and a second probe spanning intron 1. Representative results of four independent experiments are shown. (B) Top: WB of endogenous IRF9 in patient F-SV40 cells; GAPDH was used as a loading control. Bottom: STAT and phospho-STAT (pSTAT) levels were also assessed following stimulation with 1,000 U/ml of either IFN–α2b or –γ for 0.5 h on F-SV40 cells from two healthy controls (C1 and C2), the IRF9-deficient patient (IRF9−/−), her mother (IRF9+/−), a STAT1-deficient patient (STAT1−/−), a STAT2-deficient patient (STAT2−/−), an IFNGR2-deficient patient (IFNGR2−/−), and an IRF7-deficient patient (IRF7−/−). Representative results of five independent experiments are shown. (C) WB of IRF9 in IRF9-deficient U2A cells stably transfected with indicated variants (green: variants reported to be loss-of-function in in vitro assays, blue: variants found in-house, red: patient). GAPDH was used as loading control. Representative results of four independent experiments are shown. (D) WB of IRF9 in patient F-SV40 cells stably transfected with indicated variants. GAPDH was used as loading control. Representative results of four independent experiments are shown. (E) WB analysis of IRF9 localization in F-SV40 cells from two healthy controls (C1 and C2), the IRF9-deficient patient (IRF9−/−), her mother (IRF9+/−), a STAT1-deficient patient (STAT1−/−), a STAT2-deficient patient (STAT2−/−), an IFNGR2-deficient patient (IFNGR2−/−), and an IRF7-deficient patient (IRF7−/−). GAPDH and LaminA/C were used as loading controls. Representative results of three independent experiments are shown. (F) Reporter assays of ISRE or GAS-dependent firefly luciferase tested in U2A cells stimulated with 1,000 U/ml of either IFN-α2b or -γ for 16 h after being stably transfected with indicated variants (green: variants reported to be loss-of-function in in vitro assays, blue: variants found in-house, red: patient). The specific response to IFN stimulation was calculated by the ratio of firefly luciferase reporter gene activity to constitutively expressed renilla luciferase activity (RLU, relative luciferase ratio). Representative results of three independent experiments are shown. (G) EMSA analysis of ISRE and GAS binding by IFN-stimulated B-LCLs from three healthy controls (C1, C2, and C3), the IRF9-deficient patient (IRF9−/−), her mother (IRF9+/−), a STAT1-deficient patient (STAT1−/−), a STAT2-deficient patient (STAT2−/−), and an IRF7-deficient patient (IRF7−/−). Representative results of three independent experiments are shown.
Normal activation of STAT1 and STAT2 in cells expressing mutant IRF9
We then studied the function of the mutated IRF9 protein, first focusing on the proximal signaling events in the IFN responsive pathway. To assess the expression of P’s mRNAs, plasmids encoding IRF9-Δex7 and the other two missense mRNAs found in homozygosity in other individuals (Q127H, R292C), or WT IRF9, were first stably transfected into U2A cells, and IRF9 expression was verified by WB (Fig. 2 C). The K81R and R292Q alleles were included throughout the study since they have been reported to be loss-of-function in in vitro studies due to impaired interaction with IFNAR2 (R292Q) or impaired ISG up-regulation (K81R; Tang et al., 2007). All IRF9 alleles were expressed at comparable levels, indicating that these variants can be normally transcribed and translated, and that the encoded mRNAs and proteins are stable in these conditions (Fig. 2 C). We found that STAT1 and STAT2 phosphorylation was normal in cells transfected with all alleles following stimulation with type I IFN (not shown). Stable transfection of WT IRF9 in P’s F-SV40 cells restored normal expression of IRF9, suggesting that the poor expression of IRF9 in her cells was due to the homozygous Δex7 mutation in IRF9 (Fig. 2 D). STAT1 and STAT2 were normally phosphorylated in P’s cells immediately following stimulation with type I IFN, suggesting that IRF9 is dispensable for this process (Fig. 2 B). These data are consistent with previous reports and our own findings that STAT1 and STAT2 phosphorylation occurs immediately following IFN stimulation of U2A cells (Jaworska et al., 2010; Arimoto et al., 2017). Thus, the Δex7 IRF9 allele produces a truncated IRF9 protein that does not affect IFNAR-proximal signaling, in terms of STAT1 and STAT2 phosphorylation, at least in SV40 fibroblasts and U2A fibrosarcoma cells.
Abolished activation of ISGF3 in cells expressing mutant IRF9
IRF9 participates in the formation of the ISGF3 transcription factor by binding with STAT1 and STAT2 via its IAD. IRF9 also enables binding of the trimer to DNA, after its translocation to the nucleus (Veals et al., 1993; Darnell et al., 1994; Taniguchi et al., 2001; Stark and Darnell, 2012). Because P’s IRF9 protein deletion occurs in the IAD region (Fig. 1 D), we hypothesized that IRF9-Δex7 may be unable to engage with STAT1 and STAT2 to form a functional ISGF3, consistent with a recent report of this domain’s function (Rengachari et al., 2018). To test this, we first fractionated nuclear and cytoplasmic compartments of P’s F-SV40s under basal conditions to determine the localization of P’s IRF9 protein (Fig. 2 E). Consistent with the presence of a nuclear localization sequence in the N-terminus of IRF9, a small amount of IRF9 was visible in the nucleus even under basal conditions. IRF9-Δex7, although expressed at substantially lower levels than the full-length protein, is also visible in the nucleus. Consistently, U2A cells transfected with IRF9-Δex7 exhibited abolished luciferase transcription when luciferase production was driven from an IFN-stimulated response element (ISRE) promoter, but normal transcriptional activity when it was driven by a gamma activation sequence (GAS) promoter (Fig. 2 F). In contrast, the Q127H and WT variants were able to induce transcription of luciferase from both ISRE and GAS promoters. Both R292Q and R292C variants displayed an intermediate phenotype with roughly 50% activity compared with the WT allele, suggesting that they are hypomorphic. Taken together, these data strongly suggest that P’s IRF9 variant, Δex7, is unable to encode an IRF9 protein that forms a functional ISGF3 transcription factor, while R292C is hypomorphic and Q127H is neutral. Electrophoretic mobility shift assays (EMSAs) confirmed that P’s cells were not able to form a functional ISGF3 complex following IFN-α2b stimulation, as evidenced by abolished ISRE binding in vitro, although her cells were able to form gamma-activated factor complexes and bind GAS elements under the same conditions (Fig. 2 G). The patient is therefore most likely homozygous for a null IRF9 allele, as IRF9-Δex7 was the only transcript detected in the cells tested. However, one cannot exclude the remote possibility that the splicing defect is leaky in untested cells, where residual D331N expression might result in a severe but not complete form of IRF9 deficiency.
IRF9 deficiency impairs induction of ISGs
Due to the inability of P’s cells to form functional ISGF3, we hypothesized that her cells would display a profound inability to up-regulate ISGs following IFN stimulation. In contrast to control cells, P’s F-SV40s and B-LCLs did not substantially up-regulate MX1, IFIT1, or OAS1 expression in response to type I IFN, as assessed by quantitative RT-PCR (qRT-PCR) and WB (Fig. 3, A–D). Stable transfection of P’s F-SV40 cells with WT IRF9 was able to rescue the impaired induction of ISGs (Fig. 3 E). Similarly, U2A cells stably transfected with Δex7 were not able to induce transcription of either classic ISG, compared with WT IRF9 (Fig. 3 F). CXCL9 expression was used throughout these qRT-PCR experiments to demonstrate the ability of all cells to respond to type II IFN. However, the response to type I IFN is substantially broader than four or five genes, with potentially hundreds of genes displaying altered expression levels (Mostafavi et al., 2016). To characterize fully the transcriptional responses of P’s cells to type I IFN, we performed mRNA sequencing (mRNA-seq) analysis of P’s B-LCL cells and primary fibroblasts following in vitro stimulation with IFN-α2b. P’s B-LCL cells (Fig. 4, B and D–G) and primary fibroblasts (Fig. 4, A, C, and E–G) demonstrated an inability to regulate a broad array of genes, including ISGs but also negatively regulated transcripts (data not shown). However, the response in P's cells was not abolished altogether. Indeed, out of all transcripts that passed our filter criteria for being IFN-α2b–regulated in P's B-LCL cells and primary fibroblasts, 118 and 267 transcripts, respectively, were also found to be regulated by IFN in the Interferome database v2.01 (Fig. 4, C and D). Furthermore, many of the up-regulated ISGs were significantly less induced than those same ISGs were in healthy control cells (Fig. 4, E and F). Interestingly, however, some ISGs actually were induced at higher levels in IRF9-deficient cells when compared with healthy controls (Fig. 4, E and F). Furthermore, these difference were not necessarily due to a reduction in basal transcript levels, since, of those genes that are more highly induced in IRF9-deficient cells, roughly 40% exhibit higher absolute transcript levels in the IRF9-deficient cells (data not shown). To further dissect the ISG network and dysregulation in P's B-LCL cells, we selected a subset of 84 transcripts that were strongly induced (greater than fivefold linear scale) by IFN-α2b among the healthy control subjects in our study. Of these transcripts, 37 were found to be induced (>1.5-fold) and 47 were nonresponsive in P's B-LCL cells. The transcripts were then queried for ISGs in the Interferome v2.01 database, again using a 1.5-fold cut-off to differentiate between IFN-responsive and nonresponsive genes. Of these 37 responsive and 47 nonresponsive transcripts in P's cells, 24 and 39 transcripts, respectively, were identified as ISGs in the Interferome v2.01 database. These were used for a ISG response network analysis using GeneMANIA (Fig. 4 G). Notable ISGs that were not induced in P’s cells include genes encoding IFIT family proteins, ISG15, and components of the ISGylation system (USP18, HERC5). Consistent with the presence of ISGF3-independent transcription factors such as phosphorylated STAT1 homodimers downstream of the IFN receptor, however, mRNA of other important ISGs, such as DDX58, those for guanylate-binding proteins (GBP1, GBP4, and GBP5), ISG20, and STAT2, were induced by IFN-α2b in P’s cells, albeit for most genes to lower levels when compared with control subjects. The type III IFN responsive pathway cannot easily be studied in F-SV40s and B-LCLs (Kreins et al., 2015). Collectively, our studies indicate that P’s cells express a loss-of-function IRF9 allele that severely narrows the transcriptional responses to type I IFN in multiple cell types, with functional gamma-activated factor but a lack of ISGF3 complexes resulting in the induction of a small subset of ISGs.
Figure 3.
Impaired ISG induction in IRF9-deficient cells. (A) Transcription levels of MX1, IFIT1, IFIT3, and CXCL9 assessed by qRT-PCR on F-SV40 cells treated with 1,000 U/ml of IFN–α2b, -β, or –γ for 2 h. Cells were from three healthy controls (C1, C2, and C3), an IRF9-deficient patient (IRF9−/−), her mother (IRF9+/−), and STAT1-deficient (STAT1−/−), STAT2-deficient (STAT2−/−), IRF7-deficient (IRF7−/−), and IFNGR1-deficient (IFNGR1−/−) patients. Representative results of four independent experiments are shown. (B and C) WB of MX1 and IFIT3 on F-SV40 (B) or B-LCL (C) cells treated with 1,000 U/ml of IFN–α2b for various time points. GAPDH was used as a loading control. Representative results of three independent experiments are shown. (D) Transcription levels of MX1, IFIT1, IFIT3, and CXCL9 assessed by qRT-PCR of B-LCL cells treated with 1,000 U/ml of IFN–α2b, -β, or –γ for 2 h. Cells were from three healthy controls (T1, T2, and T3), an IRF9-deficient patient (IRF9−/−), her mother (IRF9+/−), and STAT1-deficient (STAT1−/−), STAT2-deficient (STAT2−/−), IRF7-deficient (IRF7−/−), and IFNGR2-deficient (IFNGR2−/−) patients. Representative results four independent experiments are shown. (E) Transcription levels of MX1, IFIT1, and CXCL9 assessed by qRT-PCR in F-SV40 cells from a healthy control (C1), P’s mother (IRF9+/−), and P (IRF9−/−) stably transfected with luciferase as a control (Luc) or indicated IRF9 variants (WT: WT IRF9, green: reported loss-of-function variants, blue: variants found in-house, red: patient variant). Cells were stimulated with 1,000 U/ml of IFN-α2b, -β, or -γ for 2 or 8 h. Representative results of four independent experiments are shown. (F) Similar to E, qRT-PCR analysis of MX1, IFIT1, and CXCL9 expression in parental HT1080 cells and U2A cells. Cells were stimulated with 1,000 U/ml of IFN-α2b, -β, or -γ for 2 or 8 h. Representative results of three independent experiments are shown.
Figure 4.
Transcriptomic analysis of ISGs in IRF9-deficient cells. mRNA-seq analysis of primary fibroblasts (A and C) and B-LCLs (B and D) from three healthy controls (C1, C2, and C3), the IRF9-deficient patient (IRF9−/−). Cells were treated with 1,000 U/ml IFN–α2b for 2 h. Heatmaps (A and B) show log2 FC values of all ISGs that were found to be differentially regulated (≥1.5-fold) in all three control subjects relative to unstimulated cells. Bar graphs (C and D) quantify the number of ISGs that were differentially regulated (≥1.5-fold) compared with unstimulated cells in healthy controls or the IRF9-deficient patient. (E) Shown are Δ log2 fold change values of a subset of ISGs that were found to be induced ≥1.5-fold (linear scale) in B-LCL cells (upper panels) or primary fibroblasts (lower panels) of the IRF9-deficient patient upon in vitro stimulation with IFN-α. To select this subset of ISGs, the IFN-α2b–induced genes in the healthy controls identified in the mRNA-seq analysis were used. In the IRF9-deficient patient, these genes were first passed through a filter by querying the gene identifiers against the interferome database and by retaining genes that were responsive to in vitro IFN stimulation. ISGs that failed to be induced at least 1.5-fold (linear scale) in patient cells were excluded. The retained ISGs were stratified in three groups of less (Δ less than −0.585), similar (−0.585 < Δ < 0.585), and higher (Δ > 0.585) induced genes relative to the average responses in the healthy control subjects. The numbers of genes in each group are shown in brackets. ***, significant differences at P < 0.0001 by the Kruskal-Wallis test. (F) Log2 FC of induced ISGs in IRF9-deficient B-LCLs (upper panels) or primary fibroblasts (lower panels) and their corresponding values in healthy donors. (G) Network analysis of a subset of highly inducible (> fivefold linear scale) ISGs among control subjects’ B-LCLs and their responsiveness in the IRF9-deficient patient. Biological pathway and physical interactions are depicted as blue and red lines, respectively. 1.5 FC was used as the cut-off to distinguish responsive (red circle) and nonresponsive (blue circle) ISGs. The highly inducible ISGs that were used for query are shaded in yellow.
Impaired anti-viral immunity in the patient’s fibroblasts
We investigated the impact of IRF9 deficiency on cell-intrinsic, nonhematopoietic immunity, using P’s F-SV40 cells. We observed an ∼10-fold difference in IAV titer at 48 h after infection when compared with healthy controls and P’s heterozygous mother (Fig. 5 A). Pre-treatment with IFN-α2b substantially reduced viral replication in healthy control cells but not P’s cells, which produced roughly 100-fold higher IAV titers than cells with an intact IFN signaling pathway (Fig. 5 A). Similarly, P’s F-SV40s were substantially more susceptible to vesicular stomatitis virus (VSV) at 12 h after infection in the absence of pretreatment with IFN-α2b, or at all time points beyond 8 h in cells that were pretreated with IFN, with roughly a 1,000-fold difference in titer between healthy controls and P (Fig. 5 B). To demonstrate that these defects in IAV and VSV immunity were IRF9 dependent, we stably transfected P’s F-SV40 cells with various IRF9 alleles or an empty vector and analyzed the ability of the transfected cells to control VSV and IAV infection (Fig. 5, C and D). Expression of WT IRF9 was able to rescue the inability of P’s cells to control of infection with either pathogen, while expression of K81R and IRF9-Δex7 alleles was unable to do so. Significantly, cells that expressed the Q127H mutation were also able to control infection with VSV or IAV, suggesting that this allele may not be deleterious in the context of type I IFN signaling and anti-viral immunity, consistent with the lack of overt viral infections experienced by the individual harboring this mutation. R292Q and R292C alleles were also somewhat able to control infection with VSV or IAV, although not at the level of the WT IRF9 allele, suggesting that a partial form of IRF9 deficiency may have contributed to the viral infections of the HOIL1-deficient patient (Boisson et al., 2012). We further showed that cell-intrinsic anti-viral immunity was broadly impaired, by testing PIV and RSV (Fig. 5, E–H). To further demonstrate that IRF9 mediates susceptibility to viral infection in vitro, we also knocked down IRF9 or mitochondrial anti-viral signaling protein (MAVS) expression in primary dermal fibroblasts from healthy individuals using siRNA, or transfected these cells with a nontargeting siRNA (siNeg) instead (Fig. 6 A). Knockdown of either IRF9 or MAVS resulted in increased viral transcripts compared with siNeg cells upon infection with HRV, suggesting that IRF9 is nonredundant for immunity to positive-sense RNA viruses in addition to the negative-sense viruses already tested. Similarly, knockdown of IRF9 increased the number of infected cells and the relative number of viral transcripts produced to a similar extent as did MAVS knockdown when compared with siNeg cells, for both RSV and PIV (Fig. 6, C–F). Collectively, these data indicate that IRF9 deficiency leads to a broad defect in cell-intrinsic immunity to viruses in vitro. Altogether, these data strongly suggest that inherited IRF9 deficiency underlies P’s impaired immunity to influenza infection and her subsequent hospitalization for ARDS.
Figure 5.
Crippled control of IAV and other viral infections in IRF9-deficient cells. (A) IAV titers in F-SV40 unstimulated (left) or pretreated (right) with 1,000 U/ml IFN-α2 for 16 h, followed by infection with (A/H1N1/CA/2009) IAV at MOI = 1. Mean ± SD (n = 3) is shown. Cells from three healthy controls were included (C1, C2, and C3), as well as those from the IRF9-deficient patient (IRF9−/−), her mother (IRF9+/−), and STAT1-deficient (STAT1−/−), STAT2-deficient (STAT2−/−), and IRF7-deficient (IRF7−/−) patients. Four independent experiments (mean ± SD) are shown. (B) VSV titers in F-SV40 cells unstimulated (left) or pretreated (right) with 1,000 U/ml IFN-α2 for 16 h, followed by infection with VSV at MOI = 3. Four independent experiments (mean ± SD) are shown. (C) IAV titers in stably transfected F-SV40 cells unstimulated (left) or pretreated (right) with 1,000 U/ml IFN-α2 for 16 h, followed by infection with IAV at MOI = 1. Cells were from a healthy control (C1), a STAT1-deficient patient (STAT1−/−), P (IRF9−/−), and P’s cells stably transfected with luciferase or WT IRF9 (gray), variants reported to be loss-of-function in in vitro assays (green), variants found in-house (blue), or the patient’s variant (red). Three independent experiments (mean ± SD) are shown. (D) VSV titers in stably transfected F-SV40 cells unstimulated (left) or pretreated (right) with 1,000 U/ml IFN-α2 for 16 h, followed by infection with VSV at MOI = 3. Four independent experiments (mean ± SD) are shown. (E) Percentage of RSV-infected (GFP+) F-SV40 cells at 24 and 48 h after infection. Cells from three healthy controls were included (C1, C4, and C5, black), as well as those from the IRF9-deficient patient (IRF9−/−, red), and cells from STAT1-deficient (STAT1−/−), STAT2-deficient (STAT2−/−), and IRF7-deficient (IRF7−/−) patients. Three independent experiments (mean ± SD) are shown. (F) Mean fluorescence intensity (MFI) of RSV-infected (GFP+) F-SV40 cells at 24 and 48 h after infection. Three independent experiments (mean ± SD) are shown. (G) Percentage of PIV-infected (GFP+) F-SV40 cells at 24 and 48 h after infection. Three independent experiments (mean ± SD) are shown. (H) MFI of PIV-infected (GFP+) F-SV40 cells at 24 and 48 h after infection. Three independent experiments (mean ± SD) are shown. MFI of GFP+ cells in individual samples were normalized to the averaged MFI of the three healthy controls at 24 h after infection in F and H.
Figure 6.
IRF9 is required for optimal control of viral infections. (A) WB confirms the efficiency of RNAi of IRF9 or MAVS in primary dermal fibroblasts. (B–F) Primary human dermal fibroblasts previously transfected with the indicated siRNA (negative control, IRF9, MAVS) were tested for control of HRV, RSV, and PIV. Cells were infected with HRV-A16 at MOI of 10 (B), RSV at MOI of 0.5 (C and D), or PIV3 at MOI of 0.1 (E and F). Relative HRV transcripts (B) were measured by qRT-PCR, and values were normalized to the siNeg control. Percentage of infected cells (C and E) and relative virus per infected cell (D and F) were measured by flow cytometric analysis of GFP+ cells. MFI of GFP+ cells in individual samples were normalized to negative control at 24 h (D and F). Shown are the mean ± SD of six (B–F) experiments. *, P < 0.05; **, P < 0.01, by Kruskal-Wallis test.
Discussion
We herein describe AR, complete IRF9 deficiency in a child with life-threatening influenza. Our data establish a causal relationship between IRF9 deficiency and this patient’s severe pulmonary influenza (Casanova et al., 2014). Following autosomal dominant GATA2 deficiency and AR IRF7 deficiency, AR IRF9 deficiency is the third described single-gene inborn error of immunity that can underlie life-threatening influenza pneumonitis (Casanova, 2015a,b; Ciancanelli et al., 2016). In contrast to IRF7 deficiency, which disrupts the amplification of antiviral IFNs, especially in pDCs, which are the only cells expressing high constitutive levels of IRF7 (Ciancanelli et al., 2015), IRF9 deficiency results in an inability of all cell types to respond fully and effectively to type I IFNs, and by inference type III IFNs. The mechanistic connection between IRF7 and IRF9 deficiency in humans suggests that the type I and III IFN responses driven by IRF7 amplification and mediated by IRF9 signaling are essential for defense against influenza virus. However, patients with other defects in type I and/or III IFN signaling pathway, including deficiency of STAT1, STAT2, JAK1, TYK2, IFNAR2, and IL10RB, have not been reported to have suffered from severe influenza (Boisson-Dupuis et al., 2012; Hambleton et al., 2013; Engelhardt and Grimbacher, 2014; Duncan et al., 2015; Kreins et al., 2015; Shahni et al., 2015; Eletto et al., 2016; Moens et al., 2017). This is perhaps because (1) they died of other severe infections before exposure to influenza virus, (2) the virus can be controlled independently of these signaling proteins, (3) at least some type I IFNs can signal without these proteins (de Weerd et al., 2013), or (4) the defect is partial, as opposed to complete, due to the hypomorphic nature of some mutant alleles (Chapgier et al., 2006; Hambleton et al., 2013; Duncan et al., 2015). Alternatively, IRF9 and IRF7, unlike the other five genes, might regulate transcription of a key subset of ISGs that specifically mediate immunity to influenza virus following type I and III IFN stimulation. A patient with IFNAR1 deficiency was recently reported, yet his additional defect of IFNGR2 prevents conclusions to be drawn as to the mechanism of infectious phenotypes (Hoyos-Bachiloglu et al., 2017). A clearer picture of the contributions of these genes to human protective immunity to influenza virus will emerge as more inborn errors are discovered and as more patients are diagnosed.
Significantly, our IRF9-deficient patient was not hospitalized in the ICU following infection with RSV or HRV, both of which were confirmed by serology and VirScan, indicating that IRF9 deficiency may not result in an extensive susceptibility to pulmonary viral infections. This contrasts with an MDA5-deficient child who was prone to severe, recurrent rhinovirus infections (Lamborn et al., 2017), and other MDA5-mutated children who were prone to RSV and other severe viral infections of the respiratory tract but not influenza (Asgari et al., 2017; Zaki et al., 2017). Human MDA5 and IRF9 thus seem to control different sets of respiratory viruses in vivo. Interestingly, IRF7 deficiency may also result in a narrow infectious phenotype, as the only reported case in humans was uniquely susceptible to influenza infection at last follow-up, aged 9 yr, and without any prophylaxis other than annual influenza vaccination (Ciancanelli et al., 2015; unpublished data). IRF9 deficiency results in a slightly broader infectious phenotype, including infection with influenza virus and likely an MMR vaccine-strain virus. The phenotype might have been broader if the patient had not been started on IgG i.v. at 2.5 yr of age. However, IRF9 deficiency is clinically closer to IRF7 deficiency when compared with GATA2 deficiency. Indeed, all known patients with the latter defect suffered from many other severe infections, including mycobacterial infections, papilloma virus infections, blastomycosis, and repeated viral infections of the respiratory tract (Collin et al., 2015). Severe influenza was the presenting feature in only one patient, who quickly thereafter displayed other phenotypes, infectious and otherwise (Sologuren et al., 2018). This broad infectious phenotype is consistent with the broad immunological phenotype of GATA2 deficiency, which is not restricted to a lack of development of type I and III IFN-producing pDCs. With the reservations listed above, it is striking that both IRF7 and IRF9 deficiency underlie such a narrow range of severe infections, almost limited to severe influenza pneumonitis. Significantly, only single cases of IRF7 and IRF9 deficiency have been reported thus far; further studies are needed to more comprehensively define the viral susceptibility resulting from these deficiencies, to determine to what extent IRF7- and IRF9-dependent type I and III IFN immunity is redundant for protective immunity against viral infections, and to suggest what other factors may compensate for deficiencies in this pathway.
Among patients with defects in the type I and/or III IFN signaling pathways, complications from MMR vaccine have been reported in patients with IFNAR2, STAT1, or STAT2 bi-allelic mutations (Hambleton et al., 2013; Duncan et al., 2015; Burns et al., 2016; Moens et al., 2017). Although relatively rare in STAT1-deficient individuals, a vaccine-strain MMR infection was reported in the only individual with AR complete STAT1 deficiency who was vaccinated (Burns et al., 2016). It is unclear whether patients with STAT2 and IFNAR2 deficiency have a complete or partial deficiency (Hambleton et al., 2013; Duncan et al., 2015; Moens et al., 2017). Interestingly, MMR vaccine complications were not observed in patients with AR partial STAT1 deficiency, nor have they been reported in patients with partial JAK1 or complete TYK2 deficiency who were vaccinated (Boisson-Dupuis et al., 2012; Kreins et al., 2015; Eletto et al., 2016). This suggests a possible divergence from mice, where STAT1 is dispensable but STAT2 is not, for protection against measles virus (Hahm et al., 2005; O’Donnell et al., 2012). The most parsimonious theory explaining these observations is that STAT2 and IRF9 mediate a physiologically relevant branch of the type I and III IFN signaling pathway in mice independently of STAT1, while some subset of this branch is dependent on STAT1 signaling in humans. STAT2-IRF9 complexes have been described in both mice and humans, but it is unclear if they could drive the cellular changes required for defense against viral infection (Lou et al., 2009; Abdul-Sater et al., 2015). Of note, in our mRNA-seq analysis, we observed up-regulation of STAT2 in P's primary fibroblasts and B-LCL in response to IFN-α2b stimulation in vitro, perhaps not coincidentally and consistent with nonredundant roles for STAT2 and IRF9 in immunity to measles infection (Hambleton et al., 2013). These results add to evidence in mice that the different components of the ISGF3 complex regulate distinct, nonredundant subsets of ISGs in addition to a jointly regulated core of genes (Li et al., 2017). Alternatively, the different MMR phenotypes in patients with various inborn errors of type I and III IFN immunity may merely reflect the impact of modifiers or of the environment itself, including the MMR inoculum. Ultimately, the experiment of nature deciphered in this report reveals the nonredundant roles of human IRF9 in the type I and/or III IFN signaling pathways for cell-intrinsic immunity against influenza virus and probably also the MMR vaccine.
Materials and methods
Patients
Whole blood, serum, and skin biopsies were obtained from the patient, her relatives, or paid healthy volunteers. These individuals gave written informed consent to participate in research protocols approved by the Institutional Review Boards at The Rockefeller University or the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Case report
We report the case of a girl born in 2013 to consanguineous parents (first cousins, f = 1/16) of Algerian descent, living in France, who was admitted to intensive care with acute respiratory distress secondary to infection with IAV at the age of 23 mo. According to her medical history, she was born after 39 wk of gestation with a birth weight of 2,910 g, height of 47 cm, and Apgar score 10/10. Her family history was negative for previous severe infectious diseases.
Since the age of 5 mo, the patient had a history of recurrent fevers (3–5 d/mo) associated with digestive features including diarrhea, abdominal pain without any vomiting, transient cutaneous rash, and intermittent urticaria, initially suggestive of an auto-inflammatory syndrome since no infectious agent was identified. Prior to any supportive treatment, IL-1β and TNFα were positive, respectively, at 17.9 pg/ml (0–15) and 25.1 pg/ml (0–20), whereas IL-6 and IL-10 were normal and negative. MEFV, MVK, CIAS1, and TRAPS were sequenced and found without any deleterious variation. Anti c1q antibodies were consistently positive, but sequencing of DNASE1L3 was normal.
She experienced five episodes of bronchiolitis revealing severe asthma (for which she was prescribed montelukast, albuterol, and ipratropium bromide), gingivostomatitis and adenitis, and four episodes of prolonged gastroenteritis. She was hospitalized in the pediatric ICU (PICU) due to idiopathic biliary perforation 14 d after MMR vaccination at 16 mo old; disseminated intravascular coagulation was suspected secondary to thrombopenia (27,000/mm3) and low prothrombin time (40%), associated with elevated liver enzymes (SGOT 274 UI/l and SGPT 128 UI/l). She was readmitted to the PICU 2 mo later with a suspicion of septic shock in the absence of a detectable pathogen, associated with elevated C-reactive protein (180 mg/l), lymphopenia (830/mm3), low platelet count (14,000/mm3), and low prothrombin time (66%).
At 23 mo of age, she was admitted to the PICU with a severe influenza A infection resulting in ARDS requiring mechanical ventilation for 6 d. Because increased oxygen was required with continuous positive airway pressure (FiO2 100%), she was intubated. She developed severe ARDS, with bilateral infiltrates on chest radiography. She was placed under mechanical ventilation in a volume support mode, with a frequency of 35 per minute and 100% FiO2. All bacteriological and viral sputum cultures were negative, except IAV. She was administered salbutamol, ipratropium bromide, ketamine, theophylline, magnesium sulfate, ceftriaxone, and oseltamivir. The oseltamivir treatment was stopped after 5 d. On the third day, bilateral parotidis and cervical adenitis appeared and were treated by amoxicillin and clavulanic acid. She was discharged from the PICU after 8 d, and her condition improved enough to be discharged after 22 d of hospitalization.
Standard immunological tests were performed. Serum levels of IgG, IgA, and IgM were unremarkable. T and natural killer lymphocyte count were conserved, whereas a modest B lymphocytosis was measured (Table S1). Lymphocyte proliferation was normal for PHA, tuberculin, and candidin (n > 50), but low in the presence of tetanus, CMV, and HSV antigens (n > 10; Table S2). T and B cell counts were within normal ranges. The patient was IgG seropositive for tetanus, diphtheria, haemophilus, measles, mumps, rubella, and HBV (Ag HBs negative, anti-HBs positive, anti-HBc negative) following routine vaccination. She remained seronegative for HIV1/2, HAV, HCV, VZV, Parvovirus B19, EBV, CMV, and pneumococcus despite proper vaccination for the latter.
Intravenous Igs at 0.4 g/kg/3 wk were begun at 2 yr 8 mo of age because of recurrent viral infections, resulting in an improvement of her recurrent fever profile, digestive symptoms, and susceptibility to viral infection.
Since i.v. Igs were started, the patient has been hospitalized on two occasions: once for acute respiratory distress secondary to metapneumovirus and once for febrile pseudo-occlusive syndrome with free fluid in Morrisson’s pouch detected by sonogram secondary to symptoms of gastroenteritis. At that time, RNA of parainfluenza 1,2,3,4 group was detected by PCR in the sputum, but no pathogen was detected known to cause gastroenteritis (also negative were rhinovirus/enterovirus, metapneumovirus, influenza A and B, coronavirus, and RSV A and B by PCR in the sputum and adenovirus, norovirus, enterovirus, and rotavirus by PCR in the stool). During neither episode was she admitted to the PICU.
WES
Exome capture was performed with the SureSelect Human All Exon 50 Mb kit (Agilent Technologies). Paired-end sequencing was performed on a HiSeq 2000 (Illumina), generating 100-base reads. We aligned the sequences with the GRCh39 reference build of the human genome using the BWA aligner (Li and Durbin, 2009). Downstream processing and variant calling were performed with the Genome Analysis Toolkit, SAMtools, and Picard (Li et al., 2009; McKenna et al., 2010). Substitution and InDel calls were made with the GATK Unified Genotyper. All variants were annotated using an annotation software system that was developed in-house (Ng and Henikoff, 2001; Adzhubei et al., 2010; Kircher et al., 2014).
Genetics
WES analysis of the patient revealed a total of 15,807 variants in P. We filtered out all variations found in a 1,000-genome database, the GnomAD, our own database of 4,892 exomes for infectious diseases other than influenza at a frequency of >1%, leaving 44 nonsynonymous coding variants in the patient’s exome: 20 homozygous and 24 heterozygous (Table S3). The heterozygous variants inherited from one parent were not considered in a model of complete penetrance. Excepting four heterozygous and two homozygous variants, none of the variants affected genes related to immunity. The mutated IRF9 allele was not found in 1,046 genomic DNA samples from 52 ethnic groups (including French) from the HGDP-CEPH Human Diversity Panel. The missense c.991G>A mutation had a CADD score of 32.21. A genomic measure of individual homozygosity was plotted for P, two European individuals from consanguineous families, and 37 individuals from nonconsanguineous families from our in-house WES database. Homozygosity was computed as the proportion of the autosomal genome belonging to runs of homozygosity (ROHs). The ROHs were defined as ranging at least 1 Mb of length and containing at least 100 single nucleotide polymorphisms (SNPs), and were estimated using the homozyg option of the PLINK software (Purcell et al., 2007). The centromeres were excluded because they are long genomic stretches devoid of SNPs and their inclusion might inflate estimates of homozygosity if both flanking SNPs are homozygous. The length of the autosomal genome was fixed at 2,673,768 kbs as previously described (McQuillan et al., 2008). We estimated the selective pressure acting on IRF9 to be 0.088 (indicative of purifying selection) by estimating the neutrality index (Stoletzki and Eyre-Walker, 2011) at the population level (PN/PS)/(DN/DS), where PN and PS are the number of nonsynonymous and synonymous alleles, respectively, at population level (1000 Genomes Project) and DN and DS are the number of nonsynonymous and synonymous fixed sites, respectively, for the coding sequence of IRF9.
Cells
PBMCs were isolated by Ficoll-Paque density gradient (Lymphoprep; Proteogenix) from the blood of the patient, the mother, and a healthy donor. SV40-immortalized dermal fibroblasts, U2A, Madin-Darby canine kidney, and Vero E6 cells were maintained in DMEM supplemented with 10% FBS. B-LCLs were grown in RPMI 1640 medium supplemented with 10% FBS. Stably transfected dermal fibroblasts were obtained by transfecting pTRIPIRF9iresRFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Stable transfectants were selected by puromycin treatment (0.3 μg/ml) and subsequent fluorescence-activated cell sorting for RFP-expressing cells yielding >80% RFP+ fibroblasts.
Plasmids
The cDNA of IRF9 was cloned into pGEMT cloning vector (Promega). Site-directed mutagenesis was performed to obtain the indicated mutant constructs. All IRF9 constructs were then subcloned into the pTRIP.CMV.IVSb.iresTagRFP_Dest vector using Gateway cloning technology (Life Technologies) and resequenced to ensure no advantitious mutations were generated during the cloning.
Western blotting
Fibroblasts were pretreated with IFN-α2b (Schering) or IFN-γ (Imukin; Boehringer Ingelheim) for the specified times before lysis. Cells were lysed in NP-40 lysis buffer (280 mM NaCl, 50 mM Tris, pH 8, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 0.5% NP-40) supplemented with 1 mM dithiothreitol, PhosSTOP (Roche), and Complete protease inhibitor cocktail (Roche). 20 μg protein lysate per lane was resolved by SDS-PAGE and transferred to polyvinylidene fluoride membrane, which was probed with unconjugated primary antibodies and HRP-conjugated secondary antibodies. Anti-GAPDH (Santa Cruz Biotechnology) or anti-Lamin A/C (Santa Cruz Biotechnology) antibodies were used as loading controls. IRF9 was probed with antibody recognizing the C-terminus at a dilution of 1:1,000 (ISGF-3γ p48 [H10]; Santa Cruz Biotechnology). SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific) was used to visualize HRP activity, and this signal was detected by an Amersham Imager 600 (GE Life Sciences).
Knockdown efficiency was determined by immunoblotting. 30 μg of total protein loaded per lane were separated on 4–12% NuPAGE Bis-Tris SDS-PAGE gels (Invitrogen) and then transferred onto nitrocellulose membranes using the Trans-Blot Turbo Transfer System (Bio-Rad Laboratories). After blocking with 5% nonfat dry milk (Bio-Rad Laboratories) in PBS containing 0.1% Tween 20 (Sigma-Aldrich), membranes were blotted with the IRF9 antibody (ISGF-3γ p48 [H10]), MAVS (clone 3993; Cell Signaling Technologies), or HSP90 (clone 68; BD Biosciences), followed by incubation with appropriate HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and SuperSignal West Pico Chemiluminescent substrate or SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific). Signal was detected by ChemiDocTM Touch Imaging System (Bio-Rad Laboratories).
EMSAs
EMSA was performed as previously described (Chapgier et al., 2006). In brief, cell activation was blocked by incubation with cold 1× PBS, and the cells were gently lysed to remove cytoplasmic proteins while keeping the nucleus intact. Nuclear lysis buffer was then added, and the recovered nuclear extracts were subjected to nondenaturing electrophoresis with Alexa 647–labeled GAS (from the FCγR1 promoter: 5′-ATGTATTTCCCAGAAA-3′) and ISRE (from the ISG15 promoter: 5′-GATCGGGAAAGGGAAACCGAAACTGAA-3′) probes. Gels were then imaged on an Amersham Imager 600 (GE Life Sciences) in the Cy5 channel.
Reporter assays
U2A cells were transfected with the indicated expression plasmids, firefly luciferase plasmids under the control of ISRE or GAS promoters, and a constitutively expressing Renilla firefly luciferase plasmid for normalization. 24 h after transfection, the cells were stimulated with IFN-α2b, -β, or -γ for 18 h or were left unstimulated. Luciferase levels were measured using Dual-Glo reagent according to the manufacturer’s protocol (Promega). Firefly luciferase values are normalized to Renilla luciferase values, and fold induction is expressed relative to unstimulated samples.
Exon trapping assays
U2A cells were transfected with pSPL3 plasmids containing a genomic region encompassing IRF9 exons 6 through 8 (WT or c.991G>A mutation). RNA was then purified from these cells, and the IRF9 splice products were amplified using flanking HIV-TAT sequences in the pSPL3 vector. RNA was then reverse-transcribed and ligated into the pGEM-T vector (Promega). DH5α cells (Thermo Fischer Scientific) were transformed with the resulting plasmids, and colony PCR followed by sequencing of IRF9 was performed to determine the splicing products produced by the mutated and WT alleles.
qRT-PCR
Total RNA from PBMCs was extracted using RNAqueous-Micro Kit (Ambio). Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Messenger RNAs were quantified with IRF9 probes Hs00960976-m1 (exon 1–2) and Hs00196051-m1 (exon 7–8; Thermo Fischer Scientific) by qRT-PCR using the Taqman Gene Expression Assay (Applied Biosystems) and normalized to the expression level of HPRT1. RNA was isolated from SV40-fibroblasts, and B-LCL cells were stimulated with 1,000 IU/ml IFN-α2b, IFN-β, or IFN-γ for 2 or 8 h with RNA lysis buffer, treated with DNase, and purified according to the manufacturer’s protocol (Zymo Research). RT-PCR was performed using random hexamers and the Superscript III reverse strand synthesis kit according to the manufacturer’s instructions (Thermo Fisher Scientific). Quantitative real-time PCR was performed with Applied Biosystems Taqman assays using the β-glucuronidase housekeeping gene for normalization. Results are expressed using the ΔΔCt method, as described by the manufacturer.
Virus assays
IAV and VSV infections
SV40 fibroblasts were infected with influenza virus (A/California/4/2009) at a multiplicity of infection (MOI) of 0.5 (MOI = 0.5). The inoculum was absorbed onto the cells for 30 min at 25°C. Cells were washed twice with Hank’s balanced salt solution and cultured at 37°C in the presence of 0.1 µg/ml TPCK-treated trypsin (Sigma-Aldrich). Virus samples were collected at the indicated times after infection. Influenza titers were determined by plaque assay on Madin-Darby canine kidney cells. Fibroblasts were infected with VSV at MOI = 3. The inoculum was absorbed onto the cells for 30 min at 25°C, washed twice with PBS, and cultured in DMEM 10% FBS at 37°C. Virus samples were collected at indicated time points. Viral titers were determined by endpoint dilution on Vero cells using the Reed and Muench calculation. Where indicated, cells were pretreated with IFN-α2b for 16 h before virus infection.
RSV, PIV, and HRV infections
HRV-A16 and recombinant RSV derived from the A2 strain and containing the enhanced GFP were used as previously described (Lamborn et al., 2017). rgPIV3, a recombinant human PIV type 3 derived from the JS strain and containing the enhanced GFP, was a gift from Dr. P. Collins, National Institute of Allergy and Infectious Diseases (Zhang et al., 2005). SV40-transformed fibroblasts were seeded at 100,000 cells per well in 12-well tissue culture plates in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin (all from Invitrogen), and 55 µM 2-ME (Sigma-Aldrich). After overnight culture, cells were infected with RSV-GFP (MOI = 0.5) or PIV3-GFP (MOI = 0.1). At 24 and 48 h after infection, cells were harvested, and flow cytometric detection of GFP in RSV-infected viable cells was performed as previously described (Lamborn et al., 2017). Flow cytometric detection of GFP in PIV3-infected viable cells was performed under similar conditions. For IRF9 siRNA knockdown experiments, human primary dermal fibroblasts were transfected with negative siRNA control, Stealth siRNA to IRF9 (HSS173591; Thermo Fisher Scientific), or siRNA to MAVS (HSS148537; Thermo Fisher Scientific), using the P3 Primary Cell 96-well Nucleofector kit (Lonza) as described (Lamborn et al., 2017). 3 d after transfection, cells were infected with RSV (MOI = 0.5), PIV (MOI = 0.1), or HRV16 (MOI = 10). RSV, PIV, and HRV replication was quantitated as described above.
VirScan analysis
Patient serum was analyzed by VirScan in two independent experiments as previously described (Xu et al., 2015). Briefly, an oligonucleotide library encoding 56 amino acid peptides tiling across the genomes of 206 viral species was synthesized on a releasable DNA microarray and cloned into T7 phage. Patient serum containing 2 μg of IgG was added to the phage library, and immunoprecipitation was performed with Protein A and G beads. Enriched peptides were identified by PCR and Illumina sequencing of the peptide cassette from the immunoprecipitated phage.
Statistical analyses
For viral replication, the Prism 7 software (GraphPad) was used to calculate P values using one-way ANOVA with Dunnett’s multiple comparisons, Kruskal-Wallis test with Dunn’s multiple comparisons.
mRNA-seq analysis
Total RNA from IFN-α2b–stimulated B-LCL and primary fibroblast cells was isolated (RNeasy kit; QIAGEN), and RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Poly (A)mRNA enrichment and cDNA library preparation was performed using a TruSeq Stranded mRNA Library Prep Kit (Illumina) in accordance with the manufacturer’s recommendations. Paired-end sequencing was performed on a HiSeq4000 (Illumina) with 150 cycles. Quality control of the raw Fastq files was performed using FastQC (Andrews, 2010). The samples were then aligned to the reference genome (Ensembl GRCh37) using a STAR aligner (Dobin et al., 2013). Aligned output was then converted to BAM files using SAMtools (Li et al., 2009). The resulting BAM files were used to quantify the read counts using the HTseq-count tool (Anders et al., 2015). The output of technical repeats (patient samples that had been stimulated in duplicate) was found to have a high correlation coefficient (not shown). Subsequent downstream analysis was performed in the R statistical programming language (Team, 2008). Read counts of technical repeats were averaged, and IFN-α2b–responsive transcripts were identified by comparing stimulated against unstimulated conditions in the three healthy control subjects, using a minimal 1.5-fold change in expression (up-regulated or down-regulated) as the cut-off. The response gene lists only those that had satisfied the filter criteria in all three controls. This resulted in a total of 1,064 and 1,576 genes that were responsive in the IFN-α2b–stimulated B-LCL and primary fibroblasts, respectively. The gene lists were then used to identify, again using a 1.5-fold change as cut-off, IFN-α2b–responsive and unresponsive genes in the cells of IRF9-, STAT1-, and STAT2-deficient patients. The overall residual responses in the patient’s cells were computed by counting the responsive genes in each patient and cell type and by calculating the percentage out of the total number of response genes in each cell type. The response in the healthy control subject was set at 100%. We used MS Excel to filter for response genes that are common or unique to specific patients for subsequent analysis.
For B-LCL network analysis, a greater than fivefold cut-off was applied to the control groups to generate the target list of IFN-α2b–induced genes. From the 749 up-regulated genes in the healthy controls, 84 were induced at > fivefold. This gene was then used to filter the IRF9-deficient patient, using the same >1.5-fold cut-off applied in RNA-seq analysis. In the IRF9-deficient patient, a total of 37 genes were found to be induced at >1.5-fold, and 47 were nonresponsive. These two lists were then entered into Interferome v2.01 and queried for ISGs, using similar experimental criteria as the current study (Rusinova et al., 2013). Of the 37 genes found to be responsive in the IRF9-deficient patient, 24 were identified as ISGs. Of the 47 nonresponsive genes, 39 were identified as ISGs. The combined ISG gene lists for the IRF9-deficient patient was used to conduct network analysis in GeneMANIA, using biological process based Gene Ontology weighting method and the maximum resultant genes set to 20 (Warde-Farley et al., 2010). The network was examined using both biological pathway (4.35%) and physical interactions (67.64%).
Data availability
mRNA-seq data have been deposited to GEO and are available under accession no. GSE117637.
Online supplemental material
Fig. S1 shows antibodies against viruses that were detected by VirScan analysis of the IRF9-deficient patient’s sera. Fig. S2 displays the PCR scheme used in exon trapping experiments. Table S1 presents the immunophenotyping results of the IRF9-deficient patient. Table S2 demonstrates the proliferation of the IRF9-deficient patient’s CD4+ T cells in response to various stimuli. Table S3 catalogs the rare, biallelic mutations present in the IRF9-deficient patient.
Supplementary Material
Acknowledgments
We thank our patient and her family. We thank S. Boucherit, T. Kochetkov, M. Fenner, M. Duchamp, A. Belkadi, L. Shang, Y. Liang, D. Papandrea, and Y. Nemirovskaya for their contributions and all members of the laboratory for fruitful discussions. We thank members of the French Memo-Flu-ARDS study group (principle investigators: B. Autran, Institut National de la Santé et de la Recherche Médicale U945, and C.-E. Luyt, Service de réanimation médicale, Hôpital Pitié-Salpêtrière, Paris) for recruiting adult severe flu patients. We thank Sandra Pellegrini for generously providing U2A fibrosarcoma cells for this study. We also thank Sara Tomei, Rebecca Mathew, and other members of Sidra Medicine's genomics core facility team for their contribution to the transcriptomics analysis, including the mRNA-seq library preparation, quality control, and Illumina sequencing.
Supported by National Center for Research Resources and National Center for Advancing Translational Sciences grant UL1TR001866; the National Institute of Allergy and Infectious Diseases for Cooperative Center on Human Immunology grant U19AI111825 at The Rockefeller University; National Institute of Allergy and Infectious Diseases grant R21AI137371; the St. Giles Foundation; the French National Research Agency under the “Investments for the Future” program grant ANR-10-IAHU-01, the Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases grant ANR-10-LABX-62-IBEID, Institut National de la Santé et de la Recherche Médicale, and Paris Descartes University. Supported in part by the Intramural Research Program at the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, and Qatar National Research Fund grant NPRP9-251-3-045. I. Melki was supported by a grant from the Fondation Bettencourt Schueller through the “Programme Santé-Sciences MD/PhD” of the Imagine Institute. N. Hernandez was supported by the Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. Y.J. Crow acknowledges European Research Council grant GA 309449. S.S.Y. Huang was supported by the Qatar National Research Fund’s Paths Toward Personalized Medicine Program grant PPM1-1220-150017.
The authors declare no competing financial interests.
Author contributions: I. Melki, A. Boulai, N. Kitabayashi, and Y.J. Crow sequenced patient cells and analyzed SNP microarray data, identifying the IRF9 mutation. N. Hernandez, M.J. Ciancanelli, and L. Abel analyzed WES data. V. Rattina assisted with bioinformatics analysis of homozygosity in patient genomic DNA. N. Hernandez, H. Jing, J. Danielson, I. Melki and A. Boulai assessed IRF9 expression and performed WB analysis. N. Hernandez and I. Melki performed qRT-PCR experiments. N. Hernandez performed luciferase reporter assays, and IAV and VSV infections. H. Jing and J. Danielson performed PIV, RSV, and HRV infections, as well as IRF9 silencing in dermal fibroblasts. S.S.Y. Huang, T. Habib, and N. Marr performed mRNA-seq analysis, while M.C. Leung and L.S. Mathew assisted with library preparation and Illumina sequencing. T. Kula and S. Elledge performed VirScan analysis of patient and control sera. Y. Rose and I. Melki assisted in the generation of F-SV40 and B-LCL cell lines. I. Melki, M.-N. Lebras, C. Dumaine, and S. Blanche cared for the patient, and collected and analyzed clinical data. J.L. Casanova, Y.J. Crow, I. Melki, and L. Lorenzo-Diaz coordinated clinical study protocol and sample collection. J.L. Casanova planned the experimental work and supervised the data analysis. H.C. Su, L.D. Notarangelo, S. Giliani, Y.J. Crow, D. Chaussabel, B. Boisson, S. Boisson-Dupuis, S.-Y. Zhang, M.P. Rodero, S. Drutman, and S. Belkaya provided advice and assisted in supervising the experimental work. N. Hernandez, M.J. Ciancanelli, Q. Zhang, and J.L. Casanova prepared the manuscript. All authors discussed and revised the manuscript.
References
- Abdul-Sater A.A., Majoros A., Plumlee C.R., Perry S., Gu A.D., Lee C., Shresta S., Decker T., and Schindler C.. 2015. Different STAT Transcription Complexes Drive Early and Delayed Responses to Type I IFNs. J. Immunol. 195:210–216. 10.4049/jimmunol.1401139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., and Sunyaev S.R.. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.2010. FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Arimoto K.I., Löchte S., Stoner S.A., Burkart C., Zhang Y., Miyauchi S., Wilmes S., Fan J.B., Heinisch J.J., Li Z., et al. 2017. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat. Struct. Mol. Biol. 24:279–289. 10.1038/nsmb.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S., Schlapbach L.J., Anchisi S., Hammer C., Bartha I., Junier T., Mottet-Osman G., Posfay-Barbe K.M., Longchamp D., Stocker M., et al. 2017. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc. Natl. Acad. Sci. USA. 114:8342–8347. 10.1073/pnas.1704259114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Paturel C., Brizard G., Chemin K., Boonstra A., O’Garra A., Vicari A., and Trinchieri G.. 2005. Type I interferon dependence of plasmacytoid dendritic cell activation and migration. J. Exp. Med. 201:1157–1167. 10.1084/jem.20041930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begue B., Verdier J., Rieux-Laucat F., Goulet O., Morali A., Canioni D., Hugot J.P., Daussy C., Verkarre V., Pigneur B., et al. 2011. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am. J. Gastroenterol. 106:1544–1555. 10.1038/ajg.2011.112 [DOI] [PubMed] [Google Scholar]
- Belkadi A., Pedergnana V., Cobat A., Itan Y., Vincent Q.B., Abhyankar A., Shang L., El Baghdadi J., Bousfiha A., Alcais A., et al. Exome/Array Consortium . 2016. Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc. Natl. Acad. Sci. USA. 113:6713–6718. 10.1073/pnas.1606460113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B., Laplantine E., Prando C., Giliani S., Israelsson E., Xu Z., Abhyankar A., Israël L., Trevejo-Nunez G., Bogunovic D., et al. 2012. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13:1178–1186. 10.1038/ni.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dupuis S., Kong X.F., Okada S., Cypowyj S., Puel A., Abel L., and Casanova J.L.. 2012. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr. Opin. Immunol. 24:364–378. 10.1016/j.coi.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousfiha A., Jeddane L., Picard C., Ailal F., Bobby Gaspar H., Al-Herz W., Chatila T., Crow Y.J., Cunningham-Rundles C., Etzioni A., et al. 2018. The 2017 IUIS Phenotypic Classification for Primary Immunodeficiencies. J. Clin. Immunol. 38:129–143. 10.1007/s10875-017-0465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C., Cheung A., Stark Z., Choo S., Downie L., White S., Conyers R., and Cole T.. 2016. A novel presentation of homozygous loss-of-function STAT-1 mutation in an infant with hyperinflammation-A case report and review of the literature. J. Allergy Clin. Immunol. Pract. 4:777–779. 10.1016/j.jaip.2016.02.015 [DOI] [PubMed] [Google Scholar]
- Casanova J.-L. 2015a Human genetic basis of interindividual variability in the course of infection. Proc. Natl. Acad. Sci. USA. 112:E7118–E7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.-L. 2015b Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc. Natl. Acad. Sci. USA. 112:E7128–E7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Conley M.E., Seligman S.J., Abel L., and Notarangelo L.D.. 2014. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J. Exp. Med. 211:2137–2149. 10.1084/jem.20140520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 2013. FluView: Influenza Associated Hospitalization Surveillance Network. Available at: http://gis.cdc.gov/grasp/fluview/FluHospChars.html.
- Chapgier A., Boisson-Dupuis S., Jouanguy E., Vogt G., Feinberg J., Prochnicka-Chalufour A., Casrouge A., Yang K., Soudais C., Fieschi C., et al. 2006. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2:e131 10.1371/journal.pgen.0020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M.J., Huang S.X.L., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A., et al. 2015. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 348:448–453. 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M.J., Abel L., Zhang S.-Y., and Casanova J.-L.. 2016. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr. Opin. Immunol. 38:109–120. 10.1016/j.coi.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia E.M., Severa M., Giacomini E., Monneron D., Remoli M.E., Julkunen I., Cella M., Lande R., and Uzé G.. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34:796–805. 10.1002/eji.200324610 [DOI] [PubMed] [Google Scholar]
- Collin M., Dickinson R., and Bigley V.. 2015. Haematopoietic and immune defects associated with GATA2 mutation. Br. J. Haematol. 169:173–187. 10.1111/bjh.13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E. Jr., Kerr I.M., and Stark G.R.. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 264:1415–1421. 10.1126/science.8197455 [DOI] [PubMed] [Google Scholar]
- Dawood F.S., Kamimoto L., D’Mello T.A., Reingold A., Gershman K., Meek J., Arnold K.E., Farley M., Ryan P., Lynfield R., et al. Emerging Infections Program Network . 2011. Children with asthma hospitalized with seasonal or pandemic influenza, 2003-2009. Pediatrics. 128:e27–e32. 10.1542/peds.2010-3343 [DOI] [PubMed] [Google Scholar]
- de Weerd N.A., Vivian J.P., Nguyen T.K., Mangan N.E., Gould J.A., Braniff S.J., Zaker-Tabrizi L., Fung K.Y., Forster S.C., Beddoe T., et al. 2013. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 14:901–907. 10.1038/ni.2667 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C.J., Mohamad S.M., Young D.F., Skelton A.J., Leahy T.R., Munday D.C., Butler K.M., Morfopoulou S., Brown J.R., Hubank M., et al. 2015. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med. 7:307ra154 10.1126/scitranslmed.aac4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D., Burns S.O., Angulo I., Plagnol V., Gilmour K.C., Henriquez F., Curtis J., Gaspar M., Nowak K., Daza-Cajigal V., et al. 2016. Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat. Commun. 7:13992 10.1038/ncomms13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt K.R., and Grimbacher B.. 2014. IL-10 in humans: lessons from the gut, IL-10/IL-10 receptor deficiencies, and IL-10 polymorphisms. Curr. Top. Microbiol. Immunol. 380:1–18. [DOI] [PubMed] [Google Scholar]
- Fu X.Y., Kessler D.S., Veals S.A., Levy D.E., and Darnell J.E. Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA. 87:8555–8559. 10.1073/pnas.87.21.8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker E.O., Kotlarz D., Boztug K., Gertz E.M., Schäffer A.A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., et al. 2009. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361:2033–2045. 10.1056/NEJMoa0907206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm B., Trifilo M.J., Zuniga E.I., and Oldstone M.B.. 2005. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 22:247–257. 10.1016/j.immuni.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Hambleton S., Goodbourn S., Young D.F., Dickinson P., Mohamad S.M.B., Valappil M., McGovern N., Cant A.J., Hackett S.J., Ghazal P., et al. 2013. STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. USA. 110:3053–3058. 10.1073/pnas.1220098110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T.. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Hoyos-Bachiloglu R., Chou J., Sodroski C.N., Beano A., Bainter W., Angelova M., Al Idrissi E., Habazi M.K., Alghamdi H.A., Almanjomi F., et al. 2017. A digenic human immunodeficiency characterized by IFNAR1 and IFNGR2 mutations. J. Clin. Invest. 127:4415–4420. 10.1172/JCI93486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Zhang S.Y., Vogt G., Abhyankar A., Herman M., Nitschke P., Fried D., Quintana-Murci L., Abel L., and Casanova J.L.. 2013. The human gene connectome as a map of short cuts for morbid allele discovery. Proc. Natl. Acad. Sci. USA. 110:5558–5563. 10.1073/pnas.1218167110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Vélez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G., et al. 2015. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA. 112:13615–13620. 10.1073/pnas.1518646112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Shang L., Boisson B., Ciancanelli M.J., Markle J.G., Martinez-Barricarte R., Scott E., Shah I., Stenson P.D., Gleeson J., et al. 2016. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods. 13:109–110. 10.1038/nmeth.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., and Pillai P.S.. 2014. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14:315–328. 10.1038/nri3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska J., Gravel A., and Flamand L.. 2010. Divergent susceptibilities of human herpesvirus 6 variants to type I interferons. Proc. Natl. Acad. Sci. USA. 107:8369–8374. 10.1073/pnas.0909951107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J., McKendry R., Pellegrini S., Flavell D., Kerr I.M., and Stark G.R.. 1991. Isolation and characterization of a new mutant human cell line unresponsive to alpha and beta interferons. Mol. Cell. Biol. 11:4189–4195. 10.1128/MCB.11.8.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.D., Hsu A.P., Ryu M.J., Wang J., Gao X., Boyer M.E., Liu Y., Lee Y., Calvo K.R., Keles S., et al. 2012. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J. Clin. Invest. 122:3692–3704. 10.1172/JCI61623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M.M., Ohnemus A., Cornitescu M., and Staeheli P.. 2012. Plasmacytoid dendritic cells and Toll-like receptor 7-dependent signalling promote efficient protection of mice against highly virulent influenza A virus. J. Gen. Virol. 93:555–559. 10.1099/vir.0.039065-0 [DOI] [PubMed] [Google Scholar]
- Kimura T., Kadokawa Y., Harada H., Matsumoto M., Sato M., Kashiwazaki Y., Tarutani M., Tan R.S., Takasugi T., Matsuyama T., et al. 1996. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1:115–124. 10.1046/j.1365-2443.1996.08008.x [DOI] [PubMed] [Google Scholar]
- Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner I., Kochs G., Kalinke U., Weiss S., and Staeheli P.. 2007. Protective role of beta interferon in host defense against influenza A virus. J. Virol. 81:2025–2030. 10.1128/JVI.01718-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreins A.Y., Ciancanelli M.J., Okada S., Kong X.F., Ramírez-Alejo N., Kilic S.S., El Baghdadi J., Nonoyama S., Mahdaviani S.A., Ailal F., et al. 2015. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212:1641–1662. 10.1084/jem.20140280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamborn I.T., Jing H., Zhang Y., Drutman S.B., Abbott J.K., Munir S., Bade S., Murdock H.M., Santos C.P., Brock L.G., et al. 2017. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 214:1949–1972. 10.1084/jem.20161759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach H., Bathke B., Gilles S., Traidl-Hoffmann C., Luber C.A., Fejer G., Freudenberg M.A., Davey G.M., Vremec D., Kallies A., et al. 2010. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J. Exp. Med. 207:2703–2717. 10.1084/jem.20092720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hofer M.J., Songkhunawej P., Jung S.R., Hancock D., Denyer G., and Campbell I.L.. 2017. Type I interferon-regulated gene expression and signaling in murine mixed glial cells lacking signal transducers and activators of transcription 1 or 2 or interferon regulatory factor 9. J. Biol. Chem. 292:5845–5859. 10.1074/jbc.M116.756510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y.J., Pan X.R., Jia P.M., Li D., Xiao S., Zhang Z.L., Chen S.J., Chen Z., and Tong J.H.. 2009. IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 69:3673–3680. 10.1158/0008-5472.CAN-08-4922 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R., Leutenegger A.L., Abdel-Rahman R., Franklin C.S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A., et al. 2008. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83:359–372. 10.1016/j.ajhg.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens L., Van Eyck L., Jochmans D., Mitera T., Frans G., Bossuyt X., Matthys P., Neyts J., Ciancanelli M., Zhang S.Y., et al. 2017. A novel kindred with inherited STAT2 deficiency and severe viral illness. J. Allergy Clin. Immunol. 139:1995–1997.e9. 10.1016/j.jaci.2016.10.033 [DOI] [PubMed] [Google Scholar]
- Mordstein M., Kochs G., Dumoutier L., Renauld J.C., Paludan S.R., Klucher K., and Staeheli P.. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4:e1000151 10.1371/journal.ppat.1000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S., Yoshida H., Moodley D., LeBoité H., Rothamel K., Raj T., Ye C.J., Chevrier N., Zhang S.Y., Feng T., et al. Immunological Genome Project Consortium . 2016. Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell. 164:564–578. 10.1016/j.cell.2015.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., and Henikoff S.. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11:863–874. 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S., Pagano J.S., and Barber G.N.. 2011. IRF7: activation, regulation, modification and function. Genes Immun. 12:399–414. 10.1038/gene.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L.A., Conway S., Rose R.W., Nicolas E., Slifker M., Balachandran S., and Rall G.F.. 2012. STAT1-independent control of a neurotropic measles virus challenge in primary neurons and infected mice. J. Immunol. 188:1915–1923. 10.4049/jimmunol.1101356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera K., Fujiwara T., Onishi Y., Itoh-Nakadai A., Okitsu Y., Fukuhara N., Ishizawa K., Shimizu R., Yamamoto M., and Harigae H.. 2016. GATA2 regulates dendritic cell differentiation. Blood. 128:508–518. 10.1182/blood-2016-02-698118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund P.I., Pietilä T.E., Veckman V., Kotenko S.V., and Julkunen I.. 2007. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol. 179:3434–3442. 10.4049/jimmunol.179.6.3434 [DOI] [PubMed] [Google Scholar]
- Palese P., and Shaw M.L.. 2007. Fields Virology. Lippincott Williams & Wilkins, Philadephia. [Google Scholar]
- Pasquet M., Bellanné-Chantelot C., Tavitian S., Prade N., Beaupain B., Larochelle O., Petit A., Rohrlich P., Ferrand C., Van Den Neste E., et al. 2013. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 121:822–829. 10.1182/blood-2012-08-447367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C., Bobby Gaspar H., Al-Herz W., Bousfiha A., Casanova J.L., Chatila T., Crow Y.J., Cunningham-Rundles C., Etzioni A., Franco J.L., et al. 2018. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 38:96–128. 10.1007/s10875-017-0464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., and Sham P.C.. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengachari S., Groiss S., Devos J.M., Caron E., Grandvaux N., and Panne D.. 2018. Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. Proc. Natl. Acad. Sci. USA. 115:E601–E609. 10.1073/pnas.1718426115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., and Hertzog P.J.. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41(Database issue, D1):D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Hata N., Asagiri M., Nakaya T., Taniguchi T., and Tanaka N.. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106–110. 10.1016/S0014-5793(98)01514-2 [DOI] [PubMed] [Google Scholar]
- Shahni R., Cale C.M., Anderson G., Osellame L.D., Hambleton S., Jacques T.S., Wedatilake Y., Taanman J.W., Chan E., Qasim W., et al. 2015. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain. 138:2834–2846. 10.1093/brain/awv182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh W.J., Blau D.M., Denison A.M., Deleon-Carnes M., Adem P., Bhatnagar J., Sumner J., Liu L., Patel M., Batten B., et al. 2010. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 177:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K.R., Kroeze E.J.B.V., Fouchier R.A.M., and Kuiken T.. 2014. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect. Dis. 14:57–69. 10.1016/S1473-3099(13)70286-X [DOI] [PubMed] [Google Scholar]
- Sole-Violan J., Sologuren I., Betancor E., Zhang S., Pérez C., Herrera-Ramos E., Martínez-Saavedra M., López-Rodriguez M., Pestano J., Ruiz-Hernández J., et al. 2013. Lethal influenza virus A H1N1 infection in two relatives with autosomal dominant GATA-2 deficiency. Crit. Care. 17(Suppl 2):15 10.1186/cc11953 [DOI] [Google Scholar]
- Sologuren I., Martinez-Saavedra M.T., Sole-Violan J., de Borges de Oliveira E. Jr., Betancor E., Casas I., Oleaga-Quintas C., Martinez-Gallo M., Zhang S.Y., Pestano J., et al. 2018. Lethal Influenza in Two Related Adults with Inherited GATA2 Deficiency. J. Clin. Immunol. [DOI] [PMC free article] [PubMed]
- Staeheli P., Grob R., Meier E., Sutcliffe J.G., and Haller O.. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8:4518–4523. 10.1128/MCB.8.10.4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., and Darnell J.E. Jr. 2012. The JAK-STAT pathway at twenty. Immunity. 36:503–514. 10.1016/j.immuni.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N., and Eyre-Walker A.. 2011. Estimation of the neutrality index. Mol. Biol. Evol. 28:63–70. 10.1093/molbev/msq249 [DOI] [PubMed] [Google Scholar]
- Tang X., Gao J.S., Guan Y.J., McLane K.E., Yuan Z.L., Ramratnam B., and Chin Y.E.. 2007. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 131:93–105. 10.1016/j.cell.2007.07.034 [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara K., Takaoka A., and Tanaka N.. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623–655. 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- Taubenberger J.K., and Morens D.M.. 2006. 1918 Influenza: the mother of all pandemics. Emerg. Infect. Dis. 12:15–22. 10.3201/eid1209.05-0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J.K., and Morens D.M.. 2008. The pathology of influenza virus infections. Annu. Rev. Pathol. 3:499–522. 10.1146/annurev.pathmechdis.3.121806.154316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.D.C. 2008. R: A language and environment for statistical computing. R Foundation for Statistical Comuting, Vienna, Austria. [Google Scholar]
- Veals S.A., Schindler C., Leonard D., Fu X.Y., Aebersold R., Darnell J.E. Jr., and Levy D.E.. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12:3315–3324. 10.1128/MCB.12.8.3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veals S.A., Santa Maria T., and Levy D.E.. 1993. Two domains of ISGF3 gamma that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol. Cell. Biol. 13:196–206. 10.1128/MCB.13.1.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(Web Server issue, suppl_2):W214-20 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk E., Pandey A.K., Leist S.R., Hatesuer B., Preusse M., Pommerenke C., Wang J., and Schughart K.. 2015. RNAseq expression analysis of resistant and susceptible mice after influenza A virus infection identifies novel genes associated with virus replication and important for host resistance to infection. BMC Genomics. 16:655 10.1186/s12864-015-1867-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza, Bautista E., Chotpitayasunondh T., Gao Z., Harper S.A., Shaw M., Uyeki T.M., Zaki S.R., Hayden F.G., Hui D.S., Kettner J.D., et al. 2010. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 362:1708–1719. [DOI] [PubMed] [Google Scholar]
- Xu G.J., Kula T., Xu Q., Li M.Z., Vernon S.D., Ndung’u T., Ruxrungtham K., Sanchez J., Brander C., Chung R.T., et al. 2015. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 348:aaa0698 10.1126/science.aaa0698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M., Thoenes M., Kawalia A., Nürnberg P., Kaiser R., Heller R., and Bolz H.J.. 2017. Recurrent and Prolonged Infections in a Child with a Homozygous IFIH1 Nonsense Mutation. Front. Genet. 8:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Cai Z., Kim Y.C., Kumar R., Yuan F., Shi P.Y., Kao C., and Luo G.. 2005. Stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3) helicase activity by the NS3 protease domain and by HCV RNA-dependent RNA polymerase. J. Virol. 79:8687–8697. 10.1128/JVI.79.14.8687-8697.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., et al. 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 317:1522–1527. 10.1126/science.1139522 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
mRNA-seq data have been deposited to GEO and are available under accession no. GSE117637.