Summary
The mammosphere assay has become widely employed to quantify stem-like cells in a population. However, the problem is there is no standard protocol employed by the field. Cell seeding densities of 1,000 to 100,000 cells/mL have been reported. These high densities lead to cellular aggregation. To address this, we have individually tracked 1,127 single MCF-7 and 696 single T47D human breast tumor cells by eye over the course of 14 days. This tracking has given us detailed information for the commonly used endpoints of 5, 7, and 14 days that is unclouded by cellular aggregation. This includes mean sphere sizes, sphere-forming efficiencies, and a well-defined minimum size for both lines. Importantly, we have correlated early cell division with eventual sphere formation. At 24 hr post seeding, we can predict the total spheres on day 14 with 98% accuracy in both lines. This approach removes cell aggregation and potentially shortens a 5- to 14-day assay to a 24 hours.
Subject Areas: Biology Experimental Methods, Cancer
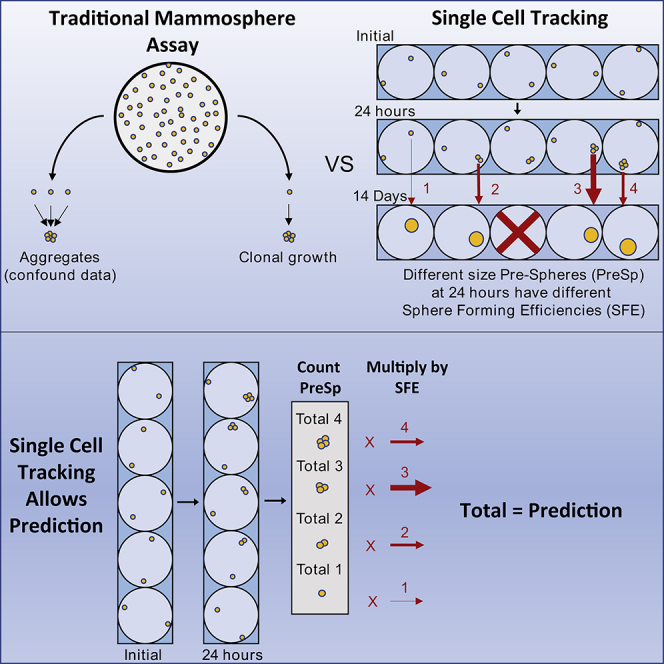
Graphical Abstract
Highlights
-
•
Single-cell tracking removes confounding aggregation from the mammosphere assay
-
•
Tracking reveals sphere-forming efficiencies much higher than commonly reported
-
•
True clonal spheres are smaller than commonly reported
-
•
At 24 hours, tracking can predict total day 14 spheres with 98% accuracy
Biology Experimental Methods; Cancer
Introduction
The cancer stem cell theory holds that there is a small sub-population of cells within a tumor that drives tumorigenesis and is capable of repopulating the tumor bulk from one cell (Al-Hajj et al., 2003, Kim and Ryu, 2017, Medema, 2013, Velasco-Velazquez et al., 2012). Although this theory remains controversial (Liu et al., 2014, Nakshatri et al., 2009), it has become widely accepted and there are numerous assays for elucidating the stem-like character of cancer cells. One property of breast cancer stem cells is that they can survive detachment from the extracellular matrix (Kim et al., 2012, Kruyt and Schuringa, 2010). Detached stem cells also retain the capability of proliferating (Alimperti et al., 2014, Dontu and Wicha, 2005). As such, the sphere formation of suspended cells is hypothesized to be a method to assay for the self-renewal potential in vitro (Ponti et al., 2005). This property, first investigated in neural cells, was further adapted for mammary epithelial cells and termed the mammosphere assay (Dontu et al., 2003). In brief, suspended cells are cultured in serum-free media containing growth factors. The fraction that survives to form spheroid colonies (mammospheres) is deemed more stem-like. This is often followed by monitoring the alterations in sphere formation following treatment (Gupta et al., 2009, Lu et al., 2014, Ma et al., 2017, Reynolds et al., 2017). Treatments that lower the sphere-forming efficiency (SFE, [spheres/cells seeded]*100) of a population are hypothesized to have reduced the stem-like sub-population of the cells.
MCF-7 human breast carcinoma cells are widely used in the mammosphere assay (Akrap et al., 2016, Fu et al., 2016, Grimshaw et al., 2008, Guttilla et al., 2012, Hinohara et al., 2012, Manuel Iglesias et al., 2013, Zhang et al., 2011). These luminal-type cells have been observed to make very cohesive, easily defined spheres. However, the problem is SFEs are quite disparate between reports and have ranged from 1% to 20% depending on the conditions (de la Mare et al., 2013, Montales et al., 2012, Morrison et al., 2012). Many factors could be contributing to these discrepancies, including growth media composition, counting procedures, and variability between different human operators performing the same assay. Of utmost importance, however, is the seeding density (Shaw et al., 2012). Due to the mobile nature of cells in suspension, cells drift and collide, leading to an aggregation tendency that is proportional to the cell density (Tolbert et al., 1980). This is problematic because clonality is an integral concept to the mammosphere assay (Shaw et al., 2012). Mammospheres should arise from a single cell to effectively measure stem-like propagation.
Attempts to address aggregation have been reported (Manuel Iglesias et al., 2013, Patel and Rameshwar, 2013, Rota et al., 2012, Shaw et al., 2012). There is no common protocol, however, and seeding densities up to 100,000 cells/mL have been reported. Varying densities can lead to large differences in SFE (Shaw et al., 2012) and beg the question of how to interpret results. If a drug treatment lowers the sphere count in an experiment, can that result be interpreted as an effect on SFE or aggregation? To completely remove results confounded by aggregates we visually tracked 1,823 verified single cells over the course of 14 days, monitoring the cell count, sphere size, and morphology.
Results
Initially Plated Single Cells and Two-Cell Clusters Have Significantly Different Sphere-Forming Efficiency
To reduce the effect of aggregation we tracked individual cells to ensure the clonality of resulting spheres. We used MCF-7, a weakly tumorigenic, luminal breast cancer cell line that has the propensity to form well-rounded, easily identifiable spheres (Manuel Iglesias et al., 2013). We initially sought to use fluorescence-activated cell sorting (FACS) to sort single cells into a 96-well plate and to subsequently track and image these sorted cells, but there were numerous technical inaccuracies involved in this procedure. The most quantifiable of these inaccuracies arose during the imaging step. Sphere formation was completely ablated after cells spent over 2 hours a day at room temperature (RT) while imaging an entire 96-well plate (Figure S1). We therefore moved to a dilution-based assay.
It is logistically impossible to achieve exactly 1 cell/well using only dilution. An average of 1 cell/well may be attained, but in practice, each well contains a different number of cells and many wells will have none. To ensure cells in nearly every well, and to investigate if aggregations had a higher chance of forming spheres, we chose a dilution of 2 cells/well. To minimize the time spent out of incubation we plated only 12 wells per 96-well plate. With 8 plates total, this combined to a full 96-wells. This was considered one full experimental replicate. This technique resulted in plates at RT in less than 15 min at a time.
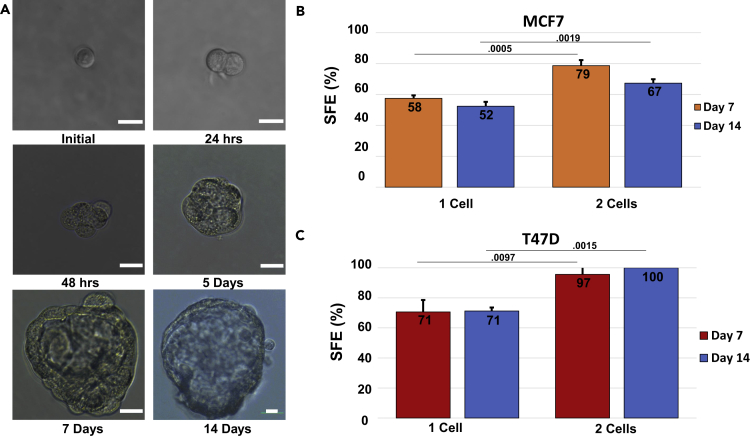
Figure 1A shows the tracking of a single cell over 14 days in suspension. Each tracked clonal outgrowth was termed a Pre-Sphere (PreSp) until day 5. Surprisingly, MCF-7 cells seeded at this low density undergo little initial movement over time in the smaller wells of a 96-well plate. Nevertheless, there were 22 instances of collision, 21 of which (95%) eventually formed spheres, indicating that aggregations have a higher propensity for sphere formation. Although these aggregates were tracked, they were removed from any subsequent single-cell calculations. In fact, any cell that moved so much as to cast doubt to its identity was removed from data for single-cell quantitative calculations as well (a total of only 6 of 1,127 MCF-7 cells and no T47Ds were “lost”).
Figure 1.
Initially Plated Single Cells and Two-Cell Clusters Have Significantly Different Sphere-Forming Efficiency (SFE)
(A) A single MCF-7 cell tracked and imaged for 14 days. Scale bars, 20 μm.
(B) Single-cell and two-cell cluster SFEs of the MCF-7 cell line. Orange bars indicate the SFEs of 1 and 2 cells at day 7. Blue bars indicate day 14. SFEs are calculated as (total spheres/total cells tracked)*100. MCF-7 spheres were considered growths over 50 μm. Statistics formulated using Student's two-tailed t test (N = 3). p Values displayed over bars.
(C) Same as (B) for the T47D cell line. Growths over 35 μm at day 7 and 40 μm at day 14 were counted as spheres.
We also observed that despite rigorous procedures to ensure a single-cell suspension, including passage through a microfilter, two-cell clusters remained at the time of initial seeding (Figure S2). These clusters showed a statistically significant increase in sphere-forming capacity of nearly 20% compared with single cells (Figure 1B). Surprisingly, for true clonal spheres originating from a single cell, we saw SFEs of 55% and 52% for days 7 and 14, respectively (Figure 1B).
To increase the robustness of our experiments, an additional cell line was tracked. T47Ds were chosen because of their ability to form spheres over numerous passages. T47Ds showed a higher propensity for movement within wells than MCF7s. In spite of manual manipulation with a pipette to reduce collisions there were 23 instances, 100% of which formed spheres by day 14. Astonishingly, the weighted average SFE of single T47D cells recorded at day 14 was 71% (Figure 1C). Two-cell clusters also remained in suspensions of T47Ds despite the use of a microfilter and passage through a needle (13 of these were removed from wells due to close proximity to single cells). It was observed that 100% of the remaining two-cell clusters formed spheres at day 14 (Figure 1C).
High Seeding Densities Are Confounded by Aggregation
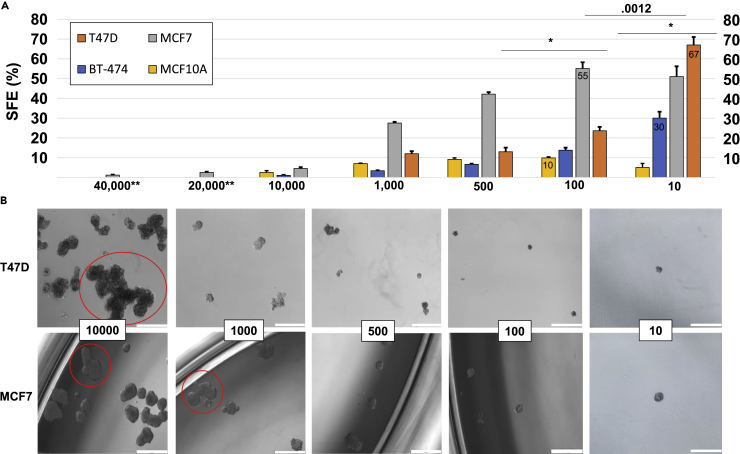
SFEs for the MCF-7 line vary greatly from one article to another and are most often reported as less than 5% (Klopp et al., 2010, Wang et al., 2011). However, our independent calculations on the raw data of Figure 1B of Morrison et al. show an SFE of ∼20% when cell density is low for MCF-7 cells (Morrison et al., 2012). Given the high efficiency we observed with single-cell tracking we sought to validate our results with an orthogonal technique. Our group and others have observed that SFE decreases with increasing seeding density (Shaw et al., 2012). We therefore performed a seeding density dilution starting at 40,000 cells/well and ending at 10 cells/well. At higher seeding densities, we also observed the lower efficiencies most often reported (Figure 2A). However, our single-cell studies were confirmed at lower seeding densities. SFEs increased significantly at densities below 1,000 cells/mL and peaked at 100 cells/mL for MCF-7 cells (Figure 2A). We observed that higher seeding densities also contained what we considered to be large aggregates (Figure 2B, red circles). The problem of whether to include these, exclude them, or estimate the number of spheres contained within the aggregate was not trivial. We found that there was a significant difference in the results depending on how these aggregations were tallied. (Statistics not reported due to the highly subjective nature of choosing aggregates. For our figure counts, all obvious aggregations were excluded.) Figure 2B illustrates the visual differences between seeding densities.
Figure 2.
High Seeding Densities Are Confounded by Aggregation
(A and B) (A) A density dilution assay was performed on MCF-7, BT-474, T47D, and MCF10A cells in 24-well low-attach plates. The cell seeding density (cells/mL) is displayed on the x axis, and the SFE is displayed on the y axis. Efficiencies were calculated as (spheres/seeded cells)*100. Spheres were counted on day 7. Statistics calculated using two-tailed Student's t test (N = 3 for T47D, BT-474, and MCF10A. N = 6 for MCF-7). p Values displayed above bars. *p < .0001. ** Indicates only performed with MCF-7 cells. (B) Representative images of MCF-7and T47D spheres. Red circles indicate examples of obvious aggregations excluded from counts. Scale bar, 500 μm.
SFE also increased significantly with lower dilution in the T47D cell line, although a dilution of 10 cells/mL was required to reach the efficiency seen in single-cell tracking. The cells had a strong tendency to move to the edge of the plate, increasing aggregation. At 10,000 cells/well the problem of aggregation was so severe that accurate counting was impossible. The BT-474 (a HER2+ cell line) and MCF10A (negative control) lines were also tested in this manner. MCF10A reached a peak of 10% SFE at 100 cells/mL, whereas BT-474 reached a maximum efficiency of 30% at 10 cells/mL.
Size Information of Spheres
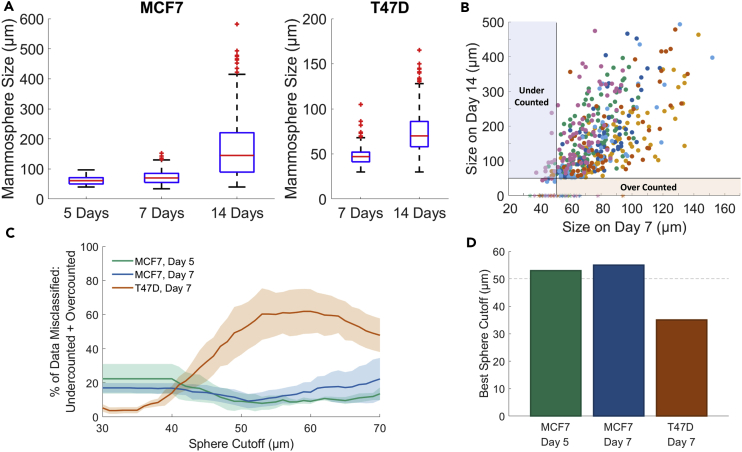
Most information on sphere size (diameter) comes from scale bars in images (Morrison et al., 2012, Wang et al., 2014, Wang et al., 2011, Wolf et al., 2013). We sought to gain a more robust measurement of clonal sphere sizes at various time points. We chose days 5, 7, and 14 for MCF-7 as these are well represented in the literature (Ji et al., 2016, Montales et al., 2012, Wang et al., 2014). The median sphere sizes of true, clonal spheres were 61, 70, and 144 μm for days 5, 7, and 14, respectively. The maximum day 5 size was 97 μm, but the mammospheres within the 25th–75th quartiles ranged from 50 to 71 μm. The day 7 size was 130 μm with an outlier as high as 152 μm, but the 25th–75th quartiles spanned a much narrower range from only 55 to 85 μm (Figure 3A). Interestingly, day 14 had a wide range of sizes from 50 to 400 μm with an outlier as large as 660 μm.
Figure 3.
Size Information of Spheres
(A) Average mammosphere size of MCF-7 and T47D cell lines. Red bar indicates mean size, and blue box indicates the 25th–75th quartiles. Whiskers represent the range of sizes, whereas red dots are outliers (for MCF-7: day 5, N = 3; days 7 and 14, N = 6. For T47D: N = 6 for days 7 and 14). Note that the y axis scale is different for T47D than MCF-7.
(B) Explanatory figure using size after 7 days compared with size on day 14 (N = 6, MCF7). Solid lines represent a sphere cutoff value of 50 μm. If this value is chosen the lavender box illustrates those individual clonal growths (termed Pre-spheres or PreSP) that were under 50 μm but grew to be spheres and were thus undercounted. The beige box illustrates those that were over 50 μm on day 7 but either died or shrunk by day 14 and were therefore overcounted.
(C and D) (C) Data from scatter charts were utilized to calculate the percent of misclassified spheres ([undercounted + overcounted]/total spheres). This was calculated for a range of cutoff values and plotted as a graph. Solid line is the weighted average, and the error is ±weighted SD. (D) The cutoff value with the smallest percent of misclassified data was calculated for each day using the graph in (C). For MCF-7 the results were 53 and 55 μm for days 5 and 7, respectively. For T47D the value was 35 μm.
Clonal T47D spheres were much smaller overall. Spheres more than 5 cells big do not generally appear until day 7. We therefore chose to eschew size measurement at day 5. Median size at day 7 was 48 μm with the 25th to 75th quartiles covering a range of 35 to 56 μm. The largest outlier was 110 μm. Median size at day 14 was moderately larger at 66 μm, ranging from 70 to 85 μm in the 25th–75th quartiles. The largest outlier was 165 μm.
This information raised the important question: “What is a sphere?” Spheres have been traditionally defined and categorized by size. The minimum size most often employed for MCF-7 cells in the literature is > 50 μm (Choi et al., 2017, Grimshaw et al., 2008, Simoes et al., 2015), but minimums ranging from 14 to 100 μm have also been reported (Gupta et al., 2009, Montales et al., 2012, Wang et al., 2011, Wolf et al., 2013). We sought to avoid ad hoc definitions and sought to find an empirical basis for categorization. We reasoned that by comparing the sizes of spheres on days 5 and 7 with sizes on day 14, we could formulate the percentage of spheres that were misclassified (Figure 3B). This would include spheres that were under the minimum size at day 5 or 7 but grew to be spheres on day 14 (undercounted), as well as those that were above the cutoff but later either died or shrunk below the cutoff values (overcounted). By comparing days 5 and 7 with day 14, we constructed a curve showing the percent of misclassified cells for values ranging from 40 to 70 μm (Figure 3C). Our only caveat was that spheres must grow at least 5 μm between measurements. We determined that MCF-7 sphere sizes of 53 and 55 μm correlated with the least misclassification for days 5 and 7, respectively (Figure 3D). As the percent of misclassification was very similar, in the range of 50 to 60 μm, we chose to adopt the widely employed value of 50 μm for the remainder of the analysis of MCF-7 cells.
T47D sphere classification required further scrutiny. PreSp as small as 30 μm and containing only 3 cells at day 7 went on to form spheres at day 14. The number of spheres that died or shrunk after day 7 (overcounted) was also negligible. The bulk of classification error arose from undercounting smaller PreSp, which went on to eventually form spheres (overcounting). The minimum size correlated with least misclassification for day 7 was 32 μm. We were attempting to avoid an ad hoc definition of how many cells a sphere contained, but a cutoff value of 32 μm is only 2–3 cells. To avoid the possibility that we were including small clusters of cells that had died, we added the caveat that spheres must grow at least 5 μm between measurements. This resulted in a minimum size of 35 μm on day 7 and 40 μm on day 14.
Size was the only parameter utilized for classifying mammospheres of either cell line. Most MCF-7 spheres were well rounded with clearly defined edges, but there were many examples of unique morphologies as well as clonal growths (still termed spheres), which would appear to be aggregations if they had not been tracked from single cells (Figure S3A). This was even more apparent in the T47D cell line. Although many T47D spheres were rounded and spherical, the majority were either dome-like mounds or actual sheets of cells. Video S1 shows a 10-μm pipette pulling a very spherical T47D growth back and forth. Video S2 shows the manipulation of a slightly curled sheet of cells (both mounds and sheets were confirmed as unattached to the plate). The overall morphologies of clonal T47D growths were even more wide ranging than MCF7s (Figure S3B) and included “spheres” of loosely associated cells that would split apart and rejoin on a daily basis. Puzzlingly, some T47D PreSp have the ability to split apart completely and continue to grow. In most cases the proximity of these secondary PreSp leads to collision with the parent sphere, but in some cases these spheres drift away to divide on their own (10 instances of splitting resulted in re-collision with parent, and 8 times the daughter sphere grew on its own.) Spheres that split away were not used in predictive calculations, but they were rigorously tracked.
A clonal, flat sheet of T47D cells curled at the edges is manually manipulated with a 10-μm pipette.
Sphere Formation can be Predicted with 98% Accuracy in 24 Hours
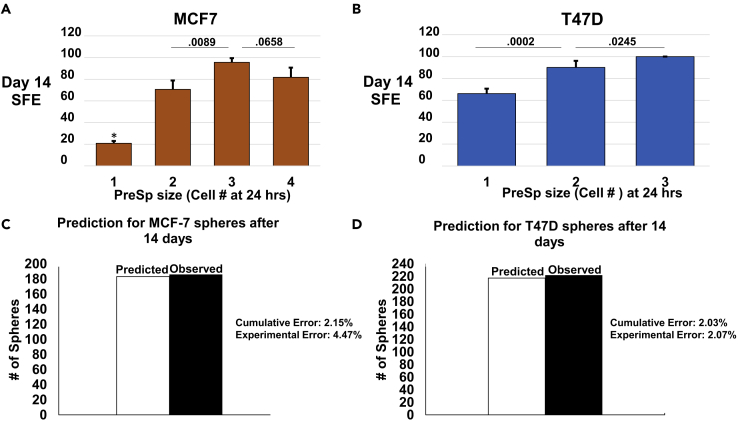
The initial impetus for this study was the hypothesis that it should be possible to predict sphere formation from an early time point using only doubling information. We set out to accomplish this by carefully tracking the fates of single cells cultured in suspension. We marked the position of each initial cell and either counted or measured the cells in resultant PreSp over 14 days. We then correlated early cell counts with eventual sphere formation. As expected, larger early PreSp grew to full spheres at a higher rate (Figure S4). Importantly, we found that 24 hr had fewer cell counts to choose from and less ambiguity in cell count (Figure S5A). PreSp of 1, 2, 3, and 4 cells were well defined and unambiguous, whereas higher counts were often estimated or found by focusing up and down through the cells (Figure S5B). As it was also the earliest time point, we chose to use 24 hr for further experimentation. We found that PreSp of size 1, 2, 3, and 4 cells each had a distinct SFE at 24 hr. Although statistical significance between the sphere-forming capacity of each PreSp size was not our goal, we found that there was significant difference between all PreSp sizes sequentially, but sizes of 2 and 4 cells were not significantly different in a direct comparison (Figure 4A).
Figure 4.
Sphere Formation can be Predicted with 98% Accuracy in 24 Hours
For a Figure360 author presentation of Figure 4, see https://doi.org/10.1016/j.isci.2018.08.015.
(A and B) Single cells were seeded and tracked. At 24 hr PreSp were counted and categorized by the number of cells in each. Each count was correlated with eventual sphere formation on days 7 and 14 (for example, there were a total of 153 single-cell MCF-7 PreSp at 24 hr. Of these, 32 eventually formed spheres on day 14, yielding an SFE of 21%). SFE for each size of PreSp is displayed on the y axis. Error bars are weighted SD. (A) Objects over 50 μm were considered spheres for the MCF-7 cell line. (B) Growths over 35 μm on day 7 and 40 μm on day 14 were considered spheres for the T47D cell line. p Values were calculated by Student's two-tailed t test and are displayed above the bars on the graph (N = 3). * Indicates significantly different from all other PreSp sizes with p <.0001.
(C and D) Single cells were again tracked over 14 days in the same manner as in (A) and (B). At 24 hr PreSp sizes were counted. SFEs from (A) were then used in a predictive capacity on these counts to determine how many spheres would form at 14 days. [For example, in one experiment there were 82 two-cell MCF-7 PreSp at 24 hr. The SFE from (A) would predict 62.76 spheres on day 14. The actual spheres formed on day 14 were then compared with these predictions.] The y axis shows the total spheres from three experiments. The white bar is the total prediction calculated using SFEs from previous experiments. The black bar is the total spheres actually observed. Difference between prediction and observation (cumulative error) at 14 days was 2.15% for MCF-7 cells (C) and 2.03% for T47Ds (D). Average error for MCF-7s was 4.47 (±.5) (C) and 2.07 (± 1.71) for T47Ds (D).
We then set up three new experiments in the same manner as above. Individual cells were tracked, and counts were made of all 1-, 2-, 3-, and 4-cell PreSp at 24 hr. The mean sphere-forming percentages from our initial experiments were then used to determine whether it was possible to predict how many spheres formed on day 14 of the newly plated experiments. Strikingly, we found that just these early 24-hr counts could predict sphere formation for day 14 with 98% accuracy (Figure 4C).
We next sought to validate these results in an additional cell line. We chose to work with T47D cells as we found their SFE to be high enough to gather meaningful data without tracking thousands of cells. T47D mammospheres are also able to be passaged over subsequent generations for long term (Manuel Iglesias et al., 2013). We repeated the methodology employed with MCF-7s insofar as three experiments were used to establish SFEs for PreSp at 24 hr (Figure 4B). These SFEs were then employed to predict the day 14 sphere formation of a further three experiments. T47Ds were slower to form spheres than MCF-7s overall, with only three instances of 3-cell PreSp at 24 hr recorded over six experiments. Again, PreSp as small as 3 cells on day 7 went on to form spheres at day 14. For this reason, we determined to focus mainly on day 14 for subsequent calculations. For T47D cells, it is also possible to predict sphere formation at day 14 with 98% accuracy using only PreSp counts at 24 hr (Figure 4D).
Sphere Formation in MCF7 and T47D Lines Does Not Depend on CD44hi/CD24- Populations
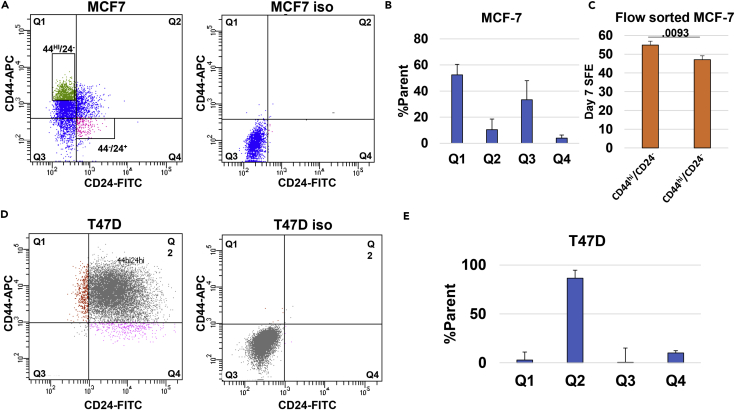
As mammosphere formation is thought to represent the portion of stem-like cells in a population, we sought to discover whether established stem cell markers could account for the high SFE revealed by single-cell tracking. Breast cancer cell populations containing the marker profile CD44+/CD24- have been reported to enrich for mammosphere formation (Gu et al., 2015, Ponti et al., 2005) and tumorigenesis in mice (Al-Hajj et al., 2003) and have been well studied in the literature. We therefore used flow cytometry to assay for the CD44/CD24 profile of the two cell lines we utilized for single-cell tracking (Figures 5A and 5D).
Figure 5.
CD44HI/CD24- Populations Do Not Play A Large Role in the Sphere-Forming Efficiency of MCF-7 or T47D Cells
(A) A representative image of MCF-7 cells sorted by flow cytometry. Q1 represents the CD44+/CD24- population, whereas Q4 contains the CD44-/CD24+ population. Box gates indicate populations sorted by FACS for further analysis. Quadrants were gated using an iso control.
(B and C) (B) The percent of parent population for each quadrant of MCF-7 cells. (N = 3) (C) MCF-7 cells were FACS sorted for CD44hi/CD24- and CD44-/CD24+ populations, and mammosphere assays were performed at a density of 10 cells/mL for each population. The graph shows the day 7 SFE for each population on the y axis. Statistics performed with Student's two-tailed t test. p Value displayed above the bar.
(D) A representative image of T47D cells sorted by flow cytometry. Q1 represents the CD44+/CD24- population, whereas Q4 contains the CD44-/CD24+ population.
(E) The percent of parent population for each quadrant of T47D cells. (N = 3).
Our MCF-7 cells exhibited a higher portion of CD44+/CD24- cells compared with the isotype control than has been commonly reported (Olsson et al., 2011, Sheridan et al., 2006, Yan et al., 2013). The average population of CD44+ cells over three experiments was over 50% (Figure 5B). As this value was very similar to the SFE of MCF-7 cells as determined by single-cell tracking, we hypothesized that this population could be responsible for that effect. To address this possibility, we employed FACS to isolate a portion of CD44hi/CD24- cells to further analyze using the mammosphere assay. We chose to use the cells expressing the highest levels of CD44 to further enrich for putative stem-like cells. These cells were then cultured in mammosphere media at a density of 10 cells/well, and the SFE at day 7 was calculated. We next proceeded to compare this population to the fraction of MCF-7 cells expressing a CD44-/CD24+ profile. We found a difference in SFE of only 10% between the two populations (Figure 5C). T47D cells expressed a CD44+/CD24+ profile almost exclusively (Figure 5D). As the CD44+/CD24- population comprised less than 5% of the cells (Figure 5E), we did not sort T47D cells for further analysis.
Discussion
The mammosphere assay has become a staple of breast cancer stem cell research. By providing a platform in which the properties of anoikis resistance and suspended proliferation can be monitored, the assay proposes to give researchers a way of quantifying the portion of putative stem cells in a population in vitro (Chang et al., 2014, Dontu and Wicha, 2005). Drug treatments or genetic modifications can then be employed, and reductions in sphere formation are hypothesized to have decreased the stem population of a sample (Fu et al., 2016, Grudzien et al., 2010). Unfortunately, there is no standard protocol employed in the field, and reported SFEs can vary significantly from publication to publication. We address here the problem of cellular aggregation as a possible cause for such discrepancies. Suspended cells can collide and form aggregations. This can lead to confounded results through an artificial lowering of SFE (Shaw et al., 2012). If seeding densities are too high, it will remain unclear if treatment causes a change in sphere formation or in aggregation. Although the idea of aggregation itself is not novel, the issue remains largely unaddressed in the field, and we illustrate clearly here the dramatic effects it can have on the experimental outcome.
We addressed the problem of aggregation by tracking individual cells from initial seeding to eventual sphere formation over the course of 14 days. We used the MCF-7 and T47D cell lines due to their common use and ready sphere-forming ability. At the very low seeding densities we employed, we lost track of a few cells due to movement or aggregation within wells (less than 3% in the MCF-7 line, 5.5% for T47Ds). What aggregations did occur, however, formed spheres at a rate of 95% in MCF-7s and 100% in T47Ds. This could be due to enhanced survival from cell-cell attachment signals (Niit et al., 2015) or the simple probability of larger PreSp forming spheres at a higher rate. Any explanation for an aggregation's enhanced sphere-forming ability, however, leads to confounded SFEs in the final calculations. In addition, cells that begin the assay as initial clusters of 2 cells also have a significantly higher chance to form eventual spheres. These confounding factors will persist in all sphere-forming assays that utilize high-density seeding.
At high seeding densities, cells and spheres have less room to move and more aggregations are formed. These aggregations can create irregular formations that also present challenges for counting. We found that using different counting procedures can result in different results. There are also non-quantifiable differences between aggregations and true clonal spheres. Single cells produce spheres containing a wide variety of morphologies ranging from very spherical to what appear to be loose aggregations of cells to shapes that are unique and non-classifiable. Methods that make an attempt to visually distinguish between spheres and aggregations have been reported (Smart et al., 2013), but the morphologies we observed underscore that spheres cannot be identified as such by the eye or by any technology that counts rounded objects. Morphologies are too wide ranging to classify spheres as clonal based on appearance alone. Single-cell tracking avoids these pitfalls.
There is also a clear need to define a standard minimum sphere size for each cell line. A size too small inevitably includes PreSp that will die off. A size too large excludes smaller PreSp that will eventually succeed in forming spheres. Our single-cell tracking allowed us to find values that minimize both these factors. The values we calculated for MCF-7 cells conform very closely to what many in the field have chosen to employ (Choi et al., 2017, Feng et al., 2017, Morata-Tarifa et al., 2016). T47D spheres proved to be much smaller and slower growing. Clonal growths as small as 3 cells continued to grow and divide until day 14. The minimum size of 35 μm we calculated for day 7 illustrates that utilizing a single blanket cutoff value for each cell line under study can exclude valuable data. A value of 50 μm applied to T47D spheres would result in over 50% of data being misclassified and a resultant underestimation of the sphere content in a population. With regard to mean sizes, clonal spheres are also demonstrably smaller than most aggregates. High-density seeding with numerous aggregations have larger spheres, and we quantified the expected distribution of clonal sphere sizes. Mean sizes as high as 100 μm have been reported on day 5 for MCF-7 spheres (Montales et al., 2012). Our data indicate that anything above 100 μm on day 5 or 150 μm on day 7 is likely the result of aggregations. T47Ds are demonstrably smaller, and the largest sphere we recorded was 165 μm at day 14. One problem is that clonal T47D growths are rarely spherical. The majority of these growths were either dome shaped or flat. This underscores the necessity to treat each cell line as unique in its morphology.
Strikingly, single-cell tracking of the MCF-7 cell line reveals an SFE of 55% at day 7 and 52% at day 14, which we confirmed using the orthogonal technique of density dilution. Most of the literature reports SFE values under 10% (de la Mare et al., 2013, Montales et al., 2012, Gupta et al., 2009, Montales et al., 2012, Wang et al., 2011, Wolf et al., 2013). However, we wish to emphasize that our data show that the SFE for MCF-7 cells is much higher than commonly reported and that all results obtained from high seeding densities are clearly confounded by aggregation. Emphasizing this are the results for T47D cells. Astonishingly, when single cells from this line are rigorously tracked over time their SFE is 71%. Even if a higher cutoff value of 40 μm is employed at day 7 the SFE is still 61%. We confirmed the results of our single-cell tracking with established cell seeding density dilutions in multiple cell lines where even a “negative control” cell line had a maximum SFE of 10%. It has been observed that SFE decreases with increasing cellular seeding density (Shaw et al., 2012). This is explained by cellular aggregation. Theoretically, SFE should remain constant if spheres are clonal. In practice, however, aggregation will reduce the number of spheres and result in lower SFE. For the mammosphere assay to give meaningful results, dilutions much smaller than those currently employed are more effective. Following single cells can also provide the additional advantage of cell fate tracking. This can provide information on the effects of drug treatments on single cells, as well as provide a means to follow cell marker persistence/progression and correlation to sphere formation.
Flow cytometry and subsequent tracking after cell sorting for stem cell markers confirmed the work of Iglesias and colleagues insofar as CD44 is not responsible for the sphere-forming capabilities of these cell lines (Manuel Iglesias et al., 2013). We tracked MCF-7 cells sorted for CD44hi/CD24- and CD44-/CE24+ populations at a density of 10 cells/mL and found that although there is a small but significant 10% difference in SFE between the two, it is not large enough to account for the high overall SFE of the entire population. The T47D cell line did not contain a CD44+/CD24- population larger than 5% and was thus ruled out for further stem cell marker study as a population that small could not contribute enough cells to account for a 71% SFE.
Finally, the mammosphere assay is also time consuming. Experiments run in triplicate can take months. We reasoned that this time could be shortened if we could generate a method of prediction based on information from early time points. We hypothesized that if cells were tracked, having their early doubling information recorded, this could be correlated to eventual sphere formation. Surprisingly, this method worked at a much earlier time point than expected and provided a wealth of biological information. After having garnered correlative information from early division and later sphere formation, we were able to apply this information to another set of experiments to predict the generation of spheres. We found that early doubling at 24 hr can predict the eventual sphere formation in a population with 98% accuracy at day 14. This has the potential to shorten sphere formation assays by a factor of 7–10, effectively reducing the traditional time frame of 7–14 days to only 24 hr.
Limitations of the Study
Although this method shortens a 7- to 14-day assay to 24 hr, it is labor intensive. Monitoring thousands of cells by eye requires substantial effort. We are currently in the process of developing a procedure to automate this process and allow high-throughput analysis.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the Kahlert Foundation and grants from the National Institutes of Health to S.S.M. (R01-CA124704, R01-CA154624), R.M.L. (T32CA154274, FA9550-16-1-0052), S.J.P.P. (5T32GM008181-30, 1F31CA232393-01), and M.I.V. (K01CA166576).
Author Contributions
P.C.B. and S.S.M. designed the study; P.C.B. performed the experiments; P.C.B. and R.M.L. analyzed the data; P.C.B. wrote the manuscript; S.S.M., R.M.L., M.I.V., S.J.P.P., K.C., K.N.T., C.J.L., and E.O. helped prepare the manuscript.
Declaration of Interests
The authors wish to declare no competing interests.
Published: October 26, 2018
Footnotes
Supplemental Information includes Transparent Methods, five figures, and two videos and can be found with this article online at https://doi.org/10.1016/j.isci.2018.08.015.
Supplemental Information
References
- Akrap N., Andersson D., Bom E., Gregersson P., Stahlberg A., Landberg G. Identification of distinct breast cancer stem cell populations based on single-cell analyses of functionally enriched stem and progenitor pools. Stem Cell Rep. 2016;6:121–136. doi: 10.1016/j.stemcr.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimperti S., Lei P., Wen Y., Tian J., Campbell A.M., Andreadis S.T. Serum-free spheroid suspension culture maintains mesenchymal stem cell proliferation and differentiation potential. Biotechnol. Prog. 2014;30:974–983. doi: 10.1002/btpr.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.W., Wang H.C., Chen C.Y., Hung T.W., Hou M.F., Yuan S.S., Huang C.J., Tseng C.N. 5-azacytidine induces anoikis, inhibits mammosphere formation and reduces metalloproteinase 9 activity in MCF-7 human breast cancer cells. Molecules. 2014;19:3149–3159. doi: 10.3390/molecules19033149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.S., Kim D.A., Chung H., Park I.H., Kim B.H., Oh E.S., Kang D.H. Screening of breast cancer stem cell inhibitors using a protein kinase inhibitor library. Cancer Cell Int. 2017;17:25. doi: 10.1186/s12935-017-0392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mare J.A., Sterrenberg J.N., Sukhthankar M.G., Chiwakata M.T., Beukes D.R., Blatch G.L., Edkins A.L. Assessment of potential anti-cancer stem cell activity of marine algal compounds using an in vitro mammosphere assay. Cancer Cell Int. 2013;13:39. doi: 10.1186/1475-2867-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G., Abdallah W.M., Foley J.M., Jackson K.W., Clarke M.F., Kawamura M.J., Wicha M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G., Wicha M.S. Survival of mammary stem cells in suspension culture: implications for stem cell biology and neoplasia. J. Mammary Gland Biol. Neoplasia. 2005;10:75–86. doi: 10.1007/s10911-005-2542-5. [DOI] [PubMed] [Google Scholar]
- Feng X., Xu X., Xiao X., Zou K., Yu W., Wu J., Tang R., Gao Y., Hao J., Zhao X. NMI inhibits cancer stem cell traits by downregulating hTERT in breast cancer. Cell Death Dis. 2017;8:e2783. doi: 10.1038/cddis.2017.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.Z., Yan Y.Y., He M., Xiao Q.H., Yao W.F., Zhao L., Wu H.Z., Yu Z.J., Zhou M.Y., Lv M.T., Zhang S.S., Chen J.J. Salinomycin induces selective cytotoxicity to MCF-7 mammosphere cells through targeting the Hedgehog signaling pathway. Oncol. Rep. 2016;35:912–922. doi: 10.3892/or.2015.4434. [DOI] [PubMed] [Google Scholar]
- Grimshaw M.J., Cooper L., Papazisis K., Coleman J.A., Bohnenkamp H.R., Chiapero-Stanke L., Taylor-Papadimitriou J., Burchell J.M. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien P., Lo S., Albain K.S., Robinson P., Rajan P., Strack P.R., Golde T.E., Miele L., Foreman K.E. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 2010;30:3853–3867. [PubMed] [Google Scholar]
- Gu W., Prasadam I., Yu M., Zhang F., Ling P., Xiao Y., Yu C. Gamma tocotrienol targets tyrosine phosphatase SHP2 in mammospheres resulting in cell death through RAS/ERK pathway. BMC Cancer. 2015;15:609. doi: 10.1186/s12885-015-1614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.B., Onder T.T., Jiang G., Tao K., Kuperwasser C., Weinberg R.A., Lander E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla I.K., Phoenix K.N., Hong X., Tirnauer J.S., Claffey K.P., White B.A. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res. Treat. 2012;132:75–85. doi: 10.1007/s10549-011-1534-y. [DOI] [PubMed] [Google Scholar]
- Hinohara K., Kobayashi S., Kanauchi H., Shimizu S., Nishioka K., Tsuji E., Tada K., Umezawa K., Mori M., Ogawa T. ErbB receptor tyrosine kinase/NF-kappaB signaling controls mammosphere formation in human breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:6584–6589. doi: 10.1073/pnas.1113271109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Zhang Y., Wang S.J., Ge H.L., Zhao G.P., Xu Y.C., Wang Y. CD44hiCD24lo mammosphere-forming cells from primary breast cancer display resistance to multiple chemotherapeutic drugs. Oncol. Rep. 2016;35:3293–3302. doi: 10.3892/or.2016.4739. [DOI] [PubMed] [Google Scholar]
- Kim W.T., Ryu C.J. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017;50:285–298. doi: 10.5483/BMBRep.2017.50.6.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.N., Koo K.H., Sung J.Y., Yun U.J., Kim H. Anoikis resistance: an essential prerequisite for tumor metastasis. Int. J. Cell Biol. 2012;2012:306879. doi: 10.1155/2012/306879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopp A.H., Lacerda L., Gupta A., Debeb B.G., Solley T., Li L., Spaeth E., Xu W., Zhang X., Lewis M.T. Mesenchymal stem cells promote mammosphere formation and decrease E-cadherin in normal and malignant breast cells. PLoS One. 2010;5:e12180. doi: 10.1371/journal.pone.0012180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruyt F.A.E., Schuringa J.J. Apoptosis and cancer stem cells: implications for apoptosis targeted therapy. Biochem. Pharmacol. 2010;80:423–430. doi: 10.1016/j.bcp.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Liu Y., Nenutil R., Appleyard M.V., Murray K., Boylan M., Thompson A.M., Coates P.J. Lack of correlation of stem cell markers in breast cancer stem cells. Br. J. Cancer. 2014;110:2063–2071. doi: 10.1038/bjc.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Clauser K.R., Tam W.L., Fröse J., Ye X., Eaton E.N., Reinhardt F., Donnenberg V.S., Bhargava R. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell. Biol. 2014;16:1105–1117. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R., Karthik G.M., Lövrot J., Haglund F., Rosin G., Katchy A., Zhang X., Viberg L., Frisell J., Williams C. Estrogen receptor beta as a therapeutic target in breast cancer stem cells. J. Natl. Cancer Inst. 2017;109:1–14. doi: 10.1093/jnci/djw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel Iglesias J., Beloqui I., Garcia-Garcia F., Leis O., Vazquez-Martin A., Eguiara A., Cufi S., Pavon A., Menendez J.A., Dopazo J. Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PLoS One. 2013;8:e77281. doi: 10.1371/journal.pone.0077281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema J.P. Cancer stem cells: the challenges ahead. Nat. Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- Montales M.T., Rahal O.M., Kang J., Rogers T.J., Prior R.L., Wu X., Simmen R.C. Repression of mammosphere formation of human breast cancer cells by soy isoflavone genistein and blueberry polyphenolic acids suggests diet-mediated targeting of cancer stem-like/progenitor cells. Carcinogenesis. 2012;33:652–660. doi: 10.1093/carcin/bgr317. [DOI] [PubMed] [Google Scholar]
- Morata-Tarifa C., Jimenez G., Garcia M.A., Entrena J.M., Grinan-Lison C., Aguilera M., Picon-Ruiz M., Marchal J.A. Low adherent cancer cell subpopulations are enriched in tumorigenic and metastatic epithelial-to-mesenchymal transition-induced cancer stem-like cells. Sci. Rep. 2016;6:18772. doi: 10.1038/srep18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison B.J., Hastie M.L., Grewal Y.S., Bruce Z.C., Schmidt C., Reynolds B.A., Gorman J.J., Lopez J.A. Proteomic comparison of MCF-7 tumoursphere and monolayer cultures. PLoS One. 2012;7:e52692. doi: 10.1371/journal.pone.0052692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H., Srour E.F., Badve S. Breast cancer stem cells and intrinsic subtypes: controversies rage on. Curr. Stem Cell Res. Ther. 2009;4:50–60. doi: 10.2174/157488809787169110. [DOI] [PubMed] [Google Scholar]
- Niit M., Hoskin V., Carefoot E., Geletu M., Arulanandam R., Elliott B., Raptis L. Cell-cell and cell-matrix adhesion in survival and metastasis: stat3 versus Akt. Biomol. Concepts. 2015;6:383–399. doi: 10.1515/bmc-2015-0022. [DOI] [PubMed] [Google Scholar]
- Olsson E., Honeth G., Bendahl P.O., Saal L.H., Gruvberger-Saal S., Ringnér M., Vallon-Christersson J., Jönsson G., Holm K., Lövgren K. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer. 2011;11:418. doi: 10.1186/1471-2407-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Rameshwar P. Tumorsphere passage for breast cancer stem cells. Protoc. Exch. 2013 [Google Scholar]
- Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M.A., Daidone M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Reynolds D.S., Tevis K.M., Blessing W.A., Colson Y.L., Zaman M.H., Grinstaff M.W. Breast cancer spheroids reveal a differential cancer stem cell response to chemotherapeutic treatment. Sci. Rep. 2017;7:10382. doi: 10.1038/s41598-017-10863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota L.M., Lazzarino D.A., Ziegler A.N., LeRoith D., Wood T.L. Determining mammosphere-forming potential: application of the limiting dilution analysis. J. Mammary Gland Biol. Neoplasia. 2012;17:119–123. doi: 10.1007/s10911-012-9258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw F.L., Harrison H., Spence K., Ablett M.P., Simoes B.M., Farnie G., Clarke R.B. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J. Mammary Gland Biol. Neoplasia. 2012;17:111–117. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- Sheridan C., Kishimoto H., Fuchs R.K., Mehrotra S., Bhat-Nakshatri P., Turner C.H., Goulet R., Jr., Badve S., Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes B.M., O'Brien C.S., Eyre R., Silva A., Yu L., Sarmiento-Castro A., Alférez D.G., Spence K., Santiago-Gómez A., Chemi F. Anti-estrogen resistance in human breast tumors is driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell Rep. 2015;12:1968–1977. doi: 10.1016/j.celrep.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart C.E., Morrison B.J., Saunus J.M., Vargas A.C., Keith P., Reid L., Wockner L., Askarian-Amiri M., Sarkar D., Simpson P.T. In vitro analysis of breast cancer cell line tumourspheres and primary human breast epithelia mammospheres demonstrates inter- and intrasphere heterogeneity. PLoS One. 2013;8:e64388. doi: 10.1371/journal.pone.0064388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert W.R., Hitt M.M., Feder J. Cell aggregate suspension culture for large-scale production of biomolecules. In Vitro. 1980;16:486–490. doi: 10.1007/BF02626461. [DOI] [PubMed] [Google Scholar]
- Velasco-Velazquez M.A., Homsi N., De La Fuente M., Pestell R.G. Breast cancer stem cells. Int. J. Biochem. Cell Biol. 2012;44:573–577. doi: 10.1016/j.biocel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Lv Q., Meng W., Tan Q., Zhang S., Mo X., Yang X. Comparison of mammosphere formation from breast cancer cell lines and primary breast tumors. J. Thorac. Dis. 2014;6:829–837. doi: 10.3978/j.issn.2072-1439.2014.03.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yu Y., Tsuyada A., Ren X., Wu X., Stubblefield K., Rankin-Gee E.K., Wang S.E. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470–1480. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J., Dewi D.L., Fredebohm J., Muller-Decker K., Flechtenmacher C., Hoheisel J.D., Boettcher M. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 2013;15:R109. doi: 10.1186/bcr3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Chen Y., Yao Y., Zhang H., Wang T. Increased invasion and tumorigenicity capacity of CD44+/CD24- breast cancer MCF7 cells in vitro and in nude mice. Cancer Cell Int. 2013;13:62. doi: 10.1186/1475-2867-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Song C., Ma Y., Tang L., Xu Y., Wang H. Effect of fibroblasts on breast cancer cell mammosphere formation and regulation of stem cell-related gene expression. Int. J. Mol. Med. 2011;28:365–371. doi: 10.3892/ijmm.2011.700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A clonal, flat sheet of T47D cells curled at the edges is manually manipulated with a 10-μm pipette.