Abstract
Cancer is the most serious disease worldwide, and ovarian cancer (OvCa) is the second most common type of gynecological cancer. There is consequently an urgent need for early-stage detection of OvCa, which requires affinity reagent biomarkers for OvCa. Systematic evolution of ligands by exponential enrichment (SELEX) and phage display technology are two powerful technologies for identifying affinity reagent biomarkers. However, the benchtop protocols for both screening technologies are relatively lengthy and require well-trained personnel. We therefore developed a novel, integrated microfluidic system capable of automating SELEX and phage display technology. Instead of using cancer cell lines, it is the first work which used tissue slides as screening targets, which possess more complicated and uncovered information for affinity reagents to recognize. This allowed for the identification of aptamer (nucleic acid) and peptide probes specific to OvCa cells and tissues. Furthermore, this developed system could be readily modified to uncover affinity reagents for diagnostics or even target therapy of other cancer cell types in the future.
I. INTRODUCTION
Cancer is responsible for tens of millions of deaths each year on a global scale.1 Although clinicians have made considerable strides in cancer diagnostics in recent years, much work remains to be done in terms of identifying small quantities of abnormally growing/dividing cells.2 The second most common type of gynecological cancer, ovarian cancer (OvCa), affects about 200,000 women each year worldwide.2,3 Unlike the deadlier, though easier to diagnose, cervical cancer, OvCa is difficult to detect in its earliest stages; biomarkers for OvCa diagnosis are therefore urgently needed since early diagnosis is generally associated with better prognosis.4,5 However, finding a biomarker specific to a single cancer still remains difficult.
Most cancer biomarkers are identified by comparing protein or gene expression profiles between normal and cancer cells.6 However, even if a specific cancer biomarker is identified, a suitable affinity reagent or probe must be designed to target, isolate, and detect it. In clinical diagnostics, antibodies are the most commonly used biomarker probes. However, antibodies are produced from the immune systems of experimental animals; they cannot be raised against toxins and small molecules that do not elicit an immune response. There are several in vitro screening techniques that can be used to identify single chain antibodies (scFvs);7 for instance, phage display technology is a subtractive proteomic technique based on (1) the cloning of foreign DNA into a filamentous phage and (2) the consequent presentation of recombinant peptide variants fused to the outer surface proteins of the phages.5,8
As an example of another biomarker identification approach, systematic evolution of ligands by exponential enrichment (SELEX) has been commonly used to select for aptamers (small, single-stranded oligonucleotides) specific to target molecules,9 and cell-SELEX can be used to bind cancer cells directly.9–11 In the latter technique, a random single-stranded DNA (ssDNA) library is used to select aptamers that (1) are specific to cancer cells (“positive selection”) and (2) lack affinity toward healthy cells (“negative selection”). Both phage display technology and SELEX require well-trained personnel and are error-prone even with such highly experienced technicians.
Microfluidic systems have been developed in recent years to screen for affinity reagents for cancer cells or biomarkers, allowing for rapid, point-of-care diagnostics.12,13 Integrated microfluidic systems have rapidly advanced and miniaturized molecular diagnostics. Amongst other advantages, they are cheaper to operate than their large-scale counterparts due to their small reaction volumes. Integrated microfluidic systems with the capacity for performing cell-SELEX workflow were first developed in 2013,14 and such systems have further been designed to target lung cancer cells, cancer stem-like cells, multiple OvCa cell lines, and even colorectal cancer cells.14–16 Specifically, microfluidic systems comprising microfluidic control modules, magnetic bead-based aptamer extraction modules, and temperature control modules have been automated to perform the entire cell-SELEX process on a single chip over the course of only three days (in comparison to one week for benchtop protocols).17 Phage display-based microfluidic systems can identify multiple protein targets simultaneously,18 and they have been used to screen for affinity reagents that bind OvCa cells.19 For instance, a micromixer was used to increase the chance for a target protein to interact with a phage within the incubation chamber, and a micropump was used to control the transport of reagents and samples. Finally, a micro-cell culture module for Escherichia coli (E. coli) was used to clonally amplify bound phages.18 With this approach, the integrated microfluidic chip could automatically perform one round of biopanning, including both positive and negative control steps, in as short as 5–6 h; this represents a significant improvement over conventional phage display (2–3 days). This chip has been further used to identify affinity probes for colon cancer cells.20 However, these studies were only focused on cultured cancer cell lines and not those cancer cells or tissues taken from human biopsies. Therefore, we developed a microfluidic platform herein for the first time capable of automatically performing both tissue-SELEX and phage display technology with OvCa tissues in order to identify cancer biomarker probes by using tissue slides. This represents the first integrated microfluidic system featuring two screening processes on one chip, which is beneficial to prepare tissue slides with a standard operation protocol suitable for microfluidic-based screening and therefore simplify future applications (such as high-throughput screening and commercialization of this platform) by using similar operation procedures. Furthermore, studying molecules interaction in different types of affinity reagents based on consistent assay systems could also avoid the bias caused by different systems. Therefore, this platform could be useful for the identification of not only OvCa biomarkers but also those diagnostic of other human cancers.
II. MATERIALS AND METHODS
A. Experimental procedures
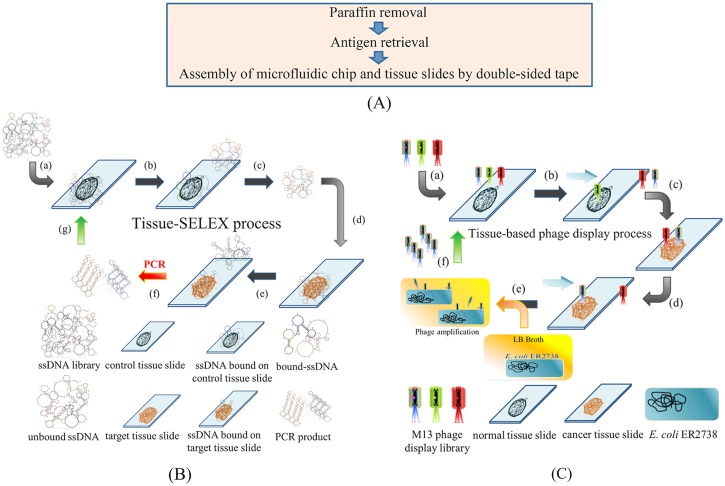
An overview of the experimental procedures on the developed system is schematically shown in Fig. 1.21 In brief, the first part is to prepare tissue slides which were covered by a paraffin layer using a 65 °C oven and organic solvents (xylene and ethanol). Then, the antigens on the tissue samples were retrieved by 95 °C citrate buffer. After antigen retrieval, paraffin-covered tissue slides were treated to expose the tissues to the reaction components, including the antigens [Fig. 1(a)]. The tissue slides and the integrated microfluidic chip were then assembled by a laser-engraved double-sided tape. Afterwards, tissue slides were attached to the microfluidic chip with a double-sided tape. In the tissue-SELEX step [Fig. 1(b)], the ssDNA library was incubated with normal ovarian tissues (“negative selection” step), which made un-wanted ssDNA bound with the normal ovarian tissue and potential aptamer candidates would suspend in buffer. In the next “positive selection” step, the ssDNA that did not bind to the normal tissues were incubated with cancer tissues, which made aptamer candidates bound with the cancer tissue and the unbound ssDNA were washed away. During the washing step, the applied gauge pressure was gradually decreased in each successive screening round from −39.9 kPa to −53.3 kPa and then to −66.6 kPa (i.e., to increase the shear force) to wash away unbound ssDNA. The washing step was repeated for six times, and the wash wastes were collected for further analysis. In the fourth step, the bound ssDNA were released by heat treatment at 95 °C for 5 min and then amplified by on-chip polymerase chain reaction (PCR). The PCR products (double-stranded DNA, dsDNA) were then used in the next round of tissue-SELEX screening after the standard ssDNA re-generation process.
FIG. 1.
Working principles of the tissue-based cancer biomarker screening process. (a) The paraffin layer was removed, and tissue antigens were boiled in citrate buffer. After antigen retrieval, tissue slides were attached to the microfluidic system and they were assembled with a laser-engraved, double-sided tape. (b) The tissue-SELEX process: (a) the ssDNA library was first incubated with normal ovarian tissues (negative selection); (b) unbound ssDNA were separated, and [(c) and (d)] incubated with ovarian cancer tissues (positive selection). (e) Aptamer candidates bound to cancer tissues, and unbound ssDNA were washed away. (f) The bound aptamer candidates were PCR amplified, and (g) PCR products were used in the next round of tissue-SELEX. (c) In the phage display process, (a) an M13 phage library was incubated with normal tissues (negative selection). [(b) and (c)] Then, unbound phages were incubated with cancer tissues (positive selection), and (d) unbound phases were washed away. (e) Positive phages were amplified in E. coli. (f) These amplified phage candidates were used in the next round of phage display.
In the phage display step [Fig. 1(c)], an M13 phage library was incubated with normal tissues (“negative selection” step), and then the unbound M13 phages were incubated with cancer tissues (“positive selection” step). In the washing step, the applied gauge pressure was modified as described above, and the wash wastes were collected for further analysis. The major difference between tissue-SELEX and phage display process was the amplification steps. For the phage display process, the amplification step was achieved by infection to its host cell, E. coli (strain R2738). E. coli cells were transported to incubate with target M13 phages for phage amplification. The amplified phage candidates were then used in the next round of phage display. The detailed experimental procedures of tissue-SELEX and phage display can be found in Tables SI and SII in the supplementary material, respectively. Note that all experimental steps, including incubation, transport, and washing, were automated by the integrated microfluidic system. However, the buffer, ssDNA library (or PCR products for all but the first round of tissue-SELEX), and E. coli were loaded manually prior to the screening process.15
B. Chip design and fabrication
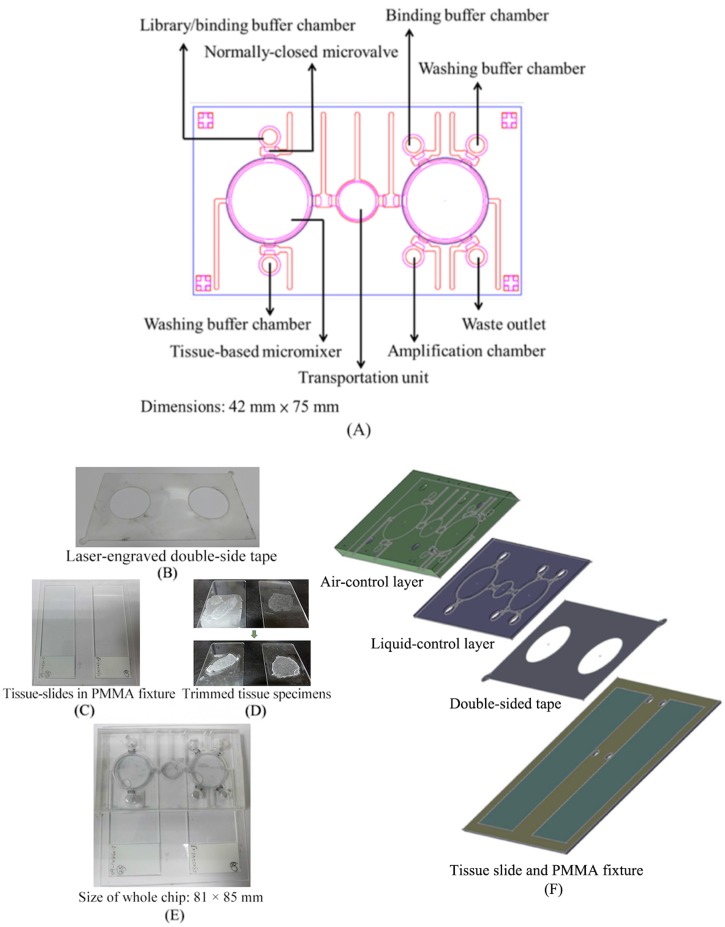
The microfluidic platform was composed of two polydimethylsiloxane (PDMS, Sylgad 184A/B, Dow Corning, USA) layers, one double-sided tape layer, and a layer containing the polymethylmethacrylate (PMMA) glass slide fixture attached to two clinical tissue slides. PMMA master molds were engraved by a computer-numerical-control (CNC) machine to produce the two PDMS layers.15 Then, the two PDMS layers were bonded together by oxygen plasma treatment. Finally, the PDMS layers were bonded with the tissue slides with the laser-engraved double-sided tape [Fig. 2(b)]. The microfluidic platform [Fig. 2(a)] was equipped with two tissue-based micromixers,22 four reagent loading chambers (for the ssDNA library, washing buffer, and binding buffer), normally-closed microvalves,23 a waste outlet, and one amplification chamber (for PCR in the tissue-SELEX module and E. coli growth for the phage display module). A custom-made, dual temperature control module was used, which was composed of two Peltier cooling/heating units that independently regulated the heating and cooling rates.
FIG. 2.
Schematic layout of the integrated microfluidic chip. (a) The microfluidic chip was equipped with two tissue-based micromixers, one transport unit, two binding buffer chambers, two washing buffer chambers, one amplification chamber, normally-closed microvalves, and one waste outlet. (b) The laser-engraved, double-sided tape was used to bind the microfluidic system with the tissue slides. (c) A tissue slide fixture was designed to hold the tissue slides. (d) Trimmed tissues were suitable for analysis. (e) The final assembled microfluidic system. (f) An exposed view of the chip.
C. Clinical tissue slide treatment, antigen retrieval, and histological staining
All clinical tissue slides were provided by the Department of Obstetrics and Gynecology of the National Cheng Kung University Hospital (NCKUH), and their dissemination was approved by the Institutional Review Board (IRB) of NCKUH (#A-ER-104-014). First, paraffin-covered tissue slides were heated at 65 °C to melt the paraffin. Then, the slides were incubated in 1× phosphate buffered saline (PBS; Sigma-Aldrich, USA) followed by xylene (Sigma-Aldrich) for 5 min, 99.8% ethanol for 3 min (Sigma-Aldrich; repeated twice in separate tanks), 90% ethanol for 3 min (repeated twice in separate tanks), 80% ethanol for 3 min (repeated twice in separate tanks), and 70% ethanol for 3 min (repeated twice in separate tanks). The tissue slides were then returned to tanks containing 1× PBS. Then, the tissue slides were immersed in 1× citrate buffer (Sigma-Aldrich) and incubated at 95 °C for 10 min for the antigen retrieval step [Fig. 1(a)].17,24
A modified hematoxylin and eosin (HE) stain was performed in a fume hood using the following sequence of solutions:24 tap water for 5 min, Gill's hematoxylin (Muto Pure Chemicals, Japan) for 5 min, tap water for 5 min, 95% ethanol for 15 s (twice in different tanks), eosin Y solution (Muto Pure Chemicals) for 1 min, 95% ethanol for 15 s (twice in different tanks), 95% ethanol for 15 s (twice in different tanks), 100% ethanol for 15 s (thrice in different tanks), and xylene for 1 min (thrice in different tanks). Coverslips were then placed on the slides. Tissue slides were stained, examined for signs of cancer, and compared with the results obtained from the carboxyfluorescein (FAM)-labeled affinity reagents.
D. ssDNA library, PCR, real-time PCR, and cloning
Molecules of the ssDNA library each contained a central region of 40 random nucleotides flanked by two 16-base-pair (bp) sequences that functioned as primer binding sites for the subsequent PCR (a total of 72 bp; 5′-GGCAGGAAGACAAACA-N40-GGTCTGTGGTGCTGT-3′). The ssDNA library was synthesized (Medclub Scientific Inc., Taiwan) and purified to a final concentration of 1 μM. A washing buffer containing 1 l of Dulbecco's phosphate-buffered saline (DPBS; Invitrogen, USA), 4.5 g of glucose, and 5 ml of 1M MgCl2 was stored at 4 °C prior to use. The binding buffer containing 1 l of DPBS, 4.5 g of glucose, 100 mg of transfer RNA, 1 g of bovine serum albumin (BSA), and 5 ml of 1M MgCl2 was used to maintain the structure of ssDNA library.10 Each selection round required 600 μl of washing buffer and 600 μl of binding buffer for incubation of the ssDNA with the tissue samples.
After three rounds of on-chip in vitro screening for tissue-SELEX or phage display, the final screened pools were amplified by PCR for confirmation. The PCRs comprised 2 μl of either the screened ssDNA pool or screened phage pool (0.1 μM), forward primers (0.5 μl, 5′-GGCAGGAAGACAAACA-3′; 0.5 μM), reverse primers (0.5 μl, 5′-ACAGCACCACAGACCA-3′; 0.5 μM), deoxynucleotide triphosphates (dNTPs, 0.5 μl, 0.2 mM), 0.125 μl of Taq DNA polymerase (New England Biolabs, USA), and double-distilled water (ddH2O) to a total volume of 25 μl. All primers used herein were synthesized by Mission Biotech, Inc. (Taiwan). The PCR was initiated with a 10-min denaturation at 94 °C, followed by 94 °C (denaturation) for 30 s, 63 °C (annealing) for 15 s, and 72 °C (extension) for 30 s for 20 cycles; a final extension step was performed at 72 °C for 10 min. The PCR products were heated to 95 °C to denature the dsDNA and then cooled rapidly to maintain the denatured DNA in the single-stranded form.
PCR products after three screening rounds were further purified and cloned using the TOPO® TA cloning kit (Invitrogen). Twenty colonies from tissue-SELEX and phage display assays were randomly selected and purified with a plasmid miniprep kit (Invitrogen). These 40 colonies were then sequenced and their folding structure and free energy were analyzed.15 Two aptamers and two peptides (Table I) were synthesized (Medclub Scientific) and used for the tissue fluorescence staining and cell lysate binding experiments to confirm their binding specificities and affinities to the target cancer tissue
TABLE I.
Aptamer and peptide sequences identified from clinical ovarian cancer tissue slides and their dissociation constants. Error terms represent standard deviation (n = 3).
| Molecule | Aptamer (5′-3′) or peptide sequence (N-C) | Kd (nM) |
|---|---|---|
| Aptamer (72 base pairs) | ||
| Tx-01 | ACAGCACCACAGACCA TCAAATTACGGAAAATCATGACGGGGTGGAACCGAGGGGG TGTTTGTCTTCCTGCC |
53.8 ± 14.9 |
| Tx-02 | ACAGCACCACAGACCA GCAAACAGCTCTGAGACGAATTCCATGTGAAAACATTCGG TGTTTGTCTTCCTGCC |
2.9 ± 0.8 |
| Peptide (12 amino acids) | ||
| Tp-01 | KRLTPLSPDHYD | 4.8 ± 1.0 |
| Tp-02 | QLKLHNKGLISS | 3.2 ± 1.5 |
E. M13 phage library, phage amplification, plaque formation assay, and phage sequencing
The M13 phage display library kit (Ph.D.™-12, New England Biolabs, USA, Cat. E8111L) was used to screen specific peptides against OvCa tissues. The library contained 1013 pfu/ml of M13 phages containing 12-mer random peptide sequences inserted into the N-terminal region of the minor coat protein (cpIII). There were three rounds of both positive and negative panning involved for the entire screening process. Between each round, the selected phages were incubated with the E. coli strain ER2738 (sold as part of phage display library kit) in Luria-Bertani (LB) broth at 37 °C for 4 h to amplify the phages.
For the plaque formation assay, phages were serially diluted from 1013 to 109 cells/ml and transfected into E. coli in LB medium at room temperature (RT) for 1-5 min. The phage-infected E. coli cells were mixed with 10 g/ml bacto-tryptone, 5 g/ml yeast extract, 5 g/ml NaCl, and 7 g/ml bacto-agar and immediately poured onto agar plates containing 50 μg/ml isopropyl-β-d-thiogalactoside (IPTG; Promega, USA) and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactoside (Xgal; Promega). Blue (positive) plaques were identified by the eye after overnight culture at 37 °C. To determine the peptide sequences of the selected phages, the inserted DNA sequences were PCR amplified with forward (5′-CCTTTAGTGGTACCTTTCTA-3′) and reverse (5′-CTTTCAACAGTTTCGGCCGA-3′) cloning primers. The amplified 95-bp PCR products were cloned with a pGEM® easy TA cloning vector (Promega), transformed into E. coli (TOP10 strain) cells, and grown on LB-agar plates containing 100 μg/ml of ampicillin (Sigma-Aldrich). In certain cases, selected phages were instead cloned as described previously.
DNAs were extracted from positive bacterial clones and sequenced in one direction with the 96 gIII sequencing primer: 5′-CCCTCATAGTTAGCGTAACG-3′. The DNA sequences were translated to 12-mer peptide sequences by Expasy (http://www.expasy.org), and the putative two-dimensional amino acid structures were predicted by PEP-FOLD2 (http://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#forms::PEP-FOLD) and PeptiMap (http://peptimap.bu.edu/). The DNA sequences were compared to those of the NCBI database with BLAST 3D, and corresponding binding sites were predicted by Uniprot (http://www.uniprot.org/) and PyMOL (http://www.pymol.org).
F. Cell culture, fluorescence staining, and the determination of aptamer dissociation constants via flow cytometry
OVCAR-3 cell lines and normal cervical cell line were provided by the Department of Obstetrics and Gynecology (NCKUH) and were used as the target cells for cell-based fluorescence staining and dissociation constant analysis. Normal cervical cell line was used as the control cells for cell-based fluorescence staining. Cells were cultured in RPMI-1640 media (Invitrogen) and culture media was supplemented with 10% fetal bovine serum (Invitrogen) and 100 U/ml of penicillin-streptomycin (Invitrogen). Cells were cultured at 37 °C at 5% CO2. 1 × 105 cells of the OVCAR-3 and normal cervical cells were harvested and then washed with 1 × PBS and then re-suspended in 200 μl binding buffer before adding 50 μl of 1 μM FAM-labeled Tx-01 aptamer or Tp-01 peptide for 30 min of incubation. After incubation, the labeled cells were washed twice with 1 × PBS and then the cells were fixed on microscope slides with ProLong® Gold Antifade Reagent (Invitrogen). The antigen-retrieved tissue slides with cancer tissues and normal tissues were stained under the same condition. The stained slides were observed and analyzed by a set of fluorescence microscopy including a collimation lens, an objective lens (Nikon LU Plan 10×/0.30 A, Nikon, Japan), three fluorescence filters (Nikon G-2A, Nikon), and a mercury lamp (MODEL C-SHG1, Nikon). Images of Tx-01 aptamer or Tp-01 peptide fluorescence staining results were captured by a DS-Qi1Mc camera (1.5 megapixels, equipped a Peltier cooling device and a programmable gain amplifier, Nikon) equipped with one digital signal controlling module.25
Aptamers (Tx-01 and Tx-02), peptides (Tp-01 and Tp-02), and ssDNA library were synthesized (>98% purity) and conjugated with FAM (excitation wavelength = 490 nm, emission wavelength = 525 nm) at the 5′- and carboxyl termini, respectively (Protech Co., Ltd., Taiwan). These fluorescently-labeled aptamers and peptides were then used for fluorescence microscopy analysis and flow cytometric assays (BD Accuri™ C6, Becton, Dickinson and Company, USA). To determine the binding affinities of the selected affinity reagents toward OVCAR-3 cells, concentrations of FAM-labeled aptamers ranging from 0.1 to 500 nM were incubated with 105 OVCAR-3 cells for 15 min in 50 μl of binding buffer. Cells (n = 3 aliquots) were then washed six times with 50 μl of washing buffer and suspended in 50 μl of binding buffer for analysis via flow cytometry. Cells without aptamer labeling were used as negative controls, and the specific binding intensity was calculated by subtracting the background intensity from the mean fluorescence intensity. A fluorescently-labeled ssDNA library was also used as a negative control to explore nonspecific binding.26 The resulting fluorescence intensity of the affinity reagents was further used to calculate the equilibrium dissociation constant (Kd) with Prism software (GraphPad Software, USA), which fitted a plot of the mean fluorescence intensity of the specific binding intensity (Y) versus the aptamer concentration (X) as follows: Y = B max X/(Kd + X).11,15
G. In vitro binding analysis by fluorescence microscopy and confocal imaging
For immunofluorescence staining, cells were seeded at a density of 1.5 × 104 cells/cm2 on a NUNC™ Lab-Tek™ II chamber slide™ (ThermoFisher Scientific, USA) and cultured at 37 °C for 18 h in a 5% CO2 incubator. Then, cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) at RT for 5 min, permeabilized with 0.01% Triton X-100 (Sigma-Aldrich) for 2 min, blocked with 1× PBS containing 3% bovine serum albumin (BSA, Sigma-Aldrich) for 60 min, and incubated with 100 nM-1 μM FAM-conjugated aptamer and anti-BRCA2 primary antibody at RT for 30 and 60 min, respectively. The anti-BRCA2 antibody (Abcam, ab9143, 1:500) was used to verify the binding site of the screened aptamer. Nuclei were stained with Hoechst 33259 (Life Technologies). Finally, cells were mounted with mounting solution (ibidi, Germany), and fluorescent images were acquired with either (1) an Olympus BX51 microscope (Japan) coupled with a DS-Qi1Mc camera (1.5 MP) equipped with a Peltier cooling device and a programmable gain amplifier (Nikon) or (2) a Carl Zeiss LSM780 confocal microscope. BX51 and confocal images were analyzed with Nikon and ZEN 2011 software (Zeiss), respectively.
H. Liquid chromatography/mass spectrometry analysis
1. Purification of target molecules of selected aptamers/peptides
Aptamers (Tx-01 and Tx-02) were modified with the amino group (–NH2) at their 5′-end and peptides (Tp-01 and Tp-02) were tagged with the biotin group at their carboxyl termini by Protech Inc. The modified aptamers can be conjugated onto the magnetic beads through the amino group (MyOne™ Carboxylic Acid, Cat. #65011, Thermo Fisher Scientific). Therefore, the target molecules of aptamers Tx-01/02 and peptides Tp-01/02 can be purified by either aptamer-conjugated magnetic beads or streptavidin resin (G Bioscience, USA, Cat. #786-555). In order to identify target molecules of aptamer Tx-01/02 and peptide Tp-01/02, protein samples from total cell lysate and membrane fractions were prepared. For protein samples from total cell lysate, cells were harvested using cell scrapers and placed into 1 ml of ice-cold 1× PBS (pH 7.4), centrifuged at 1200 rpm for 5 min at 4 °C, and washed at least twice with ice-cold 1× PBS. Finally, the cell pellets were lysed on ice with a sonicator (UP-50H, Hielscher). For the cell membrane protein samples, they were prepared by using the plasma membrane protein extraction kit following the manufacturer's instructions (ab65400, Abcam, UK).
The aptamer-coated beads (for Tx-01 and Tx-02) were incubated with approximately 100 μg of cell membrane protein extracts or total cell lysates in a final volume of 350 μl for 1 h at room temperature. The protein/aptamer-bead complexes were washed with PBS three times to remove the non-specific binding proteins. Finally, protein/aptamer-bead complexes were re-suspended in ddH2O containing 4× protein loading dye (Protech Inc) and heated at 95 °C for 10 min followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation. To purify target molecules recognized by peptide Tp-01 and Tp-02, cell membrane protein extracts or total cell lysates were incubated with 1 μM of the biotinylated peptide at 4 °C for 60 min and then incubated with streptavidin resin at room temperature for 10 min. After incubation, the resins were washed thrice with streptavidin binding buffer (20 mM NaPO4, 0.15M NaCl, pH 7.5). Target proteins were co-eluted with protein/biotinylated-peptide complexes from streptavidin resin by 8M guanidine hydrochloride (HCl) (pH 1.5). Eluted target proteins were desalted by a 5-fold dilution in ddH2O and re-concentrated to 100 μl using Amicon® Ultra-0.5 centrifugal filters (0.5 ml, 3-kDa cutoff; Millipore, Cat. #UFC500396). The re-concentrated protein samples were heated at 95 °C for 5 min in ddH2O containing 4× protein loading dye (Protech Inc.). The protein concentrations were determined by the DC protein assay kit (BioRad, Cat. #5000112). Here, the OVCAR3 cells, a serious type of ovarian cancer cell lines, were used for identification of target molecules of aptamers Tx-01/02 and peptides Tp-01/02. In addition, Hela (cervical cancer), A549 (lung cancer), MD-MBA-231 (breast cancer), and HCT8 (colon cancer) cells were used as the control cell lines for negative selection of target molecules.
2. Identification of target molecules of selected aptamers/peptide
Protein samples were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and stained with Coomassie brilliant blue. Protein samples on gels were subjected to trypsin digestion and the tryptic peptides were collected and de-salted by C18 ZipTip® before liquid chromatography-mass spectrometry (LC-MS) analysis. The tryptic peptides were analyzed by using nanoflow LC-MS/MS. Mass spectral data were acquired using a Thermo LTQ-Orbitrap Discovery Hybrid mass spectrometer (ThermoFisher, USA) equipped with a nanoelectrospray ion source (New Objective, Woburn, MA) and an Agilent 1200 Series binary high-performance liquid chromatography pump (Agilent Technologies, USA). The experiment RAW files were converted to the mgf format and subjected to Mascot analysis (ver 2.3, Matrix Science, USA) for MS/MS ion search in order to identify the target molecules of aptamer Tx-01/02 and peptide Tp-01/02.
I. Western blotting
In order to confirm the binding sites (BRCA2 and GPM6a) of screened aptamers, Western blotting was used. Total cell lysate was prepared by scraping cells from a 100-mm dish and lysing cell using a sonicator. The commercialized recombinant human BRCA2 protein (∼37 kDa, 3319 a.a.-3418 a.a., ab112253, Abcam) was purchased as a purified sample of target proteins. The human serum albumin (HAS, ∼67 kDa, A7223, Sigma-Aldrich co.) was purchased from Sigma-Aldrich as a negative control. For detecting BRCA2 protein, which is a potential binding site for the screened aptamer, cell lysates or recombinant BRCA2 proteins were denatured at 55 °C in SDS/2-ME sample buffer for 10 min. Protein samples were separated by either 10% or 6% SDS-PAGE gels and were transferred to polyvinylidene difluoride membranes (PVDF, Immobilon, Millipore) at 100 V for 120 min. The blot was blocked with 5% skim milk in PBS with 0.05% Tween 20 (PBST, pH 7.4) at RT for 1 h. To perform the aptamer-based western blot probed by the selected Tx-01 aptamer, the transferred PVDF membrane was hybridized with 100 nM Tx-01 aptamer in PBS containing 1 mM MgCl2. After thrice washing with PBS, the PVDF membrane was exposed to ethidium bromide (EtBr, 0.5 μg/ml, Cat. #15585011, ThermoFisher Scientific) for revealing the signal recognized by the Tx-01 aptamer. Antibodies used in this study are described as follows: GPM6a antibody (Cat. #ab79128, Abcam) and horseradish peroxidase (HRP)-conjugated goat-anti-rabbit antibody (Cat. #A16035, Thermo Fisher Scientific) were purchased from Abcam and Thermo Fisher Scientific, respectively. HRP-conjugated goat-anti-mouse (GTX213111) and the enhanced chemiluminescence substrate (GTX14698) were purchased from GeneTex, and western blot images were acquired with the ImageQuant LAS-4000 mini imaging system (GE Healthcare Life Sciences).
III. RESULTS AND DISCUSSIONS
A. Characterization of microfluidic devices
In this study, two different in vitro affinity reagent screening assays could work in one integrated microfluidic system. The dual functions of the microfluidic platform are beneficial to provide a standard operation protocol and less human intervention is therefore required during the tissue slide preparation. The working process is illustrated in Fig. 1. The detailed experimental procedures of tissue-SELEX and phage display technology are listed in Tables SI and SII in the supplementary material. In order to automatically perform tissue-SELEX and phage display to identify cancer-specific aptamers and peptides, respectively, a tissue-based microfluidic system was designed (Fig. 2). This microfluidic chip was equipped with two micromixers, one transport unit, two binding buffer chambers, two washing buffer chambers, one amplification chamber, and one waste outlet. The micromixer could transport 217.5 μl of liquid in one second, and the mixing index was measured to be 95.5% in 4 s under a 0.5-Hz working frequency and −39.9 kPa applied gauge pressure (Fig. S1 in the supplementary material). The shear force caused by the micromixer increased with decreasing gauge pressure such that it could be used to control the stringency of the screening (Table SIII in the supplementary material).20 It is worth noting that this micromixer was properly designed to fit the dimensions of the tissue samples (around 2 cm in diameter) while it could still perform reasonable liquid transportation and mixing under an optimal working frequency and vacuum force. The operating conditions of −39.9 kPa and 0.5 Hz were chosen for the subsequent tests.
A laser-engraved double-sided tape [Fig. 2(b)] was used to bind the chip to the tissue slides. Furthermore, to make the binding surface smoother, a tissue slide fixture [Fig. 2(c)] was created to hold the normal and cancer tissue slides together. The setup allowed for the processing of irregularly shaped tissue samples [Fig. 2(d)]. The chip [Fig. 2(e)] was 81 mm in width and 85 mm in length, and it was composed of four layers [Fig. 2(f)]: an air-control layer, a liquid-control layer, a double-sided tape layer, and a tissue slide layer. In order to make this microfluidic chip suitable for most clinical tissue slides, which are mostly irregularly shaped and large (∼2 cm), a large tissue-based micromixer (2 cm in diameter) was incorporated to fit the tissue sample. It is worth noting that this novel microfluidic platform can also be applied for other kinds of tissue slide samples.
B. Optimization of incubation times
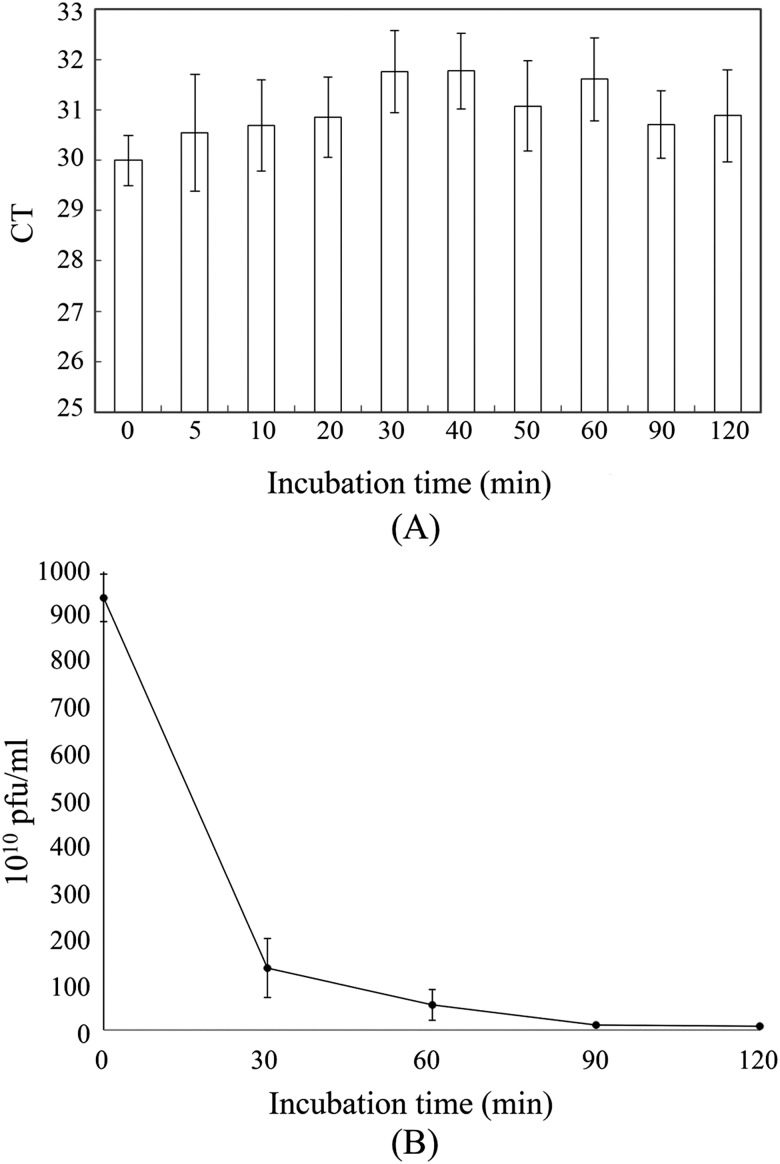
The incubation time between ssDNA library and tissue samples was first optimized. PCR was used to analyze the amount of ssDNA in the supernatant at various incubation times [Fig. 3(a)], and little ssDNA was experimentally found in the supernatant of samples incubated with reagents for 30–40 min. Therefore, 30 min was used as the optimal incubation time for all subsequent on-chip tissue-SELEX procedures. Similarly, the incubation time between M13 phages and tissue samples was optimized and a plaque formation assay was used to quantify the number of M13 phages in the supernatant after 0, 30, 60, 90, and 120 min [Fig. 3(b)]. After a 30-min, on-chip incubation, only phages with a concentration of 1010 pfu/ml were quantified, indicating that ∼90% of the phages were bound to the tissue slides. Therefore, 30 min was chosen as the optimal incubation time for on-chip phage display as well. Besides, according to the screening results of aptamers/peptides, which will be shown in Secs. III C and III D, only a few selected candidates were found and some of the screened candidates showed a high affinity and specificity on tissue samples after 30 min. It was then concluded that 30 min could be enough for our screening process. When compared with the previous bench-top tissue-based affinity reagent screening process,17 the incubation time could be dramatically reduced from 2 h to 30 min. Furthermore, under the optimized operating conditions (−39.9 kPa and 0.5 Hz) of the microfluidic system, it could provide a gentle mixing for ssDNA/phages to interact and bind with target cancer tissues. These results indicate that this integrated microfluidic system could be applied to shorten the screening process of cancer affinity reagents.
FIG. 3.
Optimization of incubation times. (a) Real-time PCR analysis of tissue-SELEX supernatants collected after varying incubation times. (b) Plaque formation assay with phage display supernatants collected at different incubation times. In panel (a), error bars represent standard deviation (N = 3). Ct = threshold cycle.
C. Identification of aptamers and peptides specific to OvCa tissues
After determining the proper incubation times, the screening procedure was enacted to identify aptamers and peptides that specifically bind OvCa tissues. After three rounds of screening, gel electrophoresis was used to detect tissue-SELEX and phage display PCR products (72 and 95 bp, respectively; Figs. S2 and S3 in the supplementary material). We also found that, upon increasing the shear force in each screening round (from 206.7 nN to 342.2 nN, as shown in Table SIII in the supplementary material), more and more weakly bound ssDNA and phages were removed. These were demonstrated by collecting the washing wastes, which were amplified by PCR and analyzed by gel electrophoresis, as shown in Figs. S2 and S3 in the supplementary material, respectively. After three rounds of on-chip tissue-SELEX, we collected the screened ssDNA pools, PCR amplified them, cloned the PCR products, and sequenced random clones; two aptamer candidates (Tx-01 and Tx-02) were identified (Table I). The ssDNA folding structures of these selected aptamers (25 °C) were predicted using MFOLD software (version 3.5; Fig. S4 in the supplementary material). We also identified two peptides candidates (Tp-01 and Tp-02) via a similar PCR + cloning + sequencing pipeline (see Table SI and Fig. S4 in the supplementary material). The sequences of the other selected aptamers and peptides are also provided in Tables SIV and SV in the supplementary material. When compared with the previous bench-top tissue-based affinity reagent screening process,17 the screening rounds could be dramatically reduced from 15 rounds to 3 rounds. Furthermore, recent next-generation sequencing technology could provide a deep analysis for the selection results, which provided a more complete diagram of the post-selection library and could observe the population dynamics during each round of the selection process.27 Because the tunable shear force of this tissue-based micromixer could wash out the weakly bound ssDNA and phages efficiently, the selected aptamers and peptides exhibited Kd values ranging from 2.9 nM to 53.8 nM (Fig. S5 in the supplementary material), which is comparable or even superior to most of the antibodies (from 10−7 to 10−9M).28
A synthesized fluorophore-labeled unselected ssDNA library was also used to be stained on OVCAR3 cells for nonspecific binding tests. The results showed that the fluorescence intensity of background (cells did not incubate with any fluorophore-labeled aptamer), FAM-labeled ssDNA library, and FAM-labeled Tx-01 was measured to be 8498, 11 545, and 18 310 (a.u.), respectively (Fig. S6 in the supplementary material). Again, the results of fluorescent images showed that ssDNA random library was non-specifically stained on OVCAR3 and Tx-01 was detected with relatively strong signals at the edge of cell membranes and nucleus in part of cells (Fig. S6 in the supplementary material). These results demonstrated that this integrated microfluidic system could be used to select high-affinity aptamers and peptides to cancer tissue slides.
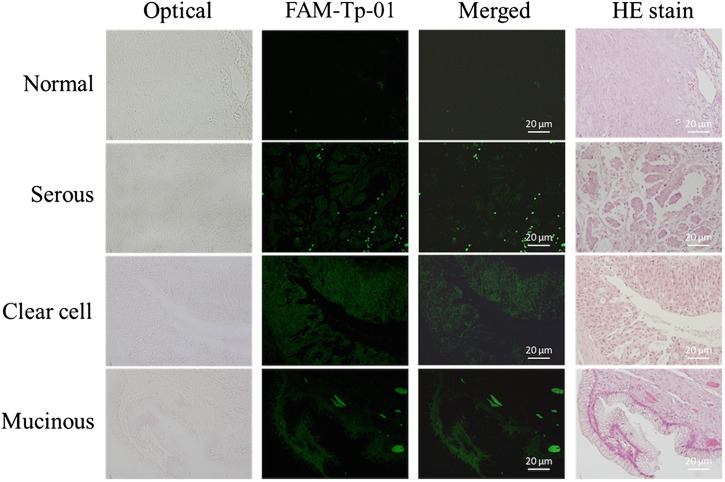
D. Specificity of the selected affinity reagents
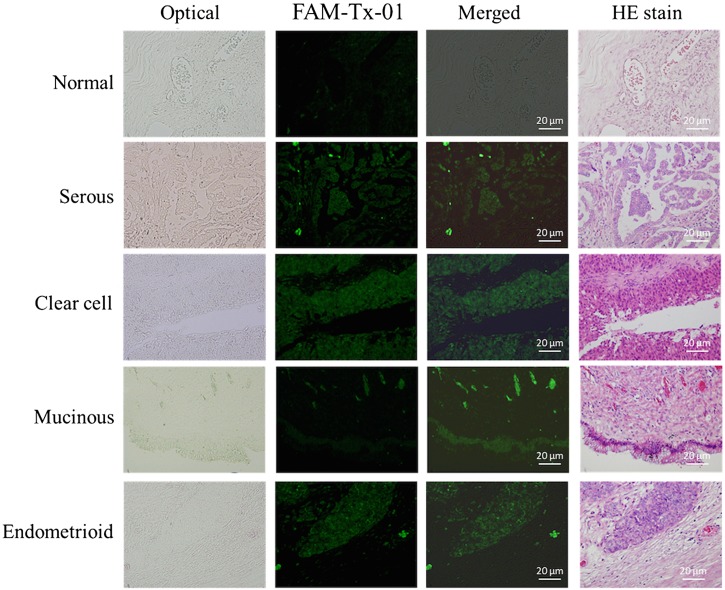
In this study, the affinity reagents screened from this novel microfluidic platform were tested with tissue slides (normal ovarian and ovarian cancer tissues) and cultured cells (OVCAR-3 ovarian cancer cell line and normal cervical cells) to demonstrate their specific binding ability. First, the on-chip screened affinity reagents modified with FAM green fluorescence dye were used to stain normal ovarian tissue slides and ovarian cancer tissue slides to explore their specificity. Aptamers and peptides were first incubated with cancer cells and tissue samples, and the results were compared to those of HE staining. Only weak signals were emitted from normal tissue slides stained with the FAM-labeled Tx-01 aptamer (Fig. 4), though significantly bright green fluorescence signals were observed upon incubation with cancer tissues in the same regions as documented upon HE staining. The staining results using the other screened aptamers (Tx-02, Tx-03, and Tx-06) were also presented in Fig. S7 in the supplementary material.
FIG. 4.
FAM-labeled aptamer staining of OvCa tissue slides. Normal OvCa tissue served as the negative control. Scale bars are 20 μm.
Similarly, with the Tp-01 peptide (Fig. 5), there were weak and bright FAM green fluorescent signals when incubated with normal and cancerous tissues, respectively (Fig. S7 in the supplementary material also presents the staining data for peptides Tp-02 and Tp-13). When compared with other ovarian cancer slide samples, clear green fluorescence signals in the cancer cells of tissue regions were observed, which were the same cancer cell regions found in the HE staining process.
FIG. 5.
FAM-labeled peptide (Tp-01) staining of OvCa tissues. Normal OvCa tissue served as the negative control. Scale bars are 20 μm.
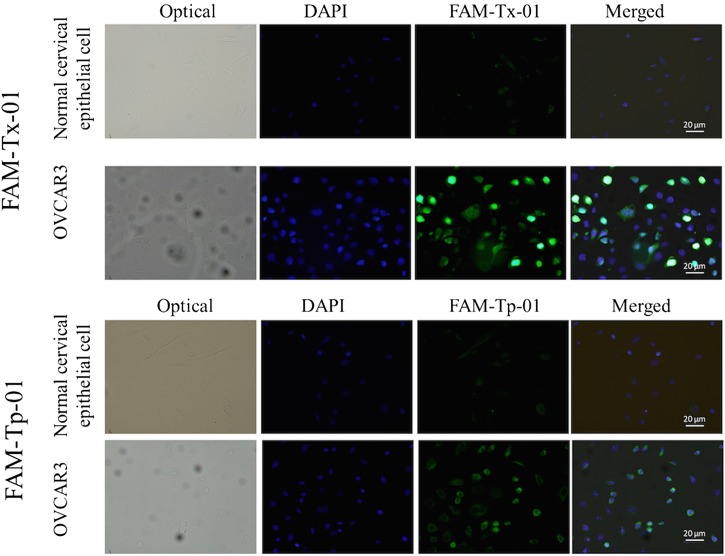
In order to test the fluorescence staining feature and capability of these selected affinity reagents, a cultured ovarian cancer cell line, OVCAR-3, and a cultured normal cervical epithelial cell line were further applied to demonstrate binding affinity toward the Tx-01 aptamer and the Tp-01 peptide (Fig. 6). Similarly, normal cervical epithelial cells presented weak green FAM fluorescent signals after the Tx-01 aptamer and the Tp-01 peptide staining. In contrast, green FAM fluorescent signals were evident when these affinity reagents were incubated with OVCAR-3 cells, which is an OvCa cell line (Fig. 6). The semi-quantitative analysis of fluorescence intensities of Fig. 6 is found in Fig. S8 in the supplementary material, indicating that the selected peptide exhibited high binding affinity toward the target tissue. Aptamer Tx-01 and oligopeptide Tp-01 toward OVCAR-3 showed clear and strong green fluorescence signals, indicating that these two screened affinity reagents may be promising to be also applied for cell-based assays as well. When compared with previous cell-based screening assays,14,16 tissue slide-based assays could result in differences in thickness of the tissues analyzed herein (0.35 to 5 μm), which would expose various cellular antigens possessing more complicated and uncovered information for affinity reagents to recognize. Furthermore, our protocol allowed for reagents and probes to interact with molecules in the cytosol and even the nucleus; such an approach could actually be advantageous in cases in which cancer biomarkers localize to these locations. Selection of affinity reagents specific to tissue samples may be also used as new probes in pathological diagnosis. Finally, tissue slides from individuals are advantageous to discover the personalized affinity reagents for cancer diagnosis/prognosis or targeted drug delivery in the future. In contrast, when coating cells, rather than tissues, on the slides, probes tend to bind only to the cell surface and the culture cell lines cannot be used for personalized medicine; if the probes are not specific to cell surface markers, the capture rate may then be markedly low.
FIG. 6.
Aptamer and peptide binding to normal (cervical epithelial cells) and OvCa cells. A cultured ovarian cancer cell line, OVCAR-3, was incubated with the FAM-labeled aptamer Tx-01 or the FAM-labeled peptide Tp-01 as described in the text. Normal cervical epithelial cells were used as the negative control cell line. Scale bars are 20 μm.
E. Binding target analysis
In order to identify the binding target molecules of aptamer Tx-01 which exhibited the best specific signal from tissue stained samples, the LC-MS/MS experiment coupled with bioinformatics analysis including Uniprot (http://www.uniprot.org/) and PyMOL (http://www.pymol.org) and literature survey was performed in this study. By using an aptamer-protein pull down assay followed by LC-MS/MS analysis, both GPM6a and BRCA2 were found to be the Tx-01 binding targets (Figs. 7 and 8). To further investigate the BRCA2 and Tx-01 interaction, we applied an aptamer-based western blot in which the transferred PVDF membrane was directly probed with Tx-01 aptamer and immersed in EtBr followed by ultraviolet (UV) exposure for detection. Figure 7 shows that GPM6a could be specifically bound with Tx-01.
FIG. 7.
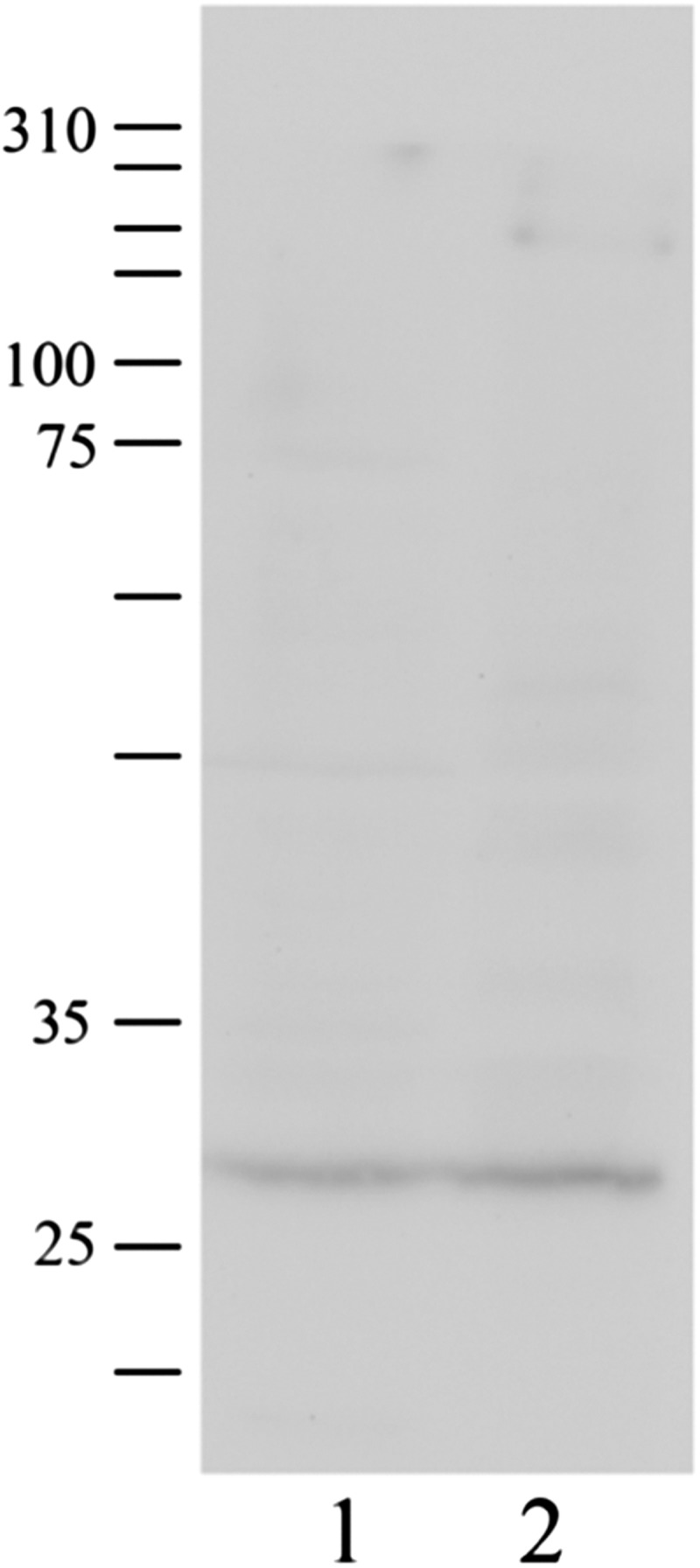
Pull down assay for the possible binding target of the Tx-01 aptamer, GPM6a. Total cell lysates were incubated with Tx-01 aptamer-coated magnetic beads and confirmed the interaction between Tx-01 aptamer and GPM6a by Western blot. Lane 1: OVCAR3 total cell lysate; lane 2: Proteins captured by Tx-01 aptamer-coated magnetic beads. (The size of GPM6a was 31 kDa; protein marker.)
FIG. 8.
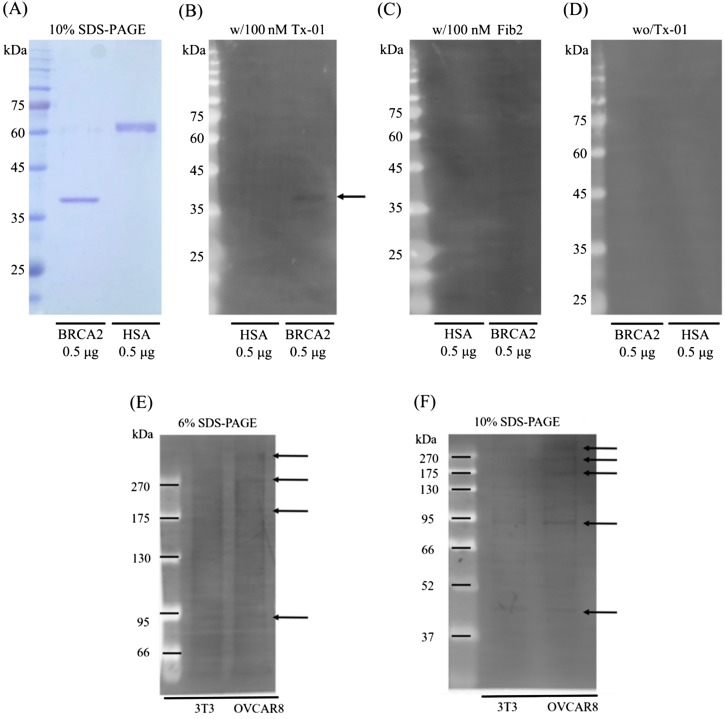
Western blot analysis of the possible binding target of the Tx-01 aptamer, BRCA2. (a) The protein size of the recombinant human BRCA2 and human serum albumin (HSA). [(b) and (c)] 0.5 μg of recombinant human BRCA2 and HSA proteins on the PVDF membrane and hybridized with Tx-01 and Fib2 aptamers, respectively. The arrow points to the protein size of recombinant human BRCA2. (d) PVDF without aptamer hybridization. [(e) and (f)] 20 μg of proteins from total cell lysates of NIH/3T3 and OVCAR8 cell lines were denatured at 55 °C and separated by 10% (b) and 6% (c) SDS-PAGE, respectively. Arrows indicate proteins of different molecular weight recognized by the Tx-01 aptamer. The leftmost lane is the molecular weight marker indicating the protein size. Protein marker (Cat. #PM007-0500, GeneDireX).
Similarly, as shown in Fig. 8, Tx-01 aptamer was able to significantly distinguish recombinant human BRCA2 protein and human serum albumin [Fig 8(b)]. Besides, a Fib2 aptamer, which recognized the human fibrinogen, was only hybridized to the recombinant human BRCA2 and hence we could exclude the non-specific binding of ssDNA aptamer on this small peptide of BRCA2 protein. In addition, the confocal images of BRCA2 and Tx-01 provided a solid evidence of the interaction between BRCA2 and Tx-01 aptamer because the fluorescence signals were co-localized at the nucleus and cell surface (Fig. S9 in the supplementary material). Furthermore, both GPM6a and BRCA2 have been reported to be highly correlated with occurrences of ovarian cancer.29–32 According to these data, we therefore conclude that both GPM6a and BRCA2 may be binding targets for Tx-01 aptamer.
IV. CONCLUSION
In this study, we have presented a novel microfluidic platform for automatically and robotically screening two different classes of affinity reagents, the DNA aptamers and peptides, by using tissues for cancer biomarker probes. This microfluidic chip carried out all transport and reaction steps and also reduced the time required to obtain data; it also reduced the likelihood of human intervention. We identified two aptamers and two peptide probes specific to OvCa tissues. One of the selected probes, Tx-01 aptamer, showed a promising specificity toward the ovarian cancer tissue. Two binding target proteins of Tx-01 aptamer, GPM6a and BRCA2 proteins, have also been identified based on liquid chromatography-mass spectrometry (LC/MS), bioinformatics, confocal microscopic imaging, and Western blotting-based analyses, which could be promising for further study and applications in cancer diagnosis and therapy. Not just could this chip be used for OvCa, but it could be readily modified to identify biomarker probes specific to other cancer cells.
Biomarker discovery is a very critical study area for disease prevention and therapy. The typical research strategy to find a biomarker is carried out by the bottom-up approach, mainly by digestion with a protease (typically trypsin) and analysis in a high-resolution LC/MS. The major advantages of these approaches are direct quantitation and high detection ranges, but the throughput and reproducibility are limited. We are here to provide a new method for the top-down approach, starting from the clinical tissue samples to affinity reagents specifically bound with cancer biomarkers. By further combining with LC/MS analysis and the bottom-up approach, the specific cancer biomarkers could be identified. As a consequence, we have screened a cancer tissue-specific aptamer, Tx-01, and then found that GPM6a is one of the major binding targets for Tx-01 aptamer, and BRCA2 may be the minor one, which provides possibilities to further study the role of these proteins in ovarian cancer and their diagnostic and therapeutic applications.
SUPPLEMENTARY MATERIAL
See supplementary material for the experimental procedures for tissue-SELEX and phage display and some additional data.
ACKNOWLEDGMENTS
The authors thank the Ministry of Science and Technology (MOST) of Taiwan for funding this work (MOST 105-2119-M-007-009 to G.B.L.). Partial financial support from the “Higher Education Support Project” of Taiwan’s Ministry of Education (Grant No. 107Q2713E1) is also greatly appreciated.
Contributor Information
Keng-Fu Hsu, Email: .
Gwo-Bin Lee, Email: .
References
- 1.Siegel R. L., Miller K. D., and Jemal A., CA Cancer J. Clin. 67(1), 7 (2017). 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., and Jemal A., CA Cancer J. Clin. 65(1), 87 (2015). 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Siegel R. L., Miller K. D., and Jemal A., CA Cancer J. Clin. 66(1), 7 (2016). 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 4.Badgwell D. and Bast R. C. Jr., Dis. Markers 23(5–6), 397 (2007). 10.1155/2007/309382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menon U., Nat. Clin. Pract. Oncol. 4, 498 (2007). 10.1038/ncponc0906 [DOI] [PubMed] [Google Scholar]
- 6.McDermott U., Downing J. R., and Stratton M. R., N. Engl. J. Med. 364(4), 340 (2011). 10.1056/NEJMra0907178 [DOI] [PubMed] [Google Scholar]
- 7.Pasqualini R. and Ruoslahti E., Nature 380(6572), 364 (1996). 10.1038/380364a0 [DOI] [PubMed] [Google Scholar]
- 8.Smith G. P., Science 228(4605), 1315 (1985). 10.1126/science.4001944 [DOI] [PubMed] [Google Scholar]
- 9.Tuerk C. and Gold L., Science 249(4968), 505 (1990). 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- 10.Sefah K., Shangguan D., Xiong X. L., O’Donoghue M. B., and Tan W. H., Nat. Protoc. 5, 1169 (2010). 10.1038/nprot.2010.66 [DOI] [PubMed] [Google Scholar]
- 11.Sefah K., Meng L., Lopez-Colon D., Jimenez E., Liu C., and Tan W., PLoS One 5(6), e14269 (2010). 10.1371/journal.pone.0014269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marx V., Nat. Methods 10(9), 829 (2013). 10.1038/nmeth.2607 [DOI] [PubMed] [Google Scholar]
- 13.Hung L. Y., Wang C. H., Fu C. Y., Gopinathan P., and Lee G. B., Lab Chip 16(15), 2759 (2016). 10.1039/C6LC00662K [DOI] [PubMed] [Google Scholar]
- 14.Weng C. H., Hsieh I. S., Hung L. Y., Lin H. I., Shiesh S. C., Chen Y. L., and Lee G. B., Microfluid. Nanofluidics 14(3–4), 753 (2012). 10.1007/s10404-012-1095-3 [DOI] [Google Scholar]
- 15.Hung L. Y., Wang C. H., Hsu K. F., Chou C. Y., and Lee G. B., Lab Chip 14(20), 4017 (2014). 10.1039/C4LC00587B [DOI] [PubMed] [Google Scholar]
- 16.Hung L. Y., Wang C. H., Che Y. J., Fu C. Y., Chang H. Y., Wang K., and Lee G. B., Sci. Rep. 5, 10326 (2015). 10.1038/srep10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Xu H., Ding H., Huang Y., Cao X., Yang G., Li J., Xie Z., Meng Y., Li X., Zhao Q., Shen B., and Shao N., J. Pathol. 218(3), 327 (2009). 10.1002/path.2543 [DOI] [PubMed] [Google Scholar]
- 18.Cung K., Slater R. L., Cui Y., Jones S. E., Ahmad H., Naik R. R., and McAlpine M. C., Lab Chip 12(3), 562 (2012). 10.1039/C2LC21129G [DOI] [PubMed] [Google Scholar]
- 19.Wang C. H., Weng C. H., Che Y. J., Wang K., and Lee G. B., Theranostics 5(4), 431 (2015). 10.7150/thno.10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Che Y. J., Wu H. W., Hung L. Y., Liu C. A., Chang H. Y., Wang K., and Lee G. B., Biomicrofluidics 9(5), 054121 (2015). 10.1063/1.4933067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung L. Y., Fu C. Y., Wang C. H., Chuang Y. J., Tsai Y. C., Lo Y. L, Lee W. B., Shiesh S. C., Chang H. Y., Hsu K. F., and Lee G. B., in Proceedings of the 19th International Conference on Solid-State Sensors, Actuators and Microsystems (TRANSDUCERS) (Institute of Electrical and Electronics Engineers, IEEE, 2017), pp. 568–571. [Google Scholar]
- 22.Weng C. H., Huang T. B., Huang C. C., Yeh C. S., Lei H. Y., and Lee G. B., Biomed. Microdevices 13(3), 585–595 (2011). 10.1007/s10544-011-9529-3 [DOI] [PubMed] [Google Scholar]
- 23.Yang Y. N., Hsiung S. K., and Lee G. B., Microfluid. Nanofluidics 6(6), 823 (2008). 10.1007/s10404-008-0356-7 [DOI] [Google Scholar]
- 24.Schmitz N., Laverty S., Kraus V. B., and Aigner T., Osteoarthr. Cartil. 18(Suppl. 3), S113 (2010). 10.1016/j.joca.2010.05.026 [DOI] [PubMed] [Google Scholar]
- 25.Hung L. Y., Chuang Y. H., Kuo H. T., Wang C. H., Hsu K. F., Chou C. Y., and Lee G. B., Biomed. Microdevices 15(2), 339 (2013). 10.1007/s10544-013-9739-y [DOI] [PubMed] [Google Scholar]
- 26.Shangguan D., Li Y., Tang Z., Cao Z. C., Chen H. W., Mallikaratchy P., Sefah K., Yang C. J., and Tan W., Proc. Natl. Acad. Sci. U.S.A. 103(32), 11838 (2006). 10.1073/pnas.0602615103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schütze T., Wilhelm B., Greiner N., Braun H., Peter F., Mörl M., Erdmann V. A., Lehrach H., Konthur Z., Menger M., Arndt P. F., and Glökler J., PLoS One 6(12), e29604 (2011). 10.1371/journal.pone.0029604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Liu Y., Teesalu T., Sugahara K. N., Kotamrajua V. R., Adams J. D., Ferguson B. S., Gong Q., Oh S. S., Csordas A. T., Cho M., Ruoslahti E., Xiao Y., and Soh H. T., Proc. Natl. Acad. Sci. U.S.A. 108(17), 6909 (2011). 10.1073/pnas.1014753108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santin A. D., Zhan F., Bellone S., Palmieri M., Cane S., Bignotti E., Anfossi S., Gokden M., Dunn D., Roman J. J., O’Brien T. J., Tian E., Cannon M. J., Shaughnessy J. Jr., and Pecorelli S., Int. J. Cancer 112(1), 14 (2004). 10.1002/ijc.20408 [DOI] [PubMed] [Google Scholar]
- 30.Antoniou A., Pharoah P. D., Narod S., Risch H. A., Eyfjord J. E., Hopper J. L., Loman N., Olsson H., Johannsson O., Borg A., Pasini B., Radice P., Manoukian S., Eccles D. M., Tang N., Olah E., Anton-Culver H., Warner E., Lubinski J., Gronwald J., Gorski B., Tulinius H., Thorlacius S., Eerola H., Nevanlinna H., Syrjakoski K., Kallioniemi O. P., Thompson D., Evans C., Peto J., Lalloo F., Evans D. G., and Easton D. F., Am. J. Hum. Genet. 72(5), 1117 (2003). 10.1086/375033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakhani S. R., Manek S., Penault-Llorca F., Flanagan A., Arnout L., Merrett S., McGuffog L., Steele D., Devilee P., Klijn J. G., Meijers-Heijboer H., Radice P., Pilotti S., Nevanlinna H., Butzow R., Sobol H., Jacquemier J., Lyonet D. S., Neuhausen S. L., Weber B., Wagner T., Winqvist R., Bignon Y. J., Monti F., Schmitt F., Lenoir G., Seitz S., Hamman U., Pharoah P., Lane G., Ponder B., Bishop D. T., and Easton D. F., Clin. Cancer Res. 10(7), 2473 (2004). 10.1158/1078-0432.CCR-1029-3 [DOI] [PubMed] [Google Scholar]
- 32.Sun Y., Shukla G. S., Kennedy G. G., Warshaw D. M., Weaver D. L., Pero S. C., Floyd L., and Krag D. N., J. Biotech. Res. 1, 55 (2009), https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3149830/. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the experimental procedures for tissue-SELEX and phage display and some additional data.