Abstract
Long non-coding RNAs (lncRNAs) are increasingly implicated in oncogenesis. Here, it is determined that LINC00152/CYTOR is upregulated in glioblastoma multiforme (GBM) and aggressive wild-type IDH1/2 grade II/III gliomas and upregulation associates with poor patient outcomes. LINC00152 is similarly upregulated in over 10 other cancer types and associates with a poor prognosis in 7 other cancer types. Inhibition of the mostly cytoplasmic LINC00152 decreases, and overexpression increases cellular invasion. LINC00152 knockdown alters the transcription of genes important to epithelial-to-mesenchymal transition (EMT). PARIS and Ribo-seq data, together with secondary structure prediction, identified a protein bound 121bp stem-loop structure at the 3′ end of LINC00152 whose overexpression is sufficient to increase invasion of GBM cells. Point mutations in the stem-loop suggest that stem formation in the hairpin is essential for LINC00152 function. LINC00152 has a nearly identical homolog, MIR4435-2HG, which encodes a near identical hairpin, is equally expressed in low-grade glioma (LGG) and GBM, predicts poor patient survival in these tumors and is also reduced by LINC00152 knockdown. Together, these data reveal that LINC00152 and its homolog MIR4435-2HG associate with aggressive tumors and promote cellular invasion through a mechanism that requires the structural integrity of a hairpin structure.
Implications
Frequent upregulation of the lncRNA, LINC00152, in glioblastoma and other tumor types combined with its prognostic potential and ability to promote invasion suggests LINC00152 as a potential biomarker and therapeutic target.
Introduction
GBM (glioblastoma) are highly aggressive grade IV gliomas and are the most common type of malignant glioma, with 10,000 new diagnoses each year [1]. GBMs are a heterogeneous group of tumors that can be separated into four different subtypes, mesenchymal, classical, proneural and neural, based on their transcriptional profile. Most of the focus on understanding glioma tumor biology has been on studying protein coding genes and microRNAs [2]. These efforts have identified commonly altered signaling pathways in GBMs, including mutations in EGFR, p53 and mTOR signaling [3,4]. Furthermore, microRNAs have been shown to play a role in many of the oncogenic phenotypes of GBMs, such as invasiveness and stemness of GBM stem cells [5,6]. Although there has been much effort on creating new targeted therapies for GBMs focusing on some of the aforementioned pathways, most have not been effective and the standard of care therapy, a combination of surgical resection, radiotherapy and Temozolomide, still leaves patients with a 5-year survival rate of roughly 10% [7].
High throughput sequencing revealed that a majority of the human genome, long thought to be transcriptionally silent, is actually expressed. Indeed, when surveyed across many different cell types it was found that nearly 80% of the human genome is actually transcribed [8]. Many of these newly discovered transcripts are lncRNAs (long noncoding RNAs). LncRNAs are a class of ncRNAs that are longer than 200 bases in length and can be further subdivided into subclasses based on chromosomal position relative to other genes, enhancers or other genomic regulatory elements. LncRNAs have been shown to play many different functional roles in the cell, in part through regulation of transcription, mRNA stability and mRNA translational efficiency [9,10]. Most of the research into the role of ncRNA in GBMs has been on microRNAs, with relatively few studies on lncRNAs. This leaves a crucial gap in our understanding of glioma pathogenesis. Indeed, lncRNAs have been shown to function in critical roles in a variety of tumor types, e.g. HOTAIR in breast cancer, SChLAP1 in aggressive prostate cancer, MALAT1 in lung cancer and DRAIC in prostate cancer [11–13].
LINC00152 is a lncRNA that was first identified as being hypomethylated during hepatocellular carcinoma tumorigenesis [14]. It is also dysregulated in gastric cancer and esophageal squamous cell carcinoma [15,16]. However, there are conflicting reports on exactly how LINC00152 functions to promote the invasive phenotype. One study has argued that LINC00152 directly interacts with EGFR and affects AKT signaling while others have suggested that LINC00152 acts as a competing endogenous RNA (ceRNA) through titrating microRNAs [5,6,17–20]. Recently, we identified LINC00152 through an in-depth genomic analysis of gliomas as being highly expressed in GBMs [21]. In this study we characterize LINC00152’s association with GBM clinical features and with tumor cell invasion and begin to functionally characterize LINC00152 structurally. Furthermore, we find that LINC00152 is overexpressed in 10 other tumor types compared to matched normal tissue and high LINC00152 expression is associated with a poor prognosis in 7 of these tumors.
Materials and Methods
Cell culture, knockdown and overexpression of LINC00152
U87 cells were maintained in MEM supplemented with 1% non-essential amino acids solution (cat # 11140-050, Gibco), 1mM sodium pyruvate (cat # 11360070, Gibco), 0.15% sodium bicarbonate (cat # 25080094, Gibco), 10% FBS and 1% P/S.
For knockdown, U87 cells were transfected during two rounds of transfection. First, cells were reverse transfected with 40 ηM of siLINC00152_II (5′-UGACACACUUGAUCGAAUA-3′), siLINC00152_III (5′-CCGGAAUGCAGCUGAAAGA-3′) or a nonspecific siGL2 control siRNA (5′-CGUACGCGGAAUACUUCGA-3′) and 9 μL of Lipofectamine RNAiMAX transfection reagent (Thermo Fisher). 24 hours later, a second round of transfection was performed using the same quantities of reagents. 24 hours after the final transfection, cells were harvested and used for subsequent analysis.
500ηg of LINC00152 or LINC00152 mutants pCDNA3-flag vectors were transfected into U87 cells using 2μL of Lipofectamine 2000 (Thermo Fisher). Cells were harvested after 48 hours for downstream analysis.
RNA isolation, cDNA synthesis, qPCR and Western blotting
Total RNA and nuclear/cytoplasmic RNAs were extracted using TRIzol total RNA isolation reagent (Thermo Fisher), Protein and RNA Isolation System (ThermoFisher), respectively. RNA samples were treated with RQ1 RNase-Free DNase (Promega) according to according to manufacturer’s instructions. cDNA was produced from 1μg RNA using Superscript III kit (Thermo Fisher) according to manufacturer’s instructions. qPCR and Western blotting were performed according to standard protocols.
LINC00152 subcellular fractionation and in situ hybridization
LINC00152 subcellular fractionation was performed using Protein and RNA Isolation System (ThermoFisher) according to manufacturer instructions.
For in situ hybridization 3×105 U87 and U251 cells were plated on the top of a cover glass in a 6-well plate. In the next day, cells were washed once with PBS and fixed for 10min with 2% paraformaldehyde. Then, cells were washed 3 times with PBS and incubated with 1mL of permeabilization buffer (1× PBS/0.5% Triton X-100) for 10min at 4°C. Cells were again washed 3 times with PBS. Next, cells were blocked with 1mL of prehybridization buffer (3% BSA in 4× SSC) for 20min at 55°C. 10ηg of LINC00152 or negative control probe were added to 2mL of hybridization buffer (10% dextran sulfate in 4× SSC) and cells were incubated overnight at 55°C. On the next day, cells were washed 3 times for 5min using washing buffer I (4× SSC, 0.1% Tween-20), 3 times for 5min using washing buffer II (2× SSC), and 3 times for 5min using washing buffer III (1× SSC). Subsequently, cells were blocked for 15min at room temperature using 2mL of blocking solution (4% BSA/1× PBS). Next, cells were incubated with 300μL of antibody solution [2% of BSA/1× PBS and digoxigenin (1:250)] for 1h. Cells were washed with 0.1% Tween-20/PBS 3 times and with alkaline Tris buffer for 5min at room temperature. Finally, the signal was developed by adding 400μL of BCP/NBT solution until the signal was visible.
MTT and matrigel invasion assays
For measuring cell growth, 1,000 cells were plated in quadruplets in 96 well plates and cell growth was measured using standard MTT reagent (Promega). To measure invasion, 2×105 U87 cells in serum free media were seeded into 24-well Matrigel Invasion Chambers (BD Biosciences) and the bottom was filled with media and 10% FBS as the chemoattractant. Cells were allowed to invade for 8 hours and then fixed and stained with crystal violet/methanol and invaded cells were counted.
Expression of LINC00152 in TCGA datasets and survival analysis
The expression of LINC00152 in GBMs and LGGs compared to normal brain and tumor subtypes was performed as previously described [21]. Expression of LINC00152 in all other TCGA tumors was determined by comparing expression data of only those tumors that had a matched normal tissue sample. Statistical significance was determined using a paired t-test. TCGA patient survival data for GBMs and LGGs were retrieved from cBioPortal (www.cbioportal.org) and survival data for the remaining tumor types were retrieved from OncoLnc (www.oncolnc.org) on 12/2016 [22–24]. The expression threshold used to separate patients are outlined in the main text. Kaplan Meier plots, hazard ratios and p-values, based on these separations were generated using the ‘survminer’ package for R.
RNA-seq analysis
U87 cells were treated with a combination of the two siRNA as mentioned earlier and total cell RNA was isolated using TRIzol and subsequently purified using RNeasy Isolation kit (Qiagen). Sequencing libraries were generated using NEB NEXT Ultra directional RNA Library prep kit and samplers were barcoded with NEBNext Multiplexing oligos per standard manufacturer protocols. Libraries were sequenced with 75 bp paired-end reads NextSeq500 instrument, in the Biomolecular Analysis Facility, University of Virginia School of Medicine. Sequencing reads were aligned to the hg38 reference genome using HISAT [25]. Gene abundances and identification of differentially expressed genes were performed using HTSeq and DESeq2 [26,27]. An adjusted P-value (obtained by DESeq2) cut-off of 0.05 and Log 2-Fold change of 2 was used to define differentially expressed genes. GSEA analysis was performed on preranked gene list based on fold change (siLINC00152/siGL2) against 50 hallmark gene sets [28]. For plotting enrichment score obtained by GSEA analysis (as shown in Fig 4B), we have included only the genes which are either induced or repressed 1.5-fold suupon siLINC00152. The raw and processed data were deposited in Gene Expression Omnibus (GEO) under accession number GSE111652.
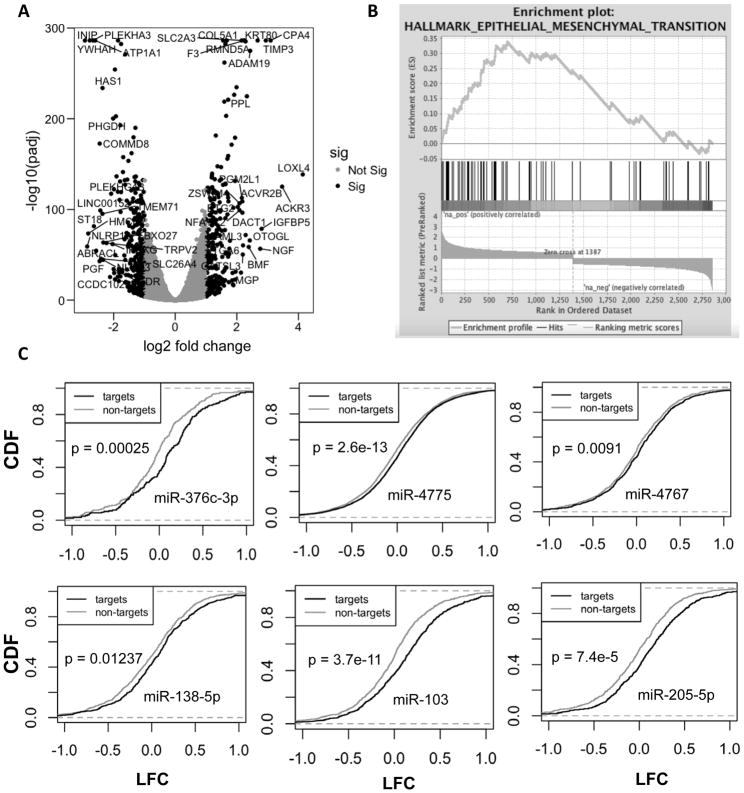
Figure 4. LINC00152 regulates genes involved in invasion in U87 cells.
A) Volcano plot of statistical significance against fold-change highlighting differentially regulated genes in black color upon siLINC00152 in U87 cells. B) Plot from gene set enrichment analysis (GSEA) showing the gene set involved in epithelial-to-mesenchymal transition (EMT) enriched among upregulated genes (left black end of spectrum) after LINC00152 knockdown in U87 cells. C) Cumulative distribution frequency plots of miRNA target mRNAs (as predicted by TargetScan: black line) or non-targets (grey line) showing fraction of genes with fold change less than that indicated on the X-axis after LINC00152 knockdown. None of the miRNAs previously proposed to be sponged by LINC00152 are released as evident from the fact that their targets are not repressed upon LINC00152 knockdown.
LINC00152 structure predictions
Secondary structure predictions of LINC00152 were determined using mfold [29]. The two structures with the lowest predicted free energies were selected for comparisons with PARIS and Ribo-seq. For PARIS data analysis of LINC00152, raw sequencing data from Lu et. al. was aligned to the hg19 genome using STAR (spliced transcripts alignment to a reference) with the alignment parameters outlines in Lu et. al. [30,31]. Aligned reads were then processed to identify gapped mapping to LINC00152 and visualized with IGV [32]. We used ribosome profiling data from Gonzalez et. al. and aligned reads to the hg19 genome using HISAT2 [33]. We then examined reads that mapped to LINC00152 for their distribution along the message to ensure that they were not legitimate ribosome footprints using IGV [32]. The predicted secondary structure elements and protein bound region were then compared to the in silico secondary structure predictions.
Results
LINC00152 is a lncRNA overexpressed in aggressive gliomas
We first identified LINC00152 from a comprehensive analysis of lncRNAs in gliomas [21]. LINC00152 was one of the most differentially expressed lncRNAs in GBMs compared to normal brain tissue, however it is not upregulated in grade II and III gliomas (Fig 1A and Sup Fig 1A). We have validated the upregulation of LINC00152 in an independent set of GBM patients compared to normal FFPE brain tissue [21].
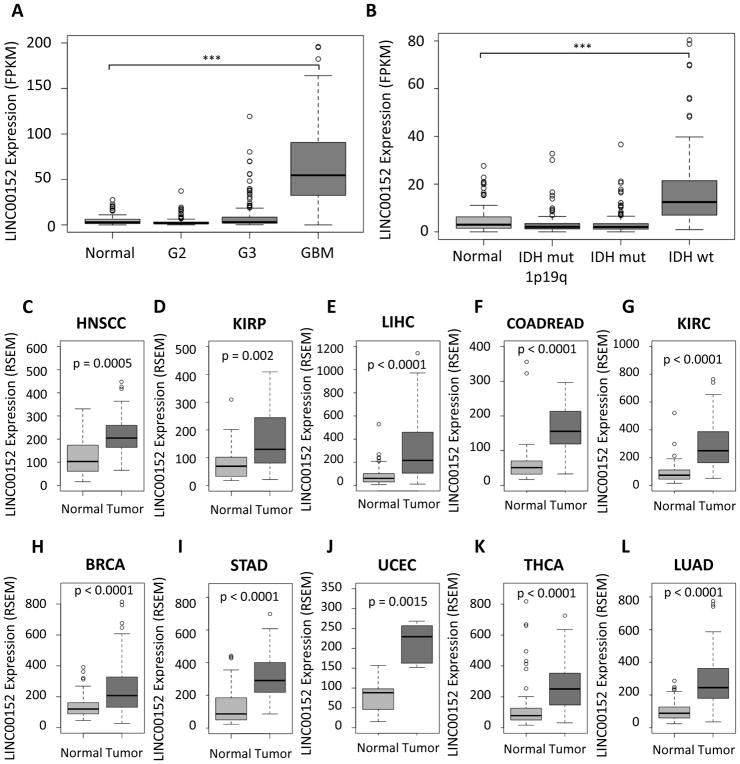
Figure 1. LINC00152 is upregulated in aggressive gliomas and in many cancer types.
A) Boxplot of LINC00152 expression in normal brain tissue, G2 (grade 2 glioma), G3 (grade 3 glioma) and GBM. B) Boxplot of LINC00152 expression in LGG subtypes and normal brain tissue. C – L) Expression (RSEM) of LINC00152 in tumors and matched normal tissue from the TCGA in head and neck squamous carcinoma, renal papillary tumor, hepatocellular carcinoma, colorectal carcinoma, renal clear cell carcinoma, breast invasive carcinoma, stomach adenocarcinoma, uterine carcinoma, thyroid carcinoma and lung adenocarcinoma, respectively.
We tested whether LINC00152 is preferentially expressed in a particular GBM subtype, but that did not appear to be the case. The differences in LINC00152 expression between the subtypes were not statistically significant, although the median expression of LINC00152 is lowest in the proneural GBM subtype (p<0.1) (Sup Fig 1B). Even though LINC00152 is not upregulated in LGGs as a whole, the IDHwt LGG subtype expresses 4 times as much LINC00152 as normal brains (p < 0.00001) (Fig 1B). This is interesting, because IDHwt LGGs are far more aggressive than the other LGG subtypes and display clinical properties similar to GBMs [34].
LINC00152 expression predicts survival in GBMs and LGGs
Since LINC00152 is upregulated in brain tumors compared to normal brain tissue, we next elucidated the association of LINC00152 association with survival of GBM and LGG patients. To do this, we assessed the survival difference of patients expressing high (top 33% highest expressing LINC00152 cohort) and low level of LINC00152 expression from the TCGA for both GBM and LGG. In GBMs, patients who had high expression of LINC00152 had a poor prognosis (p = 0.02) compared to the patients expressing low level of LINC00152, with a median survival of 11.9 and 15.4 months, respectively (Fig 2A). Furthermore, LINC00152 expression was also able to separate patients into two distinct prognostic groups in LGGs. LGG patients with high expression of LINC00152 had a median survival of 62.1 months, while the low expressing group had a median survival of 98.2 months (p < 0.0001) (Fig 2B). These results demonstrate that not only is LINC00152 overexpressed in gliomas, but that this overexpression is associated with poor patient outcome.
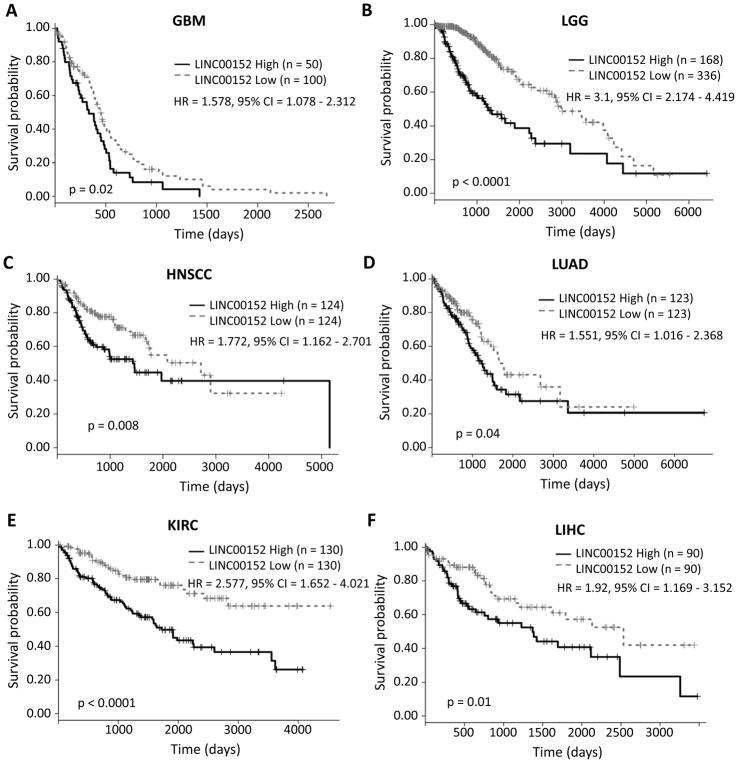
Figure 2. High level of LINC00152 expression is associated with poor patient prognosis in GBMs, LGGs and many other tumors.
A) Kaplan Meier of GBM patients separated into the top 33% highest expressing LINC00152 cohort and lower expressing cohort. B) Kaplan Meier of LGG patients separated into the 33% highest expressing LINC00152 cohort and lower expressing cohort. C–F) Kaplan Meier plots of the highest LINC00152 expressing quartile and lowest LINC00152 expressing quartiles for head and neck squamous carcinoma, lung adenocarcinoma, renal clear cell carcinoma, and hepatocellular carcinoma, respectively. Hazard ratio is indicated as “HR” and the 95% confidence interval is indicated as “CI”.
LINC00152 in other cancers
It was intriguing to examine LINC00152 expression in other cancers compared to their respective normal tissues. We compared the expression of LINC00152 in all TCGA tumor samples with paired normal and tumor RNA-seq data. Surprisingly, LINC00152 is upregulated in nearly every tumor type we analyzed, including head and neck squamous carcinoma, renal papillary tumor, hepatocellular carcinoma, colorectal carcinoma, renal clear cell carcinoma, breast invasive carcinoma, stomach adenocarcinoma, uterine carcinoma, thyroid carcinoma and lung adenocarcinoma (Fig 1C–L).
Since LINC00152 is overexpressed in the majority of tumors that we have analyzed, we next wanted to determine whether LINC00152 expression is associated with patient survival in the TCGA tumors that had higher levels of LINC00152 compared to the paired normal samples. To do this, we performed Kaplan Meier analysis for each tumor type by separating patients into two groups, the top quartile LINC00152 expressing tumors and the lowest quartile LINC00152 expressing tumors. From the original list of tumors, LINC00152 expression was associated with poor patient outcome in head and neck squamous cell carcinoma, lung adenocarcinoma, renal clear cell carcinoma and hepatocellular carcinoma (Fig 2C–F). The poor outcome of patients with renal papillary carcinoma was not statistically significant comparing the top and bottom quartiles of LINC00152 expression (p = 0.1), but the poor outcome was statistically significant (p = 0.015) when we compared patients in the top third and bottom third based on LINC00152 expression (Sup Fig 1C).
Although LINC00152 was not overexpressed in LGGs relative to normal brain, it was upregulated in an aggressive subpopulation of LGGs (those with IDH wild type) and was associated with poor patient outcome. This made us realize that even if a tumor type does not overexpress LINC00152 globally relative to normal tissue, overexpression of the lncRNA in specific tumors may still be associated with poor outcome. We therefore examined other TCGA tumors which did not show a global increase of LINC00152 expression in the cancers relative to normal tissue for the predictive value of the expression of this lncRNA. Interestingly, even among these tumors, LINC00152 expression was associated with poor patient outcome in pancreatic adenocarcinoma when we compare the tumors in the top third and bottom third (Sup Fig 1D), and acute myeloid leukemia, with the top quartile and bottom quartile for LINC00152 expression (Sup Fig 1E). These results highlight the fact that in nine tumor types (GBMs, LGGs, head and neck squamous cell carcinoma, renal clear cell carcinoma, hepatocellular carcinoma, lung adenocarcinoma, renal papillary carcinoma, pancreatic adenocarcinoma and acute myeloid leukemia) LINC00152 appears to function as unfavorable gene whose expression is associated with a poor patient outcome.
LINC00152 expression controls GBM cell invasion
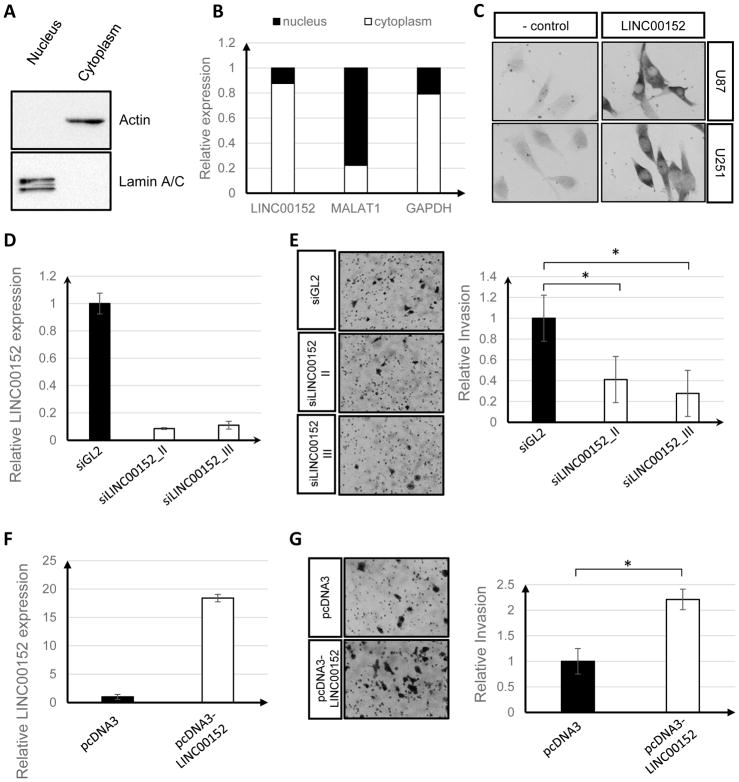
Subcellular fractionation (Fig 3A and B) and in-situ hybridization (Fig 3C) revealed that LINC00152 is primarily localized in the cytoplasm of U87 cells. We next sought to determine whether the upregulation of LINC00152 seen in GBMs is associated with any cancer phenotypes in GBM cell lines. LINC00152 has previously been shown to affect multiple cellular phenotypes, including cell growth, migration, invasion and epithelial-to-mesenchymal transition (EMT) [35,36]. We knocked down LINC00152 expression using two separate siRNAs or overexpressed the lncRNA and found that LINC00152 knockdown or overexpression did not affect cell proliferation for a period of 10 days (Sup Fig 2B and D). We next assayed whether LINC00152 expression was associated with tumor cell invasion using a transwell migration assay. Knockdown of LINC00152 in U87 cell lines led to a statistically significant reduction in cell invasion with both siRNAs targeting LINC00152 (Fig 3D and E). Conversely, overexpression of LINC00152 led to an increase of over 2-fold in the number of invaded cells (Fig 3F and G). These findings suggest that LINC00152 knockdown decreases invasion of GBM cells, while upregulation in GBMs promotes the invasive phenotype that is commonly seen in patient tumors.
Figure 3. LINC00152 is a cytoplasmic lncRNA that promotes cell invasion in U87 cells.
A) Western blot of Lamin A/C and Actin, markers of the nucleus and cytoplasm, respectively. B) qRT-PCR of LINC00152 and a cytoplasmic RNA marker, GAPDH, and a nuclear RNA marker, MALAT1. C) In-situ hybridization of LINC00152 in U87 and U251 cell lines; DRAIC lncRNA probes were used as negative control (“− control”). Purple color: positive signal. D) qRT-PCR showing knockdown of LINC00152 after treatment with two different siRNAs. E) Invasion assay with U87 cells after treatment with two different siRNAs against LINC00152; * p-value < 0.05. Pictures were adjusted by −20% in brightness and +40% in contrast. F) qRT-PCR showing overexpression of LINC00152 after transient overexpression. G) Invasion assay with U87 cells overexpressing LINC00152; * p-value < 0.05. Pictures were adjusted by +40% in contrast.
LINC00152 knockdown decreases expression of pro-invasive genes
In order to better understand how LINC00152 affects cellular invasion we performed RNA-seq on U87 following knockdown of LINC00152 using a combination of two different siRNAs. Knockdown of LINC00152 leads to large changes in gene expression, with 259 genes significantly up-regulated and 295 down-regulated at least 2-fold (Fig 4A). Thus, to determine the most significant molecular pathways regulated by LINC00152, we performed GSEA (gene set enrichment analysis), a method that can identify pathway enrichment from fold change based pre-ranked gene list from RNA-seq [28]. This analysis showed a significant enrichment of up-regulated genes upon siLINC00152 involved in Epithelial to Mesenchymal transition (EMT) (Fig 4B). Among the differentially expressed genes involved in EMT, the changes were validated by qPCR on 12 out of 13 genes after siLIN00152 treatment (Sup Table 1). More interestingly, six of the genes that were downregulated by LINC00152 knockdown were conversely upregulated by overexpression of the lncRNA: TPM2 (Tropomyosin 2), PTX3 (Pentraxin 3), IGFBP4 (Insulin growth factor binding protein 4), TGM2 (Transglutaminase 2), SPP1 (Secreted phosphoprotein 1) and LUM (Lumican)] (Sup Table 1). Moreover, overexpression of the siRNA-resistant M8 was sufficient to upregulate these genes even after knockdown of endogenous LINC00152 (Sup Fig 3). These results indicate that LINC00152 may induce U87 cells invasion by regulating the expression of at least these six genes.
LINC00152 is not involved in sponging of miRNAs
Several previous studies have suggested that LINC00152 acts as a microRNA sponge by titrating different microRNAs (miR-376c-3p, miR-4775, miR-4767, miR-138-5p, miR-103 and miR-205) in different types of tumors, including GBMs [5,6,17–20]. However, suggestions that an lncRNA acts as a microRNA sponge are sometimes questioned because the abundance of the lncRNA is often far less than that of the targets of the microRNAs and of the microRNAs themselves. If LINC00152 acts as a miRNA sponge in U87 cells we would expect that the targets of these microRNAs would be repressed upon knockdown of the lncRNA and the subsequent release of the microRNAs from interaction with the lncRNA. However, we find that there is a statistically significant up-regulation of the targets of these six microRNAs compared with non-targets when LINC00152 is knocked down ruling out the possibility of LINC00152 acting as a ceRNA for these miRNAs (Fig 4C).
Secondary structure components of LINC00152
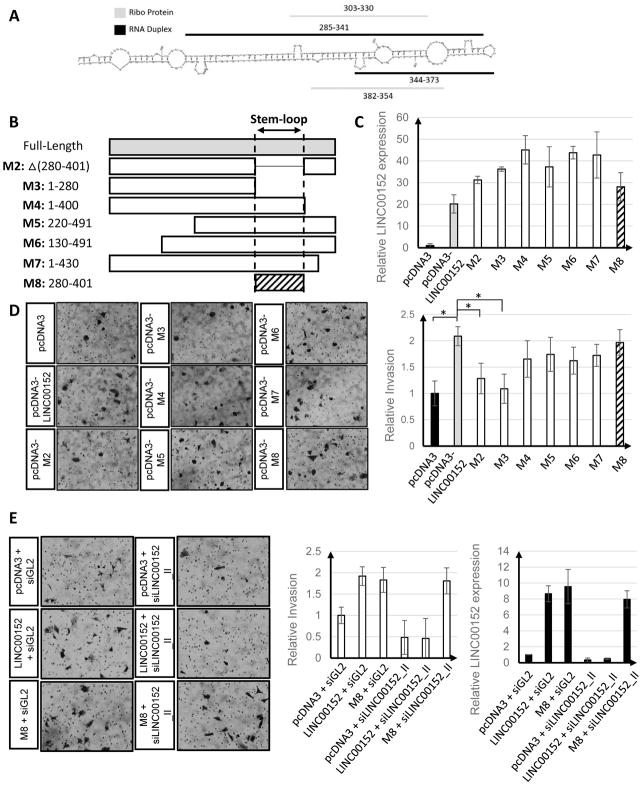
Over the past decade several new technologies have been developed to examine the secondary structures of lncRNAs on a global basis, one such technique is PARIS (psoralen analysis of RNA interactions and structures) [30]. PARIS is based on reversibly crosslinking RNA duplexes (stems of stem-loops) and gentle digestion with a single-strand RNase, S1 nuclease, to cut looped single stranded portions of an RNA’s secondary structure. The surviving RNA duplexes from the stems are then ligated to each other and subjected to high throughput sequencing. RNAs containing stem-loops will have sequencing reads corresponding to the stems with gaps (corresponding to the loops) that do not overlap with a splice site. We analyzed publicly available PARIS data from HeLa cells to determine whether LINC00152 contains any secondary structure elements that could be detected by PARIS. Following alignment, we identified reads with a 2-nt gap that were present in the PARIS libraries (Sup Fig 4C). These reads are positioned from position 285 to 373 of the 496 nt long LINC00152, with a small 2 base gap starting at position 342 (Fig 5A). Sequence analysis of this region revealed some complementarity, suggesting that this region might in fact form a stem-loop structure (Fig 5A).
Figure 5. A 120 nucleotide hairpin at the 3′ end of LINC00152 (M8) is sufficient for promoting cell invasion in U87 cells.
A) Predicted secondary structure of LINC00152 and the stem loop and protein bound regions identified by PARIS (RNA Duplex) and Ribo-seq (Sup Fig 4). B) Schematics of LINC00152 deletion mutants. C) LINC00152 qRT-PCR confirming overexpression levels of the different constructs. D) Invasion of U87 cells after overexpressing the different LINC00152 deletion mutants; * p-value < 0.05. Pictures were adjusted by −10% in brightness and +40% in contrast. E) Invasion of U87 cells decreases after treatment with si00152_II but is rescued by LINC00152 m8 overexpression; * p-value < 0.05. Pictures were adjusted by +20% in brightness and +40% in contrast.
To get a better understanding of overall LINC00152 secondary structure, we used publicly available RNA secondary structure prediction tool, mfold, to identify secondary structure predictions for LINC00152 that are consistent with a stem-loop being present from 285–373 [29]. The top 2 secondary structures with the lowest free energy differed in their exact base-pairing, but the overall stem-loop structure was largely the same. Importantly, both structures were consistent with a stem-loop being present from position 285 to 373 (Fig 5A and Sup Fig 4A and B). Furthermore, the resulting loop from the stem formation is rather small, 4 nt, which is consistent with the small 2 nt gap seen by PARIS.
We next asked if we could use a separate method to independently validate the hairpin formation in LINC00152. Ribo-seq (Ribosome profiling) is a technique that has been used to identify RNAs that interact with the ribosome and how the ribosome is distributed across those RNAs [37]. This information has also been used to ascertain that some lncRNAs are associated with ribosomes, but not translating ribosomes [38]. Recently it was determined that the polysomes isolated for Ribo-seq are contaminated with other ribonucleoprotein (RBP) complexes. As a result RNA footprints from RBPs that are not ribosome proteins can be detected in Ribo-seq data [33]. To determine if we could identify RBP-RNA footprints from LINC00152 we analyzed publicly available Ribo-seq data from normal brain samples [39]. In two out of the three normal brain Ribo-seq samples we detected a RBP footprint at positions 303–330 of LINC00152. In addition, in one of the samples there was an RBP footprint from 354–382 (Fig 5A and Sup Fig 4D). These two footprinted areas are located on opposite strands of the same stem-loop that was detected by PARIS, providing additional evidence of the existence of this stem-loop and suggesting that this stem is bound by a protein in an RBP (Fig 5A).
LINC00152 stem-loop, M8, is sufficient to promote cell invasion
In order to determine whether this newly identified, potentially protein bound, stem-loop plays a role in LINC00152 function, we created a series of LINC00152 deletion mutants (Fig 5B and Sup Fig 5A). The sites of the deletions were chosen based on PARIS and Ribo-seq analysis as well as two in silico predicted structures of LINC00152 (Fig 5A and Sup Fig 4C and D). We assessed whether independent overexpression of the mutants was able to stimulate U87 cell invasion. Overexpression of M2 (which removed the minimal amount of the protein bound stem-loop, nucleotides 280–401) or M3 (which removed the stem-loop and the remaining 3′ end) led to a decreased cell invasion significantly compared to full-length LINC00152 (p < 0.05) (Fig 5D). On the other hand, the mutant M4 (which removed the 3′ end but preserved the stem-loop) or M7 (which removed the extreme 3′ end, and also preserved the stem-loop) increased U87 cell invasion. Other deletion mutants that removed regions of LINC00152 5′ to the stem loop (M5 or M6) stimulated cellular invasion to a similar extent as full-length LINC00152. Finally, overexpression of M8, containing only the protein bound stem-loop (nucleotides 280–401) was sufficient to stimulate invasion of U87 cells (Fig 5D). These results suggest that M8 stem-loop is necessary and sufficient for stimulation of cell invasion.
Consistent with this conclusion, overexpression of the stem-loop also induced the six genes involved in EMT to the same extent as the full length LINC00152 (Sup Table 1). In addition, knockdown of LINC00152 by siLINC00152_II (a siRNA that targets a region on LINC00152 outside of M8) decreased cell invasion while the siRNA-resistant M8 was sufficient to rescue cell invasion (Fig 5E).
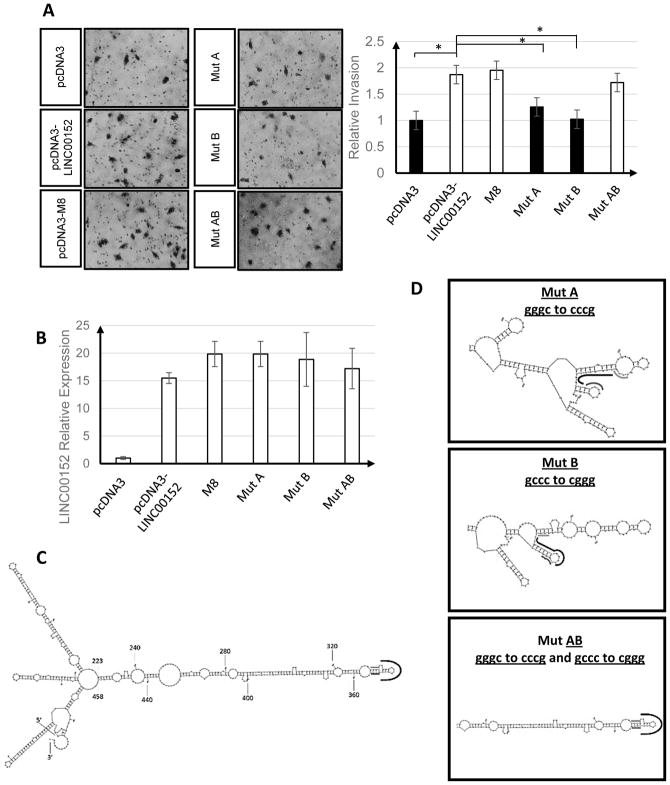
The M8 stem-loop structure is important for stimulating invasion
We next tested with point mutations whether the ability to stimulate invasion of U87 cells depends on the LINC00152 stem-loop structure. Two mutants on opposite side of the stem disrupt the stem-loop (mutA: changes bases 333–336 and mutB: changes bases 349–352) (Fig 6D). Neither mutA nor mutB stimulated the invasion of U87 cells as well as full length LINC00152 or M8 (Fig 6A). In contrast, when the two mutations were combined in mutAB, the stem-loop structure was reconstructed and this promoted invasion to the same extent as full length LINC00152 or M8 (Fig 6A). Therefore, we can conclude that the stem-loop structure itself is essential for LINC00152 to stimulate cellular invasion.
Figure 6. Point-mutation of nucleotides 333–336 or 349–352 of LINC00152 shows the importance of M8 hairpin for stimulating invasion.
A) Invasion of U87 cells after the different LINC00152 deletion mutants are overexpressed. Mut A or mut B are incapable of inducing invasion in U87 cells. Combining the two mutants (mut AB) restores the hairpin and induces invasion to the same level as full length LINC00152; * p-value < 0.05. Pictures were adjusted by +20% in brightness and +40% in contrast. B) qRT-PCR confirming overexpression of the different LINC00152 constructs. C) Predicted secondary structure of full length LINC00152 with the black line marking the sequence at the tip of the hairpin and the light grey and dark grey lines marking the residues that are mutated in Mut A or B, respectively. D) Predicted secondary structures of LINC00152 mutants A, B and AB. The black, light grey and dark grey lines mark the corresponding residues as in Fig. 6C.
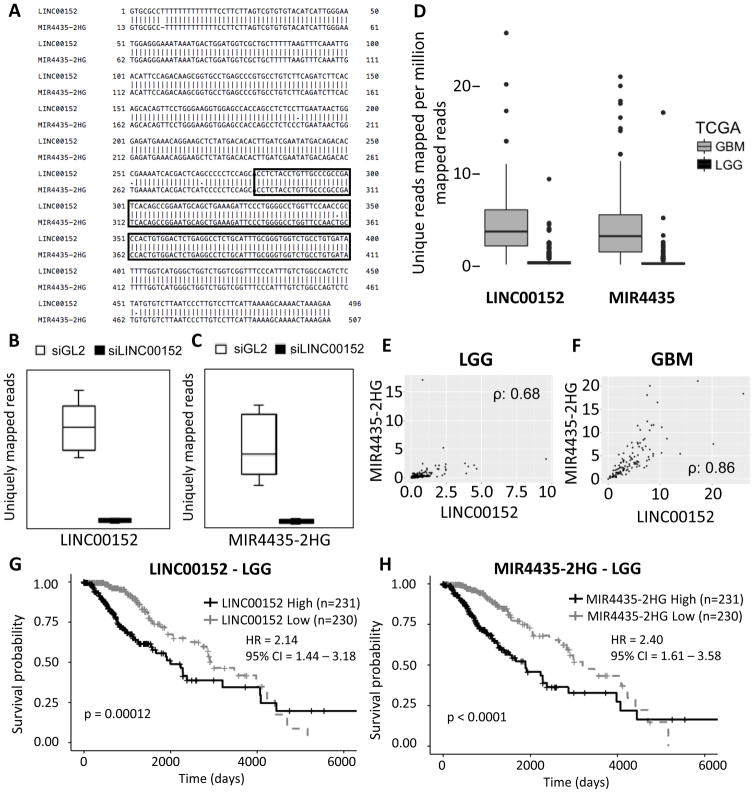
MIR4435-2HG, a homolog of LINC00152
As previously reported [40], LINC00152 is a close homolog of another lncRNA on chromosome 2, MIR4435-2HG, both have nearly identical sequences (with only 6 base mismatches) and both contain M8 sequence (Fig 7A). LINC00152 and MIR4435-2HG are both transcribed from chromosome 2, LINC00152 is located at chr2: 87455476-87606739 and MIR4435-2HG is transcribed from chr2:111196350-111495115. In order to estimate the expression level of these two RNAs, we considered RNA-seq reads that uniquely mapped without any mismatch to either LINC00152 or MIR4435-2HG. This analysis showed that LINC00152 and MIR4435-2HG are expressed at the same level in U87 cells, and both of the RNAs are knocked down upon treatment of siLINC00152 to a similar extent (Fig 7B and C). Thus, the phenotype that we observe with siRNA directed towards LINC00152 is also likely through knocking down the highly similar MIR4435-2HG. Moreover, given the high similarity between the two transcripts, and the fact that, from nucleotides 382 to 478, MIR4435-2HG forms a 97 nucleotides long stem-loop in the same position as LINC00152, it is likely that MIR4435-2HG overexpression phenocopies the effects of LINC00152 on cell invasion. However, we see an increase in cell invasion when we exogenously express LINC00152, saying that LINC00152 by itself can promote cell invasion. Again, upon considering uniquely mapped reads in TCGA RNA-seq data, we found that both LINC00152 and MIR4435-2HG are equally expressed in LGG and GBM (Fig 7D). Analysis of TCGA RNA-seq data also revealed a positive correlation between the expression of the two RNAs in GBM and LGG (Fig 7E and G), suggesting that these two RNAs may be co-regulated. Moreover, the Kaplan Meier plot to estimate survival showed that expression of either RNA is associated with poor patient survival (Fig 7G and H).
Figure 7. LINC00152 is highly similar to the lncRNA MIR4435-2HG.
A) Sequence alignment of LINC00152 and MIR4435-2HG. M8 is highlighted in the boxed area. B) LINC00152 specific RNA-seq reads in cells treated with siGL2 or siLINC00152. C) MIR4435-2HG specific RNA-seq reads in cells treated with siGL2 or siLINC00152. D) LINC00152 or MIR4435-2HG specific reads in TCGA RNA-seq data for LGG and GBM. E) Correlation of expression of LINC00152 and MIR4435-2HG in LGGs (spearman correlation 0.68 p value < 2.2 e-16). F) Correlation of expression of LINC00152 and MIR4435-2HG in GBMs (spearman correlation: 0.86, P value < 2.2e-16). G) Kaplan Meier Plot of LGG patients separated into the 50% highest expressing LINC00152 cohort and the lowest 50% expressing cohort. H) Kaplan Meier Plot of LGG patients separated into the 50% highest expressing MIR4435-2HG cohort and the lowest 50% expressing cohort. Hazard ratio is indicated as “HR” and the 95% confidence interval is indicated as “CI”.
Discussion
The human genome was once thought to be mainly dormant and that most of the transcription was devoted in producing protein coding genes. We now know that the genome is transcriptionally vibrant and only a small fraction of the expressed genome, roughly 2%, encodes for protein coding genes. GWAS and high throughput sequencing studies have found that many of the genomic lesions and expression alterations seen in cancer and other pathologies fall within non-protein coding regions of the genome and may lead to dysregulation of ncRNAs [41–43]. Furthermore, there is a growing body of evidence implicating lncRNAs in playing a direct role in normal cellular physiology, as well as driving pathogenesis in a variety of disorders, including cancer [43–46]. Indeed, recent work has illustrated the critical role that lncRNAs play in cancer, including iconic examples such as HOTAIR in breast cancer and HULC in hepatocellular carcinoma and DRAIC in prostate cancer [11,14,47].
In this study we have shown that LINC00152 is a lncRNA that is upregulated in many different cancer types and is highly upregulated in GBMs. Although LINC00152 is not upregulated in all LGGs relative to normal brain tissue, it is upregulated in the highly malignant IDHwt LGG subtype, further supporting LINC00152’s association with aggressive tumors. This raised the interesting possibility that in tumors where LINC00152 is not differentially over-expressed or is moderately upregulated in the tumor population relative to normal tissue, LINC00152 could still be highly upregulated in a more aggressive subgroup of the tumors. This was indeed found to be true in Pancreatic Adenocarcinomas and Acute Myeloid Leukemias. LINC00152 expression is associated with patient survival in nine different cancer types, including GBMs and LGGs. LINC00152 expression promotes cell invasion, which is consistent with its association with poor patient outcomes.
To assess the coding probability of LINC00152 we used the Coding Potential Assessment Tool (CPAT) and found a score of 0.0289, suggesting that this lncRNA has no coding potential, since a CPAT score of < 0.5242 is considered non-coding [48]. In addition, ExPasy [49] predicts the first LINC00152 ORF of 42 amino acids (126 nucleotides) from nucleotides 64 to 189. This ORF is not similar to any ORF predicted for the mouse transcript Gm14005. Moreover, blastx of the translated 126 nucleotide ORF did not show any hits in mouse protein database. In a different frame there is a longer ORF of 92 amino acids (which also does not have a homolog in the mouse transcript) that is preceded by a short ORF that has three stop codons. So, the longer ORF is also unlikely to be translated.
Previous studies have shown that LINC00152 is an oncogenic lncRNA involved in regulating invasion in different types of tumors [5,6,18–20], including gliomas [5,6]. Mingjun Yu and collaborators [5] have reported an in vivo tumor xenograft study, downregulation of LINC00152 produced smaller tumors and increased survival rates when compared to control. Thus, our findings reinforce the idea that LINC00152 is an oncogenic lncRNA that is associated with aggressive tumors by promoting cell invasion. Moreover, through analysis of global RNA structure mapping and RNA-protein interaction data, we identified a protein bound stem-loop in the 3′ region of LINC00152. The structure-function analysis demonstrated that this stem-loop is necessary and sufficient for stimulating invasion of U87 cells, that it can rescue the loss of invasion seen after knockdown of LINC00152 and that the base-pairing of the opposite strands of the stem-loop, rather than the sequence at the mutated sites, is more important for stimulating invasion of U87 cells. However, it is likely that there are specific sequences along the M8 stem-loop that are important for LINC00152 function that will be examined in a future study.
GSEA of RNA-seq from LINC00152 knocked down cells also supports the idea that LINC00152 is involved in promoting invasion. More specifically, TPM2, PTX3, IGFBP4, TGM2, SPP1 and LUM were downregulated by siLINC00152 and upregulated after LINC00152 overexpression. Since we did not compare the global gene-expression changes with LINC00152 overexpression and knockdown by RNA-seq, there are likely to be many other genes that will be regulated similarly to the six genes we tested in this study. Because siLINC00152 decreased invasion, we focused on genes in the RNA-seq data whose change (up or down) will decrease invasion. Of these 13 transcripts qRT-PCR after siLINC00152 validated the changes in 12. Out of these 12, 6 genes were changed in the opposite direction when LINC00152 full length or M8 was overexpressed (Sup Table 1).
In addition, despite previous suggestions, analysis of the RNA-seq showed us that LINC00152 is not acting as a ceRNA that sponges miRNAs.
LINC00152 is expressed from a syntenic location from mouse transcript. Mouse has a single gene, MIR4435-2HG (Gm14005 or MORRBID), but humans have two closely related genes, LINC00152 and MIR4435-2HG.
MIR4435-2HG is a host gene for a miRNA, as miR-4435 is transcribed from an intron of MIR4435-2HG. However, none of the effects observed by overexpression of LINC00152 are due to miR-4435, since the functional assays were done using the cDNA of LINC00152. Furthermore, miR-4435 is not detected in any of the short RNA-seq libraries (such as miRGator v3.0 and miRmine) from brain, glial cell lines and gliomas.
Mouse MIR4435-2HG has been proposed to be a pro-survival lncRNA that represses a gene in cis, the proapoptic gene BCL2L11 (BIM) by recruiting the polycomb repressive complex, PRC2, to the BCL2L11 promoter [50]. We considered the possibility that LINC00152, although cytoplasmic, is acting as a pro-oncogenic RNA by similarly suppressing BCL2L11. siLINC00152 increases BCL2L11 RNA (Sup Fig 6B–D), but this is not expected to decrease cell invasion. Second, overexpression of LINC00152 from a heterologous site or the M8 hairpin of the lncRNA did not decrease BCL2L11 (Sup Fig 6E–G) and yet increased cell invasion. Third, analyzing previously published mouse PAR-CLIP data, we determined that EZH2 from the polycomb complex associates with an intronic region of MORRBID (mouse MIR4435-2HG/MORRBID) which is not near the M8 region. Fourth mouse MIR4435-2HG encodes only the first third of the M8 hairpin that we have found to be functions in human LINC00152 or human MIR4435-2HG. Finally, LINC00152 is predominantly cytoplasmic, arguing against any role in recruiting any factors to the genome. Collectively, these results suggest that interaction with PRC2 is not necessary for the stimulation of invasion seen upon overexpression of the LINC00152 or the M8 hairpin RNA.
In conclusion, LINC00152/CYTOR and its homolog MIR4435-2HG functions as an oncogenic lncRNA in GBMs through the action of a protein-bound stem-loop and potentially plays a critical oncogenic role in a wide variety of cancer types. The results rule out a mechanism of action involving the sponging of miRNAs as proposed in the literature, or interaction with the Polycomb complex proposed for the mouse MIR4435-2HG/MORRBID RNA. LINC00152 could also serve as a tumor biomarker or a target for future cancer therapeutics.
Supplementary Material
Acknowledgments
We thank Dutta lab members for advice and helpful discussions. This work was supported by grants from the NIH R01 CA166054, AR067712 and a V foundation award D2018-002 to AD. BTRK was partly supported by CAPES BEX 0320-13-7. MK was partly supported by a DOD award PC151085.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro-Oncology. 2015;17(suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–31. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 3.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The Somatic Genomic Landscape of Glioblastoma. Cell. 2013;155(2):462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu M, Xue Y, Zheng J, Liu X, Yu H, Liu L, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16(1):110. doi: 10.1186/s12943-017-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Zhu Z, Dai J, Liao Y, Ma J, Zhou W. Knockdown of Long Noncoding RNA LINC0000125 Suppresses Cellular Proliferation and Invasion in Glioma Cells by Regulating MiR-4775. Oncol Res. 2017 doi: 10.3727/096504017X15016337254597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 8.Consortium TEP. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–8. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, et al. The non-coding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013 Feb 1;73(3):1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakurai K, Reon BJ, Anaya J, Dutta A. The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol Cancer Res. 2015;13(5):828–38. doi: 10.1158/1541-7786.MCR-15-0016-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann O, Kesselmeier M, Geffers R, Pellegrino R, Radlwimmer B, Hoffmann K, et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology. 2012;56(5):1817–27. doi: 10.1002/hep.25870. [DOI] [PubMed] [Google Scholar]
- 15.Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19(23):3658–64. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S, Ning Q, Zhang G, Sun H, Wang Z, Li Y. Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget. 2016;7(52):85728–85740. doi: 10.18632/oncotarget.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8(12):5286–5297. [PMC free article] [PubMed] [Google Scholar]
- 18.Teng W, Qiu C, He Z, Wang G, Xue Y, Hui X. Linc00152 suppresses apoptosis and promotes migration by sponging miR-4767 in vascular endothelial cells. Oncotarget. 2017;8(49):85014–85023. doi: 10.18632/oncotarget.18777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li C, et al. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2017;7(1) doi: 10.1098/rsob.160247. pii: 160247. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Wang Y, Liu J, Bai H, Dang Y, Lv P, Wu S. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am J Cancer Res. 2017;7(2):312–322. [PMC free article] [PubMed] [Google Scholar]
- 21.Reon BJ, Anaya J, Zhang Y, Mandell J, Purow B, Abounader R, et al. Expression of lncRNAs in Low-Grade Gliomas and Glioblastoma Multiforme: An In Silico Analysis. PLOS Med. 2016;13(12):e1002192. doi: 10.1371/journal.pmed.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6(269):pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2(e67) [Google Scholar]
- 25.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Meth. 2015;12(4):357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016;165(5):1267–79. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotech. 2011;29(1):24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji Z, Song R, Huang H, Regev A, Struhl K. Transcriptome-scale RNase-footprinting of RNA-protein complexes. Nat Biotech. 2016;34(4):410–3. doi: 10.1038/nbt.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Network CGAR. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372(26):2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6(40):42813–24. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14(19):3112–23. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science (80- ) 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell. 2013;154(1):240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez C, Sims JS, Hornstein N, Mela A, Garcia F, Lei L, et al. Ribosome Profiling Reveals a Cell-Type-Specific Translational Landscape in Brain Tumors. J Neurosci. 2014;34(33):10924–36. doi: 10.1523/JNEUROSCI.0084-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nötzold L, Frank L, Gandhi M, Polycarpou-Schwarz M, Groß M, Gunkel M, Beil N, Erfle H, Harder N, Rohr K, Trendel J, Krijgsveld J, Longerich T, Schirmacher P, Boutros M, Erhardt S, Diederichs S. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci Rep. 2017;7(1):2265. doi: 10.1038/s41598-017-02357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirza AH, Kaur S, Brorsson CA, Pociot F. Effects of GWAS-Associated Genetic Variants on lncRNAs within IBD and T1D Candidate Loci. PLoS One. 2014;9(8):e105723. doi: 10.1371/journal.pone.0105723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 43.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grote P, Wittler L, Währisch S, Hendrix D, Beisaw A, Macura K, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22(6):1006–14. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Grembergen O, Bizet M, de Bony EJ, Calonne E, Putmans P, Brohée S, et al. Portraying breast cancers with long noncoding RNAs. Sci Adv. 2016;2(9):e1600220. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panzitt K, Tschernatsch MMO, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a Novel Gene With Striking Up-Regulation in Hepatocellular Carcinoma, as Noncoding RNA. Gastroenterology. 2007;132(1):330–42. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013 Apr 1;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003 Jul 1;31(13):3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537(7619):239–243. doi: 10.1038/nature19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.