Abstract
Double minutes (dmin), homogeneously staining regions, and ring chromosomes are vehicles of gene amplification in cancer. The underlying mechanism leading to their formation as well as their structure and function in acute myeloid leukemia (AML) remain mysterious. We combined a range of high-resolution genomic methods to investigate the architecture and expression pattern of amplicons involving chromosome band 8q24 in 23 cases of AML (AML-amp). This revealed that different MYC-dmin architectures can coexist within the same leukemic cell population, indicating a step-wise evolution rather than a single event origin, such as through chromothripsis. This was supported also by the analysis of the chromothripsis criteria, that poorly matched the model in our samples. Furthermore, we found that dmin could evolve toward ring chromosomes stabilized by neocentromeres. Surprisingly, amplified genes (mainly PVT1) frequently participated in fusion transcripts lacking a corresponding DNA template. We also detected a significant overexpression of the circular RNA of PVT1 (circPVT1) in AML-amp cases versus AML with a normal karyotype. Our results show that 8q24 amplicons in AML are surprisingly plastic DNA structures with an unexpected association to novel fusion transcripts and circular RNAs.
Introduction
Genomic amplifications in the form of double minutes (dmin), homogeneously staining regions (hsr), and ring chromosomes are frequent in cancer [1], although rare in leukemia (<1% of cytogenetically abnormal hematological malignancies) [2], and associated with an elderly age and poor prognosis [3, 4]. Despite this clinical impact, the underlying mechanisms of amplicon formation in acute myeloid leukemia (AML) are unclear, and so are their internal structures. Chromothripsis was described as a potential mechanism involved in their genesis [5, 6]. However, the evidence we previously obtained in seven tumor cell lines harboring 8q24 amplifications clearly excluded this model in our samples [7], supporting alternative mechanisms such as the episome model [8–10]. Another crucial aspect of amplicons is their impact on the transcriptome. Amplified cancer-associated genes are often upregulated as a direct consequence of their copy number (CN) gain [11]. Nevertheless, MYC, as well as other 8q24 co-amplified genes and long noncoding RNAs (lncRNAs) (e.g., TRIB1, FAM84B, POU5F1B, PVT1, and TMEM75), represents a clear exception to this rule in myeloid malignancies [9, 12, 13]. These findings suggest that none of them could be considered as the real target for this type of amplification [14].
We investigated, at both genomic and transcriptomic levels, a large cohort of AML cases harboring MYC amplifications in the form of dmin, hsr, or ring chromosomes (AML-amp), in order to unravel the molecular mechanisms as well as potential target genes behind their genesis. To this end, we combined fluorescence in situ hybridization (FISH), immuno-FISH, PCR, single-nucleotide polymorphism (SNP) array, whole-genome sequencing (WGS), and RNA sequencing (RNA-seq). Overall, our study provides a full overview of the 8q24 amplicon organization in dmin/hsr, as well as on ring chromosomes, and sheds light on the extraordinarily high transcriptome plasticity associated with AML-amp.
Materials and methods
Sample collection
This study included a total of 24 AML-amp cases (Supplementary Table S1), two remission samples from cases that were AML-amp at diagnosis, 29 AML cases with a normal karyotype (AML-NK), 10 AML cases with chromosomal abnormalities not involving chromosome 8, 23 AML cell lines with or without 8q24 gain, and one normal peripheral blood (PB) and one bone marrow (BM) sample as controls (Supplementary Table S1 and Supplementary Methods).
Neocentromere detection on ring chromosomes
AML-amp cases MLL_11929 and MLL_11933, as well as two additional ones (#18 and #33) included in a previously described cohort [9], with ring chromosomes were investigated for the presence of neocentromere using FISH co-hybridization experiments with an anti-CENP-C polyclonal antibody (Cat. no. ABIN3072962 Eurogentec polyclonal antibody service, Aachen, Germany) for functional centromeres [15], a pan-centromeric probe detecting all types of alphoid subset sequences [16], and a BAC probe (RP11-440N18) specific for MYC.
SNP array analysis
SNP array analysis was performed on 24 AML-amp cases using the Affymetrix CytoScan HD platform (Affymetrix, Santa Clara, CA, USA) following the manufacturer’s protocol.
WGS and identification of structural variants
WGS was performed on 23 AML-amp cases and two remission samples (Supplementary Table S1) using the X Ten platform (Illumina, San Diego, CA, USA), in a paired-end 2 × 150-cycle run, with a mean coverage of 30× per sample. Candidate structural variants (SVs) were identified using the DELLY software (v.0.7.1) [17] (details are provided in Supplementary Methods).
Chromothripsis analysis
To infer chromothripsis, WGS and SNP array profiles of each case were evaluated according to published criteria [18] (Supplementary Methods). We focused on the region 8q22.3–24.1 involved in the amplifications.
RNA-seq analysis
RNA-seq was performed on 20 AML-amp cases and two remission samples (Supplementary Table S1). RNA-seq libraries were prepared using the TruSeq RNA Sample Preparation Kit v2 (Illumina) and sequenced on the HiSeq2000 platform (Illumina) in paired-end 2 × 101-cycle runs (166 Mreads/sample). To detect fusion transcripts involving amplified genes, we used both ChimeraScan (CS) [19] and FusionMap (FM) [20] (Supplementary Methods). A custom bioinformatics pipeline was implemented to combine SVs with chimeric transcript data and evaluate the occurrence of post-transcriptional events in the genesis of chimeras.
Differential gene expression analysis
To ascertain genes specifically expressed only in AML-amp, six cases harboring 8q24 amplicons with no other chromosomal changes were compared with six AML-NK cases from The Cancer Genome Atlas (TCGA) (Supplementary Table S1). The DESeq2 Bioconductor package (v1.8.1) in R environment (v3.2.2) was used for Differential gene expression analysis (DEA) of global gene expression profiles in cases with/without 8q24 amplicons as well as for explorations of 8q24 genes in the AML-amp data set (Supplementary Methods). The ToppGene suite was used to perform gene enrichment analysis (https://toppgene.cchmc.org/).
Mutation analysis of 8q24-amplified genes
Genome and transcriptome data of 20 AML-amp cases (Supplementary Table S1) were analyzed in parallel for detection of non-synonymous single-nucleotide variants (SNVs) and insertions/deletions (indels). Variants were called using GATK HaplotypeCaller [21] and VarScan2 (VarScan 2.3.7) [22] on RNA-seq and WGS data, respectively, and then filtered according to specific criteria (Supplementary Methods). Pathway analysis was performed using the MSigDB [23], GSEA [24], and DAVID [25] tools to identify pathways associated with the mutated genes.
Assessment of circular PVT1 (circPVT1) expression level
The expression of both linear and circPVT1 in eight AML-amp cases, 12 AML cell lines, nine AML-NK cases, and in one normal PB and one BM was analyzed by qPCR using properly designed primers [26]. The nine AML-NK samples were pooled and used as a calibrator; beta-2-microglobulin (B2M) was used as a reference gene. Statistical significance was evaluated using the relative expression software tool REST [27].
Results
8q24 dmin/hsr/rings amplify a 2246 kbp genomic segment encompassing 11 lncRNAs and four coding genes
By combining SNP array, WGS and FISH analyses, we defined the 8q24 amplicon organization in 23 AML-amp cases. We identified a total of 373,585 raw SVs, of which 4630 were within the 8q24-amplified regions (Supplementary Table S2) and 84 of this subset (2%) were considered as crucial for amplicon assembly, since they corresponded to CN switches. Their validation by PCR and Sanger sequencing revealed the occurrence of 30 microhomology sequences, 36 anonymous insertions, and 18 blunt-end joining events.
All AML-amp cases harbored amplified sequences derived only from chromosome 8. The 8q24 amplifications were mainly present on dmin or ring chromosomes, and only rarely on hsr (Supplementary Table S3). The proximal and distal breakpoints of the amplicons clustered in two regions: the former at chr8:125,754,780–126,442,227 (687 kbp) in 20/23 (87%) cases and the latter at chr8:130,690,866–130,950,708 (260 kbp) in 21/23 (91%) cases. The minimal commonly amplified segment was 2,245,660 bp (Supplementary Figure S1). It comprises 15 RefSeq loci, including 11 lncRNAs (LINC00861, LOC101927657, PCAT1, PCAT2, PRNCR1, CASC19, CCAT1, CASC21, CASC8, CCAT2, and CASC11) and four coding genes (TRIB1, FAM84B, POU5F1B, and MYC). In addition, the lncRNAs PVT1 and CCDC26, just outside the commonly amplified region, were amplified in 22/24 (92%) cases.
8q24 amplicon structures suggest different levels of molecular heterogeneity
All 23 cases investigated by WGS were classified according to the heterogeneity of 8q24 amplicons. In 21 out of 23 cases, we identified a most represented “ancestral” amplicon, which was repeated in a head-to-tail fashion.
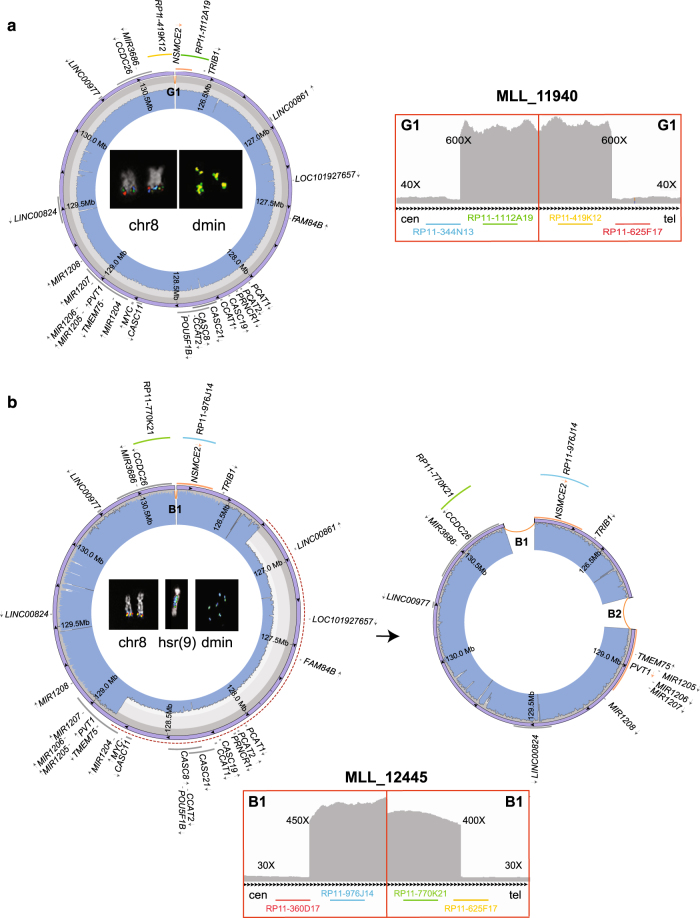
Eight cases (35%) displayed a single ancestral amplicon, ranging from 3216 kbp to 5196 kbp, without remarkable switches in their CN profiles. These cases were considered to harbor non-heterogeneous amplicons (Fig. 1a, Supplementary Figure S2a-g). A duplication SV joined the distal and the proximal breakpoints of two adjacent amplified segments in all cases except MLL_11932 and MLL_11937 (Supplementary Figure S2a-g).
Fig. 1.
Circular amplicons with no or low heterogeneity of structure. Images show the internal structure of amplified segments in cases MLL_11940 (as an example of a patient with no heterogeneous amplicons joined by a duplication) (a) and MLL_12445 (as an example of patient with two coexisting amplicons, one of which derived from the other by a deletion) (b). For each patient: IGV plot of WGS read depth at proximal and distal breakpoints of ancestral amplicons; Circos plots (outer violet circular panel) and WGS read depth (light blue inner panel) of recurrent amplicons, with internal arrows indicating amplicon orientation and genes in dark gray, or in orange if interrupted by a breakpoint (arrows represent the transcriptional orientation). Inside the Circos plots, FISH pseudocolor images showing co-localizing amplified probes, consistently colored as in the Circos plot. Probes mapping respectively upstream and downstream of the amplicons proximal and distal breakpoints are not amplified on dmin and display signal only on normal chromosome 8. The red-dashed semicircles in b represent sequence deletion on the ancestral amplicon to obtain the secondary one
The remaining 15 of the 23 cases showed different levels of amplicon heterogeneity, which we dichotomized into low-grade heterogeneity (LH) and high-grade heterogeneity (HH). In total, 12/23 (52%) cases were classified as LH based on the co-existence of heterogeneous amplicons related to, and derived from, the ancestral amplified segment through the occurrence of internal SVs (Fig. 1b, Supplementary Figure S2h-r). Notably, all LH amplicons shared proximal and distal breakpoints in the same patient (Fig. 1b).
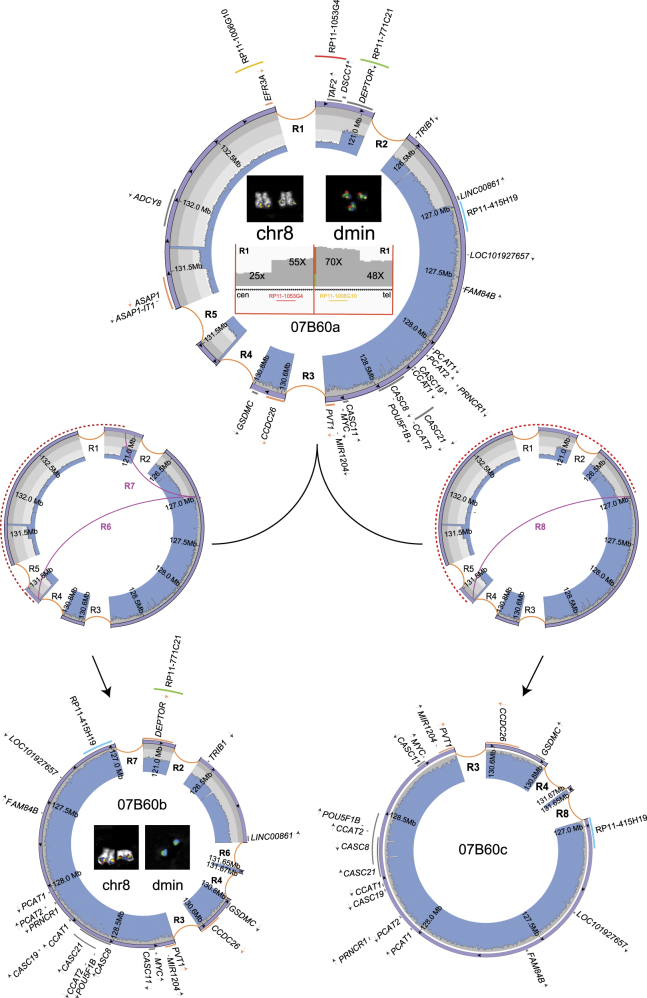
Three (13%) of the 23 cases were considered HH because they comprised distinct cell subpopulations carrying amplicons with different internal structures arranged in a noncontiguous, noncollinear array (Fig. 2, Supplementary Figure S2s-t, Supplementary Methods).
Fig. 2.
Circular amplicons with high heterogeneity of structure. Images show the internal organization and inferred evolutionary path of amplicons in case 07B60, as an example of a patient with clonal amplicon heterogeneity. The ancestral amplicon (07B60a, occurring in the 26.4% (14/53) of cells), at the top of the figure, underwent two independent rearrangement types, indicated as R6/R7 (middle left) or R8 (middle right), originating the secondary amplicons 07B60b (bottom left), observed in 73.6% (39/53) of cells, and 07B60c (bottom right), without cell specificity at the FISH level. Circos plots (in violet) represent all reconstructed amplified structures, with WGS read depth (in light blue) and genes in dark gray, or in orange if interrupted by a breakpoint (arrows represent the transcriptional orientation). Red-dashed semicircles and purple curved lines respectively represent sequence deletions and newly originated SVs labeled by appropriated codes. Inside the 07B60a Circos plot, IGV plots of WGS read depth at amplicon borders (bottom) and FISH pseudocolor images show co-amplification of delimiting amplicons proximal and distal breakpoints (top). The red-dashed semicircles represent sequence deletions on the ancestral amplicon to obtain both secondary ones. It is worth noting that dmin-harboring 07B60a amplicons showed the co-localization of all four indicated probes in the FISH experiments, contrary to those containing 07B60b, which displayed signals only for the green and blue probes, indicating a cell specificity of sequence amplifications
Complex structures at the deletions junctions on one chromosome 8 homolog
Of the 23 cases analyzed by SNP array, WGS, and FISH, 11 (48%) had heterozygous 8q24 deletions, corresponding to the amplified regions on the dmin, ring chromosomes, and hsr, on one of the chromosome 8 homologs. In four of the 11 cases, the deletion junctions were more complex than the ones in the other deleted cases, by displaying inversions with or without duplications, or reciprocal translocations at the junctions.
Chromothripsis is not involved in 8q24 amplicon genesis
The criteria defined to infer chromothripsis were evaluated across the 8q22.3–24.1 region in each AML-amp case (Table 1). According to the criterion “clustering of breakpoints”, we searched for 5–10 breakpoints within 50 kbp genomic segments. This criterion was fulfilled in 22/23 (95.7%) cases (Supplementary Figure S3a). Regarding the “regularity of oscillating copy number states”, we did not observe more than 10 CN switches in 16/23 (69.6%) cases (Supplementary Figure S3b). In the remaining seven (30.4%) cases, the CN values ranged between more than three CN states, thus exceeding the limit for chromothripsis. Interestingly, we observed loss of heterozygosity in not-deleted regions, indicating the occurrence of multi-step rearrangement events (Supplementary Figure S3b). Concerning the “randomness of DNA segments order and fragment joins”, the results were conflicting. On one hand, the order of the breakpoints was not conserved, indicating the presence of randomly joined chromosome fragments as in chromothripsis (Supplementary Figure S3c). On the other hand, there was not an equal distribution of rearrangement types, as occurs in the step-wise rearrangement model (Supplementary Figure S3d). Therefore, we considered this criterion as not validated for the assessment of chromothripsis in our samples. Finally, the criterion of “ability to walk the derivative chromosome” was not fulfilled because of the presence of nested amplified fragments (Supplementary Figure S3e); this does not fit the chromothripsis model in which no “reuse” of rearranged fragments is admitted. Taken together, the findings exclude the involvement of chromothripsis in the genesis of 8q24 amplicons.
Table 1.
Overall results of the chromothripsis analysis in 8q22.3–24.1 regions
| Sample | Clustering of breakpoints | Randomness of fragment joins/randomness of segment order | Ability to walk the derivative chromosome | Regularity of oscillating copy number state | Interspersed regions with loss and retention of heterozygosity |
|---|---|---|---|---|---|
| MLL_11929 | Y | N/Y | N | Not oscillating, >3 states | Unmatching LOH and CN loss |
| MLL_10598 | Y | N/Y | N | Oscillating, >3 states | Unmatching LOH and CN loss |
| MLL_11930 | Y | N/Y | N | Not oscillating, >3 states | Unmatching LOH and CN loss |
| MLL_11931 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| MLL_12445 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| MLL_10619 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| MLL_11932 | Y | N/Y | N | Not oscillating, <3 states | Unmatching LOH and CN loss |
| MLL_11933 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| MLL_11934 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| MLL_11935 | Y | N/Y | N | Oscillating, >3 states | Unmatching LOH and CN loss |
| MLL_11936 | Y | N/Y | N | Not oscillating, >3 states | Unmatching LOH and CN loss |
| MLL_11937 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| MLL_11938 | Y | N/Y | N | Oscillating, >3 states | Unmatching LOH and CN loss |
| MLL_11939 | Y | N/Y | N | Oscillating, >3 states | Unmatching LOH and CN loss |
| MLL_11940 | Y | N/Y | N | Not oscillating, 2 states | Unmatching LOH and CN loss |
| MLL_11941 | Y | N/Y | N | Not oscillating, >3 states | Unmatching LOH and CN loss |
| 07B60 | N | N/Y | N | Oscillating, >3 states | Unmatching LOH and CN loss |
| 1262-09 | Y | N/Y | N | Oscillating, >3 states | Unmatching LOH and CN loss |
| 757-02 | Y | N/Y | N | Not oscillating, >3 states | Unmatching LOH and CN loss |
| 756-04 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| 1-07KM6821 | Y | N/Y | N | Oscillating, >3 states | Unmatching LOH and CN loss |
| 3-06PB4872 | Y | N/Y | N | Not oscillating, 3 states | Unmatching LOH and CN loss |
| 4-09KM0664 | Y | N/Y | N | Not oscillating, >3 states | Unmatching LOH and CN loss |
Neocentromeres stabilize tandemly amplified sequences within ring chromosomes
FISH analyses of four AML-amp cases with amplicon-positive chromosomes revealed that the the primary constrictions on the ring chromosomes lacked alphoid satellite sequences but were positive for CENP-C (Fig. 3), strongly indicating the presence of functional ectopic centromeres, which are essential for stabilizing ring chromosomes during cell divisions.
Fig. 3.
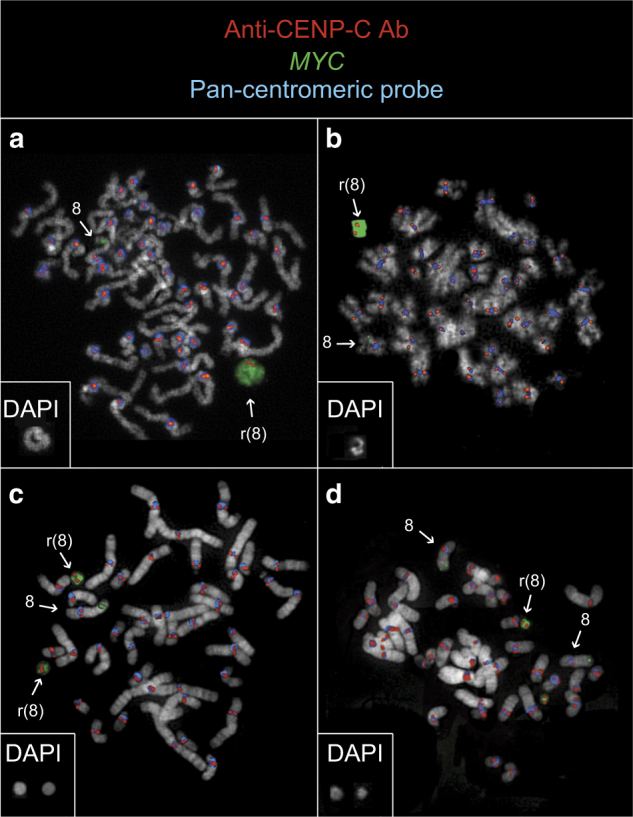
Neocentromere at ring chromosomes harboring 8q24 amplicons. Immuno-FISH co-hybridization results in cases MLL_11929 (a), MLL_11933 (b), #18 (c), and #30 (d) display ring chromosomes with MYC amplifications (green) positive for CENP-A (red) but negative for alpha-satellite DNA (blue). Square boxes at the bottom left of each panel show the DAPI image of the rings
Amplified genes/lncRNAs are recurrently involved in fusion transcripts
RNA-seq of AML-amp cases identified 12,468 and 58,032 raw chimeric transcripts by CS and FM, respectively, among which 38 and 429 involved 8q24-amplified genes (Supplementary Table S4A-B). Using RT-PCR and Sanger sequencing, we validated 35 of these 8q24 fusion transcripts (Table 2), 26 of which involved PVT1 as either a 5′ or 3′ partner (Supplementary Figure S4a). Notably, 15 chimeras with PVT1 as a 5′ partner shared exactly the same breakpoint position (at chr8:128,806,980). Other genes/lncRNAs recurrently involved in chimeras were MYC, FAM49B, RP11-89K10, CCDC26, CASC11, and CASC8. Of these, MYC and CCDC26 each showed a recurrent breakpoint position (at chr8:128,750,494 and chr8:130,365,226, respectively) in three chimeras, where they are involved as 3′ partners. Remarkably, 15 out of the 35 (43%) validated chimeras were generated by the fusion of two lncRNAs. Some fusion transcripts were common to two or more cases, were present as multiple transcript isoforms in the same case or in different cases, or together with their reciprocal fusion product (Table 2, Supplementary Table S4a).
Table 2.
List of validated chimeric transcripts involving 8q24-amplified partners
| Chimera | 5′ breakpoint | 3′ breakpoint | Tool | Recurrent | Case number | In silico translation | GenBanK accession number |
|---|---|---|---|---|---|---|---|
| 5′PVT1/3′IRF2BP2 | chr8:128806980 | chr1:234743598 | CS | Y | MLL_11929, MLL_11935, 07B60, MLL_11941, 1262/06 | Promoter swapping | MF125224 |
| 5′PVT1/3′SPRY2 | chr8:128806980 | chr13:80911891 | CS | N | MLL_11932 | Promoter swapping | MF125228 |
| 5′PVT1/3′PTP4A1 | chr8:128806980 | chr6:64286341 | FM | Y | MLL_11932, 757/02, 756/04 | Promoter swapping | MF125229 |
| 5′PVT1/3′CASC8 (LOC727677) | chr8:128806980 | chr8:128433215 | CS | Y | MLL_11938, MLL_11935 | NA | MF125263 |
| 5′PVT1/3′LINC00964 (LOC157381) | chr8:128806980 | chr8:125953705 | CS | N | MLL_11935 | NA | MF125258 |
| 5′PVT1/3′CASC11 a (BC042052) | chr8:128806980 | chr8:128701668 | CS | Y | MLL_11935, MLL_11941, 757/02, 756/04, 1262/06 | NA | MF125264 |
| 5′PVT1/3′IFRD1 | chr8:128806980 | chr7:112095818 | CS | N | MLL_11935 | Promoter swapping | MF125230 |
| 5′PVT1/3′UBB | chr8:128806980 | chr17:16285216 | FM | Y | 07B60, 756/04, MLL_11935 | Promoter swapping | MF125231 |
| 5′PVT1/3′NSMCE2 ba | chr8:128806980 | chr8:126194345 | FM | Y | MLL_11935, MLL_11932 | N-term truncated NSMCE2 | MF125232 |
| 5′PVT1/3′CXCR4 b | chr8:128806980 | chr2:136873482 | CS | N | MLL_11932 | Promoter swapping | MF125227 |
| 5′PVT1/3′CASC21 | chr8:128806980 | chr8:128351526 | FM | N | MLL_11938 | NA | MF125255 |
| 5′PVT1/3′KB-1568E2 | chr8:128806980 | chr8:131667944 | FM | N | 07B60 | NA | MF125257 |
| 5′PVT1/3′RP11-89K10 b | chr8:128806980 | chr8:127725531 | FM | N | MLL_11935 | NA | MF125259 |
| 5′PVT1/3′MYC | chr8:128806980 | chr8:128750494 | FM | Y | MLL_11932, MLL_11941 | N-term truncated MYC | MF125236 |
| 5′PVT1/3′NFIL3 c | chr8:128806980 | chr9:94173188 | FM | N | MLL_11935 | Promoter swapping | MF125234 |
| 5′PVT1/3′NFIL3 c | chr8:128903244 | chr9:94173188 | FM | Y | MLL_11932, 756/04 | Promoter swapping | MF125235 |
| 5′PVT1/3′CCDC26 c | chr8:128903244 | chr8:130365226 | FM | N | MLL_11929 | NA | MF125225 |
| 5′PVT1/3′CCDC26 c | chr8:129001537 | chr8:130365226 | CS | N | 1-07KM6821 | NA | MF125226 |
| 5′PVT1/3′LINC00824 (RP11-89M16) | chr8:129001537 | chr8:129440630 | FM | N | 1-07KM6821 | NA | MF125256 |
| 5′HIST1H2BD/3′PVT1 | chr6:26158788 | chr8:128902835 | CS | Y | MLL_11932, MLL_11935, MLL_11940 | 3′UTR swapping | MF125239 |
| 5′CXCR4/3′PVT1 ab | chr2:136875615 | chr8:128902835 | FM | N | MLL_11932 | 3′UTR swapping | MF125242 |
| 5′RP11-89K10/3′PVT1 bc | chr8:127601535 | chr8:128951754 | FM | N | MLL_11935 | NA | MF125259 |
| 5′RP11-89K10/3′PVT1 bc | chr8:127570176 | chr8:129108764 | FM | N | MLL_11932 | NA | MF125260 |
| 5′NSMCE2/3′PVT1 bc | chr8:126114729 | chr8:129108764 | FM | N | MLL_11935 | C-term truncated NSMCE2 | MF125241 |
| 5′NSMCE2/3′PVT1 bc | chr8:126194499 | chr8:128944711 | FM | Y | MLL_11929, MLL_11941, MLL_11932, 757/02 | C-term truncated NSMCE2 | MF125240 |
| 5′MDM2/3′PVT1 a | chr12:69203073 | chr8:128867408 | CS | N | MLL_11935 | C-term truncated MDM2 | MF125237 |
| 5′HNRNPA2B1/3′MYC | chr7:26240192 | chr8:128750494 | FM | N | 6-08KM2421 | promoter swapping | MF125246 |
| 5′FXR1/3′MYC | chr3:180630524 | chr8:128750494 | FM | N | 4-09KM0664 | promoter swapping | MF125244 |
| 5′FAM104A/3′MYC | chr17:71203525 | chr8:128751053 | FM | N | 6-08KM2421 | 3′UTR swapping | MF125245 |
| 5′MTSS1/3′FAM49B b | chr8:125711764 | chr8:130891718 | FM | Y | MLL_11939, MLL_11941, 756/04, 757/02, 1262/06 | C-term truncated MTSS1 | MF125248 |
| 5′FAM49B/3′MTSS1 b | chr8:130915557 | chr8:125716434 | CS | N | MLL_11936 | N-term truncated MTSS1 | MF125250 |
| 5′MPO/3′TRIB1 | chr17:56347272 | chr8:126450621 | CS | N | MLL_11935 | 3′UTR swapping | MF125251 |
| 5′CASC11/3′RP11-89K10 | chr8:127629630 | chr8:128701469 | FM | N | MLL_11935 | NA | MF125261 |
| 5′CASC8 (LOC727677)/3′CCDC26 a | chr8:128433074 | chr8:130365226 | CS | N | MLL_11935 | NA | MF125252 |
| 5′PCAT1/3′CASC21 | chr8:127890035 | chr8:128404747 | FM | N | MLL_11938 | NA | MF125262 |
Bold: chimera with genomic alteration
NA Not analyzed due to the fusion of two lncRNA
aMultiple transcript variants
bChimera showing the reciprocal fusion product in the same case
cTranscript variants in different cases
In silico translation of the 5′ PVT1 chimeras predicted the dysregulation of the protein product of their 3′ partner transcripts due to promoter swapping or N-terminus truncation (Table 2, Supplementary Figure S4b). Conversely, 3′ PVT1 fusions were predicted to display either a 3′ untranslated region substitution of their 5′ partner transcripts or a C-terminus truncation of the encoded protein. A truncation effect on the MTSS1 protein was also predicted for 5′MTSS1/3′FAM49B fusion and its reciprocal 5′FAM49B/3′MTSS1.
Interestingly, the 5′PVT1/3′CCDC26 chimera was the only fusion we detected that was also in a panel of 23 AML cell lines (Supplementary Table S4C-D). Similarly, neither the two remission samples of AML-amp cases nor the AML-NK cases harbored any of the 8q24 fusions detected in AML-amp cases.
Fusion transcripts often do not correspond to genomic rearrangements
By combining WGS and RNA-seq data, only 0.1% (12/12,468) and 0.07% (43/58,032) of the chimeric transcripts identified in our cohort by CS and FM, respectively, were detected at the genomic level. After zooming into the 8q24 chromosome segment, their percentages increased to 10.5% (4/38) and 7.2% (31/428) by CS and FM, respectively. These results suggest that the chimeric transcripts might be mostly generated by post-transcriptional events (Supplementary Table S4E). Moreover, our AML-amp cases showed a statistically significant higher number of 8q24 chimeras (mean 2.5 and 41.8 for CS and FM tools, respectively) as compared to AML-NK control cases from TCGA [0.7 and 0.2 for CS and FM, respectively; U-test p = 0.004792 (CS) and p = 1.116e−08 (FM)] (Supplementary Figure S4c-d).
AML-amp cases show a peculiar gene signature associated with 8q24 chimeras
To investigate genes differentially expressed in AML-amp cases, we performed a DEA by comparing six AML-amp cases with six AML-NK samples from TCGA. Globally, we found 853 differentially expressed genes among which only two mapped in the 8q24 region, namely, MTSS1 and RP11-532M24.1 (Supplementary Table S5A). Gene enrichment analysis showed that differentially expressed genes upregulated in AML-amp versus control cases displayed functions related to myeloid cell differentiation and homeostasis as well as chromatin assembly, nucleosome organization, histone methylation, and gene silencing (Supplementary Table S5B-C).
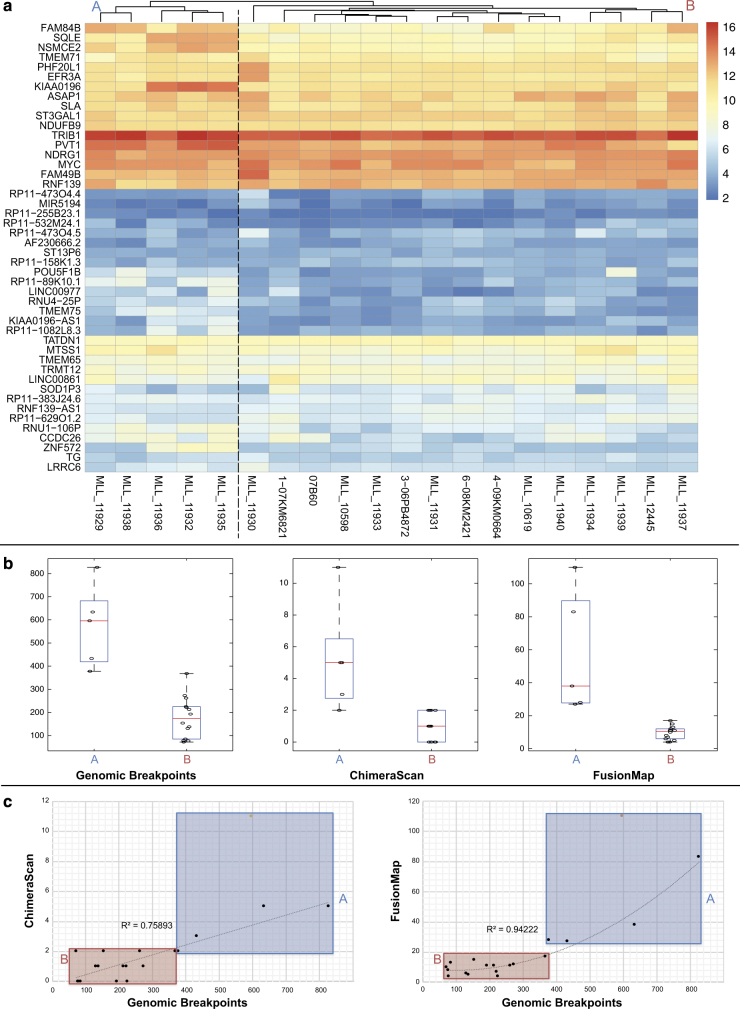
Unsupervised hierarchical clustering analysis of all the 8q24 genes in the 20 AML-amp cases revealed a high level of sample heterogeneity and clearly showed the presence of two major subgroups, denoted Group A (5 cases) and Group B (15 cases) (Fig. 4a). To ascertain the reason(s) for these two groups, we compared age, gender, cytogenetic features, and number of chimeras and genomic breakpoints involving the 8q24 region between the groups. This revealed a significant difference in the number of chimeras and genomic breakpoints (Fig. 4b). In addition, there was a positive correlation between the number of genomic breakpoints and the number of chimeras (identified by both CS and FM), indicating a close association between genomic complexity and transcriptomic plasticity of fusion transcripts (Fig. 4b-c). The DEA of Group A versus Group B samples identified nine differentially expressed genes mapping in the 8q24 region, which were upregulated in Group A (Supplementary Table S5D). Among these, we found the lncRNA PVT1 and the coding NSMCE2, which was the most significantly overexpressed gene in Group A (FDR = 10−18).
Fig. 4.
Chimeric transcripts significantly associated with a peculiar gene expression profile at amplified regions and breakpoints. a Heatmap of 8q24 gene expression levels in our AML-amp cohort. b Box and Whisker plots illustrating differences in the number of breakpoints and chimeras in 8q24 between Groups A and B. c Correlation analysis between the number of genomic breakpoints and chimeras in the 8q24 region identified by CS (left) and FM (right)
Mutations in AML-amp cases affect genes involved in cell division and chromatin modifications
We identified 467 SNVs/indels affecting 432 genes (Supplementary Table S6A), of which 19 were involved in two or more samples (Supplementary Table S6B), and 95 grouped into 31 gene families (Supplementary Table S6C). The most frequently mutated genes in the AML-amp cases were TET2 (50% of the cases), DNMT3A (25%), TP53 (15%), and KMT2C (alias MLL3, 15%). Pathway analysis grouped the 432 mutated genes into four main pathways: cell cycle, TP53 downstream pathway, chromatin modifications, and kinases (Supplementary Table S6D). These results indicate that most of the mutated genes are involved in cell division and epigenetic chromatin modifications.
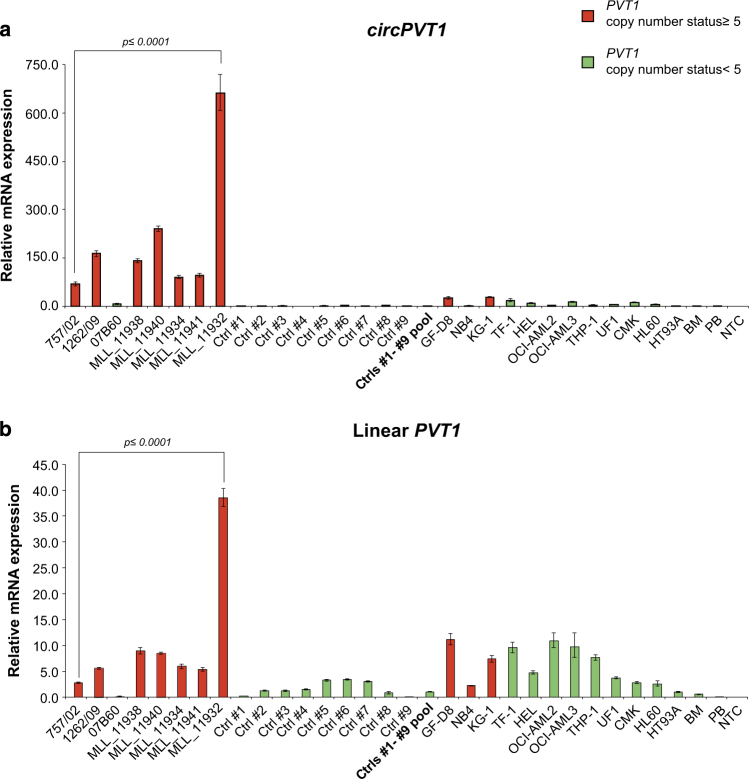
Overexpression of circular PVT1 is associated with high PVT1 CN amplification
By qPCR, we demonstrated the strong upregulation of circPVT1, generated by the circularization of PVT1 exon 2, in AML-amp cases with amplicons including PVT1 compared with AML-amp cases in which PVT1 was not amplified (as 07B60) or AML-NK samples (Fig. 5a). In AML cell lines, a slight upregulation of circPVT1 was observed only in the two cell lines carrying more than five copies of PVT1 (GF-D8 and KG-1). The PVT1 linear transcript also showed an upregulation in PVT1-amplified versus not amplified AML cases, but this difference disappeared in AML cell lines where it was similarly expressed regardless of PVT1 CN state (Fig. 5b).
Fig. 5.
Circular and linear PVT1 expression. qPCR results obtained for circPVT1 (a) and linear PVT1 (b) in AML cases and cell lines. In red and green, respectively, samples with and without PVT1 exon 2 gain. A pool of nine AML-NK cases was used as calibrator. NTC no template control
Discussion
Our study focused on the comprehensive genomic and transcriptomic characterization of 8q24 amplifications in AML. Overall, 65% of cases showed evidence of amplicon heterogeneity. We previously observed the occurrence of distinct cell subpopulations harboring different MYC amplifications [9] and here confirmed at the single-nucleotide level.
Except for that of breakpoint clustering, we found that none of the valuable chromothripsis criteria is verified in our AML-amp cases. This is in accordance with our previous results on seven tumor cell lines with 8q24 amplicon heterogeneity, which excluded the chromothripsis model as involved in dmin/hsr genesis [7], in contrast with previous literature [5, 6]. As a matter of fact, breakpoint clustering might also result from events occurring at separate temporal stages of the amplicon evolution [18]. Additionally, in 48% of cases we observed deletions corresponding to the amplified regions, not accompanied by any other rearrangement of the del(8) chromosome. Overall, these findings suggested a multi-step pathway in dmin/hsr genesis, indicating the episome model as the most reliable mechanism underlying 8q24 dmin genesis in AML, as we already proposed [9, 14].
8q24 amplifications also occurred as hsr and/or ring chromosomes in some of our cases, confirming that 8q24 amplicon architecture is shared among all these cytogenetic structures [9, 28]. Interestingly, the discovery of neocentromeres on ring chromosomes is an unprecedented result, since ectopic centromeres emerging at chromosomes composed by genomic amplifications were observed only in well-differentiated liposarcomas [29] and lung cancer [30] thus far. Centric ring chromosomes might represent an evolutionary step of scattered acentric dmin as well as hsr towards mitotic stabilization, providing an adaptive advantage to leukemia cells harboring 8q24 amplifications. With the availability of vital frozen cells, further investigation into the genomic architectures of these neocentromeres will help to better understand the evolutionary path of these amplified sequences.
In this study, we focused on the identification of chimeric transcripts that originated from the 8q24-amplified genes. To date, 8q24 amplifications have already been associated with two fusion genes, namely PVT1-NSMCE2 and BF104016-NSMCE2, found respectively in an AML case and in the HL60 cell line [31]. Although the latter was not identified by neither WGS [7] nor RNA-seq in our research, we confirmed the occurrence of the 5′PVT1/3′NSMCE2 fusion in our data set, along with its reciprocal 5′NSMCE2/3′PVT1. The lncRNA PVT1 was described as a hotspot target of genomic breakpoints in 8q24 amplifications [7, 29, 32–34]. Notably, gain of PVT1 expression is required for high MYC protein levels in 8q24-amplified human cancer cells [35]. Indeed, 26 out of 35 validated chimeras in our AML-amp cases involved this lncRNA, and most of them were recurrent in more cases and shared the same breakpoint position. Many of the 8q24 chimeras we identified lacked genomic support and were associated with dmin/hsr amplification. Overall, the 8q24-amplified genes were shown to be prone to originate chimeric transcripts, often with not-amplified genes as partners. Our evidence indicates that they might be originated by alternative mechanisms to those based on genomic rearrangements. Post-transcriptional events, such as trans-splicing or cis-splicing between adjacent genes (cis-SAGe) [36–40], might be an intriguing possibility that, however, needs further experimental support. The potential oncogenic role of these chimeras specifically accompanying 8q24 amplifications will require further investigation.
Notably, we found that AML-amp cases with the highest numbers of chimeras showed a peculiar 8q24 gene expression signature characterized by the overexpression of both PVT1 and NSMCE2, frequently found in our 8q24 chimeric transcripts. Unfortunately, such patient clustering cannot be directly correlated to disease prognostic indications, since most of Group A patients were lost during follow-up and lacked information about treatment and overall survival. However, we could envisage a possible prognostic implication given that Group A comprised three out the five cases with complex karyotype and TP53 mutation/17p13 loss, which are notoriously associated with a poorer outcome in AML [3]. Interestingly, NSMCE2, which was overexpressed in Group A cases, is required for DNA double-strand break repair and genomic integrity maintenance [41, 42]. Its depletion was shown to sensitize HeLa cells to DNA damage-induced apoptosis [43] suggesting that, alternatively, its overexpression could make tumor cells more resistant to the DNA damage induced by their wide genomic rearrangements.
Moreover, we characterized the gene signature and pathways specifically associated with the presence of 8q24 amplifications and found that the biological processes affected by these dysregulated genes are related to cell division and chromatin architecture. In particular, we observed the upregulation of genes involved in myeloid and erythroid cell development and differentiation, such as CEBPA [44] and the key erythroid transcription factors GATA1 [45], TAL1, and KLF1 [46]. According to the literature [47], KLF1 overexpression may be related to the simultaneous presence of TET2 and DNMT3A mutations in the two AML-amp cases investigated by our DEA. DNMT3A and TP53 frequencies are in line with those already reported in AML without 8q24 amplifications (30% and 13%, respectively) [48]. Conversely, TET2 and KMT2C mutations were more frequent in our cohort than in AML without amplifications (23% and 0.5%, respectively) [48, 49], although with no dysregulation at the expression level with respect to AML-NK samples. Interestingly, AML-amp cases also showed upregulated genes involved in chromatin assembly, nucleosome positioning, and H3K27 histone trimethylation, such as the replication-dependent histone genes HIST1H1B, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H2BG, HIST1H2BF, HIST1H3E (all mapping within the HIST1 cluster at 6p22-p21.3), and the replication independent H1FX (3q21.3). Considering that histone H1 and related linker histones are important for maintenance of higher-order chromatin structure and gene expression regulation [50], their significant upregulation in our cohort is of interest and might shed light on the chromatin compaction within dmin chromosomes.
Finally, the 8q24 commonly amplified genes, including MYC, PVT1, and TRIB1, showed a variegated expression pattern, consistent with previous reports [3, 9, 13, 14], thus confirming that they are not the real targets of this amplification type.
Remarkably, the transcriptional plasticity observed in our AML-amp cohort also includes circRNAs originated by back-splicing events [51], resistant to exonucleolytic degradation [52], and accumulating at relatively high levels within cells [47]. They have been proposed to act as microRNA sponges, platforms for protein interactions, transcriptional regulators of their linear counterparts, or, as recently demonstrated, as protein translators [53–57]. Concerning genes in 8q24 region, circRNAs from MTSS1, NSMCE2, FAM84B, POU5F1B, MYC, PVT1, and FAM49B have already been cataloged in the CircInteractome database (http://circinteractome.nia.nih.gov) [58]. circPVT1, generated from exon 2 of PVT1, was found as highly expressed in HeLa cervical cancer cells, human proliferating fibroblasts [59], and gastric cancer cells often accompanied by 8q24 amplifications [53]. Notably, we found a strict correlation between PVT1 CN gain and the upregulation of this circPVT1 in both AML-amp cases and cell lines. The NB4 cell line was an exception, since it did not show overexpression of circPVT1 despite its gene amplification; nonetheless, here, as well as in case 07B60, we cannot exclude the presence of additional PVT1 alterations preventing the detection of the circRNA. circPVT1 was proposed to act as a molecular sponge for tumor suppressor microRNAs, such as those belonging to let-7 [59] or miR-125 families [53]. Further investigation into its role in AML-amp cases will therefore be required.
In summary, this work adds an important piece in the puzzle of the genomic and transcriptomic impact of 8q24 amplifications in AML. The amplicon molecular heterogeneity and the deletion of the corresponding amplified regions observed in the majority of cases ruled out chromothripsis as a potential mechanism behind their origin. Moreover, the neocentromeric seeding observed on ring chromosomes, harboring tandemly amplified sequences, would be engaged to stabilize collapsed acentric dmin, providing a selective advantage to leukemia cells. Finally, the finding of post-transcriptional chimeras and circRNAs involving 8q24-amplified genes in AML-amp cases could open new scenarios in the understanding of the biological mechanisms behind MYC amplifications in leukemia.
Data availability
SNP array data are available at EMBL-EBI ArrayExpress repository, under accession number E-MTAB-5372 (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-5372/). WGS and RNA-seq data are available at NCBI Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA386992. Validated SV sequences and RT-PCR validated fusion junction sequences were submitted to GenBank repository (http://www.ncbi.nlm.nih.gov/genbank/), under accession numbers MF084292–MF084375, and MF125224–MF125264 accession numbers, respectively.
Electronic supplementary material
Supplementary Figure S1. Map of amplified 8q24 genomic segments in AML-amp cases
Supplementary Figure S3. Evaluation of the Chromothripsis criteria
Supplementary Figure S4. Fusion transcripts involving 8q24 amplified partners,
Supplementary Table S2: DELLY SVs call in AML-amp cases
Supplementary Table S3: Amplicons features in AML-amp cases
Supplementary Table S4A: 8q24 chimeras identified by ChimeraScan in AML-amp cases
Supplementary Table S5A: Differentially expressed genes in AML-amp vs AML-NK cases
Supplementary Table S6A: Mutations found in AML-amp cases
Acknowledgements
This work was supported by the AIRC (Associazione Italiana per la Ricerca sul Cancro; AIRC IG no. 14078 and 15413 to GM and CTS, respectively), the Italian Ministry of Health (Young researchers Call GR-2011-02352026 to IC), and the “5 × 1000” voluntary contributions to GM. We thank Prof. Bertil Johansson and Prof. David Gisselsson for critical review of the manuscript.
Author contributions
ALA and CTS conceived the study and wrote the manuscript, with assistance by DTo, IC, MS, GMe, and BJ. ALA, PDA, IC, MS, and GDB analyzed sequencing data and performed and reviewed bioinformatics and statistical analyses. AL, AT, BA, MP, GMe, and DTr designed and performed PCR experiments. AT, DTr, MP, and DTo performed Sanger sequencing and FISH validation of data. OP and MC performed and analyzed SNP array data. DTo and DTr performed immuno-FISH experiments. AD and AT performed mutational analysis under the supervision of LB. GD and AL performed circRNA RT-qPCR and pull-down assays. GMa, CH, LB, DM, JS, and NVR provided biological samples and clinical data. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41375-018-0033-0) contains supplementary material, which is available to authorized users.
References
- 1.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–5. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens FE Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2017.
- 3.Huh YO, Tang G, Talwalkar SS, Khoury JD, Ohanian M, Bueso-Ramos CE, et al. Double minute chromosomes in acute myeloid leukemia, myelodysplastic syndromes, and chronic myelomonocytic leukemia are associated with micronuclei, MYC or MLL amplification, and complex karyotype. Cancer Genet. 2016;209:313–20. doi: 10.1016/j.cancergen.2016.05.072. [DOI] [PubMed] [Google Scholar]
- 4.Sait SN, Qadir MU, Conroy JM, Matsui S, Nowak NJ, Baer MR. Double minute chromosomes in acute myeloid leukemia and myelodysplastic syndrome: identification of new amplification regions by fluorescence in situ hybridization and spectral karyotyping. Genes Chromosomes Cancer. 2002;34:42–47. doi: 10.1002/gcc.10038. [DOI] [PubMed] [Google Scholar]
- 5.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.L’Abbate A, Macchia G, D’Addabbo P, Lonoce A, Tolomeo D, Trombetta D, et al. Genomic organization and evolution of double minutes/homogeneously staining regions with MYC amplification in human cancer. Nucleic Acids Res. 2014;42:9131–45. doi: 10.1093/nar/gku590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwab M. Oncogene amplification in solid tumors. Semin Cancer Biol. 1999;9:319–25. doi: 10.1006/scbi.1999.0126. [DOI] [PubMed] [Google Scholar]
- 9.Storlazzi CT, Fioretos T, Surace C, Lonoce A, Mastrorilli A, Strombeck B, et al. MYC-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of MYC as the target gene. Hum Mol Genet. 2006;15:933–42. doi: 10.1093/hmg/ddl010. [DOI] [PubMed] [Google Scholar]
- 10.Storlazzi CT, Lonoce A, Guastadisegni MC, Trombetta D, D’Addabbo P, Daniele G, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20:1198–206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 12.Paulsson K, Lassen C, Kuric N, Billstrom R, Fioretos T, Tanke HJ, et al. MYC is not overexpressed in a case of chronic myelomonocytic leukemia with MYC-containing double minutes. Leukemia. 2003;17:813–5. doi: 10.1038/sj.leu.2402841. [DOI] [PubMed] [Google Scholar]
- 13.Storlazzi CT, Fioretos T, Paulsson K, Strombeck B, Lassen C, Ahlgren T, et al. Identification of a commonly amplified 4.3 Mb region with overexpression of C8FW, but not MYC in MYC-containing double minutes in myeloid malignancies. Hum Mol Genet. 2004;13:1479–85. doi: 10.1093/hmg/ddh164. [DOI] [PubMed] [Google Scholar]
- 14.Micale L, Augello B, Daniele G, Macchia G, L’Abbate A, Muehlematter D, et al. Amplification of the G allele at SNP rs6983267 in 8q24 amplicons in myeloid malignancies as cause of the lack of MYC overexpression? Blood Cells Mol Dis. 2011;47:259–61. doi: 10.1016/j.bcmd.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Trazzi S, Perini G, Bernardoni R, Zoli M, Reese JC, Musacchio A, et al. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS ONE. 2009;4:e5832. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macchia G, Nord KH, Zoli M, Purgato S, D’Addabbo P, Whelan CW, et al. Ring chromosomes, breakpoint clusters, and neocentromeres in sarcomas. Genes Chromosomes Cancer. 2015;54:156–67. doi: 10.1002/gcc.22228. [DOI] [PubMed] [Google Scholar]
- 17.Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–36. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 19.Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–4. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge H, Liu K, Juan T, Fang F, Newman M, Hoeck W. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–8. doi: 10.1093/bioinformatics/btr310. [DOI] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–5. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2016;388:208–19. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy KS. Double minutes (dmin) and homogeneously staining regions (hsr) in myeloid disorders: a new case suggesting that dmin form hsr in vivo. Cytogenet Genome Res. 2007;119:53–59. doi: 10.1159/000109619. [DOI] [PubMed] [Google Scholar]
- 29.Garsed DW, Marshall OJ, Corbin VD, Hsu A, Di Stefano L, Schroder J, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–67. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Italiano A, Attias R, Aurias A, Perot G, Burel-Vandenbos F, Otto J, et al. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23-p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–30. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Chinen Y, Sakamoto N, Nagoshi H, Taki T, Maegawa S, Tatekawa S, et al. 8q24 amplified segments involve novel fusion genes between NSMCE2 and long noncoding RNAs in acute myelogenous leukemia. J Hematol Oncol. 2014;7:68. doi: 10.1186/s13045-014-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwakawa R, Takenaka M, Kohno T, Shimada Y, Totoki Y, Shibata T, et al. Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes Chromosomes Cancer. 2013;52:802–16. doi: 10.1002/gcc.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, et al. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012;72:4954–62. doi: 10.1158/0008-5472.CAN-12-0213. [DOI] [PubMed] [Google Scholar]
- 34.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babiceanu M, Qin F, Xie Z, Jia Y, Lopez K, Janus N, et al. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016;44:2859–72. doi: 10.1093/nar/gkw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin F, Song Z, Babiceanu M, Song Y, Facemire L, Singh R, et al. Discovery of CTCF-sensitive Cis-spliced fusion RNAs between adjacent genes in human prostate cells. PLoS Genet. 2015;11:e1005001. doi: 10.1371/journal.pgen.1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaphiropoulos PG. Trans-splicing in higher eukaryotes: implications for cancer development? Front Genet. 2011;2:92. doi: 10.3389/fgene.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Gong M, Yuan H, Park HG, Frierson HF, Li H. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012;2:598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 41.Stephan AK, Kliszczak M, Morrison CG. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 2011;585:2907–13. doi: 10.1016/j.febslet.2011.04.067. [DOI] [PubMed] [Google Scholar]
- 42.Verver DE, Zheng Y, Speijer D, Hoebe R, Dekker HL, Repping S, et al. Non-SMC element 2 (NSMCE2) of the SMC5/6 complex helps to resolve topological stress. Int J Mol Sci. 2016;17:1782. doi: 10.3390/ijms17111782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–32. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohlsson E, Schuster MB, Hasemann M, Porse BT. The multifaceted functions of C/EBPalpha in normal and malignant haematopoiesis. Leukemia. 2016;30:767–75. doi: 10.1038/leu.2015.324. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu R, Engel JD, Yamamoto M. GATA1-related leukaemias. Nat Rev Cancer. 2008;8:279–87. doi: 10.1038/nrc2348. [DOI] [PubMed] [Google Scholar]
- 46.Tallack MR, Magor GW, Dartigues B, Sun L, Huang S, Fittock JM, et al. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 2012;22:2385–98. doi: 10.1101/gr.135707.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, et al. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–24. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 48.Rose D, Haferlach T, Schnittger S, Perglerova K, Kern W, Haferlach C. Subtype-specific patterns of molecular mutations in acute myeloid leukemia. Leukemia. 2017;31:11–17. doi: 10.1038/leu.2016.163. [DOI] [PubMed] [Google Scholar]
- 49.Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood. 2015;126:2491–501. doi: 10.1182/blood-2015-05-646240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sibley CR, Blazquez L, Ule J. Lessons from non-canonical splicing. Nat Rev Genet. 2016;17:407–21. doi: 10.1038/nrg.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–83. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–11. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 54.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–16. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–41. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panda AC, Grammatikakis I, Kim KM, De S, Martindale JL, Munk R, et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2016;45:4021–35. doi: 10.1093/nar/gkw1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Map of amplified 8q24 genomic segments in AML-amp cases
Supplementary Figure S3. Evaluation of the Chromothripsis criteria
Supplementary Figure S4. Fusion transcripts involving 8q24 amplified partners,
Supplementary Table S2: DELLY SVs call in AML-amp cases
Supplementary Table S3: Amplicons features in AML-amp cases
Supplementary Table S4A: 8q24 chimeras identified by ChimeraScan in AML-amp cases
Supplementary Table S5A: Differentially expressed genes in AML-amp vs AML-NK cases
Supplementary Table S6A: Mutations found in AML-amp cases
Data Availability Statement
SNP array data are available at EMBL-EBI ArrayExpress repository, under accession number E-MTAB-5372 (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-5372/). WGS and RNA-seq data are available at NCBI Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA386992. Validated SV sequences and RT-PCR validated fusion junction sequences were submitted to GenBank repository (http://www.ncbi.nlm.nih.gov/genbank/), under accession numbers MF084292–MF084375, and MF125224–MF125264 accession numbers, respectively.