Abstract
Age-dependent neurodegenerative diseases are associated with a decline in protein quality control systems including autophagy. Amyotrophic lateral sclerosis (ALS) is a motor neuron degenerative disease of complex etiology with increasing connections to other neurodegenerative conditions such as frontotemporal dementia. Among the diverse genetic causes for ALS, a striking feature is the common connection to autophagy and its associated pathways. There is a recurring theme of protein misfolding as in other neurodegenerative diseases, but importantly there is a distinct common thread among ALS genes that connects them to the cascade of autophagy. However, the roles of autophagy in ALS remain enigmatic and it is still unclear whether activation or inhibition of autophagy would be a reliable avenue to ameliorate the disease. The main evidence that links autophagy to different genetic forms of ALS is discussed.
Keywords: ALS, alsin, autophagy, C9orf72, CHMP2B, dynactin, FIG4, FTD, FUS, optineurin, p62, profilin, SOD1, TBK1, TDP-43, VAPB, VCP, UBQLN2
Introduction
ALS is an age-dependent neurodegenerative disease characterized by progressive degeneration of motor neurons (MNs), with ~10% of all cases being familial. To date, a large number of genes have been linked to ALS, including Cu/Zn superoxide dismutase (SOD1) [1], TAR DNA-binding protein (TDP-43) [2,3], fused in sarcoma (FUS) [1,4], sequestosome1 (SQSTM1/p62) [5,6], optineurin (OPTN) [7], TANK binding kinase 1 (TBK1) [8], VAMP-associated protein B (VAPB) [9,10], valosin-containing protein (VCP) [11], ubiquilin 2 (UBQLN2) [12], alsin (ALS2) [13,14], charged multivesicular body protein 2B (CHMP2B) [15–17], dynactin (DCTN) [18], profilin 1 (PFN1) [19], factor-induced gene 4 (FIG4) [20], and a hexanucleotide repeat expansion in the gene C9orf72 [21,22]. In recent years, increasing evidence has suggested that ALS and frontotemporal dementia (FTD) are two related diseases in a continuous clinical spectrum [23]. FTD is the second most common cause of dementia after Alzheimer’s disease in patients younger than 65 [24]. Biochemical and genetic studies have identified the involvement of TDP-43, FUS, and C9orf72 in both ALS and FTD, as part of evidence linking these two neurodegenerative disorders.
The toxicity caused by protein misfolding or aggregation, or proteotoxicity, is a common theme in many forms of ALS. For example, ubiquitin-positive inclusions have been reported as an almost universal pathologic feature of ALS, including those linked to SOD1, TDP-43, FUS, and C9orf72 [25]. Not only has the proteinopathy of the abnormal proteins been identified as a major pathological feature in familial ALS (fALS), but the abnormal accumulation of the wild-type forms of some proteins, such as TDP-43, has also been observed in a large fraction of the patients with sporadic ALS (sALS) [2,26]. However, the involvement of proteotoxicity in ALS is not limited to proteinaceous inclusions but is also affirmed by strong evidence of impairment in protein quality control systems, particularly autophagy, in ALS. Although proteotoxicity has been implicated in many neurodegenerative diseases, there are a multitude of ALS genes with functions related to the autophagic system, including p62, optineurin, VCP, ubiquilin 2, and TBK1. The increasingly strong connection between ALS and autophagy suggests that there are shared mechanisms underlying different forms of ALS (Figure 1 & Table 1). In this review, we summarize recent findings concerning a few select ALS genes with regard to their connections to autophagy, as part of a discussion on our current understanding of the basic disease pathways.
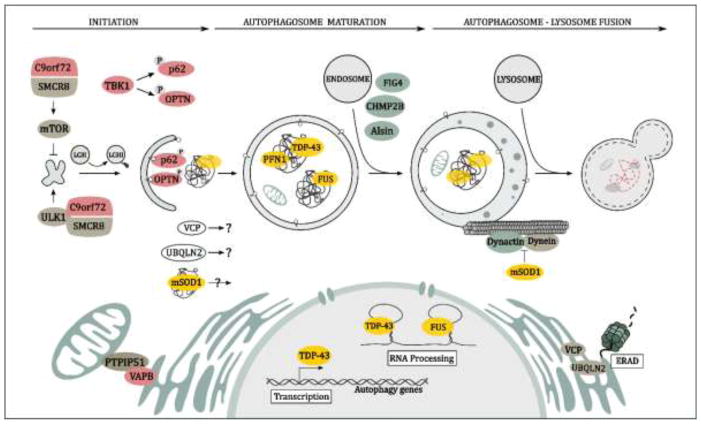
Figure 1. Schematic representation of ALS-related genes and their modulatory effects on autophagy.
The spectrum of ALS-associated genes and their various modulatory effects upon autophagy entailing the stages of autophagy initiation, autophagosome maturation, and autophagosome-lysosome/endosome fusion are depicted herein. Proteins prone to aggregation are highlighted in yellow, those involved in autophagic initiation and autophagosome formation in red, those involved in endosome trafficking and the autophagosome-lysosome fusion in green, and those with less defined roles in autophagy are signified in white.
Table 1.
Summary of ALS genes involved in autophagy impairment
| Gene | Role in Autophagy | References |
|---|---|---|
| SOD1 | Mutants induce autophagy through undefined mechanisms and interfere with autophagosome maturation by disrupting the microtubule-localized dynein-dynactin complex integral to retrograde transport. | [38–40,44,47,49] |
| TDP-43 | Gains toxicity via the formation of proteinaceous inclusions, known substrates of autophagy. Loss of nuclear functions caused by chronic sequestration in RNP granules results in reduced expression of autophagic factors that function at various events within the autophagy-lysosomal pathway. | [88–90,97–99] |
| FUS | Gains toxicity via formation of proteinaceous inclusions, known substrates of autophagy. Possible direct disruption of autophagy regulation. | [83,90,93] |
| P62 | Functions as an autophagy receptor. ALS-linked mutations may impede its association with ubiquitinated substrates or block its association with autophagic membranes via LC3-II. | [32,104] |
| OPTN | Autophagy adaptor that binds ubiquitinated cargo via UBA and LC3 via LIR; also regulates autophagosome formation and maturation. | [124–127] |
| TBK1 | Phosphorylates the UBA domain of autophagy adaptors p62 and OPTN; ALS-associated mutants induce dysfunctions in C-terminal CCD2 and serine/threonine kinase domains that may inhibit p62/OPTN activation; also involved in autophagy initiation via phosphorylation of the C9orf72 binding partner SMCR8; phosphorylation of microtubule-binding protein CEP170 underscores involvement in autophagosome maturation. | [132,134,140–143] |
| VAPB | ER resident that tethers with mitochondrial protein PTPIP51, regulating autophagosome formation; tightening of the tether represses autophagosome formation, while loosening stimulates formation; modulatory effects in autophagic induction vary depending on autophagic stimuli (chemical vs. starvation-induced); ALS-associated mutations induce oligomerization and aggregation of VAPB that overburdens ERAD, hastening ALS pathology. | [89,149–152] |
| VCP | Precise roles are uncertain, but speculated to diversely affect autophagy; ALS-associated mutations reduce mTOR activity, aberrantly induce autophagy; mutations also impede autophagosome-autolysosome maturation, and potentially impair autophagic clearance following autophagosome-lysosome fusion; mutants repress TDP-43-containing RNP complex disassembly. | [156,157,163] |
| C9orf72 | A DENN domain protein, suggested to possess multifunctional roles in autophagy initiation, nutrient sensing, and lysosomal pathways. | [137–140,186–189] |
| UBQLN2 | Recruits autophagosomes to polyubiquitinated aggregates via UBA domain interactions; mutants may hinder the endosomal pathway | [128,190,191,240] |
| ALS2 | Functions as GEF of Rab5, mediating endosome trafficking and fusion. Loss-of-function mutations lead to impeded maturation and clearance of autophagic vesicles. | [13,119,205] |
| CHMP2B | Carries out ESCRT-III dissociation from endosomal membranes, forming MVBs that transition to amphisomes, which fuse with lysosomes; toxic gain-of-function ALS-FTD-associated mutations are C-terminal truncations that disrupt late-stage endosome-lysosome fusion. | [212–214] |
| DCTN | Involved in retrograde transport of autophagic substrates; ALS-associated mutations exhibit impaired intracellular trafficking, immature autophagosome accumulation, and hastened MN degeneration. | [218–221] |
| PFN1 | Regulates actin dynamics, membrane trafficking, and synaptic structure; ALS-associated gain-of- function mutants may disrupt TDP-43 binding partner interactions, inducing deleterious conformational changes. | [229–231] |
| FIG4 | A required phosphatase for IP(3,5)P2 biosynthesis. Loss of function mutations result in a reduction in IP(3,5)P2 level, subsequently leading to enlarged endosomes and disrupted autolysosome clearance. | [235,236,239] |
SOD1
Superoxide dismutase 1 (SOD1) is a key enzyme involved in quenching toxic superoxide radicals within the cell. More than 100 different mutations of SOD1 have been linked to fALS, with approximately 20% of fALS cases and 1% of sALS cases being attributable to SOD1 mutations [1,27–29] Although the mechanism by which mutant SOD1 (mSOD1) causes ALS is not fully understood, many studies suggest that the misfolding or aggregation of mSOD1 is a key event in the pathogenic process. The role of autophagy in mSOD1 clearance was initially implicated by the observation that autophagic inhibition resulted in the accumulation of insoluble SOD1 aggregates in MNs [30]. On the basis of this finding, dysregulations in protein quality control systems such as autophagy has been suspected in ALS pathophysiology associated with mSOD1.
Pathogenic mSOD1 is characterized by the misfolding and aggregation of the protein. Autophagy adaptor p62 interacts with polyubquitinated misfolded mSOD1, and enhances the association between LC3-II and mSOD1, sequestering mSOD1 into protein inclusions [31–33]. Studies suggest that mSOD1 aggregation can affect multiple early steps in autophagy, with evidence from mSOD1 models pointing to the occurrence of hyperactive induction of autophagy [34]. Consistent with this evidence, levels of Ser757-phosphorylated ULK1, a target of mTOR, was significantly reduced in MNs of SOD1G85R mice, suggestive of AMPK activation and mTOR repression, resulting in enhanced initiation of autophagy [35,36]. Further indicating autophagic induction, amplified proteolytic processing of LC3-I to LC3-II and increased LC3-II turnover was observed in MN cell cultures expressing SOD1G93A [37]. Equivalent results were also seen in vivo, with upregulated LC3-II levels and autophagic vacuoles consistently being found in spinal cord and brainstem fractions of SOD1G93A mice [38–40]. Moreover, in spinal cords of SOD1G93A mice, there was persistent upregulation of the autophagy regulators TFEB (a coordinator of autophagic induction and lysosome biogenesis) [41], and beclin1 (an initiator of autophagy involved in autophagosome production) [42], with a corresponding increase in LC3-II [43].
Autophagosome maturation and subsequent lysosomal fusion are downstream events in autophagy at which mSOD1 is also thought to exert its deleterious effects. Autophagosomes mature during retrograde transport and are shuttled by way of the dynein-dynactin complex, acidifying prior to fusion with lysosomes located in the soma [44]. In retrograde transport, processive movement of dynein along the microtubule enables transport of autophagosomes for endosomal or lysosomal fusion [45,46]. It was found that in ALS-linked mSOD1-expressing neurons and transgenic mice, there was colocalization between dynein, and mSOD1. In mice, misfolded mSOD1 acquired a gain-of-interaction with dynein, overloading dynein-mediated retrograde axonal transport, impairing autophagosome transport [47]. Similar defects in retrograde transport were observed in another SOD1G93A murine model, where mSOD1 closely associated with the dynein-dynactin complex in MNs, sequestering dynein into mSOD1 aggregates and impairing retrograde transport efficiency at an early symptomatic stage [48]. Interestingly, it was shown in cells that overexpression of SNAPIN, a vesicle docking protein, can compete with mSOD1 for binding to dynein, thereby rescuing the effects of impaired retrograde transport via recruitment of dynein motors to late endosomes for transport [49]. Overall, mSOD1-induced dysfunctions in retrograde transport appear to contribute to a failure of late-stage fusion steps in autophagy.
Given the important role of autophagy in cleansing the cell of misfolded proteins, promoting autophagy in mSOD1 models has been thought to be beneficial [50]. To date, however, the assessment of autophagic enhancement in ALS SOD1G93A mice has uncovered conflicting results. In several studies, when SOD1G93A mice were administered with autophagy inducer lithium carbonate, they experienced delayed disease progression and prolonged lifespan [51,52], contrasting with later findings reporting worsened neuropathology in SOD1G93A mice treated with lithium [53]. Furthermore, when SOD1G93A mice were treated with autophagy enhancers including trehalose and resveratrol, animals displayed delayed symptom onset, prolonged survival, and diminished MN loss [35,54–58]. However, when SOD1G93A mice were treated with another autophagy enhancer, rapamycin, they showed accelerated MN degeneration and shortened lifespan [59]. On the other hand, in several other studies, autophagic inhibition by n-butylidenephtalide (n-BP) has yielded positive effects in ameliorating MN pathology in mSOD1 models. Specifically, n-BP treatment in SOD1G93A mice attenuated MN loss by preventing the abnormal accumulation of autophagosomes in MNs, as evidenced by the inhibition of the expression of a number of proteins: LC3-II, p62, and beclin1, as well as other autophagy-related proteins (Atg1/5) [60,61].
It appears that autophagic disturbances related to mSOD1 vary depending on the phases of disease development, further complicating the association between mSOD1 and autophagic dysregulation. In a recent study where the essential autophagy gene ATG7 was conditionally knocked out in MNs of SOD1G93A mice, autophagic inhibition resulted in an earlier onset of disease phenotypes, suggesting that autophagy fulfills a neuroprotective role at the early stage of disease. Later, however, inhibition of autophagy extended the lifespan of mice and slowed disease progression, suggesting that autophagy plays a deleterious role at an advanced disease state [62]. This stage-dependent action of autophagy has also been demonstrated in another study, which indicated that starvation-induced autophagy markedly reduced neurotoxic mSOD1 aggregation at an early pathological stage, yet dramatically increased mSOD1 inclusions at a later point of disease in SOD1G93A mice [63]. Further studies are needed to resolve the precise effects of autophagic modulation in mSOD1-associated ALS pathology.
TDP-43 and FUS
A subset of ALS-associated genes encodes RNA-binding proteins (RBPs), including TAR DNA-binding protein 43 (TDP-43) and fused in sarcoma (FUS) [64–66]. Mutations in each of these genes are accountable for about 3% of fALS cases and less than 1% of sALS cases [67]. TDP-43 mutations are also identified in approximately 2% of FTD cases [68]. The connection between these RBPs and autophagy is in part indicated by their shared tendency to exhibit misfolding and aggregation [69,70] that burden the autophagic system. Also, loss-of-function of these proteins contributes to dysregulations in autophagy [71–73]. Notwithstanding the many advances that have been made from studies of these RBPs in ALS, how the gain- and loss-of-function mechanisms of these RBPs contribute to ALS pathogenesis remains a complicated area of investigation.
Gain of toxicity
The gain of toxicity of RBPs is directly linked to their role as constituents of the ubiquitinated protein inclusions observed in most forms of ALS. To date, TDP-43-positive inclusion buildup has been described as one of the most prevalent pathological hallmarks of ALS [1,4,65,74]. The protein structures of TDP-43 and FUS share characteristics of low sequence complexity domains (LCs) [75,76], which are regions where the majority of ALS-related mutations reside [65,76]. The link between the function of LCs and RBP-positive inclusion formation has been recently studied, where mutations in LCs of both TDP-43 and FUS was shown to enhance inclusion formation [69,77,78]. LCs was reported to promote the sequestration of soluble proteins into membrane-less protein-rich liquid droplets, a phase transition event that was proposed to be vital for their normal functions but also can render them prone to aggregation [75,77,78]. As part of a normal cellular stress response, TDP-43 and FUS are transiently recruited at stress granules (SGs) for translation repression. Under chronic stress, however, their prolonged cytosolic deposition can disrupt the structural homeostasis of SGs, leading to the formation of persistent inclusions [79–81].
As a mechanism for maintaining protein homeostasis, autophagy is known as an important pathway for the clearance of TDP-43 and FUS-positive protein inclusions [82–84]. Several studies have demonstrated that pharmacological induction of autophagy enhances the TDP-43 turnover rate as well as the rate of FUS-positive SG’s clearance, in certain cases rescuing the ALS-related pathology [82,85–87]. Rapamycin treatment reduced cell death in neurons expressing FUSR521C, an ALS-linked mutation [83]. In Drosophila and mouse models of TDP-43 proteinopathy, rapamycin-induced autophagy enhanced the clearance of TDP-43 aggregates while lessening ALS/FTD-like symptoms [85,86].
RBPs and their inclusions can influence autophagy beyond being autophagic substrates. Overexpression of soluble TDP-43 in the cytoplasm was suggested to disrupt the VAPB-PTPIP51 tether, loosening ER-mitochondrial contact, subsequently enabling the recruitment of the early autophagic membrane and inducing autophagy [88,89]. Several mutations have been reported to cause direct disruption of autophagy initiation. Mutations in the C-terminal domains of TDP-43 and FUS inhibited ER-Golgi vesicle transport through depletion of the vesicle-trafficking regulator Rab1, a small GTPase required for phagophore formation [90–92]. Additional data indicated that accumulation of FUS mutants inhibited autophagy, while overexpression of Rab1 reversed this disruption [93].
Loss of function
TDP-43 and FUS are primarily localized to the nucleus, where they carry out important functions that include RNA splicing, transcriptional regulation, and the maintenance of genome integrity [72,73,94–96]. Both LC and RNA recognition motif domains are essential for their native functions. For instance, LCs allow these proteins to cluster at DNA breaks, forming scaffolds for the repair complex [75,96]. As part of the cell’s normal stress response, temporary translocation of TDP-43 to the cytoplasm can exert a protective effect through autophagic upregulation, as part of adaptation to maintain cellular homeostasis under stress. Transient discharge of TDP-43 from the nucleus was shown to relieve the repression of the FOXO transcription factors, inducers of autophagy [97]. However, under conditions of prolonged cellular stress such as those present in neurodegeneration, the protective effect of TDP-43 cytosolic sequestration is thought to be compromised, contributing to the overburdening of protein quality control [97].
In the case of ALS, chronic cytoplasmic sequestration of TDP-43 at ribonucleoprotein (RNP) granules constitutes a loss-of-function in mRNA stabilization [72], directly influencing the expression of TDP-43’s target genes, among which are important players in autophagy. Nevertheless, the overall impact of this loss-of-function for autophagic clearance remains unclear. In several instances, TDP-43 ablation inhibits the expression of positive regulators of autophagy. TDP-43 depletion was shown to downregulate the expression of the autophagic factor Atg7, an E1-like enzyme essential for the lipidation of LC3-I and autophagosome formation [98]. TDP-43 depletion also inhibited autophagosome-lysosome fusion by depleting dynactin, a motor protein essential for retrograde transport that enables vesicular fusion events in the pathway [99]. However, in other cases, the loss of TDP-43 function was reported to downregulate expression of mTOR, a negative regulator of autophagy. In particular, TDP-43 nuclear depletion resulted in reduced expression of Raptor, an mTOR adaptor protein. Downregulation of Raptor inhibits the phosphorylation of transcription factor TFEB, resulting in its nuclear translocation and a corresponding increase in the expression of autophagy and lysosome biosynthesis genes [99]. These concurrent events ultimately lead to the accumulation of autophagic vesicles observed in TDP-43 knockdown cells [99].
In sum, it is clear that the loss of TDP-43 function leads to a complex array of gene expression dysregulations that result in various aberrations to the events of autophagy; however, the combined effect of these changes on autophagic clearance remains uncertain.
SQSTM1, OPTN, and TBK1
Macroautophagy was thought to be a nonselective process, until the identification of autophagy receptors, integral components of the autophagic machinery that enable selective recruitment of specific cargo for degradation [100]. Mutations in SQSTM1/p62 and OPTN, two well-defined autophagy receptors, and in their activating kinase, TBK1, have been identified in multiple forms of ALS [7,8,101]. Specifically, mutations of these multi-functional proteins has been implicated in the impairment of the clearance of neurotoxic aggregates, as well as the disruption of inflammation regulation, conventional hallmarks of ALS [8,102,103]. p62 and OPTN share two common domains that are important for their role as autophagy receptors, the ubiquitin-associated domain (UBA) and the LC3-interacting region (LIR). The UBA domain allows these receptors to bind ubiquitinated cargo marked for degradation, and the LIR domain interacts with LC3, allowing the recruitment of receptors and cargo to the phagophore [100,104,105]. Distinctly, p62 contains another domain, PB1, which enables the oligomerization of p62 that is essential for the function of this protein in autophagy [106].
SQSTM1/p62
Mutations in sequestosome 1, or p62, account for approximately 1% of ALS cases [101,107]. Furthermore, about 2% of FTD patients in a French cohort were reported to carry p62 mutations [108]. p62 is known to be involved not only in selective autophagy but also in degradation through the ubiquitin-proteasome system (UPS) via its UBA domain [109–111]. p62 was first suggested to play a role in neurodegeneration due its enrichment in misfolded proteinaceous inclusions in many neuronal disorders including Alzheimer’s disease and Parkinson’s disease [112]. Accordingly, p62 has been widely used as a pathological marker to study these disorders. Notably, p62-positive inclusions have also been detected consistently in C9orf72-related ALS (C9ALS), the most common form of ALS [113–115].
Mutations in p62 were most commonly identified in Paget disease of bone (PDB), a skeletal disorder, occurring at a frequency of 20–50% in familial and 5–10% in sporadic PDB cases [102] with the majority of PDB-associated p62 mutations residing in the UBA domain. Similarly, p62 missense mutations of UBA were also identified in patients with ALS-FTD [5,108], suggesting that disruptions in the interactions between p62 and ubiquitinated cargo are shared between ALS-FTD and PDB. However, it remains unclear whether these shared mutations affect autophagy in both diseases. For example, expression of the p62P392L mutant impaired basal autophagy in murine osteoclasts, implicating this mutation to impeded autophagic clearance in PBD [102]. By comparison, the implications of p62 UBA domain mutations on autophagy in the context of CNS disorders such as ALS-FTD remains to be studied.
Beyond the UBA domain, additional relatively rare ALS-associated p62 mutations are diffusely expressed in other regions of p62, including the LIR domain and p62 promoter region [6,101]. Assessment of mutations in the LIR domain of p62 have particularly important implications for their possible contribution to disturbed autophagy in ALS, as this domain enables interactions with LC3, a central component in the recruitment of substrates to the autophagic membrane. A recent study reported that the ALS-associated mutation L341V in the LIR domain resulted in significantly reduced affinity of p62 for LC3-II, hindering its functional involvement with autophagic vesicles [116]. Furthermore, mutations detected in the promoter region of p62 of ALS/FTD patients suggest that reduced p62 expression could play role in the autophagy dysfunction found in this pathological spectrum [117,118].
In addition to canonical interactions between p62 and ubiquitinated inclusions, p62 also associates with mSOD1, offering insights as to how p62 may contribute to SOD1-related ALS. Early evidence of this interaction includes the observation that p62 expression and accumulation correlate with an increase in aggregation of mutant (G93A), but not wildtype, SOD1 [106]. This association was later reported to be independent of the UBA domain, instead interacting via a distinct SOD1 mutant interacting region (SMIR) to enable sequestration of mSOD1 without the presence of the ubiquitin signal [32]. In murine models, depletion of p62 was reported to shorten the lifespan of SOD1H46R and SOD1G93A mice [119]. In the SOD1H46R model, p62 loss aggravated ALS symptoms, provoking accelerated weight loss, motor dysfunction, and MN degeneration. Furthermore, a link between p62 and alsin, a causative gene in ALS, has also been suggested. Depletion of alsin exacerbated the aforementioned contribution of p62 depletion to the neuropathology of SOD1H46R mice [119].
Optineurin
Optineurin (OPTN) is a protein involved in a diverse range of cellular processes, including vesicular trafficking, maintenance of the Golgi, NF-κB modulation, and antibacterial signaling [120]. In addition, it has a well-characterized role as an autophagy receptor. Genetic analyses have uncovered OPTN mutations in cohorts of ALS patients, which account for approximately 3% of fALS and about 1% of sALS [64], raising questions as to the potential associations between OPTN dysfunction, autophagic impairment, and ALS neuropathology. Rare ALS-associated pathogenic variants of OPTN were initially reported, including deletion of OPTN exon 5, a Q398X nonsense mutation, and an E478G missense mutation [7]. Later, insertion R96L, nonsense Q165X, and truncation (c.1320delA) mutations of OPTN were detected, expanding the mutational spectrum associated with ALS [121]. Interestingly, the majority of deleterious OPTN mutations are found in the UBA region and occur in a heterozygous manner, potentially implicating haploinsufficiency in the case of a loss-of-function mechanism in pathogenesis [105]. Despite these findings, however, definitive mechanistic causes accounting for OPTN dysregulation and consequent autophagic impediments are yet to be identified.
Several pathways involving mutant OPTN-induced autophagic impairment have been proposed. In cell cultures, wildtype OPTN colocalizes with httx1 Q103 and SOD1G93C inclusion bodies, toxic protein models of Huntington’s disease and ALS, respectively [122]. Deletion of OPTN induces increases in both httx1 Q103 and SOD1G93C inclusions, hinting at the involvement of OPTN in regulating the turnover of aggregation-prone proteins [122]. Specifically, OPTN recognition of misfolded proteins is mediated by the C-terminal coiled-coiled UBA domain, which enables K63-linked polyubiquitin-mediated autophagy. Furthermore, OPTN UBA mutations, including ALS-associated E478G, reduce degradation of polyubiquitinated inclusion bodies [122,123]. In another study, OPTNE478G overexpression resulted in perturbed autophagic degradation, as shown by enhanced levels of lipidated LC3-II, with possible impeded autophagosome transport or fusion events occurring [124]. The proposed model to account for these findings posits that OPTN UBA mutants interact with wildtype OPTN, forming hybrid wildtype-mutant OPTN complexes that repress autophagosomal maturation [124]. Further in vitro studies in neuronal cultures have expanded these findings by demonstrating that the ALS-linked OPTN mutants E478G and Q398X failed to bind myosin VI (MYO6), an essential protein involved in regulating the intracellular transport of autophagosomes and endocytic/secretory vesicles [125,126]. Diminished OPTN-MYO6 binding resulted in the accumulation of immature autophagosomes and conditions of ER stress, Golgi fragmentation, and impaired secretory protein trafficking in mutant-expressing cells. In contrast, wildtype OPTN linked sequestered autophagosomes to endosomes via recruitment of MYO6 and its cargo adaptor TOM1, allowing for efficient autophagosome maturation [126]. These results have been further supported by analyses of spinal cord fractions from ALS patients, in which OPTN-MYO6 interactions were absent [126]. Thus, the compromised binding of mutant OPTN to MYO6 may represent a late-stage inhibition of autophagosome fusion with lysosomes or endosomes.
There is additional evidence to suggest OPTN’s involvement at more upstream nodes of autophagic regulation. It was shown that Ypt1, the yeast homolog of mammalian Rab1a, a GTPase involved in autophagosome formation, is a binding partner of OPTN [91,127]. The Rab1a-OPTN association was dependent upon the GTP-binding capacity of Rab1a, along with the Rab1a zinc finger domain of OPTN [127]. After siRNA-mediated silencing of Rab1a, autophagosome formation is inhibited in neuroblastoma cells, suggesting that intact Rab1a is a necessary component of autophagosome formation undertaken by OPTN [127]. Given that previous findings in cells expressing ALS-associated OPTNE478G have indicated an abolition of vesicle and autophagosome formation [128], dysfunction in Rab1a binding may underlie this pathology.
TBK1
TBK1 is a key player in various cellular processes including innate immunity, nucleic acid sensing pathways, and autophagy [129,130]. Regarding autophagy, TBK1 operates primarily by phosphorylating the autophagic adaptors p62 and OPTN. TBK1 comprises four distinct domains, an N-terminal serine/threonine kinase domain, two coiled-coiled domains (CCD1/2), and a ubiquitin-like domain [131]. TBK’s modulatory properties in autophagy were initially shown in cultured cells, with TBK1 phosphorylation of the UBA domain of OPTN on Ser-177 enhancing LC3 binding affinity and promoting autophagic clearance of cytosolic Salmonella in HeLa cells [132]. It has also been reported that TBK1 coordinates the autophagic machinery via phosphorylating Ser-403 on the UBA domain of p62, enhancing the affinity of UBA for K48/K63-linked ubiquitin chains, resulting in the autophagic clearance of mycobacteria in macrophages [133].
In ALS patients, exome sequencing has revealed that novel genetic aberrations of TBK1, encompassing loss-of-function mutations, are causal in pathogenesis. About 4% of fALS patients carry loss-of-function TBK1 mutations, however, these mutations are relatively rare (<0.1%) in sALS cases. Within a Belgian cohort, TBK1 mutations also account for about 1% of FTD patients [8,134,135]. A particularly pathogenic variant results in a (p.690–713) deletion in the C-terminal CCD2 domain of TBK1, with in vitro models exhibiting an inability of truncated TBK1 to associate with OPTN [134]. In addition, other loss-of-function mutations occurring in the serine/threonine kinase domain have been reported in a cohort of French ALS and FTD patients. The pathogenicity of these variants is considered to be a result of haploinsufficiency, since they are heterozygous missense in nature [136]. Given the essential catalytic role of the serine/threonine domain, mutations in this region may repress the activation of p62 and OPTN. Nevertheless, since the majority of ALS-associated TBK1 mutations are missense mutations with largely unknown domain-specific interactions, further analyses are necessary to deduce exactly how the constituents of the broad spectrum of mutations contribute to the neurodegenerative state.
Elucidating the mechanisms by which TBK1 regulates autophagy may provide insights into how dysregulations of the autophagic pathway contribute to ALS pathology. TBK1 potentially plays a role in autophagic initiation, since its kinase activity has recently been reported to be necessary for the proper functioning of the C9orf72 complex, whose interactions with conventional autophagic initiation factors, including ULK1 and mTOR, were also shown [137–139]. TBK1 activates C9orf72 activity via phosphorylation of its binding partner, SMCR8. In cells, depletion of SMCR8 alters autophagy, causing an accumulation of p62-positive aggregates, whereas mimicking TBK1 phosphorylation corrects this autophagic dysfunction [140]. Furthermore, in a glaucoma-associated variant of OPTN, M98K, premature retinal cell death has been attributed to amplified TBK1 activity when compared to controls. Specifically, TBK1 phosphorylation at Ser-177 of OPTN induces autophagosome formation, increasing autophagic flux and causing retinal cell death [141]. Mutations in TBK1 may also impede later autophagic events, such as dynein-dependent autophagosome maturation along microtubules. TBK1 phosphorylation of the microtubule-binding protein CEP170 is necessary for centrosomal localization and binding to the microtubule depolymerase Kif2b, which regulates the association of NuMA with dynein that is required for microtubule activity [142]. Hence, loss of TBK1 function may contribute to ALS by disrupting the microtubular retrograde transport of autophagosomes, preventing autolysosome formation [131]. In addition, TBK1 activity plays an important regulatory role in the autophagy of damaged mitochondria, since constitutive interactions of TBK1 with OPTN induces the retention of OPTN/TBK1 on damaged mitochondria and result in an enhanced binding capacity of the UBA domain of OPTN with polyubiquitin chains, promoting mitophagy in cells by facilitating LC3- mediated linkage with phagophores [143,144].
In sum, mutations in p62, OPTN, and TBK1 significantly impair autophagy through various mechanisms, resulting in ALS-relevant pathological features such as the accumulation of autophagic substrates and damaged organelles in MNs. A comprehensive understanding of the interplay between TBK1 and its effectors p62 and OPTN in autophagy may allow for targeted approaches aimed towards correcting aberrations resulting from alterations in the functions of these proteins.
VAPB
Mutations in vesicle-associated membrane protein-associated protein B (VAPB) are relatively rare, with a mutation frequency of less than 0.6% in ALS patients [145,146]. VAPB and VAPA together comprise the VAP protein family of ER tail-anchored transmembrane proteins. VAPB is associated with the membrane surrounding the ER and operates as a receptor for cytosolic protein recruitment to the ER surface. Interacting with FFAT-motif-containing polypeptides, VAPB plays diverse roles, being involved in such cellular functions as membrane trafficking, lipid transport and metabolism, membrane contact site formation, and neurotransmitter release. A rare missense mutation (P56S) in VAPB has been found to cause autosomal dominant ALS in a Brazilian cohort [10], with P56S transgenic mice later recapitulating this ALS-like pathology [147]. Another pathogenic mutation of VAPB (T46I) has been found in individuals with fALS, with this variant being speculated to impede the interaction of VAPB with cytosolic proteins by inducing dysfunctions in lipid metabolism [9]. Accumulating evidence has linked VAPB to autophagic regulation, raising the prospect that dysfunctions in VAPB may potentially underlie autophagic impairment in ALS pathogenesis.
Interactions between the ER and mitochondria are involved in mediating a range of physiological processes, including autophagy. These interactions arise as a result of ER-resident VAPB closely associating with mitochondrial protein PTPIP51, enabling ER membrane recruitment to mitochondria [88]. Autophagosomes are known to form at this ER-mitochondria interface, with the pre-autophagosome marker ATG14 and autophagosome formation marker ATG5 becoming localized to this site [148]. It was recently reported that the VAPB-PTPIP51 tether regulates autophagosome formation. Overexpression of VAPB and PTPIP51 induces a tightening of the tether and concomitant impairment of autophagosome formation. Conversely, siRNA-mediated loss of VAPB or PTPIP51 loosens the tether, stimulating autophagosome formation [89]. Interestingly, after chemically induced autophagy by rapamycin and torin 1, VAPB and PTPIP51 overexpression results in a reduction in the autophagic substrates LC3 and DFCP1, yet no such effect has been reported for cells subjected to starvation-induced autophagy [89]. Thus, the effects of VAPB-PTPIP51 in modulating autophagy appear to be dependent on the particular signaling mechanism by which stimuli induce autophagy. In addition, another means by which VAPB participates in autophagosome formation has been identified from co-immunoprecipitation assays in cells, which exhibited binding interactions between VAPB and Rab3GAP1, a protein influencing autophagosome biogenesis [149,150].
In various models assessing the effects of ALS-associated P56S, persistent intracellular inclusions and alterations in ER structure are observed, perhaps providing the trigger for downstream autophagic dysregulations. Cells overexpressing P56S displayed significant ER restructuring, accompanied by inclusions continuous with the ER [151]. In a further characterization of these results in mice, P56S induced rapid oligomerization and aggregation of mutant VAPB [152]. These effects can be explained by the buildup of inclusions positive for mutant VAPB exceeding the capacity for ER-associated degradation, suggesting that VAPB dysfunctions contribute to impairments in ER quality control, in turn, leading to conditions of ER stress. Another study employing P56S VAPB knock-in mice reported that ER stress in murine models was coupled with evidence of impaired autophagy preceding the development of ALS-like pathology. This was shown by the significant colocalization of early autophagosome markers p62 and LC3 in cytoplasmic inclusions, along with accumulation of VAPB P56S within the cytosol of MNs, indicating VAPB mislocalization from the ER to cytosolic regions [153]. There is therefore reason to speculate that perturbed autophagy may be the result of ER stress.
VCP
Valosin-containing protein (VCP/p97) is an essential AAA+-ATPase involved in a range of cellular processes that include DNA replication and repair, cell cycle regulation, and protein clearance [154]. Regarding the latter, VCP is integral to ERAD, a process in which unwanted proteins are extracted from the ER for proteasomal degradation [155]. Expanding VCP’s role in proteostasis, emerging findings now point to the involvement of VCP in autophagy [156,157].
Mutations of VCP are causal in inclusion body myopathy, Paget’s disease, and frontotemporal dementia (IBMPFD) [158]. Broadening the phenotypic spectrum of VCP mutations to ALS, whole-exome sequencing has revealed the existence of several VCP mutations (R155H, D592N, R191Q, R159G), which account for approximately 1–2% of fALS cases [11,159]. Initial evidence suggesting VCP’s involvement in autophagy has come from muscle lysates of IBMPFD patients expressing IBMPFD/ALS-associated R155H. Analysis of patient tissue revealed the accumulation of autophagosomes colocalized with p62 and LC3, coalescing at rimmed vacuoles [156]. Since then, additional studies in mouse models expressing pathogenic VCP variants have elicited similar observations [157,160,161]. Nevertheless, the precise mode of involvement of VCP in autophagy is a subject of debate.
There are several proposed mechanisms by which VCP interacts with the autophagic machinery. With regard to autophagic initiation, in neuronal cell cultures subjected to insulin withdrawal-induced induction of autophagy, VCP expression repressed the negative autophagy regulators calpain 2 and mTOR, while upregulating GSK-3β, a positive autophagic regulator [162]. Moreover, two direct targets of mTOR, EIF4EBP1 and RPS6KB1/p70S6, were found to be decreased in a murine VCPR155H model. Reduced mTOR activity was consistent with the enhanced autophagic activation and autophagosome biogenesis apparent in the model [163]. Additional studies have suggested a role for VCP in downstream autophagosome maturation. Proteomic analysis in IBMPFD murine muscle lysates and neuronal cells revealed that VCP loss in addition to presence of VCPR155H resulted in the failure of autophagosomes to mature into autolysosomes, causing accumulation of non-degradative autophagosomes, ubiquitinated aggregates, and p62-positive inclusions [156,157]. Other evidence suggests that mutant VCP interferes with processes following autophagosome-lysosome fusion, since cells expressing ALS/IBMPFD-linked VCPR155H and VCPA232E mutations exhibit accumulations of irregular, enlarged autophagic vacuoles possessing acidified autophagosomes, implying that autophagic failure occurrs at a later point, following acidification and delivery of lysosomal hydrolases [157]. Stress granule clearance represents yet another juncture in autophagy that VCP is suspected to influence. This possibility was recognized in yeast models in which the VCP ortholog Cdc48 promoted targeting of SGs for autophagic degradation [164]. These results have also been noted in HeLa cells, in which cell cultures expressing ALS/IBMPFD-associated VCP mutations were found to constitutively accumulate pathogenic TDP-43-positive SGs; in contrast, in wildtype cells, VCP regulated the amount of TDP-43-containing RNP complexes, facilitating their disassembly and targeting to autophagosomes [164].
Based on the assessment of ALS-related VCP dysfunctions, it is evident that autophagic impairment is a common theme underlying neuropathology. Extensive evidence suggests that VCP’s role in autophagy is multifaceted and varied, necessitating clarification as to which autophagic processes are directly regulated by VCP and what the specific consequences of dysregulation are.
C9orf72
A GGGGCC hexanucleotide repeat expansion in C9orf72 is the most common cause of ALS [21,22]. Notably, the frequency of C9orf72 repeat expansion varies between ALS cohorts, with very high prevalence in European cohorts (~33% in fALS, ~5% in sALS) and lower prevalence in Asian cohorts (~2% in fALS, ~0.3% in sALS). In FTD patients, the frequency of C9orf72 repeat expansion also varies within a range of 3% to 12%, depending on the studied cohort [67,165]. To date, there are three major mechanistic components that have been reported to contribute to the pathogenesis of C9ALS: (1) the loss or reduction in expression of the C9of72 protein [21,166], (2) the toxicity mediated by C9orf72 repeat expansion sense/antisense RNA transcripts [167,168] , and (3) the generation of repeat dipeptides from non-ATG translation of C9orf72 repeat expansion [169–171]. The sensitivity of patients’ cells to inhibition of autophagy was exacerbated as a result of the presence of C9orf72 repeat expansion, although the mechanism is unknown [172]. Regarding the role of C9orf72 repeat expansion in autophagy, gain-of-function components (2) and (3) may exert indirect influences that perturb the autophagy-lysosomal pathway. By sequestering important RBPs such as TDP-43 in nuclear RNA foci and preventing nuclear import via disruption of nucleocytoplasmic transport, the native function of these proteins can be compromised and the autophagy-lysosomal pathway influenced as a result [79,167,173–175]. In this section, however, we will focus on the direct influence of the repeat expansion on autophagy via (1) or through C9orf72 protein reduction or depletion.
Since the discovery of C9orf72 repeat expansion, the transcript level of the C9orf72 protein has been recognized as being reduced in the frontal and cerebellar cortices of C9ALS patients [21,166], with later studies reporting a reduction in the protein levels in patients’ brains [176]. Homology analysis of the C9orf72 protein sequence revealed a DENN-like domain in its structure suggesting that C9orf72 may play a role as a guanine exchange factor (GEF) for Rab GTPases key players in vesicular trafficking [177–179] that are extensively involved at different events of macroautophagy [92]. This role of C9orf72 in Rab-mediated events in the autophagy–lysosomal pathway was supported by the observation that C9orf72 interacts with Rab1, Rab5, Rab7, and Rab11, small GTPases known to be involved in autophagy [180,181]. In addition, a role for C9orf72 in modulating filamentous actin (F-actin) assembly was recently suggested [182]. F-actin is known to play central roles in the structure and dynamics of the cytoskeleton, the maintenance of which may be critical in the autophagy-lysosomal pathway[183–185], raising the possibility of another mechanism through which C9orf72 may affect autophagy.
In an interaction essential to its known functions, C9orf72 forms complexes with SMCR8, another DENN domain protein [137–140,186]. Multiple studies have described the many events that this complex could potentially influence within the cascade of autophagy including autophagic induction, nutrient-sensing pathways, and lysosome biogenesis/clearance [137–139,186].
Depletion of C9orf72 protein appears to inhibit autophagy initiation via the ULK1-mediated pathway [137,187]. C9orf72 was reported to interact with the ULK1/FIP100/ATG13/ATG101 complex in multiple studies [137,186–188]. Loss of C9orf72 expressionmay disrupt the induction of ULK1-mediated autophagy via two events: Firstly, C9orf72 was suggested to act as an effector protein for Rab1a, a GTPase known to be recruited for phagophore formation [92]. Through its interaction with the ULK1 complex, C9orf72 could regulate the Rab1a-dependent trafficking of ULK1 to the autophagic initiation site [187]. Secondly, C9orf72 may be essential for relaying signals from the ULK1 complex to promote downstream Rab-mediated phagophore maturation. The C9orf72-SMCR8 complex was shown to act as a GEF toward Rab8a and Rab39b [140], with expression of phosphomimetic Rab39b being found to rescue the inhibition of autophagic induction caused by C9orf72 depletion. This finding suggested that Rab39b could act immediately downstream of ULK1, whose phosphorylation via C9orf72 is required for autophagy initiation [137,140]. Notably, a recent study reported that the loss of SMCR8, C9orf72’s binding partner, resulted in an upregulation of ULK1 expression, characterizing SMCR8 as a negative regulator of autophagy induction [188]. In the same study, however, SMCR8 also exhibited another independent function in promoting autophagosome maturation [188]. The multitude of roles in autophagy, exhibited by C9orf72 and SMCR8 that results in various disturbance to the events of the autophagy cascade further demonstrate the complexity in function of the C9orf72 complex.
Autophagy is a crucial mechanism by which the cell adapts to its environment by managing and recycling its metabolism according to the availability of nutrients. The autophagy-lysosomal pathway, therefore, is directly influenced by nutrient-sensing signaling pathways, of which the mTOR-mediated pathway is a major regulator. Loss of C9orf72 function was reported to cause a defect in mTOR signaling pathway [138,139]. In the absence of C9orf72, mTOR, a negative regulator of autophagy, was inactivated [139]. In two separate studies, it was observed that mTOR inactivation leaded to the nuclear translocation of transcription factor TFEB, a master inducer of autophagy and lysosome biogenesis genes, resulting in an increase in autophagic flux [137,138]. Notably, this increase in autophagic flux was not observed in torin 1-induced autophagy [187], suggesting that there are two distinct pathways in starvation and in chemical-induced autophagy where C9orf72 may play different roles.
The potential role of C9orf72 in the lysosome pathway has been suggested by its localization to this organelle [180]. This localization was reported to be negatively regulated by amino acid availability, suggesting potentially unknown roles of the protein at the lysosome. Interestingly, inhibition of mTOR via torin 1 treatment did not result in the localization of C9orf72 to lysosomes, implicating that the lysosomal function of the protein is associated with starvation-induced, but not chemically-induced autophagy [139]. Furthermore, based on evidence from multiple studies, lysosome biogenesis has been suggested to be upregulated as a result of C9orf72 loss. Supporting this in cell models, ablation of C9orf72 resulted in swollen lysosomes that cluster near the nucleus [139]. Also, in liver lysates from C9orf72-knockout mice, a buildup of lysosomal markers, including LC3-I, LAMP1, and PSAP, was reported [138,186]. In another study, aberrantly enlarged lysosomes were observed in bone marrow-derived macrophages and microglia from C9orf72-knockout mice [189]. Upregulation of TFEB, an inducer of lysosome biogenesis via inactivation of the mTOR pathway, has been suggested to contribute to the aforementioned abnormalities in lysosomal morphology and lysosomal marker levels in C9orf72-knockout models [138].
In summary, C9orf72’s role in autophagy is multifaceted, and the ultimate consequence of C9orf72 downregulation related to the repeat expansion remains undefined. On one hand, C9orf72 has been shown to mediate autophagic induction in conjunction with the ULK1 complex, yet on the other hand, C9orf72 modulates mTOR and lysosomal functions. Together, the function of C9orf72 in the autophagy-lysosome pathways needs to be further studies.
Other ALS genes
UBQLN2
Ubiquilin 2 (UBQLN2) is part of the ubiquilin family of proteins involved in proteasome-mediated protein degradation via activity of ubiquitin-associated (UBA) and ubiquitin-like (UBL) domains. UBQLN2 may also operate as an autophagic receptor, with reported interactions between UBQLN2 and LC3-II [190]. Deletion of UBQLN2’s UBL domain abrogates its association with LC3-II [191]. According to studies in French/French–Canadian populations, UBQLN2 mutations are relatively rare and mostly found in sALS patients (~0.4%) [192,193]. UBQLN2 mutations were also found in about 0.3% of FTD cases in an extended Belgian cohort [194]. Most ALS/FTD-associated UBQLN2 mutations are clustered in UBQLN2’s proline repeat region [12], although they are also found elsewhere [192,195]. This then raises the question if autophagic dysfunction due to mutant UBQLN2 may contribute to ALS pathogenesis.
In vitro studies have conferred limited support for disturbed autophagic pathways by UBQLN2 mutations. Cells overexpressing mutant UBQLN2 showed diminished colocalization between UBQLN2 and ATG9/ATG16L1, essential factors in autophagosome-lysosome fusion [196]. Also, although wildtype UBQLN2 is found in endosomal vesicles colocalizing with p62 and autophagy initiator ULK1, ALS-linked UBQLN2 variant E478 abolished vesicle formation, inhibited endosomal trafficking, and increased inclusion body formation [128]. Similar findings have been recapitulated in animal models, where overexpression of ALS-associated UBQLN2P497H accelerated MN loss in rats via possible endosomal pathway impairment, evidenced by reduced early endosome antigen 1 [197], a nessary component of autophagosome maturation [198]. Aberrant accumulation of autophagy substrates p62 and LC3-II are also present in neural tissue of rats overexpressing UBQLN2P497H [197,199]. Despite these findings, the role of UBQLN2 in the context of autophagy remains controversial, and it is unclear by which precise mechanisms UBQLN2 may regulate autophagic flux.
Alsin
Mutations in ALS2, which encodes the protein alsin, have been identified in a rare juvenile-onset form of ALS [14]. Alsin is constitutively expressed and is localized to the cytosolic side of the endosomal membrane [29,200]. Its protein structure includes a Vps9 domain, which is crucial for its GEF role with regard to Rab5, a regulator of endosomal vesicle transport and fusion [200–204]. Alsin-mediated endosome trafficking and vesicular fusion were evidenced to be important events within the autophagy cascade that particularly influence the maturation of autophagosomes and the formation of amphisomes, two events preceding fusion with the lysosome and vesicular clearance [205]. Consistently, loss of alsin was reported to exacerbate the neurotoxicity of protein aggregates in murine models of ALS, including SQSTM1/p62-null and mSOD1 mice models. Alsin ablation leaded to elevated SOD1H46R protein buildup and toxicity by reducing autophagic clearance, which was a result of impaired endosomal trafficking and disrupted maturation of autophagosomes [206]. More recently, loss of alsin was also reported to intensify the effect of p62 deletion in SOD1H46R transgenic mice [119]. This modifier effect of alsin on p62 depletion further suggests that intact alsin-mediated endosomal functions are required for proper autophagic clearance.
CHMP2B
Charged multivesicular protein 2B (CHMP2B) is a subunit of the endosomal sorting complex required for transport (ESCRT-III), whose mutations were identified in less than 1% of a large cohort of ALS patients in North England [17]. In autophagy, dissociation of ESCRT-III from the endosomal membranes generates multivesicular bodies (MVBs) that fuse with autophagosomes, producing amphisomes that later fuse with lysosomes [207,208]. CHMP2B recycles ESCRT-III and mediates its dissociation via the recruitment of Vps4, an ATPase that facilitates membrane abscission and MVB formation. Sequencing of CHMP2B in FTD patients has revealed a splice-site mutation that produces two aberrant transcripts (CHMP2BIntron5, CHMP2BΔ10) that result in C-terminal domain truncations and significant p62 and ubiquitin deposition in patient brains [15]. CHMP2B mutations (T104N, I29V, Q206H) have also been found in ALS cohorts, with patients’ brains exhibiting immature autophagosome accumulation and upregulated LC3-II and p62 levels, implicating autophagic impairment as a common neuropathological pathway [16,17].
Various studies of autophagic disruption in CHMP2B models have recapitulated the observations made in clinical ALS-FTD cases marked by CHMP2B dysfunctions. In cells, CHMP2BIntron5 overexpression inhibited autophagic clearance, inducing the formation of ubiquitin- and p62-positive inclusions [209,210]. Similar results were reported in a CHMP2BIntron5 murine model [211]. C-terminal CHMBP2B truncations may underlie autophagic aberrations, given the role of the autoinhibitory C-terminal region in preventing self- or abnormal interactions with other ESCRT components [212]. Consistent with this explanation, truncated CHMP2BIntron5 in cells is avidly associated with mSnf7-2, a key constituent of ESCRT-III, and together they form abnormal complexes that prevent ESCRT-III dissociation from the endosomal membranes and downstream autolysosomal fusion [184,210]. In addition, in cell models, CHMP2BIntron5 displays impaired endosomal recruitment of Rab7, a GTPase required for endosome-lysosome fusion [213]. Similarly, another C-terminal truncation (ALS-associated CHMP2BT104N), accumulates in Rab5- and Rab7- positive endosomes, while also closely complexing with mSnf7-2 in cortical neurons, indicating defective endocytic trafficking [212]. There is also evidence suggesting a more upstream consequence of CHMP2B mutations in autophagy, with experiments in a Drosophila model indicating that CHMP2BIntron5 inhibits autophagosome maturation by repressing binding interactions between Vti1a and STX13, an endosomal SNARE involved in the maturation of phagophores into closed autophagosomes [214]. Thus, C-terminal CHMP2B truncations appear to exert deleterious effects that disrupt various steps in autophagy, primarily at the late-stage endosome-lysosome fusion. Further analysis is needed to decipher the role this disruption may play in ALS pathogenesis.
Dynactin
Dynactin (DCTN) is a protein found in complex with molecular motor dynein, both of which are required for retrograde transport of autophagic substrates. Mutations of DCTN are infrequent in fALS, but estimated to vary from 0.5–3% in different sALS cohorts [215]. Hinting at the involvement of dynactin mutations in the motor neuron disease, a G59S mutation in the largest subunit of dynactin, p150Glued, has been detected in patients with lower MN pathology, with patient neural tissue displaying abnormal dynein and dynactin inclusions and significant MN degeneration [18,216]. Additional ALS-associated DCTN mutations include T1249I and M751T [217].
Neuropathological features of dynactin mutations have been associated with autophagic impairment resulting from repressed retrograde transport. The G59S mutation of p150Glued inhibits dynein-dynactin-mediated retrograde transport by reducing the affinity of p150Glued for microtubules and microtubule plus-end binding protein (EB1) in transfected MNs, thereby decreasing the activity of the dynein complex. This effect results in G59S aggregation, perhaps as the result of insufficient molecular chaperone recruitment [218]. Further expanding these findings, a study of G59S transgenic mice revealed that these mice exhibit dysfunctional intracellular vesicular trafficking, since G59S p150Glued impedes the targeting of cargo to its intended destination and causes inclusion formation (specifically, autophagosomes enriched in LC3-II and ubiquitin) [219]. Also supporting the role of dynactin in autophagy, DCTN knockdown in C. elegans markedly decreased the transport of autophagosomes and significantly reduced both the speed and distance of retrograde transport. These effects induce the accumulation of immature autophagosomes and subsequent MN degeneration [220], modeling pathological features seen after DCTN silencing in ALS patients [220,221].
Interestingly, studies employing various models have suggested that the autophagy-related pathological features of dynactin mutations may be ameliorated by autophagic enhancement. In G59S p150Glued transgenic mice, consumption of a low-protein diet largely alleviates MN pathology, as a result of diet-induced autophagic induction and balancing of autophagic flux, indicated by normalized levels of LC3-II, decreased immature autophagosome formation and reduced p62-positive inclusions [222]. Similarly, the autophagic inducer rapamycin attenuated neurodegeneration and locomotor dysfunction in cultured MNs [220]. Taken together, these data suggest that autophagic modulators may compensate for retrograde transport deficits brought on by dynactin mutations.
Profilin 1
Profilin 1 (PFN1) is an actin-binding protein that facilitates the binding and conversion of actin-ADP monomers into actin-ATP, enabling its addition to growing actin filaments. PFN1 also binds to phosphoinositides and polyproline-rich ligands [223]. Collectively, these binding interactions underscore PFN1’s role in regulating cytoskeletal dynamics, membrane trafficking, and neuronal synaptic activity [224]. Emerging evidence has revealed a link between mutations of PFN1 and ALS, with whole-exome sequencing of individuals with fALS uncovering C71G, M114T, and G118V mutants of PFN1 [19]. Since then, other ALS-related pathogenic variants (R136W, T109M, A20T, Q139L) of PFN1 have been found [225–227]. Overall, mutations of PFN1 were found in 1–2% of fALS patients. In addition, they were also identified in less than 1% of FTD cases from an extended Flanders-Belgian cohort [194,228]. Various models assessing the effects of mutant PFN1 have revealed interactions of PFN1 with autophagic substrates, raising the possibility that autophagic dysregulations may be at play in mutant PFN1-related neuropathology. In transfected cells and primary mouse neurons, mutant PFN1 aggregates are reported to be positive for p62, ubiquitin, and TDP-43 [19]. Also, autophagy adaptor LC3 is sequestered in PFN1 aggregates, repressing the conversion of LC3-I to LC3-II in a potentially p62-dependent manner [229]. In transgenic mice expressing the PFN1-C71G mutant, which has been shown to possess a highly disordered non-native helical conformation [230], severe MN loss and elevated p62 levels are observed [231]. Although these studies based on overexpression models suggest a gain-of-function mechanism, the involvement of loss-of-function in disease, as suggested by the observation that mutant PFN1 displays lower levels of bound actin and inhibited axonal outgrowth would also be possible [19].
FIG4
Factor-induced gene 4 (FIG4) encodes the protein Fig4, the phosphatase that modifies phosphatidylinositol-3,5,-bisphosphate (PI(3,5)P2), thereby regulating the level of this important signaling lipid molecule of the endosomal pathway. Located on the cytosolic side of the endosomal membrane, (PI(3,5)P2) mediates retrograde trafficking of endosomes to the Golgi apparatus [232,233]. Mutations in FIG4 have been identified in 1–3% of ALS patients from central Europe, thereby qualifying as contributing factors for ALS [20,234]. These mutations are often deleterious, causing loss-of-function of the protein. Loss of Fig4 function is known to cause dysfunction in the activity of the PI(3,5)P2 biosynthesis complex, thereby resulting in a decrease in PI(3,5)P2 levels [235,236]. PI(3,5)P2 depletion, in turn, results in enlarged endosomal vesicles and defects in the endo-lysosomal pathway [235,237,238].
There have been reports of Fig4 playing a regulatory role in the autophagy–lysosomal pathway. In Drosophila models, Fig4 ablation induced lysosomal enlargement and abnormal level of lysosomal markers, resulting in a shorter lifespan and mobility defects [206]. The effect of Fig4 depletion on autophagic clearance has been further substantiated in a mouse model, in which p62/SQSTM1 and LC3-II were observed to accumulate in neurons and astrocytes of FIG4-knockout mice. A defect in autolysosome clearance was also suggested by an abnormal accumulation of LAMP-2-positive vesicles in cultured FIG4-knockout neurons and astrocytes [239]. Together with other endosome-associated proteins such as CHMP2B and alsin, the linkage of Fig4 to ALS further underscores the importance of endocytosis and autophagy in neurodegeneration.
Perspective
The common theme of autophagy defects presents an opportunity to develop a broad-spectrum strategy that could be effective for many forms of ALS (Table 1). Here, we have reviewed the main evidence linking autophagy to ALS suggesting that autophagy is a stress response whose balance may be a key to pathogenesis and treatment. Elucidating a clear connection between apparent dysregulations in autophagy and ALS pathophysiology presents numerous challenges, given that ALS is governed by a wide array of causative genes. ALS-related mutations within a subset of autophagic genes deleteriously affect different junctures of autophagy, encompassing autophagic initiation, autophagosome maturation and transport, endosome trafficking, and autophagosome-lysosome fusion. Autophagy destabilization likewise can arise from mutations in genes not canonically involved in autophagy, by way of diminished autophagic gene expression, retrograde transport impairment, or aggregation-induced overburdening of autophagic clearance. Indeed, the precise sequence of events resulting in autophagic failure depends largely on the particular context of the disease state. Both excessive and inadequate autophagic flux have been connected to ALS pathology, and autophagy may either alleviate or exacerbate the disease process at different stages. Consequently, accurately defining the nature of autophagic defects and the cell’s response to these alterations becomes increasingly important because it enables an integrated understanding of the autophagic dysregulation and proteostatic imbalance observed in ALS. Therefore, further studies are needed to provide a systematic analysis of the range of genetic players involved in autophagy and associated with ALS. These efforts may not only resolve current gaps in the understanding of autophagy in ALS but also suggest mechanism-based therapeutic measures.
Highlights.
>Many ALS-causing genetic factors share a common link to autophagy
>Diverse mechanisms in ALS impair or overactivate autophagy
>Autophagic activation or repression has variable effects in ALS models
>The nodes of autophagy amenable to therapeutic intervention remain uncertain
Acknowledgments
We are grateful to Ernest Wang and Elizabeth Alexander for their contributions in the preparation of the manuscript. This work was supported by NIH (NS074324, NS089616), the Robert Packard Center for ALS Research at Johns Hopkins, the ALS Association, The Muscular Dystrophy Association, Department of Defense, and The Judith & Jean Page Adams Chritable Foundation.
Footnotes
Author Contributions
D.N., R.T., and J.W. wrote the paper. D.N. and R.T. designed the figure with suggestions from J.W.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng H-X, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung W-Y, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak–Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar Ha, Trojanowski JQ, Lee VM-Y. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and–Amyotrophic Lateral Sclerosis. Science (80- ) 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 3.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science (80- ) 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwiatkowski TJ, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH. Mutations in the FUS/TLS Gene on Chromosome 16 Cause Familial Amyotrophic Lateral Sclerosis. Science (80- ) 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 5.Fecto F, Siddique T. UBQLN2/P62 cellular recycling pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Muscle and Nerve. 2012;45:157–162. doi: 10.1002/mus.23278. [DOI] [PubMed] [Google Scholar]
- 6.Teyssou E, Takeda T, Lebon V, Boillée S, Doukouré B, Bataillon G, Sazdovitch V, Cazeneuve C, Meininger V, Leguern E, Salachas F, Seilhean D, Millecamps S. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: Genetics and neuropathology. Acta Neuropathol. 2013;125:511–522. doi: 10.1007/s00401-013-1090-0. [DOI] [PubMed] [Google Scholar]
- 7.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 8.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu Y-F, Wang Q, Krueger BJ, Ren Z, Keebler J, Han Y, Levy SE, Boone BE, Wimbish JR, Waite LL, Jones AL, Carulli JP, Day-Williams AG, Staropoli JF, Xin WW, Chesi A, Raphael AR, McKenna-Yasek D, Cady J, Vianney de Jong JMB, Kenna KP, Smith BN, Topp S, Miller J, Gkazi A, Al-Chalabi A, van den Berg LH, Veldink J, Silani V, Ticozzi N, Shaw CE, Baloh RH, Appel S, Simpson E, Lagier-Tourenne C, Pulst SM, Gibson S, Trojanowski JQ, Elman L, McCluskey L, Grossman M, Shneider NA, Chung WK, Ravits JM, Glass JD, Sims KB, Van Deerlin VM, Maniatis T, Hayes SD, Ordureau A, Swarup S, Landers J, Baas F, Allen AS, Bedlack RS, Harper JW, Gitler AD, Rouleau GA, Brown R, Harms MB, Cooper GM, Harris T, Myers RM, Goldstein DB. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–41. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H-J, Anagnostou G, Chai A, Withers J, Morris A, Adhikaree J, Pennetta G, de Belleroche JS. Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J Biol Chem. 2010;285:40266–81. doi: 10.1074/jbc.M110.161398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura AL, Mitne-Neto M, Silva HCA, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JRM, Gillingwater T, Webb J, Skehel P, Zatz M. A Mutation in the Vesicle-Trafficking Protein VAPB Causes Late-Onset Spinal Muscular Atrophy and Amyotrophic Lateral Sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurrò MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chiò A, Traynor BJ. Exome Sequencing Reveals VCP Mutations as a Cause of Familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz Da, Kwiatkowski T, Hosler Ba, Sagie T, Skaug J, Nasir J, Brown RH, Scherer SW, Rouleau Ga, Hayden MR, Ikeda JE. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 15.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, Brandner S, Brun A, Rossor MN, Gade A, Johannsen P, Sørensen SA, Gydesen S, Fisher EM, Collinge J. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, Andersen PM, Morrison KE, Pall HS, Hardiman O, Collinge J, Shaw PJ, Fisher EMC. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B) Neurology. 2006;67:1074–1077. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- 17.Cox LE, Ferraiuolo L, Goodall EF, Heath PR, Higginbottom A, Mortiboys H, Hollinger HC, Hartley JA, Brockington A, Burness CE, Morrison KE, Wharton SB, Grierson AJ, Ince PG, Kirby J, Shaw PJ. Mutations in CHMP2B in Lower Motor Neuron Predominant Amyotrophic Lateral Sclerosis (ALS) PLoS One. 2010;5:e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 19.Wu C-H, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, Kost JE, Gonzalez-Perez P, Fox AD, Adams J, Taroni F, Tiloca C, Leclerc AL, Chafe SC, Mangroo D, Moore MJ, Zitzewitz JA, Xu Z-S, van den Berg LH, Glass JD, Siciliano G, Cirulli ET, Goldstein DB, Salachas F, Meininger V, Rossoll W, Ratti A, Gellera C, Bosco DA, Bassell GJ, Silani V, Drory VE, Brown RH, Jr, Landers JE. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, Figlewicz D, Brown RH, Meisler MH. Deleterious Variants of FIG4, a Phosphoinositide Phosphatase, in Patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NCA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GYR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Langenhove T, van der Zee J, van Broeckhoven C. The molecular basis of the frontotemporal lobar degeneration amyotrophic lateral sclerosis spectrum. Ann Med. 2012;44:817–828. doi: 10.3109/07853890.2012.665471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arvanitakis Z. Update on frontotemporal dementia. Neurologist. 2010;16:16–22. doi: 10.1097/NRL.0b013e3181b1d5c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters OM, Ghasemi M, Brown RH. Emerging mechanisms of molecular pathology in ALS. J Clin Invest. 2015;125:1767–1779. doi: 10.1172/JCI71601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosco DA, Morfini G, Karabacak NM, Song Y, Gros-Louis F, Pasinelli P, Goolsby H, Fontaine BA, Lemay N, McKenna-Yasek D, Frosch MP, Agar JN, Julien J-P, Brady ST, Brown RH. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Sayana P, Zhang X, Le W. Genetics of amyotrophic lateral sclerosis: an update. Mol Neurodegener. 2013;8:28. doi: 10.1186/1750-1326-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur SJ, McKeown SR, Rashid S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene. 2016;577:109–118. doi: 10.1016/j.gene.2015.11.049. [DOI] [PubMed] [Google Scholar]
- 29.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 30.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 31.Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 32.Gal J, Ström A-L, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW, Zhu H. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian F, Morimoto N, Liu W, Ohta Y, Deguchi K, Miyazaki K, Abe K. In vivo optical imaging of motor neuron autophagy in a mouse model of amyotrophic lateral sclerosis. Autophagy. 2014;7:985–992. doi: 10.4161/auto.7.9.16012. [DOI] [PubMed] [Google Scholar]
- 34.Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bandyopadhyay U, Nagy M, Fenton WA, Horwich AL. Absence of lipofuscin in motor neurons of SOD1-linked ALS mice. Proc Natl Acad Sci. 2014;111:11055–11060. doi: 10.1073/pnas.1409314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y. Autophagic induction of amyotrophic lateral sclerosislinked Cu/Zn superoxide dismutase 1 G93A mutant in NSC34 cells. Neural Regen Res. 2014;9:16–24. doi: 10.4103/1673-5374.125325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, Murakami T, Takehisa Y, Ikeda Y, Kamiya T, Abe K. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 39.Li A, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4:290–293. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- 40.An T, Shi P, Duan W, Zhang S, Yuan P, Li Z, Wu D, Xu Z, Li C, Guo Y. Oxidative stress and autophagic alteration in brainstem of SOD1-G93A mouse model of ALS. Mol Neurobiol. 2014;49:1435–1448. doi: 10.1007/s12035-013-8623-3. [DOI] [PubMed] [Google Scholar]
- 41.Settembre C, Ballabio A. TFEB regulates autophagy: An integrated coordination of cellular degradation and recycling processes. Autophagy. 2011;7:1379–1381. doi: 10.4161/auto.7.11.17166. [DOI] [PubMed] [Google Scholar]
- 42.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Liu H, Guan Y, Wang Q, Zhou F, Jie L, Ju J, Pu L, Du H, Wang X. The altered autophagy mediated by TFEB in animal and cell models of amyotrophic lateral sclerosis. Am J Transl Res. 2015;7:1574–1587. [PMC free article] [PubMed] [Google Scholar]
- 44.Katsumata K, Nishiyama J, Inoue T, Mizushima N, Takeda J, Yuzaki M. Dynein- and activity-dependent retrograde transport of autophagosomes in neuronal axons. Autophagy. 2010;6:378–385. doi: 10.4161/auto.6.3.11262. [DOI] [PubMed] [Google Scholar]
- 45.Johnston JA, Illing ME, Kopito RR. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton. 2002;53:26–38. doi: 10.1002/cm.10057. [DOI] [PubMed] [Google Scholar]
- 46.Ström A-L, Gal J, Shi P, Kasarskis EJ, Hayward LJ, Zhu H. Retrograde axonal transport and motor neuron disease. J Neurochem. 2008;106:495–505. doi: 10.1111/j.1471-4159.2008.05393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F, Strom AL, Fukada K, Lee S, Hayward LJ, Zhu H. Interaction between familial Amyotrophic Lateral Sclerosis (ALS)-linked SOD1 mutants and the dynein complex. J Biol Chem. 2007;282:16691–16699. doi: 10.1074/jbc.M609743200. [DOI] [PubMed] [Google Scholar]
- 48.Bilsland LG, Sahai E, Kelly G, Golding M, Greensmith L, Schiavo G. Deficits in axonal transport precede ALS symptoms in vivo. Proc Natl Acad Sci. 2010;107:20523–20528. doi: 10.1073/pnas.1006869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie Y, Zhou B, Lin MY, Wang S, Foust KD, Sheng ZH. Endolysosomal Deficits Augment Mitochondria Pathology in Spinal Motor Neurons of Asymptomatic fALS Mice. Neuron. 2015;87:355–371. doi: 10.1016/j.neuron.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 51.Feng HL, Leng Y, Ma CH, Zhang J, Ren M, Chuang DM. Combined lithium and valproate treatment delays disease onset, reduces neurological deficits and prolongs survival in an amyotrophic lateral sclerosis mouse model. Neuroscience. 2008;155:567–572. doi: 10.1016/j.neuroscience.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML, Lazzeri G, Spalloni A, Bellio N, Lenzi P, Modugno N, Siciliano G, Isidoro C, Murri L, Ruggieri S, Paparelli A. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2008;105:2052–7. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pizzasegola C, Caron I, Daleno C, Ronchi A, Minoia C, Carrì MT, Bendotti C. Treatment with lithium carbonate does not improve disease progression in two different strains of SOD1 mutant mice. Amyotroph Lateral Scler. 2009;10:221–228. doi: 10.1080/17482960902803440. [DOI] [PubMed] [Google Scholar]
- 54.Watanabe S, Komine O, Endo F, Wakasugi K, Yamanaka K. Intracerebroventricular administration of Cystatin C ameliorates disease in SOD1-linked amyotrophic lateral sclerosis mice. J Neurochem. 2017 doi: 10.1111/jnc.14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, Court FA, van Zundert B, Hetz C. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–20. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Chen S, Song L, Tang Y, Shen Y, Jia L, Le W. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy. 2014;10:588–602. doi: 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeong JK, Moon MH, Lee YJ, Seol JW, Park SY. Autophagy induced by the class III histone deacetylase Sirt1 prevents prion peptide neurotoxicity. Neurobiol Aging. 2013;34:146–156. doi: 10.1016/j.neurobiolaging.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Mancuso R, del Valle J, Modol L, Martinez A, Granado-Serrano AB, Ramirez-Núñez O, Pallás M, Portero-Otin M, Osta R, Navarro X. Resveratrol Improves Motoneuron Function and Extends Survival in SOD1G93A ALS Mice. Neurotherapeutics. 2014;11:419–432. doi: 10.1007/s13311-013-0253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Li L, Chen S, Yang D, Wang Y, Zhang X, Wang Z, Le W. Rapamycin treatment augments motor neuron degeneration in SOD1 G93A mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 60.Hsueh K-W, Chiou T-W, Chiang S-F, Yamashita T, Abe K, Borlongan CV, Sanberg PR, Yu H, Huang A, Lin S-Z, Harn H-J. Autophagic down-regulation in motor neurons remarkably prolongs the survival of ALS mice. Neuropharmacology. 2016;108:152–160. doi: 10.1016/j.neuropharm.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 61.Zhou QM, Zhang JJ, Li S, Chen S, Le WD. n-butylidenephthalide treatment prolongs life span and attenuates motor neuron loss in SOD1G93A mouse model of amyotrophic lateral sclerosis. CNS Neurosci Ther. 2017;23:375–385. doi: 10.1111/cns.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rudnick ND, Griffey CJ, Guarnieri P, Gerbino V, Wang X, Piersaint JA, Tapia JC, Rich MM, Maniatis T. Distinct roles for motor neuron autophagy early and late in the SOD1 G93A mouse model of ALS. Proc Natl Acad Sci. 2017;114:E8294–E8303. doi: 10.1073/pnas.1704294114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang K, Shi P, An T, Wang Q, Wang J, Li Z, Duan W, Li C, Guo Y. Food restriction-induced autophagy modulates degradation of mutant SOD1 in an amyotrophic lateral sclerosis mouse model. Brain Res. 2013;1519:112–119. doi: 10.1016/j.brainres.2013.04.050. [DOI] [PubMed] [Google Scholar]
- 64.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in als and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: The FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito D, Suzuki N. Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNA-binding proteins TDP-43 and FUS. Neurology. 2011;77:1636–1643. doi: 10.1212/WNL.0b013e3182343365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou Z-Y, Zhou Z-R, Che C-H, Liu C-Y, He R-L, Huang H-P. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2017;88:540–549. doi: 10.1136/jnnp-2016-315018. [DOI] [PubMed] [Google Scholar]
- 68.Borroni B, Archetti S, Del Bo R, Papetti A, Buratti E, Bonvicini C, Agosti C, Cosseddu M, Turla M, Di Lorenzo D, Pietro Comi G, Gennarelli M, Padovani A. TARDBP Mutations in Frontotemporal Lobar Degeneration: Frequency, Clinical Features, and Disease Course. Rejuvenation Res. 2010;13:509–517. doi: 10.1089/rej.2010.1017. [DOI] [PubMed] [Google Scholar]
- 69.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the als disease protein fus/tls. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fiesel FC, Kahle PJ. TDP-43 and FUS/TLS: Cellular functions and implications for neurodegeneration. FEBS J. 2011;278:3550–3568. doi: 10.1111/j.1742-4658.2011.08258.x. [DOI] [PubMed] [Google Scholar]
- 72.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46 R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling S-C, Liang TY, Mazur C, Wancewicz E, Kim AS, Watt A, Freier S, Hicks GG, Donohue JP, Shiue L, Bennett CF, Ravits J, Cleveland DW, Yeo GW. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 75.March ZM, King OD, Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Udan M, Baloh RH. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 2011;5:1–5. doi: 10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 78.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Monahan Z, Shewmaker F, Pandey UB. Stress granules at the intersection of autophagy and ALS. Brain Res. 2016;1649:189–200. doi: 10.1016/j.brainres.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lo Bello M, Di Fini F, Notaro A, Spataro R, Conforti FL, La Bella V. ALS-Related Mutant FUS Protein Is Mislocalized to Cytoplasm and Is Recruited into Stress Granules of Fibroblasts from Asymptomatic FUS P525L Mutation Carriers. Neurodegener Dis. 2017;17:292–303. doi: 10.1159/000480085. [DOI] [PubMed] [Google Scholar]
- 81.Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderwyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B. Tar DNA binding protein-43 (TDP-43) associates with stress granules: Analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X, Fan H, Ying Z, Li B, Wang H, Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci Lett. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 83.Ryu HH, Jun MH, Min KJ, Jang DJ, Lee YS, Kim HK, Lee JA. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging. 2014;35:2822–2831. doi: 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 84.Brady OA, Meng P, Zheng Y, Mao Y, Hu F. Regulation of TDP-43 aggregation by phosphorylation andp62/SQSTM1. J Neurochem. 2011;116:248–259. doi: 10.1111/j.1471-4159.2010.07098.x. [DOI] [PubMed] [Google Scholar]
- 85.Wang I-F, Guo B-S, Liu Y-C, Wu C-C, Yang C-H, Tsai K-J, Shen C-KJ. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci. 2012;109:15024–15029. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng C-W, Lin M-J, Shen C-KJ. Rapamycin alleviates pathogenesis of a new Drosophila model of ALS-TDP. J Neurogenet. 2015;29:59–68. doi: 10.3109/01677063.2015.1077832. [DOI] [PubMed] [Google Scholar]
- 87.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, Shaw C, Chandran S, Finkbeiner S. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stoica R, De Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau K-F, Vizcay-Barrena G, Lin W-L, Xu Y-F, Lewis J, Dickson DW, Petrucelli L, Mitchell JC, Shaw CE, Miller CCJ. ER mitochondria associations are regulated by the VAPB PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gomez-Suaga P, Paillusson S, Stoica R, Noble W, Hanger DP, Miller CCJ. The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy. Curr Biol. 2017;27:371–385. doi: 10.1016/j.cub.2016.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soo KY, Halloran M, Sundaramoorthy V, Parakh S, Toth RP, Southam KA, McLean CA, Lock P, King A, Farg MA, Atkin JD. Rab1-dependent ER Golgi transport dysfunction is a common pathogenic mechanism in SOD1, TDP-43 and FUS-associated ALS. Acta Neuropathol. 2015;130:679–697. doi: 10.1007/s00401-015-1468-2. [DOI] [PubMed] [Google Scholar]
- 91.Zoppino FCM, Militello RD, Slavin I, Álvarez C, Colombo MI. Autophagosome formation depends on the small GTPase rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 92.Szatmári Z, Sass M. The autophagic roles of Rab small GTPases and their upstream regulators: A review. Autophagy. 2014;10:1154–1166. doi: 10.4161/auto.29395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Soo KY, Sultana J, King AE, Atkinson R, Warraich ST, Sundaramoorthy V, Blair I, Farg MA, Atkin JD. ALS-associated mutant FUS inhibits macroautophagy which is restored by overexpression of Rab1. Cell Death Discov. 2015;1:15030. doi: 10.1038/cddiscovery.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science (80- ) 2015;349:650–655. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arnold ES, Ling S-C, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, Da Cruz S, Clutario KM, Swing D, Tessarollo L, Marsala M, Shaw CE, Yeo GW, Cleveland DW. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang T, Baldie G, Periz G, Wang J. RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response. PLoS Genet. 2014;10:e1004693. doi: 10.1371/journal.pgen.1004693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bose JK, Huang CC, Shen CKJ. Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem. 2011;286:44441–44448. doi: 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Imhaus A-F, Duménil G. The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J. 2014;33:1767–1783. doi: 10.15252/embj. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Majcher V, Goode A, James V, Layfield R. Autophagy receptor defects and ALS-FTLD. Mol Cell Neurosci. 2015;66:43–52. doi: 10.1016/j.mcn.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 101.Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, Donkervoort S, Ajroud-Driss S, Sufit RL, Heller SL, Deng H-X, Siddique T. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 102.Rea SL, Majcher V, Searle MS, Layfield R. SQSTM1 mutations–Bridging Paget disease of bone and ALS/FTLD. Exp Cell Res. 2014;325:27–37. doi: 10.1016/j.yexcr.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 103.Maruyama H, Kawakami H. Optineurin and amyotrophic lateral sclerosis. Geriatr Gerontol Int. 2013;13:528–32. doi: 10.1111/ggi.12022. [DOI] [PubMed] [Google Scholar]
- 104.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy [S] J Biol Chem. 2007 doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 105.Slowicka K, Vereecke L, van Loo G. Cellular Functions of Optineurin in Health and Disease. Trends Immunol. 2016;37:621–633. doi: 10.1016/j.it.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 106.Gal J, Ström A-L, Kilty R, Zhang F, Zhu H. p62 Accumulates and Enhances Aggregate Formation in Model Systems of Familial Amyotrophic Lateral Sclerosis. J Biol Chem. 2007;282:11068–11077. doi: 10.1074/jbc.M608787200. [DOI] [PubMed] [Google Scholar]
- 107.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Le Ber I, Camuzat A, Guerreiro R, Bouya-Ahmed K, Bras J, Nicolas G, Gabelle A, Didic M, De Septenville A, Millecamps S, Lenglet T, Latouche M, Kabashi E, Campion D, Hannequin D, Hardy J, Brice A French Clinical and Genetic Research Network on FTD/FTD-ALS. SQSTM1 mutations in French patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol. 2013;70:1403–10. doi: 10.1001/jamaneurol.2013.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 Is a Polyubiquitin Chain Binding Protein Involved in Ubiquitin Proteasome Degradation. Mol Cell Biol. 2004 doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005 doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 111.Ciani B, Layfield R, Cavey JR, Sheppard PW, Searle MS. Structure of the ubiquitin-associated domain of p62 (SQSTM1) and implications for mutations that cause Paget’s disease of bone. J Biol Chem. 2003;278:37409–37412. doi: 10.1074/jbc.M307416200. [DOI] [PubMed] [Google Scholar]
- 112.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a Common Component of Cytoplasmic Inclusions in Protein Aggregation Diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.King A, Al-Sarraj S, Troakes C, Smith BN, Maekawa S, Iovino M, Spillantini MG, Shaw CE. Mixed tau, TDP-43 and p62 pathology in FTLD associated with a C9ORF72 repeat expansion and p.Ala239Thr MAPT (tau) variant. Acta Neuropathol. 2013;125:303–310. doi: 10.1007/s00401-012-1050-0. [DOI] [PubMed] [Google Scholar]
- 114.Troakes C, Maekawa S, Wijesekera L, Rogelj B, Siklós L, Bell C, Smith B, Newhouse S, Vance C, Johnson L, Hortobágyi T, Shatunov A, Al-Chalabi A, Leigh N, Shaw CE, King A, Al-Sarraj S. An MND/ALS phenotype associated with C9orf72 repeat expansion: Abundant p62-positive, TDP-43-negative inclusions in cerebral cortex, hippocampus and cerebellum but without associated cognitive decline. Neuropathology. 2012;32:505–514. doi: 10.1111/j.1440-1789.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- 115.Andrew SA, Claire K, Bradley T, Satomi S, Shaw CE. p62 positive , TDP-43 negative , neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72 -linked FTLD and MND /ALS. Acta Neuropathol. 2011:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 116.Goode A, Butler K, Long J, Cavey J, Scott D, Shaw B, Sollenberger J, Gell C, Johansen T, Oldham NJ, Searle MS, Layfield R. Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy. 2016;12:1094–1104. doi: 10.1080/15548627.2016.1170257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rubino E, Rainero I, Chio A, Rogaeva E, Galimberti D, Fenoglio P, Grinberg Y, Isaia G, Calvo A, Gentile S, Bruni AC, St George-Hyslop PH, Scarpini E, Gallone S, Pinessi L. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Du Y, Wooten MC, Wooten MW. Oxidative damage to the promoter region of SQSTM1/p62 is common to neurodegenerative disease. Neurobiol Dis. 2009;35:302–310. doi: 10.1016/j.nbd.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hadano S, Mitsui S, Pan L, Otomo A, Kubo M, Sato K, Ono S, Onodera W, Abe K, Chen XP, Koike M, Uchiyama Y, Aoki M, Warabi E, Yamamoto M, Ishii T, Yanagawa T, Shang HF, Yoshii F. Functional links between SQSTM1 and ALS2 in the pathogenesis of ALS: Cumulative impact on the protection against mutant SOD1-mediated motor dysfunction in mice. Hum Mol Genet. 2015;25:3321–3340. doi: 10.1093/hmg/ddw180. [DOI] [PubMed] [Google Scholar]
- 120.Kachaner D, Génin P, Laplantine E, Weil R. Toward an integrative view of Optineurin functions. Cell Cycle. 2012;11:2808–2818. doi: 10.4161/cc.20946. [DOI] [PubMed] [Google Scholar]
- 121.Weishaupt JH, Waibel S, Birve A, Volk AE, Mayer B, Meyer T, Ludolph AC, Andersen PM. A novel optineurin truncating mutation and three glaucoma-associated missense variants in patients with familial amyotrophic lateral sclerosis in Germany. Neurobiol Aging. 2013;34 doi: 10.1016/j.neurobiolaging.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 122.Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013;126:580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 124.Shen WC, Li HY, Chen GC, Chern Y, Tu PH. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy- mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy. 2015;11:685–700. doi: 10.4161/auto.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kruppa AJ, Kendrick-Jones J, Buss F. Myosins, Actin and Autophagy. Traffic. 2016;17:878–890. doi: 10.1111/tra.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sundaramoorthy V, Walker AK, Tan V, Fifita JA, McCann EP, Williams KL, Blair IP, Guillemin GJ, Farg MA, Atkin JD. Defects in optineurin- and myosin VI-mediated cellular trafficking in amyotrophic lateral sclerosis. Hum Mol Genet. 2015;24:3830–3846. doi: 10.1093/hmg/ddv126. [DOI] [PubMed] [Google Scholar]
- 127.Song GJ, Jeon H, Seo M, Jo M, Suk K. Interaction between optineurin and Rab1a regulates autophagosome formation in neuroblastoma cells. J Neurosci Res. 2017 doi: 10.1002/jnr.24143. [DOI] [PubMed] [Google Scholar]
- 128.Osaka M, Ito D, Yagi T, Nihei Y, Suzuki N. Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum Mol Genet. 2015;24:1617–1629. doi: 10.1093/hmg/ddu575. [DOI] [PubMed] [Google Scholar]
- 129.Pedros C, Zhang Y, Hu JK, Choi YS, Canonigo-Balancio AJ, Yates JR, Altman A, Crotty S, Kong KF. A TRAF-like motif of the inducible costimulator ICOS controls development of germinal center TFHcells via the kinase TBK1. Nat Immunol. 2016;17:825–833. doi: 10.1038/ni.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang Q, Meng F, Chen S, Plouffe SW, Wu S, Liu S, Li X, Zhou R, Wang J, Zhao B, Liu J, Qin J, Zou J, Feng XH, Guan KL, Xu P. Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat Cell Biol. 2017;19:362–374. doi: 10.1038/ncb3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oakes JA, Davies MC, Collins MO. TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol Brain. 2017;10:5. doi: 10.1186/s13041-017-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the Autophagy Receptor Optineurin Restricts Salmonella Growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun JA, Hansen TE, Johansen T, Deretic V. TBK-1 Promotes Autophagy-Mediated Antimicrobial Defense by Controlling Autophagosome Maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, Marroquin N, Nordin F, Hübers A, Weydt P, Pinto S, Press R, Millecamps S, Molko N, Bernard E, Desnuelle C, Soriani M-H, Dorst J, Graf E, Nordström U, Feiler MS, Putz S, Boeckers TM, Meyer T, Winkler AS, Winkelman J, de Carvalho M, Thal DR, Otto M, Brännström T, Volk AE, Kursula P, Danzer KM, Lichtner P, Dikic I, Meitinger T, Ludolph AC, Strom TM, Andersen PM, Weishaupt JH. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 135.Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Philtjens S, Heeman B, Engelborghs S, Vandenbulcke M, De Baets G, Bäumer V, Cuijt I, Van den Broeck M, Peeters K, Mattheijssens M, Rousseau F, Vandenberghe R, De Jonghe P, Cras P, De Deyn PP, Martin J-J, Cruts M, Van Broeckhoven C BELNEU Consortium. Loss of TBK1 is a frequent cause of frontotemporal dementia in a Belgian cohort. Neurology. 2015;85:2116–25. doi: 10.1212/WNL.0000000000002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Le Ber I, De Septenville A, Millecamps S, Camuzat A, Caroppo P, Couratier P, Blanc F, Lacomblez L, Sellal F, Fleury MC, Meininger V, Cazeneuve C, Clot F, Flabeau O, LeGuern E, Brice A, Didic M, Dubois B, Golfier V, Hannequin D, Levy R, Michel BF, Pasquier F, Thomas-Anterion C, Puel M, Salachas F, Vercelletto M. TBK1 mutation frequencies in French frontotemporal dementia and amyotrophic lateral sclerosis cohorts. Neurobiol Aging. 2015;36:3116.e5–3116.e8. doi: 10.1016/j.neurobiolaging.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 137.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, Chen J-F. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2016;2:e1601167–e1601167. doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, Brady NR, Hamacher-Brady A, Wang J. Loss of C9orf72 Enhances Autophagic Activity via Deregulated mTOR and TFEB Signaling. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Amick J, Roczniak-Ferguson A, Ferguson SM. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell. 2016;27:3040–3051. doi: 10.1091/mbc.E16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sellier C, Campanari M, Julie Corbier C, Gaucherot A, Kolb−Cheynel I, Oulad−Abdelghani M, Ruffenach F, Page A, Ciura S, Kabashi E, Charlet−Berguerand N. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin−2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35:1276–1297. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sirohi K, Kumari A, Radha V, Swarup G. A Glaucoma-Associated variant of optineurin, M98K, activates Tbk1 to enhance autophagosome formation and retinal cell death dependent on Ser177 phosphorylation of optineurin. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pillai S, Nguyen J, Johnson J, Haura E, Coppola D, Chellappan S. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat Commun. 2015;6:10072. doi: 10.1038/ncomms10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ, Dikic I. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Millecamps S, Salachas F, Cazeneuve C, Gordon P, Bricka B, Camuzat A, Guillot-Noël L, Russaouen O, Bruneteau G, Pradat P-F, Le Forestier N, Vandenberghe N, Danel-Brunaud V, Guy N, Thauvin-Robinet C, Lacomblez L, Couratier P, Hannequin D, Seilhean D, Le Ber I, Corcia P, Camu W, Brice A, Rouleau G, LeGuern E, Meininger V. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype-phenotype correlations. J Med Genet. 2010;47:554–60. doi: 10.1136/jmg.2010.077180. [DOI] [PubMed] [Google Scholar]
- 146.Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: What do we really know? Nat Rev Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 147.Aliaga L, Lai C, Yu J, Chub N, Shim H, Sun L, Xie C, Yang WJ, Lin X, O’Donovan MJ, Cai H. Amyotrophic lateral sclerosis-related VAPB P56S mutation differentially affects the function and survival of corticospinal and spinal motor neurons. Hum Mol Genet. 2013;22:4293–4305. doi: 10.1093/hmg/ddt279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER–mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 149.Hantan D, Yamamoto Y, Sakisaka T. VAP-B binds to Rab3GAP1 at the ER: Its implication in nuclear envelope formation through the ER-Golgi intermediate compartment. Kobe J Med Sci. 2014;60:E48–E56. [PubMed] [Google Scholar]
- 150.Spang N, Feldmann A, Huesmann H, Bekbulat F, Schmitt V, Hiebel C, Koziollek-Drechsler I, Clement AM, Moosmann B, Jung J, Behrends C, Dikic I, Kern A, Behl C. RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy. Autophagy. 2014;10:2297–2309. doi: 10.4161/15548627.2014.994359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fasana E, Fossati M, Ruggiano A, Brambillasca S, Hoogenraad CC, Navone F, Francolini M, Borgese N. A VAPB mutant linked to amyotrophic lateral sclerosis generates a novel form of organized smooth endoplasmic reticulum. FASEB J. 2010;24:1419–1430. doi: 10.1096/fj.09-147850. [DOI] [PubMed] [Google Scholar]
- 152.Kuijpers M, van Dis V, Haasdijk ED, Harterink M, Vocking K, Post Ja, Scheper W, Hoogenraad CC, Jaarsma D. Amyotrophic lateral sclerosis (ALS)-associated VAPB-P56S inclusions represent an ER quality control compartment. Acta Neuropathol Commun. 2013;1:24. doi: 10.1186/2051-5960-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Larroquette F, Seto L, Gaub PL, Kamal B, Wallis D, Larivière R, Vallée J, Robitaille R, Tsuda H. Vapb/Amyotrophic lateral sclerosis 8 knock-in mice display slowly progressive motor behavior defects accompanying ER stress and autophagic response. Hum Mol Genet. 2015;24:6515–6529. doi: 10.1093/hmg/ddv360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kakizuka A. Roles of VCP in human neurodegenerative disorders. Biochem Soc Trans. 2008;36:105–108. doi: 10.1042/BST0360105. [DOI] [PubMed] [Google Scholar]
- 155.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central Pore Residues Mediate the p97/VCP Activity Required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 156.Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Watts GDG, Thomasova D, Ramdeen SK, Fulchiero EC, Mehta SG, Drachman DA, Weihl CC, Jamrozik Z, Kwiecinski H, Kaminska A, Kimonis VE. Novel VCP mutations in inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Clin Genet. 2007;72:420–426. doi: 10.1111/j.1399-0004.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- 159.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2013;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Custer SK, Neumann M, Lu H, Wright AC, Taylor JP. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum Mol Genet. 2010;19:1741–1755. doi: 10.1093/hmg/ddq050. [DOI] [PubMed] [Google Scholar]
- 161.Nalbandian A, Llewellyn KJ, Kitazawa M, Yin HZ, Badadani M, Khanlou N, Edwards R, Nguyen C, Mukherjee J, Mozaffar T, Watts G, Weiss J, Kimonis VE. The Homozygote VCPR155H/R155H Mouse Model Exhibits Accelerated Human VCP-Associated Disease Pathology. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yeo BK, Hong CJ, Chung KM, Woo H, Kim K, Jung S, Kim E-K, Yu S-W. Valosin-containing protein is a key mediator between autophagic cell death and apoptosis in adult hippocampal neural stem cells following insulin withdrawal. Mol Brain. 2016;9:31. doi: 10.1186/s13041-016-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ching JK, Weihl CC. Rapamycin-induced autophagy aggravates pathology and weakness in a mouse model of VCP-associated myopathy. Autophagy. 2013;9:799–800. doi: 10.4161/auto.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fong JC, Karydas AM, Goldman JS. Genetic counseling for FTD/ALS caused by the C9ORF72 hexanucleotide expansion. Alzheimers Res Ther. 2012;4:27. doi: 10.1186/alzrt130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, Boylan K, Patel TC, Dickson DW, Petrucelli L. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD. RNA Toxicity from the ALS/FTD C9ORF72 Expansion Is Mitigated by Antisense Intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Haeusler AR, Donnelly CJ, Periz G, Simko EAJ, Shaw PG, Kim M-S, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, Van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, Cleverley K, Nicoll AJ, Pickering-Brown S, Dols J, Cabecinha M, Hendrich O, Fratta P, Fisher EMC, Partridge L, Isaacs AM. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science (80- ) 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, Xu G, Wang G, Ying Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet. 2015;24:2426–2441. doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- 172.Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, Seeley WW, Boxer AL, Petrucelli L, Miller BL, Gao FB. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu S-L, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, Rothstein JD. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, Sun S, Herdy JR, Bieri G, Kramer NJ, Gage FH, Van Den Bosch L, Robberecht W, Gitler AD. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee K-H, Badders N, Valentine M, Miller BL, Wong PC, Petrucelli L, Kim HJ, Gao F-B, Taylor JP. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Waite AJ, Bäumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35:1779.e5–1779.e13. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang D, Iyer LM, He F, Aravind L. Discovery of novel DENN proteins: Implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 2012;3 doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Marat AL, Dokainish H, McPherson PS. DENN domain proteins: Regulators of Rab GTPases. J Biol Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RAK, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, King AE, Atkin JD. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Aoki Y, Manzano R, Lee Y, Dafinca R, Aoki M, Douglas AGL, Varela MA, Sathyaprakash C, Scaber J, Barbagallo P, Vader I, Mäger P, Ezzat K, Turner MR, Ito N, Gasco S, Ohbayashi N, El Andaloussi S, Takeda S, Fukuda M, Talbot K, Wood MJA. C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain. 2017;140:887–897. doi: 10.1093/brain/awx024. [DOI] [PubMed] [Google Scholar]
- 182.Sivadasan R, Hornburg D, Drepper C, Frank N, Jablonka S, Hansel A, Lojewski X, Sterneckert J, Hermann A, Shaw PJ, Ince PG, Mann M, Meissner F, Sendtner M. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat Neurosci. 2016;19:1610–1618. doi: 10.1038/nn.4407. [DOI] [PubMed] [Google Scholar]
- 183.Aguilera MO, Berón W, Colombo MI. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy. 2012;8:1590–1603. doi: 10.4161/auto.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee J-Y, Koga H, Kawaguchi Y, Tang W, Wong E, Gao Y-S, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao T-P. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhuo C, Ji Y, Chen Z, Kitazato K, Xiang Y, Zhong M, Wang Q, Pei Y, Ju H, Wang Y. Proteomics analysis of autophagy-deficient Atg7−/− MEFs reveals a close relationship between F-actin and autophagy. Biochem Biophys Res Commun. 2013;437:482–488. doi: 10.1016/j.bbrc.2013.06.111. [DOI] [PubMed] [Google Scholar]
- 186.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, Hu F. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4:51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, Myszczynska MA, Higginbottom A, Walsh MJ, Whitworth AJ, Kaspar BK, Meyer K, Shaw PJ, Grierson AJ, De Vos KJ. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016;35:1656–1676. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jung J, Nayak A, Schaeffer V, Starzetz T, Kirsch AK, Müller S, Dikic I, Mittelbronn M, Behrends C. Multiplex image-based autophagy RNAi screening identifies SMCR8 as ULK1 kinase activity and gene expression regulator. Elife. 2017;6 doi: 10.7554/eLife.23063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.ORourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AKMG, Ho R, Carmona S, Vit JP, Zarrow J, Kim KJ, Bell S, Harms MB, Miller TM, Dangler CA, Underhill DM, Goodridge HS, Lutz CM, Baloh RH. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.N’Diaye E-N, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Daoud H, Suhail H, Szuto A, Camu W, Salachas F, Meininger V, Bouchard JP, Dupr?? N, Dion PA, Rouleau GA. UBQLN2 mutations are rare in French and French-Canadian amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33 doi: 10.1016/j.neurobiolaging.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 193.Millecamps S, Corcia P, Cazeneuve C, Boillée S, Seilhean D, Danel-Brunaud V, Vandenberghe N, Pradat PF, Le Forestier N, Lacomblez L, Bruneteau G, Camu W, Brice A, Meininger V, LeGuern E, Salachas F. Mutations in UBQLN2 are rare in French amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:839.e1–839.e3. doi: 10.1016/j.neurobiolaging.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 194.Dillen L, Van Langenhove T, Engelborghs S, Vandenbulcke M, Sarafov S, Tournev I, Merlin C, Cras P, Vandenberghe R, De Deyn PP, Jordanova A, Cruts M, Van Broeckhoven C, van der Zee J. Explorative genetic study of UBQLN2 and PFN1 in an extended Flanders-Belgian cohort of frontotemporal lobar degeneration patients. Neurobiol Aging. 2017;34:1711.e1–1711.e5. doi: 10.1016/j.neurobiolaging.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 195.Synofzik M, Maetzler W, Grehl T, Prudlo J, vom Hagen JM, Haack T, Rebassoo P, Munz M, Schöls L, Biskup S. Screening in ALS and FTD patients reveals 3 novel UBQLN2 mutations outside the PXX domain and a pure FTD phenotype. Neurobiol Aging. 2012;33 doi: 10.1016/j.neurobiolaging.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 196.Osaka M, Ito D, Suzuki N. Disturbance of proteasomal and autophagic protein degradation pathways by amyotrophic lateral sclerosis-linked mutations in ubiquilin 2. Biochem Biophys Res Commun. 2016;472:324–331. doi: 10.1016/j.bbrc.2016.02.107. [DOI] [PubMed] [Google Scholar]
- 197.Wu Q, Liu M, Huang C, Liu X, Huang B, Li N, Zhou H, Xia XG. Pathogenic Ubqln2 gains toxic properties to induce neuron death. Acta Neuropathol. 2014;129:417–428. doi: 10.1007/s00401-014-1367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Razi M, Chan EYW, Tooze SA. Early endosomes and endosomal coatomer are required for Autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Huang B, Wu Q, Zhou H, Huang C, Xia XG. Increased Ubqln2 expression causes neuron death in transgenic rats. J Neurochem. 2016:285–293. doi: 10.1111/jnc.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Topp JD, Gray NW, Gerard RD, Horazdovsky BF. Alsin is a Rab5 and Rac1 guanine nucleotide exchange factor. J Biol Chem. 2004;279:24612–24623. doi: 10.1074/jbc.M313504200. [DOI] [PubMed] [Google Scholar]
- 201.Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 202.Otomo A, Hadano S, Okada T, Mizumura H, Kunita R, Nishijima H, Showguchi-Miyata J, Yanagisawa Y, Kohiki E, Suga E, Yasuda M, Osuga H, Nishimoto T, Narumiya S, Ikeda JE. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12:1671–1687. doi: 10.1093/hmg/ddg184. [DOI] [PubMed] [Google Scholar]
- 203.Devon RS, Orban PC, Gerrow K, Barbieri Ma, Schwab C, Cao LP, Helm JR, Bissada N, Cruz-Aguado R, Davidson T-L, Witmer J, Metzler M, Lam CK, Tetzlaff W, Simpson EM, McCaffery JM, El-Husseini aE, Leavitt BR, Hayden MR. Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc Natl Acad Sci U S A. 2006;103:9595–600. doi: 10.1073/pnas.0510197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hadano S, Kunita R, Otomo A, Suzuki-Utsunomiya K, Ikeda JE. Molecular and cellular function of ALS2/alsin: Implication of membrane dynamics in neuronal development and degeneration. Neurochem Int. 2007;51:74–84. doi: 10.1016/j.neuint.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 205.Otomo A, Kunita R, Suzuki-Utsunomiya K, Ikeda JE, Hadano S. Defective relocalization of ALS2/alsin missense mutants to Rac1-induced macropinosomes accounts for loss of their cellular function and leads to disturbed amphisome formation. FEBS Lett. 2011;585:730–736. doi: 10.1016/j.febslet.2011.01.045. [DOI] [PubMed] [Google Scholar]
- 206.Kyotani A, Azuma Y, Yamamoto I, Yoshida H, Mizuta I, Mizuno T, Nakagawa M, Tokuda T, Yamaguchi M. Knockdown of the Drosophila FIG4 induces deficient locomotive behavior, shortening of motor neuron, axonal targeting aberration, reduction of life span and defects in eye development. Exp Neurol. 2016;277:86–95. doi: 10.1016/j.expneurol.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 207.Rusten TE, Simonsen A. ESCRT functions in autophagy and associated disease. Cell Cycle. 2008;7:1166–1172. doi: 10.4161/cc.7.9.5784. [DOI] [PubMed] [Google Scholar]
- 208.Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Hulsik DL, Belly A, Goldberg Y, Chatellard-Causse C, Blot B, Schoehn G, Weissenhorn W, Sadoul R. Charged Multivesicular Body Protein 2B (CHMP2B) of the Endosomal Sorting Complex Required for Transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EMC, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III Dysfunction Causes Autophagosome Accumulation and Neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 211.Ghazi-Noori S, Froud KE, Mizielinska S, Powell C, Smidak M, Fernandez De Marco M, O’Malley C, Farmer M, Parkinson N, Fisher EMC, Asante EA, Brandner S, Collinge J, Isaacs AM. Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain. 2012;135:819–832. doi: 10.1093/brain/aws006. [DOI] [PubMed] [Google Scholar]
- 212.Han JH, Ryu HH, Jun MH, Jang DJ, Lee JA. The functional analysis of the CHMP2B missense mutation associated with neurodegenerative diseases in the endo-lysosomal pathway. Biochem Biophys Res Commun. 2012;421:544–549. doi: 10.1016/j.bbrc.2012.04.041. [DOI] [PubMed] [Google Scholar]
- 213.Urwin H, Authier A, Nielsen JE, Metcalf D, Powell C, Froud K, Malcolm DS, Holm I, Johannsen P, Brown J, Fisher EMC, van der Zee J, Bruyland M, Van Broeckhoven C, Collinge J, Brandner S, Futter C, Isaacs AM. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19:2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lu Y, Zhang Z, Sun D, Sweeney ST, Gao FB. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol Cell. 2013;52:264–271. doi: 10.1016/j.molcel.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Liu X, Yang L, Tang L, Chen L, Liu X, Fan D. DCTN1 gene analysis in Chinese patients with sporadic amyotrophic lateral sclerosis. PLoS One. 2017;12:1–8. doi: 10.1371/journal.pone.0182572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, Kennedy WR, Wendelschafer-Crabb G, Vortmeyer A, Powers R, Finnegan K, Holzbaur ELF, Fischbeck KH, Ludlow CL. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, Ludolph AC. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.WNL.0000134608.83927.B1. [DOI] [PubMed] [Google Scholar]
- 218.Levy JR, Sumner CJ, Caviston JP, Tokito MK, Ranganathan S, Ligon LA, Wallace KE, LaMonte BH, Harmison GG, Puls I, Fischbeck KH, Holzbaur ELF. A motor neuron disease-associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. J Cell Biol. 2006;172:733–745. doi: 10.1083/jcb.200511068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Laird FM, Farah MH, Ackerley S, Hoke A, Maragakis N, Rothstein JD, Griffin J, Price DL, Martin LJ, Wong PC. Motor Neuron Disease Occurring in a Mutant Dynactin Mouse Model Is Characterized by Defects in Vesicular Trafficking. J Neurosci. 2008;28:1997–2005. doi: 10.1523/JNEUROSCI.4231-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ikenaka K, Kawai K, Katsuno M, Huang Z, Jiang Y-M, Iguchi Y, Kobayashi K, Kimata T, Waza M, Tanaka F, Mori I, Sobue G. dnc-1/dynactin 1 Knockdown Disrupts Transport of Autophagosomes and Induces Motor Neuron Degeneration. PLoS One. 2013;8:e54511. doi: 10.1371/journal.pone.0054511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jiang Y-M, Yamamoto M, Tanaka F, Ishigaki S, Katsuno M, Adachi H, Niwa J, Doyu M, Yoshida M, Hashizume Y, Sobue G. Gene Expressions Specifically Detected in Motor Neurons (Dynactin 1, Early Growth Response 3, Acetyl-CoA Transporter, Death Receptor 5, and Cyclin C) Differentially Correlate to Pathologic Markers in Sporadic Amyotrophic Lateral Sclerosis. J Neuropathol Exp Neurol. 2007;66:617–627. doi: 10.1097/nen.0b013e318093ece3. [DOI] [PubMed] [Google Scholar]
- 222.Wiesner D, Sinniger J, Henriques A, Dieterlé S, Müller HP, Rasche V, Ferger B, Dirrig-Grosch S, Soylu-Kucharz R, Petersén A, Walther P, Linkus B, Kassubek J, Wong PC, Ludolph AC, Dupuis L. Low dietary protein content alleviates motor symptoms in mice with mutant dynactin/dynein-mediated neurodegeneration. Hum Mol Genet. 2015;24:2228–2240. doi: 10.1093/hmg/ddu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gibbon BC, Zonia LE, Kovar DR, Hussey PJ, Staiger CJ. Pollen profilin function depends on interaction with proline-rich motifs. Plant Cell. 1998;10:981–993. doi: 10.1105/tpc.10.6.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 225.Chen Y, Zheng Z-Z, Huang R, Chen K, Song W, Zhao B, Chen X, Yang Y, Yuan L, Shang H-F. PFN1 mutations are rare in Han Chinese populations with amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34:1922.e1–1922.e5. doi: 10.1016/j.neurobiolaging.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 226.Ingre C, Landers JE, Rizik N, Volk AE, Akimoto C, Birve A, Hübers A, Keagle PJ, Piotrowska K, Press R, Andersen PM, Ludolph AC, Weishaupt JH. A novel phosphorylation site mutation in profilin 1 revealed in a large screen of US, Nordic, and German amyotrophic lateral sclerosis/frontotemporal dementia cohorts. Neurobiol Aging. 2013;34:1708.e1–1708.e6. doi: 10.1016/j.neurobiolaging.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Smith BN, Vance C, Scotter EL, Troakes C, Wong CH, Topp S, Maekawa S, King A, Mitchell JC, Lund K, Al-Chalabi A, Ticozzi N, Silani V, Sapp P, Brown RH, Landers JE, Al-Sarraj S, Shaw CE. Novel mutations support a role for Profilin 1 in the pathogenesis of ALS. Neurobiol Aging. 2015;36:1602.e17–e27. doi: 10.1016/j.neurobiolaging.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Daoud H, Dobrzeniecka S, Camu W, Meininger V, Dupré N, Dion PA, Rouleau GA. Mutation analysis of PFN1 in familial amyotrophic lateral sclerosis patients. Neurobiol Aging. 2013;34:1311.e1–1311.e2. doi: 10.1016/j.neurobiolaging.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 229.Tanaka Y, Nonaka T, Suzuki G, Kametani F, Hasegawa M. Gain-of-function profilin 1 mutations linked to familial amyotrophic lateral sclerosis cause seed-dependent intracellular TDP-43 aggregation. Hum Mol Genet. 2016;25:1420–1433. doi: 10.1093/hmg/ddw024. [DOI] [PubMed] [Google Scholar]
- 230.Lim L, Kang J, Song J. ALS-causing profilin-1-mutant forms a non-native helical structure in membrane environments. Biochim Biophys Acta - Biomembr. 2017;1859:2161–2170. doi: 10.1016/j.bbamem.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 231.Yang C, Danielson EW, Qiao T, Metterville J, Brown RH, Landers JE, Xu Z. Mutant PFN1 causes ALS phenotypes and progressive motor neuron degeneration in mice by a gain of toxicity. Proc Natl Acad Sci. 2016;113:E6209–E6218. doi: 10.1073/pnas.1605964113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, Shisheva A. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport: Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J Biol Chem. 2007;282:23878–23891. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- 233.Rutherford AC. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci. 2006;119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Osmanovic A, Rangnau I, Kosfeld A, Abdulla S, Janssen C, Auber B, Raab P, Preller M, Petri S, Weber RG. FIG4 variants in central European patients with amyotrophic lateral sclerosis: A whole-exome and targeted sequencing study. Eur J Hum Genet. 2017;25:324–331. doi: 10.1038/ejhg.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bharadwaj R, Cunningham KM, Zhang K, Lloyd TE. FIG4 regulates lysosome membrane homeostasis independent of phosphatase function. Hum Mol Genet. 2016;25:681–692. doi: 10.1093/hmg/ddv505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nishiyama A. Brd4 Is Required for Recovery from Antimicrotubule Drug-induced Mitotic Arrest: Preservation of Acetylated Chromatin. Mol Biol Cell. 2005;17:814–823. doi: 10.1091/mbc.E05-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rudge SA, Anderson DM, Emr SD. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell. 2004;15:24–36. doi: 10.1091/mbc.E03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ferguson CJ, Lenk GM, Meisler MH. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum Mol Genet. 2009;18:4868–4878. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Cassel JA, Reitz AB Sequencing Consortium FALS. Ubiquilin-2 (UBQLN2) binds with high affinity to the C-terminal region of TDP-43 and modulates TDP-43 levels in H4 cells: Characterization of inhibition by nucleic acids and 4-aminoquinolines. Biochim Biophys Acta - Proteins Proteomics. 2013;1834:964–971. doi: 10.1016/j.bbapap.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]