Abstract
Vigilin, a nucleocytoplasmic shuttling protein, post-transcriptionally suppresses proto-oncogene c-fms expression (encoding CSF-1R) in breast cancer by binding to a 69 nt cis-acting 3-UTR element in CSF-1R mRNA. CSF-1R is an important mediator of breast cancer development, metastasis, and survival. We confirm that vigilin decreases in vitro reporter luciferase activity as well as the translation rate of target mRNAs. We further explore the mechanism of suppression of CSF-1R. We show that the 69 nt binding element has profound effects on translation efficiency of CSF-1R mRNA, not seen in the presence of mutation of the element. Also, mutation of the 69 nt element in the CSF-1R mRNA 3′UTR both interferes with direct vigilin binding and obviates effect of vigilin overexpression on translational repression of CSF-1R. We show that stable vigilin binding requires the full length 69 nt CSF-1R element, including the 26 nt pyrimidine-rich core. Furthermore, titration of endogenous vigilin and other proteins which bind the 69 nt element, by exogenously introduced CSF-1R mRNA 3′UTR containing the pyrimidine-rich sequence, increases the adhesion, motility, and invasion of breast cancer cells. This phenotypic effect is not seen when the 69 nt element is deleted. Lastly, we are the first to show that human breast tissues exhibit strong vigilin expression in normal breast epithelium. Our pilot data suggest decreased vigilin protein expression, along with shift from the nucleus to the cytoplasmic location, in the transition to ductal carcinoma in situ.
Introduction
Proto-oncogene c-fms (encoding a receptor tyrosine kinase, CSF-1R) is expressed in breast cancer [1], and serves as an important mediator of breast cancer development [2], invasion [3], [4], metastasis [5], [6], recurrence [7] and survival [8]. The binding of colony stimulating factor-1 (CSF-1) to CSF-1R activates downstream signal transduction pathways involved in these breast cancer related biological phenotypes.
We previously reported that vigilin binds a 69 nt pyrimidine-rich element in the CSF-1R mRNA 3′UTR (3499–3567) and suppresses CSF-1R mRNA expression in breast cancer cells [9]. The binding of mRNA with RNA-binding proteins can form an mRNP spatial structure whose formation is crucial in determining mRNA stability and translation efficiency [10].Vigilin suppresses the expression of c-fms mRNA by down-regulating mRNA stability as well as inhibiting translation [9].
While vigilin, a large nucleo-cytoplasmic RNA binding protein, was first described decades ago, renewed interest has focused on its role in cardiovascular disease [11]. Data on its role in cancer and the underlying mechanism is still limited. Further, it is recognized that while many of vigilin's known actions occur in the cytoplasm, vigilin's nuclear functions are less well understood [12].
In this report, we confirm and further explore the function of vigilin in the posttranscriptional suppression of CSF-1R mRNA by focusing on the role of the 69 nt binding element in the CSF-1R 3′UTR. Using both the MS2cp tethering system and a tetracycline-inducible system together, we confirm that vigilin suppresses translation of target mRNAs. We show that stable vigilin binding of CSF-1R mRNA requires the full length 69 nt element, containing a 26 nt pyrimidine-rich core. We show that mutation of the 69 nt element has profound effects on CSF-1R translation efficiency, not seen in the presence of the 69 nt wild-type. This 69 nt mutation also leads to interference of direct binding with vigilin along with obviating translational repression of CSF-1R by vigilin.
Next, we explored the in vitro phenotypes of vigilin expressing breast cancer cells by competition of protein binding to CSF-1R mRNA by excess 3′UTR CSF-1R sequences containing the vigilin binding site, as well as by use of a deletional mutant of the 69 nt element. Lastly, we are the first to show vigilin expression in human breast tissues, and study in particular the expression and cellular localization in its transition to ductal carcinoma-in-situ.
Materials and Methods
DNA Constructs
For the tethering assay, the MS2cp sequence (gifted from Dr. Ann Bin Shyu, University of Texas) was fused to the full length vigilin coding region (gifted from Dr. David Shapiro, University of Illinois) and was cloned into the pcDNA3.1(−). The luciferase (Luc) reporter contains two repeats of MS2cp binding sequences (Luc-2xMS2cp-BS) after the Luc coding sequence, and was cloned into the Tet-ON 3G inducible expression plasmid (Clontech). Chromosomally integrated Luc-2xMS2cp-BS was generated in SKOV3 ovarian cancer cells.
For the cloning of the tetracycline-inducible system of the luciferase-CSF-1R mRNA UTRs, CSF-1R mRNA 5′ and 3′UTRs were ligated to both ends of the luciferase mRNA and cloned into the Tet-ON 3G inducible expression plasmid (Clontech).
Mutation of 69 nt wild-type element was performed by replacing the pyrimidines with purines as was described previously [9].
For overexpression of the pyrimidine-rich sequence, both the 69 nt (3499–3567) and 26 nt (3499–3524) sequences from the CSF-1R mRNA 3′UTR were cloned into the pcDNA3.1(−).
Translation Induction Assay and Ribosome Profile
SKOV3 cells with a chromosomally integrated Tet-ON 3G inducible system were transfected by the indicated plasmid constructs. After the 24 h transfection, the cells were treated with 20 nM doxycycline for 4 h which was then removed with a PBS wash. After doxycycline removal, the cells were harvested every hour for RNA isolation, or medium was collected for the assay of luciferase activity. The ribosome profile is then generated, as previously described [9]. Three independent experiments were performed and the mean ± SD is shown.
Western Blotting
Endogenous proteins were detected with mouse monoclonal (CSF-1R, 1:100) or rabbit polyclonal (vigilin, 1:200) antibodies. All Western blots were developed with the Supersignal West Pico detection system (Thermoscientific) according to the manufacturer's recommendations, and the membrane was imaged using the Syngene G:Box ChemixT4 machine.
UV-Crosslinking and Label Transfer of Vigilin with CSF-1R RNA
The UV crosslinking of vigilin was performed as described previously [9], [13]. The synthesis of the Biotin-11-UTP labeled riboprobe was performed according to manufacturer's instructions (Ambion). The riboprobe was incubated with 1 μg recombinant vigilin (gift from David Shapiro, University of Illinois) [9]. To analyze protein-RNA crosslinks, reaction mixtures were electrophoresed on a SDS-PAGE gel, and bands were detected by streptavidin-HRP antibody.
Adhesion, Motility, and Invasion Assay
The wild-type terminal 578 nt of CSF-1R mRNA 3′UTR (3217-3992 nt; 3′UTR) or the deletional mutant missing the 69 nt pyrimidine-rich sequence (3′UTRΔ69) cDNA was subcloned into pcDNA 3.1 (+) (Invitrogen) [13].
The Membrane Invasion Culture System was used for assay of in vitro adhesion, motility, and invasion of BT20 breast cancer cells. Procedures were followed as described previously, except that the invasion assays were performed for 72 h [4], [14]. The degree of BT20 cell adhesion onto a filter coated with human-extracellular matrix consisting of type IV collagen, laminin, and gelatin, was measured at A585 after cell staining. The directed motility assays of BT20 cells were performed using fibronectin (25 μg/ml) as a chemoattractant in the lower wells, above which uncoated 10 μm pore filters were placed. Invasiveness was measured through a filter coated with human extracellular matrix barrier. When the effect of CSF-1R monoclonal antibody was studied, this matrix was pretreated with medium alone, 20 μg/ml mouse idiotype-matched IgG in medium, or 20 μg/ml mouse monoclonal IgG to human CSF-1R (MAB3291, R&D Systems) in medium, each for 1 h. The results are reported as mean A585 for adhesion assay, mean percent motility, or mean percent invasion ± SEM. Three independent experiments were performed.
Immunohistochemical Analysis of Vigilin and CSF-1R Expression
Human breast tissues embedded in paraffin were sectioned by the TACMASR of the University of Arizona Cancer Center. Human normal pancreatic tissues served as a positive control for vigilin expression [15]. Immunohistochemical (IHC) analysis was performed using a rabbit antihuman vigilin antibody (Sigma-Aldrich, St. Louis, MO) or mouse monoclonal anti CSF-1R antibody (Santa Cruz Biotechnology, Dallas, TX). The procedure is briefly described as follows. Slides were subjected to deparaffinization, dehydration, quenching in 0.3% hydrogen peroxide, rehydration, and antigen retrieval in 0.05% sodium azide (pH 7.4) under high pressure and at high temperature. The sections were blocked with superblock (ScyTek Laboratories, Logan, UT) for 5 minutes, washed, and incubated with a primary antibody against human vigilin (Sigma-Aldrich) at a 1:200 dilution for 16 h, or CSF-1R (Santa Cruz Biotechnology) at a 1:100 dilution for 16 h. Negative controls included the absence of the primary antibody. The sections were washed, applied with anti-polyvalent HRP polymer (ScyTek laboratories), and incubated for 30 minutes at room temperature. After washing, DAB chromogen (ScyTek laboratories) was added, and slides were counterstained in hematoxylin, dehydrated, and permanently mounted. The IHC technique used was first optimized on control tissues, on review by Ray B. Nagle, MD, pathologist in TACMASR.
Normal pancreas was used as the positive control. Negative control included the absence of primary antibody. In the normal pancreas, vigilin staining was stronger in the cytoplasm than in the nucleus.
Breast slides showing the spectrum of normal to invasive ductal cancer were obtained from the TACMASR shared service of the University of Arizona Cancer Center. The pathology was reviewed for accuracy by Demaretta Rush, MD. There were a total of 31 cases: 10 normal breast tissue, 3 atypical ductal hyperplasia (ADH), 8 ductal carcinoma in situ (DCIS), and 10 invasive ductal cancer (IDC) cases. Between 1 and 5 slides per case were stained for vigilin. One IDC case with significant heterogeneity between staining of 4 slides was not included in the analysis, for a final total of 30 cases.
Scoring of all slides was performed independently by a pathologist, Demaretta Rush, MD. Intensity was scored 0–3+. Vigilin staining was largely homogenous. Note was made of the localization of the vigilin staining (nuclear, cytoplasmic, or both) for each case.
A smaller subset of 4 slides (one each of normal breast and IDC, and 2 atypical ductal hyperplasia) was used for CSF-1R staining, along with positive (chorionic villi) and negative controls. The CSF-1R staining was cytoplasmic in location.
Statistical Analysis
Paired or unpaired t-tests, or one-way ANOVA analysis of variance on ranks was applied using Sigma Stat (Jandel Scientific Corp., San Rafael, CA), as appropriate. A P value of <0.05 was considered statistically significant. Correlation analysis was performed by Pearson Product Moment Correlation using Sigma Stat (Jandel Scientific Corp., San Rafael, CA).
Results
Vigilin Inhibits the Translation of Target mRNAs
To further verify the vigilin-induced translation repression capabilities, we used a tethering assay in SKOV3 ovarian cancer cells. This assay allowed us to study vigilin intrinsic activities (i.e., those independent of any other RBPs) in a cancer cell model. The expressed vigilin protein included a peptide tag derived from the bacteriophage MS2 coat protein (MS2cp tag, Figure 1A) [16] to enable tethering to a luciferase (Luc) reporter. The Luc reporter contains two repeats of MS2cp binding sequences (2x MS2cp-BS) after the Luc coding sequence. MS2cp binds the MS2cp-BS with high affinity and thus recruits vigilin to the Luc-2xMS2cp-BS RNA. The Luc reporter was under the control of the tetracycline inducible promoter (PTre3G promoter) to allow for tight control of transcription.
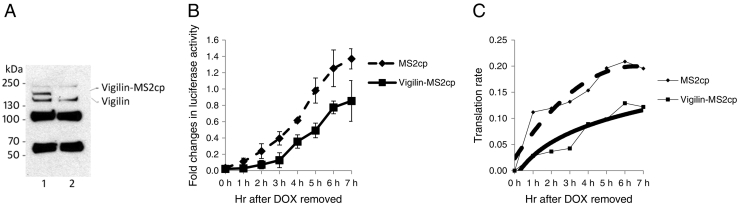
Figure 1.
(A) Lane 1: Expression of vigilin-MS2cp is detected in SKOV3 cells by vigilin antibody. Lane 2: Untransfected SKOV3 cells. (B) Linear increase of luciferase activity after doxycycline-induction in SKOV3 cells with chromosomally integrated luciferase gene fused with MS2cp-BS at the 3′ end. Cells were transfected by either vigilin-MS2cp (straight line) or MS2cp only constructs (dashed line). (C) Translation rate (Kv = L/t) is plotted in both cases.
Transfection of vigilin tagged with MS2cp (Vigilin-MS2cp, straight line) down-regulated luciferase activity when compared to the empty vector (MS2cp) transfected cells (Figure 1B, dashed line) over a 7 h period. The translation rate (Kv) is calculated by dividing luciferase activity by induction time (Kv = Luc/T). Experiments are shown measuring translation rate with a different Y axis to depict changes in translation rate more clearly (Figure 1C). In both constructs, the translation rates (Kv) reached a plateau by 7 h induction (Figure 1C). The translation rate (Kv) was 0.2 at 7 h in the empty vector (MS2cp) transfected cells. In contrast, the vigilin-tethered (Vigilin-MS2cp) cells had a translation rate (Kv) of 0.12 at 7 h. The experiment does not rule out a contribution of mRNA decay to these results. Assuming that the majority effect of this assay is on translation, there is at most a 2-fold decrease in translation rate in the vigilin-tethered cells. This data indicates that the binding of vigilin to target mRNAs represses translation.
Vigilin Requires the Full-Length 69 nt Element Including the 26 nt Pyrimidine-Rich Region in CSF-1R mRNA 3′UTR for Stable Binding
We previously reported that vigilin binds the 69 nt pyrimidine-rich element in the CSF-1R mRNA 3′UTR (3499–3567) and suppresses CSF-1R mRNA expression in breast cancer cells [9]. In the middle of the 69 nt sequence, lies a 26 nt region (3499CCUUCUUUCCUAAUCCCCUUAUCUUC3524) that is highly enriched in pyrimidines (CU = 88.5%). To further narrow down vigilin binding to the 26 nt pyrimidine-rich region in the 69 nt, the UV cross-linking and label transfer assay was performed. UV cross-linking shows that vigilin binds the 26 nt pyrimidine-rich sequence (Py26) very weakly compared to the 69 nt (Py69Wt, Figure 2A). Replacing the 26 nt pyrimidine-rich region in 69 nt by purines also interferes with vigilin binding (69Mut, Figure 2A). The competition assay with the unlabeled 26 nt pyrimidine-rich sequence competes partially with the 69 nt sequence (Figure 2B). Together, our data indicates that the 69 nt sequence including the 26 nt pyrimidine-rich region is required for stable vigilin binding.
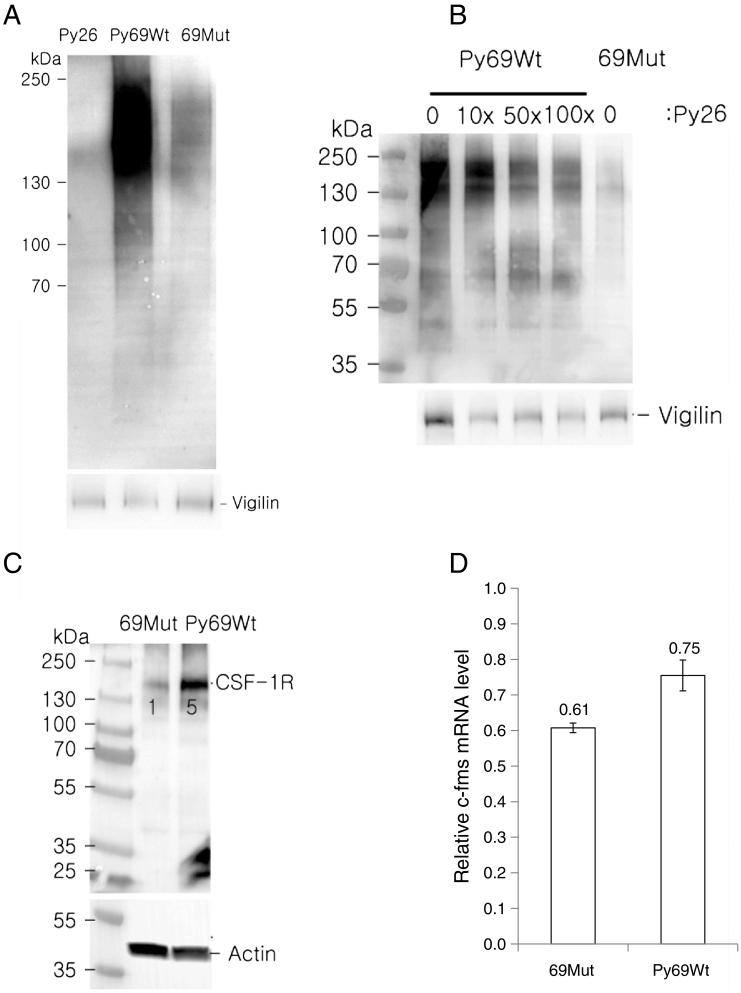
Figure 2.
(A) UV crosslinking of the pyrimidine-rich 69 nt from CSF-1R mRNA 3′UTR using recombinant vigilin. 26-Py is 26 nt pyrimidine-rich sequence only. In Py-Mut, pyrimidines are replaced by purines. Same membrane is reprobed with vigilin antibody (lower panel). (B) Competition assay between pyrimidine-rich 69 nt (Biotin-UTP labeled) and 26 nt (unlabeled) from CSF-1R mRNA 3′UTR for vigilin binding (recombinant vigilin protein) determined by UV crosslinking assay. (C) Expression of 69 nt-wild type sequence from CSF-1R mRNA 3′UTR (Py-Wt) results in increase of CSF-1R protein by 5-fold. (D) Expression of 69 nt-wild type sequence CSF-1R mRNA 3′UTR (Py-Wt) results in increase of CSF-1R mRNA by 22% compared to that in Py-Mut in BT20 breast cancer cells.
To further characterize the translation repressor activity of the pyrimidine-rich 69 nt sequence (Py69Wt), we overexpressed Py69Wt in BT20 breast cancer cells to titrate endogenous vigilin (Figure 2C). The overexpression of Py69Wt increased CSF-1R protein levels 5-fold. CSF-1R mRNA levels measured by RT-qPCR increased by <2-fold (22%, P = .005) (Figure 2D). This data indicates a major function of vigilin lies in translational repression, although it also plays a role in mRNA decay, as previously recognized.
The 69 nt RNA Binding Element Down-Regulates Efficiency of CSF-1R Translation
To study vigilin mediated translation of CSF-1R mRNA, we studied the effect of 69 nt mutation on the translation rates in the presence of 3′ and 5′ UTRs of the CSF-1R transcript. Luciferase reporter constructs fused with both the CSF-1R mRNA 5′ and 3′UTRs were made under the control of a tetracycline-inducible promoter (Figure 3A). Since tetracycline-induction assay indicates that the induction of luciferase RNA reached the maximum level by 4 h (not shown), subsequent assays were conducted using the same induction conditions. Between the wild type construct (Wt) and a construct with a mutation in the pyrimidine rich sequence in the 3′UTR which disrupts vigilin binding (Py-Mut), luciferase activity increased linearly with a 43 ± 1.1-fold increase in the pyrimidine-rich sequence mutant construct (Py-Mut, straight line) compared to a 4.4 ± 2.5-fold increase in the wild type construct (Wt, dotted line) (Figure 3B). The translation rate (Kv) is calculated by dividing luciferase activity by induction time (Kv = Luc/T). Experiments are shown measuring translation rate with a different Y axis to depict changes in translation rate more clearly (Figure 3B). The translation rate in the wild type construct (Wt) is consistent over the 7 h period (Kv = 0.63 at 7 h, dashed line) (Figure 3C). In contrast, the translation rate in the pyrimidine-rich sequence mutant (Py-Mut) increased dramatically over the 7 h period and reached the plateau by 4 h induction (Kv = 6.15 at 7 h, straight line). There is about a 10-fold increase in translation rate by 7 h induction in the pyrimidine-rich sequence mutant (Py-Mut) construct compared to the wild type construct.
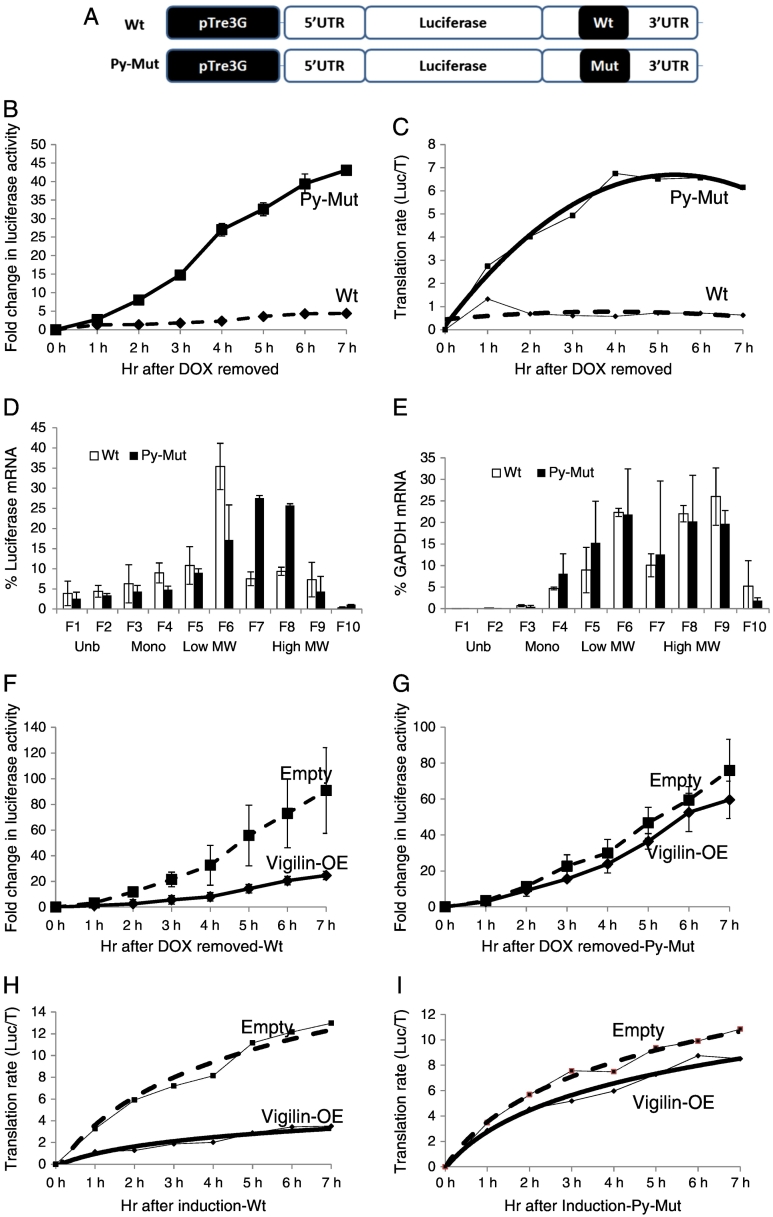
Figure 3.
(A) Luciferase gene fused with CSF-1R mRNA 5′ and 3′UTRs under the control of tetracycline-inducible pTre3G promoter. Wt = wild type 3′UTR construct. Py-Mut = pyrimidine rich sequence mutated 3′UTR construct. (B) Linear increase of luciferase activity after doxycycline-induction in SKOV3 cells with chromosomally integrated luciferase constructs. (C) Translation rate (Kv = L/t) is plotted in both Wt and Py-Mut constructs. (D) Relative distribution of luciferase mRNA in wild type (Wt) and mutant (Py-Mut) constructs in ribosome profile. Mutation of pyrimidine-rich sequence in CSF-1R mRNA 3′UTR affects the distribution of luciferase mRNA in ribosome profile, which was prepared by the sucrose density gradient ultracentrifugation. Fractions: 1–2, unbound RNPs (Unb.); 3–4, monosomes (Mono.); 5–6, low MW polysomes; 7–10, high MW polysomes. (E) Relative distribution of GAPDH mRNA in ribosome profile. (F,H) Overexpression of vigilin (straight line) decreases luciferase activity and translation rate of the wild type luciferase construct compared to the empty vector (dashed line). (G,I) Overexpression of vigilin does not significantly influence the luciferase activity and translation rate of the Py-Mut construct.
We also generated ribosome profiles to confirm the effect of the mutation in the pyrimidine-rich sequence on translation. After 4 h induction by doxycycline, treatment with cycloheximide was used to arrest ribosome migration (Figure 3D). In the wild type construct (Wt, white bar), 69.7% of luciferase mRNA cosedimented with light polyribosomes and free RNPs (Figure 3D, fractions 1–6). However, in the pyrimidine-rich sequence mutant construct (Py-Mut, black bar), 58.6% of luciferase mRNA cosedimented with heavy polyribosomes instead (Figure 3D, fractions 7–10). There was no significant correlation between the two mRNA distributions (correlation coefficient 0.396, P = .258) In contrast, no changes were observed in the control GAPDH mRNA distribution by the Py-Mut line compared to Wt (Figure 3E). The GAPDH mRNA distributions were strongly correlated with each other (correlation coefficient 0.935, P = .00007). This indicates that the binding of vigilin to the pyrimidine-rich sequence of CSF-1R mRNA has a strong effect on translational repression, in the presence of both 5′ and 3′ UTRs.
Mutation of the 69 nt Binding Element Greatly Interferes With Vigilin's Effect on CSF-1R Translation Rate
To confirm the influence of vigilin binding to the pyrimidine-rich sequence, vigilin is overexpressed either in SKOV3 with the wild type construct (Wt) (Figure 3, F and H), or in those with the pyrimidine-rich sequence mutant construct (Py-Mut) (Figure 3, G and I). The overexpression of vigilin in the Wt construct represses the translation of luciferase mRNA by 3.69-fold by 7 h induction (Figure 3F). However, the overexpression of vigilin in the Py-Mut construct does not influence the translation of luciferase mRNA (Figure 3G). The translation rate (Kv) in the Wt construct with vigilin overexpression is 3.52 at 7 h, compared to the translation rate of 12.97 at 7 h in cells transfected with empty vector (Figure 3H). In contrast, the translation rate in the Py-Mut construct with either vigilin overexpression or empty vector transfection does not show significant differences (Figure 3I).
Together, we conclude that the pyrimidine-rich 69 nt element in the mRNA 3′UTR represses the translation of CSF-1R mRNA, and that mutation of this element largely eliminates the effect of vigilin overexpression on the translation rate.
The 69 nt Pyrimidine-Rich Sequence in the CSF-1R mRNA 3′UTR Serves as a Competitor RNA for Protein Binding and Enhances the Adhesion, Motility, and Invasion of Breast Cancer Cells
The recent development of lncRNAs indicates their important role as a competitive endogenous RNA (ceRNA) in cancer development [17], [18]. To test whether exogenously introduced CSF-1R mRNA 3′UTR acts as a decoy for vigilin, we created stable clones of BT20 breast cancer cells which overexpressed the terminal 578 nt CSF-1R mRNA 3′UTR with or without the 69 nt pyrimidine-rich sequence. The overexpression of the CSF-1R mRNA 3′UTR (Wt), which contains the 69 nt pyrimidine-rich sequence, increases CSF-1R protein dramatically compared to overexpression of the CSF-1R mRNA 3′UTR with the pyrimidine-rich sequence deleted (3′UTRΔ69) (Figure 4A). This effect on CSF-1R is similar to that seen by overexpression of the 69 nt wild type sequence alone in the same cells (Figure 2C). This indicates that exogenously introduced CSF-1R mRNA 3′UTR may titrate specific binding proteins including vigilin, on the basis of its binding site within the Py69 element and as a result, endogenous CSF-1R increases.
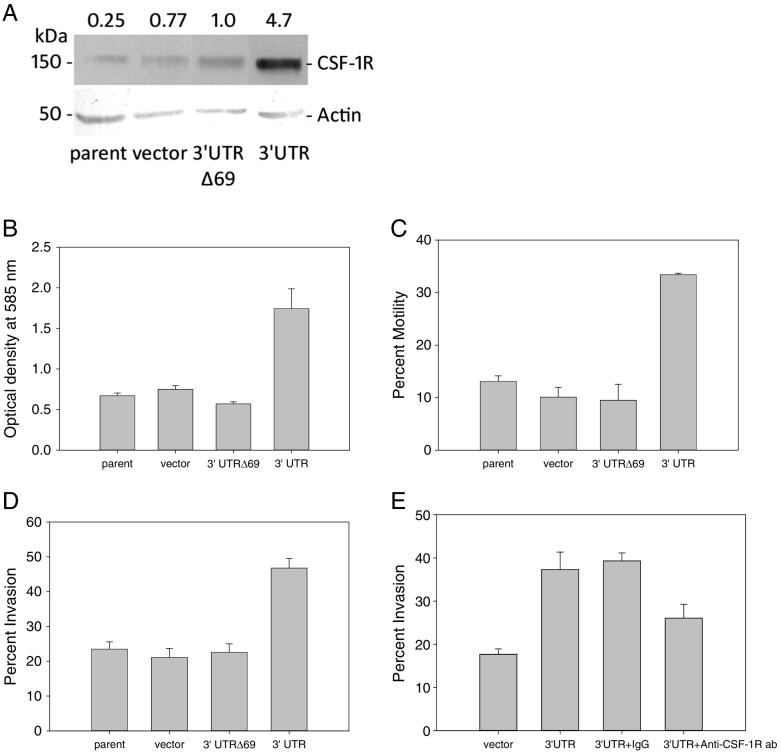
Figure 4.
(A) Stable overexpression of CSF-1R mRNA terminal 578 nt 3′UTR wild type sequence (Wt) in BT20 breast cancer cells increases CSF-1R protein level by 4.7-fold compared to that of stable overexpression of CSF-1R mRNA 3′UTR lacking the pyrimidine-rich sequence (3′UTRΔ69). (B-D) Excess CSF-1R mRNA 3′UTR wild-type, but not in the absence of the 69 nt pyrimidine-rich sequence (3′UTRΔ69), regulates in vitro adhesion, motility, and invasiveness of BT20 breast cancer cells.In vitro (B) adhesion, (C) fibronectin-directed motility, and (D) invasion of BT20 breast cancer clones through a human extracellular matrix are shown. (E) Invasion of BT20 cells with excess 3′UTR CSF-1R RNA is ~50% blocked by an anti-human CSF-1R antibody.
In order to assess whether there may be a downstream change in cellular behavior as a consequence of CSF-1R mRNA 3′UTR-induced CSF-1R up-regulation, we studied the in vitro adhesiveness, invasiveness, and motility of these stably transfected BT20 cells in vitro. Cells bearing excess CSF-1R mRNA 3′UTR wild type sequence (3′UTR) showed significantly increased adhesiveness to human extracellular matrix by 2.3-fold (P = .016, Figure 4B) compared to that of the control cells (parent, empty vector, and 3′UTRΔ69 cells). Similarly, study of fibronectin-directed motility of the 3′UTR containing cells also confirmed a 3.3-fold enhanced motility (P = .0002, Figure 4C) compared to that of the control cells (parent, empty vector, and 3′UTRΔ69). Lastly, invasiveness of the BT20 clones through human extracellular matrix demonstrated a 2.2-fold enhanced invasiveness (P = .0001, Figure 4D) of the 3′UTR containing cells compared to the 3 types of control cells. Anti-CSF-1R antibody was able to significantly inhibit by ~50% this enhanced invasiveness by excess 3′UTR CSF-1R RNA, compared to IgG control (P = .0116, Figure 4E). There was no significant difference in the in vitro behaviors studied between the parent, empty vector, and 3′UTRΔ69 cells.
Taken together, these observations show that the CSF-1R mRNA 3′UTR, via the pyrimidine-rich 69 nt sequence, regulates in vitro adhesion, motility, and invasiveness of BT20 breast cancer cells.
Vigilin Protein Expression in Human Breast Tissues
Due to the role of vigilin as a translational repressor of CSF-1R, along with CSF-1R expression in human breast cancers associated with metastasis [5], [6] and poor survival [8], we compared the expression pattern of vigilin in vivo in human normal breast, precancer, and breast cancer tissues. Prior report showed vigilin expression in rat pancreatic acini tissues [15]. We were the first to observe vigilin expression in human breast cancer cell lines [9], but expression in breast tissues has not been previously reported.
We first confirmed by immunohistochemistry that vigilin is strongly expressed in normal human pancreatic ductal cells. For comparison, staining of a pancreatic cancer sample, which is decreased, is also shown (Figure 5A). Next, we find by IHC analysis in normal breast tissue, high expression of vigilin, similar in intensity to that seen in the normal pancreas control (Figure 5B).
Figure 5.
(A) Vigilin staining in normal pancreas and in a pancreatic cancer, along with the negative controls. CSF-1R staining in chorionic villi serves as positive control, along with negative control. (B) Representative examples of immunohistochemistry of vigilin expression in normal, atypical ductal hyperplasia, DCIS, and invasive ductal cancer breast tissues. (C) Immunohistochemistry of CSF-1R expression in indicated breast tissues.
Vigilin Expression is Decreased in the Transition from Normal Breast Tissue to DCIS With Shift From Nuclear to Cytoplasmic Staining
To further characterize the vigilin expression in breast tissues, IHC was performed in a pilot set of 10 cases of normal breast tissues, 3 cases of atypical ductal hyperplasia (ADH), 8 cases of ductal carcinoma in situ (DCIS), and 9 cases of invasive ductal carcinoma (IDC) (Figure 5B). 80% or 8 of 10 normal breast tissues had 3+ intensity of vigilin staining. None of the 10 normal cases had staining in the cytoplasm which was greater than that in the nucleus (C > N). The large majority (90%) had staining that was equal in the nucleus and the cytoplasm (N=C). One case had stronger nuclear staining than in the cytoplasm (N > C).
Examination of the ADH cases showed 2 of 3 cases with altered staining from that seen in the normal breast tissue with C > N vigilin staining. The intensity of those 2 cases was 2+. The last ADH case stained 2–3+, with N=C staining of vigilin.
All 8 DCIS cases showed C > N staining. The majority (6 of 8) had ≤2+ staining of vigilin, including one case showing only 0–1+ staining. The remaining 2 cases showed 2–3+ intensity.
Taken together, there is a suggestion in this limited sample set of decline in vigilin expression during the transition from normal breast to DCIS, coupled with a shift from nuclear to more cytoplasmic staining. Interestingly, the decline of vigilin expression appears to occur prior to the ductal CIS stage, potentially seen as early as ADH.
The 9 IDC cases did not show a predominant pattern of staining. 5 of 9 had ≤2+ staining of vigilin, including one case each with only 0–1+ or 1+ staining. 4 cases had C > N and 5 C=N staining. 5 IDC cases still showed areas of 3+ staining.
CSF-1R Protein Expression Appears to Increase During Transition to Breast Carcinoma
We were able to study expression of CSF-1R protein expression in 4 tissue samples (Figure 5C), 1 each from normal or IDC, and 2 from ADH. Scoring of staining intensity suggests an increase in CSF-1R staining intensity during the transition from normal breast (1+) to invasive carcinoma, occurring as early as the ADH stage (2+). In the invasive carcinoma case (2+), there is additional CSF-1R staining whose pattern appears to be consistent with accumulation in the golgi apparatus, however, the limited number of cases limits definitive conclusion.
Discussion
In this report, we further explored the function of vigilin in the posttranscriptional repression of CSF-1R mRNA, by focusing on the 69 nt pyrimidine-rich binding element within CSF-1R 3′UTR.
We first confirmed that translational repression of target mRNAs by vigilin (Figure 1). It is known that vigilin binds a large number of mRNAs (estimated 700 mRNAs; [12]). We demonstrate that vigilin directly interacts with the pyrimidine-rich element in the CSF-1R mRNA 3′UTR, and requires the full-length 69 nt for stable binding (Figure 2). We show that mutation of the 69 nt CSF-1R element dramatically alters CSF-1R translational efficiency, and obviates vigilin effect on translational repression of CSF-1R (Figure 3).
Our data does not rule out a contribution of vigilin on mRNA decay to these experiments on translation, as the mechanisms of mRNA stability and translation are closely coupled. In fact, we have previously shown that vigilin OE can enhance CSF-1R mRNA decay by approximately 1.8-fold, while vigilin silencing had less of an effect on mRNA stability [9]. What is clear collectively is that the primary effect of vigilin is on translation, with a more minor component on mRNA decay.
This pyrimidine-rich 69 nt CSF-1R element is a target for binding by proteins other than vigilin. Translational repression and mRNA decay may be reversible by competing CSF-1R mRNA with other RNA binding proteins. We have previously characterized one of them, HuR, with a competition between vigilin and HuR demonstrated for binding this element, with opposing effects on CSF-1R expression [9], [13], [19]. The binding of vigilin to the pyrimidine-rich sequence significantly decreases CSF-1R expression (Figures 2 and 4). In contrast, the binding of HuR to this pyrimidine rich region increases the CSF-1R expression [13].
The present data indicates that the 69 nt CSF-1R binding element confers net translational repression, and that the repression effect is very significant in the presence of cellular RNA binding proteins, including vigilin and HuR (Figure 3). We have shown that the affinity of vigilin to this pyrimidine-rich region in the CSF-1R mRNA 3′UTR is at least 3-fold stronger than that of HuR in BT20 breast cancer cells by in vitro competition assay [19]. Thus, this specificity of binding of the 69 nt pyrimidine-rich region to vigilin, may explain why disruption of the pyrimidine-rich sequence appears to more significantly affect vigilin-specific actions, rather than those of HuR.
We then explored the potential roles of vigilin, along with other proteins which have affinity for the 69 nt CSF-1R element, in breast cancer cells. The sequestration of these proteins by excess 69 nt pyrimidine-rich element increases CSF-1R levels (Figures 2 and 4), and as a result, increases the degree of adhesion, motility, and invasion of breast cancer cells (Figure 4). It is notable that the degree of effect on up-regulation of CSF-1R is similar whether the terminal 578 nt CSF-1R mRNA 3′UTR containing the pyrimidine-rich sequence is overexpressed (Figure 4), or just the 69 nt pyrimidine-rich sequence (Figure 2). This underlies the impact of this small pyrimidine-rich region on post-transcriptional regulation of CSF-1R mRNA, as well as on breast cancer cellular behavior in vitro.
We have previously shown that vigilin overexpression can inhibit breast cancer cell invasiveness and motility in vitro [9]. Here, we show that deletion of the 69 nt vigilin binding element obviates effect of competition with excess CSF-1R mRNA 3′UTR on enhancement of in vitro parameters of breast cancer cellular behavior. Thus vigilin, along with other yet unidentified 69 nt binding proteins, via its binding of the CSF-1R mRNA 3′UTR pyrimidine-rich element, may have a role in the inhibition of tumor behaviors such as breast cancer invasion and metastasis. Neutralizing CSF-1R antibody inhibits the invasiveness of the excess CSF-1R mRNA 3′UTR containing breast cancer cells, suggesting the actions of the pyrimidine-rich sequence are at least in part due to the up-regulation of CSF-1R protein in these cells.
Finally, we are the first to report vigilin expression in human breast tissues and in particular in relation to transition to DCIS. CSF-1R has been well associated with metastatic spread and poor survival of breast cancer [8]. IHC analysis indicates that vigilin expression appears to be lower in transition to DCIS than in normal human breast tissues (Figure 5).
Analysis of vigilin expression along the spectrum from normal (3+) to DCIS (1–2+) showed decreasing intensity of staining, coupled with a transition from 0% of normal breast tissues demonstrating predominantly cytoplasmic based staining (C > N) to 100% of DCIS showing C > N staining of vigilin. Vigilin is well known as a nuclear cytoplasmic shuttling protein. While its cytoplasmic location has been linked to functions such as translational regulation, the role of vigilin in the nucleus is less well known (Jansen et al., 2017). It is not clear how or if Vigilin's functions related to its cytoplasmic location promote the transition from normal through ADH, to DCIS. As translational repression and usually mRNA decay are thought to be cytoplasmic processes, one may have predicted that the location of staining in the normal breast tissues would have favored a cytoplasmic location as seen in the normal pancreas, rather than a nuclear one. Once the cell is transformed into IDC it appears that vigilin's role revert to both those performed in the nucleus and the cytoplasm.
We observed in our very small subset of breast tissues stained for CSF-1R expression, that there may be an increase in CSF-1R expression which accompanies the decline in vigilin expression during the transition to DCIS.
This analysis of a pilot set of specimens for vigilin expression will need to be confirmed in a larger cohort. Interestingly, the decline of vigilin expression appears to occur prior to the ductal CIS stage. If our observation in our sample set of decline in vigilin expression during the transition from normal breast to carcinoma is confirmed, it is possible that vigilin may function as a tumor suppressor, contributing to inhibition of breast cancer development by repressing CSF-1R mRNA stability and translation.
Conclusions
CSF-1R is an important mediator of breast cancer development, metastasis, and survival. The 69 nt CSF-1R RNA binding element in its 3′UTR suppresses translation of CSF-1R. This pyrimidine-rich, non-AU-rich, 3′UTR element has been characterized to date to bind both vigilin and HuR. Vigilin, a nucleo-cytoplasmic binding protein, down-regulates mRNA stability as well as translation of target mRNAs. The full length 69 nt element is required for stable vigilin binding. This 69 nt element also mediates in vitro phenotypes of adhesion, motility, and invasion of breast cancer cells. We are the first to show that vigilin is strongly expressed in normal human breast tissue, with its expression decreasing in the transition to ductal carcinoma in situ.
If our clinical results are validated, our data may support vigilin as having a tumor suppressor function, contributing to inhibition of breast cancer development by repressing CSF-1R mRNA stability and translation.
Acknowledgments
We appreciate Steven J. Gibson and Arpan Ajit Patel for their excellent technical support. We also appreciate Women's Cancers, as well as Ray Nagle, MD, PhD, for support of this project. This work was supported by DOD grant DAMD 17-02-1-0633 (to SKC), Arizona Biomedical Research Commission grant #0802 (to SKC), and the Rodel Foundation (to SKC). Research reported in this publication was also supported by the National Cancer Institute Cancer Center Support Grant P30 CA023074 and used the Tissue Acquisition and Cellular /Molecular Analysis Shared Resource (TACMASR) for providing the breast slides.
References
- 1.Kacinski BM, Scata KA, Carter D, Yee LD, King BL, Chambers SK, Sapi E, Jones MA, Pirro MH, Stanley ER. FMS (CSF-1 receptor) and CSF-1 transcripts and protein are expressed by human breast carcinomas in vivo and in vitro. Oncogene. 1991;6:941–952. [no DOI available] [PubMed] [Google Scholar]
- 2.Lin EY, Nguyen AV, Russell RG, Jeffrey W, Pollard JW. Colony-Stimulating Factor 1 Promotes Progression of Mammary Tumors to Malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [Published March 19, 2001] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, John S, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony stimulating factor-1 receptor. Cancer Res. 2009;69:9498–9506. doi: 10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toy EP, Lamb T, Azodi M, Roy WJ, Woo HH, Chambers SK. Inhibition of the c-fms proto-oncogene autocrine loop and tumor phenotype in glucocorticoid stimulated human breast carcinoma cells. Breast Cancer Res Treat. 2011;129:411–419. doi: 10.1007/s10549-010-1247-7. [Epub 2010 Nov 10] [DOI] [PubMed] [Google Scholar]
- 5.Jeffery JJ, Lux K, Vogel JS, Herrera WD, Greco S, Woo HH, AbuShahin N, Pagel MD, Chambers SK. Autocrine inhibition of the c-fms proto-oncogene reduces breast cancer metastasis assessed with in vivo dual-modality. Exp Biol Med. 2014;239:404–413. doi: 10.1177/1535370214522588. [Epub 2014 Mar 5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toy EP, Bonafe N, Savlu A, Zeiss C, Zheng W, Flick M, Chambers SK. Correlation of tumor phenotype with c-fms proto-oncogene expression in an in vivo intraperitoneal model for experimental human breast cancer metastasis. Clin Exp Metastasis. 2005;22(1):1–9. doi: 10.1007/s10585-005-0718-4. [DOI] [PubMed] [Google Scholar]
- 7.Maher MG, Sapi E, Turner B, Gumbs A, Perotta PL, Carter D, Kacinski BM, Haffty B. Prognostic significance of colony-stimulating factor receptor expression in ipsilateral breast cancer recurrence. Clin Cancer Res. 1998;4:1851–1856. [no DOI available] [PubMed] [Google Scholar]
- 8.Kluger HM, Dolled-Filhart M, Rodov S, Kacinski BM, Camp RL, Rimm DL. Macrophage colony-stimulating factor-1 receptor expression is associated with poor outcome in breast cancer by large cohort tissue microarray analysis. Clin Cancer Res. 2004;10:173–177. doi: 10.1158/1078-0432.ccr-0699-3. [Published January 2004] [DOI] [PubMed] [Google Scholar]
- 9.Woo HH, Yi X, Lamb T, Menzl I, Baker T, Shapiro DJ, Chambers SK. Posttranscriptional suppression of proto-oncogene c-fms expression by vigilin in breast cancer. Mol Cell Biol. 2011;31:215–225. doi: 10.1128/MCB.01031-10. [Epub 2010 Oct 25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat Rev Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [Epub 2014 May 13] [DOI] [PubMed] [Google Scholar]
- 11.Mobin MB, Gerstberger S, Teupser D, Campana B, Charisse K, Heim MH, Manoharan M, Tuschl T, Stoffel M. The RNA-binding protein vigilin regulates VLDL secretion through modulation of Apob mRNA translation. Nat Commun. 2016;7 doi: 10.1038/ncomms12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng MH, Jansen RP. A jack of all trades: the RNA-binding protein vigilin. Wiley Interdiscip Rev RNA. 2017;8(6) doi: 10.1002/wrna.1448. [Epub 2017 Oct 4] [DOI] [PubMed] [Google Scholar]
- 13.Woo HH, Zhou Y, Yi X, David CL, Zheng W, Gilmore-Hebert M, Kluger HM, Ulukus EC, Baker T, Stoffer JB. Regulation of non-AU-rich element containing c-fms proto-oncogene expression by HuR in breast cancer. Oncogene. 2009;28:1176–1186. doi: 10.1038/onc.2008.469. [Epub 2009 Jan 19] [DOI] [PubMed] [Google Scholar]
- 14.Toy E.P., Azodi M., Folk N.L., Zito C., Zeiss C., Chambers S.K. Enhanced ovarian cancer tumorigenesis and metastasis by the macrophage colony stimulating factor (CSF-1) Neoplasia. 2009;11(2):126–144. doi: 10.1593/neo.81150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilgendorf I, Van de Perck M, Emmrich J, Krammer HJ, Kruse C. Vigilin and enzyme expression in isolated pancreatic acini after mellitin and gamma-interferon treatment. Pancreatology. 2003;3:336–341. doi: 10.1159/000071773. [DOI] [PubMed] [Google Scholar]
- 16.Clement SL, Lykke-Andersen J. A tethering approach to study proteins that activate mRNA turnover in human cells. Methods Mol Biol. 2008;419:121–133. doi: 10.1007/978-1-59745-033-1_8. [DOI] [PubMed] [Google Scholar]
- 17.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [Epub 2012 Jun 1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319–332. doi: 10.1016/j.cell.2015.02.043. [Epub 2015 Apr 2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo H.H., Chambers S.K. Post-transcriptional regulation of proto-oncogene c-fms in breast cancer. In: Siregar Y, editor. Oncogene and Cancer from Bench to Clinic. InTech Open Access Publishers; 2013. Invited Chapter. [chapter 13] [Google Scholar]