Abstract
Foundational studies of chloroplast genome (plastome) evolution in parasitic plants have focused on broad trends across large clades, particularly among the Orobanchaceae, a species-rich and ecologically diverse family of root parasites. However, the extent to which such patterns and processes of plastome evolution, such as stepwise gene loss following the complete loss of photosynthesis (shift to holoparasitism), are detectable at shallow evolutionary time scale is largely unknown. We used genome skimming to assemble eight chloroplast genomes representing complete taxonomic sampling of Aphyllon sect. Aphyllon, a small clade within the Orobanchaceae that evolved approximately 6 Ma, long after the origin of holoparasitism. We show substantial plastome reduction occurred in the stem lineage, but subsequent change in plastome size, gene content, and structure has been relatively minimal, albeit detectable. This lends additional fine-grained support to existing models of stepwise plastome reduction in holoparasitic plants. Additionally, we report phylogenetic evidence based on an rbcL gene tree and assembled 60+ kb fragments of the Aphyllon epigalium mitochondrial genome indicating host-to-parasite horizontal gene transfers (hpHGT) of several genes originating from the plastome of an ancient Galium host into the mitochondrial genome of a recent common ancestor of A. epigalium. Ecologically, this evidence of hpHGT suggests that the host–parasite associations between Galium and A. epigalium have been stable at least since its subspecies diverged hundreds of thousands of years ago.
Keywords: holoparasitism, horizontal gene transfer, Orobanchaceae, parasite reduction syndrome, plastome reduction, rbcL
1. Introduction
Comparative phylogenetic studies of parasitic plants are useful for understanding evolutionary changes concomitant with the major life-history shift from autotrophy to heterotrophy. At the genomic level, reduction in size and gene content of the plastome (chloroplast genome) is common in many lineages of heterotrophic angiosperms, and is associated with the shift away from photosynthesis as the primary mechanism of energy acquisition [1–4]. Using a phylogenetic framework, it is possible to reconstruct the tempo and mode of plastid genome evolution. This has stimulated the development of evolutionary models that predict convergent changes in the relaxation of selection, genome size, guanine-cytosine (GC) content, gene content, and other features with the shift to heterotrophy [2,5].
Recent foundational studies of plastome evolution in parasitic plants have generally been concentrated in lineages with high extant diversity and variation in the degree of parasitism and/or reliance on photosynthesis, such as the Orobanchaceae, Cuscuta (Convolvulaceae), Santalales, and mycoheterotrophic Ericaceae or Orchidaceae [2–4,6–8]. Many of these studies leverage a phylogenetic framework to reconstruct or infer ancestral shifts in selection on genes, and the extent of phylogenetic conservatism or homoplasy in molecular evolution. However, given the diversity of parasitic plants, the phylogenetic sampling density has traditionally been relatively coarse-grained, which limits the degree to which graduated versus punctuated hypotheses of genome loss can be distinguished, and the extent to which recent evolution can be detected. Moreover, many ecological and evolutionary processes (reviewed by [9]) relate nonlinearly, or differ altogether at contrasting scales. Therefore, it is crucial to extend this foundational research with targeted, finer-grain studies, particularly toward the tips of the phylogeny.
A recent study in mycoheterotrophic Corallorhiza orchids has begun to bridge this gap. Barrett et al. [4] found variation in plastome gene content even at the intraspecific level, providing evidence for rapid and continued genomic evolution in heterotrophic plants. However, similarly fine-scale studies have not yet been done in direct parasites (as opposed to mycoheterotrophs), and the origin of heterotrophy in Corallorhiza is several-fold more recent than it is in some lineages of parasitic plants, including the Orobanchaceae [4,10].
The primary focus of this study was to test the extent of plastome evolution of parasitic plants at shallow evolutionary time scales. Within the Orobanchaceae, Aphyllon californicum can be classified as just past Stage 1 plastome reduction (sensu [2]) because it has lost its plastid-encoded ndh genes, but still retains many primary photosynthesis genes. By contrast, the sequenced plastomes of other related holoparasites, including the sister genus Phelipanche, correspond to Stage 2 plastome reduction, lacking both ndh and photosynthesis-related genes. At the same time, plastomes of several species in Phelipanche are quite uniform, whereas the plastome of the more distantly related Orobanche gracilis was found to be considerably smaller in length than those of its Orobanche congeners and lacking atp genes (Stage 3). Motivated by these contrasting patterns among holoparasitic Orobanchaceae, we focused on the genus Aphyllon owing to its tractable size and well-elucidated fine-scale evolutionary relationships [10,11] in order to test whether plastome reduction continues at shallower evolutionary timescales in a similar fashion as it does at deeper levels in the Orobanchaceae.
Specifically, we sought to determine if the unusually large and intact plastome of A. californicum represents stochastic variation between stages of plastome reduction, or is representative of the genus Aphyllon, the latter suggesting that any level of plastome reduction may be stable. We sequenced eight plastomes representing complete taxonomic sampling of Aphyllon sect. Aphyllon, including infraspecific sampling of two widespread species: Aphyllon fasciculatum and Aphyllon ‘franciscanum', an undescribed lineage corresponding in part to the long-unrecognized Orobanche fasciculata var. franciscana but sister to Aphyllon epigalium and parasitic on a distinct suite of hosts across western North America [11]. Coupled with previous studies, this sampling allows for robust comparisons at multiple levels: within this clade, between this clade and its sister group, represented by A. californicum, and with more distantly related holoparasitic Orobanchaceae. We predicted that plastome size and gene content among species of Aphyllon sect. Aphyllon would be similar, but consistently more reduced than that of A. californicum, given that there is little reason to suspect that maintaining a miscellaneous subset of photosynthesis genes, as seen in A. californicum, is evolutionarily stable [2].
While in the course of sampling one species, A. epigalium, we serendipitously found evidence that this species contains a Galium-like copy of the plastid gene rbcL. Horizontal gene transfer (HGT) is increasingly recognized as a widespread evolutionary process and source of genetic variation in plants [12–14]. This is particularly true among parasitic plants, which have a physically and physiologically intimate connection to their host that could facilitate HGT [14,15]. However, HGT events involving the acquisition of chloroplast genes are quite rare [16]. Therefore, the study was expanded to test if our discovery was indeed the result of a host-to-parasite HGT (hpHGT) event. To do so, we screened for the rbcL xenologue in other populations of A. epigalium and its congeners, identified its genomic location by assembling large fragments (greater than 60 kb) of the A. epigalium mitochondrial genome, and finally built a robust rbcL gene tree of parasites and hosts to infer the donor lineage of the rbcL xenologue. Both subspecies of A. epigalium parasitize only perennial Galium (Rubiaceae), while congeners are exclusively found on other distantly related hosts [11,17], so we predicted that the xenologue (horizontally transmitted gene copy) would only be found in those species, and be most closely related to the plastid copies in the clade of Galium that includes most extant hosts.
2. Material and methods
(a). Aphyllon whole genome shotgun sequencing
Genomic DNA (gDNA) was extracted from floral tissue of single individuals of each of eight Aphyllon specimens using a DNeasy Plant Minikit (Qiagen, Valencia, CA, USA; see the electronic supplementary material, Methods for a description of Aphyllon habit and brief rationale for sampling floral tissue). These samples represent each of the five recognized taxa in Aphyllon sect. Aphyllon, as well as two specimens of an undescribed species currently recognized as A. fasciculatum but not parasitic on Artemisia, hereafter referred to as A. ‘franciscanum', and a second specimen of ‘true' A. fasciculatum.
The DNA was then sheared to 200–700 bp fragments using a Bioruptor sonicator (Diagenode, Denville, NJ) and gDNA libraries were prepared using a modified version of Meyer & Kircher's protocol [18]. These samples were each sequenced in 1/50 of an Illumina HiSeq 4000 DNA sequencer lane (2×150 bp paired end format) at the Vincent J. Coates Genomics Sequencing Laboratory (Berkeley, CA).
Demultiplexed sequence reads were quality filtered using Trimmomatic v.0.36 [19]. Only paired reads greater than 40 bp and with an average phred quality score of at least 20 in a 10 bp sliding window were retained. Several independent subsets of 2–2.5 million trimmed reads were assembled into contigs using the de novo assembly algorithm in Geneious v.9.1.8 (Biomatters, Auckland, New Zealand [20]). The resultant contigs that could be attributed to the plastome were then aligned and joined using the results of an independent NOVOplasty assembly (v.2.6, [21]) or in some cases Sanger sequencing with custom primers. As a final check on the assemblies, the original full read pool was reference-mapped to the de novo assembly using Geneious (see the electronic supplementary material, Methods for information on sequencing coverage of plastid genomes).
Plastome annotations were also performed in Geneious, using several closely related autotrophs and heterotrophic lamiids as references (A. californicum, Schwalbea americana and Lindenbergia philippensis [6], and Nicotiana tabacum [22]). BLASTx searches were performed to confirm open reading frames (ORFs) or pseudogenization of protein-coding regions (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and tRNAscan-SE v.2.0 was used to verify tRNA boundaries and anticodon identities [23].
Mitochondrial genome contigs of both subspecies of A. epigalium were identified based on similarity to existing GenBank mitochondrial DNA sequences and average coverage (see the electronic supplementary material). These contigs were then annotated for the presence of genes of putative plastome origin by using the Orobanchaceae and Nicotiana references above. Genes were tentatively assessed as being of host or parasite origin based on a BLASTn search. Additional annotations of mitochondrial genes were performed using MITOFY (v. 22 March 2012 [24]), tRNAscan-SE v.1.3.1, and blast+ v.2.6 [25] with default parameters.
(b). rbcL gene tree estimation
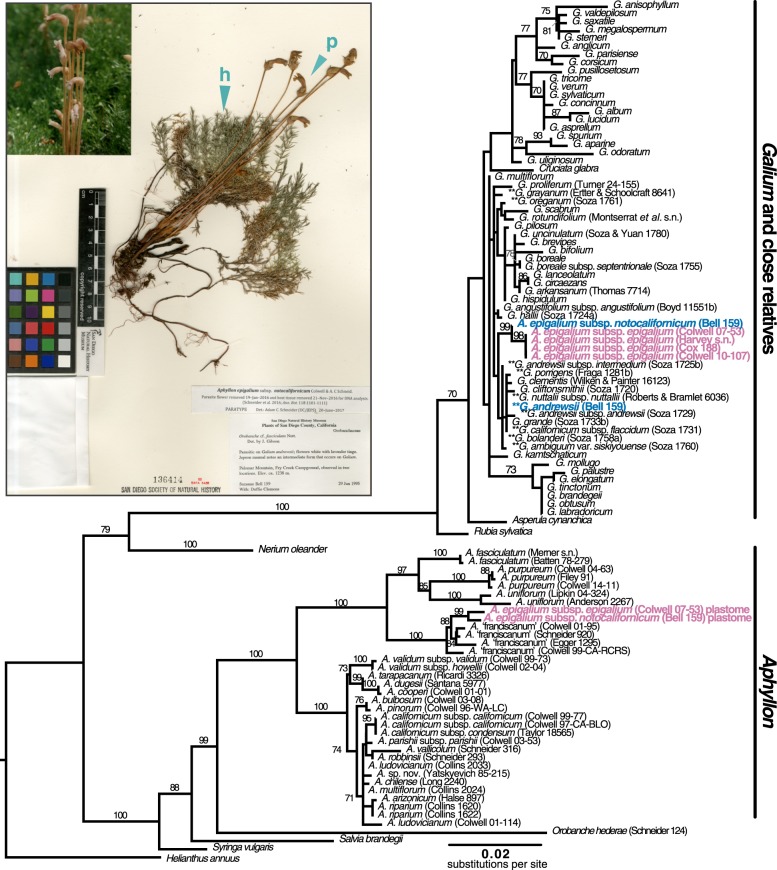
In order to verify that a putative rbcL xenologue was horizontally acquired and to better understand its phylogenetic origin, we constructed a gene tree of rbcL including both Aphyllon and Galium homologs. We included additional samples of both A. epigalium subspecies from across their geographical ranges, most other recognized Aphyllon species, many Galium species from western North America (the geographical range of A. epigalium), and representative sampling from other major Galium clades. Taxon sampling within Galium and Aphyllon were informed by previous phylogenetic studies by Soza & Olmstead [26] and Schneider et al. [11], respectively, and most DNA aliquots were generated by the authors of those studies. However, we did extract DNA from vegetative tissue of one herbarium specimen of Galium andrewsii parasitized by A. epigalium subsp. notocalifornicum using a DNeasy Plant Minikit, in order to compare sequence data from the same host/parasite pair of individuals (figure 2, inset).
Figure 2.
Maximum-likelihood rbcL gene tree of Galium and Aphyllon homologs. Bootstrap support of greater than or equal to 70% is indicated along branches. Paralogues sequenced from A. epigalium (with pink labels) form two strongly supported clades. One is nested within a clade of Galium sequences and near a subclade containing most reported host species (denoted by **), and the other is resolved within sequences of Aphyllon sect. Aphyllon, congruent with accepted infrageneric relationships [11]. Tip labels with collector names in parentheses represent sequences generated for this study; other sequences were downloaded from GenBank. Inset: collection of A. epigalium subsp. notocalifornicum parasitizing the root of its host, Galium andrewsii (Bell and Clemons 159 [SD 136414]). Blue wedges indicate approximate locations that host (h) and parasite (p) tissue were sampled from the herbarium specimen, from which differing Galium and Galium-like rbcL genes were sequenced (blue tip labels in phylogeny). Image courtesy of San Diego Natural History Museum and used with permission.
Polymerase chain reaction (PCR) amplifications of the rbcL gene were performed using AccuPower PCR PreMix kits (Bioneer, Alameda, CA, USA) and products were sequenced on both the sense and antisense strand (electronic supplementary material, Methods). We also assembled sequences of rbcL and its flanking regions for 16 additional samples of Aphyllon and Orobanche hederae from short read data generated for a separate study using the same sequencing methods as above, but using the Burrows–Wheeler Aligner [27] and a portion of the A. californicum plastome as a reference [2]. Sites with a read depth of less than 10 were converted to unknown base (N).
To test for the presence of the rbcL xenologue in close relatives of A. epigalium, the same method was used to generate a reference-guided assembly against the rbcL ORF of Galium scabrum (GenBank X81105). This technique was successful in recovering the xenologous rbcL copy in both accessions of A. epigalium. BLASTn searches were performed on the resulting assemblies, and the xenologue was assumed to be absent if the most similar sequences were each referable to Orobanchaceae, as was the case for all other taxa besides A. epigalium.
The consensus sequences were then aligned with all other rbcL sequences using default settings in Geneious v.6.1.8. Maximum-likelihood analysis was performed using RAxML-HPC2 v.8.2.10 [28] with Helianthus annuus set as the outgroup and run on the XSEDE computing cluster using the CIPRES Science Gateway v.3.3 [29]. We used the GTR+Γ model of sequence evolution, and 1000 rapid bootstrap replicates to assess branch support.
Additional methodological details are available in the electronic supplementary material. Raw Illumina reads are archived at the NCBI Short Read Archive (https://www.ncbi.nlm.nih.gov/sra), under Bioproject ID PRJNA477860. GenBank accession numbers for each rbcL sequence used are listed in the electronic supplementary material, table S1, and those generated by this study are accompanied by voucher information, the source of DNA used, and sequencing method (Sanger or Illumina).
3. Results
(a). Plastome reduction in Aphyllon
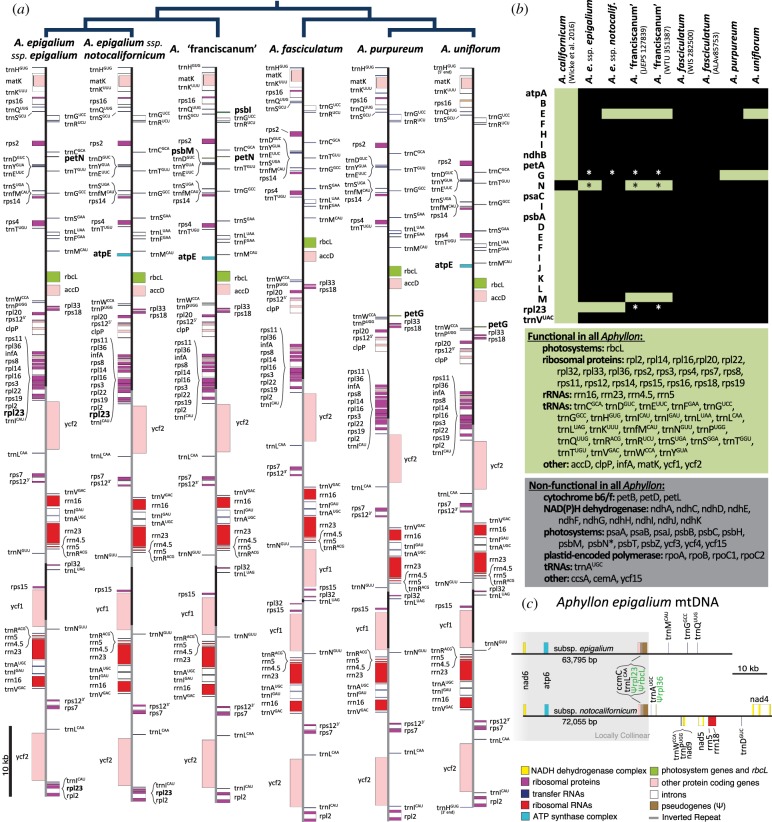
Relative to A. californicum, species of Aphyllon sect. Aphyllon have overall smaller plastomes, with a reduced GC content and disproportionally shorter single-copy regions but a similar sized inverted repeat (table 1). Collectively, all Aphyllon sect. Aphyllon species (including A. ‘franciscanum') differed in length by less than 5%, and have lost a moderate number of plastid genes (table 1 and figure 1a,b). Nearly all tRNAs and most housekeeping genes are retained. The genes that retain ORFs in some species but not others are typically small and highly divergent from the copies found in autotrophic relatives, such as petG, petN, psbM and psbN (additional detail in the electronic supplementary material, Results). In A. fasciculatum and A. ‘franciscanum', multiple plastomes sequenced from the same species differed in length by less than 1% and were identical in gene content. Structurally, all eight plastomes were nearly identical, although the inverted repeat in A. fasciculatum has dramatically expanded to encompass nearly the entire small single-copy region, including the genes ycf1, rps15, and rpl32 (figure 1a).
Table 1.
Plastid genome size and structure of Aphyllon species.
| Aphyllon speciesa | plastome (bp) | GC % | large single copy region (bp [%]) | small single copy region (bp [%]) | inverted repeat (bp [%]) | coverageb | GenBank accession number |
|---|---|---|---|---|---|---|---|
| A. epigalium subsp. epigalium | 103216 | 35.1 | 45061 (43.7%) | 8083 (7.8%) | 25036 (24.2%) | 96x | MH050785 |
| A. epigalium subsp. notocalifornicum | 103932 | 35.0 | 45566 (43.8%) | 8336 (8.0%) | 25015 (24.1%) | 132x | MH050786 |
| A. fasciculatum (WIS 282500) | 106796 | 34.7 | 43970 (41.2%) | 530 (0.5%) | 31148 (29.2%) | 56x | MH499248 |
| A. fasciculatum (ALAv85763) | 107663 | 34.6 | 44867 (41.6%) | 520 (0.5%) | 31138 (28.9%) | 153x | MH499249 |
| A. ‘franciscanum’ (JEPS 127839) | 104153 | 34.9 | 45738 (43.9%) | 8337 (8.0%) | 25039 (24.0%) | 271x | MH580292 |
| A. ‘franciscanum’ (WTU 351387) | 104135 | 34.9 | 45560 (43.8%) | 8485 (8.1%) | 25045 (24.1%) | 416x | MH580291 |
| A. purpureum | 106661 | 35.1 | 49705 (46.6%) | 7902 (7.4%) | 24527 (23.0%) | 503x | MH499250 |
| A. uniflorum | 104640 | 35.3 | 50593 (48.3%) | 7323 (7.0%) | 23362 (22.3%) | 215x | MH580290 |
| A. californicum [6] | 120840 | 36.7 | 60352 (50.0%) | 11608 (9.6%) | 24440 (20.2) | — | NC_025651 |
aMultiple samples from the same species identified by their herbarium accession number.
bCoverage is of the original read pool reference-mapped onto the assembled plastome.
Figure 1.
(a) Annotated plastomes of each species of Aphyllon sect. Aphyllon, with their phylogenetic relationships indicated above. Only one representative of A. fasciculatum (WIS282500) and A. ‘franciscanum’ (WTU 351387) is shown. Genes not shared by all samples of Aphyllon sect. Aphyllon are labelled in bold. Location of the inverted repeats are indicated by a grey backbone. (b) Differences in plastome gene content between A. californicum and eight samples of Aphyllon sect. Aphyllon. Green cells or background indicate putatively functional genes with intact ORFs; black cells or grey background indicate that the gene is lost or probably nonfunctional. An asterisk (*) indicates ambiguities in some plastomes: petG, start codon is ATT, but the ORF is otherwise intact; petN, high amino acid divergence; psbN, intact ORF in A. fasciculatum, but high sequence divergence; rpl23, no apparent stop codon, see the electronic supplementary material, appendix D for details. (c) Mitochondrial genome fragments of A. epigalium subsp. epigalium and subsp. notocalifornicum show several pseudogenes of likely host-plastid origin (Ψ, labelled in green). Grey box indicates greater than or equal to 36642 bp locally collinear block inferred using Mauve [30].
(b). Horizontal gene transfer
A 72 kb fragment of the A. epigalium subsp. epigalium mitochondrial genome, and a 64kb fragment of the A. e. subsp. notocalifornicum mitochondrial genome contain several pseudogenized genes of plastid origin, including adjacent Galium-like rbcL and rpl23 pseudogenes within a greater than 36 kb locally collinear block (figure 1c). This locally collinear region has 99.4% pairwise sequence identity between subspecies, with differences mostly owing to small, scattered indels. A Galium-like rpl36 pseudogene was also identified in subsp. notocalifornicum using a BLAST search, and, late in the revision stage of this manuscript, we discovered similar evidence of a Brassicaceae-like rbcL pseudogene in mitochondrial contigs from both samples of A. ‘franciscanum’ (data not shown).
When analysed phylogenetically, the rbcL paralogues from the A. epigalium plastid and mitochondrial genomes each formed a strongly supported clade with 99% bootstrap support (BS). However, these clades were resolved in very different places of the gene tree (figure 2, labelled in pink). Sequences of rbcL from the plastome of A. epigalium were resolved with A ‘franciscanum' as a subclade within a larger clade of Aphyllon (BS = 100%), consistent with accepted species relationships [11]. By contrast, rbcL sequences from the mitochondrial genome of A. epigalium (hereafter called the xenologous copy) were resolved within a strongly supported clade (BS = 100%) of Galium and close relatives in the Rubieae (Rubiaceae). More specifically, this mitochondrial rbcL clade from A. epigalium is sister to a western North American clade of fleshy-fruited Galium (clade 1 of [26]) that includes nearly all known host species of A. epigalium (figure 2, hosts indicated by ** as determined by Colwell et al. [17]). However, owing to low sequence variation, the precise relationships among these sequences within the Rubieae have negligible support (BS <70%).
(c). Controlling for contamination
Several lines of evidence allow us to reject host DNA contamination, a particular concern in holoparasites owing to the intimate physiological relationship with their hosts, as an explanation for the close relationship between rbcL sequences from A. epigalium and Galium. First, several independent collections of A. epigalium were sequenced to verify replicability across populations. Second, the xenologous rbcL pseudogene is phylogenetically related but not identical to the rbcL sequence from any known Galium host, or Galium species more generally. Moreover, in a known host–parasite pair of individuals (Bell and Clemons 159 (SD), we show distinct host rbcL, parasite plastid rbcL, and parasite HGT rbcL genes, indicating that the Galium-like rbcL xenologue in A. epigalium did not come from an extant host (figure 2).
4. Discussion
(a). Punctuated plastome reduction in Aphyllon
The plastomes of Aphyllon sect. Aphyllon are approximately 15% smaller than those of its sister clade (represented by A. californicum) and devoid of most photosynthesis-related genes. However, the total length (approx. 105 kb) is relatively large for holoparasitic Orobanchaceae, and considerably larger than in other genera with similar or even less gene loss [2,6]. We find support for widely recognized trends in plastome structural evolution following heterotrophy, such as reduced GC content and the disproportionate reduction of single-copy regions (table 1, [6,8,31]). Assuming that the relatively intact A. californicum plastome is characteristic of the common ancestor of all Aphyllon, we infer a rapid transition along the stem branch of Aphyllon sect. Aphyllon from Stage 1 plastome reduction (sensu [2]) to Stage 2 or early Stage 3 reduction (i.e. loss of photosystem and ATP synthase genes but retention of other housekeeping genes), followed by relative stasis during the diversification of Aphyllon sect. Aphyllon. Current divergence time estimates indicate that the length of the stem branch of Aphyllon sect. Aphyllon is approximately 2.8 Myr, whereas the crown age of the clade is 3.4–5.7 Ma [10].
More broadly, the evolution of parasitism in the Orobanchaceae and the corresponding early stages of plastome reduction occurred at least 24 Ma, with the complete loss of photosynthesis in the holoparasitic lineage including Aphyllon occurring at least 14 Ma (dates conservatively reflect lower 95% highest posterior density boundaries inferred by Schneider & Moore [10]; previous studies estimated earlier dates). Collectively, our findings support the punctuated model of Wicke et al. [2], in which large classes of plastid genes can be stable for millions of years following the evolution of parasitism, then lost in a relatively short time period (but retained in sister lineages).
Nonetheless, we do detect some evolutionary changes at shallower timescales. Structurally, gene order is identical across Aphyllon, but the major expansion of the inverted repeat boundary into the small single copy region is an autapomorphy of A. fasciculatum. Regarding gene content, we found parallel pseudogenization and loss of atpE in several lineages, including in only one of the two phylogenetically distinct subspecies of A. epigalium, which diverged between 0.3 and 0.8 Ma, more recently than any pair of heterotrophic plant taxa for which sequenced plastomes are available (figure 1a,b; [10]). Although all other ribosomal protein-coding genes remain intact, rpl23 has been repeatedly lost in Aphyllon (figure 1a,b)—a pattern that has been seen in other lineages of heterotrophic plants [2,3,5,7,32]. The remaining genes that are putatively present in some species but not others are all short subunits of larger complexes, such as petG, petN, psbM, and psbN. Barrett et al. [4] reported similar findings in the heterotrophic orchid Corallorhiza maculata. Strikingly, they found both putatively functional and nonfunctional psaJ alleles in various populations of Corallorhiza striata, and several other small photosystem subunits differentially lost between recently diverged subspecies. Given the clear loss of key genes in these complexes, their persistence may be non-adaptive. Because the genes are so short, degenerative neutral evolution will take longer to manifest by chance, even if purifying selection is relaxed and the genes are no longer being expressed [6].
Ultimately, transcriptomic, proteomic and/or physiological studies are needed to confirm hypotheses of gene function or lack thereof. For example, disruptions to promoters or untranslated regions may cause pseudogenization in spite of an intact ORF. Conversely, U to C RNA editing occurs regularly in chloroplasts [33] and could theoretically rescue the two nonsense mutations in the atpE ORF of A. epigalium subsp. notocalifornicum. However, until additional evidence becomes available, we follow convention used in similar studies of plastome evolution in parasitic plants by inferring gene presence and functionality from intact ORFs [2,4,6].
(b). Horizontal gene transfer of plastid genes and their evolutionary fate
At least three genes of Galium-plastid origin are present in the mitochondrial genome of A. epigalium: rbcL, rpl23, and rpl36 (figure 1c) As in this case, most plant-to-plant HGT has involved the mitochondrial genome as a donor and/or recipient [34]. The exact mode of HGT to parasitic plants from their hosts is still an area of active research, and probably varies among systems. HGT predominantly occurs through direct uptake of DNA, though the reverse transcription of host RNA may also occur as evidenced by the lack of intronic regions or a poly-A string at the 3′ end of the putative xenologue [16,35]. Host-plastid genes acquired by the parasite mitochondrial genome are probably the result of a two-stage process, in which intracellular gene transfer (IGT) from the plastome to the mitochondrial genome of the donor is followed by mitochondrial HGT from the donor to recipient species [16,34,36]. Insertions of a duplicate copy of rbcL into the mitochondrial genome within a species through IGT are a relatively common occurrence in angiosperms though have not yet been reported in Galium [37,38], and hpHGT events are most common between mitochondrial genomes [16]. The two-stage hypothesis could be tested in this system by surveying Galium mitochondrial DNA for the presence of related pseudogenes.
The presence of several genes of host-plastid origin (figure 1c) in mitochondrial DNA from A. epigalium suggests either multiple independent acquisitions or, more parsimoniously, the single HGT of a large genomic fragment that has since degraded. Nuclear and organellar genomes, as well as whole chloroplasts, can travel through plasmodesmata connecting grafted lines or species of tobacco [39–41]. A similar process may have occurred through the symplastic connections between holoparasitic Orobanchaceae and their hosts [42].
Though we found evidence of several mitochondrial genes of host-plastid origin, we focused on the longest, rbcL, for more detailed analysis (figure 2). In spite of the relatively low sequence divergence with its most closely related extant orthologues in the Galium plastome, this xenologue is probably not functional or even transcribed given the frameshift mutations and lack of a promoter region (electronic supplementary material, Results). In most eudicots, the plastid rbcL promoter is 156–185 nucleotides upstream of the start codon [43], but in the mitochondrial genome of A. epigalium, only approximately 100 bp separate the rbcL start codon from an upstream rpl23 pseudogene (figure 1c). Additionally, several deletions and point mutations in the mitochondrial (but not plastid) copy of rbcL appear to have disrupted stem-loop structures that are highly conserved and important for transcription termination of chloroplast genes in the 3′ untranslated region of spinach and tobacco [44]. The fact that these mutations in noncoding regions are shared by both subspecies, and that rbcL and rpl23 pseudogenes are adjacent despite being tens of kilobases apart in most intact angiosperm plastids suggest a larger HGT event was followed by substantial degradation and gene loss before the divergence of the two A. epigalium subspecies several hundred thousand years ago. On the other hand, the relatively few synapomorphic point mutations and differing frameshift mutations could be evidence for recent independent pseudogenizations in each of the two subspecies.
From an ecological perspective, the phylogenetic affinity of the rbcL xenologue to extant hosts of A. epigalium provides strong evidence that host–parasite relationships have been relatively conserved in the half-million years since the two subspecies diverged (figure 2, [10]). Low rates of rbcL molecular evolution in Galium make it difficult to identify which clade of Galium is most closely related to the rbcL xenologue with any statistical confidence. However, if the HGT did occur via a two-stage process, then rbcL pseudogenes within the Galium mitochondrial genome could provide additional phylogenetic evidence.
Conclusion
Based on eight plastomes representing exhaustive taxonomic sampling of Aphyllon sect. Aphyllon, we show that remaining primary photosynthesis genes (i.e. pet, psa and psb genes) and atp housekeeping genes were lost relatively rapidly in the stem lineage, with relatively little change since then. However, these gene losses have not been accompanied by as much of a reduction in overall plastome length as other holoparasitic Orobanchaceae with similar gene content.
Additionally, we confirm hpHGT in Aphyllon. Dozens of putative HGT events have been identified across the Orobanchaceae, and many more in other lineages of parasitic angiosperms. However, only rarely are species and gene trees of both parasite and host lineages known with sufficient precision to draw inferences about molecular evolution, historical ecology and historical biogeography, for example the sustained host–parasite association between A. epigalium and its host Galium [10,14,16,45].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank V. Soza for sharing her Galium DNA extractions, D. Kasimer, A. Seng, L. Smith and B. Wessa for assisting with laboratory work, and L. Hains and the San Diego Natural History Museum for providing access to specimens and destructive sampling permission. We also thank A. Banerjee, C. Fitzpatrick, S. Innes, M. Hetherington-Rauth, R. Rivkin, J. Santangelo, and two anonymous reviewers for providing feedback that greatly improved the manuscript.
Data accessibility
Genbank accession numbers are listed in table 1 and the electronic supplementary material, table S2. Short read archive accession numbers for raw sequence data are included in the text.
Authors' contribution
A.C.S. conceived of and conducted the research, and analysed the data. A.C.S., B.G.B., and S.S. interpreted the data. B.G.B. and S.S. provided access to laboratory facilities and some reagents. H.C. contributed data for the rbcL gene tree. A.C.S. prepared the figures and wrote the manuscript with input from all authors.
Competing interests
We declare we have no competing interests.
Funding
This work was supported in part by funding from the National Science Foundation grant DEB-1601504 to A.C.S. and B.G.B.
References
- 1.Feng Y-L, Wicke S, Li J-W, Han Y, Lin C-S, Li D-Z, Zhou T-T, Huang W-C, Huang L-Q, Jin X-H. 2016. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 8, 2164–2175. ( 10.1093/gbe/evw144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wicke S, Müller KF, de Pamphilis CW, Quandt D, Bellot S, Schneeweiss GM. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle. Proc. Natl Acad. Sci. USA 113, 9045–9050. ( 10.1073/pnas.1607576113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braukmann T, Broe MB, Stefanović S, Freudenstein JV. 2017. On the brink: the highly reduced plastomes of nonphotosynthetic Ericaceae. New Phytol. 216, 254–266. ( 10.1111/nph.14681) [DOI] [PubMed] [Google Scholar]
- 4.Barrett C, Wicke S, Sass C. 2018. Dense infraspecific sampling reveals rapid independent trajectories of plastid degradation in a heterotrophic orchid complex. New Phytol. 218, 1192–1204. ( 10.1111/nph.15072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham SW, Lam VKY, Merckx VSFT. 2017. Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 214, 48–55. ( 10.1111/nph.14398) [DOI] [PubMed] [Google Scholar]
- 6.Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25, 3711–3725. ( 10.1105/tpc.113.113373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braukmann T, Kuzmina M, Stefanovic S. 2013. Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus Grammica exhibit extensive gene loss. J. Exp. Bot. 644, 977–989. ( 10.1093/jxb/ers391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen G, Cuenca A, Seberg O. 2015. Plastome evolution in hemiparasitic mistletoes. Genome Biol. Evol. 7, 2520–2532. ( 10.1093/gbe/evv165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham CH, Storch D, Machac A. 2018. Phylogenetic scale in ecology and evolution. Global Ecol. Biogeogr. 27, 175–187. ( 10.1111/geb.12686) [DOI] [Google Scholar]
- 10.Schneider AC, Moore AJ. 2017. Parallel Pleistocene amphitropical disjunctions of a parasitic plant and its host. Am. J. Bot. 104, 1745–1755. ( 10.3732/ajb.1700181) [DOI] [PubMed] [Google Scholar]
- 11.Schneider AC, Colwell AEL, Schneeweiss GM, Baldwin BG. 2016. Extensive cryptic host-specific diversity among western hemisphere broomrapes (Orobanche s.l., Orobanchaceae). Ann. Bot. 118, 1101–1111. ( 10.1093/aob/mcw158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schönknecht G, Weber APM, Lercher MJ. 2013. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays 35, 868–875. [DOI] [PubMed] [Google Scholar]
- 13.Li F-W, et al. 2014. Horizontal transfer of an adapted chimeric photoreceptor from bryophytes to ferns. Proc. Natl Acad. Sci. USA 111, 6672–6677. ( 10.1073/pnas.1319929111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, et al. 2016. Horizontal gene transfer is more frequent with increased heterotrophy and contributes to parasite adaptation. Proc. Natl Acad. Sci. USA 113, E7010–E7019. ( 10.1073/pnas.1608765113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi Z, Bradley RK, Wurdack KJ, Wong K, Sugumaran M, Bomblies K, Rest JS, Davis CC. 2012. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genom. 13, 227 ( 10.1186/1471-2164-13-227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis CC, Xi Z. 2015. Horizontal gene transfer in parasitic plants. Curr. Opin. Plant Biol. 26, 14–19. ( 10.1016/j.pbi.2015.05.008) [DOI] [PubMed] [Google Scholar]
- 17.Colwell AEL, Watson KC, Schneider AC. 2017. A new species of Aphyllon (Orobanchaceae) parasitic on Galium in the western USA. Madroño 64, 99–107. ( 10.3120/0024-9637-64.3.99) [DOI] [Google Scholar]
- 18.Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols 2010, pdb-rot5448 ( 10.1101/pdb.prot5448) [DOI] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. ( 10.1093/bioinformatics/bts199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18 ( 10.1093/nar/gkw1060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. ( 10.1093/nar/gkw413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 27, 1436–1448. ( 10.1093/molbev/msq029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2008. BLAST+: architecture and applications. BMC Bioinform. 10, 421 ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soza V, Olmstead RG. 2010. Evolution of breeding systems and fruits of New World Galium and relatives. Am. J. Bot. 97, 1630–1646. ( 10.3732/ajb.1000130) [DOI] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post–analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, Louisiana, USA. [Google Scholar]
- 30.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. ( 10.1101/gr.2289704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C-W, Wang Y-N, Chaw S-M. 2017. Plastome evolution in the sole hemiparasitic genus laurel dodder (Cassytha) and insights into the plastid phylogenomics of Lauraceae. Genome Biol. Evol. 9, 2604–2614. ( 10.1093/gbe/evx177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braukmann T, Stefanovic S. 2012. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol. Biol. 79, 5–20. ( 10.1007/s11103-012-9884-3) [DOI] [PubMed] [Google Scholar]
- 33.Knoop V. 2011. When you can't trust the DNA: RNA editing changes transcript sequences. Cell Mol. Life Sci. 68, 567–586. ( 10.1007/s00018-010-0538-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchz-Puerta MV. 2014. Involvement of plastid, mitochondrial and nuclear genomes in plant-to-plant horizontal gene transfer. Acta Societatis Botanicorum Poloniae 83, 317–323. ( 10.5586/asbp.2014.041) [DOI] [Google Scholar]
- 35.Yoshida S, Maruyama S, Nozaki H, Shirasu K. 2010. Horizontal gene transfer by the parasitic plant Striga hermonthica. Science 328, 1128 ( 10.1126/science.1187145) [DOI] [PubMed] [Google Scholar]
- 36.Gandini CL, Sanchez-Puerta MV. 2017. Foreign plastid sequences in plant mitochondria are frequently acquired via mitochondrion-to-mitochondrion horizontal transfer. Sci. Rep. 7, 43402 ( 10.1038/srep43402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummings MP, Nugent JM, Olmstead RG, Palmer JD. 2003. Phylogenetic analysis reveals five independent transfers of the chloroplast gene rbcL to the mitochondrial genome in angiosperms. Curr. Genet. 43, 131–138. [DOI] [PubMed] [Google Scholar]
- 38.Wang XC, Chen H, Yang D, Liu C. 2018. Diversity of mitochondrial plastid DNAs (MTPTs) in seed plants. Mitochondrial DNA Part A 29, 635–642. ( 10.1080/24701394.2017.1334772) [DOI] [PubMed] [Google Scholar]
- 39.Stegemann S, Bock R. 2009. Exchange of genetic material between cells in plant tissue grafts. Science 324, 649–651. ( 10.1126/science.1170397) [DOI] [PubMed] [Google Scholar]
- 40.Stegemann S, Keuthe M, Greiner S, Bock R. 2012. Horizontal transfer of chloroplast genomes between plant species. Proc. Natl Acad. Sci. USA 109, 2434–2438. ( 10.1073/pnas.1114076109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thyssen G, Svab Z, Maliga P. 2012. Cell-to-cell movement of plastids in plants. Proc. Natl Acad. Sci. USA 109, 2439–2443. ( 10.1073/pnas.1114297109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dörr I, Kollman R. 1995. Symplastic sieve element continuity between Orobanche and its host. Plant Biol. 108, 47–55. [Google Scholar]
- 43.Manen JF, Savolainen V, Simon P. 1994. The atpB and rbcL promoters in plastid DNAs of a wide dicot range. J. Mol. Evol. 38, 577–582. ( 10.1007/BF00175877) [DOI] [PubMed] [Google Scholar]
- 44.Shinozaki K, Sugiura M. 1982. The nucleotide sequence of the tobacco chloroplast gene for the large subunit of ribulose-l,5-bisphosphate carboxylase/oxygenase. Gene 20, 91–102. ( 10.1016/0378-1119(82)90090-7) [DOI] [PubMed] [Google Scholar]
- 45.Kwolek D, Denysenko-Bennett M, Góralski G, Cygan M, Mizia P, Piwowarczyk R, Szklarczyk M, Joachimiak AJ. 2017. The first evidence of a host-to-parasite mitochondrial gene transfer in Orobanchaceae. Acta Biologica Cracoviensia Series Botanica 59, 13–22. ( 10.1515/abcsb-2016-0021) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genbank accession numbers are listed in table 1 and the electronic supplementary material, table S2. Short read archive accession numbers for raw sequence data are included in the text.