Abstract
Rapidly renewing tissues such as the intestinal epithelium critically depend on the activity of small-sized stem cell populations that continuously generate new progeny to replace lost and damaged cells. The complex and tightly regulated process of intestinal homeostasis is governed by a variety of signalling pathways that balance cell proliferation and differentiation. Accumulating evidence suggests that stem cell control and daughter cell fate determination is largely dictated by the microenvironment. Here, we review recent developments in the understanding of intestinal stem cell dynamics, focusing on the roles, mechanisms and interconnectivity of prime signalling pathways that regulate stem cell behaviour in intestinal homeostasis. Furthermore, we discuss how mutational activation of these signalling pathways endows colorectal cancer cells with niche-independent growth advantages during carcinogenesis.
Keywords: intestinal stem cells, signalling, microenvironment, colorectal cancer
1. Adult stem cells are critical for tissue homeostasis
Adult tissue homeostasis strictly depends on the balanced generation of new cells that replenish cells that are lost through natural attrition or tissue injury. This process of tissue regeneration is fuelled by small populations of stem cells that are defined by their unique ability to renew themselves persistently (self-renewal) while also giving rise to the specialized cell types of the pertinent tissue (multipotency) [1–3]. These adult stem cells are generally referred to by their tissue of origin (e.g. haematopoietic, neuronal or intestinal stem cells (ISCs)). Depending on local needs, stem cells may switch their mode of cell division from symmetric to asymmetric. Symmetric division gives rise to two identical daughter cells, both endowed with stem cell properties. Asymmetric division produces only one stem cell and a progenitor cell via signals from the microenvironment and unequal segregation of proteins or RNA, which direct distinct gene expression profiles that control the fate of the newly generated cell [4,5].
Stem cell activity is for a large part dictated externally by the microenvironment (the stem cell niche) to precisely control stem cell output and meet the homeostatic or regenerative demands of the tissue. Extracellular cues, provided by neighbouring niche cells, locally interact with stem cells to regulate their fate by activating specific signalling pathways. Here, we review current knowledge on how stem cells receive and interpret extracellular signals from their niche, focusing on the prototype model of ISCs, which undergo rapid self-renewal kinetics and give rise to the multiple specialized lineages of the intestinal epithelium [6].
2. Intestinal architecture
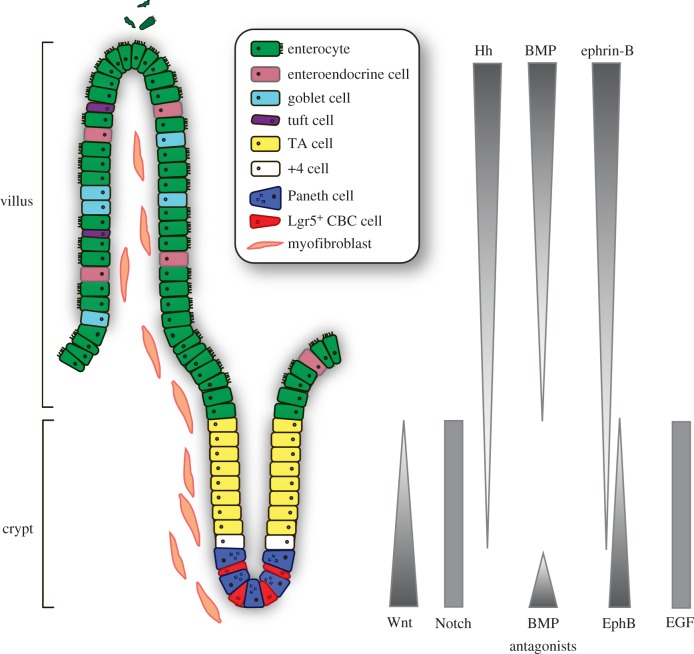
The intestinal mucosa has evolved to absorb water and nutrients while at the same time protecting the body from toxic contents of the gut lumen. The continuous renewal of the gut epithelium allows cells towards the end of their lifetime to shed off at the tip of the villus, while newly produced and differentiated cells migrate up and restock the epithelial barrier. This endless process is sustained by symmetrically dividing stem cells that reside at the crypt base (figure 1).
Figure 1.
Architecture of the small intestine and the controlling signalling pathways. Actively cycling Lgr5-positive crypt base columnar (CBC) stem cells reside at the bottom of the crypt intermingled with Paneth cells. The stem cells give rise to transit amplifying (TA) cells that terminally differentiate towards all epithelial lineages of the villus. Position 4 (+4) stem cells are mobilized upon tissue damage. Intestinal homeostasis is governed by an interconnected network of signalling pathways, regulating the balance between proliferation and differentiation.
The intestinal epithelial lining represents one of the most intensively self-replenishing organs; within 5 days the entire epithelial layer is renewed [7]. The architecture of the intestine is designed to maximize the surface for nutrient uptake and is folded into large numbers of villi and crypts in the small intestine. The colon is also folded into crypts but does not display villi. ISCs reside at the bottom of the crypts and are able to replenish the whole crypt–villus axis, generating all differentiated cell types required for the physiological function of the intestine (figure 1). Newly born cells first give rise to the rapidly proliferating subset of progenitors, also known as transit amplifying (TA) cells, that occupy the crypts and expand the population required for epithelial turnover. They migrate upwards while differentiating into one of the specialized epithelial lineages [8]. Among the differentiated cell types, nutrient absorbing enterocytes make up the majority of cells lining the villi. Other major lineages are secretory cell types such as goblet cells that produce mucus to generate a protective barrier and enteroendocrine cells that secrete various hormones that exert both local and systemic regulatory effects. Furthermore, specialized Paneth cells escape the upward flow and migrate downward to constitute the niche for ISCs at the crypt base [9,10], secreting antimicrobial peptides and essential factors for stem cell maintenance. Finally, two more rare cell types are produced, comprising secretory Tuft cells that serve as sensors for luminal contents and initiate type 2 immune responses to helminth infections [11–14], and M (microfold) cells that reside in specialized epithelium overlying Peyer's patches to communicate with the gut's immune system [15]. The continuous proliferation of crypt cells is ultimately balanced by shedding of apoptotic cells at the tip of the villus into the lumen (figure 1).
3. Intestinal stem cells
3.1. Plasticity of intestinal stem cells
At present, several populations of ISCs have been described based on their markers and localization in the crypt. Among these are the fast-cycling crypt base columnar (CBC) stem cells that are marked by leucine-rich-repeat containing G-protein coupled receptor 5 (Lgr5) [1,7,16]. In addition, a slow dividing, ‘reserve stem cell’ population was identified, also called position 4/+4 cells or label-retaining cells (LRCs) [16–19].
The Lrg5-positive CBC cells that divide every day are considered the driving force of intestinal tissue renewal. Lineage tracing experiments in mice showed that all epithelial cell types originate from the CBC cells that produced clonal ribbons of progeny with lifelong perseverance [1]. To date, Lgr5 has been validated as a bona fide stem cell marker not only in the intestine but also in the stomach pylorus [20] and corpus [21], and hair follicle [22]. Expression profiling of sorted intestinal Lgr5-positive cells provided a CBC stem cell gene expression signature [23,24], which allowed for further functional analysis of additional stem cell genes, such as Achaete-scute complex homolog 2 (Ascl2) [8,25,26], tumour necrosis factor receptor superfamily member 19 (TNFRSF19) or Troy [27], Olfactomedin 4 (Olfm4) [28] and SPARC related modulator calcium binding 2 (Smoc2) [23].
The pool of slow cycling reserve stem cells is considered to comprise quiescent stem cells that are mobilized upon tissue damage [18,19]. Several markers for these cells were identified, including polycomb protein B lymphoma Mo-MLV insertion region 1 homolog (Bmi1) [29], telomerase reverse transcriptase (Tert) [30], homeobox-only protein (Hopx) [31] and leucine-rich repeats and immunoglobulin-like domains 1 (Lrig1) [32,33].
Additionally, several secretory progenitor populations showed the ability to de-differentiate and revert to stem-like cells to replenish the crypt upon extensive tissue damage. This property was ascribed to LRCs [34] as well as to progenitors that express the Notch ligand Delta-like 1 (Dll1) [35], and to Paneth cells upon irradiation [36]. Furthermore, in addition to cells of the secretory lineage, a recent study showed that the abundant enterocyte progenitors of the absorptive lineage can dedifferentiate and replace lost ISCs upon ablation of Lgr5-expressing stem cells as well [37].
In conclusion, crypt cells display substantial plasticity, employing CBC stem cells for regular tissue renewal and reserve stem cells to act upon tissue damage. Stemness, therefore, appears extrinsically imposed on cells, placing niche signals centre stage for regulating ISC function and intestinal homeostasis.
3.2. Lgr5-positive crypt base columnar stem cells
In this review, we refer to Lgr5-positive CBCs when discussing ISCs. Lgr5-positive CBC stem cells divide once a day, generating new CBC cells that reside at the crypt base as stem cells [38]. Owing to the limited space in the crypt base, however, half of the ISCs are randomly pushed out of the niche to become committed progenitor cells, a process called ‘neutral competition’ [38,39]. In this model, all ISCs initially carry the same properties and therefore have a similar chance to persist as an ISC. Real-time intravital imaging confirmed this mechanism in vivo [39]. However, detailed quantitative analysis of individual clonal ISC lineages showed that ‘central cells’ at the crypt base have an advantage over ‘border cells’ in the upper rim of the crypt niche for long-term persistence. Border cells were more likely to be displaced into the transit-amplifying compartment, lose their stem cell properties and differentiate along the crypt–villus axis [39]. The spectrum of stem cell activity displays heterogeneity, even within the pool of cells expressing Lgr5. These cells are probably able to transit between states of variable competence, directed by niche-derived signals [39].
4. Intestinal stem cell niche
What constitutes and determines the niche for ISCs? The stem cell niche provides a nurturing and guiding environment that sustains the self-renewing, multipotent stem cell population. At the same time, the niche provides local cues for the generation and positioning of differentiated progeny. The ISC niche is constituted by neighbouring Paneth cells within the epithelial layer, and by myofibroblasts, fibroblasts, neuronal and smooth muscle cells within the subepithelial mesenchyme that tightly line the crypt base basal lamina and the extracellular matrix [10,40,41] (figure 1). The close association and direct contact of these niche cells with ISCs facilitates the supply of essential factors for ISC maintenance and proliferation. The subepithelial mesenchyme produces various Wnts and epidermal growth factor (EGF) [42–44]. Furthermore, these cells provide R-spondins, potent Wnt signalling agonists, and Noggin, gremlin 1/2 and chordin-like 1, inhibitors of bone morphogenetic protein (BMP), to repress BMP-mediated differentiation [40,42,45–47]. Recently, subepithelial telocytes were demonstrated to be a vital source of Wnt ligands, as blockage of Wnt secretion from these rare, large cells results in impaired epithelial renewal and disruption of intestinal integrity [48,49]. Similarly, subepithelial Gli1-positive mesenchymal cells provide a crucial source of Wnts, as blockage of Wnt secretion from these cells also results in stem cell loss and subsequent loss of colonic epithelium integrity, which ultimately leads to epithelial death [50]. In addition, within the epithelium, Paneth cells provide essential growth signals, including Wnt3, EGF and Notch ligands, described in detail below [10,42]. Interestingly, ablation of Paneth cells does not result in ISC depletion in vivo, but in vitro cultured mini-guts (intestinal organoids), however, lack the mesenchymal component and as such fully depend on Wnt3 production by Paneth cells for stem cell maintenance and renewal of the epithelium [10,51]. These combined findings show that both mesenchymal cells, especially telocytes and Gli1+ cells, and Paneth cells serve as important sources for growth factors in the control of tissue renewal.
Thus, ISCs and daughter cells are subjected to and directed by a broad array of signals present in their niche. Polarized gradients of these mesenchymal- and epithelial-derived signals exist both in the crypt and also along the crypt–villus axis (figure 1). The balance between the generation of new cells and their functional specialization is regulated by numerous signalling pathways, which control proper ISC maintenance and intestinal architecture. Among these are the Wnt/β-catenin, Notch, Hedgehog, BMP, EGF and Eph–ephrin signalling cascades (figure 2). Below, we review these pathways and how their interconnected circuitry governs intestinal homeostasis.
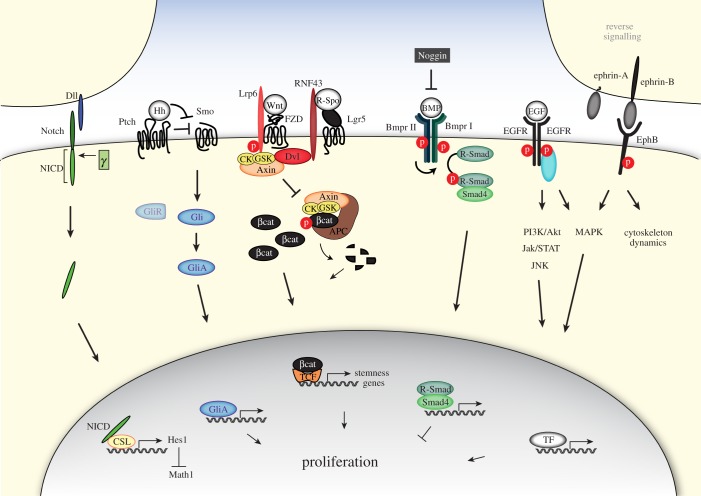
Figure 2.
Multiple key signalling pathways govern intestinal homeostasis. Representation of the principal signalling cascades of Notch, Hedgehog (Hh), Wnt, bone morphogenetic protein (BMP), epidermal growth factor (EGF) and Eph–ephrin that together control stem cell behaviour and intestinal homeostasis, see text for further details. Dll, delta-like ligand; NICD, Notch intracellular domain; γ, γ-secretase; Hh, Hedgehog; Ptch, Patched; Smo, Smoothend; Gli, glioblastoma; GliR, Gli repressor; GliA, Gli activator; Lrp6, low-densitiy lipoprotein receptor-related protein 6; FZD, Frizzled; RNF43, RING finger protein 43; Lgr5, leucine-rich-repeat containing G-protein coupled receptor 5; R-spo, R-spondin; CK, casein kinase; GSK3, glycogen synthase kinase; Dvl, Dishevelled; p, phospho-group; APC, adenomatous polyposis coli; βcat, β-catenin; TCF, T cell-specific transcription factor; BMP, bone morphogenetic protein; Bmpr I/II, BMP type I or II receptor; EGF, epidermal growth factor; EGFR, EGF receptor; PI3K, phosphoinositide 3-kinase; Jak, Janus kinase; STAT, signal transducer and activator of transcription; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; CSL, CBF1, suppressor of hairless, Lag-1; Math1, Atoh1, atonal homolog 1; Hes1, hairy and enhancer of split 1; TF, transcription factor.
5. Wnt signalling controls maintenance and size of the intestinal stem cell zone
5.1. Wnt signalling
The conserved Wnt signalling pathway determines crucial developmental processes and, importantly, controls tissue homeostasis in adult organisms. It has emerged as a pivotal player in the specification and maintenance of stem cell compartments in a wide array of tissues and organs. In the intestine, Wnt signalling is the main driving force of crypt proliferation.
Wnt ligands, produced by both Paneth and surrounding stromal cells, bind to their cognate receptors Frizzled (FZD) and low-density lipoprotein receptor-related protein 5/6 (Lrp5/6) at the surface of adjacent stem cells. Subsequent activation of the canonical Wnt pathway leads to the accumulation and nuclear entry of the transcriptional co-activator β-catenin to drive the expression of target genes involved in stem cell maintenance [6,52]. Wnt-induced stabilization of β-catenin involves the inactivation of a large multi-protein complex composed of the scaffold proteins Axin and adenomatous polyposis coli (APC) as well as the kinases glycogen synthase kinase 3β (GSK3β) and casein kinase 1 (CK1). In unstimulated cells, this destruction complex captures β-catenin and earmarks it for proteolysis through Ser/Thr phosphorylation of its flexible N-terminus [53,54]. Recognition of phospho-β-catenin by the ubiquitin ligase β-TrCP subsequently mediates its rapid ubiquitin-mediated proteasomal degradation [55,56].
Binding of Wnt to the FZD and Lrp6 receptors at the cell surface interferes with β-catenin degradation by a number of molecular rearrangements. After formation of a trimeric Wnt–FZD–Lrp5/6 complex, the cytoplasmic effector protein Dishevelled (Dvl) is recruited. Next, the activated receptor complex captures the destruction complex organiser Axin, probably through heterodimerization of the Axin and Dvl DIX domains [57–59]. The Axin-associated kinases GSK3β and CK1 turn their activity to the cytoplasmic tail of Lrp6, which, upon phosphorylation, provides a docking site for further Axin proteins. The redistribution of Axin–kinase complexes to the plasma membrane is considered a key step in the inactivation of β-catenin destruction. As a consequence, the pool of intracellular β-catenin increases and migrates to the nucleus to bind the T-cell factor (TCF) family of DNA-bound transcription factors and induces transcription of Wnt target genes (figure 2) [60]. Among the earliest target genes discovered is c-Myc, a well-known driver of proliferation of undifferentiated cells [61]. The Wnt target gene list has vastly expanded ever since, revealing multiple layers of positive and negative feedback regulation in the control of stem cell identity [23,62–65].
5.2. Wnt signalling in the intestine
The first clues for the crucial role of Wnt in the intestine originated from mouse genetic experiments. Neonatal mice deleted for TCF4, one of the main downstream effectors of Wnt, completely lack proliferative crypts, illustrating the requirement of Wnt signalling for establishment and maintenance of the stem cell compartment [66]. Maintenance of adult crypt proliferation remains dependent on Wnt signalling as conditional deletion of TCF4 in adult mice resulted in the loss of nearly all proliferating crypts, coinciding with progressive loss of Wnt target gene expression [67]. Furthermore, conditional deletion of β-catenin as well as overexpression of the diffusible Wnt inhibitor Dickkopf 1 (Dkk1) results in complete ablation of intestinal crypts in the adult mouse [68–71]. Moreover, transgenic expression of R-spondin 1 (R-Spo1), a strong Wnt agonist that acts through the Lgr4/5–Wnt receptor complex (described in more detail below), results in a massive hyperproliferation of intestinal crypts [72]. On the other hand, simultaneous deletion of both Lgr4 and Lgr5, the receptors for R-Spo, leads to the disappearance of crypts [73].
The fact that Wnt signalling plays a vital role in ISC maintenance is further illustrated by the nuclear β-catenin levels that are highest at the crypt base and gradually decrease along the crypt–villus axis [74,75]. Concordantly, the expression of various Wnt ligands (Wnt3, Wnt6 and Wnt9b) as well as their cognate receptors FZD5/7 is also highest at the crypt base [42,43,76,77]. Wnts produced by Paneth cells decorate the membranes of adjacent stem cells by binding to the highly expressed FZD receptors [78]. Through the regulation of FZD turnover and cell division, the membrane-bound reservoir of Wnts at the crypt bottom is gradually diluted, sculpting a gradient of Wnt along the crypt–villus axis [78]. Accordingly, Wnt target genes display maximum expression in the crypt base and gradually decrease moving upward along the crypt domain [40]. Interestingly, Paneth cells themselves also depend on Wnt signals [42,77,79] and require expression of the Wnt target gene Sox9 for their development and formation [80,81].
5.3. Stem cell-specific Wnt target genes
As described above, ISCs are marked by the Wnt target gene Lgr5. Transcriptome and proteome analysis of sorted Lrg5-positive cells unveiled multiple Wnt target genes among the stem cell-specific gene set [23,82]. Many of these genes have proven to be essential for stem cell maintenance and activity, regulating both positive and negative feedback signalling loops.
Among the ISC-specific Wnt target genes are the transmembrane receptor tyrosine kinase genes EphB2 and EphB3 [63]. Their expression is highly enriched on ISCs and Paneth cells, while transcription of their repulsive ligand, ephrin-B1, is concomitantly repressed in the crypts. Only upon exiting the crypt do cells start to express ephrin-B1 as a result of the decline of Wnt signals. Subsequent repulsive interactions between EphB-positive and ephrin-B-positive cells results in segregation of these cells, thereby controlling correct positioning of the cells. Indeed, EphB2/3 deficiency in mice results in a random localization of cells, including Paneth cells, along the crypt–villus axis. Hence, Wnt signalling controls architectural integrity of the stem cell zone [63,83–85].
The Wnt target gene Ascl2 is expressed in a Wnt-dependent and highly restricted fashion in ISCs [23–25,62,82,86]. Conditional knockout of Ascl2 in the adult intestinal epithelium leads to the elimination of CBC stem cells, whereas ectopic intestinal expression of Ascl2 induces hyperproliferation of crypts and de novo cryptogenesis in villi [62]. For this reason, Ascl2 is suggested to be a master regulator of the crypt stemness programme. Recent research showed that Ascl2 cooperates with β-catenin/TCF to activate the genes fundamental to the stem cell state. Ascl2 is activated when cells reach a specific Wnt/R-Spo signalling threshold and, as Ascl2 is capable of self-activation, is suggested to translate the Wnt gradient present in the crypt into a discrete ‘on’ or ‘off’ decision for stemness [25].
Another example of a stem cell-specific Wnt target gene is Tnfrsf19 or Troy, which is proposed to interact with Lgr5 and to negatively regulate Wnt/R-Spo signalling. As such, Troy is proposed to constitute a negative feedback loop to avoid over-activation of Wnt signalling, thereby preventing subsequent crypt enlargement and ultimately tumourigenesis [27].
In addition, the stem cell-specific and homologous Wnt target genes Ring finger 43 (RNF43) and zinc and ring finger 3 (ZNRF3) were shown to act in a negative feedback manner in the gut [87,88], as discussed below.
In summary, Wnt signalling constitutes the major driving force behind homeostatic self-renewal of the crypt through regulation of expression of critical regulatory genes.
5.4. Wnt pathway regulation by R-spondin and RNF43/ZNRF3
R-spondins are a group of small, secreted proteins (R-Spo1–4) that strongly potentiate Wnt/β-catenin signalling [72,89–93]. R-Spo proteins function as stem cell growth factors and can promote tissue regeneration [72,94]. Despite their biological significance there is no consensus on the exact mechanism by which R-Spo increases Wnt signalling. Various membrane proteins were proposed to act as R-Spo receptors, including Wnt receptors FZD and Lrp6 [95,96], Kremen [97], syndecan 4 [98], Lgr4/5 [73,99,100] and membrane E3 ubiquitin ligases RNF43/ZNRF3 [88,101–107].
To date, Lgrs are largely accepted as the primary receptors for R-Spo. Lgrs belong to the seven-span transmembrane receptor family and are known for their large extracellular leucine-rich repeats domain that is involved in ligand binding [108]. All four members of the R-Spo family can bind to the leucine-rich domain of Lgr4, Lgr5 and Lgr6 [73,99]. Lgr proteins have been identified as components of the Wnt receptor complex, and R-Spo binding to Lgr is hypothesized to stimulate the formation of higher order receptor complexes, thereby leading to enhanced Wnt signal transduction [73].
A new hypothesis emerged with the discovery of the two homologous genes RNF43 and ZNRF3 that display strongly enriched expression in the ISC population [87,88]. These genes encode for single-span transmembrane E3 ubiquitin ligases and operate as potent negative feedback regulators of Wnt signalling to control aberrant expansion of the crypts. Upon genetic removal of these negative regulators of Wnt signalling in the intestinal epithelium, mice showed very rapid tumour formation in the intestine. RNF43 and ZNRF3 inhibit Wnt signalling by targeting the Wnt receptors FZD and Lrp6 at the cell surface for ubiquitin-mediated endocytosis and lysosomal degradation, regulating cellular sensitivity for incoming Wnt ligands [87,88]. The underlying mechanism of RNF43/ZNRF3-mediated Wnt receptor targeting remains unknown but involves the cytoplasmic effector Dvl that was found to interact with the cytoplasmic tail of RNF43 [109]. Another regulatory layer emerged through the discovery that both RNF43 and ZNRF3 interact with R-Spo [101,104,106,110–114]. In current models, binding of R-Spo to Lgr4 recruits the RNF43/ZNRF3 receptors and induces their membrane clearance [88,105]. R-Spo-mediated removal of RNF43/ZNRF3 leads to stabilization of Wnt receptors at the cell surface, strongly enhancing cellular responses to Wnt (figure 2) [105].
A recent study demonstrates a distinct, non-redundant cooperation between Wnt and R-Spo ligands in ISC homeostasis and suggests that Wnt ligands act as priming factors that confer basal proliferative competence to ISCs by maintaining R-Spo receptor expression, which then drives the further expansion of stem cells via R-Spo ligands present in the niche [115].
In summary, the importance of Wnt signalling in collaboration with R-Spo as a major driving force of crypt proliferation is underscored by its tight regulation through multiple positive and negative regulatory feedback loops. Both R-Spo and RNF43/ZNRF3 represent prime examples of Wnt regulators with key functions in intestinal homeostasis.
6. Notch signalling regulates cell fate decisions and stemness in the crypt
The Notch signalling cascade is a highly conserved cell communication pathway that directs cell fate decisions in multicellular organisms. The mammalian Notch family comprises four single-span transmembrane Notch receptors (Notch1–4) and five single-span transmembrane Delta/Serrate/Lag2 (DSL) ligands (Jagged (Jag) 1 and 2, Delta-like (Dll) 1, 3 and 4). Notch signalling is triggered via direct cell-to-cell contact, through which the membrane-bound ligands exposed at the juxtaposed cell membrane bind and activate the Notch receptor. This ligand–receptor engagement results in the initiation of several proteolytic steps that modulate Notch receptor activity. Ultimately, the Notch intracellular domain (NICD) is released through γ-secretase protease activity (reviewed in [116,117]). Subsequently, NICD translocates to the nucleus to drive gene expression upon binding of the transcription factor CSL (CBF1, suppressor of hairless, Lag-1). CSL represses transcription of target genes in unstimulated cells but is converted into a transcriptional activator upon binding of NICD (figure 2) [118].
In the intestinal crypt, Notch signalling critically regulates the cell fate decision between absorptive and secretory cell types. This is illustrated by the use of γ-secretase inhibitors, which block Notch receptor signalling and mediate a massive conversion of proliferative cells into secretory cells [119,120]. Conversely, enhancement of Notch signalling via intestine-specific transgenic expression of NICD blocks the commitment of cells to adopt a secretory lineage fate [121].
The switch between lineages is mainly decided by two basic helix-loop-helix transcription factors. Notch signalling induces expression of the transcription factor Hes1 (hairy and enhancer of split 1) that in turn antagonizes the transcription factor Math1 (or Atoh1 (atonal homolog 1)) [122]. Math1 is the crucial regulator of the transcriptional programme for secretory lineage differentiation. Its depletion results in a complete absence of goblet, Paneth and enteroendocrine cells in the intestine [123–125], and Math1 overexpression directs progenitor cells into the secretory lineage [126]. Math1 expression is restricted to the secretory cells of the intestine, while Hes1 is expressed in most proliferative crypt cells. Inhibition of Notch signalling rapidly decreases Hes1 expression and results in upregulated Math1 expression in all crypt cells [120]. In line, deletion of Hes1 is associated with an excess of secretory cells at the expense of enterocytes [122]. Notably, similar phenotypes are observed upon simultaneous deletion of Notch1 and Notch2 genes and genetic ablation of CSL [120,127]. In summary, Notch signalling induces Hes1 levels to suppress Math1-dependent differentiation towards secretory lineages (figure 2).
How does Notch signalling affect ISCs? Lgr5-positive CBC cells predominantly express Notch1 and Notch2 receptors at the cell surface and Notch1 receptor mRNA was found enriched in these cells, signifying a regulatory role of Notch within the CBC stem cell compartment [62,128]. Neighbouring Paneth cells express Notch ligands Dll1 and Dll4, providing the ligands to activate Notch signalling in ISCs [10,129]. Notably, lineage-tracing experiments of cells undergoing active Notch signalling identified long-lived progenitors able to give rise to all the mature epithelial cell types [129,130]. Inhibition of Notch signalling results in rapid loss of CBC cells, indicating its requirement for stem cell proliferation and survival [131]. In particular, the stem cell-specific marker Olfm4 was shown to be a direct target gene of Notch signalling [131]. Thus, active Notch signalling is crucial for maintenance and activity of the ISC pool. Recently, it was shown that Notch signalling also plays a crucial role in intestinal renewal upon injury by irradiation. Interestingly, Paneth cells were shown to dedifferentiate and proliferate and start to line the crypt–villus axis. This newly gained stem cell-like capacity of Paneth cells is attributed to activated Notch signalling in the Paneth cells themselves [36].
In summary, Notch signalling regulates different aspects of intestinal homeostasis, stimulating both stem cell maintenance and cell fate determination of progenitor cells, and can induce Paneth cells to dedifferentiate upon tissue damage.
7. Hedgehog signalling regulates intestinal mesenchyme
The Hedgehog (Hh) family of secreted ligands consists of three subgroups; the Desert Hedgehog (Dhh), Indian Hedgehog (Ihh), and Sonic Hedgehog (Shh) groups [132]. Binding of Hh to the twelve-pass transmembrane receptors Patched 1 or 2 (Ptch1/2 or Ptc1–2) activates a signalling cascade that ultimately drives the activation of the zinc-finger transcription factor glioblastoma (Gli) (Gli1, Gli2 and Gli3), leading to the expression of Hh specific target genes. In the absence of Hh ligands, Ptch blocks the activity of the seven-span transmembrane protein Smoothened (Smo), and full-length Gli proteins are proteolytically processed to C-terminally truncated ‘GliR’ (Gli repressor) that actively repress a subset of Hh target genes. Binding of Hh to Ptch results in loss of Ptch activity and subsequent activation of Smo, which transduces the Hh signal to the cytoplasm, blocks the production of GliR and induces the conversion of Gli proteins into transcriptional activators (GliA) and thereby induces target gene transcription (figure 2) [132–136]. Target genes include Ptch1/2, Gli1, Hedgehog-interacting protein (Hhip) for feedback regulation [132–134,137,138], and genes that drive proliferation and survival, such as cyclin D1, myc and Bcl2 [135,139,140].
In the intestine, Ihh is the main hedgehog expressed. Low levels of Shh may be expressed at the base of the small intestinal and colonic crypts [141–144]. Ihh is secreted in a paracrine manner by epithelial cells to act on the surrounding mesenchymal cells, including smooth muscle precursor and myofibroblast cells [142,145]. In addition, intestinal macrophages and dendritic cells may directly respond to Hedgehog signalling [145]. Ihh and Shh ligands secreted by TA cells interact with Ptch receptors localized on mesenchymal cells to induce BMP production [144,146,147]. BMPs negatively regulate intestinal epithelial proliferation and restrict the number of stem cells in the crypt [40,148,149] (see paragraph below). Furthermore, (Hh downstream factor) Gli1-positive mesenchymal cells secrete Wnt ligands that are essential for stem cell renewal in the colon and in the small intestine can act as a reserve Wnt source [50].
Constitutive activation of Hh signalling, either by systemic deletion of Ptch [144] or by selective overexpression of Ihh in the intestinal epithelium (Villin-Ihh transgenic mice) [150], results in an accumulation of mesenchymal cells. Moreover, experiments in which Hh signalling was conditionally lost in the adult intestine showed that Hh not only signals for expansion of the mesenchyme but is also required to maintain smooth muscle and myofibroblast cells [150,151]. Furthermore, analysis of Ihh mutant mice showed that loss of Ihh signalling ultimately results in the loss of smooth muscle precursor cells, leading to complete loss of the villus core support structure [141]. Decreased Hh signalling in the adult intestine was also shown to enhance Wnt pathway activity, thereby compromising differentiation and driving crypt hyperplasia [143].
Thus, in current models, Hh signalling indirectly affects ISCs via (i) induction of repressive BMP signalling and (ii) modulation of adjacent stroma for supportive structure.
8. Bone morphogenetic protein signalling regulates crypt formation and terminal differentiation
BMPs were initially discovered for their ability to induce bone formation [152] but are now known to play crucial roles during organ development and tissue homeostasis [153]. BMPs comprise a class of extracellular signalling molecules that belong to the transforming growth factor-β (TGF-β) superfamily of proteins. BMPs signal via the canonical Smad-dependent pathway and can induce various non-canonical signalling pathways as well. In the canonical pathway, BMPs initiate signal transduction by binding to BMP type I and type II receptors (Bmpr1–2) that form a heterotetrameric complex. BMP receptors are single transmembrane proteins that carry serine/threonine kinase activity in their intracellular domains. Upon BMP binding, the constitutively active Bmpr2 transphosphorylates Bmpr1. Subsequently, Bmpr1 phosphorylates the receptor-bound R-Smads1/5/8 (receptor-regulated Smads). Phosphorylated Smad1/5/8 then associate with the core mediator Smad4, and the resulting Smad complex translocates to the nucleus to associate with coactivators or corepressors and regulate gene expression patterns (figure 2) [153,154]. BMP target genes include Msx homeobox genes and the proto-oncogene JunB [155,156]. Pathway activity is regulated by extracellular antagonists, such as Noggin, follistatin or gremlin, that sequester BMP ligands, thereby blocking interaction with BMP receptors [157].
In the intestine, the BMP pathway acts as a negative regulator of crypt formation and drives the terminal differentiation of mature intestinal cells [148,158,159]. BMP-2 and -4 ligands are expressed by both mesenchyme and epithelial villus cells and mainly act on the epithelial compartments that contain differentiated cells expressing BMP receptors [148,160]. Mesenchyme-to-epithelium BMP signalling promotes differentiation of progenitor cells while restraining cell proliferation [149]. BMP signalling within the crypt base stem cell niche is carefully regulated by BMP antagonists, such as gremlin1/2, chordin, Noggin and ANGPTL2, expressed by the mesenchyme surrounding the crypt [40,161,162]. Inhibition of BMP signalling in the mouse villus using transgenic expression of the BMP inhibitor Noggin, results in ectopic crypt formation [148]. Similarly, conditional deletion of Bmp receptor 1A results in hyperproliferative crypts [149]. BMP represses Wnt signalling and is expressed in an opposing gradient along the crypt–villus axis, with highest BMP signalling in the cells at the luminal surface (figure 1) [149].
9. EGF signalling is required for intestinal stem cell proliferation
EGF is an extracellular ligand that stimulates cell growth, proliferation, and differentiation by binding to its cognate receptor the epidermal growth factor receptor (EGFR). EGFR is also known as ErbB1/HER1, and is a member of the ErbB family of receptor tyrosine kinases. Upon activation by EGF binding, EGFR undergoes a transition from an inactive monomeric form to an active homodimer. EGFR dimerization stimulates its intrinsic intracellular protein-tyrosine kinase activity. As a result, several tyrosine residues in the C-terminal domain of EGFR are autophosphorylated, which drives downstream pathway activation. Downstream signalling effectors associate with phosphorylated tyrosines in the EGF receptor via their SH2 domains and initiate major cellular pro-survival and proliferation signalling cascades, including the mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K)/Akt, c-Jun N-terminal kinases (JNK), Jak/STAT and phospholipase C (PLC) pathways (figure 2) [163,164]. Of note, the small GTPase KRAS (Kirsten rat sarcoma viral oncogene homolog) acts as a crucial central relay in a number of these signalling cascades and is commonly mutated in colorectal cancers [165,166].
EGF signalling is required for proliferation and maintenance of ISCs and is produced in the niche by the surrounding Paneth cells and subepithelial mesenchyme [10,42]. In turn, EGFR is highly expressed in ISCs and in TA cells [167]. Indeed, luminally applied EGF strongly induces proliferation of the small intestinal epithelium in rats [168,169]. By contrast, blockade of EGF signalling in organoids converts actively dividing ISCs into quiescent Lgr5+ reserve stem cells that retain expression of various Wnt target genes [170]. Evidently, tight control is necessary to balance EGF-induced proliferation of ISCs. To this end, ISCs express high levels of the EGFR/ErbB inhibitor leucine-rich repeats and immunoglobulin-like domains protein 1 (Lrig1) transmembrane proteins. Lrig1 is an inducible negative feedback regulator of the ErbB receptor family that mediates the ubiquitination and subsequent degradation of canonical EGFRs [32,33,171]. Accordingly, genetic ablation of Lrig1 in mice results in enhanced EGFR/ErbB expression leading to an increase in ISC numbers and significant expansion of crypts [32,33]. This activity of EGFR signalling in intestinal maintenance is highly conserved, as shown by studies using Drosophila [172,173].
In summary, controlled expression of Lrig1 forms a negative feedback loop that allows stem cells to fine-tune their cellular response to EGF ligand-mediated signalling and ensures proper crypt size and tissue homeostasis.
Of note, a recent study demonstrated redundancy between EGF and hepatocyte growth factor (HGF) in the intestine. HGF regulates intestinal homeostasis and regeneration by engaging its receptor MET, and interestingly HGF/MET signalling can fully substitute EGFR signals in intestinal organoid cultures [174].
10. Eph–ephrin signalling directs appropriate cell positioning along the crypt–villus axis
Eph–ephrin signalling occurs via direct cell–cell contact and is involved in a wide spectrum of biological processes, including the regulation of cell positioning. Ephrin ligands are divided into two subclasses based on their structure and mode of linkage to the cell membrane (ephrin-A and ephrin-B). Ephrin-A proteins are anchored to the membrane by a glycosylphosphatidylinositol (GPI) linkage and lack a cytoplasmic domain, while ephrin-B proteins pass the membrane by their single transmembrane domain and contain a short cytoplasmic part. Eph receptors constitute the largest family of tyrosine kinase receptors and can be divided into the subclasses EphA and EphB, based on sequence similarity and binding affinities for either ephrin-A or -B [175,176].
A unique feature of Eph–ephrin signalling is that both receptor and ligand are competent to transduce signalling upon interaction. This concept of bidirectional signalling has emerged as an important mechanism by which Ephs and ephrins control cell–cell communication. Eph- and ephrin-mediated signalling are generally referred to as forward and reverse signalling, respectively [176]. Essentially, Eph signalling controls cell morphology, adhesion and migration by modifying the organization of the actin cytoskeleton and influencing the activities of integrins and intercellular adhesion molecules [85,176]. Upon binding of an ephrin ligand to the extracellular domain of an Eph receptor, intracellular tyrosine and serine residues of the receptor are auto-phosphorylated, allowing the cytoplasmic tyrosine kinase to subsequently activate and modulate downstream signalling cascades, such as MAPK, Ras and ERK signalling (figure 2) [85,177].
Although Eph–ephrins were initially studied mainly in a developmental context, their physiological functions in adult tissues are rapidly coming to light. In the intestine, high levels of Wnt signalling induce expression of EphB2 and EphB3 in the lower parts of the crypts with simultaneous transcriptional repression of their repulsive ephrin-B1 ligand [63]. Of note, Notch signalling activates the expression of ephrin-B1 in differentiated intestinal cells [178]. ISCs display high levels of EphB2 expression, while Paneth cells are EphB2 negative but express EphB3 [63,179]. The decline in Wnt cues along the crypt–villus axis results in de-repression of the repulsive ephrin-B1 ligand. At the same time, EphB2 expression progressively declines in TA cells as they migrate upwards. The gradient of repulsive EphB2/3–ephrin-B1 interaction coordinates appropriate cell positioning along the crypt–villus axis with differentiated cells moving upwards towards the villus tip [63,83,180,181]. Differentiated Paneth cells that highly express EphB3 escape this upward flow and move towards the crypt bottom. Indeed, in EphB3 null mice, Paneth cells are not restricted to crypts but spread randomly along the villi in the small intestine [63].
11. Interconnectivity of signalling pathways governs crypt–villus homeostasis
Each of the above-described signalling pathways controls intestinal homeostasis, through either a direct or indirect modulation of ISC behaviour. ISCs interpret these signals derived from the niche to ensure the balance between cell loss and cell replenishment, thereby safeguarding tissue maintenance. The interconnectivity of the signalling pathways further secures an appropriate strength and timing of signals within the stem cell niche. The Wnt pathway is the main force behind intestinal epithelium homeostasis and requires tight regulation to prevent hyperproliferation of ISCs. The expression of stem cell regulatory Wnt ligands displays a diminishing slope along the crypt–villus axis. At the +4 position, a local production of Wnt antagonists is observed, including Wnt binding secreted Frizzled-related proteins, likely to keep the reserve stem cells at that position in the quiescent state [76]. In the crypt, the Wnt pathway synergizes with Notch signalling to sustain undifferentiated and proliferative stem and progenitor cells in the crypt. Additionally, both pathways are essential for specific lineage commitment of progenitor cells along the absorptive (Notch) and secretory (Wnt) cellular differentiated states. Interestingly, inhibition of Notch signalling, using Notch blocking antibodies, caused conversion of Lgr5-expressing ISCs to secretory cells, leading to stem cell depletion. This coincided with Wnt pathway upregulation and increased secretory cell differentiation. Repression of canonical Wnt signalling rescued this phenotype, suggesting opposing and interconnected activities of Notch and Wnt signalling to guide gut homeostasis [182].
Furthermore, Wnt signalling induces high crypt-specific expression of EphB2 and EphB3, while the decreasing Wnt gradient along the crypt–villus axis generates an opposing gradient of the repulsive ephrin-B ligand, securing spatial segregation and accurate positioning of distinct cellular compartments within the crypt. In the upper part of the crypt and within the villus, the output of Wnt signalling is modulated by cooperative activity of the paracrine Hedgehog and BMP signalling cascades. As the progenitor cells move upwards from the crypt base, the Hedgehog-induced, mesenchyme-to-epithelium BMP signalling promotes differentiation while restraining cell proliferation. Importantly, at the crypt base, the pro-differentiation activity of the BMP pathway is counteracted by locally secreted mesenchyme-derived BMP antagonists. Also, Paneth cell-derived EGF-induced mitotic signalling at the crypt base is balanced by expression of EGFR inhibitor Lrig1.
Collectively, an interconnected network of developmental signalling pathways governs intestinal homeostasis by balancing the processes of cell proliferation and differentiation (figure 3).
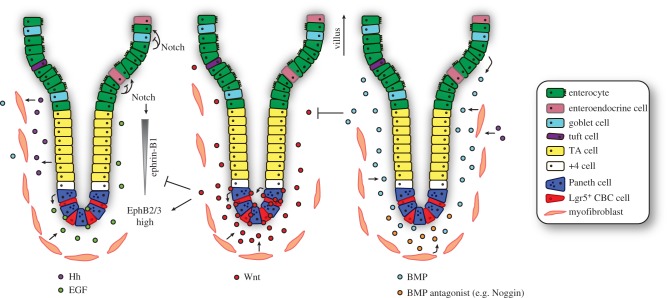
Figure 3.
Interconnectivity of signalling pathways governs crypt–villus homeostasis. Intestinal homeostasis and cell fate determination are maintained by interconnectivity of key signalling pathways between epithelial and mesenchymal cells. Wnt ligands (centre) secreted from the Paneth cells and intestinal subepithelial myofibrobasts act predominantly at the base of the crypt to maintain stem cell function and TA cell proliferation. High levels of Wnt signalling induce the expression of EphB2 (ISCs) and EphB3 (Paneth cells), and concurrently repress transcription of the repulsive ligand ephrin-B1. The decline in Wnt signals along the axis results in increased ephrin-B1 levels and proper positioning of the cells. EGF signalling (left) is required for proliferation and maintenance of ISCs and is produced in the ISC niche by the surrounding Paneth cells and subepithelial mesenchyme. Notch signalling regulates cell fate through cell-to-cell contact in the crypt (here for simplicity only visualized on the top left). Notch signalling controls the binary cell fate decision between the secretory and absorptive lineages. Hedgehog (Hh), expressed by epithelial cells in the upper part of the crypt, acts upon and maintains the myofibroblasts. This has a secondary effect on the epithelium through promotion of BMP ligand expression. BMP ligands are predominantly produced by the mesenchymal cells and partly by epithelial villus cells. Mesenchyme-to-epithelium BMP signalling promotes differentiation of progenitor cells while restraining cell proliferation. BMP signalling within the crypt stem cell niche is therefore carefully regulated by BMP antagonists expressed by the mesenchyme surrounding the crypt. Finally, BMP represses Wnt signalling and is expressed in an opposing gradient along the crypt–villus axis.
12. Sequential mutation of ‘niche-like’ signalling pathways drives intestinal carcinogenesis
Disruption of the delicate balance between proliferation and differentiation governed by key signalling pathways in the crypt can lead to hyperproliferation and ultimately tumour growth. Indeed, cancer cells inappropriately turn on a gene programme for self-renewal and survival by acquiring mutations in key components of signalling pathways that are normally provided by the external cues from the niche [183].
The majority of colorectal tumours evolve from benign to malignant lesions by acquiring a series of mutations over time: the adenoma–carcinoma sequence [184,185]. Formation of benign adenomas is initiated by activation of the Wnt signalling pathway, most commonly through inactivating mutations in APC [186]. Subsequent activating mutations in the EGFR pathway (KRAS, PIK3CA), and inactivating mutations in the TGF-β/BMP pathway (SMAD4) as well as in p53 (TP53) promote progression to an invasive and metastatic phenotype [184,185,187]. Mutations in these genes are presumed to drive colorectal carcinogenesis as they provide selective growth advantages to the mutated cells and are therefore called ‘driver’ mutations. Each human colorectal carcinoma (CRC) is regarded to harbour three to six recurrent driver mutations [187]. Recently, two elegant studies showed, using CRISPR–Cas9-engineered organoid cultures, that a combination of at least four of such driver mutations liberates the ISC from the need of niche factors, rendering gut organoid growth entirely self-sufficient [188,189]. Importantly, such mutant organoids form tumours after transplantation in mice [188,189]. Hence, the introduction of driver mutations in organoids fully copies the CRC phenotype and provides final proof that the sequential disruption of key signalling pathways governs the adenoma–carcinoma sequence.
Activation of the Wnt and the EGFR signalling pathways represent key steps in the initiation and early progression of CRC, by favouring stemness and proliferative characteristics. Subsequent blockade of BMP/TGFβ signalling suppresses differentiation and further tilts the balance towards proliferation of the cancer cells. Ultimately, inactivation of p53 results in loss of DNA damage control and, more importantly, allows CRC cells to escape apoptosis. Together, this establishes an optimal combination of events for cancer cell survival and carcinogenesis.
Interestingly, the combined inactivation of both APC and p53 appears already sufficient to induce extensive aneuploidy, a hallmark of tumour progression. Remarkably, the number of driver mutations in CRC is variable, with a fraction of CRCs carrying only a single pathway alteration [190]. In these cases, acquired microsatellite and/or chromosomal instability and epigenetic changes might alter driver pathway signalling instead.
Clearly, the sequential accumulation of driver mutations drives tumour progression towards CRC, but are the initial genetic alterations also required to maintain CRC cells at later stages of tumour progression? This question was recently addressed by Dow et al. [191] using a conditional short hairpin RNA approach to control APC levels in a subset of ISCs in the mouse. The strong reduction of APC levels initiated tumour formation along the intestine, including the colon. Upon restoration of APC expression, tumours regressed and CRC cells underwent differentiation toward normal intestinal cell types, thereby reinstating crypt–villus homeostasis. Strikingly, invasive tumours harbouring additional KRAS and TP53 mutations were also reverted to normal functioning cells after reintroduction of APC [191]. This illustrates that loss of APC is not only essential for CRC onset but remains critical for CRC maintenance even in the presence of sequential driver mutations.
13. Concluding remarks
In this review, we summarized our understanding of the unique properties and regulated activity of ISCs. A plethora of genetic studies have provided instrumental insights into the signalling networks that govern intestinal homeostasis. Crypt cells display substantial plasticity that is influenced by signals from the stem cell niche. Perturbations within these signalling pathways, most prominently the Wnt cascade, can induce tumourigenesis. A better understanding of the relationships and interconnectivity between homeostatic signalling and distinct aspects of tumour initiation and progression will be critical in the discovery of potential targets and development of strategies for therapeutic intervention.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
M.S. is supported by a Dutch Cancer Society fellowship (BUIT 2015-7526 to M.S). Work in the laboratory of B.-K.K. is supported by an ERC starting grant (Troy Stem Cells, 639050 to B.-K.K.). Work in the laboratory of M.M.M. is supported by the Netherlands Organization for Scientific Research NWO (VICI grant 91815604 and ECHO grant 711.013.012 to M.M.M.) and is part of the Oncode Institute, which is partly financed by the Dutch Cancer Society.
References
- 1.Barker N, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. ( 10.1038/nature06196) [DOI] [PubMed] [Google Scholar]
- 2.Shyh-Chang N, Daley GQ, Cantley LC. 2013. Stem cell metabolism in tissue development and aging. Development 140, 2535–2547. ( 10.1242/dev.091777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trentin A, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. 2004. Self-renewal capacity is a widespread property of various types of neural crest precursor cells. Proc. Natl Acad. Sci. USA 101, 4495–4500. ( 10.1073/pnas.0400629101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison SJ, Kimble J. 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074. ( 10.1038/nature04956) [DOI] [PubMed] [Google Scholar]
- 5.Bajaj J, Zimdahl B, Reya T. 2015. Fearful symmetry: subversion of asymmetric division in cancer development and progression. Cancer Res. 75, 792–797. ( 10.1158/0008-5472.CAN-14-2750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clevers H. 2013. The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284. ( 10.1016/j.cell.2013.07.004) [DOI] [PubMed] [Google Scholar]
- 7.Stevens CE, Leblond CP. 1947. Rate of renewal of the cells of the intestinal epithelium in the rat. Anat. Rec. 97, 373. [PubMed] [Google Scholar]
- 8.van der Flier LG, Clevers H. 2009. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260. ( 10.1146/annurev.physiol.010908.163145) [DOI] [PubMed] [Google Scholar]
- 9.Ireland H, Houghton C, Howard L, Winton DJ. 2005. Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev. Dyn. 233, 1332–1336. ( 10.1002/dvdy.20446) [DOI] [PubMed] [Google Scholar]
- 10.Sato T, et al. 2011. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. ( 10.1038/nature09637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lau W, et al. 2012. Peyer's patch M cells derived from Lgr5+ stem cells require SpiB and are induced by RankL in cultured ‘miniguts’. Mol. Cell. Biol. 32, 3639–3647. ( 10.1128/MCB.00434-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerbe F, et al. 2016. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230. ( 10.1038/nature16527) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Moltke J, Ji M, Liang HE, Locksley RM. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225. ( 10.1038/nature16161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howitt MR, et al. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333. ( 10.1126/science.aaf1648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerbe F, et al. 2011. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 192, 767–780. ( 10.1083/jcb.201010127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koo BK, Clevers H. 2014. Stem cells marked by the R-spondin receptor LGR5. Gastroenterology 147, 289–302. ( 10.1053/j.gastro.2014.05.007) [DOI] [PubMed] [Google Scholar]
- 17.Potten CS, Hume WJ, Reid P, Cairns J. 1978. The segregation of DNA in epithelial stem cells. Cell 15, 899–906. ( 10.1016/0092-8674(78)90274-X) [DOI] [PubMed] [Google Scholar]
- 18.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259. ( 10.1038/nature10408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan KS, et al. 2012. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl Acad. Sci. USA 109, 466–471. ( 10.1073/pnas.1118857109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barker N, et al. 2010. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36. ( 10.1016/j.stem.2009.11.013) [DOI] [PubMed] [Google Scholar]
- 21.Leushacke M, et al. 2017. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 19, 774–786. ( 10.1038/ncb3541) [DOI] [PubMed] [Google Scholar]
- 22.Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgård R. 2008. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291–1299. ( 10.1038/ng.239) [DOI] [PubMed] [Google Scholar]
- 23.Munoz J, et al. 2012. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 31, 3079–3091. ( 10.1038/emboj.2012.166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Flier LG, et al. 2007. The intestinal Wnt/TCF signature. Gastroenterology 132, 628–632. ( 10.1053/j.gastro.2006.08.039) [DOI] [PubMed] [Google Scholar]
- 25.Schuijers J, et al. 2015. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell 16, 158–170. ( 10.1016/j.stem.2014.12.006) [DOI] [PubMed] [Google Scholar]
- 26.Yan KS, Kuo CJ. 2015. Ascl2 reinforces intestinal stem cell identity. Cell Stem Cell 16, 105–106. ( 10.1016/j.stem.2015.01.014) [DOI] [PubMed] [Google Scholar]
- 27.Fafilek B, et al. 2013. Troy, a tumor necrosis factor receptor family member, interacts with lgr5 to inhibit Wnt signaling in intestinal stem cells. Gastroenterology 144, 381–391. ( 10.1053/j.gastro.2012.10.048) [DOI] [PubMed] [Google Scholar]
- 28.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. 2009. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17. ( 10.1053/j.gastro.2009.05.035) [DOI] [PubMed] [Google Scholar]
- 29.Sangiorgi E, Capecchi MR. 2008. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40, 915–920. ( 10.1038/ng.165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery RK, et al. 2011. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl Acad. Sci. USA 108, 179–184. ( 10.1073/pnas.1013004108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. 2011. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424. ( 10.1126/science.1213214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powell AE, et al. 2012. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158. ( 10.1016/j.cell.2012.02.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong VW, et al. 2012. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 14, 401–408. ( 10.1038/ncb2464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. 2013. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69. ( 10.1038/nature11965) [DOI] [PubMed] [Google Scholar]
- 35.van Es JH, et al. 2012. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 14, 1099–1104. ( 10.1038/ncb2581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu S, Tong K, Zhao Y, Balasubramanian I, Yap GS, Ferraris RP, Bonder EM, Verzi MP, Gao N. 2018. Paneth cell multipotency induced by Notch activation following injury. Cell Stem Cell 23, 46–59.e5. ( 10.1016/j.stem.2018.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tetteh PW, et al. 2016. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213. ( 10.1016/j.stem.2016.01.001) [DOI] [PubMed] [Google Scholar]
- 38.Snippert HJ, et al. 2010. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. ( 10.1016/j.cell.2010.09.016) [DOI] [PubMed] [Google Scholar]
- 39.Ritsma L, Ellenbroek SI, Zomer A, Snippert HJ, de Sauvage FJ, Simons BD, Clevers H, van Rheenen J. 2014. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362–365. ( 10.1038/nature12972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosinski C, et al. 2007. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl Acad. Sci. USA 104, 15 418–15 423. ( 10.1073/pnas.0707210104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meran L, Baulies A, Li VSW. 2017. Intestinal stem cell niche: the extracellular matrix and cellular components. Stem Cells Int. 2017, 7970385 ( 10.1155/2017/7970385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farin HF, Van Es JH, Clevers H. 2012. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529.e7. ( 10.1053/j.gastro.2012.08.031) [DOI] [PubMed] [Google Scholar]
- 43.Flanagan DJ, et al. 2015. Frizzled7 functions as a Wnt receptor in intestinal epithelial Lgr5+ stem cells. Stem Cell Rep. 4, 759–767. ( 10.1016/j.stemcr.2015.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valenta T, et al. 2016. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 15, 911–918. ( 10.1016/j.celrep.2016.03.088) [DOI] [PubMed] [Google Scholar]
- 45.Worthley DL, Giraud AS, Wang TC. 2010. Stromal fibroblasts in digestive cancer. Cancer Microenviron. 3, 117–125. ( 10.1007/s12307-009-0033-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabiri Z, et al. 2014. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 141, 2206–2215. ( 10.1242/dev.104976) [DOI] [PubMed] [Google Scholar]
- 47.Stzepourginski I, Nigro G, Jacob JM, Dulauroy S, Sansonetti PJ, Eberl G, Peduto L. 2017. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl Acad. Sci. USA 114, E506–E513. ( 10.1073/pnas.1620059114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Toth B, Kondo A, Massassa EE, Itzkovitz S, Kaestner KH. 2018. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246. ( 10.1038/s41586-018-0084-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoki R, et al. 2016. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol. Gastroenterol. Hepatol. 2, 175–188. ( 10.1016/j.jcmgh.2015.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, Basler K. 2018. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453. ( 10.1038/s41586-018-0190-3) [DOI] [PubMed] [Google Scholar]
- 51.Sato T, et al. 2009. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. ( 10.1038/nature07935) [DOI] [PubMed] [Google Scholar]
- 52.Krausova M, Korinek V. 2014. Wnt signaling in adult intestinal stem cells and cancer. Cell. Signal. 26, 570–579. ( 10.1016/j.cellsig.2013.11.032) [DOI] [PubMed] [Google Scholar]
- 53.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. 2002. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 16, 1066–1076. ( 10.1101/gad.230302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 17, 1371–1384. ( 10.1093/emboj/17.5.1371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. 1997. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16, 3797–3804. ( 10.1093/emboj/16.13.3797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hart M, et al. 1999. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–211. ( 10.1016/S0960-9822(99)80091-8) [DOI] [PubMed] [Google Scholar]
- 57.Cliffe A, Hamada F, Bienz M. 2003. A role of Dishevelled in relocating Axin to the plasma membrane during Wingless signaling. Curr. Biol. 13, 960–966. ( 10.1016/S0960-9822(03)00370-1) [DOI] [PubMed] [Google Scholar]
- 58.Kim SE, et al. 2013. Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science 340, 867–870. ( 10.1126/science.1232389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiedler M, Mendoza-Topaz C, Rutherford TJ, Mieszczanek J, Bienz M. 2011. Dishevelled interacts with the DIX domain polymerization interface of Axin to interfere with its function in down-regulating β-catenin. Proc. Natl Acad. Sci. USA 108, 1937–1942. ( 10.1073/pnas.1017063108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clevers H, Nusse R. 2012. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205. ( 10.1016/j.cell.2012.05.012) [DOI] [PubMed] [Google Scholar]
- 61.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. 1998. Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512. ( 10.1126/science.281.5382.1509) [DOI] [PubMed] [Google Scholar]
- 62.van der Flier LG, et al. 2009. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136, 903–912. ( 10.1016/j.cell.2009.01.031) [DOI] [PubMed] [Google Scholar]
- 63.Batlle E, et al. 2002. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111, 251–263. ( 10.1016/S0092-8674(02)01015-2) [DOI] [PubMed] [Google Scholar]
- 64.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183. ( 10.1128/MCB.22.4.1172-1183.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lustig B, et al. 2002. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 22, 1184–1193. ( 10.1128/MCB.22.4.1184-1193.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. 1998. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19, 379–383. ( 10.1038/1270) [DOI] [PubMed] [Google Scholar]
- 67.van Es JH, et al. 2012. A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol. Cell. Biol. 32, 1918–1927. ( 10.1128/MCB.06288-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fevr T, Robine S, Louvard D, Huelsken J. 2007. Wnt/β-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559. ( 10.1128/MCB.01034-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pinto D, Gregorieff A, Begthel H, Clevers H. 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17, 1709–1713. ( 10.1101/gad.267103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ, Winton DJ. 2004. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of β-catenin. Gastroenterology 126, 1236–1246. ( 10.1053/j.gastro.2004.03.020) [DOI] [PubMed] [Google Scholar]
- 71.Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. 2004. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl Acad. Sci. USA 101, 266–271. ( 10.1073/pnas.2536800100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim KA, et al. 2005. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science 309, 1256–1259. ( 10.1126/science.1112521) [DOI] [PubMed] [Google Scholar]
- 73.de Lau W, et al. 2011. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297. ( 10.1038/nature10337) [DOI] [PubMed] [Google Scholar]
- 74.Kongkanuntn R, Bubb VJ, Sansom OJ, Wyllie AH, Harrison DJ, Clarke AR. 1999. Dysregulated expression of β-catenin marks early neoplastic change in Apc mutant mice, but not all lesions arising in Msh2 deficient mice. Oncogene 18, 7219–7225. ( 10.1038/sj.onc.1203181) [DOI] [PubMed] [Google Scholar]
- 75.van de Wetering M, et al. 2002. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250. ( 10.1016/S0092-8674(02)01014-0) [DOI] [PubMed] [Google Scholar]
- 76.Gregorieff A, Pinto D, Begthel H, Destree O, Kielman M, Clevers H. 2005. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129, 626–638. ( 10.1016/j.gastro.2005.06.007) [DOI] [PubMed] [Google Scholar]
- 77.van Es JH, et al. 2005. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 7, 381–386. ( 10.1038/ncb1240) [DOI] [PubMed] [Google Scholar]
- 78.Farin HF, et al. 2016. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343. ( 10.1038/nature16937) [DOI] [PubMed] [Google Scholar]
- 79.Andreu P, et al. 2005. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development 132, 1443–1451. ( 10.1242/dev.01700) [DOI] [PubMed] [Google Scholar]
- 80.Bastide P, et al. 2007. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 178, 635–648. ( 10.1083/jcb.200704152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mori-Akiyama Y, van den Born M, van Es JH, Hamilton SR, Adams HP, Zhang J, Clevers H, de Crombrugghe B. 2007. SOX9 is required for the differentiation of Paneth cells in the intestinal epithelium. Gastroenterology 133, 539–546. ( 10.1053/j.gastro.2007.05.020) [DOI] [PubMed] [Google Scholar]
- 82.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, van Oudenaarden A. 2012. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 14, 106–114. ( 10.1038/ncb2384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cortina C, et al. 2007. EphB–ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat. Genet. 39, 1376–1383. ( 10.1038/ng.2007.11) [DOI] [PubMed] [Google Scholar]
- 84.Herath NI, Boyd AW. 2010. The role of Eph receptors and ephrin ligands in colorectal cancer. Int. J. Cancer 126, 2003–2011. ( 10.1002/ijc.25147) [DOI] [PubMed] [Google Scholar]
- 85.Pasquale EB. 2010. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180. ( 10.1038/nrc2806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jubb AM, et al. 2006. Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is upregulated in intestinal neoplasia. Oncogene 25, 3445–3457. ( 10.1038/sj.onc.1209382) [DOI] [PubMed] [Google Scholar]
- 87.Koo BK, et al. 2012. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669. ( 10.1038/nature11308) [DOI] [PubMed] [Google Scholar]
- 88.Hao HX, et al. 2012. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200. ( 10.1038/nature11019) [DOI] [PubMed] [Google Scholar]
- 89.de Lau WB, Snel B, Clevers HC. 2012. The R-spondin protein family. Genome Biol. 13, 242 ( 10.1186/gb-2012-13-3-242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kazanskaya O, Glinka A, del Barco Barrantes I, Stannek P, Niehrs C, Wu W. 2004. R-Spondin2 is a secreted activator of Wnt/β-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 7, 525–534. ( 10.1016/j.devcel.2004.07.019) [DOI] [PubMed] [Google Scholar]
- 91.Kim KA, Zhao J, Andarmani S, Kakitani M, Oshima T, Binnerts ME, Abo A, Tomizuka K, Funk WD. 2006. R-Spondin proteins: a novel link to β-catenin activation. Cell Cycle 5, 23–26. ( 10.4161/cc.5.1.2305) [DOI] [PubMed] [Google Scholar]
- 92.Li SJ, Yen TY, Endo Y, Klauzinska M, Baljinnyam B, Macher B, Callahan R, Rubin JS. 2009. Loss-of-function point mutations and two-furin domain derivatives provide insights about R-spondin2 structure and function. Cell. Signal. 21, 916–925. ( 10.1016/j.cellsig.2009.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kazanskaya O, Ohkawara B, Heroult M, Wu W, Maltry N, Augustin HG, Niehrs C. 2008. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development 135, 3655–3664. ( 10.1242/dev.027284) [DOI] [PubMed] [Google Scholar]
- 94.Ootani A, et al. 2009. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15, 701–706. ( 10.1038/nm.1951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nam JS, Turcotte TJ, Smith PF, Choi S, Yoon JK. 2006. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate β-catenin-dependent gene expression. J. Biol. Chem. 281, 13 247–13 257. ( 10.1074/jbc.M508324200) [DOI] [PubMed] [Google Scholar]
- 96.Wei Q, Yokota C, Semenov MV, Doble B, Woodgett J, He X. 2007. R-spondin1 is a high affinity ligand for LRP6 and induces LRP6 phosphorylation and β-catenin signaling. J. Biol. Chem. 282, 15 903–15 911. ( 10.1074/jbc.M701927200) [DOI] [PubMed] [Google Scholar]
- 97.Binnerts ME, et al. 2007. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc. Natl Acad. Sci. USA. 104, 14 700–14 705. ( 10.1073/pnas.0702305104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ohkawara B, Glinka A, Niehrs C. 2011. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev. Cell 20, 303–314. ( 10.1016/j.devcel.2011.01.006) [DOI] [PubMed] [Google Scholar]
- 99.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. 2011. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl Acad. Sci. USA 108, 11 452–11 457. ( 10.1073/pnas.1106083108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C. 2011. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 12, 1055–1061. ( 10.1038/embor.2011.175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen PH, Chen X, Lin Z, Fang D, He X. 2013. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 27, 1345–1350. ( 10.1101/gad.219915.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de Lau W, Peng WC, Gros P, Clevers H. 2014. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 28, 305–316. ( 10.1101/gad.235473.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moad HE, Pioszak AA. 2013. Reconstitution of R-spondin:LGR4:ZNRF3 adult stem cell growth factor signaling complexes with recombinant proteins produced in Escherichia coli. Biochemistry 52, 7295–7304. ( 10.1021/bi401090h) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng WC, de Lau W, Madoori PK, Forneris F, Granneman JC, Clevers H, Gros P. 2013. Structures of Wnt-antagonist ZNRF3 and its complex with R-spondin 1 and implications for signaling. PLoS ONE 8, e83110 ( 10.1371/journal.pone.0083110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xie Y, et al. 2013. Interaction with both ZNRF3 and LGR4 is required for the signalling activity of R-spondin. EMBO Rep. 14, 1120–1126. ( 10.1038/embor.2013.167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zebisch M, Xu Y, Krastev C, MacDonald BT, Chen M, Gilbert RJ, He X, Jones EY. 2013. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat. Commun. 4, 2787 ( 10.1038/ncomms3787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lebensohn AM, Rohatgi R. 2018. R-spondins can potentiate WNT signaling without LGRs. Elife 7, e33126 ( 10.7554/eLife.33126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barker N, Clevers H. 2010. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology 138, 1681–1696. ( 10.1053/j.gastro.2010.03.002) [DOI] [PubMed] [Google Scholar]
- 109.Jiang X, Charlat O, Zamponi R, Yang Y, Cong F. 2015. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol. Cell 58, 522–533. ( 10.1016/j.molcel.2015.03.015) [DOI] [PubMed] [Google Scholar]
- 110.Peng WC, de Lau W, Forneris F, Granneman JC, Huch M, Clevers H, Gros P. 2013. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 3, 1885–1892. ( 10.1016/j.celrep.2013.06.009) [DOI] [PubMed] [Google Scholar]
- 111.Wang D, Huang B, Zhang S, Yu X, Wu W, Wang X. 2013. Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 27, 1339–1344. ( 10.1101/gad.219360.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu K, Xu Y, Rajashankar KR, Robev D, Nikolov DB. 2013. Crystal structures of Lgr4 and its complex with R-spondin1. Structure 21, 1683–1689. ( 10.1016/j.str.2013.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zebisch M, Jones EY. 2015. ZNRF3/RNF43: a direct linkage of extracellular recognition and E3 ligase activity to modulate cell surface signalling. Prog. Biophys. Mol. Biol. 118, 112–118. ( 10.1016/j.pbiomolbio.2015.04.006) [DOI] [PubMed] [Google Scholar]
- 114.Zebisch M, Yvonne Jones E. 2015. Crystal structure of R-spondin 2 in complex with the ectodomains of its receptors LGR5 and ZNRF3. J. Struct. Biol. 191, 149–155. ( 10.1016/j.jsb.2015.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan KS, et al. 2017. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature 545, 238–242. ( 10.1038/nature22313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baron M. 2003. An overview of the Notch signalling pathway. Semin. Cell Dev. Biol. 14, 113–119. ( 10.1016/S1084-9521(02)00179-9) [DOI] [PubMed] [Google Scholar]
- 117.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. 2012. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 13, 654–666. ( 10.1038/nrg3272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pursglove SE, Mackay JP. 2005. CSL: a notch above the rest. Int. J. Biochem. Cell Biol. 37, 2472–2477. ( 10.1016/j.biocel.2005.06.013) [DOI] [PubMed] [Google Scholar]
- 119.Milano J, et al. 2004. Modulation of notch processing by γ-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82, 341–358. ( 10.1093/toxsci/kfh254) [DOI] [PubMed] [Google Scholar]
- 120.van Es JH, et al. 2005. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963. ( 10.1038/nature03659) [DOI] [PubMed] [Google Scholar]
- 121.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. 2005. Notch signals control the fate of immature progenitor cells in the intestine. Nature 435, 964–968. ( 10.1038/nature03589) [DOI] [PubMed] [Google Scholar]
- 122.Jensen J, et al. 2000. Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36–44. ( 10.1038/71657) [DOI] [PubMed] [Google Scholar]
- 123.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. 2007. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488. ( 10.1053/j.gastro.2007.03.047) [DOI] [PubMed] [Google Scholar]
- 124.van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. 2010. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat. Commun. 1, 18 ( 10.1038/ncomms1017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. 2001. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294, 2155–2158. ( 10.1126/science.1065718) [DOI] [PubMed] [Google Scholar]
- 126.VanDussen KL, Samuelson LC. 2010. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev. Biol. 346, 215–223. ( 10.1016/j.ydbio.2010.07.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Riccio O, et al. 2008. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377–383. ( 10.1038/embor.2008.7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fre S, et al. 2011. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE 6, e25785 ( 10.1371/journal.pone.0025785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pellegrinet L, et al. 2011. Dll1- and Dll4-mediated Notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240.e7. ( 10.1053/j.gastro.2011.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vooijs M, et al. 2007. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development 134, 535–544. ( 10.1242/dev.02733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.VanDussen KL, et al. 2012. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497. ( 10.1242/dev.070763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ingham PW, Nakano Y, Seger C. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406. ( 10.1038/nrg2984) [DOI] [PubMed] [Google Scholar]
- 133.Ingham PW, McMahon AP. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087. ( 10.1101/gad.938601) [DOI] [PubMed] [Google Scholar]
- 134.Jiang J, Hui CC. 2008. Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812. ( 10.1016/j.devcel.2008.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kasper M, Regl G, Frischauf AM, Aberger F. 2006. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur. J. Cancer 42, 437–445. ( 10.1016/j.ejca.2005.08.039) [DOI] [PubMed] [Google Scholar]
- 136.Hui CC, Angers S. 2011. Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537. ( 10.1146/annurev-cellbio-092910-154048) [DOI] [PubMed] [Google Scholar]
- 137.Stone DM, et al. 1996. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134. ( 10.1038/384129a0) [DOI] [PubMed] [Google Scholar]
- 138.Taipale J, Cooper MK, Maiti T, Beachy PA. 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892–897. ( 10.1038/nature00989) [DOI] [PubMed] [Google Scholar]
- 139.Zheng X, Zeng W, Gai X, Xu Q, Li C, Liang Z, Tuo H, Liu Q. 2013. Role of the Hedgehog pathway in hepatocellular carcinoma (review). Oncol. Rep. 30, 2020–2026. ( 10.3892/or.2013.2690) [DOI] [PubMed] [Google Scholar]
- 140.Kasper M, et al. 2006. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell. Biol. 26, 6283–6298. ( 10.1128/MCB.02317-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van Dop WA, et al. 2010. Loss of Indian Hedgehog activates multiple aspects of a wound healing response in the mouse intestine. Gastroenterology 139, 1665–1676.e10. ( 10.1053/j.gastro.2010.07.045) [DOI] [PubMed] [Google Scholar]
- 142.Büller NV, Rosekrans SL, Westerlund J, van den Brink GR. 2012. Hedgehog signaling and maintenance of homeostasis in the intestinal epithelium. Physiology 27, 148–155. ( 10.1152/physiol.00003.2012) [DOI] [PubMed] [Google Scholar]
- 143.van den Brink GR, et al. 2004. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat. Genet. 36, 277–282. ( 10.1038/ng1304) [DOI] [PubMed] [Google Scholar]
- 144.van Dop WA, et al. 2009. Depletion of the colonic epithelial precursor cell compartment upon conditional activation of the Hedgehog pathway. Gastroenterology 136, 2195–2203.e7. ( 10.1053/j.gastro.2009.02.068) [DOI] [PubMed] [Google Scholar]
- 145.Kolterud A, et al. 2009. Paracrine Hedgehog signaling in stomach and intestine: new roles for Hedgehog in gastrointestinal patterning. Gastroenterology 137, 618–628. ( 10.1053/j.gastro.2009.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL. 2005. Epithelial Hedgehog signals pattern the intestinal crypt-villus axis. Development 132, 279–289. ( 10.1242/dev.01576) [DOI] [PubMed] [Google Scholar]
- 147.Mao J, Kim BM, Rajurkar M, Shivdasani RA, McMahon AP. 2010. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development 137, 1721–1729. ( 10.1242/dev.044586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. 2004. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686. ( 10.1126/science.1093587) [DOI] [PubMed] [Google Scholar]
- 149.He XC, et al. 2004. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt–β-catenin signaling. Nat. Genet. 36, 1117–1121. ( 10.1038/ng1430) [DOI] [PubMed] [Google Scholar]
- 150.Zacharias WJ, Madison BB, Kretovich KE, Walton KD, Richards N, Udager AM, Li X, Gumucio DL. 2011. Hedgehog signaling controls homeostasis of adult intestinal smooth muscle. Dev. Biol. 355, 152–162. ( 10.1016/j.ydbio.2011.04.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zacharias WJ, Li X, Madison BB, Kretovich K, Kao JY, Merchant JL, Gumucio DL. 2010. Hedgehog is an anti-inflammatory epithelial signal for the intestinal lamina propria. Gastroenterology 138, 2368–2377.e4. ( 10.1053/j.gastro.2010.02.057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reddi AH. 2005. BMPs: from bone morphogenetic proteins to body morphogenetic proteins. Cytokine Growth Factor Rev. 16, 249–250. ( 10.1016/j.cytogfr.2005.04.003) [DOI] [PubMed] [Google Scholar]
- 153.Wang RN, et al. 2014. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105. ( 10.1016/j.gendis.2014.07.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Heldin CH, Miyazono K, ten Dijke P. 1997. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471. ( 10.1038/37284) [DOI] [PubMed] [Google Scholar]
- 155.Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. 1999. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem. 274, 19 838–19 845. ( 10.1074/jbc.274.28.19838) [DOI] [PubMed] [Google Scholar]
- 156.Miyazono K, Maeda S, Imamura T. 2005. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 16, 251–263. ( 10.1016/j.cytogfr.2005.01.009) [DOI] [PubMed] [Google Scholar]
- 157.Hardwick JC, Kodach LL, Offerhaus GJ, van den Brink GR. 2008. Bone morphogenetic protein signalling in colorectal cancer. Nat. Rev. Cancer 8, 806–812. ( 10.1038/nrc2467) [DOI] [PubMed] [Google Scholar]
- 158.Auclair BA, Benoit YD, Rivard N, Mishina Y, Perreault N. 2007. Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology 133, 887–896. ( 10.1053/j.gastro.2007.06.066) [DOI] [PubMed] [Google Scholar]
- 159.Qi Z, et al. 2017. BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 ( 10.1038/ncomms13824) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hardwick JC, Van Den Brink GR, Bleuming SA, Ballester I, Van Den Brande JM, Keller JJ, Offerhaus GJ, Van Deventer SJ, Peppelenbosch MP. 2004. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology 126, 111–121. ( 10.1053/j.gastro.2003.10.067) [DOI] [PubMed] [Google Scholar]
- 161.Sneddon JB, et al. 2006. Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proc. Natl Acad. Sci. USA. 103, 14 842–14 847. ( 10.1073/pnas.0606857103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Horiguchi H, Endo M, Kawane K, Kadomatsu T, Terada K, Morinaga J, Araki K, Miyata K, Oike Y. 2017. ANGPTL2 expression in the intestinal stem cell niche controls epithelial regeneration and homeostasis. EMBO J. 36, 409–424. (doi:10.15252/embj.201695690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. 2003. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp. Cell Res. 284, 31–53. ( 10.1016/S0014-4827(02)00098-8) [DOI] [PubMed] [Google Scholar]
- 164.Oda K, Matsuoka Y, Funahashi A, Kitano H. 2005. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 1, 2005. 0010 ( 10.1038/msb4100014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Malumbres M, Barbacid M. 2003. RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3, 459–465. ( 10.1038/nrc1097) [DOI] [PubMed] [Google Scholar]
- 166.Forbes SA, et al. 2015. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811. ( 10.1093/nar/gku1075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang YP, et al. 2017. A chimeric Egfr protein reporter mouse reveals Egfr localization and trafficking in vivo. Cell Rep. 19, 1257–1267. ( 10.1016/j.celrep.2017.04.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marchbank T, Goodlad RA, Lee CY, Playford RJ. 1995. Luminal epidermal growth factor is trophic to the small intestine of parenterally fed rats. Clin. Sci. 89, 117–120. ( 10.1042/cs0890117) [DOI] [PubMed] [Google Scholar]
- 169.Kitchen PA, et al. 2005. Intestinal growth in parenterally-fed rats induced by the combined effects of glucagon-like peptide 2 and epidermal growth factor. JPEN J. Parenter. Enteral Nutr. 29, 248–254. ( 10.1177/0148607105029004248) [DOI] [PubMed] [Google Scholar]
- 170.Basak O, Beumer J, Wiebrands K, Seno H, van Oudenaarden A, Clevers H. 2017. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell 20, 177–190 e174. ( 10.1016/j.stem.2016.11.001) [DOI] [PubMed] [Google Scholar]
- 171.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL III, Sweeney C. 2004. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J. Biol. Chem. 279, 47 050–47 056. ( 10.1074/jbc.M409703200) [DOI] [PubMed] [Google Scholar]
- 172.Biteau B, Jasper H. 2011. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045–1055. ( 10.1242/dev.056671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. 2011. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84–95. ( 10.1016/j.stem.2010.11.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Joosten SPJ, et al. 2017. MET signaling mediates intestinal crypt–villus development, regeneration, and adenoma formation and is promoted by stem cell CD44 isoforms. Gastroenterology 153, 1040–1053.e4. ( 10.1053/j.gastro.2017.07.008) [DOI] [PubMed] [Google Scholar]
- 175.1997. Unified nomenclature for Eph family receptors and their ligands, the ephrins. Eph Nomenclature Committee. Cell 90, 403–404. ( 10.1016/S0092-8674(00)80500-0) [DOI] [PubMed] [Google Scholar]
- 176.Kullander K, Klein R. 2002. Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 3, 475–486. ( 10.1038/nrm856) [DOI] [PubMed] [Google Scholar]
- 177.Kalo MS, Pasquale EB. 1999. Multiple in vivo tyrosine phosphorylation sites in EphB receptors. Biochemistry 38, 14 396–14 408. ( 10.1021/bi991628t) [DOI] [PubMed] [Google Scholar]
- 178.Koo BK, et al. 2009. Notch signaling promotes the generation of EphrinB1-positive intestinal epithelial cells. Gastroenterology 137, 145–155.e3. ( 10.1053/j.gastro.2009.03.046) [DOI] [PubMed] [Google Scholar]
- 179.Jung P, et al. 2011. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 17, 1225–1227. ( 10.1038/nm.2470) [DOI] [PubMed] [Google Scholar]
- 180.Merlos-Suarez A, et al. 2011. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524. ( 10.1016/j.stem.2011.02.020) [DOI] [PubMed] [Google Scholar]
- 181.Solanas G, Cortina C, Sevillano M, Batlle E. 2011. Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nat. Cell Biol. 13, 1100–1107. ( 10.1038/ncb2298) [DOI] [PubMed] [Google Scholar]
- 182.Tian H, Biehs B, Chiu C, Siebel CW, Wu Y, Costa M, de Sauvage FJ, Klein OD. 2015. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 11, 33–42. ( 10.1016/j.celrep.2015.03.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144, 646–674. ( 10.1016/j.cell.2011.02.013) [DOI] [PubMed] [Google Scholar]
- 184.Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61, 759–767. ( 10.1016/0092-8674(90)90186-I) [DOI] [PubMed] [Google Scholar]
- 185.Vogelstein B, et al. 1988. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532. ( 10.1056/NEJM198809013190901) [DOI] [PubMed] [Google Scholar]
- 186.Kinzler KW, Vogelstein B. 1997. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 386, 761–763. ( 10.1038/386761a0) [DOI] [PubMed] [Google Scholar]
- 187.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339, 1546–1558. ( 10.1126/science.1235122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Drost J, et al. 2015. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47. ( 10.1038/nature14415) [DOI] [PubMed] [Google Scholar]
- 189.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. 2015. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262. ( 10.1038/nm.3802) [DOI] [PubMed] [Google Scholar]
- 190.2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. ( 10.1038/nature11252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dow LE, O'Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, Lowe SW. 2015. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 161, 1539–1552. ( 10.1016/j.cell.2015.05.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.