Abstract
The RNA-binding protein Mei2 is crucial for meiosis in Schizosaccharomyces pombe. In mei2 mutants, pre-meiotic S-phase is blocked, along with meiosis. Mei2 binds a long non-coding RNA (lncRNA) called meiRNA, which is a ‘sponge RNA’ for the meiotic inhibitor protein Mmi1. The interaction between Mei2, meiRNA and Mmi1 protein is essential for meiosis. But mei2 mutants have stronger and different phenotypes than meiRNA mutants, since mei2Δ arrests before pre-meiotic S, while the meiRNA mutant arrests after pre-meiotic S but before meiosis. This suggests Mei2 may bind additional RNAs. To identify novel RNA targets of Mei2, which might explain how Mei2 regulates pre-meiotic S, we used RNA immunoprecipitation and cross-linking immunoprecipitation. In addition to meiRNA, we found the mRNAs for mmi1 (which encodes Mmi1) and for the S-phase transcription factor rep2. There were also three other RNAs of uncertain relevance. We suggest that at meiotic initiation, Mei2 may sequester rep2 mRNA to help allow pre-meiotic S, and then may bind both meiRNA and mmi1 mRNA to inactivate Mmi1 at two levels, the protein level (as previously known), and also the mRNA level, allowing meiosis. We call Mei2–meiRNA a ‘double sponge’ (i.e. binding both an mRNA and its encoded protein).
Keywords: Mei2, meiosis, pombe, lncRNA, Mmi1, Rep2
1. Introduction
Mei2 is an RNA-binding protein required for initiating pre-meiotic DNA synthesis and meiosis in Schizosaccharomyces pombe. Its only known target is meiRNA, a long non-coding RNA encoded by the sme2 gene [1]. Mei2 has two RNA recognition motifs (RRMs), and the C-terminal RRM is required for meiosis initiation [2]. During vegetative growth, Mei2 is inactivated and the meiotic program kept off because the Pat1 kinase phosphorylates Mei2 [3], inducing its degradation by the proteasome [4]. Signals that induce meiosis inhibit Pat1 kinase activity, allowing stabilization and accumulation of Mei2. Mutants of Mei2 that cannot be phosphorylated and degraded initiate meiosis without nitrogen starvation [3].
meiRNA is an example of a long non-coding RNA (lncRNA), which have diverse functions in eukaryotes. They range from transcriptional regulation in the nucleus, and regulation of chromatin structure [5], to posttranscriptional gene regulation [6]. lncRNAs have also been implicated in human diseases and cancer [7,8]. In fission yeast, lncRNAs play important regulatory roles in cell differentiation [9], drug tolerance [10] and phosphate metabolism [11–13].
In part—but only in part—Mei2 allows meiosis because it binds meiRNA [1,2]. The Mei2 protein localizes at a specific locus in the nucleus during meiosis, forming a ‘dot’ [1] even before the karyogamy stage of meiosis [2]. The meiRNA binds to the Mei2 protein, and is required to form the Mei2 dot. The Mei2 protein–meiRNA complex is formed at the locus of the sme2 gene, which encodes the meiRNA [14], but almost certainly not any protein [1]. Mei2 protein alone can apparently shuttle between the cytoplasm and nucleus, but binding of meiRNA traps Mei2 protein in the nucleus during meiosis [15,16]. However, adding an NLS to Mei2 protein allows it to localize to the nucleus and form a dot and promote meiosis even in the absence of meiRNA [2]. Importantly, the C-terminal RRM of Mei2 is critical for function even when the meiRNA is absent: addition of an NLS to a Mei2 mutant protein lacking this C-terminal RRM does not enable it to suppress the loss of meiRNA [2]. Thus, Mei2 protein is required in the nucleus to initiate meiosis, and this function requires its RNA-binding ability, not just nuclear localization.
The significance of the Mei2 dot was revealed when the Mei2–meiRNA complex was found to colocalize with another RNA-binding protein, Mmi1, in the nucleus during meiosis. Mmi1 is a crucial inhibitor of meiosis. During vegetative growth, Mmi1 binds to transcripts required for meiotic entry and for early meiosis, and targets them for degradation via the exosome, keeping meiosis off [17–20]. Mmi1 also recruits the Ccr4–Not complex leading to Mei2 ubiquitination by Mot2, and turnover of Mei2 protein [21]. Upon nitrogen starvation and meiotic initiation, the Mei2–meiRNA complex sequesters Mmi1 protein in the nucleus, thus allowing the mRNAs targeted by Mmi1 to escape degradation and be translated in the cytoplasm, thus initiating meiosis [17]. Because Mei2 inhibits Mmi1 protein (by sequestering it in a dot in the nucleus), while Mmi1 inhibits Mei2 (by inducing turnover), these two proteins form a double negative feedback loop, which can act as a meiotic ‘switch’.
Mmi1 binds to the hexanucleotide motif T(T/C/G)AAAC on its target RNAs [19,22]. Strikingly, the long, meiotic form of meiRNA has 23 of these Mmi1 motifs [23]. These motifs bind and titrate Mmi1 to keep it localized to the sme2 locus in the nucleus during meiosis [19,23]. Thus, meiRNA is a decoy RNA, or sponge RNA, which titrates out and effectively inactivates Mmi1 protein [23]. During vegetative growth, a shorter form of meiRNA lacking most of these Mmi1 motifs is made, and this form of meiRNA is degraded by the exosome [23]. This partly explains why meiRNA is unable to sequester Mmi1 during vegetative growth. Mei2 binds to the 5′ end of meiRNA [23].
Although the results cited above show that the Mei2–meiRNA complex promotes meiosis at least partly by titrating out Mmi1 protein, this is not the whole story. Several workers have shown that the mei2 mutant has a stronger and qualitatively different meiotic phenotype than the sme2 (meiRNA) mutant. For instance, upon nitrogen starvation, the mei2Δ mutant is defective in pre-meiotic S-phase (and, of course, in meiosis). However, the sme2Δ mutant performs pre-meiotic DNA synthesis, but cannot undertake meiosis [1]. The ts allele mei2-33, an allele where binding of the meiRNA is defective at restrictive temperature, mimics sme2Δ, and performs pre-meiotic DNA synthesis, but not meiosis [1]. Finally, the mei2-644A mutant, bearing a mutation in the C-terminal RRM, is phenotypically similar to the mei2Δ allele, in that it is defective in pre-meiotic DNA synthesis (and meiosis) [1]. This led Watanabe & Yamamoto [1] to suggest ‘Mei2 functions at least twice, prior to pre-meiotic DNA synthesis and prior to meiosis I, by coupling with different RNA partners’. That is, they suggest Mei2 must have two targets, one responsible for allowing pre-meiotic S and a second (presumably meiRNA) responsible for meiosis. Subsequent work showed that if the Mei2 protein is provided with a nuclear localization signal, it can promote meiosis even in the absence of meiRNA (i.e. it suppresses a sme2/sme2 null mutant) [2]. However, Mei2-NLS can only suppress sme2/sme2 if the C-terminal RRM of Mei2 is intact, suggesting there may be one or more additional RNA targets for promoting meiosis, and leading Yamashita et al. [2] to suggest that ‘Mei2p may have to interact with one or more RNA species in the nucleus in order to promote meiosis I.’ Shichino et al. [23] concluded that over-expression of meiRNA does not fully inactivate Mmi1, likewise suggesting Mei2 may have an additional, meiRNA-independent mechanism for inactivation of Mmi1. These various observations that the phenotype of mei2 is stronger and different from the phenotype of sme2Δ (meiRNA) all point to the possibility that Mei2 might bind additional RNA targets. Potentially there is a novel RNA target specifically involved in pre-meiotic S-phase, and possibly an additional novel target helping to inhibit Mmi1 activity, or in otherwise promoting meiosis. This idea motivated us to look for novel RNA-binding partners of Mei2, using techniques such as RIP-CHIP [24] and CLIP-Seq [25] to isolate Mei2, and identify any novel associated RNAs. Strikingly, in addition to meiRNA, we found the mmi1 mRNA as a target of Mei2, suggesting that Mei2 can inhibit Mmi1 activity independently at two levels, the mRNA level and the protein level. In addition, we found the mRNA for the S-phase transcription factor rep2, a factor important for transcription of vegetative S-phase transcripts rather than meiotic transcripts. The binding of rep2 mRNA could potentially explain why mei2Δ, but not sme2Δ, fails at pre-meiotic DNA synthesis.
2. Material and methods
2.1. Yeast strains, media and growth conditions
Schizosaccharomyces pombe culture methods were as described in [26]. When required, cells were grown in Edinburgh Minimal Media (EMM) with supplements. For transformation, cells were grown in liquid YES at 32°C overnight and then diluted in equal volume of fresh YES media. Cells were grown another 3 h before harvesting. For nitrogen starvation and sporulation, sporulation agar (SPA) and malt extract (ME) media were used [26]. For immunoprecipitation experiments using h90 strains (JLP14, JLP1760 and JLP1772), cells were grown at 32°C on YES up to OD600 = 0.6–0.8. Then they were washed and inoculated into ME media for 4–5 h at 23°C. All strains used in this study are listed in electronic supplementary material, table S4.
2.2. Construction of TAP-tagged Mei2 and Msa1 strains
The TAP tag cassette was amplified from plasmid pFA-6a-TAP-Kan with 300 nucleotides 5′ and 3′ sequences complementary to either Mei2 C-terminus or Msa1 C-terminus. This was transformed using the protocol by [27]. Kanamycin-resistant transformants were selected on YES-G418 plates, and TAP tag integration was confirmed by colony PCR and sequencing. Primers used are listed in electronic supplementary material, table S4.
2.3. Western blot
Cells were lysed according to the procedure mentioned in [26] and equal amounts of protein from each sample were resolved by 10% SDS-PAGE. Mei2-TAP and Msa1-TAP was detected by chemiluminescence using a 1 : 5000 dilution of peroxidase anti-peroxidase (PAP) antibody.
2.4. Native RNA immunoprecipitation
Cells were collected in ice, washed with cold lysis buffer, flash-frozen in liquid nitrogen and stored at −80°C. RNA immunoprecipitation (RIP) was done according to the protocol in [28] with the following modifications. All steps were performed in a cold room on ice. Protease inhibitors leupeptin, pepstatin, bestatin and aprotinin were added to a final concentration of 2 µg ml−1 of lysis buffer. PMSF was added to a final concentration of 1 mM and 100 U ml−1 of SuperaseIn (Ambion) was also added to lysis buffer. Cells were lysed by bead beating using 0.7 mm Zirconia beads in a Fast-Prep at maximum speed for 2 pulses of 20 s and incubation for 5 min on ice in between pulses. 20 µl of lysate was frozen in liquid nitrogen for western, and 50 µl was stored in liquid nitrogen as input. IgG-Agarose beads (Sigma) were used for immunoprecipitation at 4°C for 2 h. Beads were washed for a total of nine washes, three times in lysis buffer, and six times in wash buffer containing NP-40, glycerol, heparin, RNase and protease inhibitors. RNA was eluted from the beads using 1% SDS in 1X TE with SuperaseIn. RNA was purified by phenol:chloroform extraction and sodium acetate and ethanol precipitation.
2.5. Microarray
Microarray labelling and hybridization was performed using a two-colour Quick Amp Labeling Kit (Agilent). Labelled cDNAs were hybridized onto custom-made Agilent 2 × 105 K comparative genome hybridization (CGH) arrays for S. pombe (Agilent). Immunoprecipitated RNAs were labelled with Cy3. The CGH array had 50 nucleotide probes with 50 nucleotide gaps on each strand. Therefore, the genome coverage is roughly 50%. Normalized Cy5 intensities were visualized on Integrated Genome Browser (IGB) [29]. The RIP-CHIP data are available in electronic supplementary material as processed files in ‘.egr’ format for viewing in IGB. The raw data including array design are available in GEO (GSE114299).
2.6. UV crosslinking and immunoprecipitation
Cross-linking immunoprecipitation of Mei2-TAP and Msa1-TAP were done according to [30] with some modifications. Cells were UV cross-linked using Stratalinker with power 100 and default time settings. Cells were lysed exactly as in RIP (above). Lysates were sonicated using a Misonix sonicator at 80 amplitude for 30 s. Immunoprecipitation, washes and elution were done as in RIP, but with different buffers as in [30]. Following elution, 1 µl of 20 mg ml−1 proteinase K was added to the eluates and incubated for 1 h at 42°C. RNA was extracted by phenol:chloroform and precipitated using sodium acetate and ethanol.
2.7. RNA sequencing of immunoprecipitated RNA and data analysis
Immunoprecipitated RNA was DNase treated, and libraries for Illumina sequencing were made using a Nugen Ovation Universal RNA-Seq system. cDNA was made using a random-primed dUTP method to preserve strand-specificity. cDNA was sonicated using a Misonix 3000 sonicator at 60 amplitude for 10 min on ice. Libraries were PCR-amplified for 12 cycles. The quality of libraries was checked using Bioanalyzer and Qubit, and sequenced on Illumina MiSeq by paired-end sequencing. Reads were mapped to S. pombe reference transcriptome version ASM294 v. 2.26 using STAR [31]. Discordant mapping was not allowed and only annotated splice junctions were mapped. Alignments in bam format were sorted and indexed using Samtools and visualized on Integrative Genomics Viewer [32]. Deeptools ‘bamCoverage’ [33] was used to convert bam files to strand-specific bigwig files to visualize pileups at specific loci on IGV.
Peak-calling, enrichment analysis and peak annotation was done using Homer [34]. ‘Histone-mode’ was enabled in Homer to allow for wide peaks. Mei2-specific RNA targets were determined by differential peak calling against CLIP-Seq tags from untagged, and Msa1-TAP separately (electronic supplementary material, table S1). Highly significant Mei2 targets were determined by selecting the top 20 significant peaks (by p-value) from Mei2 versus untagged and Mei2 versus Msa1, and taking the intersection of the two sets. This provided us with 10 highly significant CLIP-Seq targets of Mei2 (electronic supplementary material, table S2). Msa1-specific targets were determined by differential peak calling against untagged CLIP-Seq tags (electronic supplementary material, table S3). Our CLIP-Seq data including the bigwig and peak files are available in GEO, accession code: GSE112117.
For enrichment analysis in figure 3, Homer was used to calculate the tag counts for 5 nucleotide bins over the entire length of the target RNA. These were then normalized to the percentage length of each RNA. Subsequently these were hierarchically clustered with Euclidian distance as the similarity metric and simple linkage using Cluster v. 3.0 [35]. The clusters were visualized by Java Treeview [36].
Figure 3.
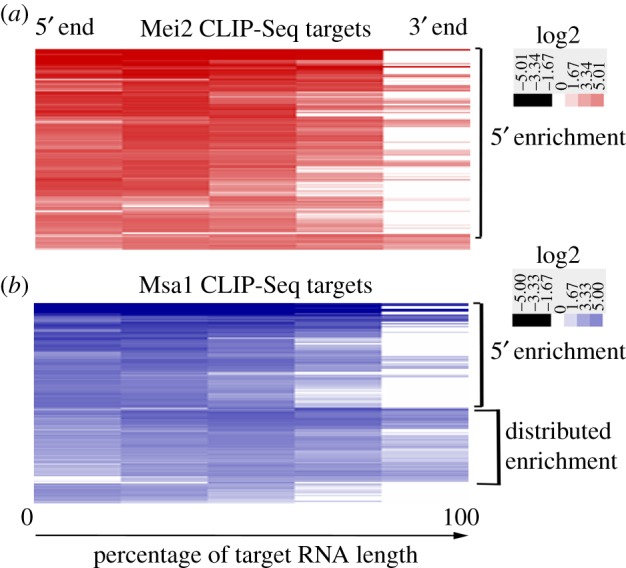
Enrichment profile along RNA length of Mei2 and Msa1 targets. (a) Enrichment profile of significant targets of Mei2 from CLIP-Seq determined by Homer. Each column in the heatmap shows enrichment that was calculated for a 5-nucleotide bin. Each row of the heatmap shows enrichment from 5′ to 3′ along the length of one particular Mei2 RNA target, normalized by length. (b) Enrichment profiles along length of Msa1 CLIP-Seq targets.
2.8. Quantitative RT-PCR
Input and co-immunoprecipitated RNAs were used to make first strand cDNA using Superscript RT III (Invitrogen Life Technologies) according to manufacturer's instructions. cDNA was used for quantitative PCR using Roche LightCycler 480 SYBR Green Master kit with gene specific primers (electronic supplementary material, table S4).
3. Results and discussion
3.1. Expression of Mei2 after nitrogen starvation and in early meiosis
We wished to identify novel RNA targets of Mei2 in meiosis, and for that purpose it was important to know at what times in meiotic time-courses Mei2 was being highly expressed. We used western blotting to assay Mei2 protein levels at various times after nitrogen starvation (which was used to initiate meiosis). We added a TAP-tag to the C-terminus of Mei2 in an h90 strain and allowed it to sporulate in nitrogen-deficient ME medium. We found that the Mei2-TAP strain sporulated as efficiently as wild-type (data not shown), indicating that addition of a TAP tag to Mei2 does not affect sporulation. Next, we did a time course and looked for Mei2 expression at different times after nitrogen starvation. We found two bands roughly corresponding to the size of full-length Mei2-TAP from 3 h to 6 h after nitrogen starvation (electronic supplementary material, figure S1A). We found by microscopy that, during these times, most cells had conjugated (data not shown). Mei2-TAP levels were higher at the 3 h and 6 h data points than at other times; this could indicate regulation of Mei2 abundance, but could also reflect asynchronous meiosis. We noticed multiple bands shorter than full-length Mei2-TAP (electronic supplementary material, figure S1A). These were probably degradation products of Mei2 protein.
3.2. Native RNA immunoprecipitation and UV cross-linking and immunoprecipitation of Mei2-TAP
To identify RNAs associated with the Mei2 RNA-binding protein during early meiosis, we used pooled samples with high Mei2 expression (4–6 h after nitrogen starvation) to immunoprecipitate Mei2-TAP with agarose-coupled IgG beads. We tested Mei2-TAP enrichment after immunoprecipitation using western blotting with a PAP antibody and found that a significant proportion of cellular Mei2-TAP was associated with the IgG beads (electronic supplementary material, figure S2A). To stabilize Mei2–RNA interactions, we used UV cross-linking and immunoprecipitation (CLIP) of Mei2-TAP according to the protocol from [37]. Many groups that study RNA–protein interactions have observed that some RNAs co-immunoprecipitate promiscuously with most RNA-binding proteins (Juan Mata 2011, personal communication). To control for such noise, we added a TAP tag to the C-terminus of another RNA-binding protein, Msa1, and found that Msa1-TAP was expressed throughout meiosis (electronic supplementary material, figure S1B). Both Mei2 and Msa1 had some level of protein degradation in meiosis, despite the addition of protease inhibitors (electronic supplementary material, figure S1C).
We performed CLIP on nitrogen starved h90 cells which were Mei2-TAP (experiment), or Msa1-TAP (control), or untagged (control). The lysates were lightly sonicated after cross-linking to break up complexes. Analysis by western blotting showed strong association of Mei2-TAP and Msa1-TAP with the IgG beads after CLIP (electronic supplementary material, figure S2B).
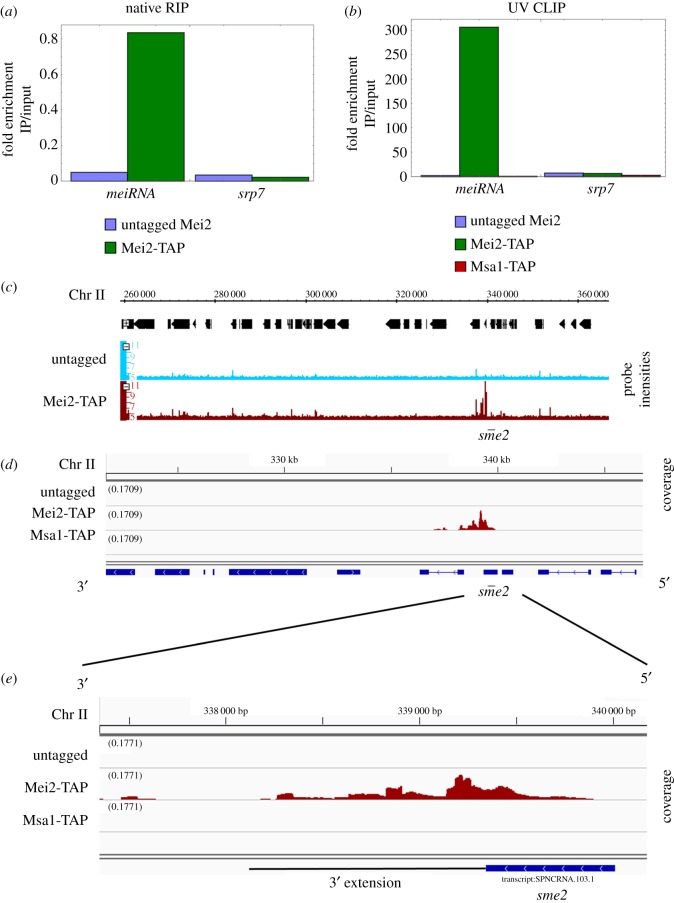
We purified RNAs that co-immunoprecipitated with Mei2-TAP after RIP, and Mei2-TAP and Msa1-TAP after CLIP. Since meiRNA is a known target of Mei2, we used qRT-PCR to test for meiRNA enrichment and found that it was highly enriched after both RIP and CLIP experiments (figure 1a,b). The fold enrichment was significantly higher after CLIP (figure 1b). To ensure specificity, we tested for enrichment of the control mRNA srp7, which should not bind Mei2, and found that it was not enriched (figure 1a,b). meiRNA was not enriched in the control CLIP experiments with Msa1-TAP, indicating that meiRNA binds specifically to Mei2 in these experiments (figure 1b).
Figure 1.
meiRNA enrichment after Mei2-TAP immunoprecipitation. (a) meiRNA enrichment after native RNA immunoprecipitation (RIP) of Mei2 shown by qRT-PCR. The y-axis shows log2 fold enrichment. srp7 is shown as a specificity control. (b) meiRNA enrichment after UV cross-linking immunoprecipitation (CLIP) by qRT-PCR is shown. srp7 is shown as a specificity control. (c) Integrated Genome Browser view of the sme2 locus after RIP-CHIP. (d) Integrative Genomics Viewer snapshot of the sme2 locus after CLIP-Seq. (e) CLIP-Seq coverage reads at the sme2 locus. The y-axis scale is normalized for all samples in (c–e).
3.3. Novel RNA-binding partners of Mei2 discovered by RIP-CHIP and CLIP-Seq
Since the specific enrichment of meiRNA suggested that the RIP experiment was successful, we looked for novel targets of Mei2 by hybridizing the extracted RNAs from RIP to a microarray (see methods). We used a tiling array chip with 50 nucleotide strand-specific probes (see Methods). We searched the genome for enriched regions using the Integrated Genome Browser and found 13 RNAs (including meiRNA) specifically enriched in the Mei2-TAP experiment versus untagged control (table 1 and figure 1c).
Table 1.
RNA targets of Mei2.
| gene | function |
|---|---|
| Mei2 targets | |
| sme2 | ncRNA acts as Mmi1 sponge |
| mmi1 | meiotic RNA turnover |
| rep2 | MBF Transcription Factor |
| mug110 | unknown |
| SPACUNK4.17 | NAD binding dehydrogenase family protein |
| SPBP23A10.11c | cell wall biosynthesis |
| RIP-CHIP only | |
| SPBC32F12.07c | ubiquitin E3 ligase |
| yox1 | MBF regulatory subunit |
| efc25 | Ras1 guanyl-nucleotide exchange factor |
| SPCC1223.14 | chorismate synthesis |
| SPCC757.12 | alpha amylase homologue |
| snoU14 | unknown |
| but2 | protein neddylation with Uba3 |
| CLIP-Seq only | |
| SPAC11D3.01c | unknown |
| rbd4 | rhomboid family protease |
| tif51 | translation elongation and termination factor eIF5A (predicted) |
| ubc7 | Hrd1 ubiquitin E2 ligase |
As an independent assay for novel RNAs, we also identified Mei2 and Msa1 targets from CLIP. For this, RNAs were assayed by reverse transcription and cDNA sequencing. We mapped the sequenced reads to the S. pombe transcriptome, visualized the alignments using IGV, and computed pileups at target loci. Exactly as in the RIP experiments (above) we found that meiRNA was highly enriched in the Mei2-TAP CLIP-Seq but not in the control Msa1-TAP CLIP-Seq (figure 1d). We used the peak-calling and annotation program Homer to analyse the genome for additional Mei2 targets (electronic supplementary material, table S1). We classified RNAs as Mei2 CLIP-Seq targets only if those RNAs immunoprecipitated in the Mei2-TAP strain but not in the control untagged strain or the control Msa1-TAP strain (electronic supplementary material, table S2, see methods).
Furthermore, to guard against the possibility that Mei2 might bind non-specifically to highly expressed meiotic RNAs, we looked at the mRNAs for many highly expressed, early meiotic genes, but none of these mRNAs was enriched in either experiment. Two examples, rec8 and mei4, are shown in electronic supplementary material, figure S3A,B.
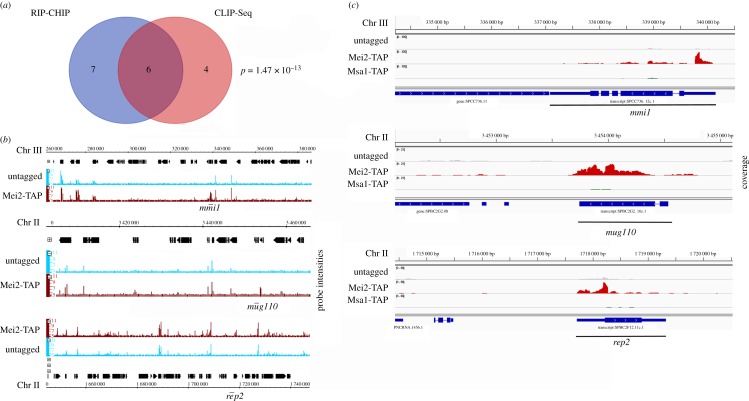
There were 13 targets of Mei2 as determined by RIP-CHIP (table 1, figures 1c and 2b; electronic supplementary material, figures S4 and S5), and 10 targets as determined by the independent CLIP-SEQ experiment (electronic supplementary material, figures S6 and S7). Strikingly, six of these targets (including meiRNA) overlapped between the two independent experimental approaches (p-value ∼ 10−13) (figure 2a). We considered these as high-confidence Mei2 targets (figure 2b,c; electronic supplementary material, figures S4 and S6). The functions of these targets are summarized in table 1. Two of the five novel targets, mmi1 and rep2 (figure 2b,c), have well characterized biological roles in controlling gene expression and meiosis.
Figure 2.
RNA partners of Mei2 in early meiosis with known roles in meiosis. (a) Venn diagram showing number of significant targets obtained by Mei2-TAP RIP-CHIP and CLIP-Seq. 6 RNA targets of Mei2 were enriched in both experiments. (b) Probe intensities from RIP-CHIP for important Mei2 targets mmi1, rep2 and mug110. (c) CLIP-Seq coverage from Mei2-TAP and Msa1 at the mmi1, rep2, and mug110 loci. The y-axis scale is normalized for (b) and (c).
The mmi1 mRNA has obvious relevance, since the Mmi1 protein inhibits meiotic entry, and since the Mmi1 protein is already known to be a target of the Mei2–meiRNA complex. Our finding here suggests that Mei2 is inhibiting Mmi1 function at two levels, the protein level (as previously shown) and also, separately and independently, the mRNA level. This could lead to a much more profound inhibition of total Mmi1 activity than if the Mei2–meiRNA complex were targeting Mmi1 protein only. Interestingly, we observe that the mmi1 primary transcript has nine consensus sequence binding sites for Mmi1 protein (T(TCG)AAAC), and the mature mRNA has seven. This suggests that there could be negative feedback of Mmi1 protein on mmi1 mRNA, or, perhaps more speculatively, that mmi1 mRNA itself is a sponge RNA for its own protein. Interestingly, CRAC analysis suggests that Mmi1 protein may bind to its own transcript during vegetative growth [38], but there is no evidence that Mmi1 targets its own transcript for destruction, somewhat supporting the view that mmi1 RNA could be a sponge for its own protein.
Rep2 is a component of the MBF transcription factor complex [39]. The MBF complex comprises Res1, Res2, Cdc10 and Rep2 in vegetative cells, and is responsible for transcription of S-phase genes during vegetative growth, including many genes needed for DNA replication [39–43]. However cellular requirements during vegetative S-phase, and during pre-meiotic S-phase, are not precisely the same, and rep2 RNA expression is reduced during nitrogen starvation [39]. Furthermore, overexpression of Rep2 blocks G1 arrest following nitrogen starvation, whereas depletion of Rep2 facilitates G1 arrest after nitrogen starvation [39]. This indicates that Rep2 promotes vegetative S-phase, and in that sense inhibits meiotic entry. During meiosis, the meiotic MBF transcription factor Rep1 probably replaces Rep2 in the MBF complex, since Rep1 is critical for transcribing a subset of pre-meiotic S-phase genes [43–46]. Sequestration of rep2 mRNA in the nucleus by the Mei2–meiRNA complex could aid in replacement of Rep2 by Rep1 in the MBF complex, thus promoting pre-meiotic S.
mug110 is a meiosis upregulated gene of unknown function. Previously mug110 mRNA was shown to bind another RNA-binding protein, Crp79 [47]. mug110 was not enriched in Msa1-TAP CLIP-Seq (figure 2c; electronic supplementary material, figure S6) suggesting that it binds specifically to Mei2 in early meiosis. mug110 is a meiosis upregulated gene, but the mug110Δ strain has no obvious meiosis or sporulation defects [48]. We made a mug110Δ mei2Δ double mutant and found that the phenotype of mug110Δ mei2Δ was indistinguishable from mei2Δ; the double mutant did not initiate meiosis (data not shown).
SPACUNK4.17 (electronic supplementary material, figure S4) is a NAD dehydrogenase family protein with an unknown function in S. pombe. In a recent screen for mutants that affect sexual reproduction, SPACUNK4.17Δ was shown to have cell-fusion defects, low mating efficiency and multi-septate cells [49]. We have noticed cell fusion defects in mei2Δ cells. The phenotypes of SPACUNK4.17Δ are either mild to moderate, but this gene could play a yet undiscovered role in early meiosis.
Finally, SPBP23A10.11c (electronic supplementary material, figure S4) is poorly characterized. It is an orthologue of S. cerevisiae TOS1, which encodes a cell wall protein under the control of the SBF/MBF transcription factor. Its RNA level goes down in response to nitrogen starvation and meiosis [50].
Some RNAs co-immunoprecipitated with Mei2 in only RIP-CHIP but not CLIP-Seq or vice versa (table 1). We did not classify these two sets of RNAs as high-confidence Mei2 targets (electronic supplementary material, figures S5 and S7). A few RNAs were enriched in both Mei2-TAP and Msa1-TAP CLIP-Seq but not in untagged controls (not shown). Both Mei2 and Msa1 are meiotic RNA-binding proteins so they may have common targets. However, some RNAs co-precipitate promiscuously in RNA IP experiments and we considered this to be because of non-specific interactions between RNAs and RNA-binding proteins.
3.4. Do the novel targets explain why mei2 mutants have stronger phenotypes than meiRNA?
Our investigation began with the observation that a mei2 mutant has a stronger and different meiotic phenotype than a meiRNA mutant. This suggested that the Mei2 protein might target additional RNAs. The five novel targets here, and in particular mmi1 mRNA and rep2 mRNA, are good candidates for such additional targets. However, the experiments required to prove this point are somewhat subtle: it is not sufficient to simply delete a candidate such as mmi1, as that removes both the mRNA and the protein, and it is already clear that an mmi1 null mutant has an extreme phenotype. Ideally, one would identify the residues in each mRNA allowing that mRNA to bind to Mei2; then mutate those residues, creating a mutant mRNA that fails to bind Mei2, and ask whether this (in combination with a sme2 deletion) phenocopies a mei2 deletion. To this end, we have searched for sequence commonalities between the RNAs that bind Mei2 (e.g. using MEME), but so far without success. At present, the sequence determinants allowing binding to Mei2 remain unknown.
In wild-type meiotic cells, Mei2 forms a dot at the sme2 locus. In a sme2Δ mutant, there is no dot. However, when an NLS is added to Mei2, then a dot is formed even in a sme2Δ mutant [2]. It will be interesting to see if this dot has any specific localization, and in particular whether it might be at any of the loci we have identified as expressing additional targets of Mei2 (mmi1, rep2, mug110, etc.)
3.5. Mei2 may have a preference for binding 5′ regions of target RNAs
meiRNA is a long non-coding RNA that is critical for meiosis. During meiosis, meiRNA has a 3′ extended form, and Mei2 binds to the 5′ end [23], while Mmi1 binds to the 3′ region. We found that the 3′ extended meiotic form of meiRNA co-immunoprecipitated with Mei2-TAP (figure 1e). However, more reads aligned to the 5′ end of meiRNA (figure 1e), so our results are consistent with a previous study [23] showing that Mei2 binds to the 5′ end of meiRNA.
Strikingly, the 5′ UTR regions of mmi1 and rep2 mRNAs were enriched in Mei2-TAP CLIP-Seq (figure 2c). This could indicate that Mei2 binds specifically to the 5′ UTR of mmi1 and rep2. It is also possible that the sequencing reads are restricted to the 5′ UTR due to an artefact of library preparation using random hexamers. However, we did not find a 5′ enrichment of other Mei2 target RNAs, e.g. mug110 and SPACUNK4.17 (figure 2c; electronic supplementary material, figure S6), suggesting that the observed 5′ enrichment is not coincidental.
To test if Mei2 has a binding preference on its RNA targets, we used Homer to calculate the enrichment of all Mei2 CLIP-Seq targets genome-wide, over the entire length of each RNA. We then clustered the global enrichment profile of Mei2 targets (figure 3a). We found that predominantly (about 60%) 5′ regions of Mei2 RNA targets were enriched, including some that had high enrichment of extreme 5′ ends (figure 3a). This strongly suggests that Mei2 has a preference for binding 5′ ends of its target RNAs, and in some cases the 5′ UTR. We also made a similar enrichment profile for Msa1 CLIP-Seq targets (electronic supplementary material, table S3), and found that about half of them had 5′ region enrichment, and the other half had enrichment distributed across the RNA (figure 3b). This confirmed that not all RNAs we recovered from CLIP-Seq had 5′ enrichment, so this enrichment was not an artefact of sequencing library preparation.
4. Conclusion
In summary, Mei2 protein binds to the meiRNA, which acts as a sponge for Mmi1 protein in the nucleus during early meiosis [23]. Mmi1 is inhibitory towards meiosis, and mmi1 mutants attempt meiosis even without nitrogen starvation [17]. We show here that in early meiosis, Mei2 not only binds to the Mmi1 protein, but also to the mmi1 mRNA. We suggest this interaction prevents mmi1 mRNA export to the cytoplasm for translation and inhibits synthesis of new Mmi1 protein. Thus, we propose a model where Mei2 simultaneously inhibits the action of mmi1 at two levels (the protein level and the transcript level), resulting in a deeper inhibition of Mmi1 activity than could be achieved by inhibition at one level alone (figure 4). Interestingly, we have also observed that the mmi1 mRNA contains multiple (7 to 9) binding sites for Mmi1, so mmi1 mRNA could act as a sponge RNA for its own protein. While Mei2 inhibits Mmi1 function at two levels, Mmi1 also inhibits Mei2 via proteolysis [21], creating a double-negative feedback loop that could act as a meiotic switch.
Figure 4.
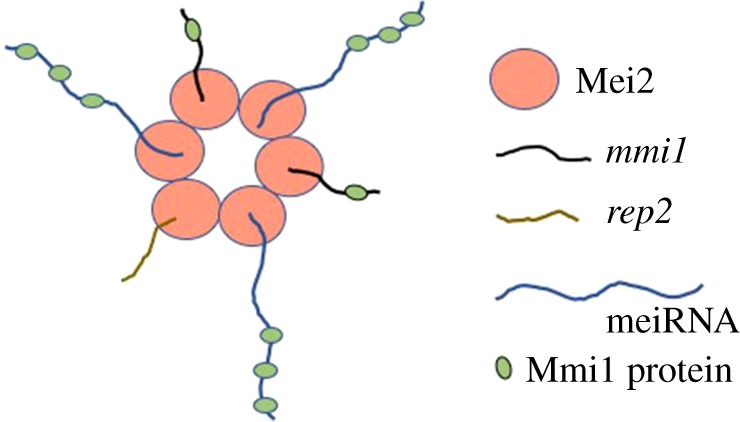
Mei2 titrates out inhibitors of meiosis. During early meiosis Mei2 binds to meiRNA, which binds Mmi1 protein, thus sequestering Mmi1. Mei2 also binds mmi1 mRNA, and probably prevents synthesis of new Mmi1 protein. Mei2 also binds rep2 mRNA, and probably prevents synthesis of new Rep2 protein. Titration of these three meiotic inhibitors (Mmi1, mmi1 mRNA, and rep2 mRNA) facilitate initiation of meiosis. The mechanism by which Mei2 forms a dot (here shown as a speculative Mei2–Mei2 protein–protein interaction) is unknown. Also unknown is whether the Mmi1 binding sites on the mmi1 mRNA participate in the titration of Mmi1 protein.
Mei2 also binds to rep2 mRNA, possibly preventing the synthesis of new Rep2 protein (figure 4). In vegetative cells, Rep2 is a component of the MBF transcription factor, but in response to nitrogen starvation, expression of Rep1 increases, and it replaces Rep2 in the pre-meiotic MBF complex. We suggest this replacement is enhanced by the inhibition of Rep2 synthesis caused by the sequestration of rep2 mRNA by Mei2. Thus, in a mei2 null mutant, at least some MBF complexes would continue to contain Rep2, and this could help explain why mei2 mutants fail to accomplish pre-meiotic DNA synthesis. Of course, the profound inhibition of Mmi1 function by Mei2 acting at two levels could also contribute to pre-meiotic S.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Hong Wang for performing microarrays and Dr Juan Mata for valuable advice on performing RIP-CHIP in fission yeast.
Data accessibility
CLIP-Seq data are available in GEO, accession code: GSE112117 (reviewer token: ‘wbyrcucolvonbsh’). RIP-CHIP data are available as raw and processed text files in GEO, accession code: GSE114299 (reviewer token: ‘mdgvaqkorfatjsz’).
Authors' contributions
K.M. carried out the molecular laboratory work, participated in data analysis, carried out sequence alignments, participated in the design of the study and drafted the manuscript; B.F. and J.L. conceived of the study, designed the study, coordinated the study and helped draft the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by NIGMS grant no. GM088297 to B.F. and J.L.
References
- 1.Watanabe Y, Yamamoto M. 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78, 487–498. ( 10.1016/0092-8674(94)90426-X) [DOI] [PubMed] [Google Scholar]
- 2.Yamashita A, Watanabe Y, Nukina N, Yamamoto M. 1998. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95, 115–123. ( 10.1016/S0092-8674(00)81787-0) [DOI] [PubMed] [Google Scholar]
- 3.Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. 1997. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386, 187–190. ( 10.1038/386187a0) [DOI] [PubMed] [Google Scholar]
- 4.Kitamura K, Katayama S, Dhut S, Sato M, Watanabe Y, Yamamoto M, Toda T. 2001. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell 1, 389–399. ( 10.1016/S1534-5807(01)00037-5) [DOI] [PubMed] [Google Scholar]
- 5.Marchese FP, Raimondi I, Huarte M. 2017. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18, 206 ( 10.1186/s13059-017-1348-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon JH, Abdelmohsen K, Gorospe M. 2013. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 425, 3723–3730. ( 10.1016/j.jmb.2012.11.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wapinski O, Chang HY. 2011. Long noncoding RNAs and human disease. Trends Cell Biol. 21, 354–361. ( 10.1016/j.tcb.2011.04.001) [DOI] [PubMed] [Google Scholar]
- 8.Huarte M. 2015. The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261. ( 10.1038/nm.3981) [DOI] [PubMed] [Google Scholar]
- 9.Touat-Todeschini L, et al. 2017. Selective termination of lncRNA transcription promotes heterochromatin silencing and cell differentiation. EMBO J. 36, 2626–2641. (doi:10.15252/embj.201796571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ard R, Tong P, Allshire RC. 2014. Long non-coding RNA-mediated transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat. Commun. 5, 5576 ( 10.1038/ncomms6576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee D, Sanchez AM, Goldgur Y, Shuman S, Schwer B. 2016. Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast. RNA 22, 1011–1025. ( 10.1261/rna.056515.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garg A, Sanchez AM, Shuman S, Schwer B. 2018. A long noncoding (lnc)RNA governs expression of the phosphate transporter Pho84 in fission yeast and has cascading effects on the flanking prt lncRNA and pho1 genes. J. Biol. Chem. 293, 4456–4467. ( 10.1074/jbc.RA117.001352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah S, Wittmann S, Kilchert C, Vasiljeva L. 2014. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 28, 231–244. ( 10.1101/gad.230177.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimada T, Yamashita A, Yamamoto M. 2003. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell 14, 2461–2469. ( 10.1091/mbc.e02-11-0738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato M, Shinozaki-Yabana S, Yamashita A, Watanabe Y, Yamamoto M. 2001. The fission yeast meiotic regulator Mei2p undergoes nucleocytoplasmic shuttling. FEBS Lett. 499, 251–255. ( 10.1016/S0014-5793(01)02567-4) [DOI] [PubMed] [Google Scholar]
- 16.Sato M, Watanabe Y, Akiyoshi Y, Yamamoto M. 2002. 14-3-3 protein interferes with the binding of RNA to the phosphorylated form of fission yeast meiotic regulator Mei2p. Curr. Biol. 12, 141–145. ( 10.1016/S0960-9822(01)00654-6) [DOI] [PubMed] [Google Scholar]
- 17.Harigaya Y, et al. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50. ( 10.1038/nature04881) [DOI] [PubMed] [Google Scholar]
- 18.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. 2010. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 29, 2173–2181. ( 10.1038/emboj.2010.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HM, Futcher B, Leatherwood J. 2011. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS ONE 6, e26804 ( 10.1371/journal.pone.0026804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. 2009. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat. Struct. Mol. Biol. 16, 255–264. ( 10.1038/nsmb.1556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonetti F, Candelli T, Leon S, Libri D, Rougemaille M. 2017. Ubiquitination-dependent control of sexual differentiation in fission yeast. Elife 6, e28046 ( 10.7554/eLife.28046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita A, et al. 2012. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol. 2, 120014 ( 10.1098/rsob.120014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shichino Y, Yamashita A, Yamamoto M. 2014. Meiotic long non-coding meiRNA accumulates as a dot at its genetic locus facilitated by Mmi1 and plays as a decoy to lure Mmi1. Open Biol. 4, 140022 ( 10.1098/rsob.140022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1, 302–307. ( 10.1038/nprot.2006.47) [DOI] [PubMed] [Google Scholar]
- 25.Licatalosi DD, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469. ( 10.1038/nature07488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsburg SL, Rhind N. 2006. Basic methods for fission yeast. Yeast 23, 173–183. ( 10.1002/yea.1347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahler J, et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- 28.Gerber AP, Herschlag D, Brown PO. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79 ( 10.1371/journal.pbio.0020079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freese NH, Norris DC, Loraine AE. 2016. Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32, 2089–2095. ( 10.1093/bioinformatics/btw069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selth LA, Gilbert C, Svejstrup JQ. 2009. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb. Protoc. 2009, pdb prot5234 ( 10.1101/pdb.prot5234) [DOI] [PubMed] [Google Scholar]
- 31.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. ( 10.1093/bioinformatics/bts635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. ( 10.1093/bib/bbs017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191. ( 10.1093/nar/gku365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, et al. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. ( 10.1016/j.molcel.2010.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20, 1453–1454. ( 10.1093/bioinformatics/bth078) [DOI] [PubMed] [Google Scholar]
- 36.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248. ( 10.1093/bioinformatics/bth349) [DOI] [PubMed] [Google Scholar]
- 37.Granneman S, Kudla G, Petfalski E, Tollervey D. 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl Acad. Sci. USA 106, 9613–9618. ( 10.1073/pnas.0901997106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilchert C, Wittmann S, Passoni M, Shah S, Granneman S, Vasiljeva L. 2015. Regulation of mRNA levels by decay-promoting introns that recruit the exosome specificity factor Mmi1. Cell Rep. 13, 2504–2515. ( 10.1016/j.celrep.2015.11.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakashima N, Tanaka K, Sturm S, Okayama H. 1995. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 14, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aves SJ, Durkacz BW, Carr A, Nurse P. 1985. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 ‘start’ gene. EMBO J. 4, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Nojima H, Okayama H. 1992. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc 10+ and SWI4 gene products. EMBO J. 11, 4923–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto M, Tanaka K, Okayama H. 1994. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 13, 1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H. 1994. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 13, 1881–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding R, Smith GR. 1998. Global control of meiotic recombination genes by Schizosaccharomyces pombe rec16 (rep1). Mol. Gen. Genet. 258, 663–670. ( 10.1007/s004380050780) [DOI] [PubMed] [Google Scholar]
- 45.Mata J, Wilbrey A, Bahler J. 2007. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 8, R217 ( 10.1186/gb-2007-8-10-r217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y, Takeda T, Nasmyth K, Jones N. 1994. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 8, 885–898. ( 10.1101/gad.8.8.885) [DOI] [PubMed] [Google Scholar]
- 47.Amorim MJ, Cotobal C, Duncan C, Mata J. 2010. Global coordination of transcriptional control and mRNA decay during cellular differentiation. Mol. Syst. Biol. 6, 380 ( 10.1038/msb.2010.38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin-Castellanos C, et al. 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 15, 2056–2062. ( 10.1016/j.cub.2005.10.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dudin O, Merlini L, Bendezu FO, Groux R, Vincenzetti V, Martin SG. 2017. A systematic screen for morphological abnormalities during fission yeast sexual reproduction identifies a mechanism of actin aster formation for cell fusion. PLoS Genet. 13, e1006721 ( 10.1371/journal.pgen.1006721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mata J, Lyne R, Burns G, Bahler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32, 143–147. ( 10.1038/ng951) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CLIP-Seq data are available in GEO, accession code: GSE112117 (reviewer token: ‘wbyrcucolvonbsh’). RIP-CHIP data are available as raw and processed text files in GEO, accession code: GSE114299 (reviewer token: ‘mdgvaqkorfatjsz’).