Abstract
The multiplicity of new therapies for breast cancer presents a challenge for treatment selection. We describe a 17-gene digital signature of breast circulating tumor cell (CTC)-derived transcripts enriched from blood, enabling high-sensitivity early monitoring of response. In a prospective cohort of localized breast cancer, an elevated CTC-Score after three cycles of neoadjuvant therapy is associated with residual disease at surgery (p=0.047). In a second prospective cohort with metastatic breast cancer, baseline CTC-Score correlates with overall survival (p= 0.02), as does persistent CTC signal after four weeks of treatment (p=0.01). In the subset with estrogen receptor (ER)-positive disease, failure to suppress ER-signaling within CTCs after three weeks of endocrine therapy predicts early progression (p=0.008). Drug-refractory ER signaling within CTCs overlaps partially with presence of ESR1 mutations, pointing to diverse mechanisms of acquired endocrine drug resistance. Thus, CTC-derived digital RNA signatures enable noninvasive pharmacodynamic measurements to inform therapy in breast cancer.
Introduction
Recent advances in therapeutics have revolutionized the management of breast cancer, with the approval of more than a dozen drugs in the past few years, including four new agents in 2017. In Hormone Receptor-positive (HR+) breast cancer, new endocrine therapies may be combined with cyclin-dependent kinase (CDK) 4/6 inhibitors 1–3, and drugs inhibiting growth factor receptors (e.g. IGFR, FGFR), oncogenic signaling pathways (e.g. PI3K, AKT, mTOR) and chromatin modifiers (e.g. HDAC) are under active clinical investigation4. Despite these increasingly effective therapeutic choices, there are few biomarkers to guide initial therapy selection and identify early responses. As a result, treatment choices are frequently empiric, and delayed clinical ascertainment of tumor response limits the ability to rapidly define an effective regimen for an individual patient.
Traditionally, assessing therapy response in metastatic breast cancer involves monitoring serum cancer antigen protein markers, such as CA15–3 and CEA, along with radiographic assessment of tumor volumes 5, 6. However in many cases, breast cancer involves bone metastases, whose responses to drug treatment are not readily assessed with radiographic imaging7 and the sensitivity and accuracy of established protein serum markers is limited4, 8. Recently, longitudinal monitoring of tumor-derived mutations detected in plasma (ctDNA) has been used as a measure of tumor response 9–11. This approach may provide a high degree of genetic information, although it typically requires initial sequencing of the primary tumor to design individualized mutational markers for each patient. Genetic heterogeneity in advanced disease may present an additional challenge in monitoring multiple subclonal mutant alleles with divergent trends following therapy 12.
While early assessment of tumor response in metastatic breast cancer presents challenges, neoadjuvant treatment of localized breast cancer is empiric and lacks early measurements of drug response, which is only ultimately evident at the time of surgical resection. In women with high risk localized breast cancer, multiple courses of neoadjuvant pre-operative chemotherapy or hormonal therapy may be administered, with the goal of reducing initial tumor burden and improve the outcome of subsequent surgical resection. ctDNA genotyping has been used to detect early relapse after neoadjuvant treatment and surgical resection, but it typically requires initial tumor sequencing and design of mutation-specific assays to measure the low fraction of mutant alleles in this minimal disease setting 10, 13. Ultra sensitive techniques to detect multiple recurrent somatic mutations in plasma without initial tumor genotyping may provide an early indication of tumor recurrence in colorectal cancer10, 13,14, but given the genetic heterogeneity of breast cancer, there is an unmet need for mutation-agnostic biomarkers for noninvasive monitoring.
CTCs are shed from tumors into the bloodstream, where a small percentage may survive, extravasate and colonize distant sites 15, 16. As such, CTCs offer a noninvasive source of whole tumor cell-derived material for serial analysis during therapy. Microscopy-based enumeration of CTCs has been established as a biomarker in metastatic breast cancer, with both the CTC number at baseline and treatment-induced changes of the number of CTCs being prognostic of progression-free (PFS) and overall survival (OS) in the context of chemotherapy treatment 17, 18. However, to date, the relatively low sensitivity and technological complexity of CTC imaging, combined with the absence of robust molecular characterization, have limited the clinical application of CTCs to guide therapeutic decision-making 19–21.
Recent advances in microfluidics have enabled the enrichment of viable and intact CTCs through the depletion of normal blood cells 22,23. Microfluidic antibody-based removal of hematopoietic cells, rather than positive capture of CTCs, enables enrichment of CTCs independent of their variable cell surface epitopes, and it also ensures CTC cellular integrity and high RNA quality 24–26. To enhance CTC detection signal following such microfluidic enrichment, we recently developed a quantitative RNA-based digital PCR scoring assay, individualized to cancer type-specific markers 27–29. The transcriptional CTC signature takes advantage of tissue lineage-associated transcripts expressed in cancer cells but absent in the normal blood cells present in the CTC-enriched product. We now describe the development of a breast cancer CTC-specific assay, providing both digital quantitation of CTC burden and intracellular ER signaling measurements, and we test its clinical utility in a prospectively monitored cohort of women receiving neoadjuvant therapy for high risk localized breast cancer and in a second prospective cohort of women with advanced metastatic breast cancer.
Results
Development of a breast cancer-specific RNA signature for CTC detection
The microfluidic CTC-iChip achieves approximately 4 log depletion of WBCs, RBCs and platelets, resulting in an output with 0.1–10% CTC purity, depending on initial CTC burden 23, 26. To develop a RNA expression signature capable of detecting breast cancer cells within the background of normal blood cells, we first analyzed RNAseq and microarray gene expression data sets derived from normal breast tissue, breast cancer and whole blood (Fig. S1, Table S1.). We ultimately selected 17 markers strongly expressed in breast-derived tissues but virtually absent in blood cells (see Methods, Fig. 1A). The markers include breast lineage-specific transcripts (PGR, SCGB2A1, PIP) and transcripts highly expressed in breast cancer (MGP, EFHD1), as well as gene products implicated in endocrine signaling (SERPINA3, WFDC2), endocrine drug resistance (AGR2), cancer growth and metastasis (MUC16, TMPRSS4), cellular signaling (FAT1, FAT2, SFRP1, SFRP2), epithelial-derived cytokines (CXCL13, CXCL14), and oncofetal antigens (PRAME). Single cell RNA sequencing confirmed the heterogenous expression of the 17 markers in 15 individual CTCs isolated from the blood of women with metastatic breast cancer; 5 similarly analyzed single WBCs have negligible expression of these genes (Fig. 1B). For maximal sensitivity and high throughput capability, we developed a droplet digital PCR assay (ddPCR) for each of the 17 markers, which was applied to the CTC-enriched product after an initial 8-cycle whole transcriptome amplification (WTA) (see Methods).
Figure 1.
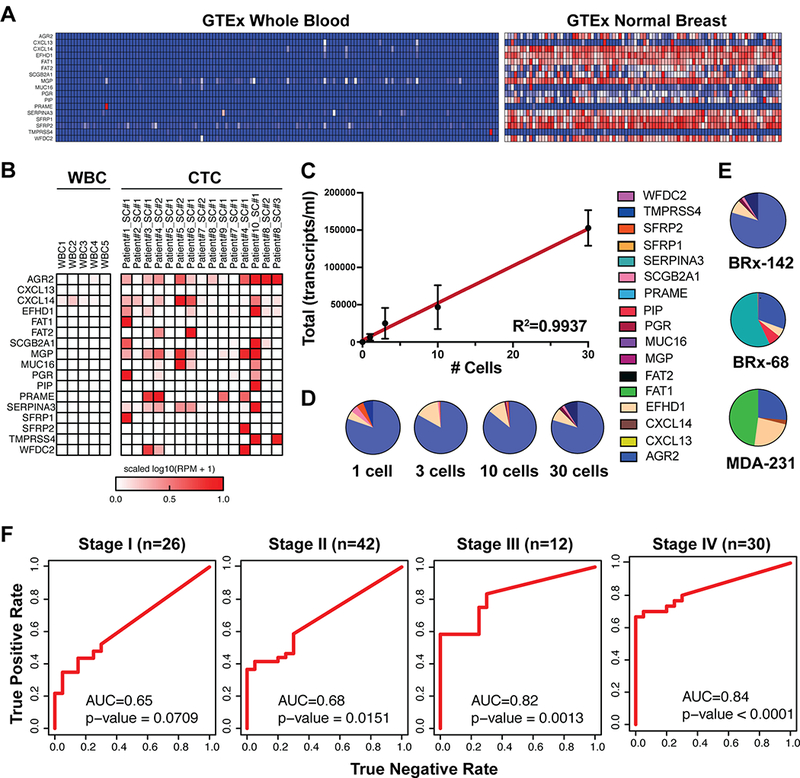
Development and validation of the breast cancer CTC-droplet digital (dd)PCR assay. A, Expression of the 17 selected breast CTC markers in whole blood versus normal breast tissue (GTeX database). B, Single-cell RNAseq-derived expression of the 17 breast CTC markers in 5 white blood cells and in 15 individually sequenced primary CTCs from women with metastatic breast cancer. C, CTC signal (total transcripts/ml of blood) from 0, 1, 3, 10 and 30 BRx-142 cells introduced into 4 ml healthy donor (HD) blood, followed by microfluidic CTC-enrichment and dd-PCR analysis (n=2, dots represent means, error bars represent SD, best fit line and R-squared statistics of the linear regression model are shown). D, Contribution of individual breast CTC markers to the signal detected from different numbers of BRx-142 cells introduced into blood. E, Contribution of individual markers to the signal detected from 30 cells each from three different cell lines (BRx-142, BRx-68 and MDA-231) introduced into blood. In all cases, cells were added to 4 ml of whole blood from HD and processed through the CTC-iChip for enrichment prior to ddPCR analysis. F, Receiver-operator characteristic (ROC) analysis of total CTC signal in healthy donors (n=20) and Stage I (N=26), Stage II (N=42), Stage III (N=12) and Stage IV (n=30) patient samples. AUC values are shown; p-values are based on Wilcoxon rank sum test.
To first test the performance of the assay in reconstitution experiments, we manually micro-manipulated 0, 1, 3, 10 or 30 cultured CTCs (cell line BRx-142 derived from a patient with metastatic breast cancer) into 4 ml healthy donor (HD) blood (approximately 20 billion cells), processed these samples through the CTC-iChip, extracted RNA from the CTC-enriched cell population, and digitally quantified the expression of the 17 markers in the product. A robust signal is observed with a single CTC, and signal increases linearly with higher numbers of spiked cells (R2=0.99) (Fig.1C; Fig. S2). Importantly, the relative expression of detected markers remains consistent from 1 to 30 spiked cells (Fig. 1D). Similar experiments performed using two other breast cancer cell lines (a second CTC-derived line, BRx-68, and the well-characterized triple negative breast cancer cell line MDA-231) demonstrate differential expression of the 17 markers illustrating the importance of using a diverse panel of cellular transcripts for optimal detection of CTCs (Fig. 1E). A remarkably similar pattern of expression is observed in the bulk RNAseq data of these cell lines (Fig. S3).
Digital CTC detection at different disease stages and in multiple breast cancer subtypes
Having benchmarked the CTC molecular signature assay in vitro, we tested its performance on healthy donors and in breast cancer patients. While we selected breast tissue lineage and breast cancer enriched transcripts for this panel, very low background expression of these markers in WBCs may still occur at levels that are detectable by a highly sensitive technique such as ddPCR. We therefore first applied the assay to an initial cohort of 33 female healthy donors, establishing normal levels of background for each marker in blood samples without CTCs (see Methods). Consequently we subtracted this background signal from all further test samples: 20 new female healthy donors (test HD cohort) and a cohort of women presenting with various stages of breast cancer, including untreated (localized pre-surgical) Stage I (N=26), Stage II (N=42), or Stage III (N=12) breast cancer, and on-treatment samples from women with Stage IV (metastatic disease; N= 30) breast cancer. We analyzed the performance of individual markers as well as the total CTC-Score using receiver-operator characteristic (ROC) analysis (Fig. S4). In patients with metastatic breast cancer, 6 genes show significant predictive value for presence of cancer as individual markers (p < 0.05, Wilcoxon Rank Sum Test; AUC range 0.62–0.75) and 4 have lower overall predictive value but their expression is completely absent in HD samples, supporting their value in a subset of heterogeneous breast cancers. In women with localized breast cancer, no single marker shows statistically significant predictive value, consistent with the generally lower signal in the blood of patients with low tumor burden. Building from these individual markers, the total CTC-Score achieves a superior, robust and statistically significant detection capability (AUC=0.84; P<0.0001 for metastatic breast cancer; AUC=0.68, P =0.0077 for localized breast cancer,Fig. S4). As expected, the performance of digital CTC detection improves with increasing stage of disease with the AUC values increasing from 0.65 (p=0.071) and 0.68 (p=0.015) in stages I and II, respectively, to 0.82 (p=0.0013) and 0.85 (p<0.0001) in stages III and IV, respectively (Fig. 1F). Sensitivity of the assay at 100% specificity was 19% for Stage I, 36% for Stage II, 58% for Stage II and 67% for Stage IV. Taken together, these results demonstrate that a panel with multiple transcripts surpasses the performance of its individual components, and highlight the benefit of multiplex assays for cancer detection in both localized and metastatic breast cancer.
Persistence of elevated CTC-Score during neo-adjuvant treatment of localized breast cancer as a predictor of residual disease
We applied the breast digital CTC-Score to a cohort of women with localized breast cancer (Stages I-III) treated with pre-operative (neo-adjuvant) therapy (BL-NEO cohort; N=54). Pretreatment baseline and monthly on-treatment blood samples were collected; of the 54 patients, 17% (n=9) had HR+ primary tumors, 46% had TNBC (n=25), and 37% (n=20) had HER2+ subtypes, consistent with the expected distribution among localized disease treated with neoadjuvant therapy (see Table 1 for baseline characteristics of patients). At a set specificity of 100%, baseline CTC-Scores are positive in 43% BL-NEO cohort patients (ROC AUC = 0.72, p = 0.0027) (Figure S5). There was no significant association between baseline CTC-Score and tumor grade, tumor diameter and nodal status within this group of patients (Figure S5).Of the 54 BL-NEO cohort patients, 37 women had their ultimate treatment response determined by both pathology analysis and clinical criteria by a clinician blinded to the CTC-Score at the time of surgical resection (after approximately 4–6 months of therapy). Patients for whom we did not have clinical or pathological assessment were not included in further analysis, but all other patients were included. Overall, a trend towards residual disease in patients with a high baseline CTC-Score (ROC AUC=0.68, p=0.055) is noted, reflecting the prognostic value of CTCs (Figure 2). However, more strikingly, an elevated CTC score during neo-adjuvant therapy (≥3 cycles) is associated with a higher probability of clinically impactful residual disease at the time of subsequent surgical resection (AUC=0.83, p=0.047) (Figure 2). Taken together this time course analysis suggests that failure to clear the digital CTC signal during neo-adjuvant therapy for localized breast cancer is associated with subsequent persistence of substantial residual tumor burden at the time of surgical resection.
Table 1.
Baseline clinico-pathological characteristics in the prospective study of locally advanced breast cancer patients starting neoadjuvant therapy (BLNEO cohort).
| Characteristic | Patients (N=54) |
| Hormone receptor status at diagnosis - no. (%) | |
| HR+ | 9 (17%) |
| TNBC | 25 (46%) |
| HER2+ | 20 (37%) |
| Tumor stage at diagnosis | |
| I | 1 (2%) |
| II | 41 (76%) |
| III | 12 (22%) |
| Tumor grade at diagnosis | |
| 1 | 3 (6%) |
| 2 | 18 (33%) |
| 3 | 33 (61%) |
| Histology at diagnosis - no. (%) | |
| Invasive Ductal Carcinoma | 54 (100%) |
| Neoadjuvant therapy - no. (%) | |
| Chemotherapy | 32 (59%) |
| Endocrine | 9 (17%) |
| Anti-HER2 | 17 (31%) |
| NA | 1 (2%) |
Figure 2.
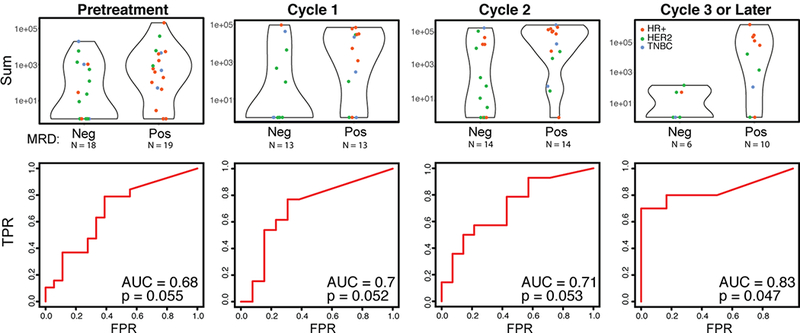
Elevated CTC-Score during presurgical neoadjuvant therapy predicts the probability of residual disease in patients with localized breast cancer at the time of surgical resection. The BL-NEO blood draws were stratified by both treatment cycle (including chemotherapy, endocrine therapy and/or anti-HER2-targeted therapy) and presence of significant residual disease upon surgery, and their CTC scores compared. Breast cancer subtypes are noted (HR+, red; HER2, green; TNBC, blue). High CTC scores in pretreatment and cycles 1 and 2 samples reveal a trend towards presence of significant residual disease, while blood draws from ≥3 cycles of therapy predict significant residual disease. ROC curves, AUC values and p-values for each of the conditions are shown. P-values were computed by comparing the performance of the CTC score to a random predictor.
Serial monitoring of CTC-Score in women with metastatic breast cancer
We tested the application of the breast digital CTC-Score for monitoring treatment response in a prospective cohort of women with metastatic breast cancer (TRACK Study; N =87), comparing measurements before initiation of new therapy and then at 3–4 weeks after start of treatment. Among the 87 patients, 60 (68%) had HR+ breast cancer, with 17 (19%) TNBC and 9 (10%) HER2+ disease. Baseline clinical characteristics of the women and treatment regimens in this cohort are shown in Table 2.
Table 2.
Baseline clinico-pathological characteristics in the prospective study of metastatic breast cancer patients starting a new line of treatment (TRACK cohort).
| Characteristic | Patients (N=87) |
| Age (years) | |
| Median | 60 |
| Range | 35–83 |
| Hormone receptor status at diagnosis - no. (%) | |
| HR+ | 60 (68%) |
| TNBC | 17 (19%) |
| HER2+ | 9 (10%) |
| Histology at diagnosis - no. (%) | |
| Ductal | 70 (79%) |
| Lobular | 7 (8%) |
| Mixed | 5 (6%) |
| Unknown | 6 (7%) |
| Current therapy - no. (%) | |
| Chemotherapy | 9 (10%) |
| Endocrine | 38 (43%) |
| Anti-HER2 | 11 (13%) |
| Other | 29 (34%) |
| Types of metastases - no. (%) | |
| Visceral | 62 (71%) |
| Bone | 57 (65%) |
| Brain | 4 (5%) |
| CA15–3 levels - no. (%) | |
| High (>30) | 52 (59%) |
| Normal (<30) | 21(24%) |
| NA | 15 (17%) |
| Number of prior therapies | |
| Median | 2 |
| Range | 0–9 |
The baseline detection rate using the CTC-Score in this prospective validation cohort, compared to a new matched set of healthy women with negative breast biopsy findings (see Methods) had an AUC= 0.86 (p<0.0001, Fig. 3A), with 68% sensitivity at a set specificity of 100%, which is comparable to that observed with the initial test metastatic cohort (AUC=0.84 p < 0.0001; Fig 1F). Having validated the assay detection rate in this prospective cohort, we dichotomized the pretreatment CTC-Scores into high (≥3000 transcripts/mL) and low (<3000 transcripts/mL) values (see Methods). Correlating the high and low CTC-scores with clinical-pathologic variables (age, hormone receptor status, histopathology, type of treatment, location of metastatic sites, number of prior therapies, CA15–3 tumor marker values) shows none of the variables analyzed to be associated with a higher CTC-Score (Table S2). We then examined the relationships between pre-treatment or week 3–4 CTC-Scores and clinical outcomes using multivariable Cox proportional hazards model of overall survival (OS) or time to progression (TTP) (see Methods). Overall survival was significantly associated with baseline CTC score, prior CDK4/6 therapy, and presence of ESR1 mutation. Patients with a high baseline CTC-Score (> 3000) had worse OS (HR: 2.70, 95% CI (1.15 to 16.7), p=0.02), compared with those with a low CTC-Score (median OS: 11.1 months versus 17.2 months) (Fig. 3B). Better OS was also seen for patients who had received CDK4/6 therapy compared to those who did not (HR: 0.23; 95% CI: 0.07 to 0.74, p=0.01) consistent with observed benefits for this class of drugs in combination with endocrine agents 1–3. Lastly, the hazard of death was reduced by two-thirds in patients who did not have ESR1 mutations (HR: 0.32; 95% CI: 0.11 to 0.95, p=0.04) (Table S3).
Figure 3.
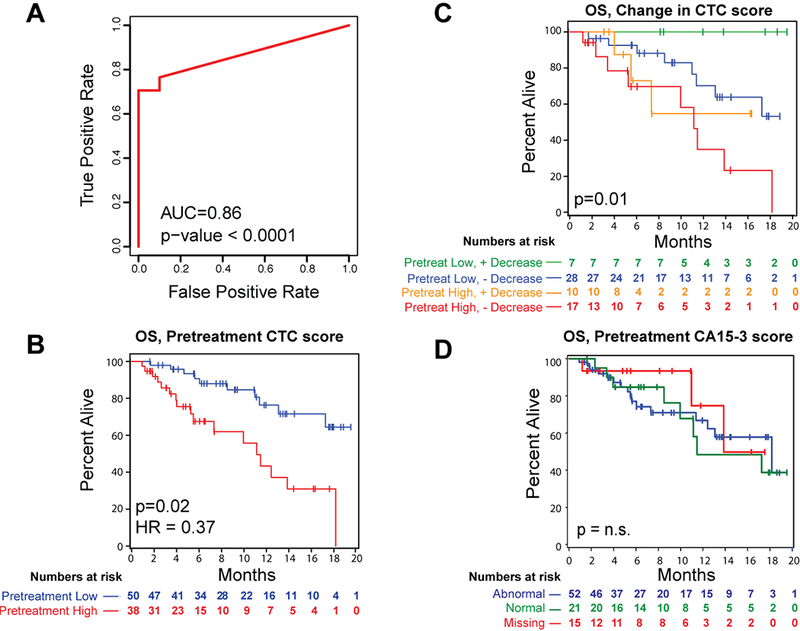
Pretreatment CTC-Score predicts overall survival in women with metastatic breast cancer receiving a new line of treatment. A, ROC analysis of total CTC signal in pretreatment samples from Stage IV patients (n= 87) from the TRACK cohort versus matched negative control (women with negative biopsies despite previously positive mammogram findings; n=10). AUC values are shown; p-values are based on Wilcoxon rank-sum test. B, Kaplan-Meier plot depicting overall survival (OS) in TRACK patients, based on pre-treatment CTC-Score. Patients were divided into two groups at a cut-off of 3000 transcripts/ml (see Methods). Patients with a high pretreatment CTC-Score (red) have a longer OS compared with those with a lower pretreatment CTC-Score (blue). Hazard ratio (HR) and p-value based on multivariable Cox proportional hazards model are shown. C, Kaplan-Meier plot depicting overall survival in TRACK patients, based on the change in CTC score between pre-treatment baseline versus 3–4 weeks on-treatment time point. Groups are defined based on low signal at pretreatment (<= 3000 transcripts/ml) with >90% reduction in signal on-treatment (green), low signal at pretreatment (<= 3000 transcripts/ml) without >90% reduction in signal on-treatment (blue), high signal at pre-treatment (>3000) with >90% reduction in signal at 3–4 weeks (orange) or high signal at pre-treatment (>3000) without >90% reduction in signal at 3–4 weeks on-treatment (red). P-value was calculated using the log-rank test. D, Kaplan-Meier plot depicting overall survival in TRACK patients, based CA15–3 levels at pretreatment. Groups are defined as abnormal CA15–3 levels (>30, blue), normal CA15–3 levels (<30, green), and missing CA15–3 levels (NA, red). P-value based on log-rank test.
Additional univariate analyses indicated that, patients who have a low CTC score at baseline that decreases further (>90%) after 3–4 weeks of treatment have significantly better OS compared to those who did not experience such decrease or started with a high pre-treatment CTC-Score (p=0.001; Fig 3C). Together, these observations suggest that both pretreatment CTC burden, as well as the early treatment-induced changes in CTCs, are important prognostic factors in predicting patient outcome. Such a correlation has been previously demonstrated using microscopic enumeration of CTCs following chemotherapy, but not endocrine treatment17, 18.
In our cohort of predominantly HR+ cancers, neither the pretreatment nor early treatment-induced changes in CTC-Score are predictive of TTP (Figure S6), a finding that is also consistent with the endocrine therapy-treated subset of patients analyzed by classical microscopic CTC enumeration 17. This difference between OS and TTP is likely related to the known heterogeneity in this patient population, variations in endocrine-based therapeutic regimens, as well as variable clinical assessment of progression in patients with HR+ breast cancer whose metastases are not readily evaluable (e.g. bone lesions). Levels of the classical protein tumor marker CA 15–3 are not predictive of either OS or TTP (Fig. 3D, Fig. S7), confirming the need for other non-invasive biomarkers in HR+ breast cancer.
Persistent estrogen signaling within CTCs identifies HR+ cases with short TTP on endocrine therapy
To identify predictive biomarkers for continued response to endocrine therapy in highly treated HR+ breast cancer (i.e. second or later lines of therapy), we focused on the subset of TRACK cohort patients who had HR+ breast cancer (n=36 at pretreatment) and were starting a new course of endocrine-based treatment (Table S4). At a set specificity of 100%, 67% had detectable CTC signal. We first performed unsupervised clustering of the 17 markers before and after initiation of treatment. Remarkably, at the 3–4 week on-treatment time point, a 6-gene subset (PIP, SERPINA3, AGR2, SCGB2A1, EFHD1 and WFDC2) identifies patients with rapid disease progression within 120 days of start of treatment (p=0.005, Fisher’s exact test) (Fig. 4A). This CTC-derived “Resistance Signature” (RS) emerges at the first on-treatment time point; while the 6 CTC transcripts also cluster together at the pretreatment time-point, their expression before initiation of therapy is not associated with clinical outcome (Fig. S8), raising the possibility that the persistence of this signature on-therapy is biologically significant.
Figure 4.
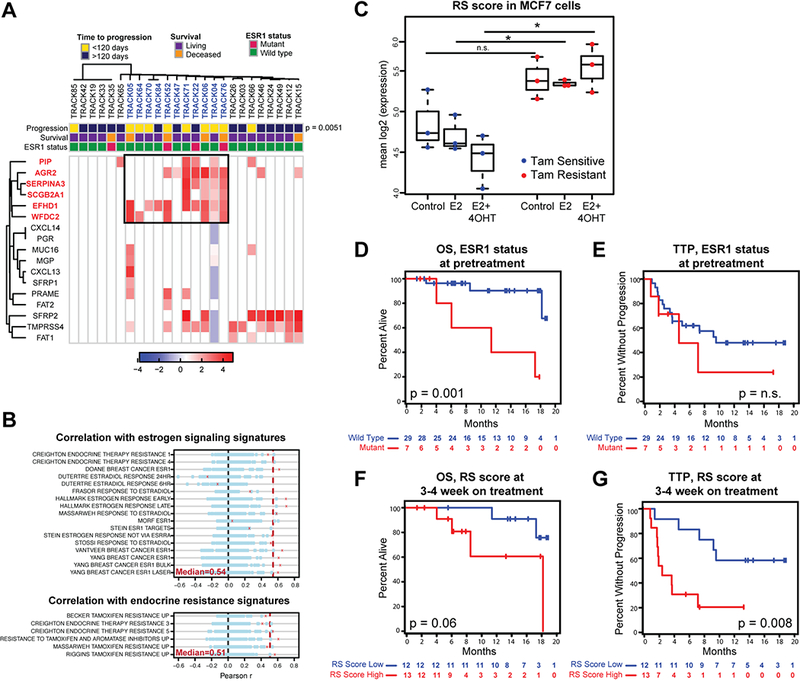
Markers associated with persistent ER signaling (RS signature) identify HR+ patients at high risk of progression on endocrine treatment. A, Unsupervised clustering of breast CTC marker expression in HR+ patients receiving endocrine therapy for 3–4 weeks. A set of markers (boxed) identifies a group of patients (colored blue) significantly enriched for progression within 120 days. P=0.0051 was calculated using Fisher’s exact test. ESR1 mutation status for each patient is shown. B, Correlations between a metascore based on the expression of the 6 high risk RS genes and GSEA signatures associated with estrogen signaling (top) and endocrine resistance (bottom) across multiple publicly available datasets are shown in red crosses. Dotted red line represents the median correlation across the multiple comparisons. Correlations with metascores based on 100 random sets of 6 genes are shown in blue circles. C, RS expression based on bulk RNAseq in tamoxifen sensitive or resistant MCF7 cells, left untreated or treated with estrogen (E2) alone or together with tamoxifen (4-OTH). Asterisks show significance of p<0.05, p-values based on two-sided t-test. D, Kaplan-Meier plots depicting OS in HR+ patients receiving endocrine therapy based on the presence of ESR1 mutations at pretreatment. Cases with ESR1 mutations (red) are compared with those with wild-type ESR1 (blue). P-values were calculated using log rank test. E, Kaplan-Meier plots depicting TTP in HR+ patients receiving endocrine therapy based on the presence of ESR1 mutations at pretreatment. Cases with ESR1 mutations (red) are compared with those with wild-type ESR1 (blue). P-values were calculated using log rank test.F, Kaplan-Meier plots of OS of HR+ patients receiving endocrine therapy based on 3–4 weeks on-treatment RS score. Groups were divided at 25 transcripts/ml. Patients with high RS CTC-Score (red) are compared with those having a low RS CTC-Score (blue). P-values were calculated using log rank test. G, Kaplan-Meier plots of OS of HR+ patients receiving endocrine therapy based on 3–4 weeks on-treatment RS score. Groups were divided at 25 transcripts/ml. Patients with high RS CTC-Score (red) are compared with those having a low RS CTC-Score(blue). P-values were calculated using log rank test.
All 6 RS transcripts are significantly enriched in ER+ tumors in the TCGA database (Fig. S9A), suggesting that their expression may be related to estrogen signaling. Indeed, a metascore based on their mean expression shows a significant correlation with the Hallmark Estrogen Receptor (Late) gene signature from the Molecular Signatures Database across multiple publicly available gene expression datasets (R = 0.70; p=1.7e-70) (Fig. S9B). The RS gene metascore is also correlated with multiple other MSigDB sets related to estrogen signaling and endocrine resistance, resulting in median correlation coefficients of 0.54 and 0.51 respectively (Fig. 4B).
To further functionally validate the association of the 6-gene RS signature with endocrine resistance, we examined an independent RNAseq dataset of MCF7 breast cancer subslones that are either sensitive or resistant to tamoxifen, with expression profiling before and after treatment with endocrine therapy30. Remarkably , the RS score at baseline was not significantly different between resistant and sensitive MCF7 cells; however after incubation with estrogen and subsequent inhibition of the pathway with tamoxifen (4OHT) the RS score appears to increase in the resistant cells, while decreasing in the sensitive cells, resulting in a significant difference between the two (Figure 4C). This result is consistent with our observations based on patient-derived CTCs, and together, they point to the RS expression signature as distinguishing breast cancer cells that are resistant to hormonal therapies following exposure both in vitro and in the clinical setting.
Activating mutations in the ESR1 gene encoding ER have been reported in breast cancers with acquired resistance to hormonal therapy, especially aromatase inhibitors (AI), and are thought to mediate persistent, ligand-independent ER signaling 13, 25, 31–33. Within our TRACK cohort, 20/36 women with HR+ metastatic breast cancer initiating a new course of endocrine treatment had previously progressed on AIs for metastatic disease, and 11/36 had undergone tumor re-biopsy during the course of clinical care, with 2 (25%) identified as having an acquired ESR1 mutation (SNaPShot genotyping, 34)(Table S5). To ascertain ESR1 mutation status in all patients, we established a digital PCR mutation-specific assay using CTC-derived RNA template, with probes specific for the hotspots L536R, Y537C, Y537N, Y537S and D538G, which together account for the majority of ESR1 mutations 13, 32. The sensitivity and accuracy of the assay was confirmed by spiking experiments introducing one or more single cells carrying an ESR1 mutation into whole blood samples followed by microfluidic CTC enrichment and digital PCR analysis, and by detection of two independent ESR1 mutations in a clinically validated patient sample (Fig. S10). Using this CTC-based assay, 5 additional patients within the TRACK HR+ cohort were found to harbor ESR1 mutations, resulting in a total mutation frequency of 7/36 (21%), a prevalence consistent with previous studies of advanced HR+ breast cancer 13, 32 (Table S5). Presence of ESR1 mutation at pretreatment is associated with worse OS, as previously reported35 (Fig. 4D), but not with a worse TTP in our patient cohort (Fig. 4E).
To correlate the CTC expression-based RS score with clinical outcomes, we used an unbiased approach to split the patients into 2 subgroups with high and low RS score (see Methods). As we previously observed for the overall CTC score, the baseline RS CTC signature is prognostic for poor overall survival (p=0.002) but not TTP (p=0.97) (Fig. S11). However, persistence of the RS signature at week 3–4 despite endocrine therapy, is associated with shorter OS (p=0.06) and worse TTP (p=0.008), consistent with it providing pharmacodynamic evidence of inadequate suppression of ER signaling by the administered endocrine therapy (Fig. 4F, Fig. 4G). Remarkably, there is limited overlap between patients with a high RS Score and those with ESR1 mutations: Of the 5 patients with ESR1 mutations for whom we had an on-treatment blood sample, only 3 have high RS scores (Table S6). Conversely, 3 of 13 patients with a high on-treatment RS Score harbor an ESR1 mutation. Thus, failure of endocrine therapy to suppress ER signaling in advanced breast cancers is only partially attributable to the acquisition of ESR1 activating mutations, suggesting distinct mechanisms that contribute to refractory disease.
Discussion
By combining initial microfluidic depletion of hematopoietic cells to enrich for intact CTCs, together with quantitative digital PCR of multiple breast-specific transcripts, we have developed a platform for high-throughput, noninvasive characterization of cancer cells in the circulation of women with breast cancer. Compared with imaging-based CTC quantitation 15,16 or analysis of individual RNA markers 36,37,38 the CTC-Score has the sensitivity and complexity to interrogate different cancer-related pathways, including estrogen receptor signaling, during the course of therapy. Indeed, failure of endocrine therapy to suppress intracellular ER signaling within cancer cells sampled in the bloodstream is associated with a very poor clinical outcome and, to our knowledge, it constitutes the first application of transcriptomic-based noninvasive pharmacodynamic monitoring in the therapy of breast cancer.
The clinical utility of CTC enumeration was first demonstrated by the observation that baseline measurements of >5 CTCs/7.5ml of blood, assayed by microscopic visualization and scoring, predict adverse overall survival in women with metastatic breast cancer treated primarily with chemotherapy 17. Women whose CTC counts failed to decline following chemotherapy also had a worse prognosis, although switching to an alternative standard chemotherapy regimen did not lead to a better outcome 18. However, since chemotherapy resistance is broadly displayed against multiple agents, and there is no predictive marker to indicate sensitivity or resistance to specific regimens, this negative result for treatment selection based on CTC monitoring is not surprising. Our digital RNA-based readout supports the prognostic value of CTC measurements at baseline and following initiation of therapy, and extends their clinical relevance to women treated primarily with endocrine-based therapies. This clinical population is particularly relevant for blood-based monitoring, since there are currently multiple therapeutic regimens available, without reliable biomarkers to direct treatment selection. It is in this context that interrogation of the ER pathways through CTC gene expression profiling holds promise for guiding treatment selection.
The 17 genes that constitute the breast CTC signature were selected to include multiple tissue-derived and cancer-related transcripts that are not expressed in contaminating blood cells. As such, the 6 genes included in the RS sub-signature do not represent canonical ER targets but their expression is nonetheless highly correlated with both ER signaling and resistance to endocrine therapy. Their persistent expression within CTCs after treatment initiation in patients with metastatic breast cancer identifies women with greatly reduced response to endocrine therapy and worse outcomes. The fact that this CTC signature emerges as a strong predictive factor 3–4 weeks after start of novel ER targeting therapy suggests that it may reflect the differential effectiveness of drug-mediated ER suppression within tumor cells. In women with highly pretreated advanced HR+ breast cancer, initiation of a new course of endocrine therapy presumably suppresses ER signaling in susceptible cancers, whereas persistent pathway activity remains evident in those where the drug fails to suppress its intended target. Given the small number of cases analyzed in this way, larger studies will be required to confirm the clinical relevance of this observation. However, it raises the possibility that ER activity continues to be an important driver of proliferation in a subset of HR+ treatment-refractory cases, and points to a need for a better understanding of underlying mechanisms that could be targeted through alternative agents. A similar rationale underlies the development of novel androgen receptor (AR) targeting agents in castrate-resistant prostate cancer39, 40.
Activating mutations in the ligand-binding domain (LBD) of the estrogen receptor gene ESR1 have been recently identified in advanced HR+ breast cancers, particularly following prolonged treatment with AIs, with reported frequencies as high as 37% 13, 25, 31–33, 41. In addition, rare ESR1 translocations, ESR1 amplifications, as well as mutations in pathways with substantial cross-talk with ER and in the regulatory regions of ER cofactors have also been reported 42–44. In our study, in the subset of patients initiating a new course of endocrine therapy, 21% had ESR1 mutations as determined by CTC-ddPCR and tissue genotyping. Thus, presence of ESR1 mutations does not account for all patients who had rapid progression (<120 days) on endocrine agents. Persistent ER signaling at 3–4 weeks of treatment with endocrine therapy, as measured by the CTC-derived RS expression signature, emerged as an independent prognostic factor in these patients. While again these observations need to be confirmed in larger clinical trials, they suggest that persistent drug-refractory ER signaling may not be solely attributable to acquired ESR1 mutations, and further supports the need to fully define the range of mechanisms driving ER activation, and potential strategies to suppress this critical signaling pathway in HR+ breast cancer.
The fact that some ESR1-mutant breast cancers had low RS Scores raises the possibility that these mutations confer constitutive ER signaling, yet at relatively lower levels of activity than cases with high on-treatment RS Scores. In addition,
ESR1 mutations are frequently subclonal11 and multiple endocrine drug resistance mechanisms may coesxist within a single patient. While our dataset is too small to allow detailed analysis of predictive power for both of these markers, it is noteworthy that patients who had a high RS Score with ESR1 mutation (3 cases) or without ESR1 mutation (10 cases) had a median TTP of 56 and 57 days, respectively; in contrast, women with an ESR1 mutation and a low RS Score (2 cases) had a median TTP of 139 days, and those with neither ESR1 mutation nor RS Score (10 cases) had a median TTP of 251 days (Table S6). Although based on small patient numbers, these data suggest that persistent expression of ER-signaling in patients treated with endocrine therapy is an independent risk factor in assessing patient’s likelihood to benefit from such treatments. Therapeutic targeting of ESR1 mutant protein through novel ER degraders is currently a major focus for drug development45, and additional strategies may be required to target breast cancers with high RS in the absence of ESR1 mutations.
Finally, we explored the application of digital CTC scoring in the neoadjuvant treatment of localized high-risk breast cancer. Setting the test specificity at a stringent level of 100%, positive CTC signal was detectable at baseline in 43% of women whose early stage breast cancer was considered sufficiently high-risk to warrant preoperative therapy (Fig. S5). A major challenge in the administration of neoadjuvant chemo and/or hormonal therapy is the absence of early markers of response, such that up to 6 months of treatment may be administered before surgical resection of the primary tumor may reveal either the desired tumor shrinkage or persistent disease.Our finding that after 3 months of neadjuvant therapy women who have higher CTC signal will have substantial residual tumor at the time of surgical resection, compared to those with lower CTC signal, suggests that CTC monitoring may help guide the presurgical evaluation of drug response, and supports previous evidence for the utility of CTC as a prognostic marker in the neoadjuvant setting46. Larger trials will be required to confirm the clinical validity of this promising blood-based predictor, and whether its clinical value varies among different histological subtypes of breast cancer subjected to different neoadjuvant treatment modalities.
The application of “liquid biopsies” to breast cancer therapeutics is rapidly evolving, in parallel with the advent of novel therapeutic agents and drugs combinations. Advances in ctDNA technology now allow for the detection of multiple somatic mutations in plasma, while pushing the limit of sensitivity to earlier stage disease11, 14. Here, we have leveraged the high specificity and signal amplification inherent in RNA-based biomarkers to provide an orthogonal assay to plasma genotyping – one that allows for the interrogation of intracellular pathways critical to understanding drug effects. ctDNA and CTCs derive from different processes within the tumor: ctDNA originates from tumor cells undergoing apoptosis or necrosis and thus likely enriches for tumor subpopulations sensitive to treatement, whereas CTCs are live cells that have intravasated into the bloodstream and are likely derived from invasive and potentially drug-resistant subclones. Thus, high throughput microfluidic enrichment of CTCs followed by multiplex digital RNA quantification may provide a novel and complementary strategy to monitor and guide therapy in both localized and advanced breast cancer.
Methods
Patients and healthy donors
All studies were conducted in accordance with Belmont Report ethical guidelines. Patient samples were collected after written informed consent through an Institutional Review Board approved protocol for CTC collection (DFHCC 05–300). 10–20ml of peripheral blood was collected. 30 on-treatment samples from 23 unique Stage IV patients were collected for the initial clinical benchmarking of the assay along with 26 pre-surgical samples from patients with newly diagnosed localized breast cancer (25 Stage I and 2 Stage II).
Pretreatment samples and samples prior to each subsequent round of neoadjuvant treatment were prospectively collected from women with newly diagnosed localized breast cancer (1 Stage I, 41 Stage II and 12 Stage III unique patients, BLNEO cohort, Table 1.) The pretreatment samples were used for establishing the performance of the assay; both pretreatment and on-treatment samples from the BLNEO cohort were used to determine if CTC monitoring of patients receiving neoadjuvant treatment is predictive of surgical outcomes. For quantitation of residual tumor burden after neoadjuvant therapy, we performed a blinded review of the surgical pathology report together with observations of the treating clinician as to whether the neoadjuvant treatment had achieved the desired goal of treatment shrinkage before surgery.
In order to determine if CTC monitoring through the breast CTC-ddPCR assay is predictive of treatment outcome and overall survival in metastatic patients, we prospectively collected pretreatment and 3–4 weeks on-treatment draws from metastatic breast cancer patients initiating a new therapy (TRACK cohort). At least one sample was collected from each of the 87 patients (baseline characteristics in Table 2). Disease progression was determined by treating physician (blinded to the CTC result) based on standard clinical and/or radiological criteria.
33 samples from female healthy donors were obtained from the blood bank (9ml average) to establish the normal expression of each marker (initial HD cohort). Additional 20 HD samples were obtained from the blood bank to establish the performance of the assay (test HD donors). To validate the detection characteristics established in the initial phase of assay development on the TRACK cohort, we also collected samples from 10 healthy women with negative breast biopsies after suspicious mammogram findings.
Marker selection
To build the breast CTC assay, we first analyzed publicly available databases, including GTEx, Oncomine, TCGA and others, and breast CTC sequencing data to identify transcripts with abundant expression in normal breast issue (lineage markers) or breast cancer (breast cancer markers) (Fig. S1). We cross-referenced potential markers to WBC gene expression data that was publicly available or generated by our lab, eliminating transcripts significantly expressed in blood. This approach identified 45 potential markers (Table S1) which we tested, first by RT-qPCR, to confirm expression in CTC cell lines and lack of expression within HD-derived WBCs; and then by ddPCR, using WTA-amplified CTC-iChip blood sample products from 10 HDs and 10 metastatic breast cancer patients. The 17 markers described in this manuscript (specific probes listed in Table S7) were chosen based on two criteria: 1) significantly higher expression in the 10 patients vs. HDs; or 2) no expression in the 10 HDs with some expression in patients, and the assay was locked in its current format and further validated in-vitro, and in patient and HD samples as described below.
Cell spike-in experiments
BRX-142 and BRx-68 CTC cell lines were derived in our lab and have been described before 25,47. MDA231 cells were obtained from ATCC, and have been authenticated using short tandem repeat profiling. All cell lines tested negative for Mycoplasma. Cells were used within 5–20 passages from thawing. For the initial in-vitro testing of the panel, and to determine the linearity of the signal, we micromanipulated increasing numbers of cultured cells into 4ml of HD blood, ran the samples through the CTC-iChip, and performed RNA extraction WTA and ddPCR as described above.
CTC-Score calculation
To normalize for differences in blood volumes among samples, all raw data were corrected for the blood-volume equivalent used in each ddPCR reaction. To further normalize the signal against HD background, the mean and twice the standard deviation of the expression of each marker within the initial cohort of 33 healthy donors was established. The product of the two values was then subtracted from every patient and healthy donor sample analyzed in this study. If the result was less than 0, it was replaced with 0. The total CTC-Score was calculated by summing the normalized expression of all markers in a sample without additional weighting and reported as transcripts/ml of blood-volume equivalent used.
ESR1 mutation detection
Probes specific for the L536R, Y537C, Y537N, Y537S and D538G ESR1 mutations have been previously published 13. Their amplification efficiency, as well as that of their respective wild-type probes, was tested on synthetic sequences (data not shown). We established the ability of Y537S to detect mutations present in cDNA from CTC-enriched IFD product by micromanipulating increasing numbers of BRx-68 cells into healthy donor blood, and then processing it as described above. 18-cycle WTA was performed using 1/3 of the extracted RNA with the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech) following manufacturer protocols; 1ul of undiluted WTA product was used per reaction. Patient samples were treated in an identical manner; probe specificity was established at 100% after testing at least 5 healthy donor samples per probe. The cut-off for the presence of ESR1 mutation was established as >3 positive droplets.
Statistical analysis
Receiver-operator curve analysis was performed to establish the specificity and sensitivity of each marker and the total CTC-Score for different cancer stages in our initial test cohort consisting of 30 on-treatment Stage IV samples, 26 pretreatment Stage I samples and the pretreatment samples from the BLNEO cohort (42 Stage II and 12 Stage III) compared to the 20 test healthy donors. The analysis was performed in R using the ROCR package. The specific script is available upon request. Wilcoxon tests were performed to establish significance of the AUC. The specificity and sensitivity in Stage IV cancer were validated using a new set of healthy donors (women with negative findings after a breast mammogram) and the pretreatment samples from the TRACK cohort. All patients, regardless of whether they were defined as CTC-potivie or negative were included in downstream analysis, as absence of CTCs is considered a meaningful biological information that has prognostic value for clinical outcomes.
For the longitudinal analysis within the BLNEO cohort, samples for which minimal residual disease or RECIST criteria data were not available were removed from the analysis resulting in 37 patients analyzed. The CTC-Score at various clinical intervals (pre-treatment and the end of cycle 1, cycle 2, or cycle 3 onward) was compared to the presence of substantial residual disease burden present at the time of surgery, as determined by a clinician blinded to CTC-Score. The R script used to perform the analysis is available upon request.
Dichotomous cut points for baseline CTC-Score, change in CTC-Score, and baseline and on-treatment RS-Score were determined to maximize the associations between high versus low scores and clinical outcomes. All patient samples were included. Cut points for the time-to-event endpoints were estimated using leave-one-out jack knife resampling of the algorithm of Contal-O’Quigley. For each score, the selected cut point was the median of the distribution of possible cut points. Comparisons of clinical variables between resulting groups are based on Fisher’s exact tests for categorical characteristics and exact Wilcoxon rank-sum tests for continuous characteristics.
To examine the relationship between pre-treatment and on-treatment clinical factors, CTC-Scores, and outcome, multivariable Cox proportional hazards models were fit for overall survival and TTP. Overall survival was defined as the interval between the date of initiation of new therapy to death from any cause. The follow-up of patients who did not die was censored at the date of last assessment of vital status. TTP was defined as the interval between the date of initiation of new therapy and first documentation of progressive disease. In the absence of documented progressive disease, follow-up was censored at date of last disease assessment. Candidate predictors in the models were factors associated with outcome based on univariate log-rank p-values of 0.2 or less: CTC-Score (either at pre-treatment or at 3–4 weeks on treatment divided at 3000 transcripts/ml), breast cancer type at diagnosis (HR+, HER2+, TNBC), prior endocrine therapy (Yes/No), prior chemotherapy (Yes/No), treatment with CDK4/6 inhibitor (Yes/No), presence of visceral metastases (Yes/No), presence of bone metastases (Yes/No), presence of brain metastases (Yes/No), CA15–3 tumor marker levels (normal defined as <30, abnormal defined as >30, missing), number of prior therapies (divided at 2), and age (divided at the median of 60 years). The model of OS was stratified by age to allow for differences in the underlying baseline hazard of death for the two different age categories. Age was a covariate in the TTP model. For on-treatment unsupervised clustering and time-to-event analyses, only patients with samples available at both pretreatment and 3–4 weeks on treatment were included. P-values for the Kaplan-Meier analyses are based on log-rank tests. Hazard ratios from the Cox models are presented with 95% confidence intervals estimated using log(-log) methods and Wald p-values. Unsupervised clustering of pretreatment and 3–4 week on-treatment samples was performed using single linkage.
Supplementary Material
Statement of Significance.
Digital analysis of RNA from CTCs interrogates treatment responses of both localized and metastatic breast cancer. Quantifying CTC-derived ER-signaling during treatment identifies patients failing to respond to ER suppression despite having functional ESR1. Thus, non-invasive scoring of CTC-RNA signatures may help guide therapeutic choices in localized and advanced breast cancer.
Acknowledgements
We thank all the patients and healthy donors who participated in this study. We acknowledge the clinical research coordinators J.B Grinnell, P.Y. Chan and G. Malvarosa for their assistance with sample and data collection. We are grateful to Massachusetts General Hospital (MGH) nurses for their help with the study; and L. Libby for invaluable technical support. This work was supported by NIH Grant 2R01CA129933, the Breast Cancer Research Foundation, Howard Hughes Medical Institute, and National Foundation for Cancer Research (to D.A.H.), NIH Quantum Grant 2U01EB012493 (to M.T. and D.A.H.), NIH grant U01CA214297 (to M.T., D.A.H and S.M), MD/PhD Training Grant T32GM007753 (to M.K.), 1-F30CA224588–01 Grant (to M.K.), ESSCO Breast Cancer Research (to S.M.), K12 5K12CA087723 (to A.B.), KL2 TR001100 (to L.M.S), ASCO Young Investigator Award (to L.M.S) and the Prostate Cancer Foundation (D.T.M.).
Footnotes
Conflict of Interest Statement: MGH has filed for patent protection for the CTC-iChip technology and molecular signatures
References
- 1.O’leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nature Reviews Clinical Oncology. 2016;13: 417–430. [DOI] [PubMed] [Google Scholar]
- 2.Hortobagyi GN, Stemmer SM, Burris HA, et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. New England Journal of Medicine. 2016;375: 1738–1748. [DOI] [PubMed] [Google Scholar]
- 3.Turner NC, Ro J, André F, et al. Palbociclib in hormone-receptor–positive advanced breast cancer. New England Journal of Medicine. 2015;373: 209–219. [DOI] [PubMed] [Google Scholar]
- 4.Hart CD, Migliaccio I, Malorni L, Guarducci C, Biganzoli L, Di Leo A. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nature Reviews Clinical Oncology. 2015;12: 541–552. [DOI] [PubMed] [Google Scholar]
- 5.Poznak CV, Somerfield MR, Bast RC, et al. Use of Biomarkers to Guide Decisions on Systemic Therapy for Women With Metastatic Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. Journal of Clinical Oncology. 2015;33: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shachar SS. Assessing Treatment Response in Metastatic Breast Cancer. American Journal of Hematology/Oncology®. 2016;12. [Google Scholar]
- 7.Lecouvet F, Talbot J, Messiou C, et al. Monitoring the response of bone metastases to treatment with Magnetic Resonance Imaging and nuclear medicine techniques: a review and position statement by the European Organisation for Research and Treatment of Cancer imaging group. European journal of cancer. 2014;50: 2519–2531. [DOI] [PubMed] [Google Scholar]
- 8.Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25: 5287–5312. [DOI] [PubMed] [Google Scholar]
- 9.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7: 302ra–133. [DOI] [PubMed] [Google Scholar]
- 11.O’Leary B, Hrebien S, Morden JP, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9: 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnedos M, Vicier C, Loi S, et al. Precision medicine for metastatic breast cancer [mdash] limitations and solutions. Nature Reviews Clinical Oncology. 2015;12: 693–704. [DOI] [PubMed] [Google Scholar]
- 13.Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7: 313ra–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells [mdash] mechanisms of immune surveillance and escape. Nature Reviews Clinical Oncology. 2017;14: 155–167. [DOI] [PubMed] [Google Scholar]
- 16.Plaks V, Koopman CD, Werb Z. Circulating tumor cells. Science. 2013;341: 1186–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. New England Journal of Medicine. 2004;351: 781–791. [DOI] [PubMed] [Google Scholar]
- 18.Smerage JB, Barlow WE, Hortobagyi GN, et al. Circulating Tumor Cells and Response to Chemotherapy in Metastatic Breast Cancer: SWOG S0500. Journal of Clinical Oncology. 2014;32: 3483–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardia A, Haber DA. Solidifying liquid biopsies: can circulating tumor cell monitoring guide treatment selection in breast cancer?: American Society of Clinical Oncology, 2014. [DOI] [PubMed] [Google Scholar]
- 20.Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14: 623–631. [DOI] [PubMed] [Google Scholar]
- 21.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11: 129–144. [DOI] [PubMed] [Google Scholar]
- 22.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. The Journal of Cell Biology. 2011;192: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karabacak NM, Spuhler PS, Fachin F, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protocols. 2014;9: 694–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu M, Bardia A, Aceto N, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science (New York, N.Y.). 2014;345: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen–dependent and–independent sorting of rare circulating tumor cells. Science translational medicine. 2013;5: 179ra147–179ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalinich M, Bhan I, Kwan TT, et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proceedings of the National Academy of Sciences. 2017;114: 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong X, Sullivan RJ, Kalinich M, et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc Natl Acad Sci U S A. 2018;115: 2467–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto DT, Lee RJ, Kalinich M, et al. An RNA-Based Digital Circulating Tumor Cell Signature Is Predictive of Drug Response and Early Dissemination in Prostate Cancer . Cancer Discov. 2018;8: 288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez-Malerva L, Park J, Zou L, et al. High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc Natl Acad Sci U S A. 2011;108: 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson DR, Wu Y-M, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nature genetics. 2013;45: 1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations [mdash] a mechanism for acquired endocrine resistance in breast cancer. Nature Reviews Clinical Oncology. 2015;12: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45: 1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2: 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016;2: 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreopoulou E, Yang LY, Rangel KM, et al. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. Int J Cancer. 2012;130: 1590–1597. [DOI] [PubMed] [Google Scholar]
- 37.Strati A, Markou A, Parisi C, et al. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer. 2011;11: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andergassen U, Kolbl AC, Hutter S, Friese K, Jeschke U. Detection of Circulating Tumour Cells from Blood of Breast Cancer Patients via RT-qPCR. Cancers (Basel). 2013;5: 1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beer TM, Tombal B. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371: 1755–1756. [DOI] [PubMed] [Google Scholar]
- 40.Ryan CJ, Smith MR, de Bono JS, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7: 11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Shen D, Shao J, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4: 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller WR, Larionov AA. Understanding the mechanisms of aromatase inhibitor resistance. Breast Cancer Res. 2012;14: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rheinbay E, Parasuraman P, Grimsby J, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanamura T, Hayashi SI. Overcoming aromatase inhibitor resistance in breast cancer: possible mechanisms and clinical applications. Breast Cancer. 2017. [DOI] [PubMed] [Google Scholar]
- 46.Bidard FC, Michiels S, Riethdorf S, et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. J Natl Cancer Inst. 2018. [DOI] [PubMed] [Google Scholar]
- 47.Jordan NV, Bardia A, Wittner BS, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


