Abstract
The roles and regulatory mechanisms of ferroptosis, a non-apoptotic form of cell death, in cancer remain unclear. The tumor suppressor BRCA1-associated protein 1 (BAP1) encodes a nuclear de-ubiquitinating (DUB) enzyme to reduce histone 2A ubiquitination (H2Aub) on chromatin. Here integrated transcriptomic, epigenomic, and cancer genomic analyses link BAP1 to metabolism-related biological processes, and identify cystine transporter SLC7A11 as a key BAP1 target gene in human cancers. Functional studies reveal that BAP1 decreases H2Aub occupancy on the SLC7A11 promoter and represses SLC7A11 expression in a DUB-dependent manner and that BAP1 inhibits cystine uptake through repressing SLC7A11 expression, leading to elevated lipid peroxidation and ferroptosis. Furthermore, we show that BAP1 inhibits tumor development partly through SLC7A11 and ferroptosis and that cancer-associated BAP1 mutants lose their abilities to repress SLC7A11 and to promote ferroptosis. Together, our results uncover a previously unappreciated epigenetic mechanism coupling ferroptosis to tumor suppression.
Keywords: BAP1, H2A ubiquitination, ferroptosis, metabolic stress, SLC7A11, tumor suppression
Introduction
To survive, cancer cells require an adequate supply of nutrients, such as amino acids, to maintain redox homeostasis and to meet their biosynthetic and bioenergetic needs 1, 2. Nutrient depletion induces metabolic stress and eventually provokes cell death, including apoptosis and other non-apoptotic forms of regulated cell death 3, 4. Cancer cells engage strategies of metabolic adaptation, including inactivation of apoptosis pathways, to survive under metabolic stress conditions and to allow tumor progression 5, 6. It is conceivable that non-apoptotic cell death pathways are also dysregulated in cancer, although the underlying mechanisms remain much less understood.
Ferroptosis is a recently identified metabolic stress-induced non-apoptotic form of regulated cell death that is caused by cystine depletion and overproduction of lipid-based reactive oxygen species (ROS), particularly lipid hydroperoxide, in an iron-dependent manner 7–11. Solute carrier family 7 member 11 (SLC7A11, also called xCT), the catalytic subunit of the cystine/glutamate antiporter system xc−, is the major transporter of extracellular cystine 12–14. Correspondingly, cystine depletion or drugs that block SLC7A11-mediated cystine uptake, such as erastin, induce ferroptosis 7. Intracellular cystine is rapidly converted to cysteine, which subsequently serves as the rate-limiting precursor for glutathione synthesis. Glutathione peroxidase 4 (GPX4) utilizes reduced glutathione (GSH) to reduce lipid hydroperoxides to lipid alcohols, to protect cells against membrane lipid peroxidation and inhibit ferroptosis 15, 16. Ferroptosis is morphologically, genetically, and biochemically distinct from other forms of regulated cell death, such as apoptosis and necroptosis 7. It is well established that cell death, most notably apoptosis, plays important roles in tumor suppression 17, 18. The roles and regulatory mechanisms of ferroptosis in tumor biology, however, still remain largely unexplored.
BRCA1-associated protein 1 (BAP1) encodes a nuclear deubiquitinating (DUB) enzyme that interacts with several transcriptional factors and chromatin-modifying enzymes, such as FOXK1/2, ASXL1/2, OGT, HCF1, and KDM1B, and plays an important role in the epigenetic regulation of gene transcription 19–25. BAP1 and its associated proteins form the polycomb repressive deubiquitinase (PR-DUB) complex, which mainly functions to remove monoubiquitin from ubiquitinated histone 2A at lysine 119 (H2Aub) 26. It has been shown that this type of histone modification regulates gene transcription through epigenetic mechanisms 27–29. BAP1 is a tumor suppressor gene with frequent inactivating mutations and deletions in a variety of sporadic human cancers, including uveal melanoma (UVM), renal cell carcinoma, mesothelioma, and cholangiocarcinoma 19, 30–33. However, the mechanisms by which BAP1 exerts its tumor suppression function, particularly the extent to which BAP1 regulation of H2Aub levels on chromatin and corresponding transcriptional targets plays a role in its tumor suppression function, remain unclear. In this study, we conduct integrative analyses to achieve a comprehensive identification of BAP1-regulated target genes and relevant biological processes in cancer cells, and identify a BAP1-mediated epigenetic mechanism that links ferroptosis to tumor suppression.
Results
Genome-wide analyses link BAP1 to metabolism-related biological processes.
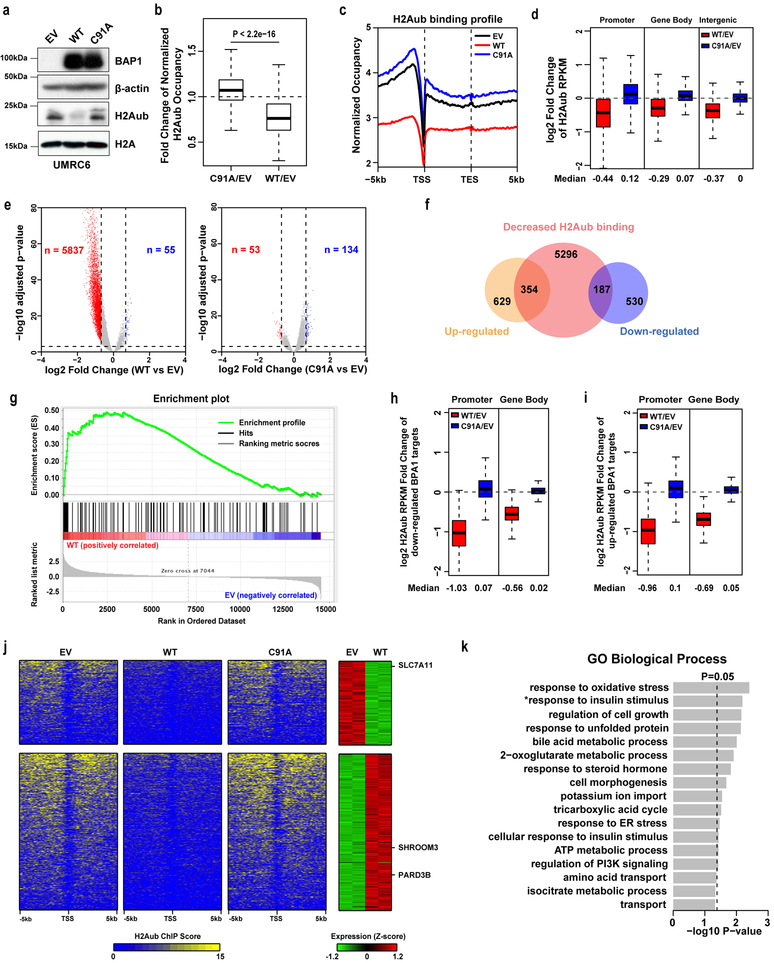
We conducted unbiased genome-wide analyses to characterize BAP1-dependent H2Aub occupancies and corresponding transcriptional alterations in the genome. To this end, we established UMRC6 cells (a BAP1-deficient renal cancer cell line) with stable expression of an empty vector (EV), BAP1 wild type (WT), and a BAP1 C91A DUB-inactive mutant 34. We confirmed that re-expression of BAP1 WT, but not its C91A mutant, in UMRC6 cells decreased global H2Aub levels (Fig. 1a). We then performed H2Aub chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) analyses in these cells. Our ChIP-seq analyses revealed that re-expression of BAP1 WT, but not its C91A mutant, resulted in significant reduction of genome-wide H2Aub occupancies in UMRC6 cells (Fig. 1b-1c). Distribution analysis showed that more than half of H2Aub bindings in EV/WT/C91A cells were detected at promoter or gene body regions (Fig. S1a). BAP1 WT, but not BAP1 C91A, cells showed decreases of H2Aub occupancies at promoter, gene body, and intergenic regions (Fig. 1d and S1b). Overall, we identified more than 5000 genes with reduced H2Aub occupancies in BAP1 WT cells compared with EV cells (Fig. 1e; FDR < 0.001).
Figure 1. Genome-wide analyses link BAP1 to metabolism-related biological processes.
a, Restoring BAP1 WT but not C91A in UMRC6 cells decreased H2Aub level. Experiment was repeated four times independently with similar results. b, Box plot showing fold changes of H2Aub occupancies in BAP1 WT or C91A compared with empty vector (EV) cells. Two-tailed unpaired Student’s t-test. n=24648 counts of promoter whose H2Aub occupancy (RPKM) is higher than 0.5 in all 3 samples. c, Average genome-wide occupancies of H2Aub in indicated cells. TSS: transcription start site; TES: transcription end site. d, Box plots of the log2 fold changes of H2Aub occupancies in promoter, gene body, and intergenic regions in BAP1 WT or C91A compared with EV cells. n=25772 for promoter and gene body, which is the total gene count in human reference. n=14237, which is the total number of intergenic regions. e, Volcano plots of H2Aub ChIP-seq data for BAP1 WT or C91A compared with EV cells. The red and blue dots represent genes with an at least 1.6-fold decrease or increase of H2Aub occupancies in BAP1 WT (left) or C91A (right) compared with EV cells. f, Venn diagram showing the overlap between 5837 genes with decreased H2Aub occupancies and 1700 differentially expressed genes (FC > 1.5, FDR < 0.05) upon restoring BAP1 in UMRC6 cells. g, GSEA showing that the 101 genes with > 2.5-fold H2Aub reduction were positively enriched in BAP1-upregulated genes. h, i, Box plots of log2 fold changes of H2Aub occupancies in promoter and gene body regions for the 187 genes (h) and 354 genes (i) as shown in Fig. 1f. j, Left 3 panels; heatmaps showing the H2Aub profile around the TSS of 187 downregulated and 354 upregulated genes (see Fig. 1f) in EV, BAP1 WT and C91A cells in decreasing order. Right panel, heatmap showing expression levels of the corresponding genes in EV and BAP1 WT cells. k, GO analysis for the 187 downregulated genes. Top annotation clusters are shown according to their enrichment scores [−log10 (p-value)]. *, positive regulation of glucose import in response to insulin stimulus. Fisher Exact test. Box and whisker plots show centre line at median, box limits at 25th/75th centiles and whiskers ±1.5× interquartile range (IQR). Unprocessed blots in Supplementary Fig. 7.
Parallel RNA sequencing (RNA-seq) analysis identified around 1,700 differentially expressed genes (983 upregulated genes and 717 downregulated genes) upon BAP1 re-expression in UMRC6 cells (Fig. S1c; fold change > 1.5, FDR < 0.05). Integration of both H2Aub ChIP-seq and RNA-seq data sets identified 354 BAP1-upregulated genes and 187 BAP1-downregulated genes with BAP1-dependent H2Aub reduction in both gene sets (Fig. 1f and Table S1). The current model proposes that H2Aub is associated with transcriptional repression 28, which would predict that BAP1-dependent H2Aub reduction correlates with BAP1-mediated transcriptional activation. To test this hypothesis, we conducted gene set enrichment analysis (GSEA) on the genes with different fold changes in BAP1-dependent H2Aub reduction (5837 genes whose H2Aub levels were reduced by BAP1 by > 1.6-fold, 1494 genes by > 2-fold, and 101 genes by > 2.5-fold). GSEA revealed that, in all three analyses, genes with BAP1-dependent H2Aub reduction were enriched with BAP1-upregulated genes; indeed, the more stringent the cutoff we employed, the more significant the correlation we observed (Fig. 1g for 101 genes, Normalized Enrichment Score = 1.61, FDR = 0; Fig. S1d for 1494 genes, Normalized Enrichment Score = 1.52, FDR = 0; Fig. S1e for 5837 genes, Normalized Enrichment Score = 1.22, FDR = 0). Thus, our analysis was consistent with the prevailing model that H2Aub correlates with transcriptional repression 28. It is also worth noting that the fold changes in H2Aub binding on promoters of the genes that were differentially regulated by BAP1 were more pronounced than those in the total set of 5837 genes (Fig. 1d, 1h, 1i and Fig. S1b, 1f, 1g; with median log2 fold change of H2Aub upon BAP1 re-expression: −0.44 for all 5837 genes vs. −1.03 for BAP1-downregulated genes vs. −0.96 for BAP1-upregulated genes). Thus, BAP1-dependent H2Aub deubiquitination correlates with BAP1-mediated transcriptional alteration. Heatmap analysis of the H2Aub profile around transcriptional start sites (TSSs) revealed that the genes that were upregulated or downregulated by BAP1 displayed reduced H2Aub signals centered on TSSs upon re-expression of BAP1 WT, but not C91A (Fig. 1j).
Gene ontology (GO) analysis of 354 BAP1-upregulated and 187 BAP1-downregulated genes revealed that, while BAP1-upregulated genes were enriched in diverse cellular processes (Fig. S1h and Table S1), the genes that were downregulated by BAP1 showed striking enrichment in metabolism-related biological processes, among which “response to oxidative stress” was the most significantly enriched (Fig. 1k and Table S1). Together, our genome-wide analyses suggested that BAP1-mediated deubiquitination of H2Aub is associated with both transcriptional activation and repression of gene targets involved in different biological processes see (Discussion), and linked BAP1-repressed genes to metabolism.
Cancer genomic analyses link SLC7A11 to BAP1-mediated tumor suppression in human cancers.
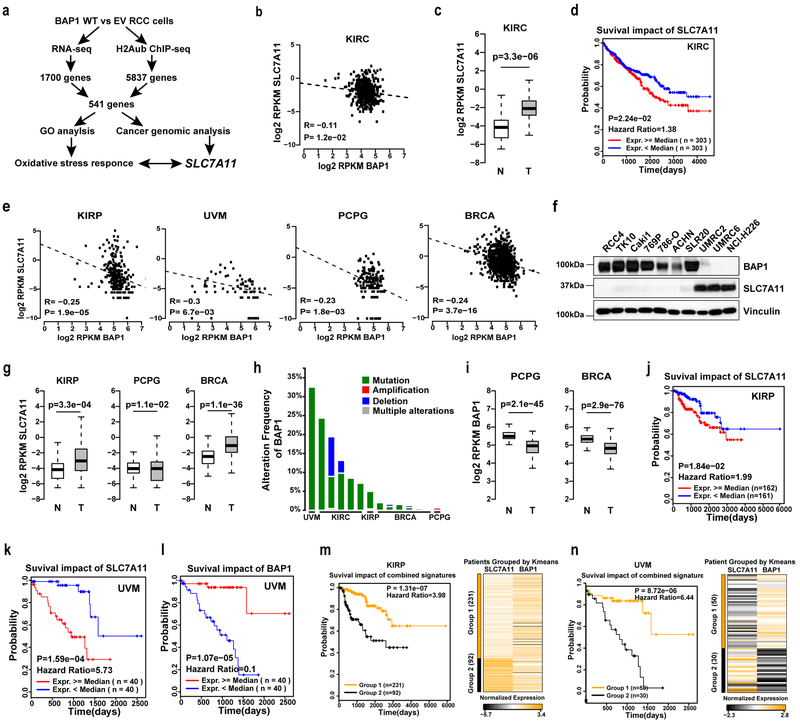
Next, we studied the potential relevance of BAP1-regulated genes to BAP1-mediated tumor suppression in human cancers through analysis of The Cancer Genome Atlas (TCGA) data sets. Since we conducted our genome-wide analyses in a renal cancer cell line, we focused on the TCGA Kidney Clear Cell Carcinoma (KIRC) data set 35 in our initial analysis. Specifically, we subjected the list of 541 genes (187 BAP1-downregulated and 354 BAP1-upregulated genes with reduced H2Aub occupancies; see Fig. 1f) to a series of computational analyses, including (i) expression correlation with BAP1 in KIRC, (ii) comparison of expression in KIRC and normal kidneys, and (iii) patient survival prediction in KIRC (Fig. 2a). Here we aimed to identify BAP1-downregulated target genes (i) whose expression is inversely correlated with that of BAP1 in KIRC, (ii) that are upregulated in KIRC compared with normal kidneys, and (iii) whose upregulation correlates with shorter patient survival, and vice versa. Such analyses identified 20 BAP1-downregulated and 26 BAP1-upregulated genes that satisfied all three criteria, among which SLC7A11 was the only downregulated gene whereas Shroom3 and PARD3B were the upregulated genes that showed statistical significance in all three analyses (Table S2). Notably, SLC7A11 was among the top genes identified in our RNA-seq and H2Aub ChIP-seq analyses (see Fig. 1j and Fig. S1c), and also among the genes involved in “response to oxidative stress,” the most significantly enriched biological process of BAP1-downregulated genes from the GO analysis (Table S1). We therefore focused on SLC7A11 in the follow-up studies.
Figure 2. Cancer genomic analyses link SLC7A11 to BAP1-mediated tumor suppression in human cancers.
a, The pipeline of identifying relevant BAP1 target genes in cancer. b, e, The scatter plots showing the inverse correlation of BAP1 and SLC7A11 expression in different cancer types. KIRC, n=606 independent samples; KIPR, n=323 independent samples, UVM, n=80 independent samples, PCPG, n=187 independent samples, BRCA, n=1218 independent samples. c, g, i, The box plots showing the comparison of SLC7A11 or BAP1 expression levels in normal tissues (N) and corresponding tumor (T) samples. KIRC, n=606 independent samples; KIRP, n=323 independent samples; Normal kidney, n=28 independent samples; PCPG, n=187 independent samples; Normal nerve, n=278 independent samples; BRCA, n=1218 independent samples; Normal breast, n=179 independent samples. d, Kaplan-Meier plots of KIRC patients stratified by SLC7A11 expression levels. f, Immunoblotting analyses of BAP1 and SLC7A11 protein levels in different cancer cell lines. Experiment was repeated twice independently with similar results. h, Bar graphs showing the frequencies of BAP1 genetic alterations in the indicated tumor types. The data are integrated from cBioPortal (http://www.cbioportal.org/), and the data sources for each column (left to right) are: TCGA, Provisional; IRC, Nat Genet 2014; TCGA, Provisional; TCGA, Nature 2013; U Tokyo, Nat Genet 2013; BGI, Nat Genet 2012; TCGA, Provisional; Sanger, Nature 2012; TCGA, Cell 2015; British Columbia, Nature 2012; TCGA, Provisional; TCGA, Nature 2012; Broad, Nature 2012; TCGA, Provisional. j, k, Kaplan-Meier plots of KIRP (j) and UVM (k) patients stratified by SLC7A11 expression levels. l, Kaplan-Meier plots of UVM patients stratified by BAP1 expression levels. m, n, Kaplan-Meier plots of KIRP (m) or UVM (n) patients stratified by unsupervised clustering on BAP1 and SLC7A11 expression. Group 1 has higher BAP1 and lower SLC7A11 expression, while Group 2 has lower BAP1 and higher SLC7A11 expression. Pearson’s correlation (two-sided) analysis was used in b and e. Two-tailed unpaired Student’s t-test was used in c, g and i. Log-rank Mantel-Cox test was used in d, j, k, l, m and n. Detailed statistical tests were described in Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 7.
As shown in Fig. 2b-2d, our analyses revealed that SLC7A11 expression inversely correlated with BAP1 expression in KIRC, that SLC7A11 exhibited higher expression in KIRC than in normal kidneys, and that higher expression of SLC7A11 predicted shorter survival in KIRC patients. We then extended the same analyses of SLC7A11 to other TCGA data sets. Such analyses revealed that BAP1 expression inversely correlated with SLC7A11 expression not only in KIRC, but also in several other human cancers, including kidney papillary cell carcinoma (KIRP), UVM, pheochromocytoma and paraganglioma (PCPG), and breast invasive carcinoma (BRCA) 36–39 (Fig. 2e). Consistent with this, we observed an inverse correlation between BAP1 and SLC7A11 levels across a panel of cancer cell lines (Fig. 2f). Further comparison of the expression levels of SLC7A11 in KIRP, PCPG, BRCA, and corresponding normal tissues showed that SLC7A11 is upregulated in these cancers (Fig. 2g). (Note that there was no normal tissue control for UVM in the TCGA or GTEx databases, so we were unable to conduct the analysis for UVM). Correspondingly, BAP1 is mutated/deleted (in UVM, KIRC, and KIRP), or downregulated (in PCPG and BRCA) in these tumor types with SLC7A11 high expression (Fig. 2h-2i). Finally, we showed that, in KIRP and UVM, patients with high SLC7A11 expression had shorter overall survival than did patients with low SLC7A11 expression, whereas UVM patients with lower BAP1 expression had significantly shorter overall survival (Fig. 2j-2l). In KIRP and UVM, the combination of high SLC7A11 and low BAP1 expression predicted even worse clinical outcomes than either parameter alone (Fig. 2m-2n). Together, our analyses identified SLC7A11 as one of the most relevant target genes in BAP1-mediated tumor suppression in human cancers and revealed that BAP1 and SLC7A11 expression levels show an inverse correlation and predict clinical outcomes in human cancers.
BAP1 suppresses SLC7A11 expression and reduces H2Aub occupancy on the SLC7A11 promoter.
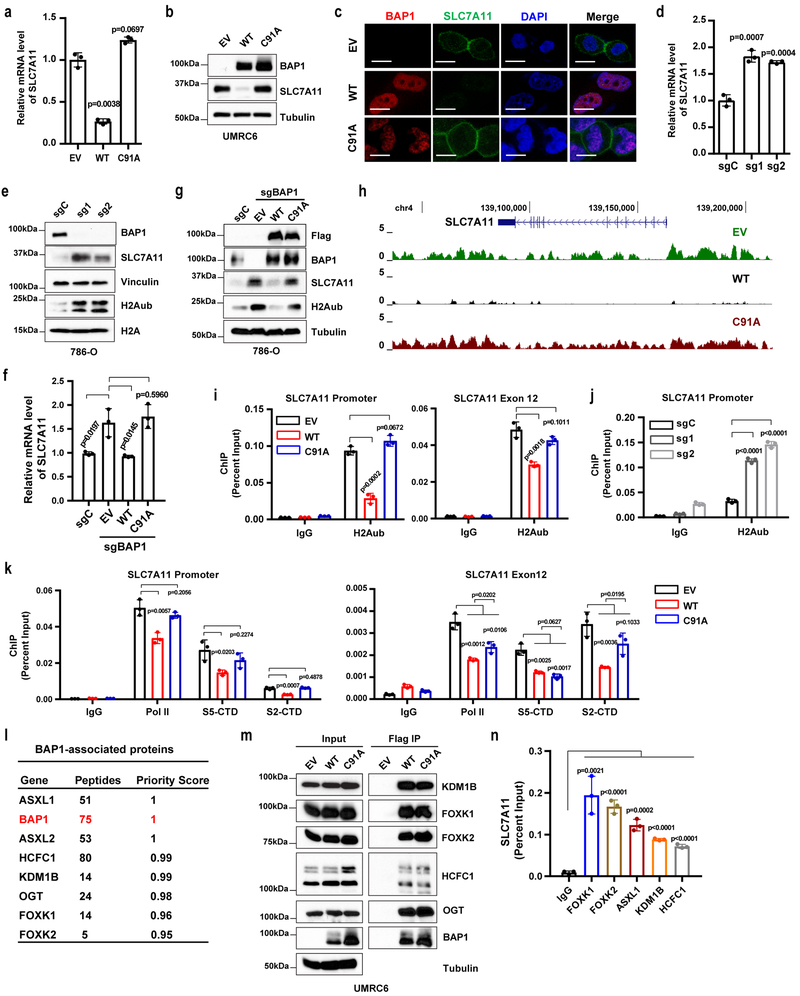
The aforementioned computational analyses prompted detailed mechanistic studies on how BAP1 regulates SLC7A11. UMRC6 cells, the BAP1-deficient cell line used in our RNA-seq and H2Aub ChIP-seq analyses, exhibited high SLC7A11 expression (see Fig. 2f). We confirmed that re-expression of BAP1 WT, but not its C91A mutant, in UMRC6 cells decreased SLC7A11 expression (Fig. 3a-3c), suggesting that BAP1-mediated repression of SLC7A11 expression requires BAP1’s DUB activity. Similarly, BAP1 re-expression in NCI-H226 cells, a BAP1-deficient mesothelioma cell line with high SLC7A11 expression (see Fig. 2f), also repressed SLC7A11 expression (Fig. S2a-2b). Conversely, BAP1 deficiency by CRISPR technology increased SLC7A11 expression in several BAP1-proficient renal cancer cells with low SLC7A11 levels, including 786-O, Caki1, and ACHN cells (Fig. 3d-3e and S2c-2f). Importantly, restoration of BAP1 WT, but not its C91A mutant, in BAP1 CRISPR knockout (KO) (sgBAP1) 786-O cells decreased SLC7A11 expression to a level similar to that in 786-O control cells (Fig. 3f-3g). In addition, Bap1 deletion in mouse embryonic fibroblasts (MEFs) 34 increased SLC7A11 expression (Fig. S2g). A luciferase assay with the SLC7A11 promoter showed that overexpression of BAP1 WT, but not its C91A mutant, decreased luciferase activity, whereas BAP1 knockdown increased it, indicating that BAP1 regulates SLC7A11 transcription (Fig. S2h-2i).
Figure 3. BAP1 suppresses SLC7A11 expression and reduces H2Aub occupancy on the SLC7A11 promoter.
a, b, SLC7A11 expression levels in indicated UMRC6 cells were measured by RT-PCR (a) and Western blotting (b). Error bars are mean ± s.d., n = 3 independent repeats (a). Experiment was repeated four times independently with similar results (d). c, BAP1 (red) and SLC7A11 (green) expression and localization in indicated cells were analyzed by immunofluorescence. Cell nuclei were labeled by DAPI (blue) staining. Scale bar, 10 µm. Experiment was repeated twice independently with similar results. d, e, mRNA and protein levels of indicated genes in indicated 786-O cells were measured by RT-PCR (d) and Western blotting (e). Error bars are mean ± s.d., n = 3 independent repeats (d). Experiment was repeated three times independently with similar results (e). f, g, mRNA and protein levels of indicated genes in indicated 786-O cells were measured by RT-PCR (f) and Western blotting (g). Error bars are mean ± s.d., n = 3 independent repeats (f). Experiment was repeated twice independently with similar results (g). h, H2Aub ChIP-Seq occupancy profiles at the SLC7A11 loci in indicated UMRC6 cells. i, ChIP-qPCR confirming the lower H2Aub binding on the SLC7A11 promoter and selected exon regions in BAP1 WT cells than in EV or BAP1 C91A cells. Error bars are mean ± s.d., n = 3 independent repeats. j, ChIP-qPCR showing the increased H2Aub binding on the SLC7A11 promoter upon BAP1 deficiency in 786-O cells. Error bars are mean ± s.d., n = 3 independent repeats. k, ChIP-qPCR analysis of RNA polymerase II (Pol II), S5-CTD, and S2-CTD binding on the SLC7A11 promoter and selected exon regions. Error bars are mean ± s.d., n = 3 independent repeats. l, Lists of BAP1 binding proteins identified by mass spectrometry. m, Interactions between indicated proteins and BAP1 were verified by Western blotting in indicated cells. Experiment was repeated twice independently with similar results. n, Bar graph showing the binding of PR-DUB proteins on the SLC7A11 promoter by ChIP-qPCR. Error bars are mean ± s.d., n = 3 independent repeats. All P values were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests were described in Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 7.
Previous studies showed that the tumor suppressor p53 represses SLC7A11 expression 40, 41. Re-expression of BAP1 in UMRC6 cells did not significantly affect p53 levels (Fig. S2j). We then studied whether p53 plays a role in BAP1 regulation of SLC7A11 expression. Consistent with previous findings in other cell lines 40, we confirmed that p53 deletion via CRISPR technology increased SLC7A11 levels in UMRC6 cells (Fig. S2k). However, restoring BAP1 in p53-deficient cells still inhibited SLC7A11 expression (Fig. S2l-2n), and the fold change in SLC7A11 expression by BAP1 restoration in p53-deficient cells was similar to that in p53-proficient cells (Fig. S2o), suggesting that BAP1 represses SLC7A11 expression independent of p53.
Analysis of our H2Aub ChIP-seq data revealed that restoration of BAP1 WT, but not BAP1 C91A, markedly decreased H2Aub occupancy at both the promoter and gene body of SLC7A11 (Fig. 3h), which was further confirmed by H2Aub ChIP assay on the SLC7A11 promoter and representative exons (Fig. 3i and S2p). Conversely, we showed that BAP1 deletion increased global H2Aub levels in 786-O, Caki1, and ACHN cells (Fig. 3e, Fig. S2c and Fig. S2e) and promoted H2Aub binding on the SLC7A11 promoter in 786-O cells (Fig. 3j). It has been shown that H2Aub regulates both transcription initiation and elongation 42, which are associated with different phosphorylation patterns of the heptad repeats of the C-terminal domain (CTD) of the largest subunit of RNA polymerase II: transcription initiation is associated with CTD serine 5 phosphorylation (S5-CTD), while transcription elongation is associated with CTD serine 2 phosphorylation (S2-CTD) 43. We found that BAP1 decreased S5-CTD binding on the SLC7A11 promoter and S2-CTD binding on exons of SLC7A11 in a DUB-dependent manner (Fig. 3k and Fig. S2q) (but BAP1 did not decrease the total protein levels of RNA polymerase II, S5-CTD, or S2-CTD; Fig. S2r). Our data thus suggested that BAP1-mediated H2Aub deubiquitination on SLC7A11 correlates with inhibition of both transcription initiation and elongation.
BAP1 interacts with several transcriptional factors and chromatin-modifying factors, forming the PR-DUB complex 19, 20. Indeed, mass spectrometry analysis identified all known protein components of the PR-DUB complex as top-ranking BAP1-interacting proteins (Fig. 3l), and some of the interactions were further confirmed by co-immunoprecipitation analysis (Fig. 3m). It should be noted that BAP1 did not appear to affect the protein levels of other PR-DUB components (Fig. S3a). It has been suggested that BAP1 is recruited to chromatin through the transcriptional factors and chromatin-modifying factors in the PR-DUB complex, such as FOXK1/2 and ASXL1/2 19. A previous study generated ChIP-seq data sets for several components in the PR-DUB complex, including FOXK1, ASXL1, and O-GlcNAC (a readout for OGT) 44. We therefore integrated these ChIP-seq data sets with our H2Aub data sets. These analyses identified around 3,000 genes that are co-occupied by FOXK1, ASXL1, and O-GlcNAC; among these, 1140 genes also exhibit BAP1-dependent decrease in H2Aub occupancy (Fig. S3b). Notably, analysis of these ChIP-seq data sets revealed striking bindings of ASXL1, FOXK1, and O-GlcNAC on the SLC7A11 promoter (Fig. S3c). The bindings of PR-DUB complex proteins on the SLC7A11 promoter were further validated by ChIP assay (Fig. 3n). Knockdown of some PR-DUB components also affected SLC7A11 expression (Fig. S3d-S3m). Together, our data suggested a model that the BAP1-containing PR-DUB complex binds on the SLC7A11 promoter, where BAP1 removes ubiquitin from H2Aub, and BAP1-dependent H2Aub reduction on SLC7A11 is associated with BAP1-mediated SLC7A11 repression.
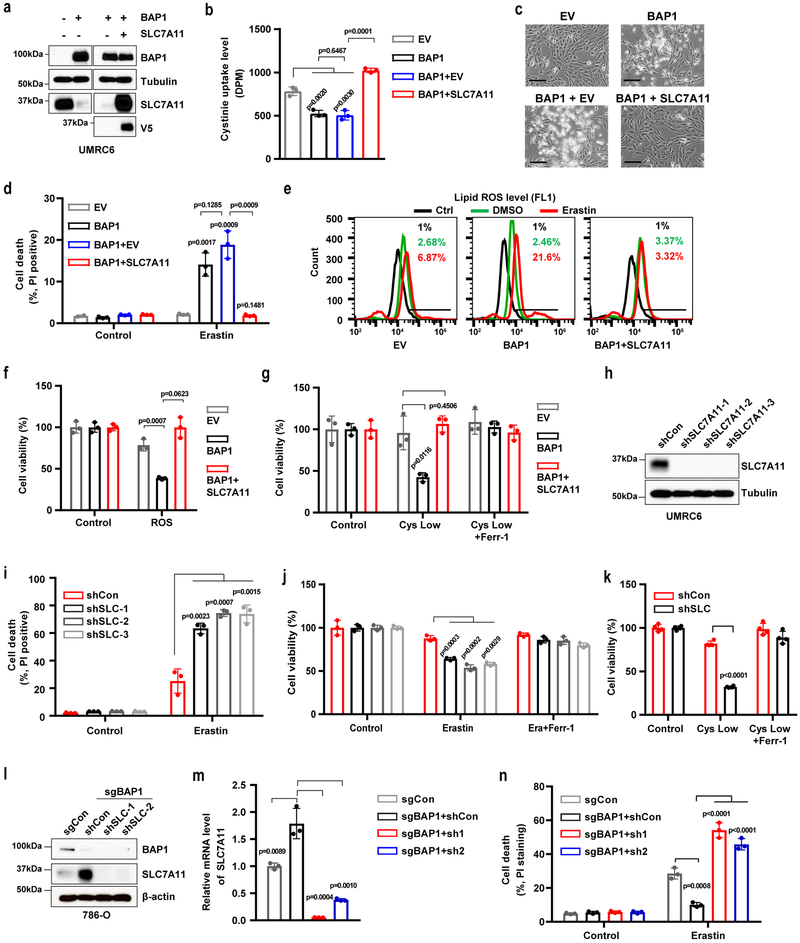
BAP1 suppresses SLC7A11-mediated cystine uptake and promotes ferroptosis.
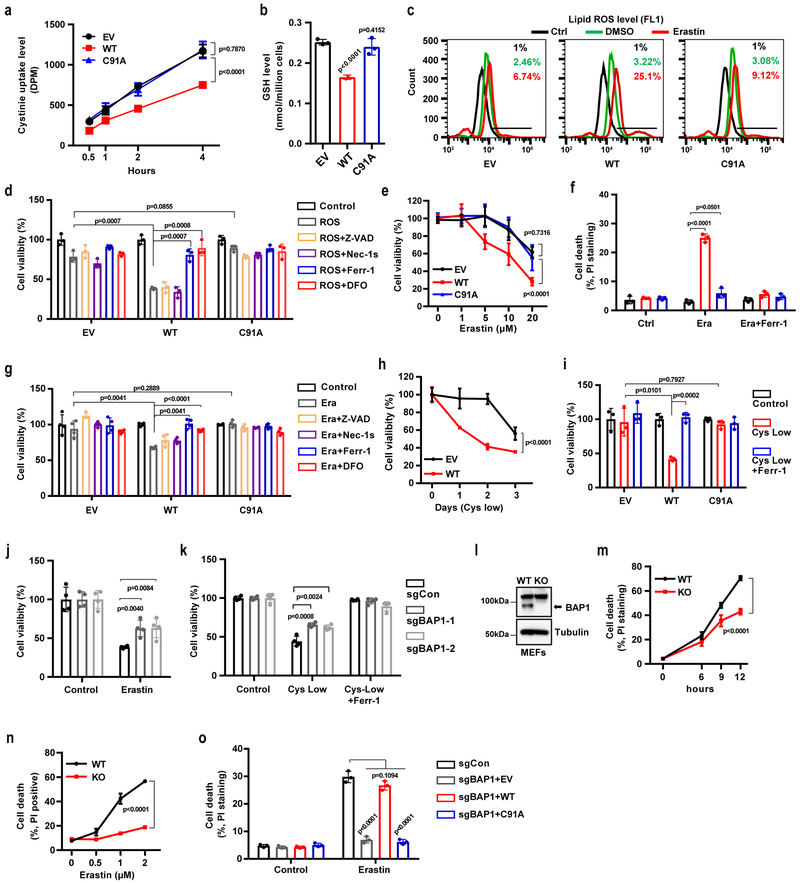
SLC7A11 mediates the uptake of extracellular cystine, a major precursor for glutathione biosynthesis 12, 14. Glutathione is then utilized by GPX4 to detoxify lipid hydroperoxide and to protect cells from ferroptosis 15. Consistently, we observed that re-expression of BAP1 WT, but not its C91A mutant, in UMRC6 cells inhibited cystine uptake (Fig. 4a), decreased GSH levels (Fig. 4b), and increased erastin-induced lipid peroxidation (Fig. 4c). Erastin treatment potently induced SLC7A11 expression without affecting BAP1 expression (Fig. S4a-4b). In addition, erastin potently induced SLC7A11 expression in BAP1-deficient cells (Fig. S4c-4d), suggesting that BAP1 represses the basal expression of SLC7A11 and that erastin induces SLC7A11 expression likely through BAP1-independent mechanisms.
Figure 4. BAP1 suppresses SLC7A11-mediated cystine uptake and promotes ferroptosis.
a, Cystine uptake levels were measured in indicated cells. b, Bar graph showing intracellular GSH levels in indicated cells. c, Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in indicated cells. Experiment was repeated three times independently with similar results. d, f, Bar graph showing cell viability in indicated cells treated with ROS (100 µM TBH) (d) or 20 µM erastin (f) combined with 5 µM Z-VAD-fmk (Z-VAD), 2 µM Necrostatin-1s (Nec-1s), 2 µM ferrostatin-1 (Ferr-1), or 100 µM deferoxamine (DFO). e, Cell viability in indicated cells was measured after treatment with different concentrations of erastin for 30 hr. g, Cell death was measured in indicated cells after treatment with Ferr-1 (2 µM) and erastin (20 µM) for 24 hr. h, Cell viability of indicated cells was measured upon culturing in cystine-low (2 µM cystine) medium for indicated durations. i, Cell viability of indicated cells was measured after culturing in cystine-low medium + Ferr-1 (2 µM) for 48 hr. j, k, Cell viability was measured in BAP1 KO 786-O cells treated with erastin (10 µM) for 24 hr (j) or cultured in cystine-low medium + Ferr-1 (2 µM) for 24 hr (k). l, Western blotting analysis of BAP1 expression in Bap1 WT and KO MEFs. Experiment was repeated twice independently with similar results. m, n, Erastin-induced cell death in Bap1 WT and KO MEFs at different time points (2 µM) (m) or at different concentrations after 8 hr of treatment (n). o, Cell death was measured in BAP1 KO 786-O cells restored with BAP1 WT or C91A mutant and treated with erastin (10 µM) for 24 hr. Error bars are mean ± s.d., n = 3 (a, b, d, f, h, j, m and n ) or 4 (e, g, i, and k) independent repeats. Two-way ANOVA analysis was performed in a, e, h, m and n. Two-tailed unpaired Student’s t-test in b, d, f, g, i, j, k and o. Detailed statistical tests were described in Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 7.
We next studied the potential role of BAP1 in ferroptosis. We showed that treatment with tert-butyl hydroperoxide (TBH), a ROS inducer 40, resulted in substantially more cell death in BAP1 WT cells than in EV or BAP1 C91A cells (Fig. 4d and S4e). Importantly, TBH-induced cell death could be largely suppressed by the ferroptosis inhibitor ferrostatin or the iron chelator deferoxamine (DFO), but not by the apoptosis inhibitor Z-VAD-fmk or the necroptosis inhibitor necrostatin-1s (Fig. 4d). Similarly, BAP1 WT, but not its C91A mutant, significantly potentiated erastin-induced cell death (Fig. 4e-4f and S4f), and erastin-induced cell death could be fully suppressed by ferrostatin or DFO, but not by Z-VAD-fmk or necrostatin-1s (Fig. 4g and S4f). Erastin treatment did not induce PARP cleavage (Fig. S4g), confirming that erastin did not induce apoptosis. Cystine depletion also induces ferroptosis 45. We observed that expression of BAP1 WT, but not the BAP1 C91A mutant, sensitized UMRC6 cells to cystine-depletion-induced cell death, which could be fully suppressed by ferrostatin (Fig. 4h-4i). Re-expression of BAP1 in NCI-H226 cells, another BAP1-deficient cell line, similarly potentiated TBH- or erastin-induced ferroptosis (Fig. S4h-4i). Conversely, BAP1 deficiency in 786-O, Caki1, and ACHN cells (Fig. 4j-4k and S4j-4k) and in MEFs (Fig. 4l-4n and Fig. S4l-4m) rendered cells more resistant to erastin- or cystine-depletion-induced ferroptosis. Importantly, re-expression of BAP1 WT, but not its C91A mutant, in BAP1 KO 786-O cells restored ferroptosis sensitivity (Fig. 4o). Finally, analysis of ferroptosis sensitivity in the panel of cell lines (see Fig. 2f) revealed that all three BAP1-low/SLC7A11-high cell lines were resistant to erastin-induced ferroptosis, whereas five of seven BAP1-high/SLC7A11-low cell lines were sensitive to erastin-induced ferroptosis (Fig. S4n). Taken together, our results convincingly showed that BAP1 promotes ferroptosis in a DUB-dependent manner.
BAP1 regulates ferroptosis through SLC7A11.
We next sought to determine whether BAP1 promotes ferroptosis by mediating repression of SLC7A11 expression. Restoration of SLC7A11 in UMRC6 BAP1 cells normalized cystine uptake, lipid peroxidation accumulation, and erastin-, ROS-, or cystine-depletion-induced ferroptosis to levels similar to those observed in UMRC6 EV cells (Fig. 5a-5g). We also restored SLC7A11 expression in NCI-H226 BAP1 cells and made similar observations (Fig. S5a-5e). Conversely, SLC7A11 knockdown in UMRC6 and NCI-H226 cells, two BAP1-deficient cancer cells that exhibited high SLC7A11 expression and were resistant to ferroptosis (see Fig. 2f and Fig. S4n), sensitized the cells to erastin- or cystine-depletion-induced ferroptosis (Fig. 5h-5k, and Fig. S5f-5i). Finally, SLC7A11 knockdown in sgBAP1 786-O cells (which exhibited increased SLC7A11 expression) also re-sensitized the cells to ferroptosis (Fig. 5l-5n). Collectively, our data strongly suggested that BAP1 promotes ferroptosis mainly through BAP1-mediated repression of SLC7A11 expression.
Figure 5. BAP1 promotes ferroptosis through SLC7A11.
a, Western blotting analysis of BAP1 and SLC7A11 in the indicated cell lines. Experiment was repeated twice independently with similar results. b, Cystine uptake levels at 2 hr were measured in the indicated cells. c, Representative phase-contrast images of the indicated cells treated with 20 μM erastin for 24 hr. Scale bar, 100 µm. d, Bar graph showing cell death in the indicated cell lines upon erastin treatment. e, Lipid peroxidation in the indicated cells after treatment with 10 µM erastin for 24 hr was assessed by flow cytometry after C11-BODIPY staining. f, Bar graph showing cell death in the indicated cells induced by ROS (100 µM TBH, 16 hr). g, Bar graph showing viability of the indicated cells cultured in cystine-low (2 µM) medium for 2 days with or without 2 µM ferrostatin-1 (Ferr-1). h, Western blotting analysis of SLC7A11 expression in SLC7A11-knockdown cell lines. Experiment was repeated twice independently with similar results. i, Bar graph showing cell death of the indicated cells upon 20 µM erastin treatment for 30 hr. j, k, Bar graph showing viability of the indicated cells treated with erastin (10 µM) with or without 2 µM Ferr-1 for 30 hr (j), or cultured in cystine-low medium with or without 2 µM Ferr-1 for 2 days (k). l, m, Western blotting (l) and RT-PCR (m) analyses of SLC7A11 in BAP1 KO 786-O cell lines with SLC7A11-knockdown. Experiment was repeated twice independently with similar results (l). n, Bar graph showing cell death in the indicated cell lines after 10 µM erastin treatment for 24 hr. Error bars are mean ± s.d., n = 3 (b, d, f, g, i, j, m and n ) or 4 (k) independent repeats. All P values were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests were described in Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 7.
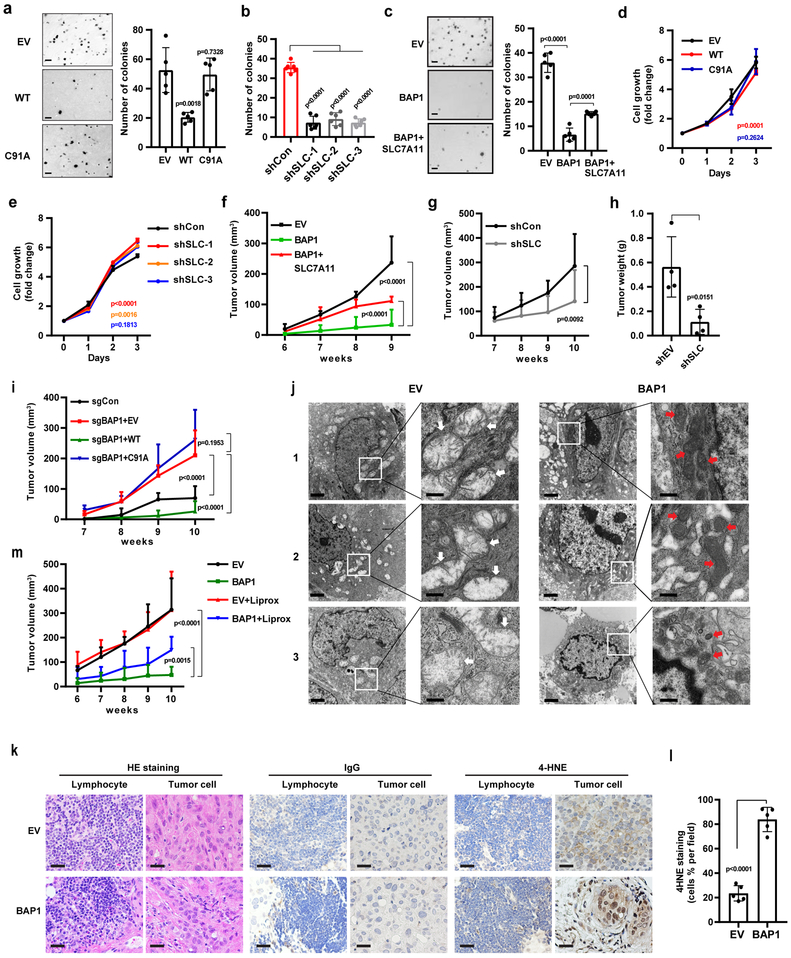
BAP1 inhibits tumor development partly through SLC7A11 and ferroptosis.
We further studied the role of BAP1 regulation of SLC7A11 in BAP1-mediated tumor suppression. BAP1 re-expression in UMRC6 cells markedly inhibited anchorage-independent growth in a DUB-dependent manner (Fig. 6a). SLC7A11 knockdown in UMRC6 cells significantly inhibited anchorage-independent growth (Fig. 6b), and importantly, restoration of SLC7A11 in UMRC6 BAP1 WT cells partially restored anchorage-independent growth compared with UMRC6 EV cells (Fig. 6c). It should be noted that BAP1 re-expression or SLC7A11 knockdown affected cell proliferation marginally (Fig. 6d and 6e). Consistently, we showed that BAP1 re-expression in a BAP1-deficient background markedly inhibited xenograft tumor development and that restoration of SLC7A11 in BAP1-expressing cells partially restored tumor development (Fig. 6f), while SLC7A11 knockdown in a BAP1-deficient background inhibited xenograft tumor development (Fig. 6g and 6h). In addition, BAP1 deletion in a BAP1-proficient background markedly increased xenograft tumor development, and restoration of BAP1 WT, but not BAP1 C91A mutant, repressed tumor development (Fig. 6i). Taken together, our data suggested that BAP1 suppresses tumor development in a DUB-dependent manner and at least partly through SLC7A11.
Figure 6. BAP1 inhibits tumor development partly through SLC7A11 and ferroptosis.
a-c, Representative images showing colonies of the indicated cell lines on soft agar. Scale bar, 500 µm. Bar graph showing the relative number of colonies formed by the indicated cells in the soft agar assay. Error bars are mean ± s.d., n = 5 (a and c) or 6 (b) independent repeats. P value was calculated using two-tailed unpaired Student’s t-test. d, e, Cell growth assays of UMRC6 cells with indicated genotypes. Error bars are mean ± s.d., n = 3 (e) or 4 (d) independent repeats. P value was determined using two-way ANOVA analysis. f, g, Volumes of xenograft tumors with the indicated genotypes at different time points (weeks) after tumor cell injection Error bars are mean ± s.d., n = 5 independent repeats. P value was determined using two-way ANOVA analysis. h, Bar graph showing the weight of tumor xenografts from the indicted genotypes. Error bars are mean ± s.d., n = 5 independent repeats. P value was calculated using two-tailed unpaired Student’s t-test. i, Tumor volumes of xenografts tumors from 786-O cell lines with indicated genotypes at different weeks. Error bars are mean ± s.d., n = 5 (sgCon and sgBAP1+EV) or 8 (sgBAP1+WT and BAP1+C91A) independent repeats. P value was determined using two-way ANOVA analysis. j, Tumor xenografts from the EV and BAP1-restored cell lines were subjected to transmission electron microscopy. White arrow: mitochondria with obvious cristae. Red arrow: shrunken mitochondria. Scale bars: left, 2 µm; right, 500 nm. Experiment was repeated twice independently with similar results. k, Hematoxylin and eosin and immunohistochemical staining of tumor xenografts from the EV and BAP1-restored cell lines. Scale bar, 50 µm. Experiment was repeated twice independently with similar results. l, Percentage of 4HNE-positive stained cells per field. Error bars are mean ± s.d., n = 5 randomly selected high-power fields. P value was calculated using two-tailed unpaired Student’s t-test. m, Volumes of xenograft tumors with the indicated genotypes and treatments at different weeks. Error bars are mean ± s.d., n = 5 independent repeats. P value was determined using two-way ANOVA analysis. Detailed statistical tests were described in Methods.
Next, we studied the potential relevance of ferroptosis in BAP1-mediated tumor suppression. Electron microscopy (EM) analysis revealed that many tumor cells from xenograft tumors with BAP1 re-expression contained shrunken mitochondria with increased membrane density, a morphologic feature of ferroptosis 7; In contrast, we never observed such cells in EV tumor samples (Fig. 6j). Since ferroptosis is characterized by overproduction of lipid peroxidation, we conducted 4-hydroxy-2-noneal (4HNE) immunohistochemistry (IHC) analysis to characterize lipid peroxidation levels 46 in EV and BAP1-re-expressing tumor samples. These studies revealed increased 4HNE staining in tumor cells, but not surrounding lymphocytes, from BAP1-re-expressing tumors compared with cells from EV tumors (Fig. 6k and 6l). Finally, we showed that treatment with the ferroptosis inhibitor liproxstatin-1 16, while did not affect EV tumor development, partially restored tumor development in BAP1-re-expressing tumors (Fig. 6m). Together, our data suggested that BAP1 promotes ferroptosis in vivo and that ferroptosis is at least partly responsible for BAP1’s tumor suppression function in vivo.
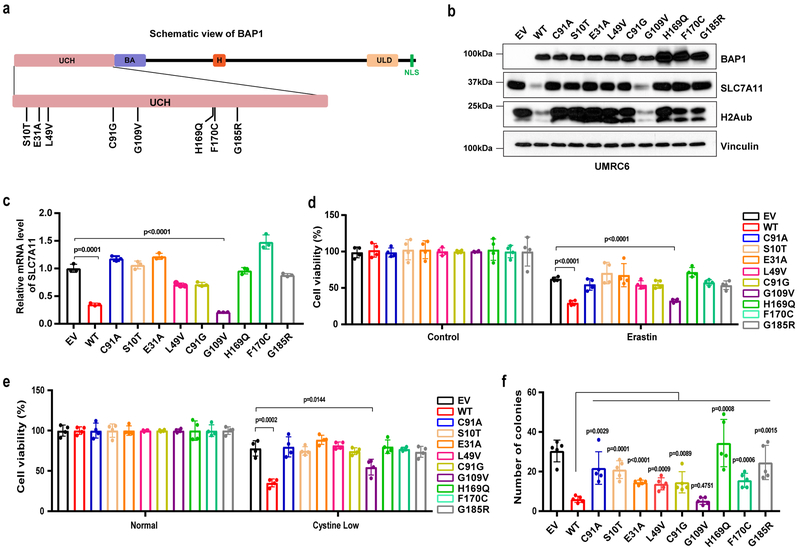
Cancer-associated BAP1 mutations are defective in regulating SLC7A11 and ferroptosis.
Many BAP1 mutations have been identified in human cancers. A survey of cancer genomics datasets from the cBioPortal for Cancer Genomics 47, 48 as of September 2017 identified about 500 somatic mutations in BAP1, including 211 nonsense (truncating) mutations and 283 missense mutations (Fig. S6a). Importantly, KIRC patients with BAP1 mutations had poor clinical outcomes (Fig. S6b). To study the effect of cancer-associated BAP1 missense mutations on ferroptosis and tumor suppression, we first utilized Mutation Assessor from cBioPortal (http://mutationassessor.org/r2/) to predict and rank the functional impact scores of all missense mutations identified in BAP1. These analyses identified eight BAP1 mutations with the highest functional impact scores (Table S3). Notably, all eight mutations are located within the UCH domain, which mediates the DUB activity of BAP1 (Fig. 7a). In fact, one mutation, C91G, is equivalent to C91A, which was used as the DUB-inactive mutant in our study. Thus, the mutation analyses strongly suggested an important role for BAP1 DUB function in human cancer. We then expressed each of these cancer-associated BAP1 mutants, as well as BAP1 WT and the C91A mutant, in UMRC6 cells. Remarkably, all BAP1 mutants, except G109V, behaved as loss-of-function mutants and were incapable of repressing SLC7A11 expression (Fig. 7b-7c) or promoting erastin- or cystine-depletion-induced ferroptosis (Fig. 7d-7e and Fig. S6c). We further showed that, while these mutants only affected cell proliferation moderately at most (Fig. S6d and S6e), all mutants except G109V at least partially lost their abilities to inhibit anchorage-independent growth (Fig. 7f).
Figure 7. Cancer-associated BAP1 mutations are defective in regulating SLC7A11 and ferroptosis.
a, Diagram showing BAP1 domain structure and mutated sites of cancer-associated BAP1 mutations generated in this study. UCH, ubiquitin C-terminal hydrolases; BA, BRCA1‑binding domain; H, HCF1‑binding domain; ULD, UCH37‑like domain; NLS, nuclear localization signal. b, Western blotting analysis of UMRC6 cells with re-expression of cancer-associated BAP1 mutants and WT BAP1 as indicated. Experiment was repeated twice independently with similar results. c, Relative mRNA levels of SLC7A11 in the indicated UMRC6 cells were determined by RT-PCR. Error bars are mean ± s.d., n = 3 independent repeats. P value was calculated using two-tailed unpaired Student’s t-test. d, e, Bar graphs showing the viability of the indicated cells treated with 20 µM erastin for 30 hr (e) or cultured in cystine-low (2 µM) medium for 2 days (e). Error bars are mean ± s.d., n = 4 independent repeats. P value was calculated using two-tailed unpaired Student’s t-test. f, Bar graph showing the relative number of colonies of the indicated cells in the soft agar assay. Error bars are mean ± s.d., n = 5 independent repeats. P value was calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests were described in Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 7.
These BAP1 mutants did not significantly affect BAP1’s interaction with other components of the PR-DUB complex (Fig. S6f). Notably, we observed that all BAP1 mutants except G109V were incapable of downregulating H2Aub (Fig. 7b), suggesting that these BAP1 mutants are at least partially DUB-inactive toward H2Aub. Overall, we observed strong correlations of these cancer-associated BAP1 mutants in terms of their effects on H2Aub levels, SLC7A11 expression, ferroptosis sensitivity, and anchorage-independent growth. Collectively, our results provided direct evidence to link BAP1’s functions in regulating H2Aub, SLC7A11 expression, and ferroptosis to its tumor suppression function in human cancer, and further suggested that BAP1’s DUB function of removing H2Aub is involved in its regulation of SLC7A11 expression, ferroptosis, and tumor suppression.
Discussion
Tumor cells require an appropriate supply of amino acids, including cystine, to survive, grow, and proliferate. Cystine plays important roles in maintaining redox balance and promoting cell survival in tumor cells; insufficient cystine supply induces metabolic stress and provokes ferroptotic cell death in tumor cells 7. Tumor cells engage different strategies to ensure an adequate supply of cystine from the tumor microenvironment. In some tumor cells with low expression of cystine transporter SLC7A11, such as chronic lymphocytic leukemia cells, stromal cells surrounding the tumor take up cystine, convert cystine to cysteine, and release cysteine into the tumor microenvironment; cysteine reuptake into tumor cells then promotes survival 49. In contrast, other types of tumor cells upregulate the expression of SLC7A11 and maintain high cystine uptake, although the underlying mechanisms by which tumor cells maintain high SLC7A11 expression remain incompletely understood. Our study revealed an important epigenetic mechanism linking SLC7A11 regulation of ferroptosis to tumor biology. Specifically, our study suggests a model in which the tumor suppressor BAP1 normally functions to repress the expression of SLC7A11 at least partly by deubiquitinating H2Aub on SLC7A11, thereby inhibiting cystine uptake into cells and rendering them more sensitive to ferroptosis. BAP1 inactivation by either mutation or expression downregulation de-represses SLC7A11 expression in tumor cells, resulting in increased cystine uptake and GSH synthesis, enhanced resistance to ferroptotic cell death, and promotion of tumor development. Notably, our integrated transcriptomic/epigenomic/cancer genomic analyses identified SLC7A11 as a BAP1 target with the most relevance to BAP1-mediated tumor suppression in human cancers, suggesting that the model described above is likely to be an important mechanism mediating BAP1’s tumor suppression function, although we do not rule out the possibility that other BAP1 targets, either transcriptional or non-transcriptional targets 50, may also play roles in BAP1-mediated tumor suppression.
Our study suggested that BAP1 represses SLC7A11 expression through BAP1-mediated H2Aub deubiquitination on SLC7A11. Other studies in mammalian cells have also shown that BAP1 represses the expression of some target genes with BAP1-dependent deubiquitination of H2Aub on the corresponding gene promoters 21, 51. However, the underlying mechanisms by which BAP1-mediated H2Aub deubiquitination represses target genes remain poorly understood, given that H2Aub generally correlates with gene repression 29. One model proposes that both H2A ubiquitination and deubiquitination need to occur in a dynamically regulated manner to maintain the repression of some target genes 52. Similarly, it has been proposed that the cycling of H2B ubiquitination and deubiquitination is critical for transcriptional activation 53. In line with this model, in Drosophila, the repression of polycomb target genes requires not only polycomb repressive complex 1 (PRC1)-mediated H2A ubiquitination, but also BAP1-mediated H2A deubiquitination, suggesting that an appropriate balance between H2A ubiquitination and deubiquitination, rather than H2A ubiquitination per se, is important for the maintenance of polycomb target gene repression 26. Thus, it is possible that BAP1, by removing ubiquitin from H2Aub on some target genes, such as SLC7A11, facilitates the dynamic cycling of H2Aub, resulting in transcriptional repression of target genes. It will be interesting to test this hypothesis in future studies.
In cell line studies, ferroptosis can be specifically induced by cystine depletion or treatment with drugs such as erastin. It can be characterized by EM analysis and cell death inhibitor rescue experiments (i.e., ferroptotic cell death can be prevented by ferroptosis inhibitors but not by inhibitors of other forms of cell death). Currently, our ability to study the clinical relevance of ferroptosis in cancer biology is hindered by a lack of established assays for characterizing ferroptosis in tissue and tumor samples equivalent to cleaved caspase-3 IHC staining for apoptosis. To our knowledge ferroptosis has not previously been identified in tumor samples in vivo. In our study, we conducted EM analysis, the gold standard assay for characterizing cell death, on tumor samples and provided definitive evidence of ferroptosis in tumors with BAP1 re-expression. Our work thus motivates further studies to develop assays that can be routinely used for characterizing ferroptosis in tissue and tumor samples.
Methods
Cell culture studies.
Cell line sources are detailed in Reporting Summary. All cell lines were free of mycoplasma contamination (tested by the vendor). Bap1 WT and KO MEFs were described in our previous publication 34. For cystine deprivation experiments, cells were cultured in DMEM with different concentrations of cystine + 10% (vol/vol) dialyzed FBS. The use of dialyzed FBS was described in our previous publication 54. To generate stable cell lines overexpressing Flag-BAP1 or mutant BAP1, HEK293T cells were transfected with either pLVX-Flag-BAP1 or mutant BAP1 constructs, together with psPAX.2 and pMD2.G third-generation lentiviral packaging systems using Lipofectamine 3000 reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. 48 hr later, lentivirus particles in the medium were collected and filtered, then the target cell lines were infected. At 24 hr post-infection, an appropriate selection antibiotic was added to obtain stable cell lines with successful transduction. To generate CRISPR KO cells, the sgRNAs were cloned into the CRISPR-V2 vector (Addgene, #52961), and lentiviral infection was conducted as described above. 72 hr later, single antibiotic selection-positive cells were sorted and plated into 96-well plates. Surviving KO clones were screened through immunoblotting with corresponding antibodies. To generate shRNA-knockdown cell lines, lentiviral transduction with shRNA vectors was conducted as described above. 72 hr later upon antibiotic selection, expression levels of target genes were determined by immunoblotting with corresponding antibodies.
Constructs and reagents.
SLC7A11 shRNAs and SLC7A11 cDNA-containing expression vectors were described in our previous publication 55. Human BAP1 cDNA was cloned into the lentivirus vector pLVX-Puro with N-terminal FLAG tag. A series of mutant pLVX-Flag-BAP1 constructs were generated by PCR mutagenesis using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) for amino acid substitutions according to the manufacturer’s instructions. All constructs were confirmed by DNA sequencing. The sequences of the primers used in PCR mutagenesis, gRNAs, and shRNAs used in this study are listed in Table S4. Erastin, ferrostatin-1, and TBH were obtained from Sigma (E7781, SML0583, and 458139). Z-VAD-fmk was obtained from R&D Systems (FMK001). Necrostatin-1s was obtained from BioVision (2263). Staurosporine was obtained from LC Laboratories (S-9300). All drugs were dissolved according to manufacturers’ instructions.
Cell death/viability/proliferation assays.
To measure cell death, cells were seeded in a 12-well plate 1 day before treatment. After treatment with appropriate drugs, cells were trypsinized and collected in a 1.5-mL tube, washed once with PBS, and stained with 2 μg/mL PI (Roche) in PBS. Dead cells (PI-positive cells) were analyzed using a BD Accuri C6 flow cytometer (BD Biosciences) 56, 57. To measure cell viability, 5000 cells per well were seeded in a 96-well plate 1 day before treatment. Upon treatment with the appropriate drugs where indicated, each well was replaced with fresh medium containing Cell Counting Kit-8 (CCK8) reagent (Sigma). After incubation for 1 hr at 37°C, the plate was analyzed using a FLUOstar Omega microplate reader (BMG Labtech), and absorbance of the wells was measured at 540 nm. To measure cell proliferation, 1500 cells per well were seeded in a 96-well plate. 24 hr after seeding, cell viability was measured using CCK8 reagent as described above. This was considered day 0. Later, cell viability was analyzed every 24 hr, and the absorbance was normalized to that measured on day 0. Cell growth was indicated by the fold change from day 0 to as long as day 4 and was graphed 58, 59.
Lipid peroxidation assay.
Lipid peroxidation levels were measured as previously described 15. Briefly, cells were incubated in a 60-mm dish containing 5 μM BODIPY 581/591 C11 dye (Invitrogen, D3861). After incubation for 30 min at 37℃, cells were washed with PBS and trypsinized, then stained with PI in PBS for 5 min. Cells were then subjected to flow cytometry analysis using an Accuri C6 flow cytometer. The FL1 channel signal in live cells was plotted as shown in the figures.
Cystine uptake assay.
Assessment of cystine uptake was conducted using modifications of a previously described protocol 49. Briefly, cells were seeded in a 12-well plate and incubated overnight. To measure cystine uptake, the medium was replaced with fresh DMEM (which contains 200 µM cystine) containing [14C] cystine (PerkinElmer) (0.04μCi), and cells were incubated for the indicated time periods. Uptake was terminated by rapidly rinsing cells twice with cold PBS, and lysed in 0.1 mM NaOH. Radioactivity (DPM) was measured using Tri-Carb® Liquid Scintillation Analyzer (PerKinElmer, Model 4810TR) in the presence of quench curve. All experiments were carried out in triplicate.
GSH assay.
GSH levels were detected using a GSH-Glo Glutathione Assay (Promega) following the manufacturer’s instructions. Briefly, 6000 cells per well were seeded in a 96-well plate 1 day before analysis. The culture medium from the wells was carefully removed, and 100 µL of prepared 1× GSH-Glo Reagent was added, followed by incubation at room temperature for 30 min. Next, 100 µL of reconstituted Luciferin Detection Reagent was added to each well and mixed briefly on a plate shaker. After 20 min, luminescence was measured using a Gen5 Microplate reader (BIOTEK). A standard curve for GSH concentration was generated, and exact GSH concentrations in different cell lines were calculated based on a GSH standard curve per the manufacturer’s instructions.
Light microscopy and immunofluorescence microscopy.
For light microscopy, cells cultured in 6-well plates were treated with reagents as indicated. Phase contrast images were obtained using an EVOSfl (Advanced Microscopy Group) microscope equipped with a 10× phase contrast objective. For trypan blue staining, cells were trypsinized and collected in a 1.5-mL tube. Cell pellets were washed once in PBS and then stained with trypan blue (Corning) for 5 min. A 20× phase contrast objective was used to acquire photographs of stained dead cells. For immunofluorescence microscopy, cells were seeded on glass coverslips and washed with PBS, then fixed in 3.7% formaldehyde. After fixation, cells were washed with PBS and permeabilized for 10 min in 0.1% TritonX-100/PBS. Blocking buffer (5% BSA in 0.1% Triton/PBS) was added for 1 hr, followed by incubation with BAP1 (1:500; Santa Cruz, sc-28383) and SLC7A11 (1:500; Cell Signaling Technology, 12691) antibodies overnight. Cells were washed with PBS three times and incubated with fluorescent secondary antibodies (ThermoFisher Scientific) for 2 h. The nuclei were labeled with 4’,6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific), and fluorescence was monitored using a confocal microscope (Leica).
Real-time PCR and ChIP-qPCR.
Real-time PCR was conducted as previously described 60, 61. ChIP experiments were performed using a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, 9003) per the manufacturer’s instructions. After chromatin digestion and sonication, antibodies against target proteins as well as a control antibody were used for ChIP. The enriched promoter fragments captured by antibodies were examined using real-time PCR. The signal relative to input was evaluated using a formula from the manufacturer’s protocol as follows: percent input = 2% × 2(C[T]2% input sample − C[T]IP sample), where C[T] = threshold cycle of PCR reaction. The sequences of primers used for real-time PCR and ChIP-qPCR are listed in Table S4. The primary antibodies used for ChIP-qPCR were: H2Aub (Cell Signaling Technology, 8240), RNA pol II CTD (Cell Signaling Technology, 2629), RNA pol II CTD phospho Ser5 antibody (Active Motif, 61085), RNA pol II CTD phospho Ser2 antibody (Active Motif, 61083), FOXK1 (Abcam, ab18196 ), FOXK2 (Bethyl Laboratories, A301–730A), HCFC1 (Bethyl Laboratories, A301–399A), OGT (Cell Signaling Technology, 5368), KDM1B (Abcam, ab193080), ASXL1 (Santa Cruz, sc-293204).
Luciferase reporter assay.
The SLC7A11 promoter was amplified using genomic DNA extracted from UMRC6 cells and cloned into pGL3 luciferase reporter vectors. Primers used to clone around 1 kb of the promoter region of SLC7A11 are listed in Table S4. The luciferase reporter assay was conducted using a Dual-Luciferase Reporter Assay System (Promega, E1910) according to the manufacturer’s instructions. Briefly, HEK293T cells were transfected with the appropriate plasmids for 48 hr, washed with PBS, and lysed for 15 min at room temperature. Cell lysates were transferred to a 384-well plate for subsequent luciferase activity measurement. Luminescence was measured using a Gen5 microplate reader (BIOTEK).
Immunoprecipitation and Western blotting.
Immunoprecipitation was conducted as previously described 62, 63. Protein extracts were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad) using standard techniques. The primary antibodies and concentrations used for Western blotting were: BAP1 (1:1000; Santa Cruz, sc-28383), SLC7A11 (1:5000; Cell Signaling Technology, 12691), tubulin (1:5000; Cell Signaling Technology, 2144), H2Aub (1:5000; Millipore, 05–678), H2A (1:5000; Millipore, ABE327), vinculin (1:10000; Sigma, V4505), FOXK1 (1:1000; Abcam, ab18196 ), FOXK2 (1:1000; Bethyl Laboratories, A301–730A), HCFC1 (1:1000; Bethyl Laboratories, A301–399A), OGT (1:1000, Cell Signaling Technology, 5368), KDM1B (1:1000; Abcam, ab193080), ASXL1 (1:1000; Santa Cruz, sc-293204), p53 (1:1000; Santa Cruz, sc-126), PARP (1:1000; Cell Signaling Technology, 9542), β-actin (1:4000; Abcam, ab8226), and V5 (1:2000; Sigma, A8012).
Tandem affinity purification of SFB-tagged protein complexes.
UMRC6 cells with stable expression of S protein, FLAG, and streptavidin-binding peptide tag (SFB)-tagged BAP1 were subjected to tandem affinity purification as previously described 64. Protein bands were excised and subjected to mass spectrometry analysis at the Taplin Mass Spectrometry Facility, Harvard Medical School. Raw mass spectrometry data were processed and further analyzed using the MUSE algorithm as previously described 64 to assign priority scores to the identified potential interacting proteins.
Xenograft model.
Xenograft model experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. The study is compliant with all relevant ethical regulations regarding animal research. Female 4- to 6-week-old athymic nude mice (Foxn1nu/Foxn1nu) were purchased from the Experimental Radiation Oncology Breeding Core Facility at MD Anderson Cancer Center and housed in the Animal Care Facility at the Department of Veterinary Medicine and Surgery at MD Anderson. Cancer cell lines were resuspended on ice with PBS and the same amount of cells were injected into mice subcutaneously. Tumor progression was monitored by bi-dimensional tumor measurements once a week until the endpoint. The tumor volume was calculated according to the equation v = length*width2*1/2. For drug treatment experiments, 10 days after tumor cell injection, nude mice were treated with PBS or 10 mg/kg liproxstatin-1 (Sigma, SML1414) through intraperitoneal injection every two days until the endpoint as indicated.
Histology and immunohistochemistry.
Xenograft tissue samples were collected and immediately fixed in 10% neutral-buffered formalin (ThermoFisher Scientific) overnight. After being washed once with PBS, samples were transferred into 70% ethanol and submitted to the Research Histology Core Laboratory at MD Anderson Cancer Center for embedding and hematoxylin and eosin staining. For IHC staining, tissue sections were processed according to methods described in our previous publications65, 66. The antibody used for IHC was anti-4HNE (1:200, Abcam, ab46545). Images were obtained at 400× magnification using a microscope (Olympus, BX43).
Transmission electron microscopy.
Xenograft tissue samples were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.3, then washed in 0.1 M sodium cacodylate buffer, treated with 0.1% Millipore-filtered cacodylate-buffered tannic acid, postfixed with 1% buffered osmium, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60℃ oven for approximately 3 days. Ultrathin sections were cut using a Leica Ultracut microtome, stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined with a JEM 1010 transmission electron microscope (JEOL USA, Inc.) at an accelerating voltage of 80 kV. Digital images were obtained using an AMT Imaging System (Advanced Microscopy Techniques Corp) at the High Resolution Electron Microscopy Facility at MD Anderson Cancer Center.
ChIP-seq.
Chromatin immunoprecipitation was performed using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, 9003) following the instructions of the manufacturer. 20 μg of chromatin was incubated overnight with 5 μL H2Aub antibody (Cell Signaling Technology, 8240). Next, 100 ng fly chromatin and 0.5 μg H2Av antibody (ActiveMotif, 61686) were added to each tube as spike-in controls. Purified ChIP-DNA was verified on an agarose gel to ensure proper fragmentation and then sent to the Sequencing and Microarray Facility at MD Anderson Cancer Center for ChIP-seq.
ChIP-seq analysis.
The raw reads (single-end, 50 bp) were aligned to human reference genome hg19 using bowtie (v1.1.0), allowing up to one mismatch. Peak calling was performed using MACS (v1.4.2) 67 with a cutoff of P ≤ 1 × 10-8. Clonal reads were automatically removed by MACS. Promoter regions were defined as regions 5 kb upstream to 5 kb downstream of the TSS. The H2Aub occupancies at promoter regions were normalized as reads per kilobase per million reads (RPKM). The fold changes and significance of H2Aub differences between conditions were determined using an MA-plot-based method with a random sampling model, which was implemented in the R package DEGseq 68; the P value was adjusted using the Benjamini-Hochberg method. For the average profile and heatmap around the TSS, the data matrix was generated using deepTools 69, and the visualization was performed using R. The enriched binding peaks of FOXK1, AXSL1, and O-GlcNAC in HEK293T cells were collected from the GEO database with accession numbers GSM1250375, GSM1250374, and GSM1250376, respectively. The binding targets of these factors were defined as the genes whose promoter regions (3 kb upstream of TSS to 3 kb downstream of TSS) were occupied by the center of a binding peak.
Gene expression profiling and survival analysis.
RNA sequencing was performed in our previous study 34. The differentially expressed genes upon restoring BAP1 in UMRC6 cells were defined by the cutoff of FDR < 0.05 with at least a 1.5-fold change. The expression data of BAP1 and SLC7A11, patient survival data, and BAP1 mutation status in 5 cancer types (UVM, KIRC, KIRP, PCPG, and BRCA) were obtained from TCGA, while the expression data for normal samples were from the GTEx database. All these data were generated using the UCSC Xena Browser (http://xena.ucsc.edu/). The survival analysis was conducted on the basis of BAP1 mutation or singular or combined expression of BAP1 and SLC7A11. For the survival analysis with combined expression of BAP1 and SLC7A11, survival was compared between two groups of patients separated by the expression of both BAP1 and SLC7A11 using unsupervised clustering; Group 1 had higher BAP1 and lower SLC7A11 expression, while Group 2 had lower BAP1 and higher SLC7A11 expression. The survival impact of different signatures was analyzed in R using the Kaplan-Meier method with a Cox proportional hazards model.
Code availability.
The software and algorithms for data analyses used in this study are all well-established from previous work. All software and custom arguments are included in Methods section. There is no unreported algorithm used in this paper. Results in Fig. 1, 2 and Supplementary Fig. 1 and 6b can be repeated using the source data in supplementary tables. The source code for data processing are available from the corresponding author on reasonable request.
Statistics and reproducibility.
Statistical analysis (two-tailed Student’s t-test) was performed using R software for Fig. 1b, 2c, 2g, 2i, S1b, S1f and S1g, and using GraphPad Prism (GraphPad Software, Inc.) for other bar graphs in this manuscript. For box plots, the upper and lower edges of the box indicate the first and third quartiles (25th and 75th percentiles) of the data, while the middle line indicates the median. The whiskers extend 1.5× interquartile range (between 25–75% ranges of data) from the edges of the boxes, respectively. The fold changes and significance of H2Aub differences between conditions were determined using an MA-plot-based method with a random sampling model, which was implemented in the R package DEGseq, and P value was adjusted using the Benjamini-Hochberg method in Fig. 1e-f. For GO biological process analysis in Fig. 1k, S1h, and Table S1, P value was determined by Fisher Exact test, and then adjusted using the Benjamini-Hochberg method. Statistical analysis (two-tailed Student’s t-test) was performed using R software for differential Expression analysis in Table S2. For survival analysis of TCGA dataset in Fig. 2d, 2j, 2k-n, S6b and Table S2, log-rank Mantel-Cox test was used by R software. Pearson’s correlation (two-sided) was performed to analyze gene correlation in Fig. 2b, 2e and Table S2. To calculate P value between groups in Fig. 4a, 4e, 4h, 4m-n, 6d, 6e-6g, 6i, 6m, S5g and S6d-e, two-way ANOVA analysis was performed. All the P values calculated above were indicated in corresponding figures. Each experiment was repeated successfully more than two times with similar results as described in figure captions. To restore BAP1, lentivirus containing BAP1 WT and C91A coding sequence prepared in multiple independent experiments were introduced to BAP1-dificient cells separately, and similar results were found across all the cell lines with successful BAP1 protein expression. To knock out or knock down genes of interest, at least two independent sgRNAs or shRNAs were employed to generate cell lines and similar results were found in all cell lines.
Data availability.
ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE101987. Previously published ChIP-seq or RNA-seq data that were re-analyzed here are available under accession code GSE95097, GSM1250375, GSM1250374, and GSM1250376. The UVM, KIRC, KIRP, PCPG and BRCA data were derived from the TCGA Research Network: http://cancergenome.nih.gov/. The data-set derived from this resource that supports the findings of this study is available in UCSC Xena Browser (http://xena.ucsc.edu/). The normal sample data were derived from the GTEx database: https://gtexportal.org/home/. The data-set derived from this resource that supports the findings of this study is available in UCSC Xena Browser (http://xena.ucsc.edu/). Source data for Fig. 1, 2 and Supplementary Fig. 1 have been provided as Supplementary Table 5 “Statistics Source Data”. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
We thank Xiaobing Shi for helpful discussion and suggestions, and Amy Ninetto from the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for manuscript editing. This research was supported by the Andrew Sabin Family Fellow Award, the Sister Institution Network Fund, and the Institutional Research Grant from The University of Texas MD Anderson Cancer Center, Anna Fuller Fund (to B.G.), and grants from the National Institutes of Health (R01CA181196 to B.G.; R01HG007538 and R01CA193466 to W.L.; and R01CA172724 to P.H.). B.G. is an Ellison Medical Foundation New Scholar and an Andrew Sabin Family Fellow. Y.Z. and P.K. are Scholars at the Center for Cancer Epigenetics at The University of Texas MD Anderson Cancer Center. P.K. is also supported by the CPRIT Research Training Grant (RP170067). This research was also supported by the National Institutes of Health Cancer Center Support Grant P30CA016672 to The University of Texas MD Anderson Cancer Center.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Pavlova NN & Thompson CB The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ & Chandel NS Fundamentals of cancer metabolism. Sci Adv 2, e1600200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green DR, Galluzzi L & Kroemer G Cell biology. Metabolic control of cell death. Science 345, 1250256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boroughs LK & DeBerardinis RJ Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 17, 351–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones RG & Thompson CB Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev 23, 537–548 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Dixon SJ et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Y et al. Ferroptosis: process and function. Cell Death Differ 23, 369–379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao JY & Dixon SJ Mechanisms of ferroptosis. Cell Mol Life Sci 73, 2195–2209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang WS & Stockwell BR Ferroptosis: Death by Lipid Peroxidation. Trends in cell biology 26, 165–176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stockwell BR et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 171, 273–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim JC & Donaldson PJ Focus on molecules: the cystine/glutamate exchanger (System x(c)(−)). Experimental eye research 92, 162–163 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Conrad M & Sato H The oxidative stress-inducible cystine/glutamate antiporter, system x (c) (−) : cystine supplier and beyond. Amino Acids 42, 231–246 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Koppula P, Zhang Y, Zhuang L & Gan B Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun (Lond) 38, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang WS et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedmann Angeli JP et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igney FH & Krammer PH Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer 2, 277–288 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Green DR & Evan GI A matter of life and death. Cancer Cell 1, 19–30 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Carbone M et al. BAP1 and cancer. Nat Rev Cancer 13, 153–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey A et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Z et al. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res 42, 6232–6242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baymaz HI et al. MBD5 and MBD6 interact with the human PR-DUB complex through their methyl-CpG-binding domain. Proteomics 14, 2179–2189 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Yu H et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol 30, 5071–5085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misaghi S et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol 29, 2181–2192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA & Dutta A The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem 284, 34179–34188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheuermann JC et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kallin EM et al. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS genetics 5, e1000506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weake VM & Workman JL Histone ubiquitination: triggering gene activity. Mol Cell 29, 653–663 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Wang H et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Harbour JW et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pena-Llopis S et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 44, 751–759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao Y et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 45, 1470–1473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bott M et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 43, 668–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai F et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc Natl Acad Sci U S A 114, 3192–3197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson AG et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 32, 204–220 e215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fishbein L et al. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell 31, 181–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research, N. et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 374, 135–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang SJ, Ou Y, Jiang L & Gu W Ferroptosis: A missing puzzle piece in the p53 blueprint? Molecular & cellular oncology 3, e1046581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP & Citterio E The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell division 3, 8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Carey M & Workman JL The role of chromatin during transcription. Cell 128, 707–719 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Hauri S et al. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep 17, 583–595 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Gao M, Monian P, Quadri N, Ramasamy R & Jiang X Glutaminolysis and Transferrin Regulate Ferroptosis. Mol Cell 59, 298–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young O, Crotty T, O’Connell R, O’Sullivan J & Curran AJ Levels of oxidative damage and lipid peroxidation in thyroid neoplasia. Head Neck 32, 750–756 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol 14, 276–286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bononi A et al. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature 546, 549–553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okino Y, Machida Y, Frankland-Searby S & Machida YJ BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J Biol Chem 290, 1580–1591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuettengruber B & Cavalli G The DUBle life of polycomb complexes. Dev Cell 18, 878–880 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Henry KW et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17, 2648–2663 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 18, 431–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koppula P, Zhang Y, Shi J, Li W & Gan B The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J Biol Chem 292, 14240–14249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X & Gan B lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle 15, 3471–3481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin A et al. The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene 33, 3183–3194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee H et al. BAF180 regulates cellular senescence and hematopoietic stem cell homeostasis through p21. Oncotarget 7, 19134–19146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin A et al. FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacological inhibition of the PI3K-AKT pathway. Cancer Res 74, 1682–1693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gan B et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468, 701–704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gan B et al. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci U S A 105, 19384–19389 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gan B, Yoo Y & Guan JL Association of focal adhesion kinase with tuberous sclerosis complex 2 in the regulation of s6 kinase activation and cell growth. J Biol Chem 281, 37321–37329 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Gan B, Melkoumian ZK, Wu X, Guan KL & Guan JL Identification of FIP200 interaction with the TSC1-TSC2 complex and its role in regulation of cell size control. J Cell Biol 170, 379–389 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X et al. Proteomic Analysis of the Human Tankyrase Protein Interaction Network Reveals Its Role in Pexophagy. Cell Rep 20, 737–749 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Gan B et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18, 472–484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gan B et al. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol 175, 121–133 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Feng Z, Wang X, Wang X & Zhang X DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Ramirez F, Dundar F, Diehl S, Gruning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE101987. Previously published ChIP-seq or RNA-seq data that were re-analyzed here are available under accession code GSE95097, GSM1250375, GSM1250374, and GSM1250376. The UVM, KIRC, KIRP, PCPG and BRCA data were derived from the TCGA Research Network: http://cancergenome.nih.gov/. The data-set derived from this resource that supports the findings of this study is available in UCSC Xena Browser (http://xena.ucsc.edu/). The normal sample data were derived from the GTEx database: https://gtexportal.org/home/. The data-set derived from this resource that supports the findings of this study is available in UCSC Xena Browser (http://xena.ucsc.edu/). Source data for Fig. 1, 2 and Supplementary Fig. 1 have been provided as Supplementary Table 5 “Statistics Source Data”. All other data supporting the findings of this study are available from the corresponding author on reasonable request.