Abstract
It has been nearly a decade since the first landmark studies implicating familial recessive Parkinson’s disease genes in the regulation of selective mitochondrial autophagy. The PTEN-induced kinase 1 (PINK1) and the E3 ubiquitin ligase Parkin (encoded by the PARK2 gene) act together to mark depolarized mitochondria for degradation. There is now an extensive body of literature detailing key mediators and steps in this pathway, based mostly on work in transformed cell lines. However, the degree to which PINK1-triggered mitophagy contributes to mitochondrial quality control in the mammalian brain, and the extent to which its disruption contributes to Parkinson’s disease pathogenesis remain uncertain. In recent years, it has become clear that there are multiple, potentially redundant, pathways of cargo specification for mitophagy. Important mitophagy-independent functions of PINK1 and Parkin are also emerging. This review summarizes key features of three major mitophagy cargo recognition systems: receptor-mediated, ubiquitin-mediated and cardiolipin-mediated. New animal models that may be useful for tracking the delivery of mitochondria into lysosomes in different neuronal populations will be highlighted. Combining these research tools with methods to selectively disrupt specific mitophagy pathways may lead to a better understanding of the potential role of mitophagy in modulating neuronal vulnerability in Parkinson’s spectrum (PD/PDD/DLB) and other neurodegenerative diseases.
Keywords: mitochondria, autophagy, neurodegeneration, mitophagy
A Brief History of Mitophagy in Parkinson’s Disease
Neurons maintain extended axonal and dendritic arbors, poised in readiness to rapidly communicate electrochemical signals across vast cellular distances. These bioenergetically demanding activities necessitate a high degree of metabolic dependence on mitochondria [33]. As mitochondria are damaged with usage, aging or disease, neurons rely upon multiple cellular quality control mechanisms to identify and either repair or replace damaged mitochondrial segments [16, 75]. Mitophagy, short for mitochondrial macroautophagy, represents the process by which mitochondrial are selectively sequestered and delivered for lysosomal degradation [43]. Mitochondrial turnover by this mechanism is believed to operate at basal levels to maintain neuronal health. In addition, mitophagy can be up-regulated in response to mitochondrial injury, cellular differentiation or stress-induced metabolic adaptation.
One of the first clues highlighting an important role for mitophagy in Parkinson’s and related diseases was an ultrastructural study delineating mitochondria marked by activated kinases within autophagosomes in neurons of patients with Parkinson’s Disease (PD) and Lewy Body Dementia (LBD) [85]. Subsequently, altered mitophagy was observed in numerous experimental models of toxic-environmental and genetic forms of PD [11, 12, 14, 16, 19, 58, 86]. In 2008, a landmark study from the Youle lab showed that Parkin, an E3 ubiquitin ligase encoded by the PARK2 gene, plays an integral role in marking severely depolarized mitochondria for lysosomal clearance [55]. This was followed by simultaneous work by several groups showing that the mitochondrial kinase encoded by a second recessive PD gene, PINK1, accumulated on depolarized mitochondria to recruit Parkin [35, 48, 56, 79]. In the intervening decade, extensive progress has been made concerning PINK1 stabilization on the surface of depolarized mitochondria, its role in phosphorylating both ubiquitin and Parkin, mitophagy adapters that link ubiquitinated mitochondria to the autophagic machinery, and other mitochondrial fusion or transport proteins whose proteasomal degradation facilitates mitochondrial sequestration [Reviewed in [25, 51, 82]]. PINK1 and Parkin regulated mitophagy is triggered by CCCP or FCCP in a wide variety of cell types, and represents the most studied pathway of cargo specification.
Unresolved Questions
Despite all this progress, the relative importance of the PINK1-Parkin mitophagy pathway in neurons, and its contribution to the pathogenesis of recessive PD and other neurodegenerative diseases remain unclear [24, 59, 62]. The majority of studies have been performed in proliferative, metabolically flexible cells using overexpressed Parkin and severe stimuli including chemical depolarization, chemical ablation of electron transport or laser-triggered ROS generation. Studies focusing on primary neurons or neurons differentiated from induced pluripotent stem cells have yielded conflicting interpretations concerning activation of the PINK1-Parkin pathway, ranging from no increase over baseline in cortical, striatal or midbrain neurons to parkin recruitment to 16% of Killer red-depolarized mitochondria in hippocampal neurons [4, 7, 62, 76]. The responses in neurons, if present are slower and/or much less robust compared to the responses observed in tumor cells.
Alternative functions for PINK1 and Parkin
While it is clear that deficiency in PINK1 or PARK2, or expression of recessive mutations linked to PD, have profound effects on mitochondrial structure and function, deciphering which of these function(s) play a key role in disease pathogenesis is not straightforward. One complicating factor in analyzing the effects of PINK1 or PARK2 deficiency, relates to growing evidence that these genes are involved in multiple aspects of mitochondrial quality control [3, 13, 64, 71]. Primary mouse Pink1−/− neurons exhibit mitochondrial fragmentation, reduced membrane potential, and lysosomal expansion [17]. Stem cell derived PINK1 RNAi neurons show increased cell death accompanied by similar membrane potential and lysosomal changes [81], and iPSC-derived neurons from PARK2- and PINK1-mutated patients clearly exhibit mitochondrial dysfunction [15]. Whether or not these mitochondrial deficits are caused by primarily by deficits in mitophagy or through dysregulation of upstream mechanisms is less clear.
An in vivo protein turnover study in Drosophila supports an important role of PINK1 and Parkin in regulating mitochondrial protein turnover in vivo [78]. However, there is a disconnect between the effects of PINK1 deficiency and autophagy (Atg7) deficiency, suggesting that PINK1 regulates respiratory complex protein stability through additional mechanisms independent of the autophagy machinery. Whether or not this could occur through mitochondrial derived vesicles [50], the effects of PINK1 on cristae structure [16] remain to be addressed experimentally.
PINK1 may also function to maintain high quality mitochondria through its ability to modulate the phosphorylation of complex I subunits, mitochondrial fission and transport proteins, mitochondrial chaperones and mitochondrial calcium transporters [39, 53, 60, 63, 69, 84]. Notably, PINK1 and Parkin may also function in the cytosol to regulate PGC-1α and mitochondrial biogenesis [42, 68, 70], suppressing autophagy [16, 20] and activating mTORC2 [54]. Additional neuron-specialized roles for PINK1 include a role in promoting dendritogenesis or maintenance of dendritic arbors [71], based on studies showing dendritic simplification in Pink1−/− neurons that can be rescued by processed, cytosolic PINK1 [18].
PINK1- and Parkin-independent mitophagy
Finally, a growing number of PINK1- and/or Parkin-independent pathways of selective mitophagy have been reported [1, 14, 30, 44, 72]. These are not mutually exclusive pathways, and there is evidence that more than one mechanism may be activated by a given stimulus. Indeed, cross-regulation, potential redundancy, and multiplication of mitophagy mechanisms relative to unicellular organisms attest to the importance of mitochondrial autophagy in mammalian cells. PINK1-independent mitophagy pathways may account for the upregulation of neuroprotective mitophagy observed in PINK1-deficient neuronal models [16, 38, 61]. While many studies have shown that PINK1 and Parkin are essential for CCCP/FCCP-triggered mitophagy, they may not be essential for mitophagy triggered by rotenone, hypoxia, ROS or iron deficiency [1, 14, 26, 44]. Given this functional redundancy, the impact of loss of function mutations on mitophagic turnover in specific neuronal populations cannot be assumed, but should be experimentally investigated keeping in mind alternative cargo-targeting mechanisms for mitophagy.
Three Mechanisms of Cargo Recognition for Selective Mitophagy
Among the stimuli that upregulate mitophagy are developmental cues, mitochondrial toxins, hypoxic-ischemic/oxidative injuries and expression of mutated proteins. Given that a given mitophagy stimuli may trigger more than one pathway, I have chosen to classify these pathways in terms of the molecules that are recognized by the autophagy machinery (Figure 1). These mitochondrial “eat-me” signals may be transcriptionally regulated, and/or post-translationally modified or exposed to stimulate mitophagy. All are recognized directly or indirectly by mammalian Atg8 homologues. While most existing studies have focused on microtubule-associated protein 1 light chain 3 (LC3), which has three human protein isoforms LC3A, LC3B and LC3C, members of the gamma-aminobutyric acid receptor-associated protein (GABARAP) family, which includes GABARAP, GABARAPL1 and GABARAPL2/Golgi-associated ATPase enhancer of 16 kDa (GATE-16), are also implicated in cargo targeting for selective autophagy [57].
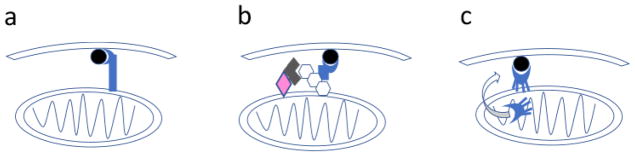
Figure 1. Three cargo targeting mechanisms for mitophagy.
There are several cargo targeting mechanisms for selective mitophagy that have been described in mammalian cells. Each pathway involves a binding interaction with the autophagy protein LC3 (black circle), or its homologs, which brings mitochondria to the phagophore as it extends to form an autophagosome. (a) Transmembrane LIR-motif receptors (purple shape) expressed on the outer mitochondrial membrane are able to directly bind LC3 for receptor-mediated mitophagy. Mitophagy in this pathway is regulated by transcriptional or post-translational mechanisms. (b) In ubiquitin-mediated mitophagy, mitochondrial depolarization results in accumulation of PINK1 (pink diamond) on the mitochondrial surface. PINK1 recruits and activates Parkin (grey notched diamond), resulting in addition of ubiquitin (white hexagons). Bifunctional LIR-domain proteins (purple shape) capable of binding ubiquitin and LC3 function as cargo adapters. (c) The phospholipid cardiolipin (purple shape) is normally sequestered inside mitochondria. Upon mitochondrial injury, cardiolipin is enzymatically externalized, allowing it to bind LC3 during cardiolipin-mediated mitophagy.
Receptor-mediated mitophagy
Selective removal of mitochondria occurs during normal developmental differentiation of several cell types, including erythrocytes. This process is mediated by transmembrane receptors that are expressed on the outer mitochondrial membrane [32] (Figure 1a). Nix, also known as BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (BNIP3L), is transcriptionally upregulated during reticulocyte maturation to erythrocytes. Nix/BNIP3L interacts directly with LC3B or GATE-16 via an LC3-interacting region (LIR), thereby mediating the sequestration of Nix-expressing mitochondria by the growing phagophore (autophagic isolation membrane) [57]. A growing number of other outer mitochondrial membrane proteins containing LIR domains have also been identified, including FKBP8 [5], with several implicated in mitophagy (Reviewed in [25].
Interestingly, hypoxia also induces expression of Nix/BNIP3L as well as a related BH3 protein BNIP3 [45], implicating similar receptor-mediated mitophagy mechanisms in injury-induced mitophagy. In addition, post-translational modifications play a key role allowing for more rapid responsiveness to hypoxic stress, as observed for the mitophagy receptor FUNDC1 [44, 46]. PINK1 and Parkin are not required for receptor-mediated mitophagy, and a recent study suggests that Nix is able to compensate for dysfunction of PINK1 or Parkin in fibroblasts from Parkinson’s disease patients [38].
Ubiquitin-mediated mitophagy
Mitochondrial depolarization, or other injuries sufficient to interrupt mitochondrial protein import, causes PINK1 protein to accumulate on the outer surface of mitochondria [35, 48, 56]. PINK1 phosphorylates and recruits parkin to the mitochondrial surface [66] and phosphorylates ubiquitin. Phospho-ubiquitin activates the E3 ligase activity of parkin [31, 36, 40], resulting in ubiquitination of mitochondrial surface proteins (Figure 1b). This leads to proteasomal degradation of outer membrane proteins and/or to mitochondrial degradation by autophagy [8, 83]. Interestingly, Parkin itself may not be essential for ubiquitin-mediated mitophagy, as other ubiquitin ligases have also been implicated [74].
A group of bifunctional LIR domain-containing cytosolic proteins with ubiquitin binding domains serve to link the ubiquitinated mitochondrion to LC3 to promote selective cargo sequestration. These adapters play a role in selective clearance of ubiquitinated protein aggregates, in xenophagy of certain bacteria, and in mitophagy [28]. Optineurin and NDP52 represent the most heavily studied mitophagy adapters, whereas the involvement of the selective autophagy adapter p62 is controversial in depolarization-mediated mitophagy [23, 41, 49, 80]. Human post-mortem studies indicate increased staining for phospho-ubiquitin in degenerating substantia nigra neurons [21], with a similar staining pattern to structures previously defined ultrastructurally as mitochondria within autophagosomes (mitophagosomes) in PD/LBD patients [85]. Taken together, these studies indicate activation of ubiquitin-mediated mitophagy in PD and related dementias.
Cardiolipin-mediated mitophagy
A third mechanism of selective mitophagy was initially reported in cortical neurons and neuroblastoma cells in response to lower dose treatments with parkinsonian toxins rotenone and 6-hydroxydopamine [14]. This pathway involves the enzymatic translocation of the inner mitochondrial membrane phospholipid cardiolipin to the outer surface of mitochondria, where it can interact directly with LC3 to mediate mitophagy (Figure 1c). The cardiolipin translocation steps require phospholipid scramblase-3 [14] and the dual functional mitochondrial nucleoside diphosphate kinase-D [29]. Like receptor-mediated mitophagy, cardiolipin-mediated mitophagy does not require PINK1 accumulation or Parkin recruitment to the mitochondria [14], but can also be triggered by staurosporine and CCCP in parallel to the PINK1-Parkin pathway. Cardiolipin interacts with multiple Atg8 family members, but only LC3 translocates to mitochondria in rotenone treated glioma cells [2]. In addition to cardiolipin, ceramide has been implicated in targeting autophagosomes to mitochondria to elicit a form of cell death involving mitophagy [65].
The composition of the four fatty acyl chains on cardiolipin may be important in regulating mitophagy, as tafazzin deficiency in MEFs results in defective mitophagosome biogenesis [27]. Tafazzin, which is mutated in Barth syndrome, functions to remodel cardiolipin. Little is currently known about the effects of cardiolipin composition on mitophagy, except that species of cardiolipin with four fatty acyl chains bind LC3 with greater affinity than lyso-cardiolipins [14]. The brain has a highly distinct cardiolipin fatty acyl profile compared to other tissues. Brain cardiolipin is composed of fewer unsaturated fatty acids, rendering it more resistant to peroxidation [6]. Given that cardiolipin lipid peroxidation is involved in apoptotic signaling [67], but not in mitophagy [14], tissue-specific differences in cardiolipin fatty acyl chain composition may regulate thresholds for mitophagy and apopotosis in different cell types.
Mitophagy Marker Mice
While much has been learned about selective cargo targeting mechanisms for mitophagy in cultured cells and neurons, demonstrating activation of mitophagy in vivo has been more elusive. Mitochondrial proteins clearly undergo turnover in vivo. As indicated above, however, this could be due to localized degradation by mitochondrial peptidases, proteasomal degradation, mitochondria derived vesicles or mitophagy. While mitophagosomes [85], or surrogate markers of activation such as phospho-ubiquitin, are observed in human PD midbrain sections [21, 85], this could reflect either increased mitophagic flux or decreased completion of mitophagy. Furthermore, beyond chemical/toxin exposures or hypoxic injuries, the physiological or pathological triggers for mitophagy remain to be clearly defined in vivo.
Within the last few years, a new pair of transgenic mouse models have been developed, which could help resolve some of these questions. Both serve to monitor the delivery of mitochondria into acidic lysosomal compartments, and thus, can be used to estimate mitophagic flux. As discussed below, studies in both model systems indicate that differences in the regulation and requirement of different cell types for mitophagy, previously delineated in culture models, are even further augmented in vivo. Although easily transfectable tumor cells have formed the basis of many advances in mitophagy research, the next frontier in neurodegeneration research will likely only be reached by studies based in neurons, astrocytes, oligodendrocytes, microglia and vascular endothelium.
mt-Keima transgenic mice
Keima is a fluorescent protein that exhibits pH dependent excitation [34]. When fused to the mitochondrial targeting sequence of COX VIII, Keima can be directed to the mitochondrial matrix, which has a pH of approximately 7.8, at which it is excited predominantly by 458-nm light. However, if the mitochondrion is present in a normally acidified lysosome (pH 4.5–5), the excitation of mt-Keima switches to 561-nm. The ratio of 561-nm:458-nm excited fluorescence intensity is often quoted as an index of mitophagic flux. However, it is important to keep in mind that this ratio would be increased whenever mitophagic delivery to lysosomes exceeds their degradation or as a result of impaired synthesis/import of new mt-Keima. Alterations in protein synthesis or lysosomal acidification are likely to occur with aging or disease states, and can be experimentally addressed through other methods.
Studies using a transgenic mouse line in which the mt-Keima reporter was inserted into the Hipp11 locus reveal that different tissues and neuron types differed greatly in levels of basal mt-Keima signal, with high levels in the heart and neural stem cells [73]. The mitophagy signal of mt-Keima in tissue sections was calculated as the number of pixels showing high red fluorescence and low green fluorescence divided by the sum of background-subtracted mt-Keima pixels irregardless of excitation wavelength. Interestingly, mt-Keima signal were highest in select populations of neurons including the small granule neurons of the dentate gyrus and large Purkinje projection neurons, but not the granule neurons in the cerebellum [73]. Similar cell type specific differences in the cerebellar autophagic response have been previously reported [22]. In the mt-Keima mice, a degree of Atg5-independent mitophagy was also observed in both dentate and Purkinje neuron populations. The dentate gyrus showed an aging related decline with marked differences observed in 21-month versus 3-month old mice [73]. Mutant Huntington has been previously reported to result in empty autophagosomes, presumably by interfering with cargo specification [47]. This correlates with diminished mt-Keima delivery to lysosomes in the dentate gyrus of mice expressing mutant Huntingtin. Interestingly, regions of the brain relevant to PD and related dementias, the cortex, striatum, and substantia nigra, showed only modest levels of basal mitophagy [73]. Given that increased mitophagy has been observed in culture models of mutant LRRK2 expression and PINK1 deficiency [9, 10, 77, 87], it would be interesting to further study how the mt-Keima mitophagy signal is altered with aging or PD-linked genetic mutations.
Outside of the brain, mt-Keima signals were augmented in the liver by high fat diet, hypoxia, expression of a proofreading-deficient form of the mitochondrial DNA polymerase POLG-γ, or by generalized cachexia [73]. This latter observation is important as these types of sensors are not designed to differentiate among the different pathways of mitophagy discussed above, or to distinguish selective mitophagy from increased mitochondrial degradation due to nonselective autophagy.
mito-QC transgenic mice
Shortly after description of the mt-Keima mouse, an alternative mouse model was reported that is based on the differential sensitivity of RFP or mCherry versus GFP to acidic environments. Called mito-QC, this mCherry-GFP-FIS1 sensor [1] is similar in principal to the commonly used RFP-GFP-LC3 tandem flux reporter [37]. Mitochondrial targeting is accomplished using the mitochondrial targeting sequence of the outer mitochondrial membrane protein FIS1. Mitochondria surrounded by neutral cytosol would emit both red and green signals, while mitochondria in the acidic lysosomal compartment would emit red only signals.
The mito-QC mouse also highlights striking tissue-specific differences in levels of basal mitophagy [52]. The highest mitophagy index was observed in the kidney and liver, with intermediate levels in the heart and skeletal muscle, and lower levels in the spleen. In the brain, red-only mitochondria, reflective of mitochondria in acidic environments, were observed in cerebellar Purkinje cells, but not cerebellar granule cells. Interestingly, the mito-QC mouse revealed a regional upregulation of mitophagy in the embryonic heart that may be related to metabolic switching to fatty acid metabolism. High resolution images are assisted by the low spectral overlap exhibited by this reporter system, and reveal subcellular concentration of mitophagolysosomes in the central multinucleated region of the tongue muscle [52]. Another advantage of this system is that the tissues can be fixed, although the molecular basis for continued quenching of the intra-lysosomal GFP signal after fixation is unclear.
Future Studies
Proper turnover of mitochondria through coordinated activities of mitophagy and mitochondrial biogenesis undoubtedly plays a key role in neuron health and function. The discovery that multiple genes implicated in familial neurodegenerative diseases play key roles in regulating ubiquitin-mediated mitophagy led to rapid research advances in understanding the molecular interactions responsible for mitochondrial cargo selection. At the same time, the existence of multiple, independent and potentially redundant pathways that serve to identify mitochondrial cargo for autophagic degradation raises additional questions. Are other mitophagy pathways able to compensate for deficits in one particular pathway? Are certain cell types or injury stimuli more dependent on one particular pathway? Does either insufficient or excessive mitophagy represent a causative, driving force behind neurodegeneration? The possibility that PINK1 and Parkin have alternative roles that are important for neurons should also be kept in mind.
Determining whether altered mitophagy plays a causative role in the pathogenesis of PD, DLB and other neurodegenerative diseases will require a more complete understanding of the level of mitophagy experienced by different populations of neurons and their support cells in the brain. Although chemicals and toxins used in culture models are likely to also trigger mitophagy in vivo, and some of these exposures are linked to human parkinsonism, for the most part in vivo triggers for brain mitophagy remain to be delineated. Both the mt-Keima and mito-QC mouse models offer new opportunities to study these processes, whether during normal development, as adaptive responses to stress, or under disease states. Delineating whether or not mitophagic flux is increased or decreased represents a first step towards understanding the potential role of mitophagy in neurodegenerative disease models. These studies can then guide development of hypothesis-driven work to delineate the relative importance of specific mitophagy pathways, and whether their stimulation or inhibition leads to neuroprotection.
Highlights.
PD-linked PINK1 and PARK2 encode proteins that regulate ubiquitin-mediated mitophagy
PINK1 and Parkin are not necessary for receptor- or cardiolipin-mediated mitophagy
Whether mitophagy is increased, decreased or unchanged in PD neurons is unclear
Mitophagy marker mice may be useful to study how mitophagy changes in neurons in vivo
Acknowledgments
Research in the Chu Laboratory is supported in part by the National Institutes of Health (AG026389, NS065789, NS101628) and the Helen Mendel Fund. Dr. Chu holds the A. Julio Martinez Chair in Neuropathology at the University of Pittsburgh.
Footnotes
The author declares no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anton Z, Landajuela A, Hervas JH, Montes LR, Hernandez-Tiedra S, Velasco G, Goni FM, Alonso A. Human Atg8-cardiolipin interactions in mitophagy: Specific properties of LC3B, GABARAPL2 and GABARAP. Autophagy. 2016;12:2386–2403. doi: 10.1080/15548627.2016.1240856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arena G, Valente EM. PINK1 in the limelight: multiple functions of an eclectic protein in human health and disease. J Pathol. 2017;241:251–263. doi: 10.1002/path.4815. [DOI] [PubMed] [Google Scholar]
- 4.Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014 doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhujabal Z, Birgisdottir AB, Sjottem E, Brenne HB, Overvatn A, Habisov S, Kirkin V, Lamark T, Johansen T. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017;18:947–961. doi: 10.15252/embr.201643147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley RM, Stark KD, Duncan RE. Influence of tissue, diet, and enzymatic remodeling on cardiolipin fatty acyl profile. Mol Nutr Food Res. 2016;60:1804–1818. doi: 10.1002/mnfr.201500966. [DOI] [PubMed] [Google Scholar]
- 7.Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherra SJ, III, Steer E, Gusdon AM, Kiselyov K, Chu CT. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. American Journal of Pathology. 2013;182:474–484. doi: 10.1016/j.ajpath.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherra SJ, 3rd, Dagda RK, Tandon A, Chu CT. Mitochondrial autophagy as a compensatory response to PINK1 deficiency. Autophagy. 2009;5:1213–1214. doi: 10.4161/auto.5.8.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherra SJ, 3rd, Steer E, Gusdon AM, Kiselyov K, Chu CT. Mutant LRRK2 Elicits Calcium Imbalance and Depletion of Dendritic Mitochondria in Neurons. Am J Pathol. 2013;182:474–484. doi: 10.1016/j.ajpath.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. Mitochondrial alpha-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. 2010;486:235–239. doi: 10.1016/j.neulet.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Human Molecular Genetics. 2010;19:R28–R37. doi: 10.1093/hmg/ddq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Qiang Wang KZ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung SY, Kishinevsky S, Mazzulli JR, Graziotto J, Mrejeru A, Mosharov EV, Puspita L, Valiulahi P, Sulzer D, Milner TA, Taldone T, Krainc D, Studer L, Shim JW. Parkin and PINK1 Patient iPSC-Derived Midbrain Dopamine Neurons Exhibit Mitochondrial Dysfunction and alpha-Synuclein Accumulation. Stem Cell Reports. 2016;7:664–677. doi: 10.1016/j.stemcr.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagda RK, Gusdon AM, Pien I, Strack S, Green S, Li C, Van Houten B, Cherra SJ, 3rd, Chu CT. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson’s disease. Cell Death Differ. 2011;18:1914–1923. doi: 10.1038/cdd.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagda RK, Pien I, Wang R, Zhu J, Wang KZ, Callio J, Banerjee TD, Dagda RY, Chu CT. Beyond the mitochondrion: cytosolic PINK1 remodels dendrites through protein kinase A. J Neurochem. 2014;128:864–877. doi: 10.1111/jnc.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dagda RK, Zhu JH, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedorowicz MA, de Vries-Schneider RL, Rub C, Becker D, Huang Y, Zhou C, Alessi Wolken DM, Voos W, Liu Y, Przedborski S. Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy. EMBO Rep. 2014;15:86–93. doi: 10.1002/embr.201337294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiesel FC, Ando M, Hudec R, Hill AR, Castanedes-Casey M, Caulfield TR, Moussaud-Lamodiere EL, Stankowski JN, Bauer PO, Lorenzo-Betancor O, Ferrer I, Arbelo JM, Siuda J, Chen L, Dawson VL, Dawson TM, Wszolek ZK, Ross OA, Dickson DW, Springer W. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep. 2015;16:1114–1130. doi: 10.15252/embr.201540514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florez-McClure ML, Linseman DA, Chu CT, Barker PA, Bouchard RJ, Le SS, Laessig TA, Heidenreich KA. The p75 neurotrophin receptor can induce autophagy and death of cerebellar Purkinje neurons. Journal of Neuroscience. 2004;24:4498–4509. doi: 10.1523/JNEUROSCI.5744-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 24.Grenier K, McLelland GL, Fon EA. Parkin- and PINK1-Dependent Mitophagy in Neurons: Will the Real Pathway Please Stand Up? Front Neurol. 2013;4:100. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamacher-Brady A, Brady NR. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci. 2016;73:775–795. doi: 10.1007/s00018-015-2087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han JY, Kang MJ, Kim KH, Han PL, Kim HS, Ha JY, Son JH. Nitric oxide induction of Parkin translocation in PTEN-induced putative kinase 1 (PINK1) deficiency: functional role of neuronal nitric oxide synthase during mitophagy. J Biol Chem. 2015;290:10325–10335. doi: 10.1074/jbc.M114.624767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu P, Liu X, Zhang J, Wang HG, Ye JM, Shi Y. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy. 2015;11:643–652. doi: 10.1080/15548627.2015.1023984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagan VE, Jiang J, Huang Z, Tyurina YY, Desbourdes C, Cottet-Rousselle C, Dar HH, Verma M, Tyurin VA, Kapralov AA, Cheikhi A, Mao G, Stolz D, St Croix CM, Watkins S, Shen Z, Li Y, Greenberg ML, Tokarska-Schlattner M, Boissan M, Lacombe ML, Epand RM, Chu CT, Mallampalli RK, Bayir H, Schlattner U. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016 doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, Gabrielson K, Kass DA, Iijima M, Sesaki H. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanki T, Klionsky DJ. The molecular mechanism of mitochondria autophagy in yeast. Mol Microbiol. 2010;75:795–800. doi: 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kann O, Kovacs R. Mitochondria and neuronal activity. Am J Physiol Cell Physiol. 2007;292:C641–657. doi: 10.1152/ajpcell.00222.2006. [DOI] [PubMed] [Google Scholar]
- 34.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010 doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 36.Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 38.Koentjoro B, Park JS, Sue CM. Nix restores mitophagy and mitochondrial function to protect against PINK1/Parkin-related Parkinson’s disease. Sci Rep. 2017;7:44373. doi: 10.1038/srep44373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostic M, Ludtmann MH, Bading H, Hershfinkel M, Steer E, Chu CT, Abramov AY, Sekler I. PKA Phosphorylation of NCLX Reverses Mitochondrial Calcium Overload and Depolarization Promoting Survival of PINK1-Deficient Dopaminergic Neurons. Cell Rep. 2015;13:376–386. doi: 10.1016/j.celrep.2015.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 41.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee Y, Stevens DA, Kang SU, Jiang H, Lee YI, Ko HS, Scarffe LA, Umanah GE, Kang H, Ham S, Kam TI, Allen K, Brahmachari S, Kim JW, Neifert S, Yun SP, Fiesel FC, Springer W, Dawson VL, Shin JH, Dawson TM. PINK1 Primes Parkin-Mediated Ubiquitination of PARIS in Dopaminergic Neuronal Survival. Cell Rep. 2017;18:918–932. doi: 10.1016/j.celrep.2016.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemasters JJ. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3) Redox Biol. 2014;2:749–754. doi: 10.1016/j.redox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lv M, Wang C, Li F, Peng J, Wen B, Gong Q, Shi Y, Tang Y. Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy. Protein Cell. 2017;8:25–38. doi: 10.1007/s13238-016-0328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 50.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McWilliams TG, Muqit MM. PINK1 and Parkin: emerging themes in mitochondrial homeostasis. Curr Opin Cell Biol. 2017;45:83–91. doi: 10.1016/j.ceb.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 52.McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, Muqit MM, Ganley IG. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–345. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morais VA, Haddad D, Craessaerts K, De Bock PJ, Swerts J, Vilain S, Aerts L, Overbergh L, Grunewald A, Seibler P, Klein C, Gevaert K, Verstreken P, De Strooper B. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344:203–207. doi: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- 54.Murata H, Sakaguchi M, Jin Y, Sakaguchi Y, Futami J, Yamada H, Kataoka K, Huh NH. A new cytosolic pathway from a Parkinson disease-associated kinase BRPK/PINK1: activation of AKT via mTORC2. J Biol Chem. 2011;286:7182–7189. doi: 10.1074/jbc.M110.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osellame LD, Duchen MR. Defective quality control mechanisms and accumulation of damaged mitochondria link Gaucher and Parkinson diseases. Autophagy. 2013;9:1633–1635. doi: 10.4161/auto.25878. [DOI] [PubMed] [Google Scholar]
- 59.Pogson JH, Ivatt RM, Sanchez-Martinez A, Tufi R, Wilson E, Mortiboys H, Whitworth AJ. The complex I subunit NDUFA10 selectively rescues Drosophila pink1 mutants through a mechanism independent of mitophagy. PLoS Genet. 2014;10:e1004815. doi: 10.1371/journal.pgen.1004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi Z, Yang W, Liu Y, Cui T, Gao H, Duan C, Lu L, Zhao C, Zhao H, Yang H. Loss of PINK1 function decreases PP2A activity and promotes autophagy in dopaminergic cells and a murine model. Neurochem Int. 2011;59:572–581. doi: 10.1016/j.neuint.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 62.Rakovic A, Shurkewitsch K, Seibler P, Grunewald A, Zanon A, Hagenah J, Krainc D, Klein C. Phosphatase and Tensin Homolog (PTEN)-induced Putative Kinase 1 (PINK1)-dependent Ubiquitination of Endogenous Parkin Attenuates Mitophagy: STUDY IN HUMAN PRIMARY FIBROBLASTS AND INDUCED PLURIPOTENT STEM CELL-DERIVED NEURONS. J Biol Chem. 2013;288:2223–2237. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandebring A, Thomas KJ, Beilina A, van der Brug M, Cleland MM, Ahmad R, Miller DW, Zambrano I, Cowburn RF, Behbahani H, Cedazo-Minguez A, Cookson MR. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS One. 2009;4:e5701. doi: 10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shidoji Y, Hayashi K, Komura S, Ohishi N, Yagi K. Loss of molecular interaction between cytochrome c and cardiolipin due to lipid peroxidation. Biochem Biophys Res Commun. 1999;264:343–347. doi: 10.1006/bbrc.1999.1410. [DOI] [PubMed] [Google Scholar]
- 68.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shlevkov E, Kramer T, Schapansky J, LaVoie MJ, Schwarz TL. Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility. Proc Natl Acad Sci U S A. 2016;113:E6097–E6106. doi: 10.1073/pnas.1612283113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siddiqui A, Bhaumik D, Chinta SJ, Rane A, Rajagopalan S, Lieu CA, Lithgow GJ, Andersen JK. Mitochondrial Quality Control via the PGC1alpha-TFEB Signaling Pathway Is Compromised by Parkin Q311X Mutation But Independently Restored by Rapamycin. J Neurosci. 2015;35:12833–12844. doi: 10.1523/JNEUROSCI.0109-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steer EK, Dail MK, Chu CT. Beyond mitophagy: cytosolic PINK1 as a messenger of mitochondrial health. Antioxid Redox Signal. 2015;22:1047–1059. doi: 10.1089/ars.2014.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Nardacci R, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun N, Yun J, Liu J, Malide D, Liu C, Rovira, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, Finkel T. Measuring In Vivo Mitophagy. Mol Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szargel R, Shani V, Abd Elghani F, Mekies LN, Liani E, Rott R, Engelender S. The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway. Hum Mol Genet. 2016;25:3476–3490. doi: 10.1093/hmg/ddw189. [DOI] [PubMed] [Google Scholar]
- 75.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. Embo J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Verma M, Callio J, Otero PA, Sekler I, Wills ZP, Chu CT. Mitochondrial calcium dysregulation contributes to dendrite degeneration mediated by PD/LBD-associated LRRK2 mutants. J Neurosci. 2017 doi: 10.1523/JNEUROSCI.3791-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wood-Kaczmar A, Gandhi S, Yao Z, Abramov AS, Miljan EA, Keen G, Stanyer L, Hargreaves I, Klupsch K, Deas E, Downward J, Mansfield L, Jat P, Taylor J, Heales S, Duchen MR, Latchman D, Tabrizi SJ, Wood NW. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS ONE. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yin XM, Ding WX. The reciprocal roles of PARK2 and mitofusins in mitophagy and mitochondrial spheroid formation. Autophagy. 2013;9:1687–1692. doi: 10.4161/auto.24871. [DOI] [PubMed] [Google Scholar]
- 83.Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L, Karsten P, Hamm S, Pogson JH, Muller-Rischart AK, Exner N, Haass C, Whitworth AJ, Winklhofer KF, Schulz JB, Voigt A. TRAP1 rescues PINK1 loss-of-function phenotypes. Hum Mol Genet. 2013;22:2829–2841. doi: 10.1093/hmg/ddt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu JH, Guo FL, Shelburne J, Watkins S, Chu CT. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathology. 2003;13:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu JH, Horbinski C, Guo FL, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. American Journal of Pathology. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu Y, Wang C, Yu M, Cui J, Liu L, Xu Z. ULK1 and JNK are involved in mitophagy incurred by LRRK2 G2019S expression. Protein Cell. 2013;4:711–721. doi: 10.1007/s13238-013-3910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]