Abstract
The present study compared the taxonomic diversity and evaluated the functional attributes of the bacterial species from Mandovi and Zuari mangrove sediments, Goa, using paired-end amplicon sequencing of 16S rDNA and culture-based analyses, respectively. 16S rDNA sequencing revealed Proteobacteria, Firmicutes, and Actinobacteria as the dominant phyla in both the sediments. However, the abundance of these phyla significantly differed between the samples. Bacteroidetes from Mandovi sediment, and Acidobacteria and Gemmatimonadetes from Zuari sediment were the other exclusive major phyla. Chloroflexi, Cyanobacteria, Nitrospirae, Planctomycetes, Verrucomicrobia, and WS3 were the minor phyla observed in both. However, a significant difference in the distribution of minor phyla and lower bacterial taxa under each phylum was noted between the sediments, indicating that the resident microbial flora completely differed between them. This was further validated by high values from distance matrix analyses between the samples. In addition, the pathogenic Vibrio sp. was recorded exclusively in Mandovi sediment, while higher abundance of ecologically important bacterial classes including Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, and Bacilli was observed in Zuari sediment. Taken together, the data indicated that Zuari sediment was taxonomically richer than Mandovi sediment, while a greater incidence of anthropogenic activities occurred in the latter. This observation was further validated by non-parametric richness estimators which were found to be higher for Zuari sediment. The cultured bacterial isolates, all identified as Firmicutes, were tested for activities related to biofertilization and production of enzymes to be used for bioremediation and chemotherapeutic applications. Higher number of bacterial isolates from Mandovi was found to produce indole-acetic-acid, tannase, xylanase, and glutaminase enzymes, and could solubilize phosphate. In contrast, higher proportion of bacterial isolates from Zuari sediment were capable of producing amylase, cellulase, gelatinase, laccase, lipase, protease, and asparaginase enzymes, emphasizing the fact that the Zuari mangrove sediment is a rich reservoir for economically and biotechnologically important bacterial species.
Keywords: Mandovi, Mangrove, Paired-end amplicon sequencing, Taxonomic diversity, Zuari
Introduction
Mangroves, one of the diverse and biologically productive ecosystems found along tropical and sub-tropical coastal regions, is estimated to cover 17.0 million hectares (ha) worldwide including 121 countries (Aizpuru et al. 2000). Asia has the largest amount [42%] of the world’s mangroves with India contributing an area of 4627.6 sq km (Giri et al. 2015). In India, Goa occupies 22 sq kms of mangrove-covered areas (http://www.iomenvis.nic.in accessed on 29th June, 2018). An area of 700 ha, 900 ha, and 200 ha of mangroves is found along the Mandovi estuary in North, Zuari estuary in South and Cumbarjua canal connecting these two estuaries, respectively (goaenvis.nic.in/mangrove.htm; accessed on 29th June, 2018).
The mangrove biome is unique due to its high salt-tolerance and adaptive capabilities in harsh coastal environment. In addition, their rhizosphere and surrounding sediments are hotspots of diverse microbial activities which contribute to nutrient assimilation in the mangrove ecosystem (Bai et al. 2013). The bacteria release nutrients into the water to be used by marine organisms through degradation of detritus produced from the fallen leaves and branches, thereby transferring the organic matter and energy from the land to marine ecosystems establishing the basis of marine food chain (Bai et al. 2013). Increasing anthropogenic activities, coupled with a lack of proper infrastructure for protection of the mangroves, has resulted in destruction of the mangroves, sometimes beyond their sustainable potential, thus imposing a threat to this important ecosystem (Cabral et al. 2016).
Identification of microorganisms that can attribute to augment plant tolerance to adverse climatic and edaphic conditions, and that can aid to increase mangrove productivity under natural conditions by enhancing nutrient absorption, water uptake, mineral utilization, salt-tolerance, and modification of plant structure and functions is very much essential. In addition, the biodiversity of soil microbes is a useful indicator of ecosystem change (Castaneda and Barbosa 2017). Though there is an upsurge in interest on bacterial diversity of mangrove sediment in recent years, most of the studies on mangroves of Goa relied on the study of individual microorganisms through culture-based assays (Poharkar et al. 2016; Kharangate and; Bhosle 2016). In modern era of scientific advancements, ‘omic’ technologies are emerging as potential tools to ensure holistic insight into environmental systems, thereby leading to in-depth understanding of the complex mechanisms in an ecosystem (Sharma and Lal 2017). It is a revolutionary tool that helps to detect microbes and to decipher their full biological activities within the environmental niche which, otherwise, would have remained unnoticed due to our inability of mimicking their growth conditions under laboratory set-ups. To the best of our knowledge, there is one report on bacterial diversity from mangrove ecosystem of Goa using 454 pyrosequencing which was limited to the diversity of denitrificants only (Fernandes et al. 2015). However, in-depth phylogenetic analysis of bacterial community from the mangrove sediments can be beneficial for developing a better perception on the ecological role of bacteria in mangrove ecosystem.
Therefore, the first objective of our study was to explore the overall bacterial composition, abundance, and diversity in the mangrove sediments of Mandovi and Zuari estuaries of Goa using 16S rDNA amplicon sequencing on illumina MiSeq platform. This method is significantly reliable in its efficiency to discriminate between similar soil samples in comparison to the other next-generation sequencing technologies [NGS] (Habtom et al. 2017). The 427 bp-long V3–V4 region of 16S rRNA was targeted for sequencing, because this region provides more accurate estimates than others for both bacteria and archaea, and has been extensively recommended to be used for the identification of microbiome on illumina Miseq platform that provides short-paired reads of 250 bp (Kim et al. 2011).
Furthermore, to translate the generated knowledge on taxonomic diversity of bacterial community of mangrove sediments for agricultural and soil remediation uses, bacteria were cultured and were studied for enzymatic activities. The previous studies from these environments mainly focused on metal mitigation and antimicrobial actions among specific cultured bacterial isolates (Kharangate and Bhosle 2016). However, many reports show the prevalence of bacteria in mangrove ecosystems throughout the world with capability of degradation of organic matter, promoting nutrient cycles and plant growth (Soares et al. 2014). Therefore, in this study, we investigated the biofertilization and bioremediation activities for the cultured isolates to derive a complete picture for the ongoing biochemical activities in these mangrove ecosystems. The isolates were also studied for the production of two important antineoplastic enzymes, asparaginase and glutaminase (Husain et al. 2016; Reda 2015). This comparison of the microbial consortia with respect to these activities will help to evaluate the importance of these mangrove ecosystems towards biotechnological applications and thereby the need for conservation of its microflora.
Methods
The investigations of this study were carried out on samples obtained at mangrove forests of Ribandar (15.4993°N and 73.8684°E) and Cortalim (15.4058°N and 73.9286°E) along Mandovi (M) and Zuari (Z) estuaries, respectively, during April–May, 2017. Avicennia officinalis and A. alba are the two true mangrove species reported from these two places constituting 80% and 40% of the total mangrove vegetation in Ribandar and Cortalim, respectively; Sonneratia alba (15%) and Acanthus illicifolius (5%) are also found at Ribander mangroves, while Cortalim mangroves are also rich in A. officinalis and A. alba, Acanthus illicifolius, Aegiceras corniculatum, and Excoecaria agallocha (http://www.forest.goa.gov.in/mgr/ accessed on 26th August, 2018). A total of six cores of sediment samples from the intertidal zone, three cores from each site, were collected from a depth of 2 cm. The sediment cores were collected using a soil-hole borer from at a depth of 2 cm, transferred to sterile plastic containers with the help of sterile spatula, and were taken to the laboratories for analysis within 1 h of collection. A part of the samples to be used for NGS analyses was stored at − 20 °C, and a part of the samples to be used for culture-based analyses was stored at 4 °C. The temperature of each site was recorded in situ, while the pH of the sediment samples was determined by suspending 1 g sediment in 10 ml sterile water. The moisture content of the samples was measured by following the standard protocol (http://www.environment.nsw.gov.au).
DNA extraction
Sediment samples were submitted to Xcelris Labs, Ahmedabad, for obtaining the NGS data. The procedure involved the extraction of genomic DNA from 1 g of composite mixture of sediments from three cores using Xcelgen Sediment gDNA kit using the indirect lysis method. For this, the sediment samples were homogenized by vortexing with glass beads, followed by precipitation of DNA using isopropanol and column purification of the precipitated DNA; the RNA-free DNA was eluted in 30 µl tris-ethylenediaminetetraacetic acid (TE) buffer. Quality of gDNA was checked on 0.8% agarose gel against Hind III marker (New England Biolabs). The quantity and the purity of DNA was measured using Qubit dsDNA high-sensitivity assay kit (Life Technologies).
MiSeq library preparation, sequence generation, and clustering
The libraries for paired-end amplicon sequencing were prepared following 16S metagenomic sequencing library preparation protocol using Nextera XT Index Kit (Illumina Inc.). The V3–V4 region from the DNA obtained was amplified using adapter-ligated primers (Pro341F; 5′-CCTACGGGNBGCASCAG-3′ and Pro805R; 5′-GACTACNVGGGTATCTAATCC-3′), followed by the addition of multiplexing indices (Takahashi et al. 2014). The amplicon library was purified on 1X AMpureXP beads (Beckman Coulter), checked on Bioanalyzer 2100 (Agilant Technologies), and quantified with fluorometer (Life Technologies). 10–20 pM of the purified amplicon was loaded onto Illumina platform for cluster generation and sequencing. Primary image analysis and base-calling for the raw FASTQ reads were performed on MiSeq instrument which included the estimation of various quality parameters, and screened using spacer, conserved region, and mismatch filters. The paired-sequence reads for each library were stitched together using a bioinformatics tool called Flash (Magoc and Salzberg 2011) and were filtered of the chimeric sequences using usearch61 algorithm (Edgar 2010). The quality-filtered reads with phred quality score (Q) > 30 were used for Operational Taxonomic Unit (OTU) clustering. The sequences with 97% similarity were grouped under a single OTU and each OTU was assigned taxonomically against Greengenes database using UCLUST algorithm v1.2.22q (Edgar 2010). The metagenomic data were deposited at Sequence Read Archive (National Center for Biotechnology Information, NCBI; https://www.ncbi.nlm.nih.gov/sra) to obtain accession number (PRJNA362281).
For culture-based studies, 1 g of soil was suspended in 10 ml of sterile water. Bacterial viable counts were obtained by serial tenfold dilution of soil suspension plated onto nutrient agar (NA) medium supplemented with 2% sea-salt and 0.01% cyclohexamide and incubated at 37 °C for 24 h. The colonies with different morphological characteristics were isolated and streaked to obtain pure colonies, and were stored at 4 °C. The pure bacterial cultures were tested for: (1) biochemical activities for use as biofertilizer: plant growth-promoting factor indole-acetic-acid [IAA] production (Ali et al. 2009) and phosphate solubilization (Chaiharn and Lumyong 2011); (2) enzyme activities for bioremediation: amylase (Gupta et al. 2016), cellulase (Kasana et al. 2008), xylanase (Nagar et al. 2012), lipase (Mobarak-Qamsari et al. 2011), protease (Gupta et al. 2016), gelatinase (Balan et al. 2012), laccase (Tekere et al. 2001), and tannase (Brahmbhatt et al. 2014); (3) enzyme activities for chemotherapeutic applications: asparaginase (Mahajan et al. 2013) and glutaminase (Gupta et al. 2016). Each test for the bacterial isolates was done in triplicates.
The bacterial isolates which showed more than two potent biochemical activities were sequenced by Sanger sequencing with universal 16S rDNA primers, namely, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) using big dye terminator v3.1 cycle sequencing kit on ABI 3730xl Genetic Analyzer [Eurofins, Goa] (Haldar and Sengupta 2015). The obtained sequences were trimmed, stitched, and used to identify the bacterial species using Basic Local Alignment Search Tool (BLAST) from NCBI database. The sequences were deposited at GenBank (http://www.ncbi.nlm.nih.gov) to obtain accession numbers (MG255961–MG255977 and MG266442–MG266474). The significant difference in the abundance of bacterial species between the samples was compared using online p value calculator (http://www.graphpad.com/quickcalcs/).
Results and discussion
The pH, temperature, and moisture content of the sediment samples from Mandovi (M) were 6.5, 35 °C, and 28% respectively, and from Zuari (Z) were 6.7, 35 °C, and 29.6% respectively.
MiSeq library sizes and the sequence data from M and Z were 605 bp and 603 bp and 262 Mb and 345 Mb respectively. The total number of stitched reads obtained from MiSeq platform was 1,051,918 and 1,381,440, respectively, from M and Z. After quality filtering, the reads obtained from M and Z were 435,958 and 583,906, respectively, which indicated that 41% and 42% of the reads were retained after quality filtering of the raw reads. The bioproject accession number for metagenome sequences is PRJNA362281 and it is accessible in the link https://www.ncbi.nlm.nih.gov/bioproject/?term. A total number of 16,760 OTUs were obtained from these reads. Out of these, 13,195 OTUs (78.72%) were assigned taxonomically.
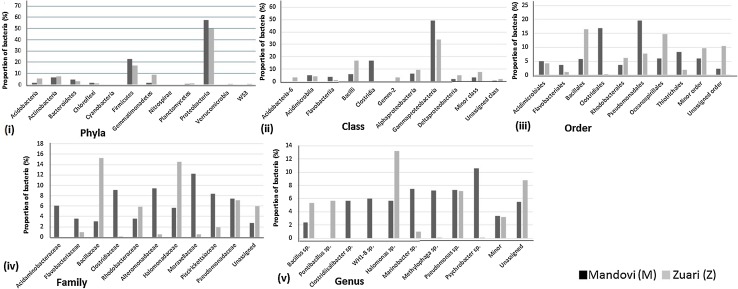
A total number of 12 bacterial phyla and 1 archaeal phylum were identified from these OTUs in both M and Z. The OTUs with low sequence reads (< 5%) were grouped together as minor phyla, while the rest (sequence reads > 5%) were marked as major phyla (Fig. 1i). Proteobacteria was the most abundant bacterial phylum in both M and Z; the proportion being 57.6 and 50%, respectively (p value < 0.005); followed by Firmicutes (23% in M and 17% in Z; p value < 0.005). The other abundant phyla were Actinobacteria (6.5%) followed by Bacteroidetes (5%) in M and Gemmatimonadetes (9%) followed by Actinobacteria (7.4%) in Z, respectively. Our findings corroborate earlier findings which showed these phyla as the major identified phyla from mangrove sediments of Goa and other tropical regions (Fernandes et al. 2015; Alzubaidy et al. 2016; Ghosh and Bhadury 2018).
Fig. 1.
Proportion of bacterial (i) phyla, (ii) class, (iii) order, (iv) family, and (v) genera in Mandovi (M) and Zuari (Z) mangrove sediment samples
Both Gemmatimonadetes (9%) and Acidobacteria (5.5%) which were observed as major phyla in Z were recorded in low proportion in M (1.5% and 2% respectively). It is noteworthy to mention that abundance of Gemmatimonadetes and Acidobacteria has been reported previously from well-preserved mangrove sediments (Jiang et al. 2013), therefore, indicating that the mangrove sediment of Z is ecologically rich than that of M.
Minor components of the bacterial assemblage were affiliated to Chloroflexi (1.5% in M and 1.1% in Z), Cyanobacteria (0.3% in M and 0.4% in Z), Nitrospirae (0.1% in M and 0.3% in Z), Planctomycetes (0.7% in M and 1.4% in Z), Verrucomicrobia (0.3% in M and 0.5% in Z), and WS3 (0.3% in M and 0.5% in Z). This is the first study which has shown the presence of these minor phyla in Goa mangrove ecosystems. However, these have been reported from other mangrove regions of the world through illumina sequencing (Hong et al. 2015; Basak et al. 2015). A recent study has not only identified Planctomycetes and Chloroflexi but has also highlighted their role in nitrogen cycling from Sundarban mangrove sediments (Ghosh and Bhadury 2018). Planctomycetes have been reported to be a component of microbial core in mangrove ecosystem taking part in methane and sulphur metabolism also (Rampadarath et al. 2018).
The difference in distribution of bacteria between M and Z became more prominent with a finer resolution (Fig. 1ii–v). However, the proportion of unassigned taxa increased with lower taxonomic classification. The major five phyla were sub-divided into 14 classes. Out of these, nine formed major classes, while the other five classes, with a proportion less than 5% in both the samples, were pooled together as one minor class; the latter constituted 3.3% and 7.9% in M and Z, respectively. The rest of the classes which could not be resolved taxonomically were grouped under unassigned class that constituted 0.7% and 1.9% in M and Z, respectively (p value < 0.005) (Fig. 1ii).
Among the Proteobacteria, the two major classes observed were Gammaproteobacteria (49.02% in M and 34.1% in Z; p value < 0.005) followed by Alphaproteobacteria (6.3% in M and 9.5% in Z; p value < 0.005). Both of these bacterial classes are active participants in carbon, nitrogen, and sulphur cycling, and metagenomic analyses have identified these classes from vast number of mangrove sediments (Dias et al. 2010; Andreote et al. 2012; Fernandes et al. 2015). In a recent study by Ghosh and Bhadury, similar proportion (68%) of Proteobacteria with persistence of two major classes, namely, Gammaproteobacteria and Alphaproteobacteria, were identified by MiSeq sequencing from Sundarban mangrove sediments. (Ghosh and Bhadury 2018). Though Deltaproteobacteria formed a major class in Z (5.2%), it was observed as a minor class in M (1.9%; p value < 0.005). It is noteworthy to mention that this group of bacteria was observed in high proportion from pristine Tuvem mangrove ecosystems of Goa previously (Fernandes et al. 2015).
The class Bacilli that has been previously reported from other mangrove sediments were significantly higher in Z than in M [17% and 6%; p value < 0.005] (Balakrishnan et al. 2015; Mishra et al. 2012). Acidobacteria-6 was also significantly higher (p value < 0.005) in Z (3.3%) than M (0.3%). It is noteworthy to mention that Proteobacteria and Firmicutes, identified from other mangrove sediments, have been previously reported to be potent sulphate reducers, nitrate reducers, and methanogens (Fernandes et al. 2015; Alzubaidy et al. 2016; Rampadarath et al. 2018). Therefore, taken together, abundance of Gammaproteobacteria and higher proportion of Alphaproteobacteria, Deltaproteobacteria, Bacilli, and Acidobacteria in Z indicated Zuari mangrove sediment to be taxonomically rich, with microorganisms showing greater functional activity than that from Mandovi mangrove sediment.
In contrast, the Clostridia were higher in M (17%) than in Z (0.3%; p value < 0.005). Flavobacteriia (3.6% in M and 1.1% in Z) and Acidimicrobiia (5% in M and 4.2% in Z) were the only two functionally important bacterial groups, significantly higher in M than Z (p value < 0.005).
The bacterial classes from both M and Z were resolved into 8 major orders, 12 orders pooled together as 1 minor order (6% in M and 9.6% in Z), and 1 unassigned order (2.2% and 10.5% in M and Z). Pseudomonadales (20%), followed by Clostridiales (17%), were the two major orders of bacteria in M, while Bacillales (16.5%) followed by Oceanospirillales (15%) formed the two major orders in Z. Pseudomonadales was the third most abundant order in Z (8%), thereby indicating a significant difference in distribution of this order between M and Z [p value < 0.005] (Fig. 1iii). Interestingly, all these groups of bacteria were able to degrade hydrocarbon pollutants and have been detected from urban mangrove forests with elevated anthropogenic inputs of hydrocarbons (Gomes et al. 2008). One recent study from the sediments of Sundarban, the largest mangrove ecosystem, has identified several sequences belonging to Sphingomonadales, Chromatiales, Alteromonadales, Oceanospirillales, and Bacteroidetes, which are known to play important roles in coastal carbon cycling (Ghosh and Bhadury 2018).
A high proportion of Rhodobacterales (6.1%) and a low proportion of nitrogen fixing Rhizobiales (1%) were identified from Z (Fig. 1iii). In contrast, a low proportion of Rhodobacterales (3.6%) was observed in M. However, Rhizobiales were completely absent in this sediment. Rhodobacterales were previously identified from the mangrove microbiome and has been reported to be involved in methane, nitrogen, and sulphur metabolism (Rampadarath et al. 2018). On the other hand, the pathogenic Vibrionales was found at a proportion of 3.4% in M, but was completely absent in Z. This observation not only indicated the presence of pathogens in mangrove ecosystems leading to contagion of native fauna, but also pointed to a greater incidence of anthropogenic activities and pollution in M. To reinforce this finding, a recent report has also identified pathogenic Vibrio spp. from ten mangrove ecosystems of Goa using culture-based techniques (Poharkar et al. 2016). Ten families were ascertained from these 18 orders. The two orders Acidimicrobiales and Gem-2 could not be resolved into families, and were, therefore, grouped as unassigned families (2.7% in M and 6% in Z; p value < 0.005). Moraxellaceae (12.2%), and Alteromonadaceae (9.4%) followed by Clostridiaceae (8.4%) were the three major families in M, while Bacillaceae (15.3%) followed by Halomonadaceae (14.5%), Pseudomonadaceae (7%), and Rhodobacteraceae (6%) formed the major families of bacteria in Z. In addition to Clostridiaceae (9.1%), M also harbored Acidaminobacteraceae (6.1%) as a major bacterial family from Firmicutes. However, in Z, Clostridiaceae (0.1%) was recorded in very low proportion and Acidaminobacteraceae was not observed at all (Fig. 1iv).
Altogether 14 genera with 9 major, 4 minor (3.4% in M and 3.2% in Z), and 1 unassigned (5.5% in M and 8.8% in Z) from M and Z were observed (Fig. 1v). Psychrobacter spp. (10.6%) and Halomonas spp. (13.19%) from Gammaproteobacteria were the two dominant genera in M and Z, respectively. Pseudomonas spp. was the second dominant genus with similar proportion in both M and Z (7.5%). Marinobacter spp. (7.3%) and Methylophaga spp. (7.2%) were the other major genera in M and were present in very low proportion in Z (0.1%). Clostridiisalibacter spp. (5.7%) and WH1-8 spp. (6.1%) were the two dominant genera in M, but were not observed in Z at all. On the other hand, Pontibacillus spp. (5.7%) and Bacillus spp. (5.3%) were dominant in Z. Though the former was not found, the latter genus was observed in low proportion in M (2.4%).
Euryarchaeota was the only phylum from Archaea observed in both M and Z. 16S rDNA-based metagenomic analyses have identified this Euryarchaeota previously from other mangrove ecosystems (Bhattacharyya et al. 2015; Mendes et al. 2012). Halobacteria was the class observed from Euryarchaeota phylum (0.6%) in Z.
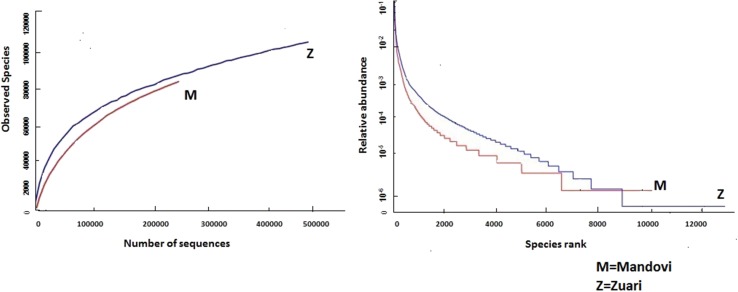
On the basis of OTUs, the alpha and beta diversity, and species richness and evenness were calculated using the software QIIME [Quantitative Insight Into Microbial Ecology] (Caporaso et al. 2010). Non-parametric richness indicators, Chao1 index, and observed species richness estimates accounted to 12172.76 and 8477.00 for M, and 16439.8 and 10,728 for Z, respectively, indicating significantly higher bacterial richness in Z (p value < 0.001). Steep gradient of rarefaction and rank-abundance curves obtained for Z depicted higher number of bacterial taxa with uneven distribution in comparison to M (Fig. 2). Low Jaccard similarity index (0.354) and high Bray–Curtis dissimilarity index (0.861) along with high Euclidean distance (72076.73) between the two sediment samples potentiated the findings, indicating that the bacterial composition between M and Z was very different.
Fig. 2.
(i) Rarefaction and (ii) rank-abundance curves obtained from the Illumina sequence reads for Mandovi (M) and Zuari (Z) mangrove sediment samples
A total number of 60 bacterial isolates: 35 from M, named from M001 to M035 and 25 from Z, named from Z001 to Z025, were isolated on NA plates. These isolates were tested for their Gram characteristics: a total of 43 isolates (71.6%) belonged to Gram-positive rods, while 17 isolates (28.3%) were Gram-positive cocci.
The bacterial isolates from M and Z were compared with respect to their abilities to be used for the biofertilization and capabilities of producing biodegradative and antineoplastic enzymes (Table 1). It was noted that only one isolate each from M and from Z exhibited the maximum number of biochemical activities, that is 7 out of 13 tests, showing IAA production > 5 µg/ml, phosphate solubilization, cellulase, xylanase, lipase, gelatinase, and laccase activities.
Table 1.
Bacterial species showing bioremediation and biofertilization activities
| Activity | Mandovi (M) | Zuari (Z) | Identified species from Mandovi (M) and Zuari (Z) (strain no) | ||
|---|---|---|---|---|---|
| Number of isolates (%) of isolates | Number of isolates (%) of isolates | ||||
| I. Biofertilizer activity | |||||
| IAA production > 5 μg/ml | 15 | 42.85% | 8 | 32.00% | Bacillus cereus (M027, Z001, Z004); B. toyonensis (M009, M016); Lysinibacillus macroides (M002, M004, M005, M010, M017, M020, M021, M024, Z010, Z019); Paenibacillus illinoisensis (Z011); P. pabuli (Z020) P. xylanilyticus (M032, M033); Staphylococcus hominis (M006, M013, Z017); S. pasteuri (Z018) |
| Phosphate solubilization | 4 | 11.42% | 1 | 4.00% | Bacillus toyonensis (M022); Lysinibacillus macroides (M002, M003, M005, Z010) |
| II. Biodegradative enzymes | |||||
| Amylase | 13 | 37.30% | 14 | 56.00% | Bacillus cereus (M028, Z001, Z004, Z005, Z015); B. toyonensis (M001, M009, M012, M016, M022, M026; Z003, Z008, Z012, Z013); Lysinibacillus macroides (M003, M004, M019, M020, M024); Paenibacillus illinoisensis (Z002); P. xylanilyticus (M033, Z014); Staphylococcus hominis (Z017) |
| Cellulase | 6 | 17.14% | 17 | 68.00% | Bacillus cereus (M027, M029, Z001, Z004, Z005, Z015); B. toyonensis (M001, M026, Z003, Z006, Z008, Z012, Z013); Lysinibacillus macroides (M002, M004, Z010, Z019); Paenibacillus illinoisensis (Z002, Z011); P. spabuli (Z020); P. xylanilyticus (Z007, Z014); Staphylococcus hominis (Z017) |
| Xylanase | 11 | 31.42% | 3 | 12% | Bacillus toyonensis (M001, M022, Z005); Lysinibacillus macroides (M002, M003, M004, M005, M008, M017, M021, M024, Z010); Staphylococcus hominis (Z017); S. pasteuri (M007) |
| Protease | 0 | – | 4 | 16% | Bacillus cereus (Z009); B. toyonensis (Z012); Lysinibacillus macroides (Z019); Paenibacillus illinoisensis (Z011) |
| Gelatinase | 5 | 14.28% | 17 | 68.00% | Bacillus cereus (Z001, Z004, Z005, Z009, Z015); B. toyonensis (M001, M016, M026, Z003, Z006, Z008, Z012, Z013); Lysinibacillus macroides (M002, Z010); Paenibacillus illinoisensis (Z002); P. pabuli (strain Z020); P. xylanilyticus (Z007, Z014) Staphylococcus hominis (Z017); S. pasteuri (M007, Z018) |
| Lipase (Tween 80) | 8 | 22.8% | 12 | 48.00% | Bacillus cereus (Z009, Z015); B. toyonensis (M022, Z008, Z012, Z013) Lysinibacillus macroides (M003, M004, M002, M008, M021, Z010, Z019); Paenibacillus illinoisensis (Z002, Z011); P. xylanilyticus (Z014); Staphylococcus hominis (M006); S. pasteuri (Z018) |
| Lipase (TBT) | 2 | 5.71% | 10 | 40% | Bacillus cereus (Z001, Z004, Z005); B. toyonensis (M016, Z003, Z006, Z013) Lysinibacillus macroides (M002, Z010); Paenibacillus illinoisensis (Z002, Z011) |
| Tannase | 15 | 42.85% | 0 | – | Bacillus toyonensis (M009, M015, M016); Lysinibacillus macroides (M002, M003, M004, M005, M008, M010, M017, M019, M020); Staphylococcus hominis (M006); S. pasteuri (M007) |
| Laccase | 3 | 8.5% | 13 | 52% | Bacillus cereus (Z001, Z004, Z005, Z015); B. toyonensis (Z003, Z006, Z008, Z012, Z013); Lysinibacillus macroides (M002, M003, M005, Z010); Paenibacillus illinoisensis (Z002); P. xylanilyticus (Z014) |
| III. Antineoplastic enzymes | |||||
| Glutaminase | 11 | 31.42% | 0 | – | Bacillus toyonensis (M016, M022, M023); Lysinibacillus macroides (M017, M019, M020, M021, M024); Staphylococcus hominis (M013); S. pasteuri (M025) |
| Asparaginase | 0 | – | 3 | 12% | Bacillus cereus (Z005); Paenibacillus xylanilyticus (Z014); P. illinoisensis (Z002) |
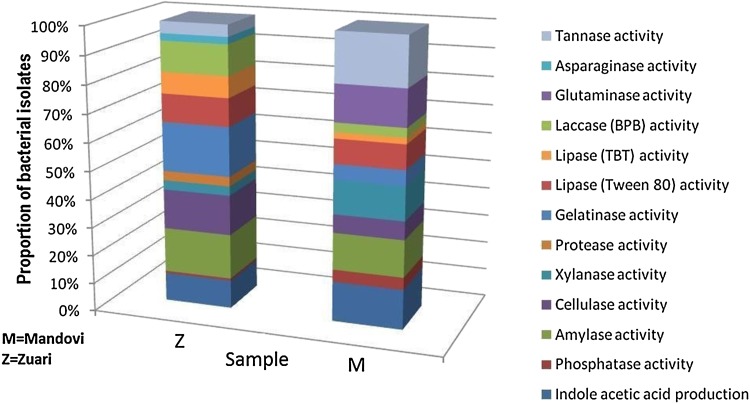
The amount of produced IAA varied from 2 to 8 µg/ml in both M and Z. However, the proportion of bacterial isolates producing ≥ 5 µg/ml IAA were more in M (42.85%) as compared to Z (32.00%). Four isolates (11.42%) from M and one isolate (4%) from Z produced a clear zone indicating phosphate solubilization. The zone diameter varied from 2.4 to 2.7 mm ± (0.22 to 0.34) in sample M, while it was 2.9 mm ± (0.44 to 0.52) for Z sample, respectively. One isolate from M (M002) produced more than 5 µg/ml IAA as well as a clearance zone of 2.5 mm ± 0.39 of phosphate solubilization, thereby indicating that this isolate could be important candidate for biofertilizer formulation.
With respect to bioremediation activities, xylanase-producing bacteria were significantly more in M (31.42%) than Z (12%; p value < 0.005). The diameter of the zone of clearance for xylanse activity varied between 1.8 and 2.2 mm ± 0.21 in both M and Z. The tannase activity was observed in M only where 45.71% isolates showed the activity with the diameter of produced zone varying from 1.1 to 2.3 mm ± 0.14.
The other five enzyme activities including amylase, cellulase, lipase, gelatinase, and laccase were more in Z than in M (Fig. 3). The clearance zone diameter produced by the bacterial isolates from Z for amylase, cellulase, and lipase activities varied from 1.6 to 2.2 mm ± 0.16. For gelatinase activity, the zone diameters recorded were around 3 to 3.4 mm ± 0.2.
Fig. 3.
Distribution of enzyme activities with respect to biofertilization, biodegradation, and chemotherapy in Mandovi (M) and Zuari (Z) mangrove sediment samples
Four (16%) bacterial isolates showed protease activity from Z, while this activity was not found among the bacterial isolates from M. Out of the four bacterial isolates which showed positive protease activity, three were also positive for gelatinase activity. Though protease activity was not recorded from isolates from M, five of the isolates (14.28%) could degrade gelatin. Eight (22.85%) and twelve (48%) isolates from M and Z could degrade tween 80, while two (5.71%) and ten (40%) isolates from M and Z could degrade tri-butyryl toluene (TBT), respectively. One isolate from M and four isolates from Z could degrade both Tween 80 and TBT.
Eleven (31.42%) isolates from M showed glutaminase activity, while 12% of the isolates (3 isolates) from Z utilized asparagine. Interestingly, asparaginase activity was not found in isolates from M and glutaminase activity was not exhibited among the isolates from Z, indicating the exclusivity of bacteria producing glutaminase in M, and of those producing asparaginase in Z (Fig. 3). It is noteworthy to mention that this is the first report on glutaminase and asparaginase activities from mangrove sediments. The previous studies have isolated bacterial species showing these activities from other soil samples (Saxena et al. 2015; Bhattacharyya et al. 2007).
A total number of 50 bacterial isolates: 31 out of 35 (88.5%) and 19 out of 25 (76%) from M and Z, respectively, which showed positive results for more than two biochemical activities, were sequenced and 1.2–1.4 kb of sequence were obtained for each of them. A correlation of the identified species with their biochemical activities is given in Table 1. The percentage of identity for all the sequenced isolates was 98%–99%. GenBank accession numbers for the identified species are MG255961–MG255977 and MG266442–MG266474, and the details are accessible by the link https://www.ncbi.nlm.nih.gov/nuccore/.
The identified bacterial isolates belonged to the phylum Firmicutes; the latter also being identified as a dominant phylum from both M and Z using Illumina sequencing. The identified Firmicutes belonged to Bacillus spp. [12 from M (38.7%) and 10 from Z (52.67%)], Lysinibacillus spp. [11 from M (35.4%) and 2 from Z (10.5%)], Paenibacillus spp. [3 from M (9.67%) and 5 from Z (26.3%)], and Staphylococcus spp. [5 from M (16.1%) and 2 from Z (10.5%)]. It is noteworthy to mention that Bacillus spp. were also observed as dominant species from Firmicutes from both M and Z by Illumina sequencing and the proportion of this species was observed to be higher in Z than M by both culture-based and NGS methods.
The two species identified from Bacillus genus were Bacillus cereus CCM 2010 (4 from M and 4 from Z) and Bacillus toyonensis BCT-7112 (8 from M and 6 from Z). Lysinibacillus macroides LMG 18,474 (11 from M and 2 from Z) and Paenibacillus xylanilyticus XIL14 (3 from M and 2 from Z) were the two common species present in both the sediments. Two Paenibacillus illinoisensis JCM 9907 and one Paenibacillus pabuli HSCC 492 species were identified in Z. Staphylococcus hominis DM 122 (2 from M and 1 from Z) and Staphylococcus pasteuri ATCC 51,121 (3 from M and 1 from Z) were the two types of species observed from Staphylococcus genus. The isolate which showed the maximum number of positive biofertilization and bioremediation activities in both M and Z was identified as Lysinibacillus macroides LMG18474 (isolate number M002 and Z010 from M and Z, respectively).
Conclusions
This is the first study that explored the bacterial diversity for two major mangrove sediments of Goa: Mandovi and Zuari, using Illumina sequencing and revealed the presence of distinct bacterial consortia in these two sediments. Higher proportion of ecologically important bacterial species from Proteobacteria and Bacillus indicated Zuari mangrove sediment to be an environmentally rich niche, while the presence of pathogenic bacteria Vibrio spp. in Mandovi sediment is indicative of a greater incidence of anthropogenic activities and pollution in the latter. Pollution, particularly in the Mandovi estuary, has risen due to anthropogenic and industrial activities, the major contribution coming from metal (Bicholkar and Nazareth 2015; Ratnaprabha and Nayak 2011), as well as of sewerage, as recorded by the Goa State Pollution Control Board (GSPCB) which has declared the River Mandovi to be the most polluted river in Goa caused by the release of raw sewerage, the off-shore casinos in the estuary forming the main contributors to sewerage contamination, and of rampant dumping of garbage (goaspcb.gov.in/). The analysis of functional attributes identified higher number of bacterial isolates capable of producing biodegradative and antineoplastic enzymes from Zuari sediment, thereby reinforcing the fact that the Zuari mangrove sediment is a rich repository for biotechnologically important bacterial species. Taken together, this study not only is valuable in evaluation of ecology of these regions but also points towards the necessity for restoration of sediment microflora in these mangrove ecosystems of Goa.
Acknowledgements
The research work was funded by National Post-Doctoral Fellowship Scheme of Science and Engineering Research Board, Department of Science and Technology, New Delhi. The authors thank Goa University, Goa, for providing the necessary infrastructural support for this research work.
Author contributions
The sample collection, culture-based isolation, and data analysis for both NGS and Sanger sequencing and preparation of manuscript was done by S. Haldar. S. Nazareth contributed in the correction/editing of the manuscript.
Compliance with ethical standards
Conflict of interest
On behalf of all the authors, the corresponding author declares that there is no conflict of interest.
Contributor Information
Shyamalina Haldar, Email: haldarshyamalina@gmail.com.
Sarita W. Nazareth, Email: sarita@unigoa.ac.in
References
- Aizpuru M, Achard F, Blasco F. EEC Research project n 15017-1999-05 FIED. Ispra: ISP FR–Joint Research Centre; 2000. Global assessment of cover change of the mangrove forests using satellite imagery at medium to high resolution. [Google Scholar]
- Ali B, Sabri AN, Ljung K, Hasnain S. Auxin production by plant associated bacteria: impact on endogenous IAA content and growth of Triticum aestivum L. Lett Appl Microbiol. 2009;48(5):542–547. doi: 10.1111/j.1472-765X.2009.02565.x. [DOI] [PubMed] [Google Scholar]
- Alzubaidy H, Essack M, Malas TB, Bokhari A, Motwalli O, Kamanu FK, Jamhor SA, Mokhtar NA, Antunes A, Simões MF, Alam I, Bougouffa S, Lafi FF, Bajic VB, Archer JA. Rhizosphere microbiome metagenomic of gray mangroves (Avicennia marina) in the Red Sea. Gene. 2016;576:626–636. doi: 10.1016/j.gene.2015.10.032. [DOI] [PubMed] [Google Scholar]
- Andreote FD, Jiménez DJ, Chaves D. The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS One. 2012;7(6):e38600. doi: 10.1371/journal.pone.0038600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Li J, He Z, Van Nostrand JD, Tian Y, Lin G, Zhou J, Zheng T. GeoChip-based analysis of the functional gene diversity and metabolic potential of soil microbial communities of mangroves. Appl Microbiol Biotechnol. 2013;97(15):7035–7048. doi: 10.1007/s00253-012-4496-z. [DOI] [PubMed] [Google Scholar]
- Balakrishnan S, Indira K, Srinivasan MJ. Mosquitocidal properties of Bacillus species isolated from mangroves of Vellar estuary, Southeast coast of India. Parasit Dis. 2015;39(3):385–392. doi: 10.1007/s12639-013-0371-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Balan SS, Nethaji R, Sankar S, Jayalakshmi S. Production of gelatinase enzyme from Bacillus spp. isolated from the sediment sample of Porto Novo Coastal sites. Asian Pac J Trop Biomed. 2012;2:S1811–S1816. doi: 10.1016/S2221-1691(12)60500-0. [DOI] [Google Scholar]
- Basak P, Majumder NS, Nag S, Bhattacharyya A, Roy D, Chakraborty A, SenGupta S, Roy A, Mukherjee A, Pattanayak R, Ghosh A, Chattopadhyay D, Bhattacharyya M. Spatiotemporal analysis of bacterial diversity in sediments of Sundarbans using parallel 16S rRNA gene tag sequencing. Microb Ecol. 2015;69(3):500–511. doi: 10.1007/s00248-014-0498-y. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya P, Chakrabarti K, Tripathy S, Chakraborty A, Kim K, Kim SH. L-asparaginase and L-glutaminase activities in submerged rice soil amended with municipal solid waste compost and decomposed cow manure. J Environ Sci Health B. 2007;42(5):593–598. doi: 10.1080/03601230701389462. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Majumder NS, Basak P, Mukherji S, Roy D, Nag S, Haldar A, Chattopadhyay D, Mitra S, Bhattacharyya M, Ghosh A. Diversity and distribution of Archaea in the Mangrove sediment of Sundarbans. Archaea. 2015;2015:968582. doi: 10.1155/2015/968582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicholker AA, Nazareth SW. A comparative study of metal tolerance and sorption capacities of varied fungal genera frommetal—polluted estuarine environments for potential in metal bioremediation. Kavaka. 2015;44:16–29. [Google Scholar]
- Brahmbhatt D, Modi HA, Jain NK. Preliminary isolation and screening of tannase producing bacteria and fungi. Int J Curr Microbiol Appl Sci. 2014;3(11):193–203. [Google Scholar]
- Cabral L, Júnior GV, Pereira de Sousa ST, Dias AC, Lira Cadete L, Andreote FD, Hess M, de Oliveira VM. Anthropogenic impact on mangrove sediments triggers differential responses in the heavy metals and antibiotic resistomes of microbial communities. Environ Pollut. 2016;216:460–469. doi: 10.1016/j.envpol.2016.05.078. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda LE, Barbosa O. Metagenomic analysis exploring taxonomic and functional diversity of soil microbial communities in Chilean vineyards and surrounding native forests. Peer J. 2017;30(5):e3098. doi: 10.7717/peerj.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiharn M, Lumyong S. Screening and optimization of indole-3-acetic acid production and phosphate solubilization from rhizobacteria aimed at improving plant growth. Curr Microbiol. 2011;62(1):173–181. doi: 10.1007/s00284-010-9674-6. [DOI] [PubMed] [Google Scholar]
- Dias AC, Andreote FD, Rigonato J, Fiore MF, Melo IS, Araújo WL. Brazilian non-disturbed mangrove sediment. Antonie Van Leeuwenhoek. 2010;98(4):541–551. doi: 10.1007/s10482-010-9471-z. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2246–2460. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fernandes SO, Kirchman DL, Michotey VD, Bonin PC, LokaBharathi PA. Bacterial diversity in relatively pristine and anthropogenically-influenced mangrove ecosystems (Goa, India) Braz J Microbiol. 2015;45(4):1161–1171. doi: 10.1590/S1517-83822014000400006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Bhadury P. Investigating monsoon and post-monsoon variabilities of bacterioplankton communities in a mangrove ecosystem. Environ Sci Pollut Res Int. 2018;25(6):5722–5739. doi: 10.1007/s11356-017-0852-y. [DOI] [PubMed] [Google Scholar]
- Giri C, Long J, Abbas S, Murali RM, Qamer FM, Pengra B, Thau D. Status and distribution of mangrove forests of the world using earth observation satellite data. Glob Ecol Biogeogr. 2015;20(1):154–159. doi: 10.1016/j.jenvman.2014.01.020. [DOI] [Google Scholar]
- Gomes NC, Borges LR, Paranhos R, Pinto FN, Mendonça-Hagler LC, Smalla K. Exploring the diversity of bacterial communities in sediments of urban mangrove forests. FEMS Microbiol Ecol. 2008;66(1):96–109. doi: 10.1111/j.1574-6941.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- Gupta S, Sharma P, Dev K, Sourirajan A. Halophilic bacteria of Lunsu produce an array of industrially important enzymes with salt tolerant activity. Biochem Res Int. 2016;2016:9237418. doi: 10.1155/2016/9237418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtom H, Demanèche S, Dawson L, Azulay C, Matan O, Robe P, Gafny R, Simonet P, Jurkevitch E, Pasternak Z. Soil characterisation by bacterial community analysis for forensic applications: a quantitative comparison of environmental technologies. Forensic Sci Int Genet. 2017;26:21–29. doi: 10.1016/j.fsigen.2016.10.005. [DOI] [PubMed] [Google Scholar]
- Haldar S, Sengupta S. Impact of plant development on the rhizobacterial population of Arachis hypogaea: a multifactorial analysis. J Basic Microbiol. 2015;55(7):922–928. doi: 10.1002/jobm.201400683. [DOI] [PubMed] [Google Scholar]
- Hong Y, Liao D, Hu A, Wang H, Chen J, Khan S, Su J, Li H. Diversity of endophytic and rhizoplane bacterial communities associated with exotic Spartina alterniflora and native mangrove using Illumina amplicon sequencing. Can J Microbiol. 2015;61(10):723–733. doi: 10.1139/cjm-2015-0079. [DOI] [PubMed] [Google Scholar]
- Husain I, Sharma A, Kumar S, Malik F. Purification and characterization of glutaminase free asparaginase from Pseudomonas otitidis: induce apoptosis in human leukemia MOLT-4 cells. Biochimie. 2016;121:38–51. doi: 10.1016/j.biochi.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Jiang XT, Peng X, Deng GH, Sheng HF, Wang Y, Zhou HW, Tam NF. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol. 2013;66(1):96–104. doi: 10.1007/s00248-013-0238-8. [DOI] [PubMed] [Google Scholar]
- Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr Microbiol. 2008;57(5):503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- Kharangate-Lad A, Bhosle S. Studies on siderophore and pigment produced by an adhered bacterial strain Halobacillus trueperi MXM-16 from the mangrove ecosystem of Goa, India. Indian J Microbiol. 2016;56(4):461–466. doi: 10.1007/s12088-016-0591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Morrison M, Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods. 2011;84(1):81–87. doi: 10.1016/j.mimet.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan RV, Saran S, Saxena RK, Srivastava AK. A rapid, efficient and sensitive plate assay for detection and screening of l-asparaginase-producing microorganisms. FEMS Microbiol Lett. 2013;341(2):122–126. doi: 10.1111/1574-6968.12100. [DOI] [PubMed] [Google Scholar]
- Mendes LW, Taketani RG, Navarrete AA, Tsai SM. Shifts in phylogenetic diversity of archaeal communities in mangrove sediments at different sites and depths in southeastern Brazil. Res Microbiol. 2012;163(5):366–377. doi: 10.1016/j.resmic.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Mishra RR, Swain MR, Dangar TK, Thatoi H. Diversity and seasonal fluctuation of predominant microbial communities in Bhitarkanika, a tropical mangrove ecosystem in India. Rev Biol Trop. 2012;60(2):909–924. doi: 10.15517/rbt.v60i2.4026. [DOI] [PubMed] [Google Scholar]
- Mobarak-Qamsari E, Kasra-Kermanshahi R, Moosavi-Nejad Z. Isolation and identification of a novel, lipase-producing bacterium, Pseudomnas aeruginosa KM110. Iran J Microbiol. 2011;3(2):92–98. [PMC free article] [PubMed] [Google Scholar]
- Nagar S, Mittal A, Kumar D, Gupta VK. Production of alkali tolerant cellulase free xylanase in high levels by Bacillus pumilus SV-205. Int J Biol Macromol. 2012;50(2):414–420. doi: 10.1016/j.ijbiomac.2011.12.026. [DOI] [PubMed] [Google Scholar]
- Poharkar KV, Kerkar S, D’Costa D, Doijad S, Barbuddhe SB. Mangrove ecosystems: an adopted habitat for pathogenic Salmonella spp. Water Environ Res. 2016;88(3):264–271. doi: 10.2175/106143016X14504669767733. [DOI] [PubMed] [Google Scholar]
- Rampadarath S, Bandhoa K, Puchooa D, Jeewon R, Bal S. Metatranscriptomics analysis of mangroves habitats around Mauritius. World J Microbiol Biotechnol. 2018;34(4):59. doi: 10.1007/s11274-018-2442-7. [DOI] [PubMed] [Google Scholar]
- Ratnaprabha S, Nayak GN. Mudflats in lowermiddle estuary as a favorable location for concentration of metals, west coast of India. Indian J Geomar Sci. 2011;40:372–385. [Google Scholar]
- Reda FM. Kinetic properties of Streptomyces canariusl-Glutaminase and its anticancer efficiency. Braz J Microbiol. 2015;46(4):957–968. doi: 10.1590/S1517-838246420130847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Upadhyay R, Kango N. Isolation and identification of actinomycetes for production of novel extracellular glutaminase free l-asparaginase. Indian J Exp Biol. 2015;53(12):786–793. [PubMed] [Google Scholar]
- Sharma A, Lal R. Survey of (Meta)genomic approaches for understanding microbial community dynamics. Indian J Microbiol. 2017;57(1):23–38. doi: 10.1007/s12088-016-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares Júnior FL, Dias AC, Fasanella CC, Taketani RG, de Souza Lima AO, Melo IS, Andreote FD. Endo- and exoglucanase activities in bacteria from mangrove sediment. Braz J Microbiol. 2014;44(3):969–976. doi: 10.1590/S1517-83822013000300048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One. 2014;9(8):e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekere M, Mswaka AY, Zvauya R, Read JS. Growth, dye degradation and ligninolytic activity studies on Zimbabwean white rot fungi. Enzyme Microb Technol. 2001;28(4–5):420–426. doi: 10.1016/S0141-0229(00)00343-4. [DOI] [PubMed] [Google Scholar]