Abstract
Anastasis is a natural cell recovery phenomenon that rescues cells from the brink of death. Programmed cell death such as apoptosis has been traditionally assumed to be an intrinsically irreversible cascade that commits cells to a rapid and massive demolition. Interestingly, recent studies have demonstrated recovery of dying cells even at the late stages generally considered immutable. Here, we examine the evidence for anastasis in cultured cells and in animals, review findings illuminating the potential mechanisms of action, discuss the challenges of studying anastasis and explore new strategies to uncover the function and regulation of anastasis, the identification of which has wide-ranging physiological, pathological and therapeutic implications.
Keywords: anastasis, apoptosis, mutagenesis, programmed cell death, reversal of apoptosis, reversal of cell death process
1. Introduction
Programmed cell death is an essential component of life. Among over 20 forms of programmed cell death that have been proposed [1–3], apoptosis is by far the most well studied for its regulatory mechanisms in cell suicide, and essential roles in embryonic development and normal homeostasis by eliminating the unwanted, injured or dangerous cells in the body [4–6]. While genetic or pharmaceutical manipulation can allow dying cells to survive that otherwise would normally die [7–9], initiation of apoptosis is generally believed to represent an irreversible commitment to cell death [10,11]. Events canonically marking this ‘point of no return’ in apoptosis include release of mitochondrial cytochrome c into the cytosol [12–17], activation of execution caspase proteases [18–20], and their associated or downstream events [1], such as DNA damage, cell surface exposure of ‘eat me’ signals, cell shrinkage and fragmentation of the cell body. However, a growing body of evidence has demonstrated that dying cells can exit the initiated death process and recover, even at late stages generally accepted as beyond the ‘point of no return’. We coined a term anastasis (Αναστάσης), which means ‘rising to life’ in Greek [21], to describe the recovery of dying cells after brink of death, using reversal of apoptosis as the first example. More recent studies expand the anastasis field that dying cells can recover following important cell death hallmarks of apoptosis and potentially other forms of cell death process (figure 1 and table 1). Removal of the death stimulus is sufficient to allow anastasis to occur in vitro and in vivo, indicating that anastasis is an intrinsic recovery phenomenon. In this review, we gather and summarize the strength of evidence supporting anastasis, discuss the possible mechanisms of anastasis at each step of the cell death process and address the key challenges of studying and furthering our understanding of anastasis. Given the novelty of this field, we also look forward to some of the potential physiological, pathological and therapeutic implications of anastasis, and propose future directions for the study of anastasis.
Figure 1.
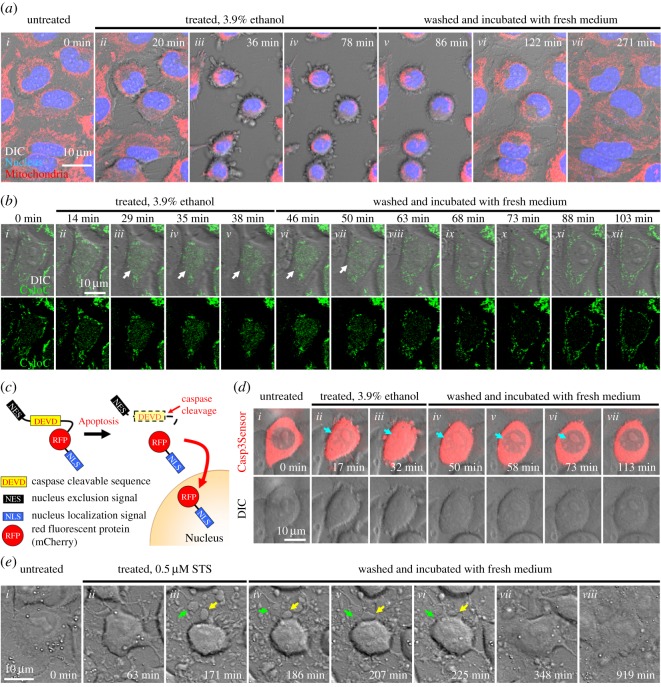
Cell recovery after attempted late apoptosis. (a) Time-lapse live cell confocal microscopy showing reversal of apoptosis in HeLa cells. To visualize mitochondria and nucleus, cells were stained with MitoTracker (red fluorescence) and Hoechst (blue fluorescence) for confocal microscopy, respectively. Cell morphology was observed with differential interference contrast (DIC) microscopy. In healthy cells, mitochondria formed a tubular network which extended throughout the cytoplasm (i). During exposure to a cell death stimulus of 3.9% ethanol, cells displayed morphological hallmarks of apoptosis including mitochondrial fragmentation, nuclear condensation, cell shrinkage, and plasma membrane blebbing (ii–iv). After washing and incubating the apoptotic dying cells with fresh culture medium, reversal of apoptosis occurred as indicated by morphological recovery of the cells (v–vii). The original movie can be viewed in electronic supplementary material. (b) Recovery of an apoptotic cell after mitochondrial cytochrome c release to cytosol. Time-lapse live cell confocal microscopy of a HeLa cell expressing a fusion protein of cytochrome c-GFP (CytoC). Before cell death induction, cytochrome c localized in tubular mitochondria (i). During apoptosis induced by 3.9% ethanol, cytochrome c was released to cytosol (ii–v). After removal of the cell death inducer, cytosolic cytochrome c was reduced in the recovered cell (vi–xii). Merged images of cytochrome c-GFP (green fluorescence) and DIC for cell morphology (top row), and images of cytochrome c-GFP only (bottom row). White arrows indicate cytosolic signal of cytochrome c-GFP. (c) Schematic diagram of a caspase-3 biosensor fusion protein NES-DEVD-RFP-NLS (Casp3Sensor). (d) Recovery of an apoptotic cell after caspase-3 activation. Time-lapse live cell confocal microscopy of a HeLa cell expressing fusion protein of Casp3Sensor. In the healthy cell, the Casp3Sensor localized in cytosol (i). During apoptotic induction of 3.9% ethanol, the caspase-3 cleaved biosensor translocated from cytosol to nucleus (ii,iii). After removal of the death stimulus, the signal of nuclear Casp3Sensor was diminished in the recovered cell (iv–vii). Merged images of Casp3Sensor (red fluorescence) and DIC for cell morphology (top row), and images of DIC only (bottom row). Blue arrows indicate nuclear signal of Casp3Sensor. (e) Apoptotic bodies fuse to cell body during cell recovery. Time-lapse live cell DIC microscopy of a healthy HeLa cell (i), the same cell after treating with a cell death stimulus of 0.5 µM staurosporine (STS; ii,iii), and then being washed and further incubated with fresh culture medium to remove the staurosporine (iv–viii). Green and yellow arrows indicate two apoptotic bodies that fused to a cell body during recovery of the cell.
Table 1.
Reversal of cell death events.
| cell death events | references |
|---|---|
| externalization of phosphatidylserine (in vitro) | [21–24] |
| externalization of phosphatidylserine (in vivo) | [25] |
| cytochrome c release | [26] |
| incomplete MOMP | [27] |
| mitochondrial fragmentation | [21,26,28,29] |
| caspase activation (in vitro) | [21,26,28–33] |
| caspase activation (in vivo) | [34,35] |
| plasma membrane blebbing | [21,26,28,29] |
| cell shrinkage | [21,24,26,28,29,31,32] |
| DNA damage | [21,27,30] |
| nuclear condensation | [21,26,28,29,32] |
| RIPK3 activation | [24,36] |
| apoptotic body formation and cell fragmentation | [26] |
2. Cell death process: considered irreversible
Apoptosis is executed by the sophisticated cellular demolition mechanisms, in which mitochondrial cytochrome c release and caspase activation are critical steps in this process of cell suicide [12–20]. During apoptosis, pro-apoptotic cell death factors translocate to and fragment mitochondria, leading to mitochondrial outer membrane permeabilization (MOMP), which releases apoptogenic factors into the cytosol [14,37–39]. These factors include cytochrome c to initiate the caspase protease cascade [40,41], Smac/DIABLO to suppress the inhibitor of apoptosis protein (IAP) for enhancing caspase activation [42,43], and specific DNases for apoptosis such as apoptosis-inducing factor (AIF) and endonuclease G (EndoG), which enzymatically cleave the genome [44–46]. Activated caspases mediate apoptosis by directly and indirectly cleaving hundreds of cellular substrates. For example, caspases activate DNA fragmentation factor/caspase-activated DNase (DFF40/CAD) that destroys the genome by cleaving its inhibitor, DFF45/ICAD [47,48], and cleave DNA-repairing enzyme Poly(ADP)-ribose polymerase-1 (PARP) that plays a critical role in maintaining genomic stability [49,50]. Activated caspases also cleave flippases at the plasma membrane, leading to cell surface exposure of phosphatidylserine, which then acts as an ‘eat me’ signal recognized by phagocytic cells [51]. Caspase cleavage of cytoskeletons and their regulators contributes to plasma membrane blebbing, cell shrinkage and fragmentation [52–60], signalling and facilitating the phagocytosis of apoptotic cells and recycling of their contents [4,61,62].
Importantly, apoptosis is a rapid and massive cellular destruction process [63]. The process to activate apoptosis is multivariate, requiring minutes to days or even longer after a death stimulus is applied. Once initiated, pro-apoptotic cell death factors such as BAX translocate to and fragment mitochondria within 15 min [64,65], leading to mitochondrial damage and release of apoptogenic factors including cytochrome c and SMAC to occur within 1–5 min [66–68], followed by rapid caspase activation and morphological features of apoptosis, including nuclear condensation, plasma membrane blebbing and cell shrinkage within 10–15 min [69–71]. While activated caspases execute cellular destruction by proteolysis of functional and structural components, apoptotic events also render mitochondria dysfunctional, disrupting cellular bioenergetics and metabolism [72–74]. Notably, mitochondrial damage or caspase activation alone is sufficient to cause cell death independently [18,39]. Therefore, apoptosis is generally considered to be irrevocable [10,11], especially at late times after these critical cell death-executing activities occur. However, recent studies reveal that recovery of dying cells is possible, even after reaching these critical cell death events.
3. Evidence and potential mechanisms of anastasis
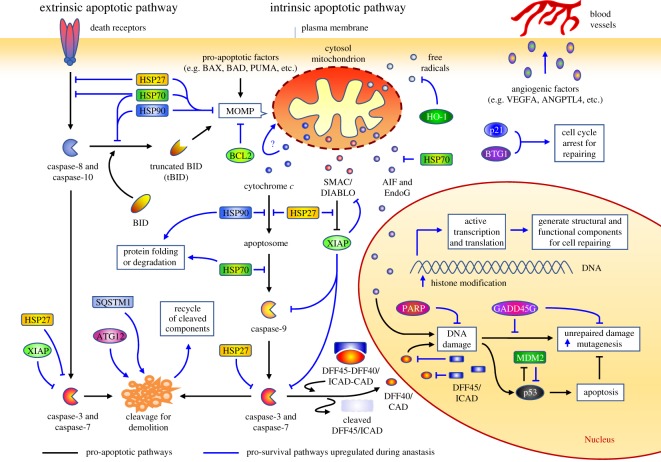
Can a dying cell recover from the brink of cell death after reaching the generally assumed ‘points of no return’? If so, how can a dying cell reverse a cell death decision? Recovery should involve arresting programmed death cascades, restoring normal cellular functions and repairing damage. While the precise mechanisms remain unclear, recent studies have demonstrated anastasis and provided new insights into the potential strategies possibly adopted by anastatic cells to halt and reverse the initiated cell death process (figure 2).
Figure 2.
Proposed mechanism of anastasis during cell recovery. Upregulation of pro-survival pathways identified during anastasis interact with the apoptosis network to suppress initiated death cascade and promote cell recovery.
3.1. Recovery after cytochrome c release
Considering that dysfunctional energy production of damaged mitochondria, initiation of the proteolytic caspase cascade and demolition of the genome can each independently cause cell death, recovery of dying cells after cytochrome c release by MOMP would seem unlikely [12–17]. Nevertheless, recent studies reveal an unexpected reversibility of apoptosis after mitochondrial fragmentation (figure 1a) and cytochrome c release (figure 1b). For example, single cell live microscopy study demonstrated that cytochrome c-releasing and mitochondria-fragmented dying cells are able to recover [26], even after plasma membrane blebbing, a downstream hallmark of effector caspase activation [57,58], is observed [26]. This suggests a reversibility of apoptosis even after these once considered irrevocable steps. Besides, recovery of dying cells after cytochrome c release could also occur in vivo. During heart failure, significant numbers of dying cardiomyocytes exhibit several hallmarks of apoptosis, including mitochondrial release of cytochrome c and activation of caspase-3, but these cells maintain normal nuclear morphology with the absence of terminal morphological features of apoptosis, indicating that the dying cells may not complete the apoptotic process [75–77]. This phenomenon, termed apoptosis interruptus, suggests possible arrest or interference with the apoptotic process [78], and therefore, could explain the clinical observation on recovery of cardiomyocytes during reversal of heart failure, in which the failing heart was unloaded with a left ventricular assist device to reduce the physical stress on the heart that can cause apoptosis [79,80].
Mitochondria serve as the powerhouses of the cell [14,81]. Release of cytochrome c from mitochondria to cytosol causes mitochondrial damage and dysfunction during apoptosis [14,82]. How can anastatic cells obtain energy to recover from apoptosis? By contrast to what was originally interpreted as an ‘all or nothing’ event [12,66,67], incomplete release of cytochrome c is observed in a subset of mitochondria in dying cells [26,83,84], possibly due to incomplete mitochondrial outer membrane permeabilization (iMOMP) [27]. This suggests that some remaining intact or partially functional mitochondria may be available to support early energy requirements of anastasis. At later times in anastasis, mitochondria show recovery, as determined by time-lapse live cell microscopy revealing fragmented mitochondria can fuse and regain their normal tubular structure (figure 1a) [21,26,28,29,85], thereby restoring the powerhouse to provide bioenergetics and metabolic support during subsequent recovery. Of note, some cell types, such as neurons and cardiomyocytes, are able to resist cell death induction when cytochrome c is introduced into the cytosol by microinjection [86,87], suggesting the existence of intrinsic mechanisms for handling the cytosolic cytochrome c without triggering inevitable cell death.
Logically, the recovering cells will need to inhibit cytochrome c release, degrade cytosolic apoptogenic factors and remove the damaged mitochondria to restore energy production, but the related molecular mechanism remains unclear. A recent time-course whole-genome gene expression microarray study provides mechanistic insights by identifying the molecular signature of anastasis in primary mouse liver cells, which reveals striking temporal changes of gene transcription during anastasis [29]. This signature included upregulation of the autophagy-related protein Atg12, autophagic adaptor protein Sqstm1, heat shock proteins Hspb1, Dnajb1, HSPa1a and Hsp90aa1 (HSP27, HSP40, HSP70 and HSP90, respectively, for human homologue), and Hmox1 for encoding for heme oxygenase (HO-1) [29]. ATG12 and SQSTM1 are key regulators of mitochondrial homeostasis and remove impaired mitochondria by mitophagy [88–91], suggesting that autophagy may be involved in degrading damaged mitochondria after cytochrome c release in anastatic cells. ATG12 also promotes the rapid degradation of cytosolic cytochrome c in the absence of caspase activation [92,93]. Besides, the chaperone proteins HSP27 and HSP70 have been shown to suppress the mitochondrial release of cytochrome c [94–98], and cooperate with HSP90 to inhibit cytochrome c-mediated caspase activation [99–105], thereby halting further caspase activation. Studies also demonstrated that HSP70 and HSP40 suppress mitochondrial translocation of pro-apoptotic BAX and caspase-8-mediated activation of pro-apoptotic BCL-2 family member BID [106,107], while HSP27 suppresses mitochondrial translocation of the cleaved BID [94], thereby preventing further MOMP-mediated leakage of apoptogenic factors. HSP70 also interacts with and suppresses AIF and EndoG, preventing DNA destruction after MOMP [108–110]. Expression of heme oxygenase could also promote cell survival by removing free radicals that are generated during apoptosis [111]. Collectively, the upregulation of this set of genes during anastasis suggests candidate regulators that degrade released cytochrome c, suppress the activated executioners and restore a functional network of healthy mitochondria that is essential for energy production.
3.2. Arrest of activated caspase cascade
During the execution phase of apoptosis, activated caspases carry out the proteolytic destruction of key functional and structural components of the cell [18,20]. This rapid and massive cellular demolition is initiated and amplified through multiple apoptotic pathways mediated by caspases (figure 2). Activation of caspases ultimately leads to fragmentation of organelles [18,20,38], destruction of the genome by activating apoptotic DNases [47,48], cell surface exposure of phagocytic signals [51] and formation of apoptotic bodies in preparation for the phagocytic clearance of dead cells [4,61,62]. Activated caspases also cleave BID, which subsequently translocates to mitochondria to further promote MOMP to accelerate the apoptotic cascade [112–116]. Given the critical role of caspases in initiating and amplifying death signals that destroy or cripple a multitude of cellular processes, apoptosis used to be considered to be inevitable after caspase activation [18–20].
Interestingly, recent studies demonstrate that reversal of apoptosis can actually occur after caspase activation in cultured cells and in live animals. Time-lapse live cell microscopy of human cervical cancer HeLa cells expressing a caspase-3 biosensor detected caspase activation after ethanol-induced apoptosis, and showed that the same cells were able to recover and eliminate the cleavage-activated biosensor after being washed and incubated with fresh medium (figure 1c,d) [21,26]. These caspase-activated cells can also recover after mitochondrial fragmentation, cytosolic and nuclear condensation, plasma membrane blebbing and cell shrinkage [21,26,85], thereby indicating that apoptosis can be reversible at the cell execution stage.
It is technically challenging to demonstrate anastasis in animals, because cells that underwent anastasis can be morphologically indistinguishable from surrounding healthy cells. To overcome this difficulty, an in vivo CaspaseTracker biosensor was developed [34], which can be activated by effector caspase to trigger permanent fluorescent protein expression within anastatic cells and their progeny, allowing the tracking of cell fate [34]. Studies done in vivo demonstrate that after transiently exposing Drosophila melanogaster transgenic for CaspaseTracker biosensor to environmental insults, such as protein starvation or cold shock that trigger apoptosis in various tissues including egg chambers (somatic and germ cells), fluorescent protein was permanently expressed in these cells of the recovered flies, but not in those of untreated control flies [34,85]. This indicates that anastasis can occur after caspase activation in animals. Notably, this biosensor is also activated both during and after development [34,35,85], suggesting a potential involvement of anastasis during embryogenesis and normal homeostasis.
Broadening studies of non-apoptotic caspase activity reveal the existence of a still poorly understood strategy for cells to handle the activated caspases without the cells being killed. These include regulation of neuronal activity [117–122], spermatid individualization [123,124], microRNA processing [125], cell proliferation [126] and cell fate patterning [127]. Remarkably, cultured cells have been shown to tolerate sublethal caspase activity without triggering apoptosis [30,128–130]. These studies indicate that caspase activation is not the ‘point of no return’. Although it is still unclear whether cells use the same or different mechanism to manage the activated caspases for anastasis and non-apoptotic caspase activity, several genes that are specifically upregulated during reversal of apoptosis could serve as suppressors of the caspases [29]. For example, HSP27 has been shown to bind procaspase-3 and inhibit the caspase-9-mediated proteolysis required for the activation of caspase-3 [100], while HSP27, HSP70 and HSP90 can suppress apoptosome formation and activation of caspase-9 [99–105], thus preventing further activation and amplification of the caspase cascade. Upregulation of murine double minute (MDM2), an inhibitor of p53 [131–133], occurs in anastatic cells [29], and that could suppress the p53-mediated pro-apoptotic DNA damage response during reversal of apoptosis. Expression of MDM2 can also activate the X-linked inhibitor of apoptosis protein (XIAP) [134], which arrests the death cascade by inhibiting the activated initiator and executioner caspases [135–140], and promoting the degradation of Smac/DIABLO [141,142]. Transcription is also active during anastasis [21,29,31], and that can generate building blocks for the recovering cells to repair damage and regain normal morphology. Interestingly, anastatic cells also display upregulation of potent angiogenic factors, such as ANGPTL4 and VEGFA [29], which promote angiogenesis and vascular permeability [143–146]. This could enhance anastasis by facilitating the nutrient supply and removal of cellular wastes such as those generated by degradation of the caspase-cleaved products, but this has not yet been confirmed experimentally.
3.3. Repairing DNA damage
DNA damage is a hallmark of apoptosis, executed by apoptotic DNases such as AIF and EndoG being released from the mitochondria [44–46], and DFF40/CAD activated by caspases that cleave its inhibitor of DFF45/ICAD [47,48]. Executioner caspases also abolish the DNA repair system in dying cells by cleaving the DNA-repair enzyme PARP [49,50].
Anastasis can occur at this late stage in the cell death process. Mouse primary liver cells and non-cancerous NIH3T3 fibroblasts that had undergone apoptosis following exposure to ethanol, a cell death inducer, and showed mitochondrial release of AIF and EndoG, cleavage of ICAD and PARP and breakage of DNA strands, were able to reverse apoptosis after removing the death inducer and restoring normal culture conditions [21]. Although recovery is possible, some surviving cells acquired new chromosomal abnormalities and underwent an oncogenic transformation, as indicated by loss of contact inhibition of growth (focus formation) and anchorage-independent growth (proliferation in soft agar) [21]. An increased frequency of cells with micronuclei, a biomarker of DNA damage [11,147], has also been observed in both primary cells and cancer cell lines that recovered from apoptosis [21,29].
DNA damage and oncogenic transformation can also be promoted by iMOMP and sublethal activation of caspase-3, presumably through apoptotic DNases, whereas suppressing caspase activation or apoptotic DNase activity has the opposite effect of inhibiting transformation [28,30]. Studies with leukaemia cell lines also showed that sublethal apoptosis induction promotes MLL gene translocations, TEL breaks and formation of TEL-AML1 fusion [148–150], which are often detected during cancer progression [151,152]. These observations support the notion that activation of apoptotic machines can promote mutagenesis by damaging the genome when the cells attempt apoptosis but do not die, and then recover [21]. Therefore, it is possible that DNA damage sustained during apoptosis may not be appropriately repaired during anastasis, leading to the acquisition of new mutations.
How can dying cells with damaged DNA survive through anastasis? One possibility is that during anastasis, upregulation of the ubiquitin ligase MDM2 occurs [29], leading to degradation of the tumour suppressor p53, thereby suppressing the pro-apoptotic p53-mediated DNA damage response [131–133]. HSP70 upregulated during anastasis may also interact with and suppress AIF and EndoG to stop DNA destruction initiated after MOMP [29,108–110]. While being cleaved by caspases during apoptosis, the expression of ICAD and PARP has been shown to return to pre-apoptosis levels during anastasis [21], enabling ICAD to inhibit the DNase activity of CAD, and PARP to repair DNA damage in anastatic cells. In addition, cell cycle arrest is important for DNA repair [153,154], and upregulation of the cell cycle arrest genes such as Btg1, Cdkn1a and Trp53inp1 occurs during anastasis [29].
3.4. Reunion after formation of apoptotic bodies
Remarkably, anastasis can still occur even after the fragmentation of the dying cells [26,85]. Time-lapse live cell microscopy has demonstrated that HeLa cell fragments resulting from the formation of apoptotic bodies can coalesce to an apparently normal morphology after removing the cell death inducer staurosporine (figure 1e) [85]. A similar coalescence of fragmented cells has also been observed with the cultured human small cell lung carcinoma H446 cell line after recovery from ethanol-induced apoptosis [26]. As mentioned above, recovered cells often display an increased number of micronuclei and chromosomal abnormalities [21,26,29], suggesting unrepaired DNA damage after anastasis. Failure of apoptotic bodies with broken chromosomes to faithfully reassociate during anastasis may lead to increased aneuploidy [21,26].
It is unclear how the pieces of fragmented apoptotic cells can coalesce back to a seemingly normal structure. Recent studies have revealed that surface exposure of phosphatidylserine may be a key requirement for different types of cell fusion, including macrophage polykaryon formation [155], myotube formation [156,157], viral infection [158] and fusion of severed axons [159]. This leads to speculation that the fragmented cells with externalization of PS could use this strategy for fusion and repair during anastasis.
Under normal physiological conditions, apoptotic body formation is followed by phagocytosis. Perhaps related to recovery after the formation of apoptotic bodies may be the intriguing question of whether engulfed cell fragments can undergo anastasis. Internalized living cells undergoing entosis have been shown to escape their cannibalized state, ultimately surviving and proliferating [160]. However, entosis is not typical phagocytosis and is distinguished as an invasion of a living cell into the cytoplasm of another cell [160], and so it may not represent how macrophages engulf apoptotic cell fragments [4,38].
3.5. Removal of externalized phosphatidylserine
Under normal conditions, phosphatidylserine (PS) is restricted to the inner leaflet of the plasma membrane. During apoptosis, however, PS becomes exposed on the outer surface of the cell and represents an ‘eat me’ signal that facilitates rapid engulfment by phagocytes [61,62]. Despite the deployment of surface PS, dying cells can undergo anastasis. For example, apoptotic BCL1.3B3 B lymphoma cells (induced by pro-apoptotic anti-immunoglobulin antibodies [22]) or mouse mammary carcinoma MOD cells (with temperature-sensitive p53 and induced by shift to permissive temperature [23]) were able to proliferate after removing the death inducer, indicating that anastasis can occur after surface exposure of PS [22,23]. Anastasis after the PS exposure has also been observed in neonatal rat primary cardiomyocytes and the mouse cardiac muscle HL1 cell line after transient cell death induction of ethanol [21,26].
It is expected that anastatic cells need to remove this ‘eat me’ signal in order to escape phagocytosis, and there is evidence that this can happen. Recovering cells have been shown to retain surface-exposed PS for only a few hours, suggesting that the removal of previously exposed PS is an active process during anastasis [21,26], although the exact molecular mechanism is unclear. Recovery after PS exposure has also been observed in live animals. PS identified by annexin V in rabbit and mouse cardiomyocytes recovering from apoptosis induced by transient ischaemic injury, was detected inside the surviving cells [25]. This suggests the internalization of PS in the anastatic cells to its pre-apoptotic plasma membrane location, rather than, for example, engulfment of dead cells by their neighbouring cells, but this interpretation requires further evaluation and corroboration.
4. Remaining key questions and challenges
Anastasis is a new field, and the current gaps in our knowledge raise fundamental questions about which kinds of cell death processes are reversible, what strategies can be developed to overcome the challenges of identifying the functions and regulatory pathways of anastasis, and how knowledge of anastasis might be translated to pharmacological intervention.
Anastasis is a term coined to describe the cell recovery phenomenon that reversal of apoptosis has been reported as the first example [21]. Interestingly, recent subsequent studies suggest that other forms of cell death may also be reversible. Apart from apoptosis, the cell death stimulus staurosporine induces necroptosis [161,162], starvation triggers autophagy [163,164] and cold shock leads to necrotic injury characterized by plasma membrane rupture [34,165–167], and interestingly recovery of these dying cells has been observed in vitro or in vivo after being returned to optimal culture conditions [28,34,85]. In addition, and although genetic manipulation is required, RIPK3-activable biosensor-induced necroptotic cells have been shown to recover when the biosensor was turned off [24], further supporting that necroptosis is reversible. Research extending the study of anastasis to different forms of cell death can be expected to provide a deeper understanding of its functions, mechanisms and consequences.
Two major challenges complicate studying the reversibility of cell death processes. First, a single cell death inducer can activate multiple cell death pathways, in which interconnections and interactions allow for complicating ‘cross talk’ among them [161–168], making it difficult to test the reversibility of one specific pathway. To solve this problem, suppression of key regulators of specific cell death pathways, such as caspases for apoptosis, MLKL for necroptosis and LC3 for autophagy, will be required to isolate pathways and determine which kinds of cell death are reversible after induction by ‘broad-spectrum’ death conditions. Second, it remains technically challenging to detect and track anastasis, especially in animals where histologically recognizable markers of apoptosis disappear after anastasis, and there is not yet a specific biomarker identified for anastasis. The recent development of the CaspaseTracker biosensor provided a novel strategy to identify apoptotic dying cells and track the fate of anastatic cells in live animals [34,35,85], but is unable to unambiguously distinguish between apoptotic and non-apoptotic caspase activation [161–167,169], making the biosensor not necessarily apoptosis-specific. Development of a trackable system to specifically follow anastatic cells recovered after different kinds of cell death will facilitate these in vivo studies.
Developing and refining pharmacological agents and other strategies to modulate anastasis requires a solid understanding of its molecular mechanism. This is an area of ongoing investigation. Whole-genome gene expression studies have been performed on anastasis in mouse primary liver cells [29] and human cervical cancer HeLa cells [31] after ethanol-induced apoptosis, as well as in mouse NIH3T3 fibroblasts after RIPK3-activated biosensor-induced necroptosis [36], but the putative master regulators of the process remain elusive. It is possible that anastasis is mediated by multiple pathways rather than by a specific master regulator. Future studies of post-translational and epigenetic influences on anastasis will provide new insights into its regulation.
The existence of a definite irreversible checkpoint for cell death is also not known. Dying cells undergoing fragmentation and membrane permeabilization are generally expected to commit demise, but as we have discussed, some cells can still recover [26,34,85], suggesting that there is a new tipping point. While the mechanism is not known, it is possible that cell-specific thresholds exist for survival; for example, cytosolic microinjection of cytochrome c induces death in the neonatal rat dermal fibroblasts but not the rat cardiomyocytes [87]. Some cells, for example neurons, can also avoid initiation of the death cascade or live with activated cell executioners such as caspases [34], although this is not well understood [119,170]. Clearly, future work is required to fully define the checkpoint of cell death and anastasis.
5. Looking forward
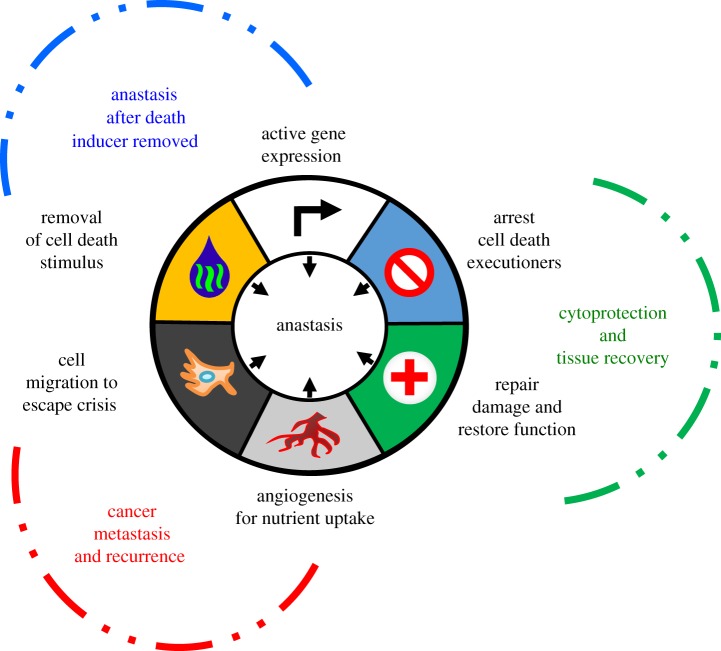
There are numerous gaps in our current knowledge of anastasis. Here, we discuss the potential biological significance of this process (figure 3), though we must stress that further research is needed in the area. Anastasis may serve as a pro-survival force to balance apoptosis, and may therefore participate in the regulation of cell death and survival during embryonic development and normal homeostasis, as well as other physiological and pathological conditions.
Figure 3.
Emerging hallmarks of anastasis. This illustration encompasses the proposed features and consequences of anastasis.
Promoting anastasis may represent as a previously unrecognized beneficial mechanism of preserving differentiated cells that are difficult to replace, such as neurons and cardiomyocytes. For example, in flies, rats and rabbits, photoreceptor cells die if they are exposed to constant light, but this retinal degeneration can be reversed within the first few days by returning the animals to normal light–dark cycles [171–173]. As the proliferation of photoreceptor cells is not expected in the retina, anastasis may protect the retinal function by aiding the survival of severely injured photoreceptor cells, thus providing a novel approach to regenerative medicine. As another example, following brain injury neuronal cell death is mediated by cell death programmes that include apoptosis, as evidenced by caspase-3 and calpain activation [174]. Treatment of such traumas with anti-apoptotic erythropoietin and therapeutic hypothermia can reduce the extent of damage and improve functional outcomes [175,176], identifying apoptosis as a therapeutic target and raising the possibility that promoting anastasis might further enhance recovery from brain injury by rescuing dying neurons. As a third example, apoptosis occurring in cardiomyocytes of failing hearts [177,178], appears possible to arrest or reverse well beyond the expected ‘point of no return,’ as discussed above. It has been proposed that damaged heart tissue may undergo repair by phagocytic removal of the apoptotic cells and replacement by therapeutically induced compensatory division of healthy cardiomyocytes [179–181]. Anastasis may have a role in this process by limiting permanent damage and promoting cell survival following transient cell death-inducing stresses to the heart [78]. Anastasis can also occur in cultured primary liver cells [21,29], suggesting that it may augment recovery after liver injury as an additional or alternative mechanism other than cell division for liver regeneration [182].
Inhibiting anastasis may also have beneficial application. For example, cancer cells may employ anastasis as an escape tactic to survive cell death-inducing anti-cancer therapy, possibly contributing to cancer recurrence. Chemotherapy and radiotherapy kill cancer cells by inducing various types of cell death including apoptosis [183,184]. Primary cancers often exhibit dramatic initial responses to such therapies [185–189]. However, most metastatic cancers, including lung, brain, skin and pancreatic cancers, inevitably recur, leading to treatment failure [185–189]. Anastasis has been observed in various cultured human cancer cell lines including cervical cancer, small cell lung carcinoma, neuroblastoma, skin cancer, testicular cancer, liver cancer, breast cancer and prostate cancer [21,26,28,29,31–33,85], thereby suggesting that this process could be a common occurrence in cancers. Furthermore, upregulation of genes involved in cell migration (MMP9, MMP10 and MMP13) and angiogenesis (ANGPTL4, ANGPT2 and VEGFA) during anastasis [29] suggests a plausible connection between anastasis and cancer metastasis during a recurrence. More work is needed to determine whether this mechanism could contribute to cancer recurrence and spread, and whether targeting anastasis could provide a new strategy to fight cancer by suppressing the ability of apoptotic cancer cells to escape anti-cancer therapy.
As mentioned, some anastatic cells acquire permanent genetic changes or undergo an oncogenic transformation at a higher frequency than control cells that did not attempt cell death [21,26,29]. Therefore, anastasis may contribute to tumourigenesis by rescuing genetically damaged cells, potentially accounting for the observation that repeated tissue injury increases cancer risk. This has been observed in various tissues, such as alcohol-damaged liver [190,191] and chronic thermal esophageal injury caused by swallowing very hot beverages [192–194]. Moreover, anti-cancer therapy can trigger apoptosis in normal cells due to off-target effects, raising the possibility that anastasis to spare those cells following anti-cancer therapy could give rise to secondary cancers. For example, anastasis in haematopoietic stem cells could underlie the appearance of acute myeloid leukaemia arising after completion of anti-cancer treatments [195–197]. In that vein, anastasis occurring between cycles of genotoxic anti-cancer therapy could allow treatment-induced mutations to be perpetuated, leading to progression and evolution of drug resistance in recurrent cancers [185–189,198]. Thus, blocking or interfering with anastasis during anti-cancer therapy may offer a novel therapeutic strategy for preventing or arresting the progression of cancer and development of anti-cancer drug resistance.
In bacteria, yeast and plants, stress-induced mutagenesis is proposed to serve as an adaptive mechanism to cope with environmental changes by introducing new mutations for natural selection [199–202]. While stress-induced mutagenesis could benefit microorganisms and plants for the fitness of survival, it could be an unrecognized mechanism to cause genetic diseases when anastasis occurs in DNA-damaged germ cells. For example, epidemiological studies reveal that individuals born at the time of famines have a higher chance of developing transgenerational inheritable diseases such as breast cancer, coronary heart diseases, diabetes and obesity [203–210]. In response to environmental stresses such as starvation or temperature shock, germ cells undergo apoptosis, but fertility can resume shortly after the stressed animals are returned to a more normal environmental condition, presumably by restoring germ cell production [211–213]. Interestingly, recent studies demonstrated the occurrence of anastasis in germ cells of Drosophila after transient exposure to physiological and environmental stresses such as starvation or cold shock [34], thereby raising an intriguing possibility that recovered germ cells might be able to acquire new mutations from unrepaired DNA damage generated during apoptosis, leading to genetic alterations in their progeny [21,34]. If true, this mutagenetic mechanism might promote deleterious disease by causing changes along beneficial survival enhancing genetic alterations for natural selection, which could promote evolution especially in response to environmental changes.
6. Conclusion
The links between anastasis and tissue recovery, evolution of diseases and cell death decision remain to be elucidated. Further work is needed to determine whether anastasis is involved in these processes. Should they turn out to be true, studies on the physiological and pathological roles of anastasis could provide new insights into multidisciplinary fields of research that enhance our understanding in the control of cell death and survival, and also offer potential to identify new therapeutic approaches for cancers, heart failure, degeneration, tissue injury and regeneration medicine by mediating reversibility of cell death processes.
Supplementary Material
Acknowledgements
We invite and encourage others studying anastasis and related pathways to contact us for further discussion and idea exchange. We are grateful to Ralph Bohlmann and James Voelz for their ideas and suggestions for the word anastasis. We thank Ming Chiu Fung, Shenandoah Robinson and Kathleen Schwarz for valuable discussion of this work, and Douglas R. Green for providing HeLa cells stably expressing cytochrome c-GFP. Ho Lam Tang is an Investigator of The Hartwell Foundation.
Data accessibility
Data are available from the cited references and the electronic supplementary material.
Authors' contributions
H.M.T and H.L.T. designed study and wrote this review article.
Competing interests
We have no competing interests.
Funding
This work was supported by funding to H.L.T. through a National Cancer Institute Transition Career Development Award (CA204458) and a Hartwell Foundation Individual Biomedical Research Award.
References
- 1.Kroemer G, et al. 2009. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16, 3–11. ( 10.1038/cdd.2008.150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L, et al. 2012. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 19, 107–120. ( 10.1038/cdd.2011.96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluzzi L, et al. 2018. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541. ( 10.1038/s41418-017-0012-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr JF, Wyllie AH, Currie AR. 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257. ( 10.1038/bjc.1972.33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobson MD, Weil M, Raff MC. 1997. Programmed cell death in animal development. Cell 88, 347–354. ( 10.1016/S0092-8674(00)81873-5) [DOI] [PubMed] [Google Scholar]
- 6.Fuchs Y, Steller H. 2011. Programmed cell death in animal development and disease. Cell 147, 742–758. ( 10.1016/j.cell.2011.10.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddien PW, Cameron S, Horvitz HR. 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature 412, 198–202. ( 10.1038/35084096) [DOI] [PubMed] [Google Scholar]
- 8.Hoeppner DJ, Hengartner MO, Schnabel R. 2001. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412, 202–206. ( 10.1038/35084103) [DOI] [PubMed] [Google Scholar]
- 9.Martinou I, Desagher S, Eskes R, Antonsson B, Andre E, Fakan S, Martinou JC. 1999. The release of cytochrome c from mitochondria during apoptosis of NGF-deprived sympathetic neurons is a reversible event. J. Cell Biol. 144, 883–889. ( 10.1083/jcb.144.5.883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. The cell cycle and programmed cell death. Molecular Biology of the Cell, 4th edn, pp. 983–1026. New York, NY: Garland Science, Taylor and Francis Group. [Google Scholar]
- 11.Holland AJ, Cleveland DW. 2012. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 18, 1630–1638. ( 10.1038/nm.2988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinou JC, Desagher S, Antonsson B. 2000. Cytochrome c release from mitochondria: all or nothing. Nat. Cell Biol. 2, E41–E43. ( 10.1038/35004069) [DOI] [PubMed] [Google Scholar]
- 13.Green DR, Kroemer G. 2004. The pathophysiology of mitochondrial cell death. Science 305, 626–629. ( 10.1126/science.1099320) [DOI] [PubMed] [Google Scholar]
- 14.Ow YP, Green DR, Hao Z, Mak TW. 2008. Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9, 532–542. ( 10.1038/nrm2434) [DOI] [PubMed] [Google Scholar]
- 15.Bhola PD, Letai A. 2016. Mitochondria—judges and executioners of cell death sentences. Mol. Cell. 61, 695–704. ( 10.1016/j.molcel.2016.02.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kale J, Osterlund EJ, Andrews DW. 2018. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 25, 65–80. ( 10.1038/cdd.2017.186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burke PJ. 2017. Mitochondria, bioenergetics and apoptosis in cancer. Trends Cancer 3, 857–870. ( 10.1016/j.trecan.2017.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riedl SJ, Shi Y. 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5, 897–907. ( 10.1038/nrm1496) [DOI] [PubMed] [Google Scholar]
- 19.Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P. 2014. Cell death. Molecular Biology of the Cell, 6th edn., pp. 1021–1034. New York, NY: Garland Science, Taylor and Francis Group. [Google Scholar]
- 20.Julien O, Wells JA. 2017. Caspases and their substrates. Cell Death Differ. 24, 1380–1389. ( 10.1038/cdd.2017.44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang HL, et al. 2012. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Biol. Cell. 23, 2240–2252. ( 10.1091/mbc.E11-11-0926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammill AK, Uhr JW, Scheuermann RH. 1999. Annexin V staining due to loss of membrane asymmetry can be reversible and precede commitment to apoptotic death. Exp. Cell Res. 251, 16–21. ( 10.1006/excr.1999.4581) [DOI] [PubMed] [Google Scholar]
- 23.Geske FJ, Lieberman R, Strange R, Gerschenson LE. 2001. Early stages of p53-induced apoptosis are reversible. Cell Death Differ. 8, 182–191. ( 10.1038/sj.cdd.4400786) [DOI] [PubMed] [Google Scholar]
- 24.Gong YN, Guy C, Olauson H, Becker JU, Yang M, Fitzgerald P, Linkermann A, Green DR. 2017. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell 169, 286–300 e216. ( 10.1016/j.cell.2017.03.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenis H, et al. 2010. Annexin A5 uptake in ischemic myocardium: demonstration of reversible phosphatidylserine externalization and feasibility of radionuclide imaging. J. Nucl. Med. 51, 259–267. ( 10.2967/jnumed.109.068429) [DOI] [PubMed] [Google Scholar]
- 26.Tang HL, Tang HM, Hardwick JM, Fung MC. 2015. Strategies for tracking anastasis, a cell survival phenomenon that reverses apoptosis. J. Vis. Exp. 96, e51964 ( 10.3791/51964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichim G, et al. 2015. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol. Cell. 57, 860–872. ( 10.1016/j.molcel.2015.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang HL, Yuen KL, Tang HM, Fung MC. 2009. Reversibility of apoptosis in cancer cells. Br. J. Cancer 100, 118–122. ( 10.1038/sj.bjc.6604802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang HM, Talbot CC Jr, Fung MC, Tang HL. 2017. Molecular signature of anastasis for reversal of apoptosis. F1000Res. 6, 43 (doi:10.12688/f1000research.10568.2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SS, Xie X, Wong CS, Choi Y, Fung MC. 2014. HepG2 cells recovered from apoptosis show altered drug responses and invasiveness. Hepatobiliary Pancreat. Dis. Int. 13, 293–300. ( 10.1016/S1499-3872(14)60042-4) [DOI] [PubMed] [Google Scholar]
- 31.Liu X, He Y, Li F, Huang Q, Kato TA, Hall RP, Li CY. 2015. Caspase-3 promotes genetic instability and carcinogenesis. Mol. Cell. 58, 284–296. ( 10.1016/j.molcel.2015.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun G, Guzman E, Balasanyan V, Conner CM, Wong K, Zhou HR, Kosik KS, Montell DJ. 2017. A molecular signature for anastasis, recovery from the brink of apoptotic cell death. J. Cell Biol. 216, 3355–3368. ( 10.1083/jcb.201706134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y, So C, Lam HM, Fung MC, Tsang SY. 2018. Apoptosis reversal promotes cancer stem cell-like cell formation. Neoplasia 20, 295–303. ( 10.1016/j.neo.2018.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang HL, Tang HM, Fung MC, Hardwick JM. 2015. In vivo caspasetracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Sci. Rep. 5, 9015 ( 10.1038/srep09015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding AX, Sun G, Argaw YG, Wong JO, Easwaran S, Montell DJ. 2016. CasExpress reveals widespread and diverse patterns of cell survival of caspase-3 activation during development in vivo. Elife 5, e10936 ( 10.7554/eLife.10936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong YN, Guy C, Crawford JC, Green DR. 2017. Biological events and molecular signaling following MLKL activation during necroptosis. Cell Cycle 16, 1748–1760. ( 10.1080/15384101.2017.1371889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15, 2922–2933. [PubMed] [Google Scholar]
- 38.Taylor RC, Cullen SP, Martin SJ. 2008. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231–241. ( 10.1038/nrm2312) [DOI] [PubMed] [Google Scholar]
- 39.Galluzzi L, Kepp O, Kroemer G. 2012. Mitochondria: master regulators of danger signalling. Nat. Rev. Mol. Cell Biol. 13, 780–788. ( 10.1038/nrm3479) [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157. ( 10.1016/S0092-8674(00)80085-9) [DOI] [PubMed] [Google Scholar]
- 41.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413. ( 10.1016/S0092-8674(00)80501-2) [DOI] [PubMed] [Google Scholar]
- 42.Du C, Fang M, Li Y, Li L, Wang X. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42. ( 10.1016/S0092-8674(00)00008-8) [DOI] [PubMed] [Google Scholar]
- 43.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102, 43–53. ( 10.1016/S0092-8674(00)00009-X) [DOI] [PubMed] [Google Scholar]
- 44.Susin SA, et al. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 441–446. ( 10.1038/17135) [DOI] [PubMed] [Google Scholar]
- 45.Joza N, et al. 2001. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature 410, 549–554. ( 10.1038/35069004) [DOI] [PubMed] [Google Scholar]
- 46.Li LY, Luo X, Wang X. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412, 95–99. ( 10.1038/35083620) [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Zou H, Slaughter C, Wang X. 1997. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 89, 175–184. ( 10.1016/S0092-8674(00)80197-X) [DOI] [PubMed] [Google Scholar]
- 48.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. 1998. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391, 43–50. ( 10.1038/34112) [DOI] [PubMed] [Google Scholar]
- 49.Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. 1994. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 371, 346–347. ( 10.1038/371346a0) [DOI] [PubMed] [Google Scholar]
- 50.Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. 1997. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11, 2347–2358. ( 10.1101/gad.11.18.2347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S. 2014. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168. ( 10.1126/science.1252809) [DOI] [PubMed] [Google Scholar]
- 52.Cotter TG, Lennon SV, Glynn JM, Green DR. 1992. Microfilament-disrupting agents prevent the formation of apoptotic bodies in tumor cells undergoing apoptosis. Cancer Res. 52, 997–1005. [PubMed] [Google Scholar]
- 53.Orth K, Chinnaiyan AM, Garg M, Froelich CJ, Dixit VM. 1996. The CED-3/ICE-like protease Mch2 is activated during apoptosis and cleaves the death substrate lamin A. J. Biol. Chem. 271, 16 443–16 446. ( 10.1074/jbc.271.28.16443) [DOI] [PubMed] [Google Scholar]
- 54.Kothakota S, et al. 1997. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science 278, 294–298. ( 10.1126/science.278.5336.294) [DOI] [PubMed] [Google Scholar]
- 55.Brancolini C, Lazarevic D, Rodriguez J, Schneider C. 1997. Dismantling cell–cell contacts during apoptosis is coupled to a caspase-dependent proteolytic cleavage of beta-catenin. J. Cell Biol. 139, 759–771. ( 10.1083/jcb.139.3.759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morishima N. 1999. Changes in nuclear morphology during apoptosis correlate with vimentin cleavage by different caspases located either upstream or downstream of Bcl-2 action. Genes Cells 4, 401–414. ( 10.1046/j.1365-2443.1999.00270.x) [DOI] [PubMed] [Google Scholar]
- 57.Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. 2001. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3, 339–345. ( 10.1038/35070009) [DOI] [PubMed] [Google Scholar]
- 58.Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J. 2001. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 3, 346–352. ( 10.1038/35070019) [DOI] [PubMed] [Google Scholar]
- 59.Communal C, Sumandea M, de Tombe P, Narula J, Solaro RJ, Hajjar RJ. 2002. Functional consequences of caspase activation in cardiac myocytes. Proc. Natl Acad. Sci. USA 99, 6252–6256. ( 10.1073/pnas.092022999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moss DK, Betin VM, Malesinski SD, Lane JD. 2006. A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. J. Cell Sci. 119, 2362–2374. ( 10.1242/jcs.02959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arandjelovic S, Ravichandran KS. 2015. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 16, 907–917. ( 10.1038/ni.3253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elliott MR, Ravichandran KS. 2016. The dynamics of apoptotic cell clearance. Dev. Cell. 38, 147–160. ( 10.1016/j.devcel.2016.06.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green DR. 2005. Apoptotic pathways: ten minutes to dead. Cell 121, 671–674. ( 10.1016/j.cell.2005.05.019) [DOI] [PubMed] [Google Scholar]
- 64.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. 1997. Movement of Bax from the cytosol to mitochondria during apoptosis. J. Cell Biol. 139, 1281–1292. ( 10.1083/jcb.139.5.1281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou L, Chang DC. 2008. Dynamics and structure of the Bax–Bak complex responsible for releasing mitochondrial proteins during apoptosis. J. Cell Sci. 121, 2186–2196. ( 10.1242/jcs.024703) [DOI] [PubMed] [Google Scholar]
- 66.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. 2000. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2, 156–162. ( 10.1038/35004029) [DOI] [PubMed] [Google Scholar]
- 67.Goldstein JC, Munoz-Pinedo C, Ricci JE, Adams SR, Kelekar A, Schuler M, Tsien RY, Green DR. 2005. Cytochrome c is released in a single step during apoptosis. Cell Death Differ. 12, 453–462. ( 10.1038/sj.cdd.4401596) [DOI] [PubMed] [Google Scholar]
- 68.Rehm M, Dussmann H, Prehn JH. 2003. Real-time single cell analysis of Smac/DIABLO release during apoptosis. J. Cell Biol. 162, 1031–1043. ( 10.1083/jcb.200303123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tyas L, Brophy VA, Pope A, Rivett AJ, Tavare JM. 2000. Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer. EMBO Rep. 1, 266–270. ( 10.1093/embo-reports/kvd050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JH. 2002. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process: role of caspase-3. J. Biol. Chem. 277, 24 506–24 514. ( 10.1074/jbc.M110789200) [DOI] [PubMed] [Google Scholar]
- 71.Takemoto K, Nagai T, Miyawaki A, Miura M. 2003. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 160, 235–243. ( 10.1083/jcb.200207111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chipuk JE, Green DR. 2005. Do inducers of apoptosis trigger caspase-independent cell death? Nat. Rev. Mol. Cell Biol. 6, 268–275. ( 10.1038/nrm1573) [DOI] [PubMed] [Google Scholar]
- 73.Kroemer G, Martin SJ. 2005. Caspase-independent cell death. Nat. Med. 11, 725–730. ( 10.1038/nm1263) [DOI] [PubMed] [Google Scholar]
- 74.Tait SW, Green DR. 2010. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632. ( 10.1038/nrm2952) [DOI] [PubMed] [Google Scholar]
- 75.Narula J, et al. 1999. Apoptosis in heart failure: release of cytochrome c from mitochondria and activation of caspase-3 in human cardiomyopathy. Proc. Natl Acad. Sci. USA 96, 8144–8149. ( 10.1073/pnas.96.14.8144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reed JC, Paternostro G. 1999. Postmitochondrial regulation of apoptosis during heart failure. Proc. Natl Acad. Sci. USA 96, 7614–7616. ( 10.1073/pnas.96.14.7614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanoh M, et al. 1999. Significance of myocytes with positive DNA in situ nick end-labeling (TUNEL) in hearts with dilated cardiomyopathy: not apoptosis but DNA repair. Circulation 99, 2757–2764. ( 10.1161/01.CIR.99.21.2757) [DOI] [PubMed] [Google Scholar]
- 78.Narula J, Haider N, Arbustini E, Chandrashekhar Y. 2006. Mechanisms of disease: apoptosis in heart failure—seeing hope in death. Nat. Clin. Pract. Cardiovasc. Med. 3, 681–688. ( 10.1038/ncpcardio0710) [DOI] [PubMed] [Google Scholar]
- 79.Dandel M, Weng Y, Siniawski H, Stepanenko A, Krabatsch T, Potapov E, Lehmkuhl HB, Knosalla C, Hetzer R. 2011. Heart failure reversal by ventricular unloading in patients with chronic cardiomyopathy: criteria for weaning from ventricular assist devices. Eur. Heart J. 32, 1148–1160. ( 10.1093/eurheartj/ehq353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Demirozu ZT, Frazier OH. 2012. Remission of chronic, advanced heart failure after left ventricular unloading with an implantable left ventricular assist device. Tex. Heart Inst. J. 39, 268–270. [PMC free article] [PubMed] [Google Scholar]
- 81.Siekevitz P. 1957. Powerhouse of the cell. Sci. Am. 197, 131 ( 10.1038/scientificamerican0757-131) [DOI] [Google Scholar]
- 82.Suen DF, Norris KL, Youle RJ. 2008. Mitochondrial dynamics and apoptosis. Genes Dev. 22, 1577–1590. ( 10.1101/gad.1658508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heiskanen KM, Bhat MB, Wang HW, Ma J, Nieminen AL. 1999. Mitochondrial depolarization accompanies cytochrome c release during apoptosis in PC6 cells. J. Biol. Chem. 274, 5654–5658. ( 10.1074/jbc.274.9.5654) [DOI] [PubMed] [Google Scholar]
- 84.Munoz-Pinedo C, Guio-Carrion A, Goldstein JC, Fitzgerald P, Newmeyer DD, Green DR. 2006. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc. Natl Acad. Sci. USA 103, 11 573–11 578. ( 10.1073/pnas.0603007103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang HM, Fung MC, Tang HL. 2018. Detecting anastasis in vivo by CaspaseTracker biosensor. J. Vis. Exp. 132, e54107 ( 10.3791/54107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neame SJ, Rubin LL, Philpott KL. 1998. Blocking cytochrome c activity within intact neurons inhibits apoptosis. J. Cell Biol. 142, 1583–1593. ( 10.1083/jcb.142.6.1583) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Potts MB, Vaughn AE, McDonough H, Patterson C, Deshmukh M. 2005. Reduced Apaf-1 levels in cardiomyocytes engage strict regulation of apoptosis by endogenous XIAP. J. Cell Biol. 171, 925–930. ( 10.1083/jcb.200504082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J. 2010. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142, 590–600. ( 10.1016/j.cell.2010.07.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Villa E, et al. 2017. Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 20, 2846–2859. ( 10.1016/j.celrep.2017.08.087) [DOI] [PubMed] [Google Scholar]
- 90.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131. ( 10.1038/ncb2012) [DOI] [PubMed] [Google Scholar]
- 91.Okatsu K, et al. 2010. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15, 887–900. ( 10.1111/j.1365-2443.2010.01426.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Colell A, et al. 2007. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129, 983–997. ( 10.1016/j.cell.2007.03.045) [DOI] [PubMed] [Google Scholar]
- 93.Gama V, et al. 2014. The E3 ligase PARC mediates the degradation of cytosolic cytochrome c to promote survival in neurons and cancer cells. Sci. Signal. 7, ra67 ( 10.1126/scisignal.2005309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Paul C, Manero F, Gonin S, Kretz-Remy C, Virot S, Arrigo AP. 2002. Hsp27 as a negative regulator of cytochrome c release. Mol. Cell. Biol. 22, 816–834. ( 10.1128/MCB.22.3.816-834.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, Morimoto RI, Massie B. 2000. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol. Cell. Biol. 20, 7146–7159. ( 10.1128/MCB.20.19.7146-7159.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Steel R, Doherty JP, Buzzard K, Clemons N, Hawkins CJ, Anderson RL. 2004. Hsp72 inhibits apoptosis upstream of the mitochondria and not through interactions with Apaf-1. J. Biol. Chem. 279, 51 490–51 499. ( 10.1074/jbc.M401314200) [DOI] [PubMed] [Google Scholar]
- 97.Tsuchiya D, et al. 2003. Overexpression of rat heat shock protein 70 is associated with reduction of early mitochondrial cytochrome c release and subsequent DNA fragmentation after permanent focal ischemia. J. Cereb. Blood Flow Metab. 23, 718–727. ( 10.1097/01.WCB.0000054756.97390.F7) [DOI] [PubMed] [Google Scholar]
- 98.Clemons NJ, Buzzard K, Steel R, Anderson RL. 2005. Hsp72 inhibits Fas-mediated apoptosis upstream of the mitochondria in type II cells. J. Biol. Chem. 280, 9005–9012. ( 10.1074/jbc.M414165200) [DOI] [PubMed] [Google Scholar]
- 99.Garrido C, Bruey JM, Fromentin A, Hammann A, Arrigo AP, Solary E. 1999. HSP27 inhibits cytochrome c-dependent activation of procaspase-9. FASEB J. 13, 2061–2070. ( 10.1096/fasebj.13.14.2061) [DOI] [PubMed] [Google Scholar]
- 100.Pandey P, Farber R, Nakazawa A, Kumar S, Bharti A, Nalin C, Weichselbaum R, Kufe D, Kharbanda S. 2000. Hsp27 functions as a negative regulator of cytochrome c-dependent activation of procaspase-3. Oncogene 19, 1975–1981. ( 10.1038/sj.onc.1203531) [DOI] [PubMed] [Google Scholar]
- 101.Bruey JM, et al. 2000. Hsp27 negatively regulates cell death by interacting with cytochrome c. Nat. Cell Biol. 2, 645–652. ( 10.1038/35023595) [DOI] [PubMed] [Google Scholar]
- 102.Beere HM, et al. 2000. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2, 469–475. ( 10.1038/35019501) [DOI] [PubMed] [Google Scholar]
- 103.Li CY, Lee JS, Ko YG, Kim JI, Seo JS. 2000. Heat shock protein 70 inhibits apoptosis downstream of cytochrome c release and upstream of caspase-3 activation. J. Biol. Chem. 275, 25 665–25 671. ( 10.1074/jbc.M906383199) [DOI] [PubMed] [Google Scholar]
- 104.Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES. 2000. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat. Cell Biol. 2, 476–483. ( 10.1038/35019510) [DOI] [PubMed] [Google Scholar]
- 105.Pandey P, et al. 2000. Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90. EMBO J. 19, 4310–4322. ( 10.1093/emboj/19.16.4310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gotoh T, Terada K, Oyadomari S, Mori M. 2004. hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 11, 390–402. ( 10.1038/sj.cdd.4401369) [DOI] [PubMed] [Google Scholar]
- 107.Gabai VL, Mabuchi K, Mosser DD, Sherman MY. 2002. Hsp72 and stress kinase c-jun N-terminal kinase regulate the bid-dependent pathway in tumor necrosis factor-induced apoptosis. Mol. Cell. Biol. 22, 3415–3424. ( 10.1128/MCB.22.10.3415-3424.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ravagnan L, et al. 2001. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 3, 839–843. ( 10.1038/ncb0901-839) [DOI] [PubMed] [Google Scholar]
- 109.Gurbuxani S, et al. 2003. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene 22, 6669–6678. ( 10.1038/sj.onc.1206794) [DOI] [PubMed] [Google Scholar]
- 110.Kalinowska M, Garncarz W, Pietrowska M, Garrard WT, Widlak P. 2005. Regulation of the human apoptotic DNase/RNase endonuclease G: involvement of Hsp70 and ATP. Apoptosis 10, 821–830. ( 10.1007/s10495-005-0410-9) [DOI] [PubMed] [Google Scholar]
- 111.Gozzelino R, Jeney V, Soares MP. 2010. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 50, 323–354. ( 10.1146/annurev.pharmtox.010909.105600) [DOI] [PubMed] [Google Scholar]
- 112.Li H, Zhu H, Xu CJ, Yuan J. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501. ( 10.1016/S0092-8674(00)81590-1) [DOI] [PubMed] [Google Scholar]
- 113.Slee EA, Keogh SA, Martin SJ. 2000. Cleavage of BID during cytotoxic drug and UV radiation-induced apoptosis occurs downstream of the point of Bcl-2 action and is catalysed by caspase-3: a potential feedback loop for amplification of apoptosis-associated mitochondrial cytochrome c release. Cell Death Differ. 7, 556–565. ( 10.1038/sj.cdd.4400689) [DOI] [PubMed] [Google Scholar]
- 114.Ferreira KS, Kreutz C, Macnelly S, Neubert K, Haber A, Bogyo M, Timmer J, Borner C. 2012. Caspase-3 feeds back on caspase-8, bid and XIAP in type I Fas signaling in primary mouse hepatocytes. Apoptosis 17, 503–515. ( 10.1007/s10495-011-0691-0) [DOI] [PubMed] [Google Scholar]
- 115.Milhas D, Cuvillier O, Therville N, Clave P, Thomsen M, Levade T, Benoist H, Segui B. 2005. Caspase-10 triggers bid cleavage and caspase cascade activation in FasL-induced apoptosis. J. Biol. Chem. 280, 19 836–19 842. ( 10.1074/jbc.M414358200) [DOI] [PubMed] [Google Scholar]
- 116.Upton JP, Austgen K, Nishino M, Coakley KM, Hagen A, Han D, Papa FR, Oakes SA. 2008. Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol. Cell. Biol. 28, 3943–3951. ( 10.1128/MCB.00013-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jonas EA, et al. 2004. Proapoptotic N-truncated BCL-xL protein activates endogenous mitochondrial channels in living synaptic terminals. Proc. Natl Acad. Sci. USA 101, 13 590–13 595. ( 10.1073/pnas.0401372101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Z, Jo J, Jia JM, Lo SC, Whitcomb DJ, Jiao S, Cho K, Sheng M. 2010. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 141, 859–871. ( 10.1016/j.cell.2010.03.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hyman BT, Yuan J. 2012. Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat. Rev. Neurosci. 13, 395–406. ( 10.1038/nrn3228) [DOI] [PubMed] [Google Scholar]
- 120.Maor-Nof M, Yaron A. 2013. Neurite pruning and neuronal cell death: spatial regulation of shared destruction programs. Curr. Opin. Neurobiol. 23, 990–996. ( 10.1016/j.conb.2013.06.007) [DOI] [PubMed] [Google Scholar]
- 121.Yu F, Schuldiner O. 2014. Axon and dendrite pruning in Drosophila. Curr. Opin. Neurobiol. 27, 192–198. ( 10.1016/j.conb.2014.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Neukomm LJ, Freeman MR. 2014. Diverse cellular and molecular modes of axon degeneration. Trends Cell Biol. 24, 515–523. ( 10.1016/j.tcb.2014.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arama E, Agapite J, Steller H. 2003. Caspase activity and a specific cytochrome c are required for sperm differentiation in Drosophila. Dev. Cell 4, 687–697. ( 10.1016/S1534-5807(03)00120-5) [DOI] [PubMed] [Google Scholar]
- 124.Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E. 2010. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev. Cell 19, 160–173. ( 10.1016/j.devcel.2010.06.009) [DOI] [PubMed] [Google Scholar]
- 125.Weaver BP, Zabinsky R, Weaver YM, Lee ES, Xue D, Han M. 2014. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. eLife 3, e04265 ( 10.7554/eLife.04265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fogarty CE, Bergmann A. 2017. Killers creating new life: caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death Differ. 24, 1390–1400. ( 10.1038/cdd.2017.47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Weaver BP, Weaver YM, Mitani S, Han M. 2017. Coupled caspase and n-end rule ligase activities allow recognition and degradation of pluripotency factor LIN-28 during non-apoptotic development. Dev. Cell 41, 665–673 e666. ( 10.1016/j.devcel.2017.05.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Orth JD, Loewer A, Lahav G, Mitchison TJ. 2012. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell 23, 567–576. ( 10.1091/mbc.E11-09-0781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hain KO, Colin DJ, Rastogi S, Allan LA, Clarke PR. 2016. Prolonged mitotic arrest induces a caspase-dependent DNA damage response at telomeres that determines cell survival. Sci. Rep. 6, 26766 ( 10.1038/srep26766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miles MA, Hawkins CJ. 2017. Executioner caspases and CAD are essential for mutagenesis induced by TRAIL or vincristine. Cell Death Dis. 8, e3062 ( 10.1038/cddis.2017.454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245. ( 10.1016/0092-8674(92)90644-R) [DOI] [PubMed] [Google Scholar]
- 132.Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299. ( 10.1038/387296a0) [DOI] [PubMed] [Google Scholar]
- 133.Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387, 299–303. ( 10.1038/387299a0) [DOI] [PubMed] [Google Scholar]
- 134.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. 2009. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 15, 363–375. ( 10.1016/j.ccr.2009.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun C, et al. 1999. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature 401, 818–822. ( 10.1038/44617) [DOI] [PubMed] [Google Scholar]
- 136.Srinivasula SM, et al. 2001. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410, 112–116. ( 10.1038/35065125) [DOI] [PubMed] [Google Scholar]
- 137.Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Shi Y. 2001. Structural basis of caspase-7 inhibition by XIAP. Cell 104, 769–780. ( 10.1016/S0092-8674(01)00272-0) [DOI] [PubMed] [Google Scholar]
- 138.Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. 2001. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell 104, 781–790. ( 10.1016/s0092-8674(02)02075-5) [DOI] [PubMed] [Google Scholar]
- 139.Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. 2001. Structural basis for the inhibition of caspase-3 by XIAP. Cell 104, 791–800. ( 10.1016/S0092-8674(01)00274-4) [DOI] [PubMed] [Google Scholar]
- 140.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. 2003. Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell 11, 519–527. ( 10.1016/S1097-2765(03)00054-6) [DOI] [PubMed] [Google Scholar]
- 141.MacFarlane M, Merrison W, Bratton SB, Cohen GM. 2002. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 277, 36 611–36 616. ( 10.1074/jbc.M200317200) [DOI] [PubMed] [Google Scholar]
- 142.Flanagan L, Sebastia J, Tuffy LP, Spring A, Lichawska A, Devocelle M, Prehn JH, Rehm M. 2010. XIAP impairs Smac release from the mitochondria during apoptosis. Cell Death Dis. 1, e49 ( 10.1038/cddis.2010.26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guo L, Li SY, Ji FY, Zhao YF, Zhong Y, Lv XJ, Wu XL, Qian GS. 2014. Role of Angptl4 in vascular permeability and inflammation. Inflamm. Res. 63, 13–22. ( 10.1007/s00011-013-0678-0) [DOI] [PubMed] [Google Scholar]
- 144.Babapoor-Farrokhran S, et al. 2015. Angiopoietin-like 4 is a potent angiogenic factor and a novel therapeutic target for patients with proliferative diabetic retinopathy. Proc. Natl Acad. Sci. USA 112, E3030–E3039. ( 10.1073/pnas.1423765112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ferrara N, Gerber HP, LeCouter J. 2003. The biology of VEGF and its receptors. Nat. Med. 9, 669–676. ( 10.1038/nm0603-669) [DOI] [PubMed] [Google Scholar]
- 146.Simons M, Gordon E, Claesson-Welsh L. 2016. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 17, 611–625. ( 10.1038/nrm.2016.87) [DOI] [PubMed] [Google Scholar]
- 147.Fenech M. 2007. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2, 1084–1104. ( 10.1038/nprot.2007.77) [DOI] [PubMed] [Google Scholar]
- 148.Stanulla M, Wang J, Chervinsky DS, Thandla S, Aplan PD. 1997. DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Mol. Cell. Biol. 17, 4070–4079. ( 10.1128/MCB.17.7.4070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT. 2001. Apoptotic triggers initiate translocations within the MLL gene involving the nonhomologous end joining repair system. Cancer Res. 61, 4550–4555. [PubMed] [Google Scholar]
- 150.Eguchi-Ishimae M, Eguchi M, Ishii E, Miyazaki S, Ueda K, Kamada N, Mizutani S. 2001. Breakage and fusion of the TEL (ETV6) gene in immature B lymphocytes induced by apoptogenic signals. Blood 97, 737–743. ( 10.1182/blood.V97.3.737) [DOI] [PubMed] [Google Scholar]
- 151.Fernandez LC, Torres M, Real FX. 2016. Somatic mosaicism: on the road to cancer. Nat. Rev. Cancer 16, 43–55. ( 10.1038/nrc.2015.1) [DOI] [PubMed] [Google Scholar]
- 152.Bolouri H, et al. 2018. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 24, 103–112. ( 10.1038/nm.4439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Branzei D, Foiani M. 2008. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9, 297–308. ( 10.1038/nrm2351) [DOI] [PubMed] [Google Scholar]
- 154.Hustedt N, Durocher D. 2016. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9. ( 10.1038/ncb3452) [DOI] [PubMed] [Google Scholar]
- 155.Helming L, Winter J, Gordon S. 2009. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J. Cell Sci. 122, 453–459. ( 10.1242/jcs.037200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.van den Eijnde SM, van den Hoff MJ, Reutelingsperger CP, van Heerde WL, Henfling ME, Vermeij-Keers C, Schutte B, Borgers M, Ramaekers FC. 2001. Transient expression of phosphatidylserine at cell–cell contact areas is required for myotube formation. J. Cell Sci. 114, 3631–3642. [DOI] [PubMed] [Google Scholar]
- 157.Hochreiter-Hufford AE, et al. 2013. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497, 263–267. ( 10.1038/nature12135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zaitseva E, Zaitsev E, Melikov K, Arakelyan A, Marin M, Villasmil R, Margolis LB, Melikyan GB, Chernomordik LV. 2017. Fusion stage of HIV-1 entry depends on virus-induced cell surface exposure of phosphatidylserine. Cell Host Microbe 22, 99–110 e117. ( 10.1016/j.chom.2017.06.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Abay ZC, Wong MY, Teoh JS, Vijayaraghavan T, Hilliard MA, Neumann B. 2017. Phosphatidylserine save-me signals drive functional recovery of severed axons in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 114, E10 196–E10 205. ( 10.1073/pnas.1703807114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS. 2007. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131, 966–979. ( 10.1016/j.cell.2007.10.040) [DOI] [PubMed] [Google Scholar]
- 161.Dunai ZA, Imre G, Barna G, Korcsmaros T, Petak I, Bauer PI, Mihalik R. 2012. Staurosporine induces necroptotic cell death under caspase-compromised conditions in U937 cells. PLoS ONE 7, e41945 ( 10.1371/journal.pone.0041945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Simenc J, Lipnik-Stangelj M. 2012. Staurosporine induces different cell death forms in cultured rat astrocytes. Radiol. Oncol. 46, 312–320. ( 10.2478/v10019-012-0036-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Codogno P, Meijer AJ. 2005. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 12(Suppl. 2), 1509–1518. ( 10.1038/sj.cdd.4401751) [DOI] [PubMed] [Google Scholar]
- 164.Tsapras P, Nezis IP. 2017. Caspase involvement in autophagy. Cell Death Differ. 24, 1369–1379. ( 10.1038/cdd.2017.43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Drobnis EZ, Crowe LM, Berger T, Anchordoguy TJ, Overstreet JW, Crowe JH. 1993. Cold shock damage is due to lipid phase transitions in cell membranes: a demonstration using sperm as a model. J. Exp. Zool. 265, 432–437. ( 10.1002/jez.1402650413) [DOI] [PubMed] [Google Scholar]
- 166.Yi SX, Moore CW, Lee RE Jr. 2007. Rapid cold-hardening protects Drosophila melanogaster from cold-induced apoptosis. Apoptosis 12, 1183–1193. ( 10.1007/s10495-006-0048-2) [DOI] [PubMed] [Google Scholar]
- 167.Quinn PJ. 1985. A lipid-phase separation model of low-temperature damage to biological membranes. Cryobiology 22, 128–146. ( 10.1016/0011-2240(85)90167-1) [DOI] [PubMed] [Google Scholar]
- 168.Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. 2013. Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta 1833, 3448–3459. ( 10.1016/j.bbamcr.2013.06.001) [DOI] [PubMed] [Google Scholar]
- 169.Tang HL, Tang HM, Fung MC, Hardwick JM. 2016. In vivo biosensor tracks non-apoptotic caspase activity in Drosophila. J. Vis. Exp. 117, e53992 ( 10.3791/53992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bell RAV, Megeney LA. 2017. Evolution of caspase-mediated cell death and differentiation: twins separated at birth. Cell Death Differ. 24, 1359–1368. ( 10.1038/cdd.2017.37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McKechnie NM, Foulds WS. 1980. Recovery of the rabbit retina after light damage (preliminary observations). Albrecht von Graefes Arch. Klin. Exp. Ophthalmol. 212, 271–283. ( 10.1007/BF00410521) [DOI] [PubMed] [Google Scholar]
- 172.Milligan SC, Alb JG Jr, Elagina RB, Bankaitis VA, Hyde DR. 1997. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 139, 351–363. ( 10.1083/jcb.139.2.351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gordon WC, Casey DM, Lukiw WJ, Bazan NG. 2002. DNA damage and repair in light-induced photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 43, 3511–3521. [PubMed] [Google Scholar]
- 174.Wang KK. 2000. Calpain and caspase: can you tell the difference? Trends Neurosci. 23, 20–26. ( 10.1016/S0166-2236(99)01479-4) [DOI] [PubMed] [Google Scholar]
- 175.Jantzie LL, Corbett CJ, Firl DJ, Robinson S. 2015. Postnatal erythropoietin mitigates impaired cerebral cortical development following subplate loss from prenatal hypoxia-ischemia. Cereb. Cortex 25, 2683–2695. ( 10.1093/cercor/bhu066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shankaran S. 2014. Outcomes of hypoxic-ischemic encephalopathy in neonates treated with hypothermia. Clin. Perinatol. 41, 149–159. ( 10.1016/j.clp.2013.10.008) [DOI] [PubMed] [Google Scholar]
- 177.Narula J, et al. 1996. Apoptosis in myocytes in end-stage heart failure. N. Engl. J. Med. 335, 1182–1189. ( 10.1056/NEJM199610173351603) [DOI] [PubMed] [Google Scholar]
- 178.Arbustini E, Brega A, Narula J. 2008. Ultrastructural definition of apoptosis in heart failure. Heart Fail. Rev. 13, 121–135. ( 10.1007/s10741-007-9072-8) [DOI] [PubMed] [Google Scholar]
- 179.Leri A, Rota M, Pasqualini FS, Goichberg P, Anversa P. 2015. Origin of cardiomyocytes in the adult heart. Circ. Res. 116, 150–166. ( 10.1161/CIRCRESAHA.116.303595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Foglia MJ, Poss KD. 2016. Building and re-building the heart by cardiomyocyte proliferation. Development 143, 729–740. ( 10.1242/dev.132910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S, Srivastava D. 2018. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104–116.e112. ( 10.1016/j.cell.2018.02.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Forbes SJ, Newsome PN. 2016. Liver regeneration – mechanisms and models to clinical application. Nat. Rev. Gastroenterol. Hepatol. 13, 473–485. ( 10.1038/nrgastro.2016.97) [DOI] [PubMed] [Google Scholar]
- 183.Johnstone RW, Ruefli AA, Lowe SW. 2002. Apoptosis: a link between cancer genetics and chemotherapy. Cell 108, 153–164. ( 10.1016/S0092-8674(02)00625-6) [DOI] [PubMed] [Google Scholar]
- 184.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. 2009. Cell death. N. Engl. J. Med. 361, 1570–1583. ( 10.1056/NEJMra0901217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Davis AJ, Tannock JF. 2000. Repopulation of tumour cells between cycles of chemotherapy: a neglected factor. Lancet Oncol. 1, 86–93. ( 10.1016/S1470-2045(00)00019-X) [DOI] [PubMed] [Google Scholar]
- 186.Kim JJ, Tannock IF. 2005. Repopulation of cancer cells during therapy: an important cause of treatment failure. Nat. Rev. Cancer 5, 516–525. ( 10.1038/nrc1650) [DOI] [PubMed] [Google Scholar]
- 187.Wagle N, et al. 2011. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29, 3085–3096. ( 10.1200/JCO.2010.33.2312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Demedts IK, Vermaelen KY, van Meerbeeck JP. 2010. Treatment of extensive-stage small cell lung carcinoma: current status and future prospects. Eur. Respir. J. 35, 202–215. ( 10.1183/09031936.00105009) [DOI] [PubMed] [Google Scholar]
- 189.Manjila S, Ray A, Hu Y, Cai DX, Cohen ML, Cohen AR. 2011. Embryonal tumors with abundant neuropil and true rosettes: 2 illustrative cases and a review of the literature. Neurosurg. Focus 30, E2 ( 10.3171/2010.10.FOCUS10226) [DOI] [PubMed] [Google Scholar]
- 190.Boffetta P, Hashibe M. 2006. Alcohol and cancer. Lancet Oncol. 7, 149–156. ( 10.1016/S1470-2045(06)70577-0) [DOI] [PubMed] [Google Scholar]
- 191.McKillop IH, Schrum LW. 2005. Alcohol and liver cancer. Alcohol 35, 195–203. ( 10.1016/j.alcohol.2005.04.004) [DOI] [PubMed] [Google Scholar]
- 192.Castellsague X, Munoz N, De Stefani E, Victora CG, Castelletto R, Rolon PA. 2000. Influence of mate drinking, hot beverages and diet on esophageal cancer risk in South America. Int. J. Cancer 88, 658–664. ( 10.1002/1097-0215(20001115)88:4%3C658::AID-IJC22%3E3.0.CO;2-T) [DOI] [PubMed] [Google Scholar]
- 193.Islami F, et al. 2009. Tea drinking habits and oesophageal cancer in a high risk area in northern Iran: population based case–control study. BMJ 338, b929 ( 10.1136/bmj.b929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Loomis D, et al. 2016. Carcinogenicity of drinking coffee, mate, and very hot beverages. Lancet Oncol. 17, 877–878. ( 10.1016/S1470-2045(16)30239-X) [DOI] [PubMed] [Google Scholar]
- 195.Smith RE, Bryant J, DeCillis A, Anderson S. 2003. Acute myeloid leukemia and myelodysplastic syndrome after doxorubicin-cyclophosphamide adjuvant therapy for operable breast cancer: the National Surgical Adjuvant Breast and Bowel Project Experience. J. Clin. Oncol. 21, 1195–1204. ( 10.1200/JCO.2003.03.114) [DOI] [PubMed] [Google Scholar]
- 196.Travis LB, et al. 2005. Second cancers among 40 576 testicular cancer patients: focus on long-term survivors. J. Natl Cancer Inst. 97, 1354–1365. ( 10.1093/jnci/dji278) [DOI] [PubMed] [Google Scholar]
- 197.Chaturvedi AK, et al. 2007. Second cancers among 104 760 survivors of cervical cancer: evaluation of long-term risk. J. Natl Cancer Inst. 99, 1634–1643. ( 10.1093/jnci/djm201) [DOI] [PubMed] [Google Scholar]
- 198.Yates LR, Campbell PJ. 2012. Evolution of the cancer genome. Nat. Rev. Genet. 13, 795–806. ( 10.1038/nrg3317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.McClintock B. 1984. The significance of responses of the genome to challenge. Science 226, 792–801. ( 10.1126/science.15739260) [DOI] [PubMed] [Google Scholar]
- 200.Capy P, Gasperi G, Biemont C, Bazin C. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity (Edinb) 85, 101–106. ( 10.1046/j.1365-2540.2000.00751.x) [DOI] [PubMed] [Google Scholar]
- 201.Rosenberg SM. 2001. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2, 504–515. ( 10.1038/35080556) [DOI] [PubMed] [Google Scholar]
- 202.Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR. 2003. An active DNA transposon family in rice. Nature 421, 163–167. ( 10.1038/nature01214) [DOI] [PubMed] [Google Scholar]
- 203.Elias SG, Peeters PH, Grobbee DE, van Noord PA. 2004. Breast cancer risk after caloric restriction during the 1944–1945 Dutch famine. J. Natl Cancer Inst. 96, 539–546. ( 10.1093/jnci/djh087) [DOI] [PubMed] [Google Scholar]
- 204.Painter RC, De Rooij SR, Bossuyt PM, Osmond C, Barker DJ, Bleker OP, Roseboom TJ. 2006. A possible link between prenatal exposure to famine and breast cancer: a preliminary study. Am. J. Hum. Biol. 18, 853–856. ( 10.1002/ajhb.20564) [DOI] [PubMed] [Google Scholar]
- 205.Painter RC, de Rooij SR, Bossuyt PM, Simmers TA, Osmond C, Barker DJ, Bleker OP, Roseboom TJ. 2006. Early onset of coronary artery disease after prenatal exposure to the Dutch famine. Am. J. Clin. Nutr. 84, 322–327; quiz 466–327 ( 10.1093/ajcn/84.2.322) [DOI] [PubMed] [Google Scholar]
- 206.Roseboom TJ, van der Meulen JH, Osmond C, Barker DJ, Ravelli AC, Schroeder-Tanka JM, van Montfrans GA, Michels RP, Bleker OP. 2000. Coronary heart disease after prenatal exposure to the Dutch famine, 1944–45. Heart 84, 595–598. ( 10.1136/heart.84.6.595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.de Rooij SR, et al. 2006. Glucose tolerance at age 58 and the decline of glucose tolerance in comparison with age 50 in people prenatally exposed to the Dutch famine. Diabetologia 49, 637–643. ( 10.1007/s00125-005-0136-9) [DOI] [PubMed] [Google Scholar]
- 208.Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN, Bleker OP. 1998. Glucose tolerance in adults after prenatal exposure to famine. Lancet 351, 173–177. ( 10.1016/S0140-6736(97)07244-9) [DOI] [PubMed] [Google Scholar]
- 209.Ravelli AC, van der Meulen JH, Osmond C, Barker DJ, Bleker OP. 1999. Obesity at the age of 50 y in men and women exposed to famine prenatally. Am. J. Clin. Nutr. 70, 811–816. ( 10.1093/ajcn/70.5.811) [DOI] [PubMed] [Google Scholar]
- 210.Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ. 2008. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 115, 1243–1249. ( 10.1111/j.1471-0528.2008.01822.x) [DOI] [PubMed] [Google Scholar]
- 211.Drummond-Barbosa D, Spradling AC. 2001. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev. Biol. 231, 265–278. ( 10.1006/dbio.2000.0135) [DOI] [PubMed] [Google Scholar]
- 212.Salinas LS, Maldonado E, Navarro RE. 2006. Stress-induced germ cell apoptosis by a p53 independent pathway in Caenorhabditis elegans. Cell Death Differ. 13, 2129–2139. ( 10.1038/sj.cdd.4401976) [DOI] [PubMed] [Google Scholar]
- 213.Aitken RJ, Findlay JK, Hutt KJ, Kerr JB. 2011. Apoptosis in the germ line. Reproduction 141, 139–150. ( 10.1530/REP-10-0232) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the cited references and the electronic supplementary material.