Loyher et al. demonstrate that lung tumors are densely colonized by macrophages of various ontogenies. These distinct developmental origins dictate tumor-associated macrophages relative anatomical distributions, functions and responses to anti-cancer therapies.
Abstract
Tissue-resident macrophages can self-maintain without contribution of adult hematopoiesis. Herein we show that tissue-resident interstitial macrophages (Res-TAMs) in mouse lungs contribute to the pool of tumor-associated macrophages (TAMs) together with CCR2-dependent recruited macrophages (MoD-TAMs). Res-TAMs largely correlated with tumor cell growth in vivo, while MoD-TAMs accumulation was associated with enhanced tumor spreading. Both cell subsets were depleted after chemotherapy, but MoD-TAMs rapidly recovered and performed phagocytosis-mediated tumor clearance. Interestingly, anti-VEGF treatment combined with chemotherapy inhibited both Res and Mod-TAM reconstitution without affecting monocyte infiltration and improved its efficacy. Our results reveal that the developmental origin of TAMs dictates their relative distribution, function, and response to cancer therapies in lung tumors.
Graphical Abstract
Introduction
The tumor microenvironment (TME) can regulate malignant potential and contributes to tumor heterogeneity. Tumor-associated macrophages (TAMs) are the most abundant host cells within the TME (Qian and Pollard, 2010) and have been implicated in the promotion of invasiveness (Wyckoff et al., 2007), growth (Pollard, 2004), angiogenesis (Lewis et al., 2016), metastasis (Kitamura et al., 2015), and immunosuppression (Boissonnas et al., 2013; Broz et al., 2014). TAMs have been suggested to limit the efficacy of chemotherapeutic agents and to promote tumor relapse (Hughes et al., 2015), although they can in some cases be required for optimal therapy response (De Palma and Lewis, 2013).
It is considered that TAMs mainly arise from the differentiation of monocytic precursors (Cortez-Retamozo et al., 2012; Franklin et al., 2014). However, in many tissues, pools of resident macrophages have been identified; these originate from embryonic precursors and self-maintain independently of hematopoietic stem cells (Gomez Perdiguero et al., 2015). Distinct transcriptional programs initiated in embryonic, fetal, or adult progenitors (Mass et al., 2016) and the exposure to specific tissue environments (Gosselin et al., 2014; Lavin et al., 2014) may explain the specialization and diversity of macrophages in healthy as well as neoplastic tissues. The lung environment is densely colonized by subsets of mononuclear phagocytic cells displaying various spatial organizations, functions, and dependence for blood monocytes in their maintenance. Interstitial macrophages (IMs) represent a discrete population in the steady-state lung largely outnumbered by alveolar macrophages (AMs; Rodero et al., 2015; Gibbings et al., 2017). IMs and AMs express different surface markers, which allow their identification, and they have been described to arise from distinct developmental waves without interconverting (Guilliams et al., 2013; Tan and Krasnow, 2016).
So far, the contribution of these different resident macrophage subsets in the generation of lung TAMs has not been reported.
Herein, the TAM network in lung tumors is studied based on transgenic fluorescent reporter mice and fate-mapping models that enable the discrimination of the lung mononuclear phagocyte subsets according to their origin and localization. We showed that the TAM compartment is intermingled by both yolk sac–derived interstitial and monocyte-derived recruited macrophages, differentially represented in the TME depending on the anatomical site of tumor development in the lung. Finally, we highlight their respective implication on lung tumor development and response to various anti-cancer therapies.
Results
Lung macrophage subsets differentially accumulate during tumor development
We studied the impact of tumor growth on the different subset of lung myeloid cells after inoculating TC-1 lung carcinoma cells, which induce multifocal tumor nodules (Lin et al., 1996; Ji et al., 1998). The tumor-associated myeloid signature was monitored along tumor evolution using flow cytometry phenotyping combined with an unsupervised visual implementation of t-distributed stochastic neighbor embedding (tSNE [viSNE]) analysis. The generated tSNE plot was calculated with 12 parameters, including cell anatomical distribution between the tissue parenchyma and the vasculature. This distinction is achievable using anti-CD45 antibody injected intravenously that allows a blood/tissue partitioning of cells (see dashed gates, Fig. 1 A and Fig. S1). 10 clusters obtained using unsupervised analysis were subsequently assigned to a specific cell population according to expression level of each marker and previously described phenotypes (Fig. S1 A; Misharin et al., 2013; Gibbings et al., 2017; Sabatel et al., 2017). In brief, cluster 7 and cluster 8 were identified as classical Ly6Chigh and nonclassical Ly6Clow/- monocytes (Mo), respectively, with CD11bhighSiglec-F−Ly6G−Fc-gamma receptor 1low (CD64low) expression profile. Cluster 2 included CD11blowCD11chighSiglec-Fhigh cells, representing AMs, while cluster 1 included CD11bhighSiglec-F−Ly6G−CD64+ cells, representing a distinct subset of lung macrophages named here Ly6Clow/-CD64+ Mac. These different macrophage subsets were clearly distinguished from cluster 3, identified as CD11b−I-A[b]+CD11c+CD103+Ly6C−CD64− cells and cluster 5, identified as CD11b+I-A[b]+CD11c+Ly6C−CD64− cells, known as conventional dendritic cells cDC1 and cDC2, respectively. Clusters 6 and 9 are defined as CD11bhighLy6G+ cells (representing two subsets of neutrophils with differential expression of CD24). Cluster 10 represented CD11bhighSiglec-Fint cells identified as eosinophils (Fig. 1 A and Fig. S1, A and B). In the absence of tumor, Ly6Chigh-Mo (cluster 7) and Ly6Clow/--Mo (cluster 8) resided almost exclusively within lung vessels, whereas AMs (cluster 2) and Ly6Clow/-CD64+ Mac (cluster 1) were detected in the lung parenchyma only (Fig. 1 A and Fig. S1 C). With tumor expansion, cluster 4 including Ly6Chigh CD64+ cells appeared together with a progressive and massive accumulation of Ly6Clow/-CD64+ Mac (cluster 1; Fig. 1 A). Supervised analysis of Ly6C, CD64 expression, and intravascular cell labeling on CD11b+Siglec-F−Ly6G− gated cells suggested that Ly6Chigh-Mo progressively up-regulate CD64 and differentiate into Ly6Clow/-CD64+ Mac upon tumor infiltration (Fig. S1, C and D). In contrast, Ly6Clow/--Mo did not apparently accumulate, and they remained mostly intravascular (Fig. 1 and Fig. S1). Blood/tissue partitioning of monocyte and macrophage subsets (Fig. 1, C and D) was next performed using supervised analysis, and the observations made on tSNE plots were confirmed. While the Ly6Clow/-CD64+ Mac subset massively accumulated in the tumor parenchyma exclusively, the number of AMs per milligram of tissue strongly diminished with tumor growth, leading to 50% reduction in their number per whole lungs after 20 d (Fig. 1 B). Overall, these observations suggest that monocyte-derived macrophages (MoD-Mac) and lung-resident macrophages might differentially contribute to the TME.
Figure 1.
Lung macrophage subsets differentially accumulate during tumor development. (A) Representative tSNE dimension 1 and 2 plots of the lung myeloid compartment evolution after TC-1 cell intravenous inoculation. Upper panel delineates cell blood/tissue partitioning (dashed gates). Color clusters are represented over time (lower panels). (B) Blood/tissue partitioning monitoring of lung monocytes and macrophages during tumor growth. Dots represent mean of the absolute number ± SEM/milligram of tissue (upper panels) or absolute number per whole lung (lower panels). Statistical differences are indicated compared with D0. For all panels, n = 6–8 mice per time point out of three independent experiments, two-way ANOVA with Bonferroni multiple comparisons test was performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). See also Figs. S1.
Macrophages have distinct origins within lung tumors
We previously demonstrated that the MacBlue × Cx3cr1EGFP/+ mouse can be used to discriminate monocyte and macrophage subsets in lungs according to their relative expression of the enhanced CFP (ECFP) and enhanced GFP (EGFP) fluorescent reporters (Rodero et al., 2015). Histological analysis of TC-1tdTomato tumor-bearing mice along tumor expansion unveiled that tumor nodules were infiltrated by distinct cell subsets expressing EGFP and ECFP (Fig. 2 A and Fig. S2 A). The fluorescent signatures of monocytes and macrophages in tumor-free and tumor-bearing MacBlue × Cx3cr1EGFP/+ mice were compared using tSNE algorithm, and clusters were assigned as previously described (Fig. S2 B). In brief, classical Ly6Chigh (cluster 4) and nonclassical Ly6Clow/- (cluster 6) monocyte subsets both expressed high level of ECFP and, respectively, low and high levels of EGFP according to their relative expression of the Cx3cr1 gene reporter. AMs (cluster 1) expressed high level of ECFP, but no EGFP (Fig. S2 C). Interestingly, the Ly6Clow/-CD64+ Mac subset was distributed between clusters 2 and 3, suggesting different cell origin in this subset. In tumor-free animals, cluster 3 was dominant and was mainly composed of EGFPhigh ECFPneg (named EGFP+ macrophages, representing 87 ± 4.2% of the total Ly6Clow/-CD64+ Mac; Fig. S2 D). We previously observed that this subset typically represents IMs located in the pleura, along blood vessels and nearby large airways of the lungs (Rodero et al., 2015). Following tumor inoculation, cluster 2, including Ly6Clow/-CD64+ Mac subset, expressing high level of ECFP and EGFP (named ECFP+ macrophages), accumulated along with cluster 3, but became dominant as soon as day 10 (70.4 ± 9.8% of the Ly6Clow/-CD64+ Mac; Fig. S2).
Figure 2.
Macrophages have distinct origins within lung tumors. (A) Lung cryo-sections of tumor free and TC-1tdTomato tumor bearing MacBlue × Cx3cr1EGFP/+ mice show the distribution of ECFP+ and EGFP+ cells within tumor nodules over time. (B) Dot plots show the relative proportion of macrophage subsets in tumor-free, tumor-bearing MacBlue × Cx3cr1EGFP/+ (WT) and MacBlue × Cx3cr1EGFP/+ × Ccr2−/− (Ccr2−/−) mice at indicated time points, numbers indicate the mean percentage ± SD of ECFP+EGFP+ TAMs (left panel). Box and whisker graph shows the absolute number per milligram of tissue of indicated myeloid subsets in WT and Ccr2−/− mice (right panel; n = 10 mice out of three independent experiments, two-way ANOVA with Bonferroni multiple comparisons test was performed). (C) Lung cryo-sections show the distribution of ECFP+ and EGFP+ cells in TC-1 tumor-bearing MacBlue × Cx3cr1EGFP/+ (WT), MacBlue × Cx3cr1EGFP/+ × Ccr2−/− (Ccr2−/−) mice and C57Bl6 host parabiont with MacBlue × Cx3cr1EGFP/+ mice (left panels). Ratio of ECFP+/EGFP+ cell numbers in lung tumors. Each dot represents mean of ECFP+/EGFP+ cell ratio from different tumor nodules per mouse, red bars indicate mean, mice are pooled from at least two independent experiments (right panel). (D) Lung cryo-sections show the distribution of ECFP+ and EGFP+ cells in LLC tumor bearing MacBlue × Cx3cr1EGFP/+ mouse (day 15; left panel), and ratio of ECFP+/EGFP+ cells in LLC tumors (middle panel). Each dot represents mean of ECFP+/EGFP+ cell ratio from different tumor nodules per mouse, red bar indicates mean, mice are pooled from two independent experiments. Dot plot show the relative proportion of macrophages subsets in LLC tumor bearing MacBlue × Cx3cr1EGFP/+ mice at the indicated time point; numbers indicated the mean percentage ± SD of ECFP+EGFP+ TAMs (right panel; n = 4 mice out of two independent experiments). (E) Lung cryo-section shows the distribution of ECFP+ and EGFP+ cells in spontaneous pulmonary metastases from MacBlue × Cx3cr1EGFP/+ × PyMT-ChOVA mouse (left panel). Middle panel shows the lack of EGFP+ cells in pulmonary metastases from C57Bl6 PyMT-ChOVA host parabiont. Right panel shows the quantification based on histological analyses. Each dot represents mean of ECFP+/EGFP+ cell ratio from different tumor nodules per mouse, red bar indicates mean, all mice are independents. ANOVA with Bonferroni multiple comparisons test were performed for panel B and C and unpaired Student's t test was performed for panel E. For all panels: *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. See also Fig. S2.
The reduction of AMs was confirmed in the second tSNE signature (cluster 1; Fig. S2 B). Co-labeling of MacBlue × Cx3cr1EGFP/+ mice with Siglec-F on histological sections showed that ECFP+Siglec-F+ AMs remained exclusively localized in the healthy alveolar space, outside tumor nodules (Fig. S2 E), suggesting that AMs are progressively eliminated during tumor expansion or that they completely change their phenotype. We thus hypothesized that tumor development leads to the accumulation of lung TAMs from distinct origins. To address this, the distribution of EGFP+ or ECFP+ cells was analyzed in tumor-bearing MacBlue × Cx3cr1EGFP/+ × Ccr2−/− mice. ECFP+ macrophages were substantially reduced in Ccr2−/− mice, whereas EGFP+ macrophages and AMs were unaffected (Fig. 2 B). This suggests a monocytic origin of ECFP+ macrophages, while EGFP+ macrophage accumulation is CCR2 independent. Macrophage distribution was next compared on histological lung sections of tumor-bearing MacBlue × Cx3cr1EGFP/+ (WT), MacBlue × Cx3cr1EGFP/+ × Ccr2−/− (Ccr2−/−), and C57Bl6 host parabiont with the MacBlue × Cx3cr1EGFP/+ donor mouse. In pulmonary nodules of WT mice, the ratio of ECFP+/EGFP+ cells was 0.57 ± 0.10, whereas the corresponding ratio was 0.14 ± 0.08 in Ccr2−/− mice and 0.96 ± 0.07 in host parabiont mice (Fig. 2 C). These results support that TAMs in lung tumors are composed of both ECFP+ monocyte-derived macrophages (ECFP+-TAMs) and a CCR2-independent local accumulation of EGFP+ resident IMs (EGFP+-TAMs).
These two fluorescent subsets were also present within Lewis lung carcinoma (LLC) nodules with similar proportion. ECFP+/EGFP+ cells ratio within nodules was 0.65 ± 0.03 on histological sections, among which ECFP+-TAMs represented 58 ± 8.5% of total TAMs as depicted by flow cytometry analysis (Fig. 2 D). We next evaluated the origin of TAMs in spontaneous pulmonary metastases using the PyMT-ChOVA breast cancer model. Within spontaneous pulmonary metastases of MacBlue × Cx3cr1EGFP/+ × PyMT-ChOVA mice, the ECFP+/EGFP+ cells ratio was 0.54 ± 0.14, while EGFP+ cells were absent in nodule of parabiont mice (Fig. 2 E). These results suggest that TAMs are of dual origins both during the growth of lung carcinoma cells and metastatic cells.
Lung IMs macrophages of embryonic origin accumulate within tumors
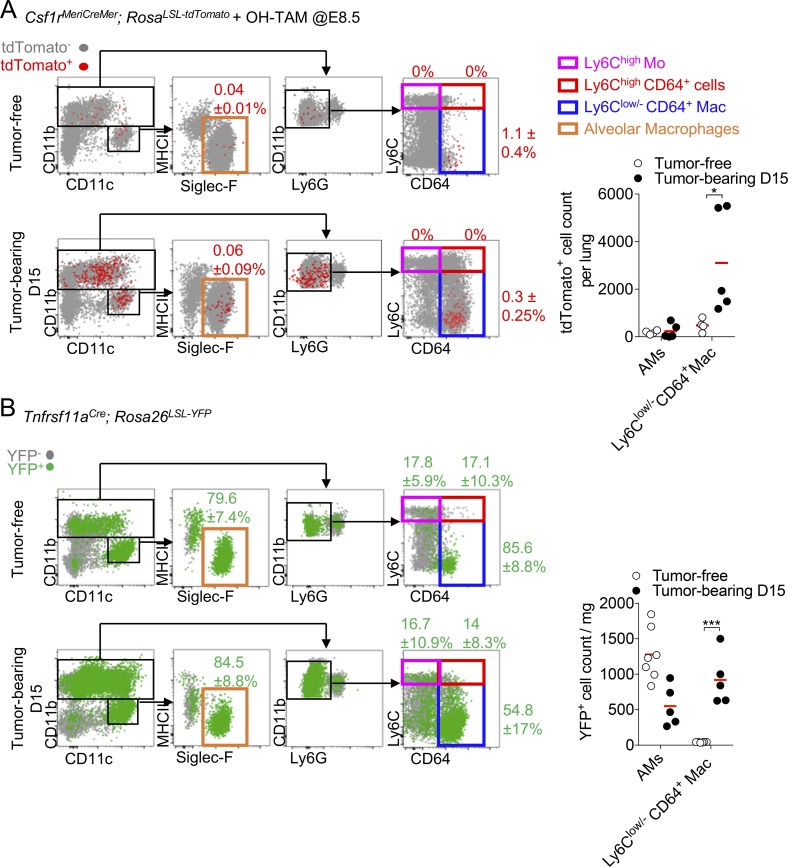
To further confirm that lung IMs contributed to the TAM compartment, we performed fate-mapping experiments using Csf1rMeriCreMer; RosaLSL-tdTomato reporter mice pulsed with 4-hydroxytamoxifen (OH-TAM) at embryonic day 8.5 (E8.5) to label cells derived from erythro-myeloid progenitors (EMP; Schulz et al., 2012; Mass et al., 2017). In this context, a small fraction of Ly6Clow/-CD64+ Mac, and to some lesser extent Siglec-F+ AMs macrophages were labeled (Fig. 3 A). In the presence of tumor, the embryonic-derived tdTomato+ Ly6Clow/-CD64+ Mac strongly expanded, but not tdTomato+ Siglec-F+ AMs (Fig. 3 A, right panel), confirming the previous observation made in the MacBlue × Cx3cr1EGFP/+ model. Expression of Tnfrsf11a during early EMP-derived macrophage differentiation allows more efficient and relatively specific lineage tracing of tissue-resident macrophages using the Tnfrsf11aCre (Mass et al., 2016). Ly6Clow/-CD64+ Mac and Siglec-F+ AMs were mostly YFP+ in the healthy lungs of Tnfrsf11aCre; Rosa26LSL-YFP mice, whereas <20% of each Mo subsets and neutrophils were labeled consistent with studies showing an embryonic origin of the former populations (Guilliams et al., 2013; Tan and Krasnow, 2016). Upon tumor development, only the proportion of YFP+ cells among the total Ly6Clow/-CD64+ Mac diminished in accordance with the appearance of a YFP− Ly6Clow/-CD64+ Mac (Fig. 3 B). YFP+ Ly6Clow/-CD64+ Mac, but not Siglec-F+ AMs, dramatically increased in absolute count confirming the expansion of the embryonic-derived interstitial subset with tumor growth (Fig. 3 B, right panel). These different fate-mapping models further confirm that interstitial resident macrophages of embryonic origin contribute to the pool of TAMs in lung tumors together with monocyte-derived macrophages.
Figure 3.
Lung IMs are of embryonic origin and accumulate within tumors. (A) Dot plots show tdTomato+ myeloid cell subsets in Csf1rMeriCreMer; RosaLSL-tdTomato after OH-TAM pulse at E8.5 in adult tumor-free mice and 15 d after TC-1 inoculation (left panels). Mean percentage ± SD of tdTomato+ cells among each subset is indicated. Right panel shows the numbers per milligram of tissue of the tdTomato+ AMs and Ly6Clow/- CD64+ Mac. Bars represent mean of four mice per group out of two independent experiments. Two-way ANOVA with Bonferroni multiple comparisons test was performed. (B) Dot plots show YFP+ macrophage subsets in Tnfrsf11aCre; Rosa26LSL-YFP mice. Mean percentage ± SD of YFP+ cells among each subset is indicated (left panels). Right panel shows the absolute number per milligram of YFP+ AMs and Ly6Clow/- CD64+ Mac. Bars represent mean of four mice per group out of two independent experiments. Two-way ANOVA with Bonferroni multiple comparisons test was performed. For all panels: *, P < 0.05; ***, P < 0.001.
Resident and MoD-TAMs harbor distinct phenotypes and distribution
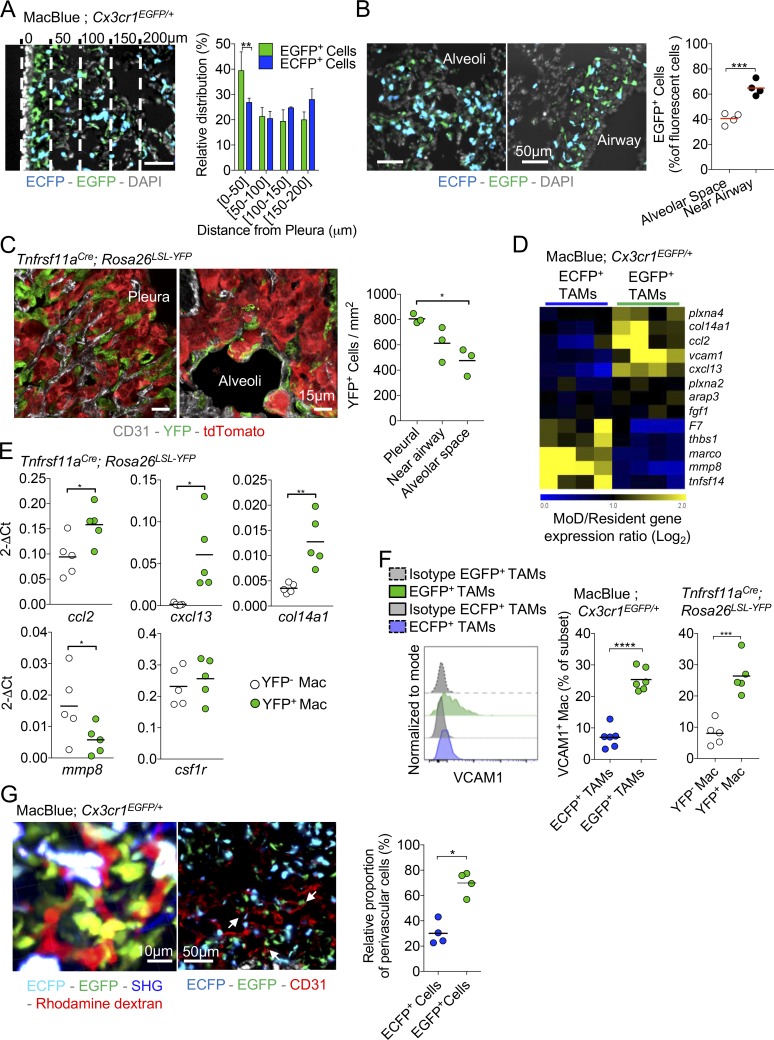
Because of their different origin, we speculated that the distribution and phenotype of EGFP+- and ECFP+-TAMs might be different. We previously demonstrated that, in tumor-free lungs of MacBlue × Cx3cr1EGFP/+, EGFP+ IMs were mostly localized in the lung pleura and in the vicinity of large airways (Rodero et al., 2015). Accordingly, in tumor nodules located nearby the lung pleura, EGFP+ cells showed a gradient of distribution falling with increase distance from the pleura, while the ECFP+ cell distribution was equal (Fig. 4 A). In tumors that developed in the central alveolar space of the lung, EGFP+ cells represented 40.5 ± 7.8% of total fluorescent cells while in tumors that developed near large airways, the ratio of EGFP+ cells was higher (65 ± 8.6%; Fig. 4 B). EGFP+ cells displayed a more stellar-like morphology compared with ECFP+ cells. EGFP+ cells were relatively sessile but interacted with each other and exhibited a highly protrusive activity across tumor cells (Fig. S3 A and Videos 1 and 2). The dynamics of ECFP+ cells were heterogeneous, likely reflecting the diversity of their composition, including monocytes or macrophages with higher displacement compared with EGFP+ cells as depicted by the relative track straightness distribution (Fig. S3 B).
Figure 4.
Resident and MoD-TAMs harbor distinct phenotypes and anatomical distribution. (A) Lung cryo-section shows the distribution of ECFP+ and EGFP+ cells in TC-1 pulmonary tumor nodule located in the vicinity of the pleura. Bar, 50 µm. Dashed lines represent each 50-µm interval, starting from the pleura. Graph represents the relative distribution of EGFP+ cells and ECFP+ MoD cells as a function of the distance from the surface (pleura; bars represent means ± SD of four mice out of two independent experiments; two-way ANOVA was performed). (B) Lung cryo-sections show the distribution of ECFP+ and EGFP+ cells in pulmonary tumor nodules located in the alveolar space or near large airways, 15 d after TC-1 inoculation. Graph represents the percentage of EGFP+ cells among fluorescent cells in each sub-anatomical compartment (dots represent the mean ratio per mouse, mice are pooled from two independent experiments, and red bars indicate mean. Unpaired Student’s t test was performed). (C) Lung cryo-sections of Tnfrsf11aCre; Rosa26LSL-YFP mice shows YFP+ cells in pulmonary tumor nodules located in the pleura or in the alveolar space 15 d after tdTomato+ TC-1 inoculation. Graph represents the number of YFP+ cells in each sub-anatomical compartment (dots represent the mean ratio per mouse, mice are pooled from two independent experiments, and bars indicate means; one-way ANOVA was performed). (D) Heat map shows a selection of transcripts involved in extracellular matrix interaction and remodeling, differentially expressed between EGFP+ and ECFP+-TAMs. (E) Expression of indicated genes, relative to GAPDH (2-ΔCt), as determined by qPCR of FACS-sorted YFP+ and YFP− TAMs in Tnfrsf11aCre; Rosa26LSL-YFP mice. Each dot represents one mouse, pooled from two independent experiments. (F) Representative histogram shows VCAM expression gated on TAM subsets from MacBlue × Cx3cr1EGFP/+ mice (left panel). Middle panel shows the quantification of VCAM1 expression by EGFP+- and ECFP+-TAMs in MacBlue × Cx3cr1EGFP/+. Right panel represents the quantification of VCAM1 expression by YFP+ and YFP− macrophages in Tnfrsf11aCre; Rosa26LSL-YFP mice. Mice are pooled from three independent experiments, bars indicate means; unpaired Student’s t test was performed. (G) TPLSM 3D reconstruction shows the perivascular location of the EGFP+ cell network within a tumor nodule (left panel). Lung cryo-section from TC-1 tumor-bearing MacBlue × Cx3cr1EGFP/+ at day 15 shows fluorescent subset distribution regarding tumor vasculature using CD31 staining (middle panel). Close interactions of EGFP+ cells with the vasculature are indicated (white arrows). Scatter plot represents the relative proportion of perivascular EGFP+ cells and ECFP+ cells in tumor nodules (right panel; dots represent the mean proportion per mouse pooled from at least two independent experiments; bars indicate means; unpaired Student’s t test was performed). For all panels: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. See also Fig. S3 and Videos 1 and 2.
Similar to EGFP+ cells, YFP+ TAMs in Tnfrsf11aCre; Rosa26LSL-YFP mice were more abundant in tumor nodules developing next to the pleura compared with nodules located in the alveolar space (Fig. 4 C). Along with tumor expansion (between days 15 and 20), accumulation of ECFP+ cells was observed at the tumor margin, whereas the proportion of EGFP+ cells remained higher in the tumor core (Fig. S3 C). This suggests that the relative composition of EGFP+-TAMs and ECFP+-TAMs in the TME is determined by the specific site of tumor development, as well as the phase of tumor evolution. Based on phenotypic surface markers (CD206, IA[b], and CD11c), we did not find any distinct expression between the two TAM subsets (Fig. S2 B), suggesting that both subsets are composed of M1/M2-like profiles. To further compare the two TAM subsets, we performed whole transcriptome microarray analysis on EGFP+- and ECFP+-TAMs sorted 20 d after TC-1 inoculation. Up to 604 differentially expressed genes (either up or down, with a P value < 0.05 by Student’s t test) were identified between the two TAM subsets (Fig. S3 D). The Ingenuity Knowledge Base identified their association with functional groups and the most relevant groups (with a cut-off value at P < 0.01, given by the score from Fisher’s Exact Test) were listed (Fig. S3 E). These functional groups were involved in cellular signaling, cell morphology and trafficking, tissue remodeling associated to cancer development. We found a set of transcripts related to extracellular matrix and vasculature interactions that were differentially expressed between EGFP+-TAMs and ECFP+-TAMs. For instance, the transcripts Marco, Mmp8, F7, Tnfsf14, and Thbs1 were found to be expressed at higher levels in ECFP+-TAMs compared with EGFP+-TAMs (Fig. 4 D). The transcripts for Col14a1, Ccl2, and Cxcl13, as well as Vcam1 and Plxna4 (involved in adhesion-dependent processes and angiogenesis; Gambardella et al., 2010; Tamagnone, 2012), were all up-regulated in EGFP+-TAMs compared with ECFP+-TAMs. Col14a Ccl2, Cxcl13 transcripts were also higher in YFP+ TAMs in the Tnfrsf11aCre; Rosa26LSL-YFP model, whereas YFP− TAMs expressed a higher level of Mmp8. YFP+ and YFP− TAMs expressed similar levels of csf1r transcripts (Fig. 4 E). VCAM1 expression was confirmed at the protein level and defined a marker mostly restricted to the EGFP+-TAM subset and was expressed accordingly in YFP+ TAMs of Tnfrsf11aCre; Rosa26LSL-YFP lungs (Fig. 4 F). Near the tumor vasculature, EGFP+ cells were more abundant than ECFP+ cells, displaying a typical perivascular-like morphology around the vessels (Fig. 4 G). We concluded that despite a similar surface marker expression profile, ECFP+-TAMs and EGFP+-TAMs are distinct subsets and we speculated they might be differentially involved in tumor growth.
Resident TAMs support tumor cell growth and MoD cells are associated with tumor spreading in the lung
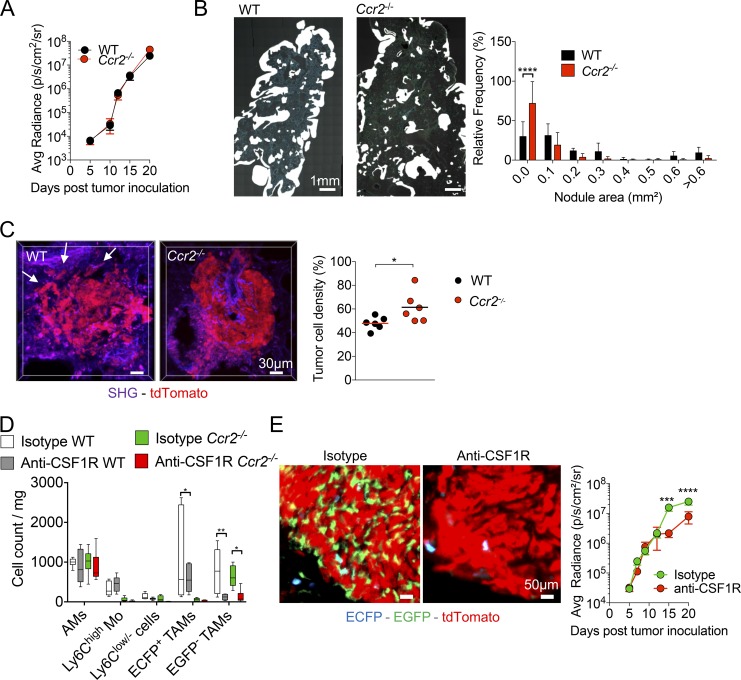
The relative contribution of the TAM subsets on tumor growth was next evaluated comparing tumor evolution in WT and CCR2-deficient mice. Tumor growth was similar in WT and Ccr2−/− mice, as monitored by bioluminescence (Fig. 5 A). However, histological analysis showed that nodule surface was smaller in CCR2-deficient mice compared with WT mice (Fig. 5 B). This discrepancy might be explained by a more disperse and lower density of tumor cells within pulmonary nodules of WT compared with Ccr2−/− (Fig. 5 C). Overall, these results confirm that even in the absence of monocyte-derived TAMs, tumor cells can efficiently grow in vivo and suggest that resident TAMs are sufficient to support tumor cell expansion, while MoD cells might contribute to tumor cell dissemination.
Figure 5.
Resident TAMs support tumor cell growth, and MoD cells are associated with tumor spreading in the lung. (A) Tumor growth was monitored in WT and Ccr2−/− mice by bioluminescence imaging (graph represents mean ± SEM of n = 10 mice per group from two independent experiments). (B) Wide field images of a whole lung cryo-section 15 d after TC-1 inoculation in WT and Ccr2−/− mice (left panels). White mask indicates nodule size and distribution. Graph shows the relative distribution of nodule areas (right panel; n = 5–7 mice in each group out of three independent experiments; two-way ANOVA comparing WT and Ccr2−/− frequency of each distribution was performed). (C) TPLSM 3D reconstructions show tumor cell density in a representative nodule from TC-1tdTomato tumor-bearing WT and Ccr2−/− mice; white arrows indicate spread tumor cells (left panels). Tumor cell density was measured using 3D reconstruction images of tdTomato+ tumor nodule (dots represent the mean of at least four different tumor nodules per mouse, pooled from two independent experiments; bars indicate means; unpaired Student’s t test was performed). (D) Box and whisker graph represents numbers per milligram of lung of each indicated myeloid subset after indicated treatment at day 15. Tumor-bearing WT and Ccr2−/− mice were treated every 2 d with anti-CSF1R between days 5 and 14 (right panel; n = 6 mice from two independent experiments. One-way ANOVA comparing each subset individually for each condition was performed). (E) Lung cryo-sections of TC-1tdTomato tumor-bearing MacBlue × Cx3cr1EGFP/+ × Ccr2−/− mice treated with anti-CSF1R or isotype control show depletion of EGFP+ cells at day 15 (left panels). Tumor burden was monitored by bioluminescence imaging after TC-1-Luc inoculation (right panel). Mice were treated every 2 d with anti-CSF1R starting day 5 (graphs represent mean ± SEM, n = 10 mice per group out of two independent experiments. Two-way ANOVA with Bonferroni multiple comparisons test was performed). For all panels: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Transient anti-CSF1R treatment is known to target mature macrophages, but does not block monocyte infiltration into tumors (Kitamura et al., 2018). Compared with other resident macrophages, AMs have been described to be uniquely dependent on GM-CSF (Guilliams et al., 2013; Schneider et al., 2014) and, as a result, should not be targeted by the treatment. Treatment of tumor-bearing WT mice with anti-CSF1R depleted ECFP+-TAMs and more profoundly EGFP+-TAMs but not monocytes and AMs (Fig. 5 D). Anti-CSF1R treatment does not allow to distinguish the relative contribution of monocytes, ECFP+-TAMs and EGFP+-TAMs on tumor growth. To investigate the contribution of resident TAMs only on tumor growth, we performed anti-CSF1R treatment on CCR2-deficient mice. This treatment strongly depleted the remaining EGFP+-TAMs in tumor nodules of CCR2-deficient mice as well and strongly reduced tumor burden (Fig. 5, D and E). Our results corroborate the role of interstitial lung macrophages as a trophic support for tumor cells, while MoD cells are associated with tumor remodeling and spreading.
Distinct sensitivity and recovery of Res-TAMs and MoD-TAMs after chemotherapy
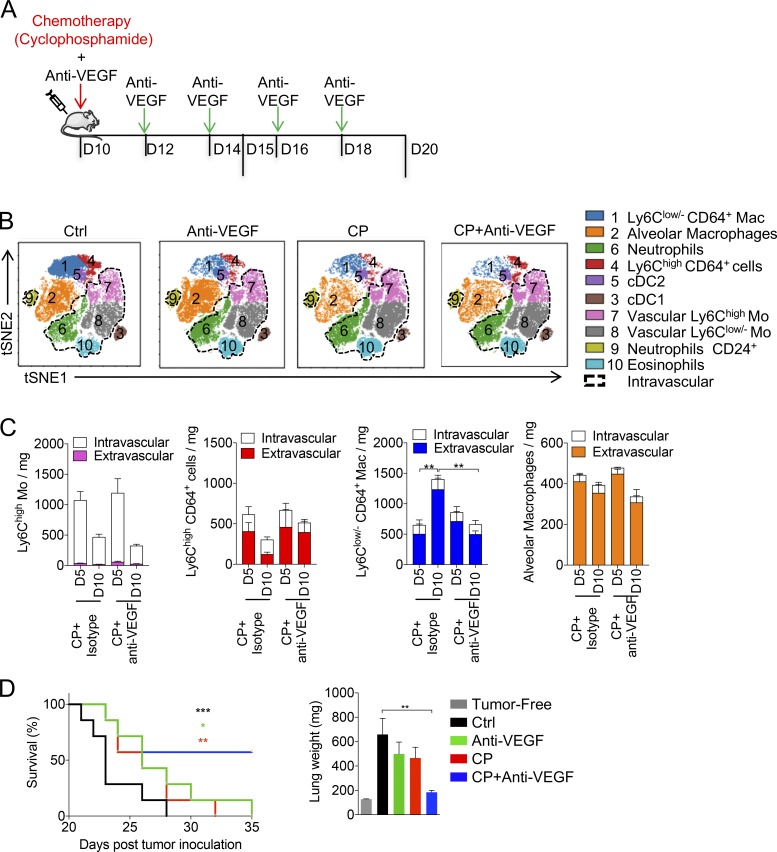
TAMs play major roles in the response to anti-cancer therapies (Mantovani and Allavena, 2015). We next addressed how the two TAM subsets respond to conventional chemotherapy. Cyclophosphamide (CP) is a classical alkylating agent with known myeloablative properties (Jacquelin et al., 2013). A single injection of CP led to a strong reduction in tumor burden, which relapsed 15 d after chemotherapy (Fig. 6 A). The number of circulating Ly6Chigh-Mo was reduced 2 d after chemotherapy, but recovered with a significant overshoot between days 5 and 10 post-therapy (Fig. S4 A). Circulating Ly6Clow/--Mo displayed a delayed recovery compared with Ly6Chigh-Mo, but the numbers of both monocyte subsets finally dropped at day 15 after CP, correlating with tumor relapse (Fig. S4 A). Intravascular CD45 staining was performed, and the recovery of myeloid cells in the lungs was monitored (Fig. 6 B). Monocyte and macrophage subsets were also depleted in both vascular and parenchymal compartments of the lungs within 2–3 d (Fig. 6 C). Monocyte subsets transiently rebounded at day 5 after CP treatment, and their accumulation was associated with macrophage recovery, peaking at day 10 (Fig. 6 C). Among macrophages, both EGFP+ and ECFP+ subsets were depleted by CP treatment, but the massive recovery at day 10 was mainly constituted by ECFP+-TAMs (Fig. 6 D). ECFP+ MoD cells accumulated in the vicinity of living tumor cells between 5 and 10 d after CP and participated in the clearance of the apoptotic debris (Fig. 6 E). The proportion of phagocytic cells among different subsets was quantified by flow cytometry between 10 and 15 d after CP (Fig. 6 F). ECFP+-TAMs represented the most abundant phagocytic subsets while EGFP+-TAMs poorly contributed to tumor clearance (Fig. 6 F). The numbers of monocytes and macrophages were lower in Ccr2−/− mice compared with WT mice 15 d after CP treatment (Fig. S4 B). This defect was associated with a reduced efficacy of chemotherapy (Fig. S4, C and D). We conclude that CP treatment targets both EGFP+-TAMs and ECFP+-TAMs, but these subsets differentially recover and contribute to tumor elimination. Because one single dose of CP was not sufficient to completely eradicate the tumor and led to tumor relapse, we next aimed at improving therapy efficacy.
Figure 6.
Distinct sensitivity and recovery of Res-TAMs and MoD-TAMs after chemotherapy. (A) Impact of CP treatment on tumor growth was monitored by bioluminescence imaging (graph represents mean ± SEM of n = 10 mice per group out of three independent experiments). (B) Dot plots show Ly6C and CD64 expressions of Siglec-F−CD11b+Ly6G− lung cells over time after CP treatment. Mean percentage ± SD of cells in each quadrant are indicated. (C) Blood/tissue partitioning monitoring of lung monocytes and macrophages during tumor growth after chemotherapy (graphs represent mean of the absolute number ± SEM/milligram of tissue, n = 6–10 mice per time point out of two to four independent experiments, two-way ANOVA with Bonferroni multiple comparisons test was performed. Only statistical differences compared with day of treatment [day 0] are indicated for each compartment). (D) Graph shows the absolute number per milligram of tissue of ECFP+-TAMs and EGFP+-TAMs from MacBlue × Cx3cr1EGFP/+ mice after CP (n = 4–10 mice per time point out of three independent experiments, two-way ANOVA with Bonferroni multiple comparisons test was performed). (E) Lung cryo-sections of TC-1tdTomato tumor-bearing MacBlue × Cx3cr1EGFP/+ mice show TAM subset distribution within tumor nodules following CP treatment. (F) Dot plots show TC-1tdTomato phagocytosis by the indicated mononuclear phagocyte subsets (red). Fluorescent background from nonfluorescent TC-1 tumor is overlaid (left panels; black). Box and whisker graphs show the relative proportion of phagocytic cells among indicated subsets at 10 and 15 d after CP treatment (right panels; n = 9 mice out of three independent experiments). For all panels: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. See also Fig. S4.
Anti-VEGF combination with CP reduces TAM recovery and enhances chemotherapy efficacy
The pro-angiogenic molecule VEGF has been implicated in vessel reconstruction and tumor relapse following chemotherapy (Hughes et al., 2015; Lewis et al., 2016). In addition, the combination of anti-VEGF with chemotherapy has shown greater efficacy than chemotherapy or targeted therapy alone in patients bearing nonsmall cell lung cancer and metastatic breast cancer (Cohen et al., 2007; Montero and Glück, 2012). Because TAMs have been shown to express VEGFR1 (FLT1; Qian et al., 2015), we speculated that the combination of anti-VEGF with CP could directly target TAMs and improve therapeutic outcome. Tumor-bearing mice were treated or not with CP in combination or not with anti-VEGF (Fig. 7 A). We determined the impact of the combined therapy on the myeloid signature of the TME using the previous unsupervised viSNE analysis based on 12 parameters including intravascular CD45 staining (dashed gates; Fig. 1 A, Fig. 7 B, and Fig. S1). The myeloid signature of the vascular compartment was similar in each condition. Anti-VEGF treatment in combination with CP induced a striking reduction of the TAM signature (cluster 1) in comparison to single treatments. Interestingly, in the combined regimen, the tumor-infiltrating Ly6Chigh CD64+ cell subset (cluster 4) was increased compared with CP or anti-VEGF treatments alone (Fig. 7 B). We thus quantified the recovery of monocytes and macrophages between days 5 and 10 after chemotherapy in mice treated with anti-VEGF or isotype control (Fig. 7 C). The combination of CP and anti-VEGF blocked TAM recovery between days 15 and 20, whereas neither the Ly6Chigh-Mo rebound nor the infiltration of Ly6Chigh CD64+ cells were affected, suggesting that TAM diminution was not a result of a reduction of monocyte infiltration. AM number remained unaffected between the two conditions (Fig. 7 C). The efficacy of the combined therapy was evaluated on advanced stages of tumor development (day 20 after tumor inoculation). Compared with both treatments alone, the combination resulted in prolonged mouse survival and normalization of the lung weight (Fig. 7 D).
Figure 7.
Anti-VEGF combination with CP reduces TAM recovery and enhances chemotherapy efficacy. (A) Tumor-bearing mice were treated with CP 10 d after tumor inoculation and treated or not every 2 d with anti-VEGF. (B) Representative tSNE dimension 1 and 2 plots show the impact of the different therapies on the myeloid signature at day 15. Cell subsets are color grouped, and dashed black gates delineate blood/tissue partitioning. (C) Blood/tissue partitioning monitoring of lung monocytes and macrophages during tumor growth at days 15 and 20 after CP or CP + anti-VEGF. (Bars represent mean of the total number of cell ± SEM. For all panels n = 6 mice per time point out of two independent experiments; two-way ANOVA with Bonferoni multiple comparisons test was performed.) (D) Survival curve shows the efficacy of combined therapy started from day 20 (n = 7 mice per group, data are representative of two experiments; log-rank [Mantel-Cox] test was performed to compare each survival curve with the one of CP + anti-VEGF). Corresponding lung weights are reported (one-way ANOVA with Bonferroni multiple comparisons test was performed). For all panels: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Anti-VEGF targets Res-TAM and MoD-TAM accumulation
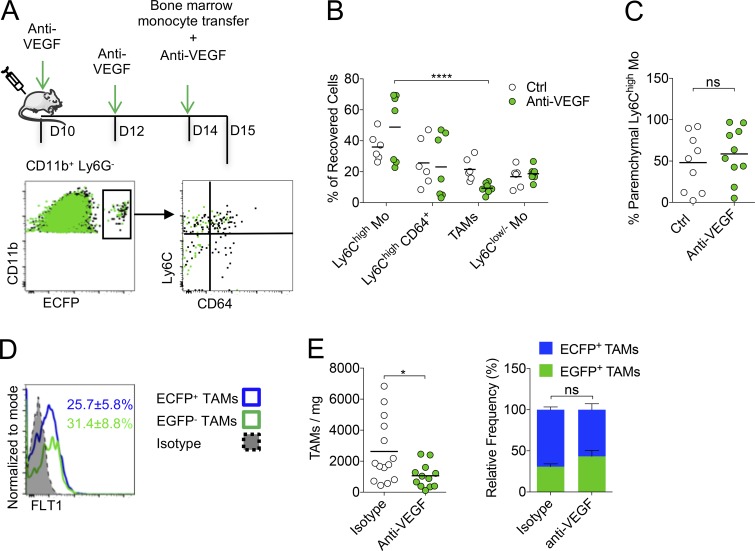
To further investigate the action of anti-VEGF on myeloid cells, we adoptively transferred bone marrow monocytes in anti-VEGF or isotype-treated WT mice (Fig. 8 A). The proportion of recovered TAMs was significantly reduced at the expense of Ly6Chigh-Mo (Fig. 8 B), while the infiltration (measured by intravascular CD45 staining) of the latter was unchanged (Fig. 8 C), indicating that anti-VEGF did not block monocyte infiltration, but rather reduced their differentiation into TAMs and/or TAM survival. FLT1 expression was already detected on Ly6Chigh CD64+ cells, but the expression on Ly6Chigh- and Ly6Clow/--Mo was low (Fig. S5 A). EGFP+-TAMs and ECFP+-TAMs harbored similar expression of FLT1 (Fig. 8 D). VEGF was found mainly along blood vessels but also in the tumor parenchyma, in proximity to TAMs without preferential colocalization with ECFP+ or EGFP+ cells (Fig. S5 B). Anti-VEGF treatment of tumor-bearing mice led to a significant reduction in the number of TAMs, but the proportions of EGFP+-TAMs and ECFP+-TAMs were similar, demonstrating that both macrophage subsets are reduced by this treatment (Fig. 8 E). Our results support that anti-VEGF targets both monocyte-derived and resident TAM accumulation through a mechanism independent of their recruitment but rather affects their survival or proliferation.
Figure 8.
Anti-VEGF targets Res-TAM and MoD-TAM accumulation. (A) Bone marrow monocytes from MacBlue mice were adoptively transferred in tumor-bearing WT mice treated or not with anti-VEGF (upper panel). Representative overlay dot plots show the phenotype of recovered MoD cells from tumor-bearing mice treated (green) or not with anti-VEGF (black) 24 h after transfer (lower panels). (B) Graph shows the relative proportion of recovered cells in each mouse pooled from two independent experiments (black bars indicate means; two-way ANOVA with Bonferroni multiple comparisons test was performed). (C) Graph shows the proportion of Ly6Chigh-Mo infiltration evaluated by blood/tissue partitioning in each mouse pooled from two independent experiments (black bars indicate means; Student’s t test was performed). (D) Representative histogram plot of FTL1 expression by ECFP+ and ECFP+-TAMs, mean percentage ± SD of FLT1+ cell in each subset out of six mice from two independent experiments are indicated. (E) Left panel shows the number of TAMs/milligram in indicated conditions. Right panel shows the relative proportion of TAMs in mice treated with anti-VEGF or isotype. Black bars indicate means of two independent experiments, Student’s t test was performed. For all panels: *, P < 0.05; ****, P < 0.0001; ns, not significant. See also Fig. S5.
Discussion
Embryonic-derived macrophages have recently been shown to contribute to the generation of TAMs in the pancreas and in the brain (Bowman et al., 2016; Zhu et al., 2017). These discoveries challenge the dogma on the origin of TAMs and raise the question whether this observation is applicable to other tissues such as the lungs, which are colonized by distinct macrophage subsets. AMs represent the main and typical resident macrophages of the lungs, maintaining immune homeostasis in the alveoli lumen (Trapnell and Whitsett, 2002). AMs acquire their unique signature and self-maintain via GM-CSF–dependent induction of PPAR-γ after birth (Guilliams et al., 2013; Schneider et al., 2014). Less is known about the functions and origin of IMs, but it has been suggested that they develop earlier than AMs in the embryo (Tan and Krasnow, 2016) and self-maintain independently of adult hematopoiesis (Rodero et al., 2015; Gibbings et al., 2017).
The implication of the chemokine receptor CCR2 in the recruitment of monocytes and on their subsequent differentiation into TAMs is well established in both primary and metastatic sites of various tumor types. This CCR2–CCL2 axis can contribute to an amplification loop of tumor progression (Franklin et al., 2014; Kitamura et al., 2015; Loyher et al., 2016). In these studies, the role of this axis on resident macrophages could not be excluded. However, lung IMs have been shown to expand independently of CCR2 and to display regulatory functions in the context of allergy (Sabatel et al., 2017).
Using the MacBlue × Cx3cr1EGFP/+ model, we unveiled the accumulation of an EGFP+-TAM subset that was unaffected by CCR2 deficiency and not reconstituted in parabiosis experiments, demonstrating that this subset originates from macrophages that were already present in the healthy lungs before tumor development. Preferential labeling of CD11b+ IMs was previously achieved using Csf1rMeriCreMer mice pulsed with OH-TAM at E8.5 (Schulz et al., 2012). Using the same approaches to trace EMP-derived macrophages, we unveiled that embryonically seeded lung-resident IMs persist and proliferate to represent a large fraction of TAMs within pulmonary tumors and confirmed our hypothesis made using the MacBlue × Cx3cr1EGFP/+ mouse system. While we could not firmly exclude that some EGFP+-TAMs arise from AM differentiation, it is unlikely that upon tumor parenchymal infiltration AMs lose the expression of Siglec-F and ECFP reporter while up-regulating CD11b and EGFP. No progressive change in the expression of these surface markers that could support this hypothesis was observed in the AM population during tumor development. Thus, resident IMs are likely to represent a unique tissue-resident subset involved in the accumulation of EGFP+-TAMs. Loss of ECFP expression from monocyte-derived macrophages could lead to overestimation of EGFP+-TAMs; however, monocyte-derived cells maintained ECFP expression in parabiosis and transfer experiments, suggesting that this phenomenon barely occurs during this time frame and would only minimally perturb our quantification of EGFP+-TAMs. Fate-mapping studies led to similar observations and strengthen the fidelity of the MacBlue × Cx3cr1EGFP/+ system to study lung macrophages. Concomitantly, we observed an increase in the number of Ly6Chigh monocytes. Infiltrating Ly6Chigh-Mo seemed to up-regulate CD64 suggesting an intermediate toward the progressive differentiation into TAMs. We subsequently identified a distinct population of monocyte-derived TAMs arising from CCR2-dependent monocyte recruitment. At later time points, these TAMs became the most abundant population. The dual origin of macrophages was also observed in TAMs of LLC lung nodules and PyMT-ChOVA spontaneous pulmonary metastases suggesting that it might occur for any neoplastic tissue development in the lung.
Within lung tumor nodules, the relative distribution and abundance of EGFP+-TAMs compared with the recruited ones were in accordance with the localization of IMs before tumor development. We previously showed in the MacBlue × Cx3cr1EGFP/+ mice that interstitial EGFP+ macrophages are abundant in the pleura, airways, and at the periphery of large blood vessels (Rodero et al., 2015). Lineage tracing of yolk sac–derived macrophages labeled mostly IMs that persisted in adults and localized in these same particular locations, but scarcely in the central lung parenchyma (Tan and Krasnow, 2016). The local environment can dictate macrophage phenotypes in vivo (Gosselin et al., 2014; Lavin et al., 2014; van de Laar et al., 2016). Despite a very close proximity between the two TAM subsets in tumor nodules, their transcriptomic profiles were distinct and were associated with different distribution depending on the anatomical localization of the tumor, further arguing that origin poises macrophages for differing functions. No typical M1 or M2 profile could be attributed to EGFP+-TAMs or ECFP+-TAMs, suggesting that this paradigm does not fully resolve the polarization process of TAMs. Nevertheless, the relative proportion and specific features of tissue-resident macrophages might contribute to the heterogeneity of different TME according to the anatomical site of tumor development. Further studies are needed to investigate whether it could serve as a prognostic factor of tumor growth and response to therapies.
Anti-CSF1R treatment depleted most of EGFP+-TAMs, but ECFP+-TAMs were only partially targeted, while Ly6Chigh monocytes and AMs were unaffected. Despite the lack of ECFP expression in the MacBlue mouse, it was previously shown that adult tissue macrophages express CSF1R. The differential utilization of the truncated CSF1R promoter of the MacBlue binary transgene in macrophages was proposed to reflect different survival dependency on CSF1. Thus, cell expressing the ECFP reporter would be CSF1-independent in contrast to EGFP+ macrophages that require the upstream depleted region of the CSF1-regulated promoter region of the MacBlue transgene (Sauter et al., 2014; Hawley et al., 2018). This was supported by the reduced impact of anti-CSF1R treatment on ECFP+-TAMs compared with EGFP+-TAMs. Depletion of EGFP+-TAMs in Ccr2−/− mice led to drastic reduction in tumor growth, which links resident TAMs more directly to tumor trophic functions. ECFP+-TAMs displayed increased motility, in accordance to enriched cellular movement associated pathways and turned up to accumulate at the tumor margin. In this regard, Mmp8 and Tnfsf14 enrichment (implicated in airway remodeling; Doherty et al., 2011) could argue for a licensing of monocyte-derived TAMs for remodeling of the surrounding environment and modification of the tumor architecture. Indeed, recruitment of MoD cells was associated with reduced tumor cell density, higher spreading, and increased invasion of pulmonary nodules. We could not fully differentiate the relative contribution of tumor-infiltrating ECFP+ monocytes versus ECFP+-TAMs. Monocyte-dependent cytotoxic activity could be suspected as observed after CP-induced monocyte rebound. Thus ECFP+-cells represent a heterogeneous population balancing between tumor destruction and remodeling, favoring spreading and invasiveness. This observation raises important questions about cancer therapies targeting TAM subsets and suggests that depleting resident macrophages but keeping the phagocytic activities of MoD cells would yield a better outcome for chemotherapies.
The targeting of VEGF in combination with chemotherapy including CP (Dellapasqua et al., 2008) has been shown to be beneficial (Motz and Coukos, 2011). Although TAMs have clearly been shown to participate in the process of angiogenesis within tumor (Lewis et al., 2016), few studies have investigated the impact of this therapeutic combination on the immune cellular composition of the TME. Moreover, FLT1 expression and signaling by pulmonary TAMs are implicated in their pro-tumor activity, partly via downstream regulation of the master macrophage regulator CSF1 (Qian et al., 2015). VEGF has been proposed to act as a chemoattractant factor for monocytes (Kaplan et al., 2005; Grunewald et al., 2006), but the beneficial effect of anti-VEGF combination was associated with a drastic reduction of both EGFP+ resident and ECFP+ monocyte-derived TAMs without affecting tumor-monocyte infiltration, suggesting that VEGF contributes to monocyte differentiation and/or TAM survival. Indeed, FLT1 expression was increased upon monocyte to macrophage differentiation, which corroborates previous studies (Barleon et al., 1996; Qian et al., 2015). This observation is in accordance with the hypothesis of a loss of anti-tumor activity of tumor-infiltrating monocytes upon differentiation into TAMs. The clinical relevance of our results lies in the fact that anti-VEGF could improve chemotherapy efficacy through functions that go beyond its main expected role on angiogenesis and leukocyte recruitment. Increasing knowledge of the impact of such molecule on the different TAM subsets according to their origin might allow for further development and improvement of anti-cancer dosing regimens and combinations.
Materials and methods
Mice and fate mapping
C57Bl6 mice were purchased from Janvier Labs. Cx3cr1EGFP/+ (Jung et al., 2000), Csf1r-Gal4VP16/UAS-ECFP (MacBlue; Ovchinnikov et al., 2008), and Ccr2−/− mice were intercrossed to generate MacBlue × Cx3cr1EGFP/+, MacBlue × Cx3cr1EGFP/+ × Ccr2−/− litermate mouse strains. These strains, and MMTV PyMT-P2A-mCherry-P2A-OVA (PyMT-ChOVA) mice (Engelhardt et al., 2012) were bred at Pitié-Salpêtrière animal facility. Csf1rMeriCreMer; Rosa26tdTomato-LSL (Qian et al., 2011) and Tnfrsf11aCre; Rosa26YFP-LSL (Maeda et al., 2012) were bred at the Memorial Sloan Kettering Research Animal Resource Center. For the labeling of the EMP lineage, pulse labeling was performed in Csf1rMeriCreMer; Rosa26tdTomato-LSL E8.5 embryos with 4-hydroxytamoxifen (OH-TAM, Sigma-Aldrich). Embryonic development was estimated considering the day of vaginal plug formation as 0.5 d after coitum. Cre recombination was induced by a single injection of 37.5 mg per kg (body weight) of OH-TAM into pregnant females. OH-TAM was supplemented with 18.75 mg per kg (body weight) progesterone (Sigma-Aldrich) to counteract the mixed estrogen agonist effects of tamoxifen, which can result in fetal abortions.
All mice were maintained under SPF conditions and used between 8 and 14 wk old except for PyMT-ChOVA that develop primary breast tumors and lung metastases at around 25 wk.
Ethical statement
All experiment protocols were approved by the French animal experimentation and ethics committee and validated by Service Protection et Santé Animales, Environnement with the number A-75-2065 for tumor experiments and A-75-1315 for parabiosis experiments. Sample sizes were chosen to assure reproducibility of the experiments and according to the 3 Rs of animal ethic regulation. Animal procedures involving Csf1rMeriCreMer; Rosa26tdTomato-LSL and Tnfrsf11aCre; Rosa26YFP-LSL were performed in adherence to project license IACUC 15-04-006, issued by Institution Review Board from Memorial Sloan Kettering Cancer Center.
Parabiosis
C57Bl6 female host parabionts were generated with MacBlue × Cx3cr1EGFP/+ females. Blood T cell chimerism was tested after 2 wk and was between 40 and 60%, whereas Ly6Chigh-Mo chimerism was 10–20%. At this time, both animals were injected with TC-1 cells and analyzed 15 d later.
Cells
The TC-1 tumor cell line was derived from primary lung epithelial cells of a C57Bl6 mouse cotransformed with HPV-16 oncoproteins E6 and E7 and c-Ha-ras oncogene (Lin et al., 1996; Ji et al., 1998). TC-1 cells expressing Luciferase (TC-1-Luc) were provided by T.-C. Wu (Johns Hopkins University, Baltimore, MD). TC-1tdTomato cell line was generated by infection of TC-1 cells with a tdTomato-lentivirus (provided by M. Lambert, Institut Cochin, Paris, France). Cells were expanded and sorted with BD Facs AriaIII (platform CyBio, Institut Cochin). TC-1tdTomato cells were >97% pure. LLC (CRL-1642) were obtained from American Type Culture Collection. All cell lines were cultured in RPMI-1640 with 10% fetal bovine serum, Na-Pyruvate, anti-biotic, and anti-mitotic (GIBCO BRL Invitrogen) and incubated at 37°C in 5% CO2. For in vivo experiments, 5 × 105 cells were injected intravenously in the tail vein to generate lung tumors.
Blood/tissue partitioning
Intravascular CD45 labeling was performed as previously described (Rodero et al., 2015; Hamon et al., 2017). Mice were injected intravenously with 1 µg of anti-CD45 (clone 30-F11). 2 min after injection, blood was drawn and mice were sacrificed. Lungs were harvested and bathed in a large volume of PBS.
Anti-cancer therapies
CP (Sigma-Aldrich) treatment was performed by a single intraperitoneal injection diluted in PBS at 175 mg/kg at 10 d after tumor inoculation or at 20 d for advanced cancer treatment protocols. The anti-VEGF antibody (B20-4.1.1) and isotype control were supplied by Genentech. Antibodies were administrated intraperitoneally at 5 mg/kg, every 2 d after chemotherapy. The anti-CSF1R antibody (AFS98, BioXcell) or rat IgG2a κ isotype controls were administrated intraperitoneally at 50 mg/kg every 2 d starting on day 5.
Flow cytometry
Blood was drawn via retro-orbital puncture with heparin and directly stained with antibodies. After staining, erythrocytes were lysed with buffer containing 0.15 M NH4Cl, 0.01 mM KHCO3, and 0.1 mM EDTA and resuspended in FACS buffer containing PBS, BSA (0.5%), and 2 mM EDTA. Lungs were harvested and digested in RPMI medium (GIBCO BRL Invitrogen) with 1 mg/ml collagenase IV (Sigma-Aldrich) for 30 min at 37°C and filtered through a 40-µm pore cell strainer (Becton Dickinson). 1/10 of the cell suspension was incubated with 1 µg/ml purified anti-CD16/32 (2.4G2, BD Biosciences) for 10 min at 4°C then surface staining was performed by additional 20-min incubation with appropriate dilution of the surface marker antibodies. Cells were then washed once in FACS buffer and analyzed directly by flow cytometry. The panel of antibodies used was anti-CD11b (clone M1/70), anti-Ly6C (clone AL-21), anti-Ly6G (clone 1A8), anti-NK1.1 (clone PK136), anti-IA[b] (clone AF6-120.1), anti-CD11c (clone HL3), anti-Siglec-F (clone E50-2440), anti-CD103 (clone M290), anti-CD45 (clone 30-F11), anti-CD24 (clone M1/69), anti-CD64 (clone X54-5/7.1; PharMingen; BD Biosciences), anti-FLT1 (clone 141522; R&D Systems), anti-CD206 (clone C068C2; Biolegend), and anti-VCAM1 (clone 429; BD Biosciences). Calculation of absolute cell number was performed by adding to each vial a fixed number (10,000) of nonfluorescent 10-µm polybead carboxylate microspheres (Polysciences) according to the formula no. of cells = (no. of acquired cells × 10,000) / (no. of acquired beads). Number of cells obtained for each sample was normalized per milligram of tissue or for the whole lungs and per milliliter of blood. Flow cytometry acquisition was performed on the flow cytometer FACS LSRFortessa X-20 (Becton Dickinson) with DIVA Flow Cytometry software. Flow cytometry data were analyzed with FlowJo software (Tree Star) and, when indicated, visualized using viSNE (Amir et al., 2013), a dimensionality reduction method, which uses the Barnes-Hut acceleration of the t-SNE algorithm. viSNE was implemented using Cytobank (Chen and Kotecha, 2014).
Multi-photon imaging
Lung experiments were performed on freshly explanted tissue according to our previously work (Rodero et al., 2015). Lungs were carefully collected and were immobilized in an imaging chamber perfused with oxygenated (95% O2 plus 5% CO2) RPMI medium containing 10% FCS. Local temperature was monitored and maintained at 37°C. To define the tumor vasculature 2MDa tetramethylrhodamine-Dextran (Invitrogen) was injected i.v. before the imaging session. The two-photon laser-scanning microscopy (TPLSM) set-up used was a 7MP (Carl Zeiss) coupled to a Ti:Sapphire Crystal multiphoton laser (ChameleonU, Coherent), which provides 140-fs pulses of near-infrared light, selectively tunable between 680 and 1050 nm and an optical parametric oscillator (OPO-MPX, Coherent) selectively tunable between 1,050 and 1,600 nm. The system included a set of external nondescanned detectors in reflection with a combination of a LP-600-nm followed by LP-462-nm and LP-500-nm dichroic mirrors to split the light and collect the second harmonic generation signal with a 417-/60-nm emission filter, ECFP with a 480-/40-nm emission filter, EGFP with a 525-/50-nm emission filter, and tdTomato with a 624-/40-nm emission filter. The excitation wavelength was 850 nm for the nonlinear optical beam and 1100 nm for the OPO beam. Cell motility was measured every 30 s by five consecutive 3-µm z-spacing stacks (total 12-µm thickness) using a plan apochromat ×20 (numerical aperture = 1) water immersion objective. Fluorescent cells were monitored over time with three-dimensional (3D) automatic tracking and manual correction with Imaris software (Bitplane). The acquisition and analysis protocols for all experimental conditions to be compared were identical. Velocity and straightness were determined using Imaris. The track straightness corresponds to the ratio of the distance between the initial and the final positions of each cell to the total distance covered by the same cell. Tumor cell density analyses were performed with ImageJ software using default threshold. Data are expressed as a percent of tdTomato signal among the total surface of the tumor nodule, recorded from a z projection of five consecutive 5-µm z-spacing stacks of the tumor nodule imaged from 40-µm-thick lung sections.
Histological analysis
In brief, organs were harvested and fixed in 4% PFA for 24 h and then incubated in 30% sucrose-PBS overnight at 4°C before being embedded in Tissue-Tek optimal cutting temperature compound (Sakura Finetek) and frozen at −80°C. Sectioning was completed on a HM550 Cryostat (Thermo Fisher) at −20°C; 5-µm or 15-µm sections were collected on Superfrost Plus Slides (Thermo Fisher Scientific) and stored at −20°C until use. Tissue sections were rehydrated with 0.5% Triton-PBS during 10 min. A first block step was performed with 3% BSA solution during 30 min, followed by 1-h incubation at 37°C with the primary antibodies rat anti-mouse CD31 PE (clone MEC 13.3; Becton Dickinson) or the rat IgG2a, κ isotype control (clone eBR2a; eBioscience), and anti-VEGF clone B20-4.1.1 or isotype control (Genentech) and rat anti-mouse Siglec-F PE-CF594 (clone E50-2440) were used at the appropriate dilution. The slides were then incubated with Avidin/Biotin Blocking kit (SP-2001; Vector Laboratories) following manufacturer protocol. Biotinylated secondary antibody binding (donkey anti-rat IgG for CD31 and Siglec-F stainings and AF647 anti-mouse IgG2a for VEGF staining [Biolegend]) was then performed during 30 min at room temperature followed by Cy3-conjugated Streptavidin staining during 30 min at room temperature (Jackson ImmunoResearch Laboratories). Slides were counterstained and mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories) and analyzed by using Axio Z1 fluorescent microscope (Carl Zeiss) using Zen software. ECFP, EGFP, DAPI, Cy3, and AF647 signals were acquired using an ExBP 475/40, EmBP 530/50 for EGFP, an ExBP 436/25, EmBP 480/40 for ECFP, an ExG365, EmBP 445/50 for DAPI, an ExBP 545/25, and EmBP 605/70 for Cy3 and an ExBP 640/30 and EmBP 690/50 for AF647 light cube filters. Acquisition settings were identical for both isotype and CD31/VEGF staining. Cell quantification was performed by counting the number of ECFP+ and EGFP+ cells using ImageJ software (National Institutes of Health).
For Tnfrsf11aCre; Rosa26LSL-YFP mice, lungs images were acquired using Zeiss LSM 880 laser-scanning confocal microscope. Cell quantification was performed by counting manually the number of ECFP+ and EGFP+ (or YFP+) cells in each field. Between four and five fields were chosen from whole lung sections of at least three mice and separated according to their anatomical location. Tumor nodules located at the surface of the lung are considered in contact with the lung pleura, tumor nodules in contact with airways are easily defined by the presence of a large autofluorescent epithelial mono-layer, and tumor in the alveolar space represent all other nodules within the tumor parenchyma. All histological quantifications are presented as a mean or relative distribution of the different tumor nodules for each individual mouse.
In vivo TC-1-Luciferase cell luminescence
In vivo bioluminescence imaging was conducted on the In Vivo Imaging System spectrum (Perkin Elmer), using the Living Image acquisition and analysis software (Perkin Elmer). Prior to imaging, mice were injected with d-luciferin i.p. (150 mg/kg, 100 µl/mouse) and anesthetized with isoflurane. The radiance from the lung region was quantified with the same software.
Transcriptomic analysis
20 d after inoculation of TC-1 cells in MacBlue × Cx3cr1EGFP/+, CD11b+CD64+Ly6C−ECFP+EGFP+ MoD-TAMs, and CD11b+CD64+Ly6C−EGFP+ resident TAMs were sorted using FACSAria (Becton Dickinson) with a purity >95%. TAMs were gated as depicted on Fig. S2 A. The two subsets were separated according to ECFP and EGFP expression. Cells were sorted from four independent biological replicates, and total RNA was extracted using RNeasy Micro kit (QIAGEN) and quality was monitored with the 2100 Bioanalyzer (Agilent Technologies). Each biological replicate was hybridized to Affimetrix PICO V2 MouseGene2.OST microarrays. Analysis of gene-expression profiles was performed using Multi Experiment Viewer (MeV), which provides bioinformatics tools for integrative data analysis (Saeed et al., 2003, 2006). After validation of the RNA quality with Bioanalyzer 2100 (using Agilent RNA6000 nano chip kit), 2 ng of total RNA was reverse transcribed following the Ovation Pico System V2 (Nugen). In brief, the resulting double-strand cDNA is used for amplification based on SPIA technology. After purification according to Nugen protocol, 3.6 µg of Sens Target DNA were fragmented, and biotin was labeled using Encore Biotin Module kit (Nugen). After control of fragmentation using Bioanalyzer 2100, cDNA was then hybridized to GeneChip Mouse Gene 2.0 ST (Affymetrix) at 45°C for 17 h. After overnight hybridization, chips are washed on the fluidic station FS450 following specific protocols (Affymetrix) and scanned using the GCS3000 7G. The scanned images are then analyzed with Expression Console software (Affymetrix) to obtain raw data (cell files) and metrics for Quality Controls. Data were normalized using RMA algorithm in Bioconductor with the custom CDF versus 21. Statistical analyses were performed with the use of Partek GS. First, variations in gene expression were analyzed using unsupervised hierarchical clustering and PCA to assess data from technical bias and outlier samples. 25,429 genes have been identified in macrophage transcriptome. Student’s t test was applied on EGFP+ and ECFP+ macrophage transcriptomic data, and we obtained 604 differentially expressed genes between these two conditions with a P value of 0.05 based on t-distribution with overall α (critical P value) and the variance assumption was Welch approximation (unequal group variances). Hierarchical clustering was performed on significant genes only with gene tree and sample tree and then with optimized gene and sample leaf order. Pearson correlation was the distance metric used to perform hierarchical clustering, with complete linkage clustering. Data have been adjusted with median center genes/rows, median center samples/column, and to the end unlog2 data transformation was applied. Within the context of biological systems Ingenuity Pathway Analysis brings powerful analysis and allowed us to determine relevant bio functions and networks about the 604 differentially expressed genes.
Quantitative real-time PCR (qRT-PCR)
15 d after inoculation of TC-1 cells in Tnfrsf11aCre; Rosa26YFP-LSL, YFP+ TAMs and YFP− TAMs were sorted using FACSAria (Becton Dickinson). TAMs were gated as depicted on Fig. 3 B. 50,000 cells were directly sorted in 1 ml Trizol LS reagent (Thermo Fisher Scientific). RNA extraction was performed using Direct-zol RNA MicroPrep plus (Zymo Research), following manufacturer’s instructions. RNA concentration was measured with nanodrop2000. cDNA preparation was performed with Quantitect Reverse transcription kit (Qiagen) as per manufacturer’s instructions. qRT-PCR was performed on 10 ng cDNA. qRT523 PCR was performed on a Quant Studio 6 Flex using TaqMan Fast Advance Mastermix, and TaqMan probes (Thermo Fisher Scientific) were used for Gapdh (Mm99999915_g1), Ccl2 (Mm00441242_m1), Cxcl13 (Mm04214185_s1), Col14a1 (Mm00805269_m1), Mmp8 (Mm00439509_m1), and Csf1r (Mm01266652_m1) transcripts.
Adoptive transfer experiment
10 d after inoculation of TC-1 cells, mice were treated with isotype control or anti-VEGF every 2 d. On day 14 after tumor inoculation, 107 bone marrow cells isolated from MacBlue mice were adoptively transferred and the recovered cells were analyzed by FACS 24 h later.
Statistical analysis
All statistical analyses were performed with Graphpad Prism. Each sample values were first tested for Gaussian distribution by D’Agostino and Pearson omnibus normality test. Accordingly, multigroup analysis of variances were performed, and one-way or two-way ANOVA tests were followed by Bonferroni post tests for Gaussian distribution or Kruskal Wallis followed by Dunn’s multiple comparisons for non-Gaussian distribution. For simple comparison analysis, Student’s t test was performed to compare parametric distribution and Mann-Whitney for nonparametric distribution. For survival curves, Log-rank (Mantel-Cox) test was performed. For all pooled experiments, individual replicated were either statistically significant or showed the same trends on their own. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Online supplemental material
Fig. S1, related to Fig. 1, presents tSNE dimension 1 and 2 plots for each phenotypic marker and full gating strategy used for the study. Fig. S2, related to Fig. 2, presents additional information associated with the characterization of the MacBlue × Cx3cr1EGFP/+ model. Fig. S3, related to Fig. 4, presents additional characterization of the dynamic and distribution of macrophages subsets and the enriched function group associated with differentially regulated transcript. Fig. S4, related to Fig. 3, show the impact of CP on blood monocytes, lung myeloid populations, and survival in WT and Ccr2−/− mice. Fig. S5, related to Fig. 8, shows the expression of FLT1 in the different lung myeloid subsets and expression of VEGF in tumor nodules. Videos 1 and 2 show the dynamic interactions and protrusive activities of EGFP+ and ECFP+ cells within tumor nodules.
Supplementary Material
Acknowledgments
The authors wish to thank the Plateforme Imagerie Pitié-Salpêtrière for assistance with the two-photon microscope, the animal facility Nouvelle Animalerie Commune for mouse-breeding assistance and Angéline Duche and Sebastien Jacques from sequencing platform GENOM’IC (Institut Cochin, Paris, France). The authors also thank M. Lambert for the tdTomato viral construct.
P.-L. Loyher is funded by Fondation ARC pour la Recherche sur le Cancer and Memorial Sloan Kettering Cancer Center Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center fellowships. P. Hamon is funded by la Ligue Contre le Cancer. This work was supported by funding from the European Community's Seventh Framework Program (FP7/2007-2013) No. 304810, RAIDs, Institut National de la Santé et de la Recherche Médicale, Roche, la Ligue Contre le Cancer, Fondation pour la Recherche Médicale équipe labélisée, Fondation ARC pour la Recherche sur le Cancer (to A. Boissonnas), National Institutes of Health (NIH) National Cancer Institute (grant P30CA008748 to F. Geissmann), Memorial Sloan Kettering Cancer Center core grant, NIH/National Institute of Allergy and Infectious Diseases grant 1R01AI130345-01 and NIH/National Heart, Lung, and Blood Institute grant 1 R01HL138090-01 (to F. Geissmann).
A. Savina is employed by Roche. The authors declare no further competing financial interests.
Author contributions: P.-L.M. Loyher and P. Hamon designed, performed the experiments, analyzed, and interpreted the data and wrote the manuscript, A. Meghraoui-Kheddar, E. Goncalves, and B. Combadière analyzed and interpreted the data; M. Laviron, S. Torstensson, C.B. de Chanville, and Z. Deng performed some experiments and wrote the manuscript. N. Bercovici provided reagent and wrote the manuscript; F. Geissmann, A. Savina, and C. Combadière provided reagent and designed research. A. Boissonnas supervised the study, designed, performed the experiments, analyzed and interpreted the data, and wrote the manuscript.
References
- Amir A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31:545–552. 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barleon B., Sozzani S., Zhou D., Weich H.A., Mantovani A., and Marmé D.. 1996. Migration of human monocytes in response to vascular endothelial growth factor (VEGF) is mediated via the VEGF receptor flt-1. Blood. 87:3336–3343. [PubMed] [Google Scholar]
- Boissonnas A., Licata F., Poupel L., Jacquelin S., Fetler L., Krumeich S., Théry C., Amigorena S., and Combadière C.. 2013. CD8+ tumor-infiltrating T cells are trapped in the tumor-dendritic cell network. Neoplasia. 15:85–94. 10.1593/neo.121572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman R.L., Klemm F., Akkari L., Pyonteck S.M., Sevenich L., Quail D.F., Dhara S., Simpson K., Gardner E.E., Iacobuzio-Donahue C.A., et al. 2016. Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies. Cell Reports. 17:2445–2459. 10.1016/j.celrep.2016.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz M.L., Binnewies M., Boldajipour B., Nelson A.E., Pollack J.L., Erle D.J., Barczak A., Rosenblum M.D., Daud A., Barber D.L., et al. 2014. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 26:638–652. 10.1016/j.ccell.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.J., and Kotecha N.. 2014. Cytobank: providing an analytics platform for community cytometry data analysis and collaboration. Curr. Top. Microbiol. Immunol. 377:127–157. [DOI] [PubMed] [Google Scholar]
- Cohen M.H., Gootenberg J., Keegan P., and Pazdur R.. 2007. FDA drug approval summary: bevacizumab (Avastin) plus Carboplatin and Paclitaxel as first-line treatment of advanced/metastatic recurrent nonsquamous non-small cell lung cancer. Oncologist. 12:713–718. 10.1634/theoncologist.12-6-713 [DOI] [PubMed] [Google Scholar]
- Cortez-Retamozo V., Etzrodt M., Newton A., Rauch P.J., Chudnovskiy A., Berger C., Ryan R.J., Iwamoto Y., Marinelli B., Gorbatov R., et al. 2012. Origins of tumor-associated macrophages and neutrophils. Proc. Natl. Acad. Sci. USA. 109:2491–2496. 10.1073/pnas.1113744109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellapasqua S., Bertolini F., Bagnardi V., Campagnoli E., Scarano E., Torrisi R., Shaked Y., Mancuso P., Goldhirsch A., Rocca A., et al. 2008. Metronomic cyclophosphamide and capecitabine combined with bevacizumab in advanced breast cancer. J. Clin. Oncol. 26:4899–4905. 10.1200/JCO.2008.17.4789 [DOI] [PubMed] [Google Scholar]
- De Palma M., and Lewis C.E.. 2013. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 23:277–286. 10.1016/j.ccr.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Doherty T.A., Soroosh P., Khorram N., Fukuyama S., Rosenthal P., Cho J.Y., Norris P.S., Choi H., Scheu S., Pfeffer K., et al. 2011. The tumor necrosis factor family member LIGHT is a target for asthmatic airway remodeling. Nat. Med. 17:596–603. 10.1038/nm.2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt J.J., Boldajipour B., Beemiller P., Pandurangi P., Sorensen C., Werb Z., Egeblad M., and Krummel M.F.. 2012. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 21:402–417. 10.1016/j.ccr.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K., Pamer E.G., and Li M.O.. 2014. The cellular and molecular origin of tumor-associated macrophages. Science. 344:921–925. 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambardella L., Hemberger M., Hughes B., Zudaire E., Andrews S., and Vermeren S.. 2010. PI3K signaling through the dual GTPase-activating protein ARAP3 is essential for developmental angiogenesis. Sci. Signal. 3:ra76 10.1126/scisignal.2001026 [DOI] [PubMed] [Google Scholar]
- Gibbings S.L., Thomas S.M., Atif S.M., McCubbrey A.L., Desch A.N., Danhorn T., Leach S.M., Bratton D.L., Henson P.M., Janssen W.J., and Jakubzick C.V.. 2017. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. Am. J. Respir. Cell Mol. Biol. 57:66–76. 10.1165/rcmb.2016-0361OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F., and Rodewald H.R.. 2015. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 518:547–551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F., and Glass C.K.. 2014. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 159:1327–1340. 10.1016/j.cell.2014.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald M., Avraham I., Dor Y., Bachar-Lustig E., Itin A., Jung S., Chimenti S., Landsman L., Abramovitch R., and Keshet E.. 2006. VEGF-induced adult neovascularization: recruitment, retention, and role of accessory cells. Cell. 124:175–189. 10.1016/j.cell.2005.10.036 [DOI] [PubMed] [Google Scholar]
- Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., Deswarte K., Malissen B., Hammad H., and Lambrecht B.N.. 2013. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210:1977–1992. 10.1084/jem.20131199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon P., Loyher P.L., Baudesson de Chanville C., Licata F., Combadiere C., and Boissonnas A.. 2017. CX3CR1-dependent endothelial margination modulates Ly6Chigh monocyte systemic deployment upon inflammation in mice. Blood. 129:1296–1307. 10.1182/blood-2016-08-732164 [DOI] [PubMed] [Google Scholar]
- Hawley C.A., Rojo R., Raper A., Sauter K.A., Lisowski Z.M., Grabert K., Bain C.C., Davis G.M., Louwe P.A., Ostrowski M.C., et al. 2018. Csf1r-mApple Transgene Expression and Ligand Binding In Vivo Reveal Dynamics of CSF1R Expression within the Mononuclear Phagocyte System. J. Immunol. 200:2209–2223. 10.4049/jimmunol.1701488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R., Qian B.Z., Rowan C., Muthana M., Keklikoglou I., Olson O.C., Tazzyman S., Danson S., Addison C., Clemons M., et al. 2015. Perivascular M2 Macrophages Stimulate Tumor Relapse after Chemotherapy. Cancer Res. 75:3479–3491. 10.1158/0008-5472.CAN-14-3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin S., Licata F., Dorgham K., Hermand P., Poupel L., Guyon E., Deterre P., Hume D.A., Combadière C., and Boissonnas A.. 2013. CX3CR1 reduces Ly6Chigh-monocyte motility within and release from the bone marrow after chemotherapy in mice. Blood. 122:674–683. 10.1182/blood-2013-01-480749 [DOI] [PubMed] [Google Scholar]
- Ji H., Chang E.Y., Lin K.Y., Kurman R.J., Pardoll D.M., and Wu T.C.. 1998. Antigen-specific immunotherapy for murine lung metastatic tumors expressing human papillomavirus type 16 E7 oncoprotein. Int. J. Cancer. 78:41–45. [DOI] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R.N., Riba R.D., Zacharoulis S., Bramley A.H., Vincent L., Costa C., MacDonald D.D., Jin D.K., Shido K., Kerns S.A., et al. 2005. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 438:820–827. 10.1038/nature04186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Qian B.Z., Soong D., Cassetta L., Noy R., Sugano G., Kato Y., Li J., and Pollard J.W.. 2015. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Exp. Med. 212:1043–1059. 10.1084/jem.20141836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Doughty-Shenton D., Cassetta L., Fragkogianni S., Brownlie D., Kato Y., Carragher N., and Pollard J.W.. 2018. Monocytes Differentiate to Immune Suppressive Precursors of Metastasis-Associated Macrophages in Mouse Models of Metastatic Breast Cancer. Front. Immunol. 8:2004 10.3389/fimmu.2017.02004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., and Amit I.. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 159:1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C.E., Harney A.S., and Pollard J.W.. 2016. The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell. 30:365 10.1016/j.ccell.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Lin K.Y., Guarnieri F.G., Staveley-O’Carroll K.F., Levitsky H.I., August J.T., Pardoll D.M., and Wu T.C.. 1996. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 56:21–26. [PubMed] [Google Scholar]
- Loyher P.L., Rochefort J., Baudesson de Chanville C., Hamon P., Lescaille G., Bertolus C., Guillot-Delost M., Krummel M.F., Lemoine F.M., Combadière C., and Boissonnas A.. 2016. CCR2 Influences T Regulatory Cell Migration to Tumors and Serves as a Biomarker of Cyclophosphamide Sensitivity. Cancer Res. 76:6483–6494. 10.1158/0008-5472.CAN-16-0984 [DOI] [PubMed] [Google Scholar]
- Maeda K., Kobayashi Y., Udagawa N., Uehara S., Ishihara A., Mizoguchi T., Kikuchi Y., Takada I., Kato S., Kani S., et al. 2012. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat. Med. 18:405–412. 10.1038/nm.2653 [DOI] [PubMed] [Google Scholar]
- Mantovani A., and Allavena P.. 2015. The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 212:435–445. 10.1084/jem.20150295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E., Ballesteros I., Farlik M., Halbritter F., Günther P., Crozet L., Jacome-Galarza C.E., Händler K., Klughammer J., Kobayashi Y., et al. 2016. Specification of tissue-resident macrophages during organogenesis. Science. 353:aaf4238 10.1126/science.aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E., Jacome-Galarza C.E., Blank T., Lazarov T., Durham B.H., Ozkaya N., Pastore A., Schwabenland M., Chung Y.R., Rosenblum M.K., et al. 2017. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. 549:389–393. 10.1038/nature23672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin A.V., Morales-Nebreda L., Mutlu G.M., Budinger G.R., and Perlman H.. 2013. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol. 49:503–510. 10.1165/rcmb.2013-0086MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero A., and Glück S.. 2012. Long-Term Complete Remission with nab-Paclitaxel, Bevacizumab, and Gemcitabine Combination Therapy in a Patient with Triple-Negative Metastatic Breast Cancer. Case Rep. Oncol. 5:687–692. 10.1159/000346345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz G.T., and Coukos G.. 2011. The parallel lives of angiogenesis and immunosuppression: cancer and other tales. Nat. Rev. Immunol. 11:702–711. 10.1038/nri3064 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D.A., van Zuylen W.J., DeBats C.E., Alexander K.A., Kellie S., and Hume D.A.. 2008. Expression of Gal4-dependent transgenes in cells of the mononuclear phagocyte system labeled with enhanced cyan fluorescent protein using Csf1r-Gal4VP16/UAS-ECFP double-transgenic mice. J. Leukoc. Biol. 83:430–433. 10.1189/jlb.0807585 [DOI] [PubMed] [Google Scholar]
- Pollard J.W. 2004. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer. 4:71–78. 10.1038/nrc1256 [DOI] [PubMed] [Google Scholar]
- Qian B.Z., and Pollard J.W.. 2010. Macrophage diversity enhances tumor progression and metastasis. Cell. 141:39–51. 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R., Kaiser E.A., Snyder L.A., and Pollard J.W.. 2011. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 475:222–225. 10.1038/nature10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B.Z., Zhang H., Li J., He T., Yeo E.J., Soong D.Y., Carragher N.O., Munro A., Chang A., Bresnick A.R., et al. 2015. FLT1 signaling in metastasis-associated macrophages activates an inflammatory signature that promotes breast cancer metastasis. J. Exp. Med. 212:1433–1448. 10.1084/jem.20141555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero M.P., Poupel L., Loyher P.L., Hamon P., Licata F., Pessel C., Hume D.A., Combadière C., and Boissonnas A.. 2015. Immune surveillance of the lung by migrating tissue monocytes. eLife. 4:e07847 10.7554/eLife.07847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatel C., Radermecker C., Fievez L., Paulissen G., Chakarov S., Fernandes C., Olivier S., Toussaint M., Pirottin D., Xiao X., et al. 2017. Exposure to Bacterial CpG DNA Protects from Airway Allergic Inflammation by Expanding Regulatory Lung Interstitial Macrophages. Immunity. 46:457–473. 10.1016/j.immuni.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 34:374–378. [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., and Quackenbush J.. 2006. TM4 microarray software suite. Methods Enzymol. 411:134–193. 10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
- Sauter K.A., Pridans C., Sehgal A., Bain C.C., Scott C., Moffat L., Rojo R., Stutchfield B.M., Davies C.L., Donaldson D.S., et al. 2014. The MacBlue binary transgene (csf1r-gal4VP16/UAS-ECFP) provides a novel marker for visualisation of subsets of monocytes, macrophages and dendritic cells and responsiveness to CSF1 administration. PLoS One. 9:e105429 10.1371/journal.pone.0105429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., and Kopf M.. 2014. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15:1026–1037. 10.1038/ni.3005 [DOI] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Tamagnone L. 2012. Emerging role of semaphorins as major regulatory signals and potential therapeutic targets in cancer. Cancer Cell. 22:145–152. 10.1016/j.ccr.2012.06.031 [DOI] [PubMed] [Google Scholar]
- Tan S.Y., and Krasnow M.A.. 2016. Developmental origin of lung macrophage diversity. Development. 143:1318–1327. 10.1242/dev.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell B.C., and Whitsett J.A.. 2002. Gm-CSF regulates pulmonary surfactant homeostasis and alveolar macrophage-mediated innate host defense. Annu. Rev. Physiol. 64:775–802. 10.1146/annurev.physiol.64.090601.113847 [DOI] [PubMed] [Google Scholar]
- van de Laar L., Saelens W., De Prijck S., Martens L., Scott C.L., Van Isterdael G., Hoffmann E., Beyaert R., Saeys Y., Lambrecht B.N., and Guilliams M.. 2016. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity. 44:755–768. 10.1016/j.immuni.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Wyckoff J.B., Wang Y., Lin E.Y., Li J.F., Goswami S., Stanley E.R., Segall J.E., Pollard J.W., and Condeelis J.. 2007. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 67:2649–2656. 10.1158/0008-5472.CAN-06-1823 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Herndon J.M., Sojka D.K., Kim K.W., Knolhoff B.L., Zuo C., Cullinan D.R., Luo J., Bearden A.R., Lavine K.J., et al. 2017. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity. 47:597 10.1016/j.immuni.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.