Abstract
Medulloblastoma is the most common malignant tumor that arises from the cerebellum of the central nervous system. Clinically, medulloblastomas are treated by surgery, radiation, and chemotherapy, all of which result in toxicity and morbidity. Recent reports have identified that DDX3, a member of the RNA helicase family, is mutated in medulloblastoma. In this study, we demonstrate the role of DDX3 in driving medulloblastoma. With the use of a small molecule inhibitor of DDX3, RK-33, we could inhibit growth and promote cell death in two medulloblastoma cell lines, DAOY and UW228, with IC50 values of 2.5 μM and 3.5 μM, respectively. Treatment of DAOY and UW228 cells with RK-33 caused a G1 arrest, resulted in reduced TCF reporter activity, and reduced mRNA expression levels of downstream target genes of the WNT pathway, such as Axin2, CCND1, MYC, and Survivin. In addition, treatment of DAOY and UW228 cells with a combination of RK-33 and radiation exhibited a synergistic effect. Importantly, the combination of RK-33 and 5 Gy radiation caused tumor regression in a mouse xenograft model of medulloblastoma. Using immunohistochemistry, we observed DDX3 expression in both pediatric (55%) and adult (66%) medulloblastoma patients. Based on these results, we conclude that RK-33 is a promising radiosensitizing agent that inhibits DDX3 activity and down-regulates WNT/β-catenin signaling and could be used as a frontline therapeutic strategy for DDX3-expressing medulloblastomas in combination with radiation.
Introduction
Medulloblastoma is the most common malignant form of pediatric brain tumor that occurs in the cerebellum of the central nervous system [1]. Despite treatment advances in recent years, current treatment strategies are associated with long-term toxicities. About 40% of patients experience recurrence of the disease and 30% of patients will eventually die [2]. Current standard treatments include surgical resection and craniospinal irradiation, followed by chemotherapy. Although these strategies have the potential to increase the survival of 70–80% of patients with medulloblastoma, they are associated with serious treatment-induced morbidity. Histologically, medulloblastoma is defined into four subtypes: classic, desmoplastic/nodular; medulloblastoma with extensive nodularity; and large cell/anaplastic [3]. Genetically, medulloblastoma is subtyped into five subgroups: WNT-activated; Sonic hedgehog (SHH)-activated and p53-mutant; SHH-activated and p53-wildtype; non-WNT/non-SHH Group 3; and non-WNT/non-SHH Group 4 [4]. More recently, medulloblastoma has been subtyped into 7 distinct groups [5], [6]. Approximately 10% of medulloblastoma show up-regulation of the WNT pathway with mutations mainly in β-catenin (CTNNB1), which is the principal effector of the WNT pathway [7], [8], [9], [10]. Less common mutations, including adenomatous polyposis coli (APC), Axin1, and Axin2, which are the key factors in the WNT signaling pathway, are also found in medulloblastoma [7], [11]. Somatic mutations in Smoothened (SMO), Suppressor of Fused (SUFU), and Patched-1 (PTCH-1) are the molecular abnormalities found in SHH-type medulloblastoma [12], [13], [14]. Group 3 and group 4 tumors are associated with genomic instability [11] and MYC amplification [15], [16].
DDX3, an RNA helicase, is involved in many biological activities that regulate multiple steps in gene expression, such as transcription [17], [18], mRNA translation [18], [19], splicing [20], and nuclear export [21], [22]. However, mutations in DDX3 have been reported in various cancers, such as chronic lymphocytic leukemia [23], head and neck squamous cell carcinomas [24], and T-cell acute lymphoblastic leukemia [25]. About 8% of medulloblastoma cases involve mutations in DDX3, of which 11% involve the pediatric WNT- and -SHH subtypes [26]. Mutations in DDX3 lead to the activation of mutated β-catenin in TCF/LEF reporter assays, suggesting the oncogenic role of DDX3 in medulloblastoma [27]. DDX3 is also involved in the potentiation of β-catenin signaling through translational regulation of Rac1 in a helicase-dependent manner [28]. In colorectal cancer, DDX3, acting as a mediator in the activation of β-catenin through the CK1ε/Dvl2 axis, is associated with tumor invasion and a worse prognosis [29]. In recent studies, we have shown that RK-33 impaired WNT signaling through the disruption of the DDX3-β-catenin pathway in both lung [30] and colorectal cancers [31]. We have also shown that RK-33, in combination with radiation, induced tumor regression in mouse models of lung cancer [30]. Moreover, RK-33 specifically promoted radiation sensitization in DDX3 over-expressing cells [30], [32].
To validate the functional utility of inhibiting DDX3 activity for tumor ablation, a small molecule that targets DDX3 has been synthesized by our laboratory, referred to as RK-33 [30]. RK-33 binds to the ATP-binding domain of DDX3 and inhibits its RNA-helicase activity [30]. Here, we show the growth inhibition of medulloblastoma cell lines by RK-33 which was associated with reduced transcript levels of WNT-regulated genes. We also demonstrate synergy between RK-33 treatment and radiation both in vitro and in a mouse model of medulloblastoma. Lastly, we also studied DDX3 expression in human medulloblastoma samples.
Materials and Methods
DAOY and UW228 cells were a kind gift from Dr. Michael Lim (Johns Hopkins University, Baltimore, MD, USA). The DAOY cell line (SHH subtype with a mutated p53 gene) was grown in DMEM/F-12 50/50, 1x (Dulbecco's Modification of Eagle's Medium/Ham's F-12 50/50 mix) with 10% fetal bovine serum, L-glutamine and 15 nM HEPES. The UW228 cell line (SHH subtype with a mutated p53 gene) was grown in MEM, 1x (Minimum Essential Medium Eagle) with Earle's salts and L-glutamine supplemented with 10% fetal bovine serum. The cell lines were maintained under sterile conditions in a humidified incubator with 5% CO2 at 37°C. Transfection was performed using the JetPrime transfection reagent (Polyplus, New York, NY, USA) and TransLT1 (Mirus, Madison, WI, USA). For the DDX3 knockdown experiments, siControl (non-targeting pool) and siDDX3 sequences were purchased from Dharmacon, Lafayette, CO, USA.
Patient Samples
A medulloblastoma tissue microarray with 80 cases was previously assembled at the Johns Hopkins Hospital Department of Pathology. Survival data are available for these de-identified patient specimens under IRB approved protocol, NA_00015113. These cases were previously assigned to a molecular subgroup by the German Cancer Research Center (DKFZ), Heidelberg, Germany, using an immunohistochemical panel to identify 4 distinct subgroups; WNT, SHH, Group 3, and Group 4 using antibodies as previously described [16], [33]. Missing cases were attributable to damaged or detached cores during cutting or staining, or to cores not containing tumor. Pathology was reviewed according to the 2007 WHO classification for central nervous system tumors [3]. Clinicopathological data were retrieved from the pathology report and patient files.
Immunohistochemistry
Four μm-thick sections were cut, mounted on Super Frost slides (Menzel & Glaeser, Brunswick, Germany), deparaffinized in xylene, and rehydrated in decreasing ethanol dilutions. Endogenous peroxidase activity was blocked with 1.5% hydrogen peroxide buffer for 15 minutes and was followed by antigen retrieval through boiling for 20 minutes in 10 mM citrate buffer (pH 6.0) for DDX3. Slides were subsequently incubated in a humidified chamber for 1 hour with anti-DDX3 (1:1000, pAb r647) [34]. After washing with PBS, slides were incubated with poly-HRP-anti-mouse/rabbit/rat IgG (BrightVision Immunologic, Duiven, The Netherlands) as a secondary antibody for 30 minutes at room temperature. Peroxidase activity was developed with diaminobenzidine and hydrogen peroxide substrate solution for 10 minutes. The slides were lightly counterstained with hematoxylin and mounted. Appropriate positive and negative controls were used throughout. Scoring of DDX3 was performed by P.V.D. The intensity of cytoplasmic DDX3 expression was scored semi-quantitatively as absent (0), low (1), moderate (2), or strong (3). Cases with a score of 0 to 1 were classified as having low DDX3 expression and evaluated against cases with strong expression.
Immunoblotting
DAOY and UW228 cells were harvested at 60–70% confluence for protein expression analysis. Standard SDS-PAGE and immunoblotting protocols were followed. The primary antibody used for DDX3 and actin was mouse monoclonal antibody and the secondary antibody used was anti-mouse antibody for both DDX3 and actin.
DDX3 Knockdown in Medulloblastoma Cell Lines
DAOY and UW228 cells were plated at 7.5 x 104 cells per well in six-well plates and incubated overnight. Cells were transfected with siDDX3 (25 nM) using the JetPrime transfection reagent. Media was refreshed 24 h after transfection. The cells were harvested after 72 h of transfection and protein concentration was estimated before Western blotting.
Cytotoxicity Assay
Cytotoxicity was determined using an MTS assay. DAOY and UW228 cells were plated in duplicates at 1 x 103 cells per well in a 96-well plate. After overnight incubation, cells were treated with various concentrations of RK-33 (1 μM - 16 μM) and DMSO as a control, for about 72 h. An MTS reagent (10%) was added and cells incubated for an additional 2 h followed by reading absorbance at 490 nm (Victor V3, Perkin-Elmer).
Immunoblotting
DAOY and UW228 cells were plated at 1.5 x 105 cells per well in a six-well plate and incubated overnight. DAOY cells were treated with DMSO or 2.5 μM RK-33, while UW228 cells were treated with 3.5 μM RK-33. After 48 h, cells were lysed in SDS-extraction buffer (100 nM Tris–HCl, 2% SDS, 12% glycerol, 10 mM EDTA, pH 6.7) with an added protease inhibitor. Protein concentration was determined and 30 μg of each sample was loaded onto a 10% SDS-PAGE gel. After protein transfer to the PVDF membrane, DDX3 (1:500 in 5% BSA, mAb AO196) and actin (1:10,000 in 5% milk, A5441, Sigma-Aldrich, St. Louis, MO, USA) primary antibodies were added. After overnight incubation at 4°C, mouse secondary antibodies were added. The blots were developed with Clarity Western ECL (BioRad, Hercules, CA, USA) and imaged with a G-Box Chemi XR5 (Syngene, Frederick, MD, USA).
Cell Cycle Analysis
DAOY and UW228 cells were plated in duplicates at 1.5 x 105 cells per well in a six-well plate. After overnight incubation, cells were treated with RK-33 (1 μM and 2 μM) or DMSO. Briefly, after 24 h of RK-33 treatment, cells were trypsinized and fixed in 70% ethanol overnight at −20°C. Fixed cells were washed with PBS and resuspended in a DNA staining solution (5 μg/ml propidium iodide, 0.5 mg/ml RNase A) for one hour at room temperature. Cell cycle acquisition was performed on a FACScan I or FACScalibur instrument (BD Biosciences, San Jose, CA, USA). Debris was gated out and 10–25,000 cells were acquired per sample. Data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR, USA).
Apoptosis Assay
DAOY and UW228 cells were plated in duplicates at 1.5 x 105 cells per well in six-well plates. After overnight incubation, cells were treated with RK-33 (2 μM) and DMSO as a control. After 24 h of drug treatment, apoptosis was measured with an Annexin V binding detection kit. Briefly, cells floating in the supernatant, along with the adherent fraction, were trypsinized and then washed. Cells were incubated with Annexin V-FITC and PI for 15 min at room temperature in the dark. Cells were immediately analyzed by flow cytometry. Debris was gated out while all other cells were studied. Viable cells excluded both Annexin V-FITC and PI. Early apoptotic cells were Annexin V-FITC-positive and PI-negative, whereas cells that were no longer viable, due to apoptotic or necrotic cell death, were positively stained by both Annexin V and PI. The amounts of stained cells in each quadrant was reported.
TCF Reporter Assays
The transcriptional activity of TCF4 was measured for DAOY and UW228 cells using the dual luciferase assay (Promega, Madison, WI, USA) according to the manufacturer's instructions. Briefly, DAOY and UW228 cells were plated at 3 x 104 cells per well in a 24-well plate. After overnight incubation, cells were transfected with 500 ng TOP-FLASH or FOP-FLASH constructs and 50 ng phRL Renilla constructs as a transfection control, using an LT1 transfection reagent. After overnight incubation, cells were treated with DMSO or 2.5 μM RK-33 for DAOY cells and 3.5 μM for UW228 cells and incubated for 24 h. The cells were lysed and the transcriptional activity of TCF4 was measured using a dual luciferase assay. For the DDX3 knockdown experiments, DAOY and UW228 cells were plated at 3 x 104 cells per well in a 24-well plate. After overnight incubation, cells were transfected with 10 nM siDDX3 for DAOY cells and 25 nM siDDX3 for UW228 cells using the JetPrime transfection reagent. The following morning, cells were transfected with TOP-FLASH and FOP-FLASH constructs using the TransLT1 transfection reagent and incubated for another 48 h, after which the cells were lysed for the luciferase assay. Luminescence was measured using a luminometer (Berthold Sirius, Oak Ridge, TN, USA). Relative TCF4-promoter activity was calculated by normalizing TOP-FLASH and FOP-FLASH readings for Renilla luciferase readings, and then, dividing normalized TOP-FLASH readings by normalized FOP-FLASH readings.
Quantitative Reverse Transcriptase Polymerase Chain Reaction
DAOY and UW228 cell lines were treated with RK-33 (2.5 μM and 3.5 μM) and siDDX3 (25 nM). After 24–48 h of treatment, cells were harvested and RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA, USA), and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA), followed by qPCR using SYBR green (Bio-Rad, Hercules, CA, USA) on a CFX96 Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA). Amplification of 36B4, a housekeeping gene, was used to normalize gene expression values. Primer sequences: DDX3F: 5′-GGAGGAAGTACAGCCAGCAAAG-3′, DDX3R: 5′-CTGCCAATGCCATCGTAATCACTC-3′, AXIN2 F: 5‘-TCAAGTGCAAACTTTCGCCA-ACC-3′, AXIN2 R: 5′-TAGCCAGAACCTATGTGATAAGG-3′, MYC F: 5′-CGTCTCCAC-ACATCAGCACAA-3′, MYC R: 5′-CACTGTCCAACTTGACCCTCTTG-3′, CCND1 F: 5′-GGCGGAGGAGAACAAACAGA-3′, CCND1 R: 5′-TGGCACAGAGGGCAACGA-3′, Survivin F: 5′-CCACCGCATCTCTACATTCA-3′, Survivin R: 5′-TATGTTCCTCTATGGG-GTCG-3′. Fold changes in mRNA expression were calculated using the 2(−ΔΔCT) method [35]. Statistical significance was calculated by performing a paired Student's t-test on the ΔCT values.
Clonogenic Assay
The radio-sensitivity of RK-33 was determined by a clonogenic survival assay. DAOY and UW228 cells at 200–700 were plated in duplicate in six-well plates and incubated overnight. The following day, cells were treated with RK-33 (1–3 μM) or DMSO. After 2.5 h of RK-33 treatment, the cells were exposed to increasing doses of radiation (0, 1, 2, and 3 Gy). After seven days of incubation, colonies consisting of more than 50 cells were scored. Colonies were washed with PBS and stained with 0.05% (w/v) crystal violet dye and then counted. The plating efficiency (PE) was calculated as “mean colony counts/cells seeded,” and the surviving fraction (SF) was calculated as “colonies / (cells seeded x plating efficiency),” and plating efficiency was defined as “mean colony counts for un-irradiated controls / cells seeded.”
In Vivo Combination of RK-33 Treatment With Radiation in Nude Mice
All mice were maintained under sterile conditions, and experiments were conducted according to the guidelines of the Johns Hopkins Animal Care and Use Committee. All animals were maintained under regulated temperature and humidity. Nude mice were injected in the flank with 1 x 106 DAOY cells mixed with matrigel. Mice were monitored until the tumor was felt by palpation. Mice were distributed randomly over four groups of three mice: DMSO as a control; RK-33 (50 mg/kg); radiation (5 Gy); and a combination of RK-33 (50 mg/kg) and radiation (5 Gy). Mice were treated intraperitoneally with RK-33 every alternate day for two weeks. A single dose of radiation (5 Gy) was given using a Small Animal Radiation Research Platform (SARRP) with a circular beam focused on the tumor site. Mice were monitored for tumor growth. The size of tumors was measured over a period of time (42 days) and tumor volume was calculated [V = 1/2(length x width2)] using Vernier calipers.
Statistical Analysis
The Cox proportional hazards analysis and Kaplan–Meier survival plots were used to evaluate differences in time to death between high and low values of DDX3 for subgroups of patient characteristics that had an adequate count of patients. The hazard ratio, 95% confidence interval and p-value for subgroups with adequate patients are provided. For subgroups where there were zero deaths for either the high or low value of DDX3 (perfect prediction) were not able to be evaluated for a hazard ratio.
All bars indicate standard deviation, except where indicated. Unpaired T-tests were used to determine significance. All experiments were performed at least twice (biological replicates) independently in replicates of 2 or 3. Graphs represent mean ± SD, *P < .05, **P < .01, ***P < .001.
Results
DDX3 Expression Increased in Medulloblastomas
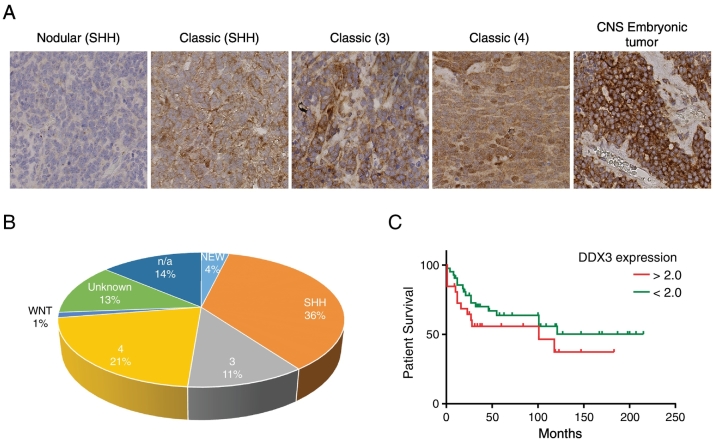
To determine the expression levels of DDX3 in medulloblastoma samples, we evaluated a cohort of 80 medulloblastoma patients (15 samples were not scorable) for DDX3 expression by means of tissue microarrays. In normal brain tissue, we saw little or no expression of DDX3 (data not shown). However, in 31 of 56 (55%) pediatric medulloblastoma samples and in 6 of 9 (67%) adult medulloblastoma samples, we detected DDX3 expression Figure 1A. In addition, high cytoplasmic DDX3 expression was observed among different histological subtypes of medulloblastoma patients Figure 1, A and B. Importantly, no significant association between high and low cytoplasmic DDX3 and survival was observed in medulloblastoma patients Figure 1C and Table 1.
Figure 1.
DDX3 expression in patients with medulloblastoma. A. DDX3 expression in the different molecular subgroup of pediatric and adult medulloblastoma as seen by immunohistochemical staining B. Sample distribution of the different molecular subgroups of medulloblastoma (n = 65) used in our study. C. Kaplan–Meier curve showing overall survival of medulloblastoma patients with low (<2) and high (>2) DDX3 expressing tumors.
Table 1.
Clinical Parameters
|
Total |
DDX3 |
HR |
95% C.I. |
P Value |
||
|---|---|---|---|---|---|---|
| Low (Alive/Dead) | High (Alive/Dead) | |||||
| Age | ||||||
| 0–18 | 49 | 24 (14/10) | 25 (14/11) | 1.09 | 0.46–2.56 | 0.85 |
| 19+ | 8 | 3 (2/1) | 5 (4/1) | 0.45 | 0.03–7.37 | 0.57 |
| Gender | ||||||
| Female | 25 | 14 (7/7) | 11 (8/3) | 0.40 | 0.10–1.56 | 0.19 |
| Male | 34 | 13 (9/4) | 21 (10/11) | 1.98 | 0.63–6.23 | 0.24 |
| Race | ||||||
| White | 30 | 15 (7/8) | 15 (9/6) | 0.77 | 0.27–2.22 | 0.63 |
| Black | 11 | 6 (4/2) | 5 (3/2) | 1.11 | 0.15–8.16 | 0.92 |
| German subgroup | ||||||
| C | 7 | 3 (0/3) | 4 (0/4) | 0.58 | 0.10–3.49 | 0.55 |
| D | 12 | 4 (3/1) | 8 (6/2) | 0.70 | 0.06–7.92 | 0.78 |
| SHH | 23 | 9 (9/0) | 14 (10/4) | NA | NA | NA |
| Subtype | ||||||
| Classic | 26 | 11 (8/3) | 15 (8/7) | 1.78 | 0.46–6.88 | 0.41 |
| Mod A | 8 | 4 (3/1) | 4 (3/1) | 1.41 | 0.09–22.64 | 0.81 |
| Nodular | 10 | 4 (4/0) | 6 (5/1) | NA | NA | NA |
| Sev A | 6 | 2 (0/2) | 4 (1/3) | 0.16 | 0.01–1.75 | 0.13 |
Correlations between DDX3 and other clinicopathological variables in pediatric medulloblastoma patients. P values calculated by Cox proportional hazard analysis. HR, Hazard Ratio; C.I., Confidence Interval.
Inhibition of DDX3 in Medulloblastoma Cell Lines Decreased Proliferation
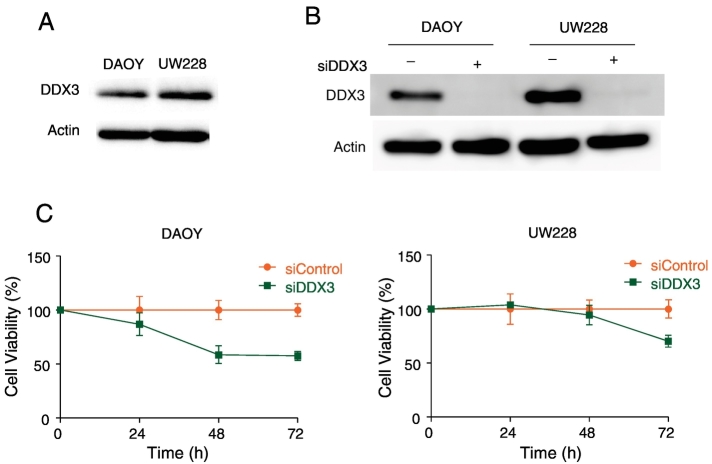
To ascertain whether DDX3 inhibition decreases proliferation, we initially assessed DDX3 expression levels in the medulloblastoma cell lines DAOY and UW228. As shown in Figure 2A, both these cell lines express high levels of DDX3. To determine the role of DDX3 on the growth of DAOY and UW228 cells, we specifically knocked down DDX3 expression using a validated siRNA [31] Figure 2B. As shown in Figure 2C, the knockdown of DDX3 resulted in reduced cellular proliferation.
Figure 2.
Inhibition of DDX3 in medulloblastoma cell lines results in a decrease of proliferation. A. Intrinsic DDX3 expression levels in DAOY and UW228 cells, as detected by immunoblotting B. Immunoblotting showing DDX3 expression levels after knockdown of DDX3 using 25 nM siDDX3. C. Proliferation assay of DAOY and UW228 cells after knockdown of DDX3 measured by MTS assay.
RK-33 Inhibited Cell Viability in Medulloblastoma Cell Lines
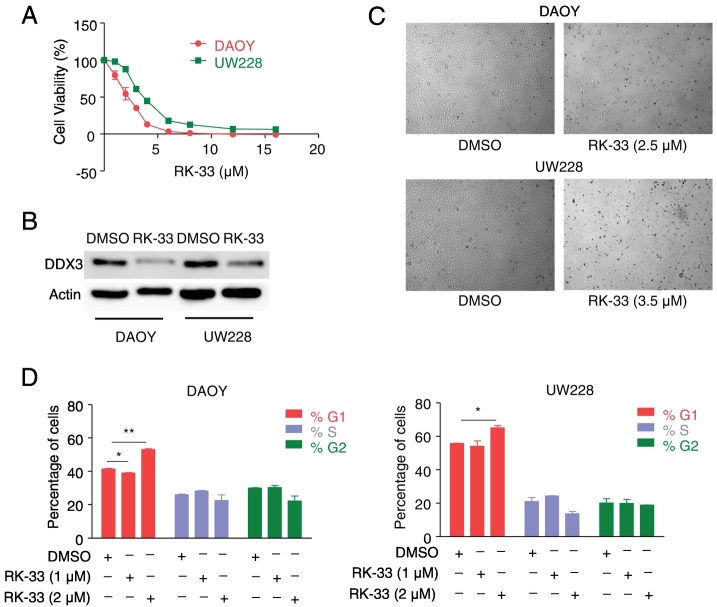
As DDX3 expression is high in medulloblastoma cell lines, we next evaluated whether these cell lines were sensitive to RK-33, a small molecule that targets DDX3. The structure of RK-33 is shown in Supplementary Figure 1. DAOY and UW228 cell lines were treated with various concentrations of RK-33, and cell viability was measured on day 3 with an MTS assay. Both the DAOY and the UW228 cell lines were sensitive to RK-33, with IC50 values of 2.5 μM and 3.5 μM, respectively Figure 3A. In order to determine the effect of RK-33 on DDX3 expression levels, we treated DAOY and UW228 cell lines with IC50 values of RK-33. After 24 h of treatment with RK-33, we scored for DDX3 expression levels by western blotting. The result showed decreased levels of DDX3 expression in both the DAOY and the UW228 cell lines, as shown in Figure 3B. Photomicrographs of cells treated with RK-33 or DMSO are shown in Figure 3C. Based on these observations, we can conclude that RK-33 can effectively reduce DDX3 levels, as a result of which cell death mechanisms are potentially enhanced.
Figure 3.
RK-33 inhibits cell viability in medulloblastoma cell lines. A. Cytotoxicity assay showing the sensitivity of DAOY and UW228 cells to various concentrations of RK-33 after 72 h incubation as measured by MTS assay. B. Immunoblots showing DDX3 expression levels in medulloblastoma cell lines after treatment with RK-33 for 48 h. C. Cells treated with RK-33 indicating cell death. D. Graph displaying changes in cell cycle phases after treating DAOY and UW228 cells with RK-33 for 24 h.
To extend the findings, we evaluated the expression level of DDX3 in two additional medulloblastoma cell lines: D425-Med and HD-MB03 cells. As shown in Supplementary Figure 2, both of these cell lines express DDX3 with D425-Med cells exhibiting somewhat higher levels then HD-MB03 cells. Furthermore, both D425-Med and HD-MB03 cells were sensitive to RK-33 treatment when grown as monolayers on adherent plates with IC50 values of 2.3 and 6.5 μM respectively Supplementary Figure 2A. Moreover, as shown in the Calcein AM green fluorescence (viable cells) photomicrographs and plots of Supplementary Figure 2, B and C. when these cell lines were grown in suspension in non-adherent plates their sensitivity to RK-33 treatment remained in a similar range of IC50 values of 4.5 and 5.8 μM for D425-Med and HD-MB03 cells respectively.
RK-33 Induced a G1-Phase Cell Cycle Arrest and Caused Apoptosis
Next, to define the effects of RK-33 on cell cycle profile, we performed flow cytometry analysis on RK-33-treated DAOY and UW228 cells. Both DAOY and UW228 cells exhibited G1 arrest within 24 h of RK-33 treatment Figure 3D. This is indicative that RK-33-induced G1 arrest in both DAOY and UW228 cells might be dependent on DDX3 levels in these two cell lines.
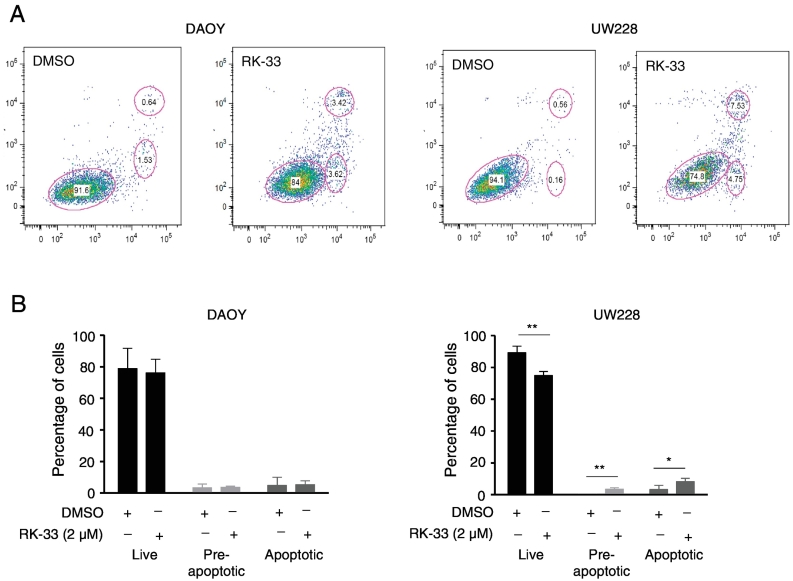
In order to delineate whether the cell cycle arrest at the G1 phase as a result of RK-33 treatment was due to apoptosis, we next performed flow cytometric analysis using Annexin V/PI staining. DAOY and UW228 cells were treated with RK-33 (2 μM), incubated for 24 h, and subjected to Annexin V/PI staining to assess the percentage of viable, early apoptotic, and late apoptotic cells Figure 4A. The analysis showed that the number of viable cells, pre-apoptotic, and apoptotic cells in DAOY was 83%, 3%, and 3%, respectively. In UW228, the number of viable cells, pre-apoptotic, and apoptotic cells was 74%, 4%, and 6%, respectively Figure 4B. From this observation, we conclude that treatment of DAOY and UW228 cells with RK-33 induced a G1-phase cell cycle arrest with minor cell death.
Figure 4.
Induction of apoptosis in DAOY and UW228 cells treated with RK-33. A. Representative histograms displaying the flow cytometric density plot of Annexin V/PI staining after DAOY and UW228 cell line treatment with RK-33 (2 μM) or DMSO (control) for 24 h. B. Histogram representing the average population of viable cells, pre-apoptotic, and apoptotic cells that were determined by estimating the stained cells with Annexin V/PI staining.
DDX3 Knockdown and RK-33 Treatment Decreased WNT/β-Catenin Signaling
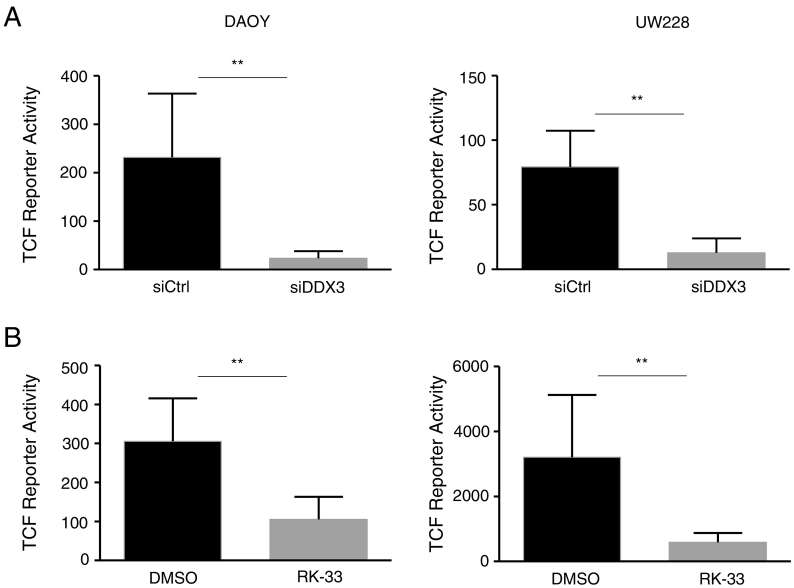
In order to investigate the relation between the interference of the oncogenic WNT/β-catenin pathway with the observed reduction in proliferation, we tested whether DDX3 inhibition causes a reduction in TCF promoter activity using a TOP/FOP luciferase assay. By transfecting DDX3 knockdown cell lines using TOP-FLASH or FOP-FLASH, the knockdown of DDX3 resulted in a significant decrease in TCF activity in both DAOY and UW228 cells. Similarly, RK-33 also significantly reduced TCF activity in both DAOY and UW228 cells Figure 5, A and B.
Figure 5.
WNT signaling activity is reduced by DDX3 inhibition in DAOY and UW228 cells. A. Histogram displaying TCF reporter activity of cell lines after treatment with 10 nM siDDX3 for DAOY cells and 25 nM for UW228 cells. B. TCF reporter activities after treatment with RK-33 (2.5 μM for DAOY cells and 3.5 μM for UW228 cells).
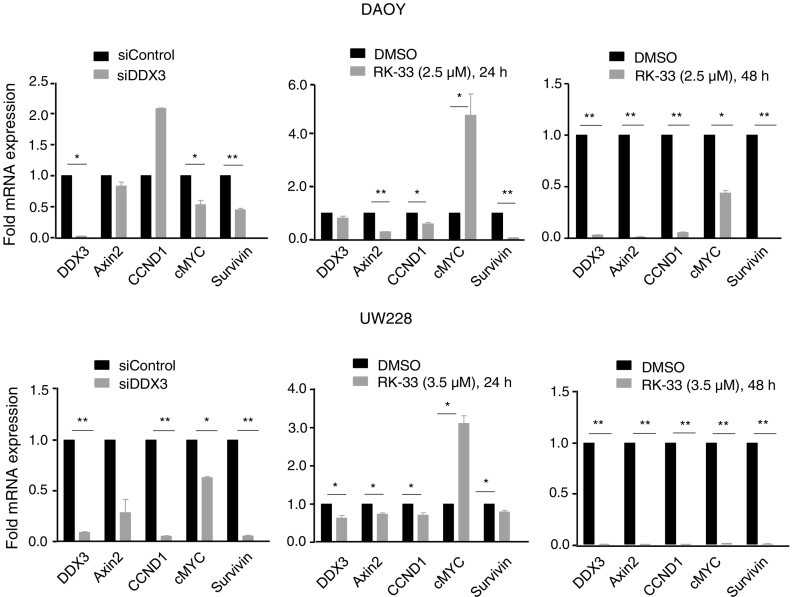
Since the WNT/β-catenin signaling pathway activates downstream target genes, such as Axin2, CCND1, MYC, and Survivin, which promote tumor progression, we next evaluated whether reduced TCF reporter activity in medulloblastoma cell lines also resulted in reduced expression of mRNA of TCF regulated genes. From this analysis, we observed that DDX3 knockdown resulted in a reduced expression of Axin2, CCND1, MYC, and Survivin in both DAOY and UW228 cell lines. RK-33 treatment also significantly reduced transcription of Axin2, CCND1, MYC, and Survivin, as shown in Figure 6. Based on these findings, we conclude that DDX3 inhibition resulted in decreased WNT/β-catenin signaling in both DAOY and UW228 cells, which further confirms the role of DDX3 in cell cycle progression and in the generation of a tumor phenotype.
Figure 6.
Quantitative reverse transcriptase polymerase chain reaction. Relative mRNA expression of WNT target genes after knockdown of DDX3 with 25 nM siDDX3 in DAOY and UW228 cell lines and RK-33 treatment (2.5 μM for DAOY cells and 3.5 μM for UW228 cells) for 24–48 h.
Combination of RK-33 and Radiation Exhibited Synergistic Cell Death Effect, Both In Vitro and in a Preclinical Model of Medulloblastoma
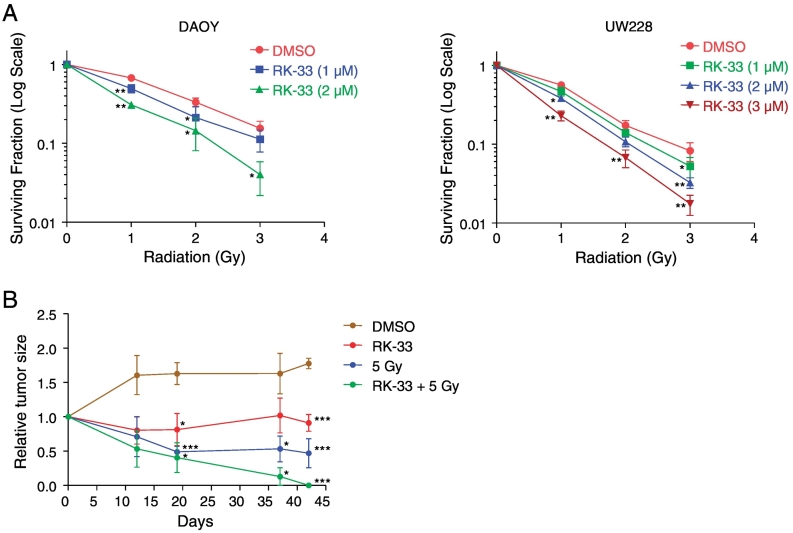
In order to evaluate the effect of RK-33 in combination with radiation, we treated DAOY and UW228 cell lines with RK-33 and different doses of radiation in a colony forming assay. Based on the data obtained, we observed that a combination of RK-33 and radiation resulted in a significantly higher inhibitory effect than using single treatment of either RK-33 or radiation Figure 7A. This indicates that RK-33 is able to radiosensitize DAOY and the UW228 cell lines, resulting in enhanced cell death.
Figure 7.
In vitro and in vivo combination of RK-33 and radiation. A. Effect of a combination of RK-33 of various concentrations (1–3 μM) and radiation of different doses (0 Gy to 3 Gy) on the survival of DAOY and UW228 cell lines. B. Graph detailing tumor size effects in DAOY-inoculated SCID mice after treatment with DMSO, RK-33, radiation (5 Gy), and a combination of RK-33 and radiation. Bars indicate SEM.
To determine whether RK-33 could be used clinically as a radio-sensitizer for medulloblastoma treatment, we treated tumors generated using DAOY cell line in nude mice, with RK-33 in combination with radiation. Following tumor generation in the flank, mice were randomly grouped and were treated with DMSO, RK-33, radiation (5 Gy), and a combination of RK-33 and radiation (5 Gy). Of the three different treatment groups, 5 Gy in combination with RK-33 demonstrated the highest tumor reduction Figure 7B. This is indicative of the use of RK-33 as a potential radiosensitizer for the treatment of medulloblastoma.
Discussion
About 10% of medulloblastomas are driven by mutations in the WNT signaling pathway, but are considered a low risk subgroup [36]. This subgroup is identified by the β-catenin nucleo-positive immunophenotype and is associated with a significantly better overall survival, thus suggesting that nuclear β-catenin accumulation is a marker of favorable outcome in medulloblastoma [37]. In a normal cell, the cytoplasmic β-catenin levels are maintained by the phosphorylation of β-catenin by a cytoplasmic destruction complex consisting of APC, axin, casein kinase 1α (CK1α), and glycogen synthase kinase (GSK3β), and further β-catenin is degraded by the ubiquitin proteasome complex [38], [39]. Mutations in the phosphorylation site lead to β-catenin stabilization, which is translocated to the nucleus where it interacts with the TCF/LEF complex and causes up-regulation of WNT target genes, such as Axin2, CCND1, MYC, and Survivin [8], [40], [41], [42], [43].
Earlier reports have shown that DDX3 is among the most mutated gene in pediatric and adult medulloblastoma [26], [44]. A positive correlation between the high expression of DDX3 and poor prognosis has been reported in human tumors [45]. DDX3 mutations often occur simultaneously along with β-catenin mutations, and this mutant DDX3 increases the activity of mutated β-catenin activity, thereby enhancing the nuclear translocation of β-catenin. The role of DDX3 in WNT/β-catenin signaling was first reported by Cruciat et al. [46]. Since a percentage of medulloblastomas are driven by mutations in the WNT signaling pathway, we investigated the possible role of DDX3 in WNT-associated medulloblastoma. Furthermore, we investigated the potent anti-tumor activity of RK-33, a small molecule inhibitor of DDX3, on medulloblastoma cell lines in vitro and in vivo. We demonstrated that DDX3 is over-expressed in medulloblastoma cell lines, and targeting DDX3 with RK-33 not only reduced WNT signaling, but also caused a G1 arrest. We also showed that the inhibition of DDX3 resulted in reduced proliferation of medulloblastoma cell lines. This demonstrates the dependence of medulloblastoma cell lines on DDX3 for their survival. However, some reports have shown a contradictory role for DDX3 as both an oncogene [28], [30], [47] as well as tumor suppressor gene [48], [49]. Several known WNT/β-catenin downstream targets, such as Axin2, CCND1, MYC, and Survivin, were decreased by RK-33 treatment, as demonstrated by RT-PCR analysis in the medulloblastoma cell lines used in this study. Earlier, it was shown that Norcantharidin impairs medulloblastoma growth by inhibiting WNT/β-catenin signaling [50]. The biflavone ginkgetin also suppresses the growth of medulloblastoma by inhibiting the WNT/β-catenin signaling pathway, inducing G2/M phase arrest, and also reducing the expression of the downstream target genes of the WNT/β-catenin signaling pathway, such as Axin2, CCNDI, and Survivin in DAOY cells [51]. Our results also parallel those, which were reported earlier.
The in vivo non-toxic effect, as well as radio-sensitization of RK-33, has been reported previously in lung cancer and prostate cancer from our laboratory [30], [32]. Based on these previous results, we evaluated the effect of RK-33 as a radio-sensitizer by performing in vitro clonogenic assays using DAOY and UW228 cells. A combination of 2 μM RK-33 and 3 Gy radiation in the DAOY cell line, and a combination of 3 μM RK-33 and 3 Gy radiation in UW228 cell line, reduced the clonogenic ability compared to RK-33 or radiation alone. Treatment of mice with DAOY flank tumors with a combination of RK-33 and a 5 Gy radiation dose showed a significant reduction in tumor size compared to RK-33 and radiation alone. Moreover, in both pediatric and adult patient medulloblastoma samples, we detected that over 55% and 66% of the samples had DDX3 expression, respectively.
Conclusions
Currently, in the field of medulloblastoma, extensive attention has been focused on the post-therapy quality of life and reducing the risk of long-term side effects. The goal is to minimize the intensity of the treatment, as well as use less toxic - and more targeted - novel agents in the treatment of medulloblastoma. In this regard, RK-33, a small molecule inhibitor of DDX3, acts as a promising novel therapeutic agent that not only can reduce the toxicities associated with the treatment, but also reduce the dose of radiation in the treatment of medulloblastoma. In conclusion, RK-33 acts as a promising small molecule inhibitor that can target and induce cell death in medulloblastomas in which DDX3/WNT-β-catenin is over-expressed.
Funding
Funding for this work was supported by Safeway (VR) and FAMRI (VR).
Acknowledgments
We would like to acknowledge support for the statistical analysis from Carol Thompson of the National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health through Grant Number 1UL1TR001079.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.09.002.
Appendix A. Supplementary Data
Supplementary Figure RK-33 inhibits viability of D425-Med and HD-MB03 medulloblastoma cell lines as monolayers and in suspension. A. Plots of viable cell numbers vs RK-33 concentration from the Calcein AM live cell assay showing that treatment of monolayers of D425-Med and HD-MB03 cells RK-33 results in cell death. Insets of immunoblots from the same gel show that both cell lines have ample amounts of DDX3. B. & C. Representative images (2x objective) of Calcein AM live (green fluorescence) assay of D425-Med and HD-MB03 grown in suspension that were treated for 72 h with the various concentrations of RK-33 (shown in the upper left corner of each image: 0 – 12 μM). The plots associated with each group of images provide the means to evaluate IC50 values.
References
- 1.Rogers HA, Sousa S, Salto C, Arenas E, Coyle B, Grundy RG. WNT/beta-catenin pathway activation in Myc immortalised cerebellar progenitor cells inhibits neuronal differentiation and generates tumours resembling medulloblastoma. Br J Cancer. 2012;107:1144–1152. doi: 10.1038/bjc.2012.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 5.Schwalbe EC, Lindsey JC, Nakjang S, Crosier S, Smith AJ, Hicks D, Rafiee G, Hill RM, Iliasova A, Stone T. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18:958–971. doi: 10.1016/S1470-2045(17)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, Grobner S, Segura-Wang M, Zichner T, Rudneva VA. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellison DW, Kocak M, Dalton J, Megahed H, Lusher ME, Ryan SL, Zhao W, Nicholson SL, Taylor RE, Bailey S. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol. 2011;29:1400–1407. doi: 10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zurawel RH, Chiappa SA, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic beta-catenin mutations. Cancer Res. 1998;58:896–899. [PubMed] [Google Scholar]
- 9.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Northcott PA, Hielscher T, Dubuc A, Mack S, Shih D, Remke M, Al-Halabi H, Albrecht S, Jabado N, Eberhart CG. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol. 2011;122:231–240. doi: 10.1007/s00401-011-0846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slade I, Murray A, Hanks S, Kumar A, Walker L, Hargrave D, Douglas J, Stiller C, Izatt L, Rahman N. Heterogeneity of familial medulloblastoma and contribution of germline PTCH1 and SUFU mutations to sporadic medulloblastoma. Fam Cancer. 2011;10:337–342. doi: 10.1007/s10689-010-9411-0. [DOI] [PubMed] [Google Scholar]
- 15.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soulat D, Burckstummer T, Westermayer S, Goncalves A, Bauch A, Stefanovic A, Hantschel O, Bennett KL, Decker T, Superti-Furga G. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 2008;27:2135–2146. doi: 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 19.Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merz C, Urlaub H, Will CL, Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 22.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandimarte L, La Starza R, Gianfelici V, Barba G, Pierini V, Di Giacomo D, Cools J, Elia L, Vitale A, Luciano L. DDX3X-MLLT10 fusion in adults with NOTCH1 positive T-cell acute lymphoblastic leukemia. Haematologica. 2014;99:64–66. doi: 10.3324/haematol.2013.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P, Taylor MD. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HH, Yu HI, Cho WC, Tarn WY. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2015;34:2790–2800. doi: 10.1038/onc.2014.190. [DOI] [PubMed] [Google Scholar]
- 29.He TY, Wu DW, Lin PL, Wang L, Huang CC, Chou MC, Lee H. DDX3 promotes tumor invasion in colorectal cancer via the CK1epsilon/Dvl2 axis. Sci Rep. 2016;6 doi: 10.1038/srep21483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, Levine A, Irving A, Korz D, Tantravedi S. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med. 2015;7:648–669. doi: 10.15252/emmm.201404368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heerma van Voss MR, Vesuna F, Trumpi K, Brilliant J, Berlinicke C, de Leng W, Kranenburg O, Offerhaus GJ, Burger H, van der Wall E. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget. 2015;6:28312–28326. doi: 10.18632/oncotarget.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie M, Vesuna F, Tantravedi S, Bol GM, Heerma van Voss MR, Nugent K, Malek R, Gabrielson KL, Van Diest PJ, Tran PT. RK-33 radiosensitizes prostate cancer cells by blocking the RNA helicase DDX3. Cancer Res. 2016;76(21):6340–6350. doi: 10.1158/0008-5472.CAN-16-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remke M, Hielscher T, Korshunov A, Northcott PA, Bender S, Kool M, Westermann F, Benner A, Cin H, Ryzhova M. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol. 2011;29:3852–3861. doi: 10.1200/JCO.2011.36.2798. [DOI] [PubMed] [Google Scholar]
- 34.Angus AG, Dalrymple D, Boulant S, McGivern DR, Clayton RF, Scott MJ, Adair R, Graham S, Owsianka AM, Targett-Adams P. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol. 2010;91:122–132. doi: 10.1099/vir.0.015909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan JS, Reed A, Chen F, Stewart CN., Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85–96. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clifford SC, Lusher ME, Lindsey JC, Langdon JA, Gilbertson RJ, Straughton D, Ellison DW. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5:2666–2670. doi: 10.4161/cc.5.22.3446. [DOI] [PubMed] [Google Scholar]
- 37.Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, Pearson AD, Clifford SC, United Kingdom Children's Cancer Study Group Brain Tumour C beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children's Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23:7951–7957. doi: 10.1200/JCO.2005.01.5479. [DOI] [PubMed] [Google Scholar]
- 38.Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25:7482–7491. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- 39.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 40.Koch A, Waha A, Tonn JC, Sorensen N, Berthold F, Wolter M, Reifenberger J, Hartmann W, Friedl W, Reifenberger G. Somatic mutations of WNT/wingless signaling pathway components in primitive neuroectodermal tumors. Int J Cancer. 2001;93:445–449. doi: 10.1002/ijc.1342. [DOI] [PubMed] [Google Scholar]
- 41.Eberhart CG, Tihan T, Burger PC. Nuclear localization and mutation of beta-catenin in medulloblastomas. J Neuropathol Exp Neurol. 2000;59:333–337. doi: 10.1093/jnen/59.4.333. [DOI] [PubMed] [Google Scholar]
- 42.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 43.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 44.Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miao X, Yang ZL, Xiong L, Zou Q, Yuan Y, Li J, Liang L, Chen M, Chen S. Nectin-2 and DDX3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. Int J Clin Exp Pathol. 2013;6:179–190. [PMC free article] [PubMed] [Google Scholar]
- 46.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 47.Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P, Jr., Mukadam S, Van Diest P, Chen JH, Farabaugh P. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27:3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 49.Wu DW, Lee MC, Wang J, Chen CY, Cheng YW, Lee H. DDX3 loss by p53 inactivation promotes tumor malignancy via the MDM2/Slug/E-cadherin pathway and poor patient outcome in non-small-cell lung cancer. Oncogene. 2014;33:1515–1526. doi: 10.1038/onc.2013.107. [DOI] [PubMed] [Google Scholar]
- 50.Cimmino F, Scoppettuolo MN, Carotenuto M, De Antonellis P, Dato VD, De Vita G, Zollo M. Norcantharidin impairs medulloblastoma growth by inhibition of Wnt/beta-catenin signaling. J Neurooncol. 2012;106:59–70. doi: 10.1007/s11060-011-0645-y. [DOI] [PubMed] [Google Scholar]
- 51.Ye ZN, Yu MY, Kong LM, Wang WH, Yang YF, Liu JQ, Qiu MH, Li Y. Biflavone ginkgetin, a novel Wnt inhibitor, suppresses the growth of medulloblastoma. Nat Prod Bioprospect. 2015;5(2):91–97. doi: 10.1007/s13659-015-0056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure RK-33 inhibits viability of D425-Med and HD-MB03 medulloblastoma cell lines as monolayers and in suspension. A. Plots of viable cell numbers vs RK-33 concentration from the Calcein AM live cell assay showing that treatment of monolayers of D425-Med and HD-MB03 cells RK-33 results in cell death. Insets of immunoblots from the same gel show that both cell lines have ample amounts of DDX3. B. & C. Representative images (2x objective) of Calcein AM live (green fluorescence) assay of D425-Med and HD-MB03 grown in suspension that were treated for 72 h with the various concentrations of RK-33 (shown in the upper left corner of each image: 0 – 12 μM). The plots associated with each group of images provide the means to evaluate IC50 values.