Tanaka et al. show that Sox12, a member of SoxC family, is induced by TCR-NFAT signaling in T cells, binds to the Foxp3 promoter and drives its transcription, and induces the differentiation of T reg cells in the periphery during colitis.
Abstract
Peripherally induced regulatory T (pT reg) cells play indispensable roles in regulating gut inflammation; however, the mechanism underling the differentiation of pT reg cells under inflammatory conditions remains largely unknown. Here, we show that the expression of Sox12, a member of SoxC family, is significantly induced in T reg cells in colitic mice. We also show that TCR–NFAT signaling induces Sox12 expression in CD4+ T cells. Although Sox12 is not required for the development of thymus-derived T reg (tT reg) cells, Sox12 is involved in the development of pT reg cells under inflammatory conditions in an adoptive transfer colitis model. Moreover, we found that enforced expression of Sox12 is sufficient to promote Foxp3 expression in CD4+ T cells even in the absence of TGF-β or IL-2 and that Sox12 binds to Foxp3 promoter and drives its transcription. These results suggest that TCR-NFAT signaling induces the development of pT reg cells in colitic mice partly through Sox12 induction.
Introduction
Regulatory T (T reg) cells, defined by the expression of Foxp3, have a central role in the protection against excessive inflammatory responses caused by infections or autoimmune diseases. T reg cells are also important for the maintenance of immune tolerance in gut where trillions of microbes and food antigens are present (Barnes and Powrie, 2009; Tanoue et al., 2016). Among T reg cells, thymus-derived T reg (tT reg) cells have an indispensable role in maintaining immune tolerance to self-antigens. However, peripherally induced T reg (pT reg) cells, which arise from Foxp3 induction during T cell differentiation in the periphery and generate the majority of gut T reg population (Ai et al., 2014), play indispensable roles in commensal microbiota composition and the suppression of mucosal allergic inflammation (Josefowicz et al., 2012b). In autoimmune colitis models, pT reg cells, in concert with tT reg cells, have been shown to act to restore immune tolerance (Haribhai et al., 2009). These findings suggest that pT reg cells have critical roles in the suppression of gut inflammation.
Regarding the mechanisms underlying the induction of Foxp3 during T cell differentiation in the periphery, strong TCR signaling with suboptimal costimulation, TGF-β, IL-2, retinoic acid, and microbial metabolites have been shown to induce the development of pT reg cells both in vivo and in vitro (Bilate and Lafaille, 2012; Josefowicz et al., 2012a; Arpaia et al., 2013; Furusawa et al., 2013). Among the downstream pathways of these signals, Smad3 and NFAT binding to conserved noncoding sequence 1 (CNS1) of Foxp3 gene plays a central role in Foxp3 induction (Josefowicz et al., 2012a). However, Levine et al. have demonstrated that TCR signaling is dispensable for the maintenance of Foxp3 expression in T reg cells (Levine et al., 2014). Another group reported that TCR stimulation leads to down-regulation of Foxp3 by degrading FOXO1 protein (Bothur et al., 2015). In this context, the roles of TCR–NFAT signaling in Foxp3 induction are still debatable.
Recently, it has been shown that T reg cells acquire an activated phenotype, enhance suppressive activity, and increase their populations under inflammatory conditions (Rosenblum et al., 2011; Shafiani et al., 2013). Regarding the mechanism underlying the maintenance of activated T reg cells, Arvey et al. have shown that Foxp3 poises its targets genes by inducing repressive chromatin formation under inflammatory conditions (Arvey et al., 2014). Although this study has uncovered the importance of Foxp3-dependent transcriptional program in activated T reg cells under inflammatory conditions, specific transcription factors which regulate Foxp3 expression under inflammatory conditions remain unknown.
We show here that Sox12, a member of SoxC family, is a transcription factor whose expression is significantly enhanced in T reg cells in dextran sulfate sodium (DSS)–induced colitis and that TCR–NFAT signaling induces Sox12 expression. We also show that although Sox12 is not required for the development of tT reg cells, Sox12 is involved in the development of pT reg cells in colitis induced by T cell transfer. Moreover, Sox12 binds to the promoter of Foxp3 gene and drives its transcription. Our data uncover a novel mechanism underlying the development of pT reg cells in colitic mice.
Results and discussion
Sox12 expression is induced in T reg cells by TCR–NFAT signaling
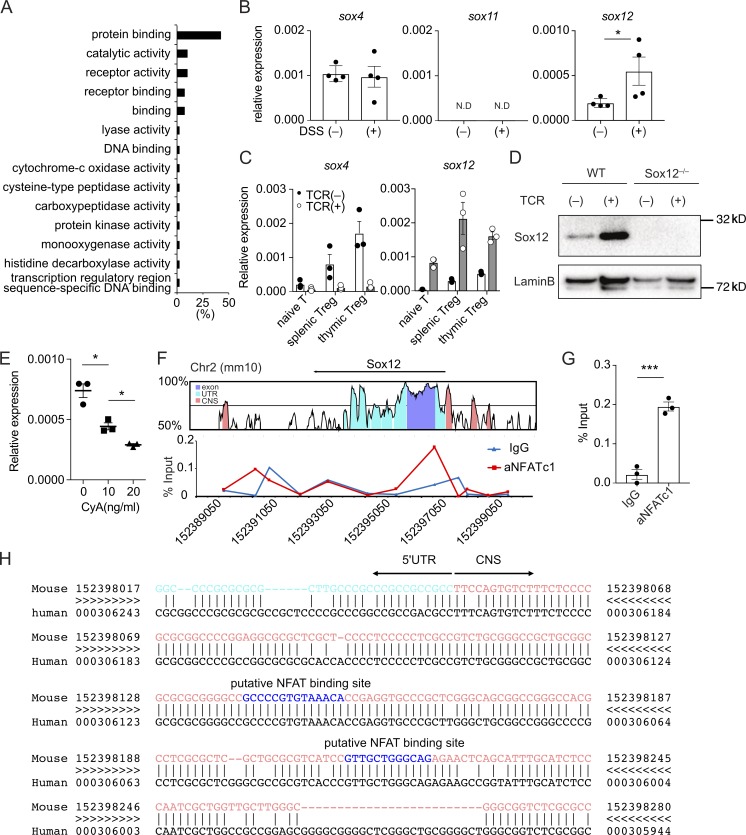
To identify genes specifically expressed in T reg cells under inflammatory conditions, we first analyzed gene expression profiles of splenic T reg cells, which were isolated as hCD2+ CD4+ cells from Foxp3-reporter Foxp3hCD2 mice, with or without the induction of DSS-induced colitis. RNA sequencing identified 56 protein-coding genes whose expression were altered more than twofold by DSS treatment (Fig. 1 A). Among the 56 genes, we focused on Sex determining region Y box 12 (Sox12) because Sox12 is the only transcription factor. Sox12 is a member of SoxC family composed of Sox4, Sox11, and Sox12, which have a high mobility group (HMG) box domain in N-terminal region and a transactivation domain in C-terminal region (Dy et al., 2008). Previous studies have revealed that SoxC family proteins play important roles in the development of the heart, nerve system, kidney, and pancreas (Penzo-Méndez, 2010). Regarding immunological aspects, while Sox4 has been shown to be involved in the differentiation of Th2 cells and survival of B cell precursor (Kuwahara et al., 2012; Sun et al., 2013; Mallampati et al., 2014), the roles of Sox11 and Sox12 have not been reported yet.
Figure 1.
T cell receptor signaling induces Sox12 expression in T reg cells. (A) Gene ontology of 56 differentially expressed transcripts in splenic T reg cells from Foxp3hCD2 mice treated with or without 3% DSS water (more than twofold). (B) Quantitative PCR analysis of SoxC family genes in splenic T reg cells in mice treated with or without DSS. Data are compiled of four independent experiments. (C) Sox4 and Sox12 expression in naive CD4+ T cells, splenic T reg cells, and thymic T reg cells cultured with or without anti-CD3ε/CD28 (TCR) stimulation for 24 h. Data are compiled from three independent experiments. (D) Nuclear proteins of WT or Sox12−/− CD4+ T cells stimulated with or without TCR for 24 h were immunoblotted with antibodies against Sox12 and LaminB1. Data are representative of three independent experiments. (E) Naive CD4+ T cells were stimulated with TCR in the presence of indicated amounts of cyclosporin A (0–20 µg/ml) for 24 h, and Sox12 expression was assessed by qPCR. Data are compiled from three independent experiments. (F and G) Naive CD4+ T cells were stimulated with TCR for 4 h. ChIP-qPCR assay for Sox12 gene locus was performed with anti-NFATc1 antibody or control mouse IgG. Shown are VISTA plot of Sox12 gene locus (GRCm38/mm10, Chr2) and representative NFATc1 binding plot to Sox12 regulatory region (F) and means ± SEM of percent input of NFATc1 or control mouse IgG binding to the promoter of Sox12 gene (G). (H) Shown are 5′UTR (light blue), upstream CNS sequences (pink), and putative NFAT binding sequences (blue) of Sox12 gene analyzed by rVISTA. (C, E, and G) *, P < 0.05; ***, P < 0.001 by unpaired t test.
We therefore examined the expression of SoxC family members in T reg cells. As shown in Fig. 1 B, Sox4 and Sox12 but not Sox11 were expressed in splenic T reg cells at steady state. Importantly, the expression levels of Sox12 were significantly elevated in splenic T reg cells isolated from DSS colitis-induced mice as compared with those from control mice, whereas the expression levels of Sox4 were similar between these cells (Fig. 1 B). Since Sox4 and Sox12 were expressed in T reg cells in colitis-induced mice, we next analyzed the effect of TCR stimulation on the expression of Sox4 and Sox12 in naive CD4+ T cells, thymic T reg cells, and splenic T reg cells. As shown in Fig. 1 C, both Sox4 and Sox12 were highly expressed in T reg cells as compared with naive CD4+ T cells in steady-state conditions. Interestingly, TCR stimulation increased Sox12 expression but decreased Sox4 expression in T reg cells (Fig. 1, C and D). These results suggest that the expression of Sox4 and Sox12 is differently regulated in T reg cells and that Sox4 and Sox12 may have a distinct role in T reg cells.
To determine the mechanism underlying TCR signaling-mediated induction of Sox12, we examined the effect of cyclosporin A (CsA), an inhibitor of calcineurin-mediated NFAT activation, on Sox12 expression in CD4+ T cells and found that CsA inhibited Sox12 expression in a dose-dependent manner (Fig. 1 E). Chromatin immunoprecipitation (ChIP) assay revealed that NFATc1 bound to the upstream of exon 1 of Sox12 gene locus where CNS is located (Fig. 1, F and G). Moreover, we found that putative NFAT binding sequences were located in this region (Fig. 1 H). These results suggest that Sox12 is induced by TCR-mediated NFAT activation in CD4+ T cells.
Sox12−/− naive CD4+ T cells fail to convert into pT reg cells in colitic mice
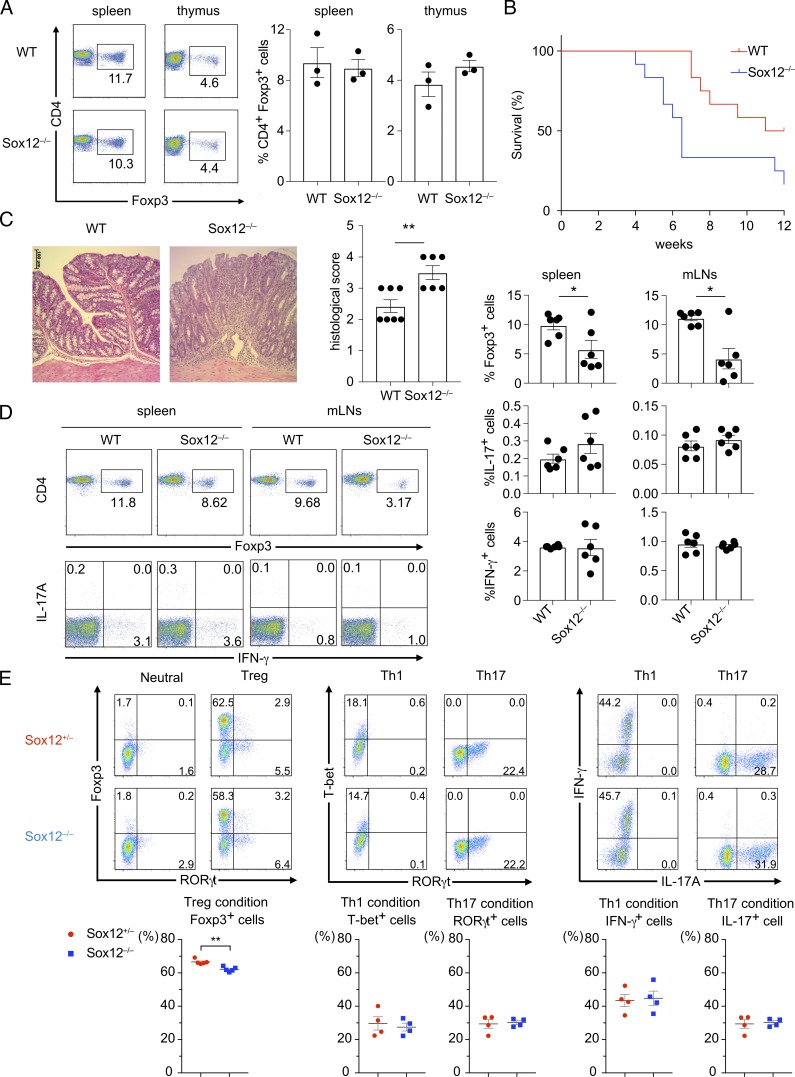
To examine whether Sox12 is involved in T reg cell differentiation, we next analyzed the development of T reg cells in Sox12−/− mice. The development of T cells in the thymus (Fig. S1, A and B) and the percentage of CD4+ Foxp3+ cells in thymus and spleen (Fig. 2 A) were similar between Sox12−/− mice and littermate WT mice at steady state. The frequency of CD4+ Foxp3+ cells in lamina propria of large intestine was also similar between Sox12−/− mice and littermate WT mice at the age of 3 and 6 wk (Fig. S1 C). Moreover, the frequency of Helios+ cells in colonic T reg cells were similar between Sox12−/− mice and WT mice (Fig. S1 C), suggesting that Sox12 does not affect T reg differentiation at steady state.
Figure 2.
Sox12−/− naive CD4+ T cells fail to convert into pT reg cells in adoptive transfer colitis. (A) Representative FACS profiles of CD4 versus Foxp3 of CD4+ T cells harvested from spleen and thymus of WT or Sox12−/− mice are shown (left). Bar plots indicate the percent of Foxp3+ cells (right). (B–E) Naive CD4+ T cells from WT or Sox12−/− mice were transferred to RAG2−/− mice to induce adoptive transfer colitis. (B) Survival plot is depicted. Data are compiled from four independent experiments (two to three mice in each group in each experiment). (C) Shown are representative photomicrographs of H&E staining at 14 d after cell transfer (left) and means ± SEM of histopathological scores (right). Bar, 100 µm. (D) CD4+ T cells were harvested from spleen and mLNs at 14 d after cell transfer and stimulated with PMA/ionomycin for 4 h. Shown are representative FACS profiles of CD4 versus Foxp3 and IFN-γ versus IL-17A and means ± SEM of the percentages of the indicated cells. Data are compiled from three independent experiments (six mice in each group). (E) Naive CD4+ T cells from Sox12−/− mice and littermate Sox12+/− mice were stimulated under neutral, iT reg, Th1, and Th17 conditions. Representative FACS profiles of indicated transcription factors and cytokines (upper panels) and means ± SEM of the percentages of the indicated cells are shown. Data are compiled from four independent experiments (n = 4 or 5). (C, D, and E) *, P < 0.05; **, P < 0.01 by unpaired t test.
Given that Sox12 expression was elevated in splenic T reg cells in DSS colitis-induced mice, we hypothesized that Sox12 is associated with the differentiation of pT reg cells in colitic mice. To test this hypothesis, we used an adoptive transfer colitis model that mimics human inflammatory bowel diseases and is frequently used for studying T reg cell function (Ai et al., 2014). When naive CD25− CD4+ T cells isolated from Sox12−/− mice or littermate WT mice were injected intraperitoneally to RAG2−/− mice, the mice transplanted with Sox12−/− naive CD4+ T cells exhibited shorter survival rate than mice transplanted with WT naive CD4+ T cells (Fig. 2 B). Histopathologic examination of the colon revealed severe colitis in mice transplanted with Sox12−/− naive CD4+ T cells (Fig. 2 C). In addition, the frequency of Foxp3+ CD4+ cells in spleen and mesenteric lymph nodes (mLNs) was significantly reduced in mice transplanted with Sox12−/− naive CD4+ T cells as compared with those transplanted with WT naive CD4+ T cells (Fig. 2 D). On the other hand, the numbers of IL-17A+ CD4+ T cells and IFN-γ+ CD4+ T cells in spleen and mLNs were comparable between mice transplanted with Sox12−/− naive CD4+ T cells and those transplanted with WT naive CD4+ T cells (Fig. 2 D).
We next analyzed in vitro differentiation of T cells in Sox12−/− CD4+ T cells (Fig. 2 E). Under iT reg conditions, although Foxp3 expression was reproducibly decreased in Sox12−/− CD4+ T cells as compared with that in control Sox12+/− CD4+ T cells (Fig. 2 E), the levels of decrease were modest. In addition, in vitro differentiation of Th1 cells under Th1 conditions and that of Th17 cells under Th17 conditions were similar between Sox12−/− CD4+ T cells and Sox12+/− CD4+ T cells (Fig. 2 E). The strongly impaired T reg cell differentiation in colitic mice (Fig. 2 D) with the modest impairment of in vitro T reg differentiation (Fig. 2 E) suggests that the environmental factor(s) in colitic mice may play critical roles in Sox12-mediated induction of pT reg cells.
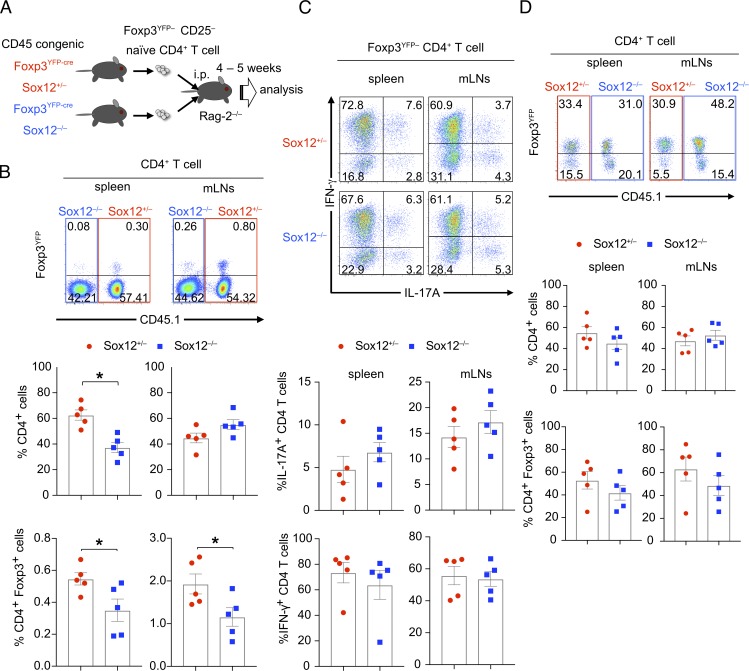
To examine whether Sox12 induces the expression of Foxp3 in a cell-intrinsic manner in vivo, we next performed a mixed cell transfer experiment. In this experiment, purified Foxp3YFP− CD25− naive CD4+ T cells isolated from Foxp3YFP-cre Sox12−/− mice and congenically marked (CD45) littermate Foxp3YFP-cre Sox12+/− mice were mixed in a 1:1 ratio, injected intraperitoneally to RAG2−/− mice, and analyzed 4–5 wk after the transfer (Fig. 3 A). As shown in Fig. 3 B, the frequency of Sox12−/− CD4+ T cells was lower in spleen, but tended to be higher in mLNs (flow cytometry profiles and upper box plots), suggesting that deficiency of Sox12 might alter tissue distribution of effector CD4+ T cells. Importantly, the frequencies of Foxp3YFP+ cells in spleen and mLNs were significantly decreased in Sox12−/− CD4+ T cells as compared with Sox12+/− CD4+ T cells (Fig. 3 B, flow cytometry profiles and bottom box plots). On the other hand, the frequencies of IL-17A+ CD4+ T cells and IFN-γ+ CD4+ T cells in Foxp3YFP− cells were similar between Sox12−/− CD4+ T cells and Sox12+/− CD4+ T cells (Fig. 3 C).
Figure 3.
Intrinsic expression of Sox12 is involved in the development of pT reg cells. (A–C) Foxp3YFP− CD25− CD62Lhi CD44lo CD4+ T cells isolated from CD45 congenic Foxp3YFP-cre Sox12−/− mice and littermate Foxp3YFP-cre Sox12+/− mice were mixed in a 1:1 ratio and injected intraperitoneally to RAG2−/− mice. 4–5 wk after the transfer, spleen and mLNs were analyzed. (B) Representative FACS profiles of CD45.1 versus Foxp3YFP of CD4+ T cells from spleen and mLNs (upper panels), the percentages of Sox12−/− CD4+ T cells and Sox12+/− CD4+ T cells among CD3+ CD4+ cells (middle panels), and the percentages of Foxp3YFP+ CD4+ T cells in Sox12−/− CD4+ T cells and Sox12+/− CD4+ T cells (lower panels) are shown. Data are compiled from three independent experiments (n = 5). *, P < 0.05 by unpaired t test. (C) Cells from spleen and mLNs were stimulated with PMA/ionomycin for 4 h. Shown are representative FACS profiles of IL-17A versus IFN-γ of Sox12+/− Foxp3YFP− CD4+ T cells and Sox12−/− Foxp3YFP− CD4+ T cells and means ± SEM of the percentages of indicated cells. (D) Foxp3YFP+ CD25+ CD4+ T cells isolated from Foxp3YFP-cre Sox12−/− mice and congenically marked littermate Foxp3YFP-cre Sox12+/− mice were mixed and injected intraperitoneally to RAG2−/− mice. Representative FACS profiles of CD45.1 versus Foxp3YFP of CD4+ T cells in spleen and mLNs at 2 wk after the transfer, frequencies of Sox12−/− CD4+ T cells and Sox12+/− CD4+ T cells in CD3+ CD4+ cells (middle panels), and frequencies of Foxp3YFP+ cells in Sox12−/− CD4+ T cells and Sox12+/− CD4+ T cells (bottom panels) are shown. Data are compiled from four independent experiments (n = 5).
To examine the role of Sox12 in the stability of T reg cells in vivo, purified Foxp3YFP+ CD25+ T reg cells from Foxp3YFP-cre Sox12−/− mice and those from congenically marked littermate Foxp3YFP-cre Sox12+/− mice were cotransferred to RAG2−/− mice, and the fate of these cells was examined 2 wk after the transfer. The number of CD4+ T cells as well as the frequency of Foxp3+ cells in CD4+ T cells was not significantly different between Sox12−/− cells and Sox12+/− cells (Fig. 3 D), suggesting that Sox12 does not significantly affect the stability and cell survival of T reg cells. Collectively, these results suggest that Sox12 is cell-intrinsically involved in the development of pT reg cells, although it is still possible that Sox12 is also involved in the function of pT reg cells.
Sox12 increases T reg cells by inducing Foxp3
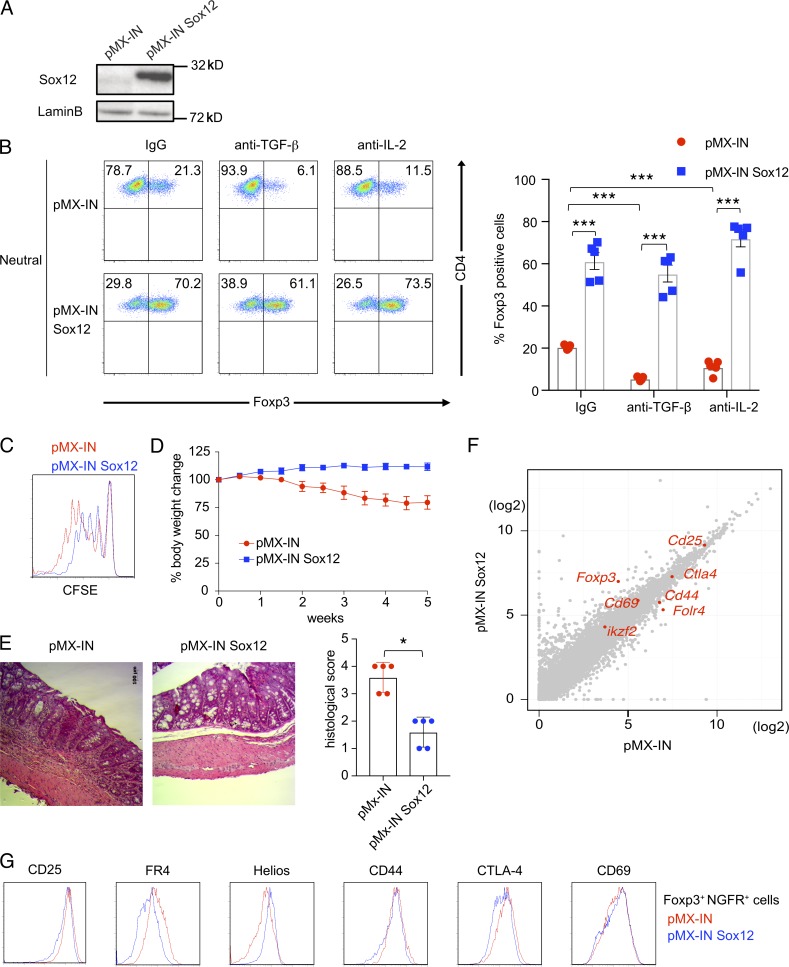
We next examined the effect of Sox12 on Foxp3 expression in CD4+ T cells by using pMX-IN-Sox12 retrovirus (Sox12 retrovirus) which induces forced expression of Sox12 protein (Fig. 4 A). When sorted Foxp3YFP− naive CD4+ T cells from Foxp3YFP-cre mice were infected with Sox12 retrovirus under neutral conditions, the numbers of Foxp3+ cells were significantly increased as compared with those infected with control retrovirus (Fig. 4 B). Sox12-mediated Foxp3 induction was still observed even when TGF-β or IL-2, important cytokines for pT reg cell differentiation (Josefowicz et al., 2012a), was neutralized by antibodies (Fig. 4 B), suggesting again that Sox12 intrinsically induces Foxp3 expression in CD4+ T cells.
Figure 4.
Sox12 induces the expression of Foxp3. (A) Naive CD4+ T cells were infected with retroviruses of pMX-IN-Sox12 or pMX-IN (as a control). 2 d after the infection, infected cells were isolated and subjected to immunoblotting with indicated antibodies. (B) Foxp3YFP− naive CD4+ T cells were stimulated with TCR under neutral conditions and infected with retroviruses of either pMX-IN-Sox12 or pMX-IN in the presence of anti–TGF-β, anti–IL-2, or control IgG, and the expression of Foxp3 was evaluated. Shown are representative FACS profiles (left) and means ± SEM of the percentages of Foxp3+ CD4+ T cells (right) compiled from four independent experiments. ***, P < 0.001 by one-way ANOVA followed by Dunnett’s test. (C) Naive CD4+ T cells were stimulated with TCR and infected with retroviruses of either pMX-IN-Sox12 or pMX-IN for 24 h. Cells were then stimulated with TCR for additional 2 d. Infected human NGFR+ cells were sorted and co-cultured with CFSE-labeled responder naive CD4+ T cells. Shown is a representative CFSE fluorescence intensity 3 d after co-culture. (D and E) Naive CD4+ cells were transferred to RAG2−/− mice to develop adoptive transfer colitis. Where indicated, pMX-IN-Sox12–infected CD4+ T cells or pMX-IN–infected CD4+ T cells (as a control) were cotransferred to the mice. (D) Shown are changes in body weight after cell transfer. Data are compiled from three independent experiments (two to three mice in each group in each experiment). (E) Shown are representative photomicrographs of colons and means ± SEM of histopathological scores. *, P < 0.05 by unpaired t test. Bar, 100 µm. (F) A scatter plot of RNA-Seq (log2 RPKM) for all coding transcripts in pMX-IN-Sox12–infected CD4+ T cells versus pMX-IN–infected CD4+ T cells. Representative T reg–associated transcripts are shown in red. (G) Representative histograms of T reg–associated molecules in Foxp3+ CD4+ NGFR+ cells in WT CD4+ T cells infected with pMX-IN-Sox12 (blue) or pMX-IN (red) viruses.
We also examined whether the enforced expression of Sox12 enhances suppressive activity in CD4+ T cells. As shown in Fig. 4 C, when CD4+ T cells infected with Sox12 retrovirus were co-cultured with CFSE-labeled naive CD4+ T cells, these cells suppressed the proliferation of responder cells more strongly than control retrovirus-infected CD4+ T cells. In addition, when CD4+ T cells infected with Sox12 retrovirus were cotransferred with naive CD4+ T cells to RAG2−/− mice, these cells significantly reduced the severity of adoptive transfer colitis as compared with control retrovirus-infected CD4+ T cells (Fig. 4, D and E). These results indicate that Sox12 induces suppressive ability in CD4+ T cells.
Sox12 induces Foxp3 expression by binding the promoter
To address the mechanism underlying Sox12-mediated induction of T reg cells, we next compared the transcriptomes of CD4+ T cells infected with Sox12 retrovirus and those with control retrovirus by RNA sequencing. Some of enriched gene ontology (GO) terms by the forced expression of Sox12 are related to immune systems, although the majority of enriched GO terms are related to cell proliferation, cell death, and organ development (Fig. S2), consistent with the fact that SoxC family members are involved in the organ development. Among several T reg–associated transcripts including Cd25, Cd44, Cd69, Ctla4, Folr4 (encoding FR4), Foxp3, and Ikzf2 (encoding Helios), Foxp3 was the most differentially expressed gene in CD4+ T cells with Sox12 induction (Fig. 4 F and Table S1). Consistently, flow cytometric analysis showed that, although some of T reg–associated molecules were differentially expressed between Sox12-expressing CD4+ Foxp3+ T cells and control CD4+ Foxp3+ T cells at 4 d after the induction, the difference was modest (Fig. 4 G), suggesting that Foxp3 is likely to be a primary target of Sox12.
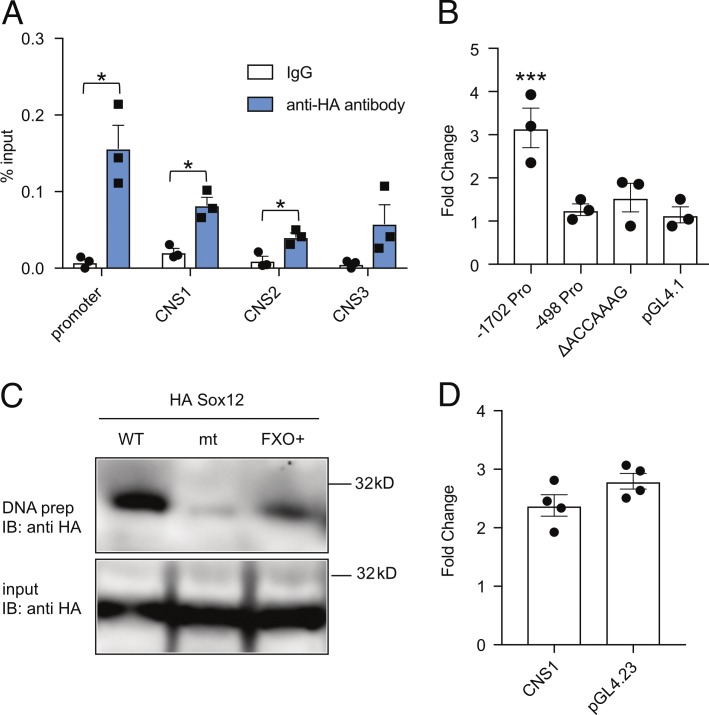
We next performed ChIP–quantitative PCR (qPCR) assay to test whether Sox12 directly binds to the regulatory elements of Foxp3 gene. Because no anti-Sox12 antibody suitable for ChIP is available, CD4+ T cells were retrovirally transduced to express HA-tagged Sox12, and the Sox12 binding was evaluated by using anti-HA antibody. As shown in Fig. 5 A, Sox12 strongly bound to the promoter region and weakly bound to CNS1 and CNS2 of Foxp3 gene. Luciferase reporter assay revealed that Sox12 activated −1702 bp Foxp3 promoter, but not −498 bp Foxp3 promoter (Fig. 5 B), suggesting that −1702 to −499 bp region of Foxp3 promoter is important for Sox12-mediated Foxp3 promoter activation. We also found that a putative Sox binding sequence (ACCAAAG) is located in −1702 to −499 bp region of Foxp3 promoter and that Sox12 did not activate −1702 bp Foxp3 promoter lacking the putative Sox12 binding sequence (ΔACCAAAG; Fig. 5 B). DNA precipitation assay confirmed that Sox12 bound to WT DNA probe that contains the putative Sox binding sequence but not to its mutant (Fig. 5 C). We also assessed whether Sox12 enhances CNS1 activity because CNS1 has been demonstrated to be critical for induced T reg cell differentiation (Josefowicz et al., 2012b). As shown in Fig. 5 D, Sox12 did not significantly enhance the reporter activity of minimal promoter plus CNS1 construct as compared with that of minimal promoter construct (pGL4.23). Collectively, these results suggest that Sox12 induces Foxp3 expression by directly binding to the Foxp3 promoter and then induces pT reg cell differentiation.
Figure 5.
Sox12 binds to and activates the promoter of Foxp3. (A) Naive CD4+ T cells were infected with retrovirus of pMX-IN-HA-Sox12, and cells were restimulated with TCR for additional 12 h. ChIP-qPCR assay for the promoter and CNSs of Foxp3 gene locus was performed with anti-HA antibody or control rabbit IgG. Data are compiled from three independent experiments. (B) Luciferase assay with indicated Foxp3 promoter Luci constructs in EL4 cells. Data are compiled from three independent experiments. (C) DNA precipitation assays of HA-Sox12 with biotinylated double-stranded DNA probes containing a portion of Foxp3 promoter or its mutant at Sox12-binding site (ΔACCAAAG) were performed. FXO+ probe was used as a positive control. Shown are representative of three independent experiments. (D) Luciferase assays with pGL4.23-Foxp3 CNS1 enhancer-Luci construct or pGL4.23 (as a control) were performed in EL4 cells. Data are compiled from four independent experiments. (A and B) *, P < 0.05; ***, P < 0.001 by unpaired t test.
Concluding remarks
We have shown that Sox12 promotes pT reg cell differentiation in colitic mice. Moreover, Sox12 could induce Foxp3 expression in CD4+ T cells even in the absence of TGF-β and IL-2. Given that antigens and inflammatory cytokines are abundant at the inflammatory site and these factors preferentially induce effector T cells, Sox12-mediated pT reg cell differentiation in the inflamed gut seems to be important to prevent excessive inflammatory responses. Although further studies are needed, our data provide new insights into the mechanisms underlying pT reg cell differentiation under inflammatory conditions to prevent autoimmune diseases such as inflammatory bowel diseases.
Materials and methods
Mice
BALB/c mice and C57BL/6 mice were purchased from Charles River Laboratories. RAG2−/− mice and Foxp3YFP-cre mice were purchased from Jackson Laboratory and crossed to CD45.1 congenic mice. Foxp3hCD2 mice (Miyao et al., 2012) were gifts from S. Hori (Institute of Physical and Chemical Research, Kanagawa, Japan). Sox12−/− mice (Bhattaram et al., 2010) were backcrossed onto C57BL/6 mice for eight generations. To generate Foxp3YFP-cre Sox12−/− mice and congenically marked littermate Foxp3YFP-cre Sox12+/− mice, Foxp3YFP-cre Sox12+/− mice (CD45.1+/CD45.2+) were crossed to Foxp3YFP-cre Sox12−/− mice (CD45.2+/CD45.2+). All mice were housed in microisolator cages under specific pathogen–free conditions. Chiba University Animal Care and Use Committee approved protocols of animal experiments.
Reagents
Antibodies to CD3ε (145-2C11), CD28 (37.51), CD11c (HL3), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3), CTLA4 (UC10-4B9), IL-4 (11B11), IL-4 (BDV4-1D11), and IFN-γ (XMG1.2), human NGFR (C40-1457), and human CD2 (RPA-2.10) were purchased from BD Biosciences. Antibodies to CD8α (53–6.7), CD16/32 (93), CD25 (PC61 and 7D4), B220 (RA3-6B2), CD45.1 (A20), CD45.2 (104), FR4 (12A5), IL-6 (MP5-20F3), and IL-17A (TC11-18H10.1) were purchased from BioLegend. Antibodies to Foxp3 (FJK-16s) and Helios (22F6) were purchased from eBioscience. Anti-TGF-β mAb (1D11) and anti–IL-2 mAb (JES6-1A12) were purchased from R&D Systems. Cyclosporin A was purchased from Sigma-Aldrich.
Plasmids
pMX-IRES-NGFR (pMX-IN) is a retrovirus vector expressing human NGFR under the regulation of an internal ribosome entry site. pMX-IN-Sox12 is a gift from M. Yamashita (Ehime University, Ehime, Japan). HA tag was fused to Sox12 by PCR amplification, and HA-Sox12 was subcloned into pMX-IN to create pMX-IN-HA-Sox12. Sox12 and HA-Sox12 were also subcloned into pcDNA3 (Invitrogen). pGL4.1 and pGL4.23 vectors were purchased from Promega Biotech Inc. pGL4.1 vector containing Foxp3 promoter fragment (−1702 to +174 bp; −1702 bp Foxp3 promoter vector) and Foxp3 CNS1 enhancer fragment (+1988 to +2738) were gifts from Y. Tone (Tone et al., 2008). CNS1 enhancer fragment was subcloned into pGL4.23 vector. Mutations were introduced on luciferase reporter vectors with a KOD plus mutagenesis kit (Toyobo). All constructs were verified by sequencing.
Cell isolation and analysis
CD62L+ CD25− TCRγδ− CD4+ T cells (naive CD4+ T cells) were isolated from lymph nodes or spleen by using a CD4+ CD62L+ T cell isolation kit II (Miltenyi Biotec) according to the manufacturer’s instruction. The resultant cells were >95% pure CD4+ CD25− CD62L+ T cells by flow cytometric analysis. Flow cytometric analyses were performed on a FACSCalibur (BD Biosciences) or a FACSCantoII (BD Biosciences) with FlowJo software (Tree Star).
Cell culture
Naive CD4+ T cells were stimulated with anti-CD3ε mAb (5 µg/ml) in the presence of irradiated splenocyte as antigen presenting cells under neutral conditions (anti–IL-4 mAb [10 µg/ml] and anti–IFN-γ mAb [10 µg/ml]), iT reg conditions (TGF-β [3 ng/ml], anti–IL-6 mAb [10 µg/ml], anti–IL-4 mAb, and anti–IFN-γ mAb), Th1 conditions (IL-12 [10 ng/ml], IL-2 [10 ng/ml], anti–IL-6 mAb, and anti–IL-4 mAb), and Th17 conditions (TGF-β [3 ng/ml], IL-6 [10 ng/ml], anti–IL-4 mAb, and anti–IFN-γ mAb).
DSS-induced colitis
Foxp3hCD2 mice (6–8 wk old) were given 3% (wt/vol) DSS solution (MP Biomedicals) orally for 7 d. hCD2+ cells were purified from spleens with a SH800 cell sorter (SONY).
Adoptive transfer colitis
6–8-wk-old RAG2−/− mice were injected intraperitoneally with CD25− CD4+ cells (5 × 105 cells/mouse) from WT or Sox12−/− mice and the mice were weighed twice per week. Moribund mice were euthanized based on Institutional Animal Care and Use Committee protocol. Histopathological scoring was performed as described previously (Asseman et al., 1999) in a blinded fashion.
In vivo development of pT reg cells in a mixed cell transfer experiment
CD45 congenic Foxp3YFP− CD25− CD62Lhi CD44lo CD4+ T cells were isolated from Foxp3YFP-cre Sox12−/− mice and congenically marked littermate Foxp3YFP-cre Sox12+/− mice by using a SH800 cell sorter. The resultant cells were >99% pure Foxp3YFP− CD25− CD62Lhi CD44lo CD4+ T cells by flow cytometric analysis. Isolated cells (3 × 105 cells, each) were mixed in a 1:1 ratio and injected intraperitoneally into CD45.2+/CD45.2+ background RAG2−/− mice. 4–5 wk after the transfer, cells from spleen and mLNs were analyzed.
Stability of T reg cells in a mixed cell transfer experiment
Foxp3YFP+ CD25+ CD4+ T cells were isolated from Foxp3YFP-cre Sox12−/− mice and congenically marked littermate Foxp3YFP-cre Sox12+/− mice by using a SH800 cell sorter. The resultant cells were >99% pure Foxp3YFP+ CD25+ CD4+ T cells by flow cytometry. Isolated cells (1.25 × 105 cells, each) were mixed in a 1:1 ratio and injected intraperitoneally into CD45.2+/CD45.2+ background RAG2−/− mice. 2 wk after the transfer, cells from spleen and mLNs were analyzed.
Suppression assay
Human NGFR+ cells were sorted by a SH800 cell sorter and suppression assay was performed as described previously (Kawashima et al., 2013).
Retrovirus-mediated gene expression
Retrovirus-mediated gene induction for naive CD4+ T cells was performed by a RetroNectin-bound virus infection method (Takara Bio) as described previously (Suto et al., 2008). In brief, Foxp3YFP− naive CD4+ T cells isolated from Foxp3YFP-cre WT mice were stimulated with anti-CD3/CD28 antibodies under neutral conditions for 24 h and infected with retroviruses of either pMX-IN-Sox12 or pMX-IN (as a control) in the presence of anti-TGF-β, anti-IL-2, or control IgG (10 µg/ml) for 24 h. Cells were then stimulated with anti-CD3/CD28 antibodies for another 4 d in the presence of antibodies.
For the analysis of the transcriptome of CD4+ T cells infected with Sox12 retrovirus, naive CD4+ T cells were stimulated with anti-CD3/CD28 antibodies under neutral conditions for 24 h, infected with retroviruses of either pMX-IN-Sox12 or pMX-IN (as a control) for 24 h, and then stimulated with anti-CD3/CD28 antibodies for 24 h. Infected human NGFR-positive cells were positively purified by PE-conjugated anti-NGFR antibody and anti-PE microbeads (Miltenyi Biotec) and then dead cells were removed by Lympholyte-M (Cedarlane Laboratories) according to the manufacturer’s instructions.
Real-time PCR analysis
Extraction of total cellular RNA, reverse transcription, and qPCR analysis were performed with a StepOnePlus real-time PCR system (Applied Biosystems) as described previously (Suto et al., 2008).
PCR primers and probes are as follows: Sox4 forward, 5′-CGGCTGCATCGTTCTCTCC-3′; Sox4 reverse, 5′-GGTAGACGTGCTTCACTTTCTTG-3′; Sox11 forward, 5′-CGACGACCTCATGTTCGACC-3′; Sox11 reverse, 5′-GACAGGGATAGGTTCCCCG-3′; Sox12 forward, 5′-CCCGAGGTTACCGAGATGATC-3′; Sox12 reverse, 5′-GCTGACGGTGGGCTCAGTAG-3′; Sox12 probe, FAM-5′-ACTGGCGCTCGTCTAGTATCGCCGA-3′-TAMRA; β-actin forward, 5′-GCTCTGGCTCCTAGCACCAT-3′; β-actin reverse, 5′-GCCACCGATCCACACAGAGT-3′; β-actin probe, FAM-5′-GTCAAGATCATTGCTCCTCCTGAGCGC-3′-TAMRA. The levels of target genes were normalized to the levels of β-actin.
RNA-seq analysis
Total RNA was prepared using a PureLink RNA mini kit (Invitrogen). RNA-seq libraries were prepared using a SureSelect Strand Specific RNA Library Preparation kit (Agilent). Sequencing was performed on an Illumina HiSeq1500 using a TruSeq Rapid SBS kit (Illumina) in a 50-base single-read mode. mRNA profiles were calculated with Cufflinks software and expressed as RPKM (reads per kilobase of exon model per million mapped fragments). For Fig. 1 A, protein-coding genes whose expression was more than 1 RPKM in either one or both conditions were selected for the analysis.
ChIP–qPCR analysis
ChIP was performed using a ChIP assay kit (Millipore) and Dynabeads Protein G (Invitrogen) as described previously (Hiramatsu et al., 2010). Anti-NFATc1 antibody (7A6; Abcam) and anti-HA antibody (ab9110; Abcam) were used for ChIP assays. DNA contents were measured by qPCR with a SYBR green reagent. Data were expressed as the percent input for each ChIP fraction.
Sequences of qPCR primers for ChIP-qPCR analyses are as follows: primer set 1 (mm10, chr2, start position: 152390018); forward, 5′-TGTAGATCATGCTGGCCTTGAA-3′; reverse, 5′-CAGCACATGCCTTTGATCACA-3′; primer set 2 (mm10, chr2, start position: 152391151); forward, 5′-TCAAGACCCTTCCTACCTGTCAGT-3′; reverse, 5′-AGGAATGGCTGACCCAGAAA-3′; primer set 3 (mm10, chr2, start position: 152391643); forward, 5′-CAGGCCTCAATCTTCCAACTG-3′; reverse, 5′-GTCCCTATGAGCTACCATGTTGAA-3′; primer set 4 (mm10, chr2, start position: 152392737); forward, 5′-CTCCACGCTAGTCAGGGCTTA-3′; reverse, 5′-TTGTTTTTGTCTAATGGACTATCCTTTG-3′; primer set 5 (mm10, chr2, start position: 152393720); forward, 5′-GGAGTCTGGTGGAAGAAATGCT-3′; reverse, 5′-CCTGCCCCAGTGTCTAAGCT-3′; primer set 6 (mm10, chr2, start position: 152395142); forward, 5′-AGAGAAATAGAGAAAGAGCGCAAGA-3′; reverse, 5′-CCAGCTGCCTCCTTTAAAGCT-3′; primer set 7 (mm10, chr2, start position: 152396149); forward, 5′-CCTTTCGGAGTTGGCAACAG-3′; reverse, 5′-GCATGCTTGATGCTTCTTGGA-3′; primer set 8 (mm10, chr2, start position: 152397512); forward, 5′-GCCACTGGTCCATGATTTTTCT-3′; reverse, 5′-CACATTAAGCGGCCGATGA-3′; primer set 9 (mm10, chr2, start position: 152398365); forward, 5′-GCGCTGCCCATCTGTTCT-3′; reverse, 5′-GCGGCAGGTGTCATTGAATA-3′; primer set 10 (mm10, chr2, start position: 152398675); forward, 5′-CGCCTGCTTTCCAAAGACA-3′; reverse, 5′-CAAGCTAGGACCGCCAGACT-3′; primer set 11 (mm10, chr2, start position: 152399419); forward, 5′-CCTGGGTGGTTCTTTGTTCAG-3′; reverse, 5′-TGCACTTGAAGATAAGCAAAAGGA-3′; primer set 12 (mm10, chr2, start position: 152400126); forward, 5′-AACCTGTCTCCAACCATTTTTTG-3′; reverse, 5′-CGGTTCAGAATGGTCCTTAAGG-3′.
Luciferase reporter assay
EL4 cells (5 × 105 cells) were resuspended in resuspension buffer R containing indicated pcDNA3 vectors (pcDNA3-Sox12 or empty pcDNA3) and pGL4 vectors with pRL-TK and electroporated by using Neon transfection system (Invitrogen). Luciferase reporter assay was performed as previously described (Tanaka et al., 2014).
Western blot
Western blot was performed as described previously (Hiramatsu et al., 2010) by using anti-Sox12 antibody (B01P; Abnova) and anti-Lamin B1 antibody (Ab16048; Abcam).
DNA precipitation assay
293T cells were transfected with pcDNA3-HA-Sox12 and DNA precipitation assay was performed as described previously (Tanaka et al., 2014).
Data analysis
Data are summarized as means ± SEM. The statistical analysis of the results was performed by either the unpaired Student’s t test or ANOVA followed by Dunnett’s test. P < 0.05 was considered significant.
Data availability
The accession no. for RNA-seq datasets in this paper is GSE115732.
Online supplemental material
Fig. S1 shows development of thymocytes and colonic T reg cells in Sox12−/− mice. Fig. S2 shows GO terms of transcripts altered by the infection of Sox12 retrovirus. Table S1 shows differentially expressed genes between CD4+ T cells infected with Sox12 retrovirus and those with mock retrovirus.
Supplementary Material
Acknowledgments
We thank Dr. S. Hori for Foxp3hCD2 mice, Dr. M. Yamashita for pMX-IN-Sox12, Dr. Y. Tone for –1702 Foxp3 promoter reporter and CNS1 fragment (+1988 to +2738), and Ms. J. Iwata for technical help.
This work was supported in part by Grants-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, the Japanese government, Japan Society for the Promotion of Science (JSPS) KAKENHI grant numbers JP23591432 and JP26461461, Leading Graduate School at Chiba University (LGS) Program, and Institute for Global Prominent Research, Chiba University, Japan.
The authors declare no competing financial interests.
Author contributions: S. Tanaka and A. Suto designed and performed experiments. T. Iwamoto, T. Kageyama, T. Tamachi, H. Takatori, K. Suzuki, K. Hirose, O. Ohara, and H. Nakajima analyzed and interpreted the data. S. Tanaka, A. Suto, and H. Nakajima wrote the manuscript. V. Lefebvre provided resources and interpreted the data.
References
- Ai T.L., Solomon B.D., and Hsieh C.S.. 2014. T-cell selection and intestinal homeostasis. Immunol. Rev. 259:60–74. 10.1111/imr.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., Liu H., Cross J.R., Pfeffer K., Coffer P.J., and Rudensky A.Y.. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 504:451–455. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A., van der Veeken J., Samstein R.M., Feng Y., Stamatoyannopoulos J.A., and Rudensky A.Y.. 2014. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 15:580–587. 10.1038/ni.2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseman C., Mauze S., Leach M.W., Coffman R.L., and Powrie F.. 1999. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J. Exp. Med. 190:995–1004. 10.1084/jem.190.7.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes M.J., and Powrie F.. 2009. Regulatory T cells reinforce intestinal homeostasis. Immunity. 31:401–411. 10.1016/j.immuni.2009.08.011 [DOI] [PubMed] [Google Scholar]
- Bhattaram P., Penzo-Méndez A., Sock E., Colmenares C., Kaneko K.J., Vassilev A., Depamphilis M.L., Wegner M., and Lefebvre V.. 2010. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 1:9 10.1038/ncomms1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilate A.M., and Lafaille J.J.. 2012. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu. Rev. Immunol. 30:733–758. 10.1146/annurev-immunol-020711-075043 [DOI] [PubMed] [Google Scholar]
- Bothur E., Raifer H., Haftmann C., Stittrich A.B., Brüstle A., Brenner D., Bollig N., Bieringer M., Kang C.H., Reinhard K., et al. 2015. Antigen receptor-mediated depletion of FOXP3 in induced regulatory T-lymphocytes via PTPN2 and FOXO1. Nat. Commun. 6:8576 10.1038/ncomms9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy P., Penzo-Méndez A., Wang H., Pedraza C.E., Macklin W.B., and Lefebvre V.. 2008. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 36:3101–3117. 10.1093/nar/gkn162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., et al. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 504:446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- Haribhai D., Lin W., Edwards B., Ziegelbauer J., Salzman N.H., Carlson M.R., Li S.H., Simpson P.M., Chatila T.A., and Williams C.B.. 2009. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J. Immunol. 182:3461–3468. 10.4049/jimmunol.0802535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu Y., Suto A., Kashiwakuma D., Kanari H., Kagami S., Ikeda K., Hirose K., Watanabe N., Grusby M.J., Iwamoto I., and Nakajima H.. 2010. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. J. Leukoc. Biol. 87:703–712. 10.1189/jlb.0909639 [DOI] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., and Rudensky A.Y.. 2012a Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Niec R.E., Kim H.Y., Treuting P., Chinen T., Zheng Y., Umetsu D.T., and Rudensky A.Y.. 2012b Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 482:395–399. 10.1038/nature10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima H., Takatori H., Suzuki K., Iwata A., Yokota M., Suto A., Minamino T., Hirose K., and Nakajima H.. 2013. Tumor suppressor p53 inhibits systemic autoimmune diseases by inducing regulatory T cells. J. Immunol. 191:3614–3623. 10.4049/jimmunol.1300509 [DOI] [PubMed] [Google Scholar]
- Kuwahara M., Yamashita M., Shinoda K., Tofukuji S., Onodera A., Shinnakasu R., Motohashi S., Hosokawa H., Tumes D., Iwamura C., et al. 2012. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat. Immunol. 13:778–786. 10.1038/ni.2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.G., Arvey A., Jin W., and Rudensky A.Y.. 2014. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15:1070–1078. 10.1038/ni.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallampati S., Sun B., Lu Y., Ma H., Gong Y., Wang D., Lee J.S., Lin K., and Sun X.. 2014. Integrated genetic approaches identify the molecular mechanisms of Sox4 in early B-cell development: intricate roles for RAG1/2 and CK1ε. Blood. 123:4064–4076. 10.1182/blood-2013-12-543801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao T., Floess S., Setoguchi R., Luche H., Fehling H.J., Waldmann H., Huehn J., and Hori S.. 2012. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 36:262–275. 10.1016/j.immuni.2011.12.012 [DOI] [PubMed] [Google Scholar]
- Penzo-Méndez A.I. 2010. Critical roles for SoxC transcription factors in development and cancer. Int. J. Biochem. Cell Biol. 42:425–428. 10.1016/j.biocel.2009.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum M.D., Gratz I.K., Paw J.S., Lee K., Marshak-Rothstein A., and Abbas A.K.. 2011. Response to self antigen imprints regulatory memory in tissues. Nature. 480:538–542. 10.1038/nature10664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiani S., Dinh C., Ertelt J.M., Moguche A.O., Siddiqui I., Smigiel K.S., Sharma P., Campbell D.J., Way S.S., and Urdahl K.B.. 2013. Pathogen-specific Treg cells expand early during mycobacterium tuberculosis infection but are later eliminated in response to Interleukin-12. Immunity. 38:1261–1270. 10.1016/j.immuni.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Mallampati S., Gong Y., Wang D., Lefebvre V., and Sun X.. 2013. Sox4 is required for the survival of pro-B cells. J. Immunol. 190:2080–2089. 10.4049/jimmunol.1202736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto A., Kashiwakuma D., Kagami S., Hirose K., Watanabe N., Yokote K., Saito Y., Nakayama T., Grusby M.J., Iwamoto I., and Nakajima H.. 2008. Development and characterization of IL-21-producing CD4+ T cells. J. Exp. Med. 205:1369–1379. 10.1084/jem.20072057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Suto A., Iwamoto T., Kashiwakuma D., Kagami S., Suzuki K., Takatori H., Tamachi T., Hirose K., Onodera A., et al. 2014. Sox5 and c-Maf cooperatively induce Th17 cell differentiation via RORγt induction as downstream targets of Stat3. J. Exp. Med. 211:1857–1874. 10.1084/jem.20130791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T., Atarashi K., and Honda K.. 2016. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 16:295–309. 10.1038/nri.2016.36 [DOI] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., and Tone M.. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202. 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession no. for RNA-seq datasets in this paper is GSE115732.