Abstract
CTLA4 is an essential negative regulator of T-cell immune responses and a key checkpoint regulating autoimmunity and antitumor responses. Genetic mutations resulting in quantitative defects in the CTLA4 pathway are also associated with the development of immune dysregulation syndromes in humans. It has been proposed that CTLA4 functions to remove its ligands CD80 and CD86 from opposing cells by a process known as transendocytosis. A quantitative characterization of CTLA4 synthesis, endocytosis, degradation, and recycling and how these affect its function is currently lacking. In a combined in vitro and in silico study, we developed a mathematical model and identified these trafficking parameters. Our model predicts optimal ligand removal in an intermediate affinity range. The intracellular CTLA4 pool as well as fast internalization, recovery of free CTLA4 from internalized complexes, and recycling is critical for sustained functionality. CD80-CTLA4 interactions are predicted to dominate over CD86-CTLA4. Implications of these findings in the context of control of antigen-presenting cells by regulatory T cells and of pathologic genetic deficiencies are discussed. The presented mathematical model can be reused in the community beyond these questions to better understand other trafficking receptors and study the impact of CTLA4 targeting drugs.
Introduction
In a healthy organism, the immune system has to maintain a balance between immune activation and inhibition. For T cells, the decision between these two outcomes is influenced by receptors that are not specific for antigen but provide context for antigen recognition. CD28 and CTLA4 are two such transmembrane receptors expressed by T lymphocytes with opposing stimulatory and inhibitory functions, respectively. These receptors bind to the same costimulatory ligands, CD80 and CD86, which are expressed by antigen-presenting cells (1, 2). Although ligation of CD28 with costimulatory ligands is essential for full T-cell activation and effector functions (3, 4, 5), CTLA4 inhibits excess costimulation by competing for CD28 ligands and thereby preventing uncontrolled T-cell activation and clonal expansion of cells specific for our own tissues. Accordingly, CTLA4 deficiency results in autoimmunity (6, 7, 8). Early data suggested that CTLA4 directly inhibits the cells that express it, referred to as the cell-intrinsic pathway of immune inhibition (9, 10). Such cell-intrinsic mechanisms predict that autoimmunity arises in a CTLA4-deficient model because of uncontrolled responses of activated T cells. However, cell-intrinsic inhibition also implies that in a chimeric model containing a mixture of CTLA4-deficient and CTLA4-sufficient cells, hyperactive CTLA4-deficient cells should still be observed. Surprisingly, a normal immune system with no hyperactivation was found when this idea was tested experimentally (11, 12, 13, 14). This observation suggests CTLA4 may act through a cell-extrinsic mechanism of inhibition in which CTLA4-expressing cells exert control on their surroundings. This fits with the demonstration that CTLA4 plays a critical role in regulatory T cells (Tregs), an immune cell population with potent cell-extrinsic function (15). Recently, it has been shown that CTLA4 molecules on T cells are able to internalize costimulatory molecules from the surface of antigen presenting cells (APCs) by a process known as transendocytosis (16), which represents a potential mechanism for cell-extrinsic inhibition of T-cell activation (17).
Unlike many immune regulatory proteins, CTLA4 molecules are mostly observed intracellularly in cytoplasmic vesicles (18, 19) as a result of their interaction with the μ2 subunit of the clathrin adaptor protein complex AP2 (20, 21, 22, 23). In contrast, CD28 is retained on the plasma membrane (24). The unusual localization of CTLA4 raises questions about how this affects its function. For example, it is not clear how the steady-state distribution of CTLA4 molecules results from dynamic trafficking between cytosol and plasma membrane. Various parameters affecting internalization, degradation, and recycling rates all have the potential to influence the cellular function. Given the number of parameters that can affect CTLA4 function (e.g., trafficking rates, ligand binding affinities, competition between ligands, etc.), mathematical models are useful in understanding and predicting the impact of changes in these parameters on CTLA4 suppressive function. We have therefore derived a mathematical model for in silico experimentation to test how various parameters can influence CTLA4 function.
In this study, we extract CTLA4 trafficking rates from a series of experimental protocols and corresponding mathematical models to understand the behavior of CTLA4 and study its role in ligand clearance. The model shows that cytoplasmic CTLA4 acts as a buffer for CTLA4 surface expression and boosts ligand uptake at early time points of ligand exposure. In silico ligand-uptake experiments suggest that stable CTLA4-ligand binding does not guarantee an efficient ligand uptake and instead ligand removal is optimally fast for an intermediate affinity range of ligand-CTLA4 interaction. We showed that the process of ligand uptake highly depends on the recycling process, and even expression of a normal amounts of total CTLA4 does not guarantee an efficient ligand uptake when the recycling process is impaired. Thus, molecular binding and trafficking properties determine the efficiency of ligand clearance. Furthermore, when CTLA4+ cells are exposed to a mixture of natural ligands, we find that CD80 is the dominant competitor and the uptake of CD86 is highly affected by the presence of CD80 but not vice versa.
Materials and Methods
CTLA4 internalization assay
Chinese hamster (Cricetulus griseus) ovary (CHO) cells, expressing the virally transduced (MP-71 retroviral-vector-containing) human CTLA4 (UniProtKB accession: CTLA4_HUMAN) were placed in fluorescence activated cell sorter (FACS) tubes (Sarstedt, Nümbrecht, Germany) at 1.5 × 106 cell/mL density (150 μL) before being placed on ice. The cells were washed in 200 μL phosphate-buffered saline buffer (PBS (pH 7.4); Life Technologies, Carlsbad, CA) and 2% w/v bovine serum albumin (ACROS Organics, Geel, Belgium) by spinning cells at 500 g for 5 min. The surface CTLA4 was stained using phycoerythrin (PE) fluorophore-conjugated mouse anti-human CTLA4 antibody (PE-CTLA4; BD Pharmingen, San Jose, CA) at 1:100 dilution for 20 min on ice. After removing the excess antibody and washing the cells with ice-cold PBS buffer, the cells were placed in a 37°C water bath for various durations, allowing CTLA4 internalization from the CHO cell surface, before being placed on ice and fixed with 3% (v/v) paraformaldehyde (PFA; Agar Scientific, Essex, UK) for 15 min. The cells were washed, and the surface CTLA4 was stained with Alexa Fluor 647 conjugated chicken anti-mouse secondary antibody (ThermoFisher Scientific, Waltham, MA) at 1:1000 dilution for 30 min on ice before being washed and run through a BD LSRFortessa (5L SORP) flow cytometer (BD Biosciences, San Jose, CA) using yellow-green 561 nm (586/15 band-pass filter) and red 640 nm (670/14 band-pass filter) lasers for PE and Alexa Fluor 647 fluorophores, respectively. The data were recorded using BD FACSDiva Software (Version 6.2), and the emission-spectra-compensated mean fluorescence intensity (MFI) of each fluorophore was obtained using FlowJo 10.0.8r1 software.
CTLA4 molecular counting
Quantum Simply Cellular anti-Mouse IgG kits (Bangs Laboratories, Fishers, IN) were used according to the manufacturer’s instructions to quantify the number of virally transduced human CTLA4 expressed in CHO cells. Briefly, beads 1–4, which are of known size and antibody-binding capacity, were stained with varying amounts of PE-CTLA4 for 30 min on ice. The beads were washed in media (Dulbecco’s modified Eagle’s medium (Life Technologies), 10% fetal bovine serum (Labtech), 2 mM L-Glutamine (Life Technologies), 1× Penicillin-Streptomycin (Life Technologies)) by spinning at 1060 g for 5 min before their mean fluorescence intensity (MFI) was obtained using the BD LSRFortessa flow cytometer using the yellow-green 561 nm (586/15 band-pass filter) laser at a set voltage. Bead saturation levels with PE-CTLA4 antibody were assessed using the FlowJo 10.0.8r1 software, and the best MFI for each bead was plotted to establish a calibration curve. In parallel to the above, various numberd of CHO cells expressing human CTLA4 were fixed with 3% (v/v) PFA (Agar Scientific) for 15 min on ice before being washed in media and permeabilized with 0.1% (v/v) saponin in media. The cells were then stained in an identical manner to the beads and run through the BD LSRFortessa flow cytometer using identical settings and voltages as the beads. The obtained CHO cell MFIs were then compared against the calibration curve to obtain the number of CTLA4 molecules per CHO cell.
Synthesis-block assay
CHO cells expressing CTLA4 were placed in a 96-well U-bottom plate (Falcon, Corning, NY) at 2 × 106 cell/mL (130 L) before adding cycloheximide at 50 ng/mL or dimethyl sulfoxide vehicle control for the indicated times. The cells were fixed as described before, then permeabilized in 150 μL PBS+ (PBS, bovine serum albumin, 0.1% saponin) for 10 min at room temperature before being stained with BNI3 clone PE-CTLA4 antibody as described before. After removing the excesses antibody and washing the cells with ice cold PBS+ twice, they were resuspended in PBS, run on the flow cytometer, and analyzed as described before.
Lysosomal block assay
A total of 1.5 × 105 cells per condition were plated in a 96-well plate and pulsed with anti-CTLA4-PE (BN13) for 1 h at 37°C. Cells were then washed twice and resuspended in medium containing dimethyl sulfoxide or NH4Cl for 2, 4, or 6 h. At each time point, cells were transferred to FACS tubes and fixed in 2% PFA on ice for 10 min. Cells were then washed three times in PBS and analyzed for CTLA4 expression by flow cytometry.
All the experiments were performed in triplicates, and the MFI values were averaged from 10,000 cells (events) for each repeat.
Mathematical models
Different mathematical models were developed according to the experimental protocols. Models were studied analytically and numerical simulations were performed by C++ (Boost.Numeric.Odeint) and MATLAB (MathWorks, Natick, MA). A differential evolution algorithm was implemented in C++ for parameter estimation. Markov chain Monte Carlo (MCMC) analysis was performed in MATLAB. The mathematical analysis is described in the Supporting Materials and Methods.
Results
Generation of the model
The rates of CTLA4 internalization, recycling, and degradation were estimated from a series of in vitro experiments in which the individual processes of CTLA4 trafficking were perturbed. These experiments were replicated by generating corresponding mathematical models. The parameters of the model were determined by simultaneously fitting all parameters to all experimental results with a combined cost function using a differential evolution algorithm (see Supporting Materials and Methods). To examine the impact of the experimental data variations on the estimated parameters, we employed an MCMC approach to obtain the distribution of the parameters (see Supporting Materials and Methods). The optimal values of the CTLA4 trafficking rates obtained by the differential evolution are within the 68% confidence interval of their distributions. We therefore used the optimal values of the parameters in our simulations. The experimental data used in this study were sufficient to determine all of the CTLA4 trafficking parameters and their distributions given in Table 1. Although the parameters were identified using data from all experiments, for the purpose of clarity, the different experiments informing the model are presented individually below, together with their respective mathematical model.
Table 1.
Ligand-Independent CTLA4 Trafficking Parameters
| Parameter | Description | Dimension | Optimal valuea | Posterior mean ± standard deviationb |
|---|---|---|---|---|
| σi | Rate of CTLA4 synthesis | # min−1 | 10,100c | 13,414 ± 6936 |
| ki | Rate of CTLA4 internalization | min−1 | 0.2988 | 0.379 ± 0.116 |
| kr | Rate of CTLA4 recycling | min−1 | 0.0243 | 0.028 ± 0.012 |
| δn | Rate of nonlysosomal degradation | min−1 | 0.0014 | 0.0016 ± 0.00057 |
| δl | Rate of lysosomal degradation | min−1 | 0.0024 | 0.0035 ± 0.0014 |
| δc | Rate of cytoplasmic degradation | min−1 | δl + δn | 0.0051 ± 0.0015 |
Optimal values were obtained by differential evolution algorithm. See Supporting Materials and Methods.
Parameter distributions were obtained by MCMC analysis. See Supporting Materials and Methods.
This value represents the rate of CTLA4 synthesis of the cell line used for the molecular counting experiment. See Supporting Materials and Methods.
CTLA4 trafficking in the absence of ligands
After synthesis, CTLA4 is transported from the Golgi to the cell surface. This transport is dependent on the enzymes phospholipase D and ADP ribosylation factor 1 (25) and occurs via the fusion of CTLA4-containing vesicles with the plasma membrane. Surface-expressed CTLA4 is then subject to constitutive endocytosis, independent of ligand binding, via interaction of the tyrosine-based motif (YVKM) in the CTLA4 cytoplasmic domain with clathrin adaptor complex AP2 (20, 21, 22, 23, 26). After internalization, CTLA4 molecules can be recycled to the plasma membrane or degraded in lysosomal compartments (24, 27, 28). These features of CTLA4 behavior are seen in primary human T cells and are preserved in model systems transfected with CTLA4 (26).
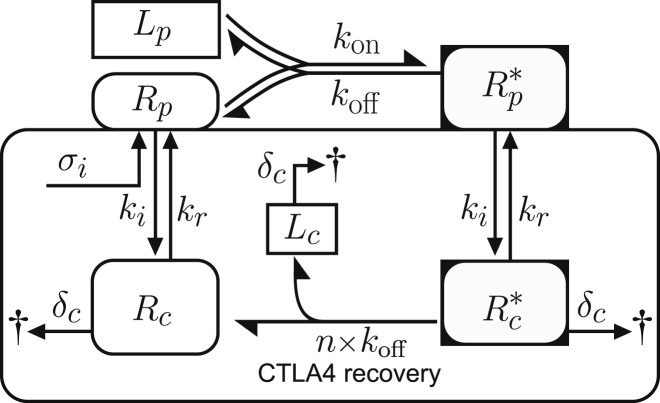
Based on the above biological observations, we constructed a trafficking model, which comprises processes of protein synthesis, recycling, internalization, and degradation. This model, depicted schematically in Fig. 1 A, relies on the following assumptions: CTLA4 molecules are synthesized by a constant rate σi with dimension of molecules (#) per minute. Newly synthesized CTLA4 molecules are incorporated to the plasma membrane (Rp(t)) and subsequently internalized into the cytoplasm (Rc(t)) with rate ki. This assumes that newly synthesized CTLA4 molecules do not directly undergo degradation in the cytoplasm but only after endocytosis. Cytoplasmic CTLA4 molecules are degraded in lysosomal and nonlysosomal pathways with constant rates δl and δn, respectively. Therefore, the total cytoplasmic degradation rate is δc = δl + δn. Nondegraded cytoplasmic CTLA4 molecules are recycled to the plasma membrane with constant rate kr. All variables and parameters represent the value for a single cell (per cell). Such a trafficking model can be represented by the ordinary differential equations (ODE), given in Eq. S1 (see Supporting Materials and Methods).
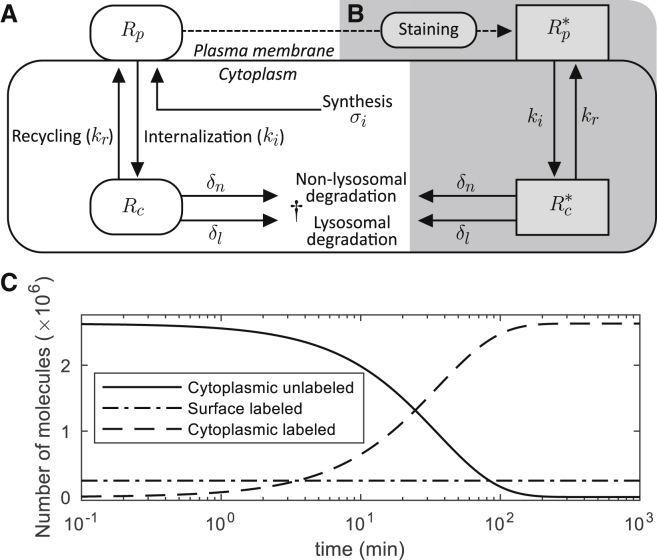
Figure 1.
Ligand-independent CTLA4 trafficking and CTLA4 staining models. (A) Natural trafficking of CTLA4 molecules. The corresponding ODE-based model is given in Eq. S1. (B) By extracellular CTLA4 staining with antibody, the CTLA4 pool is segregated into labeled (shown with gray background) and unlabeled pools. This is a simplified model of a more complex staining model described in the Supporting Materials and Methods and shown in Fig. S1. (C) CTLA4 staining simulation. Equation S12 was solved analytically, and the solution, given in Eq. S13, was plotted.
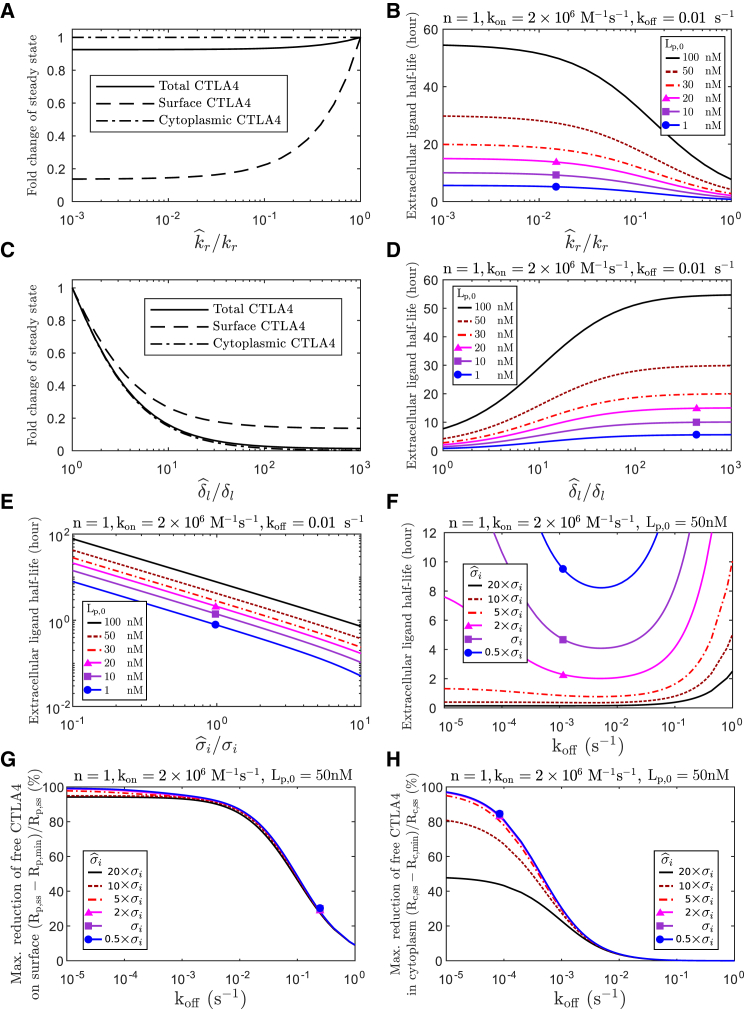
To recapitulate the experimental observation that CTLA4 molecules are mostly located in the cytoplasm, the model in its steady-state (ss) condition has to satisfy the condition ki > kr + δc (see Eq. S2, with condition Rp,ss/Rc,ss < 1). Despite the dependence of steady-state total CTLA4 values on the rate of protein synthesis, the ratio of surface/cytoplasmic CTLA4 is independent of the level of synthesis (Eq. S2). Further, the steady-state absolute number of the cytoplasmic CTLA4 molecules is independent of the rates of internalization and recycling (Eq. S2).
To inform our model, initial parameter estimates were determined using a variety of different antibody staining protocols. By staining CTLA4 with specific fluorescent antibodies at 4°C, it is assumed that all surface molecules present on the plasma membrane are labeled at a particular time point. Subsequently, trafficking of CTLA4 is controlled by changes in the temperature by which 37°C permits internalization and recycling of CTLA4 not seen at 4°C. To label a subset of the cytoplasmic CTLA4, staining was performed at 37°C for a certain duration, after which the kinetics of labeled CTLA4 could be observed. These basic principles were used in various ways to monitor internalization, recycling, and degradation of CTLA4 molecules. The general mathematical model for the dynamics of CTLA4 staining is described in the Supporting Materials and Methods (see Eq. S3; Fig. S1). The staining model is simplified based on the following assumptions: A) the concentration of staining antibodies in the medium is sufficiently large and does not drop as a result of target binding (thermic bath approximation); B) staining antibodies bind to CTLA4 molecules with a higher rate than the rate of trafficking processes (quasi-steady-state approximation); C) staining antibodies remain bound to CTLA4 molecules during the time of sample observation (negligible unbinding rate); and D) staining antibodies do not alter the natural trafficking of CTLA4 molecules and associated rates. The simplified model is shown in Fig. 1, A and B, in which and are the numbers of labeled CTLA4 molecules on the cell surface and in the cytoplasm, respectively. The corresponding ODEs are given in Eq. S12 (see Supporting Materials and Methods). Note that in the simplified staining model, the number of unlabeled surface CTLA4 during the staining is zero because all free surface CTLA4 molecules are labeled instantly when exposed to staining antibodies in the medium (assumption B). Before starting the staining experiment, the number of CTLA4 molecules is assumed at equilibrium (Eq. S2). During the staining process at 37°C, the unlabeled pool of cytosolic CTLA4 molecules becomes exhausted with a half-life of t1/2 = ln(2)/(kr + δc) ≈ 25 min as a result of transport to the plasma membrane and subsequent labeling, as well as cytosolic degradation (see Eq. S13 a). Therefore, the rates of recycling and degradation dictate the duration of the staining process required to label the majority of total cellular CTLA4 molecules.
The dynamics of the different pools of CTLA4 molecules during persistent staining is shown in Fig. 1 C. Because of instant labeling of surface molecules as well as continued exocytosis of newly synthesized CTLA4, the number of labeled CTLA4 molecules on the cell surface starts and remains at its equilibrium value, whereas unlabeled cytoplasmic CTLA4 molecules become exhausted over time.
CTLA4 internalization
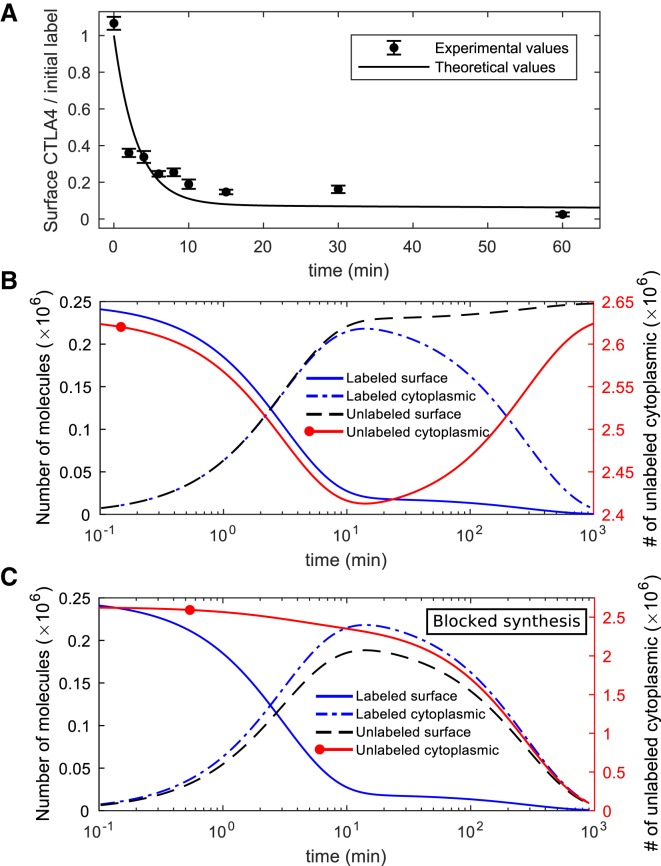
To observe the internalization of surface CTLA4 molecules, we used a CTLA4 internalization assay described in Materials and Methods. In this experiment, CTLA4-expressing cells were labeled at 4°C with a fluorescently conjugated anti-CTLA4 antibody (PE-CTLA4). Excess antibody was washed off and the temperature raised to 37°C for various durations to permit CTLA4 trafficking. After this period, a secondary antibody (Alexa Fluor 647) was applied at 4°C to detect the remaining surface antibody-labeled CTLA4. In this setting, the initial amount of surface CTLA4 is quantified by PE, and the number of molecules still at the surface after a period of trafficking is quantified by Alexa 647. The data from this experiment are shown in Fig. 2 A.
Figure 2.
In silico and in vitro internalization data. (A) The fraction of initially labeled CTLA4 molecules detected on the cell surface over time is shown. Experimental data points are scaled with the factor 1/Sint (see Table S1). (B) The kinetics of all different CTLA4 pools in the model. Unlabeled CTLA4 follows Eq. S1. Unlabeled cytoplasmic CTLA4 molecules (Rc(t)) drop transiently (right y axis). (C) An in silico internalization experiment with a block of CTLA4 synthesis. Both unlabeled CTLA4 pools clear over time; however, the surface unlabeled pool transiently reaches 76% of its steady-state value in normal cells. To see this figure in color, go online.
In the model, the number of surface CTLA4 molecules before starting the staining process is derived from the steady-state solution of the CTLA4 trafficking model given in Eq. S2. This number is assumed to be fully labeled on ice at t = 0, leaving no unlabeled molecules on the cell surface. The solution of the dynamic equations provides the time course of the number of CTLA4 molecules in different pools (see Supporting Materials and Methods; Eq. S15). Surface labeled CTLA4 molecules vanish over time monotonically, becoming cytoplasmic because of internalization (Fig. 2 B), whereas the cytoplasmic labeled CTLA4 pool diminishes after a transient increase because of degradation. Newly synthesized and recycled unlabeled CTLA4 molecules replace the labeled pool over time.
An in silico internalization experiment in which CTLA4 synthesis is stopped (σi = 0) predicts that unlabeled CTLA4 molecules on the plasma membrane are cleared after transient maintainance at 76% of its pre-experiment (normal steady-state) value (see Fig. 2 C) resulting from the utilization of the cytoplasmic CTLA4 pool. This suggests that without new CTLA4 synthesis, CTLA4 stored in the cytoplasm can temporarily buffer surface CTLA4 expression.
Modeling perturbations of the CTLA4 steady state
CTLA4 synthesis is key for long-term maintenance of surface expression
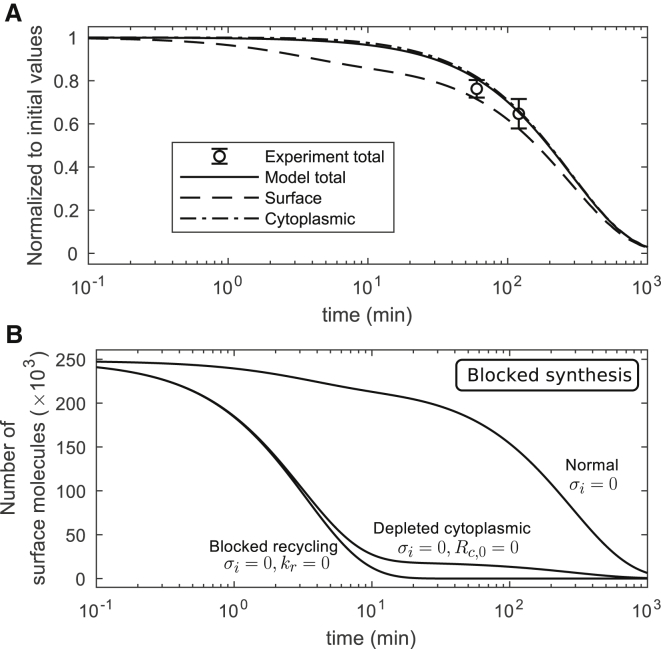
Quantification of the contribution of protein synthesis to CTLA4 homeostasis was based on the synthesis-block experiment (see Materials and Methods), in which CTLA4-expressing cells were treated with Cycloheximide (CHX) for different durations and the total number of CTLA4 molecules was measured and compared with cells without CHX treatment.
In the model, we assume that CHX treatment of cells completely blocks protein synthesis (σi = 0). The time evolution of surface and cytoplasmic CTLA4 in the presence of CHX is calculated (see Supporting Materials and Methods; Eq. S18) and depicted in Fig. 3 A. At early time points, surface CTLA4 is quickly lost because of fast internalization and recycling partially compensates surface CTLA4 loss. Total CTLA4 reduces by degradation in the cytoplasm.
Figure 3.
Block of CTLA4 synthesis. (A) A single experimental data point was compared to the theoretical value in Eq. S21. CTLA4 molecules in the cytoplasm can transiently compensate CTLA4 loss on the plasma membrane after the block of synthesis. However, both pools become exhausted with extended exposure to CHX. (B) In silico experiment of blocked synthesis with additional depletion of cytoplasmic CTLA4 (Rc,0 = 0) or block of recycling (kr = 0). These curves are obtained by numerical solution of the trafficking model in Eq. S1. In the absence of cytoplasmic CTLA4 or of the recycling process, the surface CTLA4 clears quickly, reducing by 50% within ∼2.41 and 2.32 min, respectively.
When the CTLA4 synthesis is blocked (σi = 0) and the remaining processes (internalization, recycling, and cytosolic degradation) are included (see Fig. 3 A), the surface, cytoplasmic, and total CTLA4 molecules are reduced by 50% within 160, 198, and 195 min, respectively. Surface CTLA4 is reduced by 8% within 3 min and cytoplasmic CTLA4 only by 0.4% during this time, which indicates a rapid loss of surface CTLA4 at early time points. As expected, CTLA4 synthesis is important for maintaining the homeostatic number of CTLA4 molecules.
Next, we addressed the contribution of the cytoplasmic pool to ask how internalization impacts CTLA4 surface expression. In an in silico experiment, a complete depletion of the cytoplasmic CTLA4 (by imposing a zero initial value, Rc,0 = 0) or a complete block of recycling (kr = 0) was tested in the absence of CTLA4 synthesis. This showed that surface CTLA4 molecules clear within a few minutes in comparison to the half-life of 160 min under the normal condition (σi = 0) (see Fig. 3 B). The model predicts that in the absence of new synthesis, CTLA4 can be maintained at the cell surface by recycling. Thus, the cytoplasmic reserves of CTLA4 molecules are key for maintaining the surface CTLA4 pool.
Lysosomal dominates nonlysosomal degradation
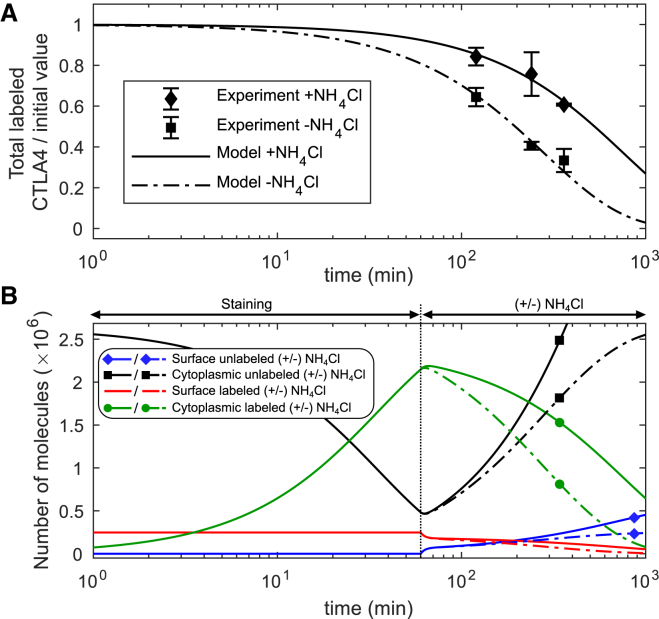
To quantify the impact of lysosomal degradation on CTLA4 trafficking, we used the lysosomal block assay, in which CTLA4 kinetics were measured in the presence or absence of ammonium chloride (NH4Cl) (see Materials and Methods). NH4Cl inhibits lysosomal degradation through lysosomal pH neutralization. In this experiment, cells were initially pulsed with a labeling antibody (anti-CTLA4 PE) at 37°C for 60 min. After washing, degradation of CTLA4 molecules was quantified by flow cytometry (see Fig. 4 A).
Figure 4.
Block of lysosomal-degradation. (A) In silico versus in vitro results with and without addition of NH4Cl. (B) In silico prediction of different CTLA4 pools for blocked lysosomal degradation (+NH4Cl, solid lines) and normal condition (−NH4Cl, dash-dotted lines). To see this figure in color, go online.
To reconstruct this experiment in silico, we used the staining model (Fig. 1, A and B) and calculated the number of labeled CTLA4 molecules after 60 min. Starting from this value as the initial condition, the time evolution of the CTLA4 pools was calculated (see Supporting Materials and Methods; Eq. S22) in the presence (+) or absence (−) of NH4Cl. It was assumed that NH4Cl does not impact on the nonlysosomal degradation pathway. A comparison of both conditions reveals the differences in the kinetics of CTLA4 molecules (see Fig. 4 B): after the initial decrease of the number of unlabeled CTLA4 molecules during the staining step, it increases to its steady-state value or higher in the absence or presence of the lysosomal inhibitor, respectively. This increase is due to ongoing CTLA4 synthesis. The rate of degradation decreased by ∼63% and the half-life of total labeled CTLA4 molecules increased from ∼3.3 to 8.8 h in the presence of the lysosomal inhibitor (see Fig. 4 A). The model predicts that the lysosomal degradation pathway is dominant.
According to the steady-state number of CTLA4 molecules in the CTLA4 trafficking model Fig. 1 A (see Rp,ss/Rc,ss in Eq. S2), reduction of the cytoplasmic degradation (δc) leads to a reduction in the fraction of the total CTLA4 molecules that appears on the cell surface.
These results suggest that the lysosomal degradation of CTLA4 is a fast process that regulates the homeostatic number of CTLA4 molecules and, when blocked, shifts the steady-state subcellular distribution of CTLA4 toward the cytoplasm.
CTLA4 recycling is slower than internalization
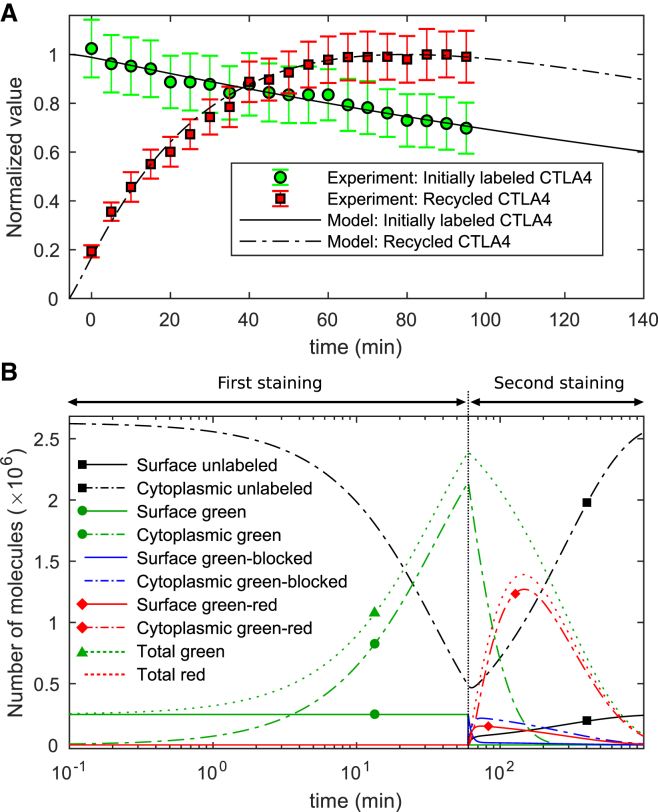
For quantification of CTLA4 recycling, we used a two-step staining protocol to label CTLA4 molecules on the plasma membrane, allow them to internalize, and then to label molecules upon their return to the surface (26). In the first step, CTLA4-expressing cells were incubated with Alexa488-conjugated anti-CTLA4 (a green antibody) for 60 min at 37°C. Then, cells were washed, and surface CTLA4 molecules were blocked at 4°C with unconjugated anti-human IgG, a secondary antibody without any color. This prevented those molecules that remained on the surface and were green labeled from binding to a secondary antibody. After washing, cells were incubated for different times with Alexa555-conjugated anti-human IgG (a red secondary antibody) at 37°C. The red antibody only binds those green-labeled CTLA4 molecules that recycled back to the plasma membrane. The red-fluorescent intensity was quantified with confocal microscopy, reflecting the time evolution of the labeled subset of recycled CTLA4. Experimental data are taken from (26) and shown in Fig. 5 A.
Figure 5.
Recycling experiment and simulation. (A) In vitro data taken from (26) and corresponding in silico values for the number of green- and red-labeled CTLA4 molecules. (B) Model prediction of different CTLA4 pools during the staining procedures (see Eq. S27). The vertical line at t = 60 min indicates the block of green-labeled CTLA4 and the starting time of the second staining step. To see this figure in color, go online.
The corresponding in silico experiment replicates the two staining steps. At first, the number of green-labeled CTLA4 molecules stained on the cell surface and subsequently internalized within 60 min is calculated from the staining model in Fig. 1, A and B. During the CTLA4 block on ice, we assumed that all green-labeled molecules on the cell surface are blocked (denoted by green-blocked in Fig. 5 B) and undergo natural trafficking at normal temperature. In the model, application of the red secondary antibody labels only those CTLA4 molecules that recycle back from the cytosol and do not belong to the blocked subset. This defines a green-red labeled pool of molecules (see Supporting Materials and Methods; CTLA4 Recycling Model).
According to the modeling results (Fig. 5 B), in the second staining step, green-blocked molecules clear monotonically from the cell surface by internalization, whereas the green-blocked cytoplasmic molecules vanish after a transient increase, reaching 88% of total initially blocked CTLA4 molecules after 14 min (t = 74 min) of trafficking. Such a transient increase is also observable for green-red labeled molecules, which originate from the green-labeled cytoplasmic pool. The number of green-red labeled molecules on the cell surface reaches its maximal value (at t = 74 min) faster than the cytoplasmic pool (at t = 147 min) because the labeling is only possible by passing through the cell surface first. The green- and red-labeled CTLA4 molecules clear over time and are replaced with the unlabeled CTLA4 molecules.
In an in silico recycling experiment in which the CTLA4 degradation is blocked (δc = 0) in the second staining step, the cytoplasmic green-labeled (not blocked) CTLA4 is exclusively cleared by recycling with a predicted half-life of ∼28 min. These results suggest that recycling of the cytoplasmic CTLA4 occurs rapidly but significantly slower than internalization (t1/2 = ln(2)/ki = 2.32 min; see Fig. 3 B).
Biological implications
In the previous section, all parameters of the model have been identified by experimental constraints in a simultaneous fitting to all staining experiments. In the following, the model is evaluated and used to generate predictions on CTLA4 dynamics and functionality.
The subcellular CTLA4 distribution is not altered by CTLA4 synthesis
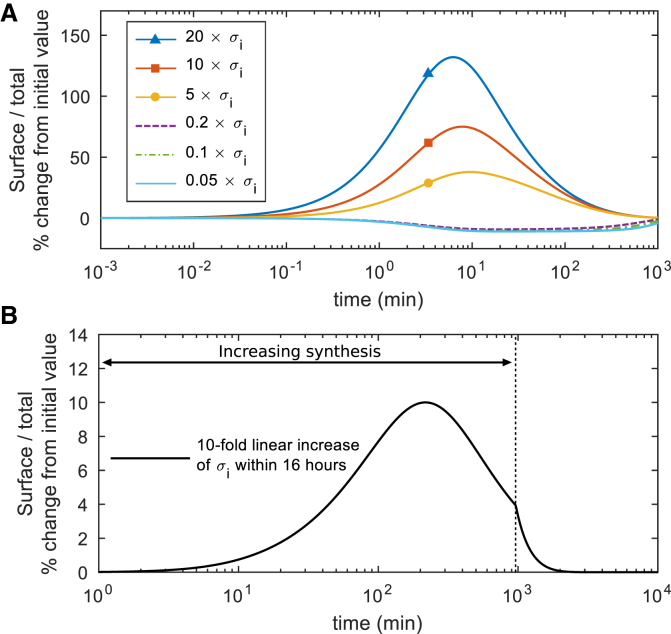
Because Tregs are known to upregulate CTLA4 synthesis upon TCR ligation, we investigated how altering this parameter in the model impacted CTLA4 cellular distribution. The ratio of surface/cytoplasmic CTLA4 in the steady-state condition is independent of the rate of CTLA4 synthesis (see Eq. S2). However, by changing synthesis, the trafficking model deviates from its equilibrium state and the absolute number of CTLA4 molecules changes to a new steady-state value. This transition induces a transient change in the subcellular distribution of CTLA4. By increasing or decreasing the rate of CTLA4 synthesis, the fraction of CTLA4 molecules on the cell surface transiently increases or decreases, respectively, but returns to the steady-state distributions over time. Fig. 6 A shows how a sharp change in the rate of protein synthesis transiently deviates the subcellular distribution, whereas a smooth change in the rate of protein synthesis has less impact on the distribution of CTLA4 molecules (Fig. 6 B). Given the fact that CTLA4 upregulation can be on the order of 10-fold within 16 h (29), these results suggest that the subcellular distribution of CTLA4 is only subtly altered by upregulation of CTLA4 synthesis.
Figure 6.
CTLA4 subcellular distribution transiently changes upon variation of CTLA4 synthesis. The rate of protein synthesis (σi) is varied (A) sharply and (B) smoothly from its original value, and the dynamic change of the fraction of CTLA4 molecules expressed on the cell surface (Rp(t)/(Rp(t) + Rc(t))) is calculated. After a transient change, the fraction converges to its original (steady-state) value (kr + δc)/(ki + kr + δc). Deviation (%) of the fraction from its steady-state value is shown. To see this figure in color, go online.
High affinity ligands exhaust cytosolic CTLA4
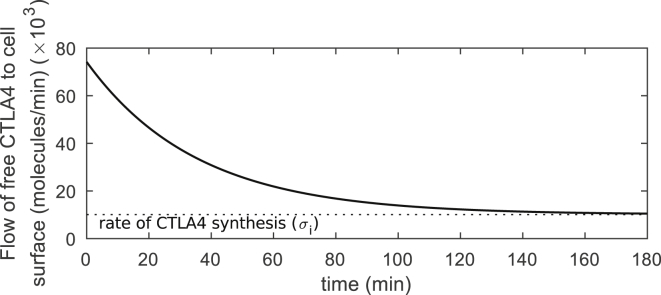
The CTLA4 staining model in Fig. 1, A and B relies on the assumption that labeling antibodies remain bound to and degrade along with the bound CTLA4 molecules. Theoretically, this is a good approximation for ligands with high affinity (high on rate and low off rate) to CTLA4 molecules, such as pharmacological ligands. The rate of ligand removal from the medium to the cytoplasm, in addition to the rate of internalization, also depends on the flow of free (unbound) CTLA4 molecules to the cell surface (Fcs), which is provided by CTLA4 synthesis (σi) and recycling (with rate kr) of the free cytoplasmic CTLA4 molecules (Rc(t))
| (1) |
Upon exposure to high affinity ligands, the free cytoplasmic pool of CTLA4 molecules clears over time (Rc(t) → 0) and reduces by 98% within 2.3 h (see Fig. 1 C, unlabeled cytoplasmic subset). Consequently, the flow of free CTLA4 molecules to the cell surface is reduced and limited to the newly synthesized CTLA4 molecules (Fcs(t) → σi) (see Fig. 7).
Figure 7.
Flow of free CTLA4 to the cell surface. The flow of free CTLA4 molecules to the cell surface is obtained from Eq. 1. This flow converges to the rate of CTLA4 synthesis by clearing the pool of free cytoplasmic CTLA4 molecules (Rc(t)). The analytic function of Rc(t) is given in Eq. S13a.
These results suggest that flow of free CTLA4, which controls the rate at which CTLA4 can internalize new ligands, is increasingly compromised by binding of high-affinity “noncompetitive” ligands, such as monoclonal antibodies. The rate of passage from the medium into the cytoplasm for high-affinity ligands is highest at the time of exposure to the ligands, when there are still many free CTLA4 molecules in the cytoplasm.
Ligand uptake is most efficient at intermediate affinities
To investigate the uptake kinetics of physiological ligands, we employed a ligand uptake model that explicitly considers CTLA4-ligand binding reactions (see Fig. 8). We relaxed the previous assumption in the staining model (Fig. 1, A and B) that ligands are abundantly available in the extracellular environment (assumption A). It is assumed that modeled ligands in the extracellular medium (with concentration Lp(t)) react with free surface CTLA4 (Rp(t)) with a constant on rate (kon) and off rate (koff). The complexes can be internalized and degraded by the rate of CTLA4 internalization (ki) and degradation (δc), respectively. Free ligands appear in the cytoplasm (Lc(t)) via unbinding of the cytoplasmic complexes to the reactants with the rate n × koff, with n ≤ 1. We assumed that free ligands cannot bind again to free CTLA4 in the cytoplasm. Because free ligands in the cytoplasm do not interact with other trafficking molecules in the model, the absolute value of their degradation rate does not impact extracellular ligand uptake. For simplicity, we assumed the same rate as for the degradation of CTLA4-ligand complexes. Unbound CTLA4 molecules directly join other free cytoplasmic CTLA4 molecules (Rc(t)), which subsequently can be recycled (with rate kr) or degraded (with rate δc). The mathematical model is described in Eq. S35 (see Supporting Materials and Methods). We performed in silico cell culture experiments with a cell count of 106 cells/mL. This gives a volume of the extracellular medium per cell (Vγ) equal to 10−9 L/cell. This setting was kept for all in silico experiments.
Figure 8.
Scheme of binding reaction and CTLA4 trafficking for limited extracellular ligand concentration. The corresponding mathematical model is given in Eq. S35.
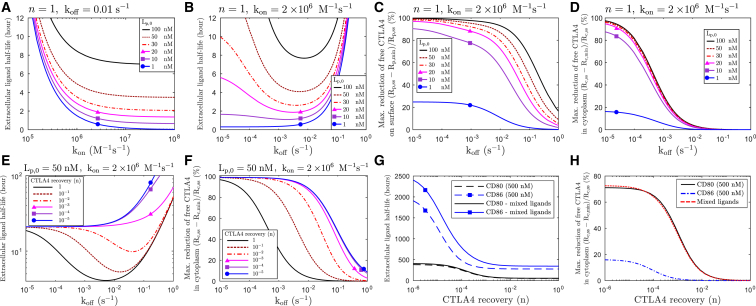
To investigate the role of ligand binding affinities in the kinetics of ligand uptake, the ligand uptake model in Fig. 8 is solved numerically for ligands with different on rates (fixed off rates) or off rates (fixed on rates). The time at which half of the initial extracellular ligand concentration is depleted (extracellular ligand half-life) was monitored for different initial ligand concentrations (see Fig. 9, A and B). These results indicate how fast modeled ligands can be depleted from the extracellular environment via CTLA4 trafficking.
Figure 9.
Extracellular ligand uptake and CTLA4 occupancy. (A and B) The kinetics of ligand uptake is obtained numerically from model Eq. S35 for ligands with different binding affinities. Affinities are varied by changing either (A) on rate (kon) or (B) off rate (koff) for various initial extracellular ligand concentration. (C and D) The maximal reduction of free CTLA4 by ligand uptake is shown for free CTLA4 (C) on the plasma membrane and (D) in the cytoplasm. (E and F) The same representation as in (B) and (D) for different CTLA4 recovery rates n is shown. (G and H) The same representation as in (B) and (D) for an in silico mixed-ligand experiment is shown. A two-ligands version of the ligand-uptake model Eq. S35 was solved with binding affinities of CD80 (kon = 2.15 × 106 M−1 s−1, koff = 0.43 s−1) and CD86 (kon = 1.96 × 106 M−1 s−1, koff = 5.1 s−1) (40), each with initial ligand concentration of 500 nM. The results are compared with single-ligand experiments with the same initial ligand concentration. To see this figure in color, go online.
According to Fig. 9 A, the uptake of the extracellular ligands with higher on rates (higher affinities) is faster. In contrast, this process is optimally fast only for an intermediate range of off-rate values (see Fig. 9 B). For low off rates (high-affinity ligands), uptake is not efficient because the recovery of CTLA4 from internalized CTLA4-ligand complex and their subsequent recycling is not efficient. For high off rates (low-affinity ligands), CTLA4-ligand complexes on the cell surface are not stable and complex internalization is limited by complex concentration, which results in an inefficient ligand uptake. We conclude that ligand removal from the medium is monotonic in dependence on on rates but exhibits a maximal efficiency at intermediate off rates.
To evaluate the capacity of modeled ligands to bind and reduce free CTLA4 molecules, the minimal number of the free CTLA4 molecules (Rp,min and Rc,min) is obtained during the process of ligand uptake. The degree of reduction from the initial (steady-state) value is depicted in Fig. 9, C and D and shows that this reduction on the plasma membrane depends on both extracellular ligand concentration and ligand binding affinity (see Fig. 9 C). CTLA4 depletion in the cytoplasm is largely independent of the ligand concentration above 10 nM (see Fig. 9 D). A similar level of reduction in the free cytoplasmic and surface CTLA4 occurs on different values of off rates, with lower off rates in the cytoplasm (see Fig. 9, C and D). This difference results from a complex interplay of different parameters in the model.
To further investigate the role of the CTLA4 recovery, we varied the factor n in the range 0 < n ≤ 1; lower n corresponds to slower CTLA4 recovery. By reducing the CTLA4 recovery (n), ligand uptake becomes slower (see Fig. 9 E) because of a reduced number of free CTLA4 molecules in the cytoplasm (see Fig. 9 F). When CTLA4 recovery (n) is highly reduced, no optimal off rate can be found at which the ligand uptake is maximum. In other words, in the absence of the CTLA4 recovery, ligand removal from the medium is monotonic in dependence on off rates. Therefore, the process of CTLA4 recovery facilitates uptake of ligands with intermediate off rates.
The presence of CD80 dominates CTLA4 function
A mixture of ligands with different binding affinities results in a binding competition for CTLA4, which may impact the trafficking of CTLA4 and the kinetics of ligand uptake. To investigate the effect of such binding competition for natural ligands (CD80/CD86), we employed an in silico mixed-ligands experiment by solving a two-ligands version of the model Eq. S35. Because the impact of binding affinity/avidity of natural ligands on the rate of CTLA4 recovery (n × koff) is unknown, we obtained ligand-uptake kinetics for different values of the CTLA4 recovery by changing the value of parameter n (0 < n ≤ 1).
The reduction of free cytoplasmic CTLA4 is compared between single-ligand and mixed-ligands experiments. Irrespective of the degree of CTLA4 recovery, CD86 uptake is affected by the presence of CD80 molecules, whereas CD80 removal is almost independent of the presence of CD86 molecules (Fig. 9 G). The extent of reduction of free cytoplasmic CTLA4 (for low to intermediate values of n) in the mixed-ligands experiment is similar to the case of exposure to CD80 alone (Fig. 9 H). The effect of ligand competition on the kinetics of ligand removal is reduced when the concentration of ligands is low (data not shown). This result suggests that CD86 removal is affected by the presence of CD80 but not vice versa.
Expression of adequate CTLA4 does not guarantee efficient ligand uptake
The process of ligand uptake relies on an efficient flow of free CTLA4 to the plasma membrane, provided by synthesis (σi) and recycling of free cytoplasmic CTLA4 (krRc(t); see Eq. 1). Therefore, alterations in the synthesis and recycling processes, as well as a reduced number of free CTLA4 molecules in the cytoplasm, can result in inefficient ligand uptake. The steady-state number of free cytoplasmic CTLA4 molecules (Rc,ss) is independent of the recycling process, directly related to the synthesis, and inversely related to the degradation process (see Eq. S2). Therefore, to quantify the impact of impairment in recycling and CTLA4 expression on the kinetics of ligand uptake, we varied separately the rates of recycling , lysosomal degradation and synthesis .
Reduced recycling does not significantly alter total free CTLA4 (∼7.5% reduction when recycling is completely blocked, i.e., ). In contrast, increased lysosomal degradation strongly reduces total CTLA4 (compare Fig. 10, A and C). Alteration in both processes can slow down the ligand-uptake process to a similar extent (compare Fig. 10, B and D). This result suggests that a normal process of ligand removal does not only rely on an adequate number of CTLA4 molecules (limited lysosomal degradation) but also on an effective recycling process. In other words, expression of an adequate number of CTLA4 molecules does not guarantee efficient ligand uptake, and efficiency of the recycling process needs to be taken into account.
Figure 10.
Alterations in recycling, degradation, and synthesis. Impact of alteration in the rate of recycling and degradation on (A and C) the steady-state number of free CTLA4 molecules and (B and D) the kinetics of ligand uptake. The fold change in the steady-state number of free CTLA4 molecules by changing the rate of (A) recycling or (C) lysosomal degradation , is calculated from Eq. S2. kr, δl, and σi denote the estimated natural rates for CTLA4 recycling, lysosomal degradation, and synthesis, respectively, given in Table 1. The ligand uptake model in Fig. 8 (Eq. S35) is solved numerically with altered rates of (B) recycling , (D) lysosomal degradation , and (E) synthesis for different initial ligand concentrations (Lp,0), and (F) the half-life of extracellular ligand concentration is shown for different CTLA4 synthesis rates and off rates. The maximal degree of reduction of free CTLA4 molecules (G) on the cell surface and (H) in the cytoplasm is obtained for different rates of CTLA4 synthesis and off rates. Rp,ss and Rc,ss are the steady-state number of free CTLA4 molecules in the absence of ligand and are evaluated for each of the considered synthesis rates. To see this figure in color, go online.
Alteration in the rate of CTLA4 synthesis directly impacts surface, cytoplasmic, and therefore total CTLA4 with a similar factor (see Eq. S2). An increased rate of synthesis results in a faster ligand uptake (see Fig. 10 E). Depending on the extracellular concentration of the ligands, an extreme increase of the synthesis rate could destroy the existence of the off-rate range in which ligand uptake is efficient (see Fig. 10 F, ). However, an intermediate increase retains this property. An excess of CTLA4 synthesis reduces the degree of reduction in the free cytoplasmic CTLA4 but has a limited impact on the free surface CTLA4 molecules (see Fig. 10, G and H). This result suggests that an increased rate of CTLA4 synthesis can reduce the extent of CTLA4 occupancy in the cytoplasm and its impact on the kinetics of ligand uptake.
Discussion
In the cell-extrinsic regulatory function of CTLA4, an efficient depletion of costimulatory ligands (CD80/CD86) via transendocytosis relies on the trafficking features of CTLA4. In this study, an intracellular trafficking model is constructed by taking into account the essential subprocesses involved in the constitutive trafficking of transmembrane proteins. Starting from a simplistic mathematical model of CTLA4 trafficking, we only added extra complexities when necessary to capture the specifics of different independent experiments used for characterizing the kinetics of CTLA4 molecules. This approach enabled us to carefully estimate all CTLA4 trafficking parameters in the original model (see Table 1) and avoid uncertainties arising from considering a complex model that suffers from unidentifiable parameters. Some aspects of CD28 and CTLA4 signaling have been investigated by mathematical modeling. Using a deterministic model, Jansson et al. (30) considered CD28 and CTLA4 interactions with their natural ligands in immunological synapse in the absence of ligand uptake. More recently, a spatially resolved stochastic model of T cell-APC interaction has been developed that assumes CTLA4 and ligand internalization without quantitative parameter estimation (31). The estimated parameters in our work can inform such theoretical attempts focusing on detailed events during T cell-APC interactions, in which CTLA4 and its quantitatively unique trafficking features are essential.
The analytic solutions derived from the mathematical models revealed the explicit dependencies of experimental measurements on each of the trafficking parameters. For example, these explicit solutions verified that the distribution (Eq. S2) and the observed kinetics of CTLA4 (Eqs. S17, S21, S25, S33, and S34) are independent of the absolute value for the rate of protein synthesis. Therefore, cells with different steady-state levels of CTLA4 synthesis would still exhibit similar CTLA4 behavior. We estimated a synthesis rate of ∼104 molecules/min/cell for CHO cells, which is quite high because of the expression on a vector construct. Given that T cells are smaller in size and the total number of CTLA4 molecules is lower compared to CHO cells (∼38-fold lower MFI in stimulated human CD45RA Tregs, data not shown), a lower rate of CTLA4 synthesis in T cells is expected and is likely to vary substantially for different cell types and conditions. Nonetheless, our estimated internalization rate of CTLA4 is within the same order of magnitude as transferrin receptor, a carrier protein of iron-bound transferrin, with 50% of surface transferrin receptor being internalized within 5 min (32, 33). This is consistent with their uptake by similar AP2 mechanisms.
Our CHO model, combined with the mathematical model, provided a setup to establish the relevant parameters of CTLA4 trafficking by which the impact of the parameter variations on CTLA4 biology can be investigated. Although we recognize that CHO cells are likely to have differences to primary T cells, they nonetheless provide a starting point for our combined modeling approach. The exact parameter values in T cells will depend on cell type (e.g., naïve or memory Treg), activation state, and the T-cell stimulus; therefore, values are bound to vary. CTLA4 molecules show an unusual distribution in both CHO and T cells, with a high fraction of total CTLA4 being present in the cytoplasm at equilibrium. The estimated parameters confirmed the expectation that this distribution results from very fast trafficking rates in which the ratio of internalization/recycling rates is high (in the range of ∼12 in our cell line). The concepts underpinning our model are largely compatible with the behavior of CTLA4 seen in primary T cells in our experience (26). Moreover, these concepts have been used to predict the function of CTLA4 on Tregs (34) as well as measure deficits in CTLA4 function in T cells derived from patients (29, 35). Taken together, these suggest that the concepts described here are generally robust and relevant to the behavior of primary human T cells. We have found that this large storage of CTLA4 in the cytoplasm, in combination with fast recycling, provides a temporary compensation for surface CTLA4 loss when protein synthesis is interrupted (see Fig. 3 and unlabeled surface CTLA4 in Fig. 2 C). Physiologically, one can therefore consider that additional CTLA4 molecules can be recruited to the plasma membrane in the absence of new protein synthesis, which fits well with the rapid increase in CTLA4 at the immune synapse that precedes new protein synthesis. The rate of CTLA4 molecules newly expressed on the cell surface depends on both CTLA4 synthesis and the number of recycled free CTLA4 molecules (see Eqs. S1 and 1). Together with the speed of internalization (ln(2)/ki = 2.3 min), these processes determine how rapidly antibodies/ligands are removed from the extracellular medium or the opposing cell. According to the model, the flow of free CTLA4 molecules to the plasma membrane is highest at the time of initial exposure to antibodies or ligands and decreases within ∼2.3 h (see Fig. 7). During this period of time, the pool of free cytoplasmic CTLA4 molecules is exhausted and replaced by an antibody-labeled pool in the case of high-affinity antibodies (see Fig. 1 C). In this situation, the rate of antibody removal from the medium is decreased and ultimately limited to the binding of newly synthesized CTLA4 molecules. Viewed in the context of the transendocytosis model, this result suggests that efficient ligand capture may be achieved when CTLA4-expressing T cells remain bound to a single APC for a short duration (less than 2.3 h). The T cell experiences a period during which the depleted free cytoplasmic CTLA4 is restored and the internalized ligands are degraded. This may limit the efficiency of CTLA4-expressing T cells in removing target molecules from multiple subsequent APCs or the same APC over extended times. In a two-photon imaging study, it was shown that Tregs, which constitutively express CTLA4 (36, 37), make transient interactions with lymph node resident dendritic cells with an average duration of 3.3 min and up to 40 min with migratory activated dendritic cells (38) suggesting rapid depletion by CTLA4 is essential. These measured contact durations, according to the model, would allow for an efficient and persistent removal of costimulatory molecules.
The above limitation induced by the depletion of the free cytoplasmic CTLA4 and an increase in bound CTLA4 can be overcome if CTLA4 molecules return to the recycling pool after ligand binding (CTLA4 recovery). Our results show that an optimal affinity must exist at which CTLA4 most efficiently takes up ligands. If affinity is too weak, then binding to ligands on the cell surface is compromised. However, the data suggest that if affinity is too high, then this may compromise the regeneration of a recycling pool of unbound CTLA4. In this scenario, CTLA4 molecules bound to their target would fail to dissociate from the target after internalization and show delayed re-entry into the pool of free cytosolic CTLA4 available for recycling (see Fig. 8). Given that the CTLA4-ligand off rate would control the rate of CTLA4 recovery, this indicates that the two ligands may differ substantially. On the one hand, a low off rate due to too-high affinity/avidity would limit CTLA4 recovery. On the other hand, a low off rate increases interaction on the cell surface and by this increases the probability of transendocytosis. Therefore, a subtle balance between ligand-binding reaction and CTLA4 trafficking is needed for maintenance of efficient transendocytosis in a stable CTLA4+ T cell-APC interaction. The model predicts how the kinetics of ligand uptake is altered by variations in binding affinities (Fig. 9) and trafficking parameters (Fig. 10).
Our simulations for ligand removal process aimed to investigate the natural ligands (CD80/CD86) as well as therapeutic ligands that block CTLA4 for therapeutic purposes. The therapeutic ligands typically bind CTLA4 with higher affinities than natural ligands, which enables their exploitative binding. For example, Ipilimumab (kD = 18.2 nM) and Tremelimumab (kD = 5.89 nM) bind CTLA4 with higher affinity compared to natural ligands (kD,CD86 = 2.6 μM, kD,CD80 = 0.2 μM) (39, 40). A mapping between affinities (off rates) to the uptake kinetics of physiological ligands in our model should be made with caution because of the factors that cannot be inferred experimentally and the impact of multivalent avidity affecting ligand binding. These processes include recovery of CTLA4 from internalized complexes. In addition to dissociation, other processes are involved in guiding recovered CTLA4 to the recycling endosomes that slow down the CTLA4 recovery. We therefore introduced the parameter n (n ≤ 1) in the rate of CTLA4 recovery (n × koff) to account for these uncertainties. By reducing the value of n, we observed the impact of CTLA4 recovery on ligand removal (see Fig. 9, E–H). Our results predict a nonlinear relationship between ligand-CTLA4 affinity and extracellular ligand half-life that is not validated experimentally and motivates further experiments.
For modeled ligands with off rates in the range of CD80/CD86 off rates, free cytoplasmic CTLA4 is not reduced irrespective of the ligand concentration (see Fig. 9 D for off rates higher than 10−2 (s−1) and Fig. 9 H for values of n higher than 10−1). This suggests that the flow of free CTLA4 to the surface is not affected by complex formation with the natural ligand and complex internalization, and consequently, ligand uptake keeps working at maximal capacity. However, this is based on monomeric affinities, whereas the interaction avidities are unknown. The CD80-CTLA4 interactions are dimer-dimer interactions with the potential for very substantial avidity enhancement (41). In contrast, CD86-CTLA4 interactions appear to be monomeric. Accordingly, this offers the clear possibility that available CTLA4 for recycling may differ substantially between the two ligands (see Fig. 9 H for low values of n that would correspond to higher avidities). In this regard, it is of considerable interest that our data show that CD80-CTLA4 interactions are dominant over those of CD86 and CD80 affects clearance of CD86 but not vice versa (see Fig. 9 G). Although the dominance of CD80 could be readily explained by its high affinity/avidity for CTLA4, our in silico results suggest that this dominance could also be partially impacted by the CTLA4 occupancy with CD80 in the cytoplasm and shortage of the recycling free CTLA4. Consistent with this, we have observed experimentally that capture of CD80 largely inhibits uptake of CD86 when both are coexpressed on CHO cells (data not shown). In the context of T-cell activation, this would suggest enhanced CD28-CD86 binding and signaling when CD80 is expressed.
In the context of T cell-APC interactions, upon cytoskeletal polarization of T cells toward APCs, cytoplasmic CTLA4 is translocated to the immunological synapse, which may facilitate CTLA4 exocytosis and increase CTLA4 surface density in the quasi-two-dimensional interaction face (24, 35). This could result in a significantly higher binding rate of ligands and CTLA4 than in solution.
Lipopolysaccharide-responsive and beige-like anchor protein (LRBA) is an intracellular protein that colocalizes with CTLA4 in endosomal vesicles. Patients with LRBA mutations exhibit a reduction in CTLA4 expression in the nonactivated state (42). However, because CTLA4 is synthesized normally, activation of T cells can still upregulate CTLA4 (29). It has been suggested that the interaction with LRBA in the endosomes rescues CTLA4 from lysosomal degradation such that LRBA deficiency results in higher degradation and lower recycling of CTLA4 (42, 43). Our model predicted that recycling defects alone cannot account for a significant reduction in the total number of CTLA4 molecules, in contrast to the impact of lysosomal degradation (see Fig. 10, A–C). Given that the defect in CTLA4 expression in some patients with LRBA mutation is not very large after activation (29), our in silico results suggest that the problem in patients is mainly the need for effective recycling to capture costimulatory ligands.
This experimentally validated model provides a framework for, to our knowledge, new insights into the control of immune T-cell responses. Having established such a model, important biological questions related to immune regulation and autoimmune disease can be addressed. We have addressed how the two natural ligands CD80 and CD86, with different binding affinities for CTLA4, might have differential impact on CTLA4 biology. We have analyzed the hypothetical impact of biologically relevant changes in parameters affecting synthesis, recycling, ligand affinity, and competition, revealing insights into behavior that can now be further explored. Moreover, it is already evident that mutations in CTLA4 (44, 45) and most likely in its ligands will occur in the context of immunological diseases. Such mutations may affect the binding affinity with unknown consequences. Accordingly, with additional experimental data, it would become possible to predict the impact of such changes on immune function with the help of this model.
Author Contributions
M.M.-H., L.S.K.W., and D.M.S. designed research. S.K. developed and analyzed mathematical models. S.K. and J.D.B.H. performed MCMC analysis. B.R., E.W., and N.H. performed experiments. All authors contributed in analyzing the data and writing the manuscript.
Acknowledgments
The authors wish to thank Sebastian Binder and Ghazal Montaseri for revising the manuscript.
S.K. acknowledges the support of the German Federal Ministry of Education and Research for the eMED project SYSIMIT. B.R. and E.W. were supported by UK Biotechnology and Biological Sciences Research Council. P.A.R. and M.M.-H. were supported by the Human Frontier Science Program (RGP0033/2015). M.M.-H was supported by iMed the Helmholtz Initiative on Personalized Medicine. L.S.K.W. was supported by the Medical Research Council. N.H. and D.M.S. were supported by the Wellcome Trust. J.D.B.H. was supported by the Helmholtz International Graduate School for Infection Research, grant number VHGS-202.
Editor: Ruth Baker.
Footnotes
Supporting Materials and Methods, three figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30973-1.
Contributor Information
David M. Sansom, Email: d.sansom@ucl.ac.uk.
Michael Meyer-Hermann, Email: mmh@theoretical-biology.de.
Supporting Material
References
- 1.Sansom D.M. CD28, CTLA-4 and their ligands: who does what and to whom? Immunology. 2000;101:169–177. doi: 10.1046/j.1365-2567.2000.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenschow D.J., Walunas T.L., Bluestone J.A. CD28/B7 system of T cell costimulation. Annu. Rev. Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- 3.Keir M.E., Sharpe A.H. The B7/CD28 costimulatory family in autoimmunity. Immunol. Rev. 2005;204:128–143. doi: 10.1111/j.0105-2896.2005.00242.x. [DOI] [PubMed] [Google Scholar]
- 4.Linsley P.S., Brady W., Ledbetter J.A. Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation. J. Exp. Med. 1991;173:721–730. doi: 10.1084/jem.173.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson C.B., Lindsten T., June C.H. CD28 activation pathway regulates the production of multiple T-cell-derived lymphokines/cytokines. Proc. Natl. Acad. Sci. USA. 1989;86:1333–1337. doi: 10.1073/pnas.86.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain N., Nguyen H., Kang J. Dual function of CTLA-4 in regulatory T cells and conventional T cells to prevent multiorgan autoimmunity. Proc. Natl. Acad. Sci. USA. 2010;107:1524–1528. doi: 10.1073/pnas.0910341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tivol E.A., Borriello F., Sharpe A.H. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 8.Waterhouse P., Penninger J.M., Mak T.W. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 9.Walunas T.L., Lenschow D.J., Bluestone J.A. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1:405–413. doi: 10.1016/1074-7613(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 10.Krummel M.F., Allison J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmann M.F., Waterhouse P., Ohashi P.S. Normal responsiveness of CTLA-4-deficient anti-viral cytotoxic T cells. J. Immunol. 1998;160:95–100. [PubMed] [Google Scholar]
- 12.Bachmann M.F., Köhler G., Kopf M. Cutting edge: lymphoproliferative disease in the absence of CTLA-4 is not T cell autonomous. J. Immunol. 1999;163:1128–1131. [PubMed] [Google Scholar]
- 13.Bachmann M.F., Gallimore A., Kopf M. Normal pathogen-specific immune responses mounted by CTLA-4-deficient T cells: a paradigm reconsidered. Eur. J. Immunol. 2001;31:450–458. doi: 10.1002/1521-4141(200102)31:2<450::aid-immu450>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 14.Homann D., Dummer W., von Herrath M.G. Lack of intrinsic CTLA-4 expression has minimal effect on regulation of antiviral T-cell immunity. J. Virol. 2006;80:270–280. doi: 10.1128/JVI.80.1.270-280.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wing K., Onishi Y., Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 16.Qureshi O.S., Zheng Y., Sansom D.M. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science. 2011;332:600–603. doi: 10.1126/science.1202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker L.S., Sansom D.M. Confusing signals: recent progress in CTLA-4 biology. Trends Immunol. 2015;36:63–70. doi: 10.1016/j.it.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linsley P.S., Bradshaw J., Mittler R.S. Intracellular trafficking of CTLA-4 and focal localization towards sites of TCR engagement. Immunity. 1996;4:535–543. doi: 10.1016/s1074-7613(00)80480-x. [DOI] [PubMed] [Google Scholar]
- 19.Leung H.T., Bradshaw J., Linsley P.S. Cytotoxic T lymphocyte-associated molecule-4, a high-avidity receptor for CD80 and CD86, contains an intracellular localization motif in its cytoplasmic tail. J. Biol. Chem. 1995;270:25107–25114. doi: 10.1074/jbc.270.42.25107. [DOI] [PubMed] [Google Scholar]
- 20.Shiratori T., Miyatake S., Saito T. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity. 1997;6:583–589. doi: 10.1016/s1074-7613(00)80346-5. [DOI] [PubMed] [Google Scholar]
- 21.Chuang E., Alegre M.L., Thompson C.B. Interaction of CTLA-4 with the clathrin-associated protein AP50 results in ligand-independent endocytosis that limits cell surface expression. J. Immunol. 1997;159:144–151. [PubMed] [Google Scholar]
- 22.Zhang Y., Allison J.P. Interaction of CTLA-4 with AP50, a clathrin-coated pit adaptor protein. Proc. Natl. Acad. Sci. USA. 1997;94:9273–9278. doi: 10.1073/pnas.94.17.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider H., Martin M., Rudd C.E. Cytolytic T lymphocyte-associated antigen-4 and the TCR zeta/CD3 complex, but not CD28, interact with clathrin adaptor complexes AP-1 and AP-2. J. Immunol. 1999;163:1868–1879. [PubMed] [Google Scholar]
- 24.Egen J.G., Allison J.P. Cytotoxic T lymphocyte antigen-4 accumulation in the immunological synapse is regulated by TCR signal strength. Immunity. 2002;16:23–35. doi: 10.1016/s1074-7613(01)00259-x. [DOI] [PubMed] [Google Scholar]
- 25.Mead K.I., Zheng Y., Sansom D.M. Exocytosis of CTLA-4 is dependent on phospholipase D and ADP ribosylation factor-1 and stimulated during activation of regulatory T cells. J. Immunol. 2005;174:4803–4811. doi: 10.4049/jimmunol.174.8.4803. [DOI] [PubMed] [Google Scholar]
- 26.Qureshi O.S., Kaur S., Sansom D.M. Constitutive clathrin-mediated endocytosis of CTLA-4 persists during T cell activation. J. Biol. Chem. 2012;287:9429–9440. doi: 10.1074/jbc.M111.304329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iida T., Ohno H., Saito T. Regulation of cell surface expression of CTLA-4 by secretion of CTLA-4-containing lysosomes upon activation of CD4+ T cells. J. Immunol. 2000;165:5062–5068. doi: 10.4049/jimmunol.165.9.5062. [DOI] [PubMed] [Google Scholar]
- 28.Catalfamo M., Tai X., Henkart P.A. TcR-induced regulated secretion leads to surface expression of CTLA-4 in CD4+CD25+ T cells. Immunology. 2008;125:70–79. doi: 10.1111/j.1365-2567.2008.02822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou T.Z., Verma N., Sansom D.M. Identifying functional defects in patients with immune dysregulation due to LRBA and CTLA-4 mutations. Blood. 2017;129:1458–1468. doi: 10.1182/blood-2016-10-745174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansson A., Barnes E., Nilsson P. A theoretical framework for quantitative analysis of the molecular basis of costimulation. J. Immunol. 2005;175:1575–1585. doi: 10.4049/jimmunol.175.3.1575. [DOI] [PubMed] [Google Scholar]
- 31.Sugár I.P., Das J., Sealfon S.C. Multiscale modeling of complex formation and CD80 depletion during immune synapse development. Biophys. J. 2017;112:997–1009. doi: 10.1016/j.bpj.2016.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciechanover A., Schwartz A.L., Lodish H.F. Kinetics of internalization and recycling of transferrin and the transferrin receptor in a human hepatoma cell line. Effect of lysosomotropic agents. J. Biol. Chem. 1983;258:9681–9689. [PubMed] [Google Scholar]
- 33.Watts C. Rapid endocytosis of the transferrin receptor in the absence of bound transferrin. J. Cell Biol. 1985;100:633–637. doi: 10.1083/jcb.100.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou T.Z., Qureshi O.S., Sansom D.M. A transendocytosis model of CTLA-4 function predicts its suppressive behavior on regulatory T cells. J. Immunol. 2015;194:2148–2159. doi: 10.4049/jimmunol.1401876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokosuka T., Kobayashi W., Saito T. Spatiotemporal basis of CTLA-4 costimulatory molecule-mediated negative regulation of T cell activation. Immunity. 2010;33:326–339. doi: 10.1016/j.immuni.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi T., Tagami T., Sakaguchi S. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J. Exp. Med. 2000;192:303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Read S., Malmström V., Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J. Exp. Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matheu M.P., Othy S., Cahalan M.D. Imaging regulatory T cell dynamics and CTLA4-mediated suppression of T cell priming. Nat. Commun. 2015;6:6219. doi: 10.1038/ncomms7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He M., Chai Y., Gao G.F. Remarkably similar CTLA-4 binding properties of therapeutic ipilimumab and tremelimumab antibodies. Oncotarget. 2017;8:67129–67139. doi: 10.18632/oncotarget.18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins A.V., Brodie D.W., Davis S.J. The interaction properties of costimulatory molecules revisited. Immunity. 2002;17:201–210. doi: 10.1016/s1074-7613(02)00362-x. [DOI] [PubMed] [Google Scholar]
- 41.Ikemizu S., Gilbert R.J., Davis S.J. Structure and dimerization of a soluble form of B7-1. Immunity. 2000;12:51–60. doi: 10.1016/s1074-7613(00)80158-2. [DOI] [PubMed] [Google Scholar]
- 42.Lo B., Zhang K., Jordan M.B. AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science. 2015;349:436–440. doi: 10.1126/science.aaa1663. [DOI] [PubMed] [Google Scholar]
- 43.Sansom D.M. IMMUNOLOGY. Moving CTLA-4 from the trash to recycling. Science. 2015;349:377–378. doi: 10.1126/science.aac7888. [DOI] [PubMed] [Google Scholar]
- 44.Schubert D., Bode C., Grimbacher B. Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat. Med. 2014;20:1410–1416. doi: 10.1038/nm.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuehn H.S., Ouyang W., Uzel G. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014;345:1623–1627. doi: 10.1126/science.1255904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.