Abstract
Lung cancers, the leading cause of cancer mortality worldwide, are characterized by a high metastatic potential. Growing evidence reveals that Spindlin 1 (SPIN1) is involved in tumor progression and carcinogenesis. However, the role of SPIN1 in non-small-cell lung cancer (NSCLC) and the molecular mechanisms underlying SPIN1 in human NSCLC remain undetermined. Here we examined the function of SPIN1 in human NSCLC and found that the expression of SPIN1 was closely correlated with the overall survival and poor prognosis of NSCLC patients. Aberrant regulation of microRNAs (miRNAs) has an important role in cancer progression. We revealed that miR-409 inhibits the expression of SPIN1 by binding directly to the 3′ UTR of SPIN1 using dual-luciferase reporter assays. Overexpression of miR-409 significantly suppressed cell migration, growth, and proliferation by inhibiting SPIN1 in vitro and in vivo. SPIN1 overexpression in miR-409-transfected NSCLC cells effectively rescued the suppression of cell migration, growth, and proliferation regulated by miR-409. miR-409 regulates the PI3K/AKT (protein kinase B) pathway in NSCLC. Moreover, clinical data showed that NSCLC patients with high levels of miR-409 experienced significantly better survival. miR-409 expression was also negatively associated with SPIN1 expression. Taken together, these findings highlight that the miR-409/SPIN1 axis is a useful pleiotropic regulatory network and could predict the metastatic potential in NSCLC patients early, indicating the possibility that miR-409 and SPIN1 might be attractive prognostic markers for treating NSCLC patients.
Keywords: non-small lung cancer, NSCLC, miR-409, SPIN1, therapy
Introduction
Non-small-cell lung cancer (NSCLC) has been implicated as the most common type of lung cancer worldwide.1, 2, 3, 4, 5, 6 The survival rate of patients with NSCLC after diagnosis is only slightly more than 15%. Despite enormous advances in radiotherapy and chemotherapy to treat NSCLC during the past decades,3, 7, 8, 9, 10, 11, 12, 13 the clinical outcome for NSCLC patients has not yet significantly improved. Therefore, identification of new therapeutic approaches and candidates for clinical treatment of NSCLC is necessary.
Spindlin 1 (SPIN1), a member of the SPIN/SSTY gene family, has emerged as an essential regulator that is specifically expressed during gametogenesis in unfertilized oocytes and embryos in mice.14 SPIN1 regulates meiotic resumption and forms a ribonucleoprotein complex during meiotic maturation in mouse oocytes.15 SPIN1 overexpression in somatic cells also causes metaphase arrest.15 Recent reports have indicated that ectopic expression of SPIN1 is closely correlated with cancer progression and metastasis. SPIN1 promotes ovarian cancer cell proliferation through activation of WNT/TCF-4 signaling,14 suggesting that SPIN1 might have key roles in the aggressiveness of tumors. In human triple-negative breast cancer (TNBC), SPIN1 has been revealed to be upregulated.16 SPIN1 is also overexpressed in glioma tissues and has been identified as a potential candidate of miR-489.17 However, the essential role of SPIN1 in human NSCLC development and carcinogenesis remains largely unknown.
MicroRNAs (miRNAs) are small, non-coding, single-stranded RNAs that have been described to function as an endogenous means of RNAi and plays critical roles in various essential biological processes, including development, metabolism, and apoptosis.18, 19, 20, 21, 22 Accumulating evidence indicates that miRNAs such as miR-138,23 miR-330-3p,24 miR-503,25 and miR-582-3p are grossly dysregulated in NSCLC pathogenesis.26 However, the molecular mechanisms by which miRNAs regulate SPIN1 in human NSCLC carcinogenesis and the possible link between SPIN1 and miRNAs in human NSCLC have not yet been fully elucidated.
In this work, we identified that SPIN1 is closely associated with the overall survival and prognosis of NSCLC patients. SPIN1 was found to be a novel direct downstream target and functional regulator of miR-409 in NSCLC. We also demonstrated that miR-409 plays an important role in controlling cell migration, growth, and proliferation by targeting SPIN1 using in vivo and in vitro approaches. Furthermore, miR-409 expression was negatively associated with SPIN1 expression in NSCLC patients. Therefore, miR-409 and SPIN1 might be promising novel prognostic and therapeutic biomarkers for treating NSCLC patients.
Results
SPIN1 in NSCLC Patients Is Closely Correlated with Clinical Prognosis
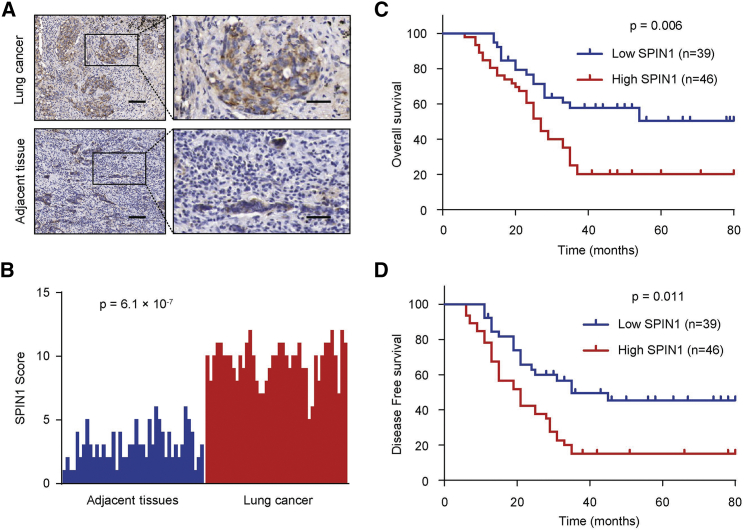
To examine the function of SPIN1 in NSCLC patients, we carried out a histopathologic assay and investigated SPIN1 expression in 85 NSCLC samples and adjacent noncancerous tissues (Figure 1A). We found that SPIN1 possessed a higher expression in human NSCLC tissues than in the corresponding nontumor tissues (p = 6.1 × 10−7) (Figure 1B). We then detected the clinical parameters of SPIN1 in NSCLC patients to identify the association between SPIN1 expression and clinicopathological characteristics. The findings showed that SPIN1 expression was closely correlated with tumor size, histological stage, pleural invasion, and metastasis (Table 1). According to the Kaplan-Meier survival analysis, we identified that NSCLC patients with low expression of SPIN1 had better overall survival (OS) (p = 0.006) and disease-free survival (DFS) (p = 0.011) than those with high SPIN1 expression (Figures 1C and 1D). Taken together, these findings reveal the important clinical significance of SPIN1 in prognosis and metastasis of NSCLC patients.
Figure 1.
SPIN1 Is Closely Correlated with Clinical Prognosis in NSCLC Patients
(A) Representative immunohistochemistry images of SPIN1 expression in human lung cancer tissues and adjacent tissues. Scale bars, 150 μm (left) and 50 μm (right). (B) SPIN1 expression scores in lung cancer tissues and matched adjacent tissues (n = 85) were compared with the Mann-Whitney U test. (C) Kaplan-Meier survival curves and log rank tests were used to compare overall survival (OS) of NSCLC patients with low and high scores for SPIN1. (D) Kaplan-Meier survival curves and log rank tests were used to compare disease-free survival (DFS) of NSCLC patients with low and high scores for SPIN1.
Table 1.
Associations between SPIN1 Expression and Clinicopathological Characteristics
| Characteristics | n | SPIN1 Expression |
p Value | |
|---|---|---|---|---|
| High (n, %) | Low (n, %) | |||
| Age | ||||
| ≥60 | 47 | 25 (53.2) | 22 (46.8) | 0.438 |
| <60 | 38 | 17 (44.7) | 21 (55.3) | |
| Gender | ||||
| Male | 62 | 30 (48.4) | 32 (51.6) | 0.505 |
| Female | 23 | 13 (56.5) | 10 (43.5) | |
| Tumor Size (cm) | ||||
| >3 | 34 | 26 (76.5) | 8 (23.5) | 5.689 × 10−7** |
| ≤3 | 51 | 11 (21.6) | 40 (78.4) | |
| TNM Stage | ||||
| I | 43 | 10 (23.3) | 33 (76.7) | 2.726 × 10−4** |
| II | 31 | 18 (58.1) | 13 (41.9) | |
| III | 11 | 9 (81.8) | 2 (18.2) | |
| Pleural Invasion | ||||
| No | 68 | 17 (25.0) | 51 (75.0) | 3.910 × 10−4** |
| Yes | 17 | 12 (70.6) | 5 (29.4) | |
| Metastasis | ||||
| No | 51 | 16 (31.4) | 35 (68.6) | 0.001** |
| Yes | 34 | 23 (67.6) | 11 (32.4) | |
The p values were calculated by Pearson’s χ2 test. *p < 0.05, **p < 0.01. TNM, tumor, node, metastasis.
miR-409 Inhibits the Expression of SPIN1 by Targeting Its 3′ UTR
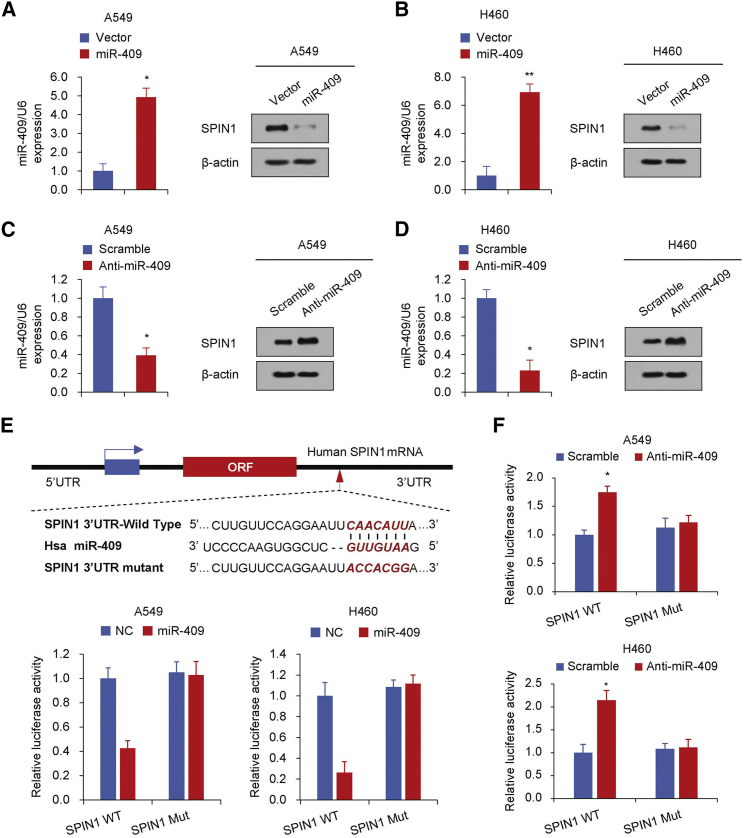
Next we used two target prediction programs, TargetScan and miRanda, to investigate the potential targeting miRNAs of SPIN1 and predict its potential functional binding site. Several miRNA candidates that target SPIN1 were selected, including miR-23-3p, miR-409, miR-431-5p, and miR-489. We then performed a western blot analysis to demonstrate the effect of the abovementioned miRNAs on the expression of SPIN1 in human NSCLC cell lines (Figure S1). Consistent with the findings seen previously in glioma,17 miR-489 also inhibited SPIN1 expression in human NSCLC cells. Importantly, miR-409 had the most pronounced inhibitory effect on SPIN1 expression. Hence, we hypothesize that miR-409 might be an important regulator in human NSCLC. According to the western blot analysis, we identified that overexpression of miR-409 significantly inhibited the production of SPIN1 in human NSCLC cell lines (Figures 2A and 2B; Figure S1). miR-409 inhibition also promoted SPIN1 expression in the same NSCLC cell lines (Figures 2C and 2D). Notably, miR-409 did not regulate the mRNA expression level of SPIN1, indicating that this regulation is posttranscriptional (Figure S2).
Figure 2.
miR-409 Inhibits SPIN1 Expression by Targeting its 3′ UTR
(A–D) Immunoblot analysis of the indicated NSCLC cell lines transfected with miR-409 (A and B) or anti-miR-409 (C and D). The histograms on the left of the immunoblots show the corresponding miR-409 mRNA expression levels. (E) miRNA luciferase reporter assays in A549 and H460 cells co-transfected with wild-type or mutated SPIN1 reporters and miR-409. Top: wild-type and mutant forms of putative miR-409 target sequences in the 3′ UTR of SPIN1. Bold and italicized fonts indicate putative miR-409-binding sites in the 3′ UTR of SPIN1. Underlining indicates mutations introduced into the 3′ UTR of SPIN1. (F) miRNA luciferase reporter assays in A549 and H460 cells co-transfected with wild-type or mutated SPIN1 reporters and anti-miR-409. Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05, **p < 0.01.
We next transiently co-transfected the A549 and H460 cell lines with luciferase reporter constructs containing the wild-type or mutated SPIN1 3′ UTR and miR-409 or anti-miR-409 to determine whether SPIN1 is a direct and specific target of miR-409. The data identified that miR-409 overexpression inhibited the 3′ UTR luciferase reporter activity of SPIN1 but did not have an effect on the luciferase activity of the reporter in which the miR-409 binding sites were mutated (Figure 2E). miR-409 inhibition also promoted the 3′ UTR reporter luciferase activity of SPIN1 (Figure 2F). Taken together, these findings show that miR-409 inhibits SPIN1 expression by directly targeting the 3′ UTR of SPIN1 in human NSCLC.
miR-409 Inhibits the NSCLC Cell Migration, Growth, and Proliferation Abilities through Inhibition of SPIN1
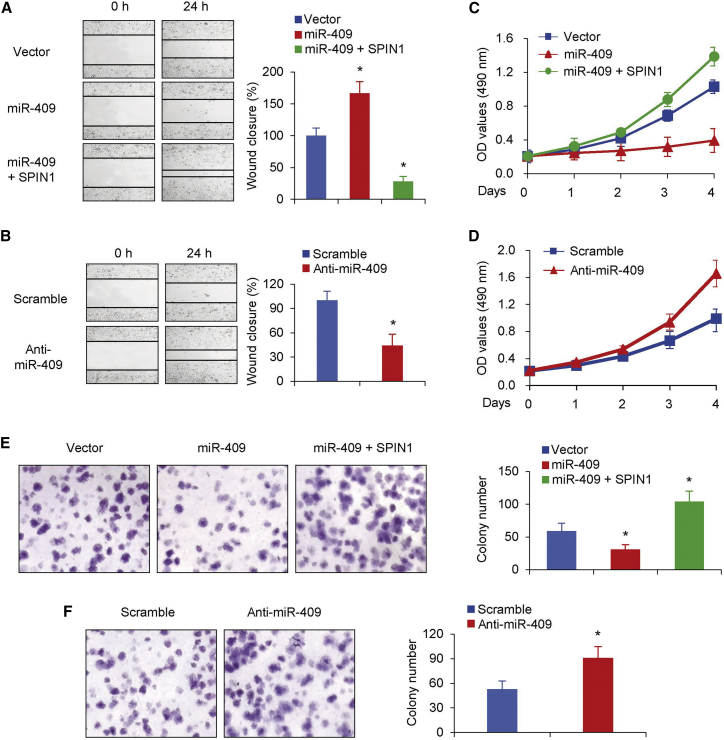
Next we used wound healing assays to find out whether miR-409 regulates the migration ability of NSCLC cell lines. The findings indicated that miR-409 overexpression significantly suppressed the migration ability of NSCLC cell lines, whereas re-expression of SPIN1 rescued the effect of miR-409 on NSCLC cells (Figure 3A). Furthermore, inhibition of miR-409 promoted the migration of human NSCLC cells compared with a control treatment (Figure 3B), which was in line with the data found above.
Figure 3.
miR-409 Inhibits NSCLC Cell Migration, Growth, and Proliferation through Inhibition of SPIN1
(A and B) Wound healing assays were conducted in A549 cells transfected with miR-409 or miR-409 and SPIN1 (A) or miR-409 inhibitor (B). Cell migration was measured 24 hr after the cell layers were scratched. (C and D) A549 cells expressing miR-409 or miR-409 and SPIN1 (C) and A549 cells transfected with miR-409 inhibitor (D) were cultured in regular medium. At the specified times, cell numbers were determined with the CCK-8 assay. The representative immunoblot shows SPIN1 expression. (E and F) A549 cells transfected with miR-409 (E) or miR-409 inhibitor (F) were plated and assayed for colony formation after 3 weeks. Representative images show colonies in plates (left). Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05.
We then performed cell growth and colony formation assays to assess whether miR-409 had an effect on the cell proliferation phenotypes of NSCLC cell lines. Cells transfected with miR-409 were used for cell growth analysis. Consistent with the data above, overexpression of miR-409 suppressed the proliferative ability and colony formation (Figures 3C and 3E). Moreover, SPIN1 restoration reversed the effect of miR-409 on cell proliferation and colony formation (Figures 3C and 3E). miR-409 inhibition also promoted NSCLC cell proliferation and colony formation (Figure 3D and 3F). Taken together, these data demonstrate that miR-409 impairs cell proliferation by suppressing SPIN1 production, suggesting that SPIN1 is an important regulator of miR-409 function in human NSCLC progression.
miR-409 Regulates Activation of the PI3K/AKT Pathway in NSCLC
Previous reports have revealed that SPIN1 mediates the phosphatidylinositol 3-kinase (PI3K)/AKT (protein kinase B) pathway in breast cancer, melanoma, and glioma.17, 27, 28 SPIN1 has been shown to regulate the process of spindle organization and chromosomal stability.15, 27, 29 The findings above demonstrate that miR-409 acts as the upstream regulator of SPIN1. We next examined whether miR-409 regulates spindle organization and chromosomal stability through activation of the PI3K/AKT pathway in NSCLC. The results indicated that miR-409 overexpression inhibited CREB1 and BCL2 expression in the PI3K/AKT signaling pathway (Figures S3A–S3C), whereas miR-409 knockdown promoted CREB1 and BCL2 expression (Figures S3D–S3F). Furthermore, overexpression of miR-409 suppressed phosphorylated AKT (p-AKT) as well as downstream targets of the PI3K/AKT pathway, including CyclinD1 (Figure S3G). Inhibition of miR-409 upregulated the PI3K/AKT pathway with increased expression levels of p-AKT and CyclinD1 (Figure S3H). Taken together, these data indicate that miR-409 regulates the PI3K/AKT pathway in NSCLC.
miR-409 Inhibits the Initiation of NSCLC
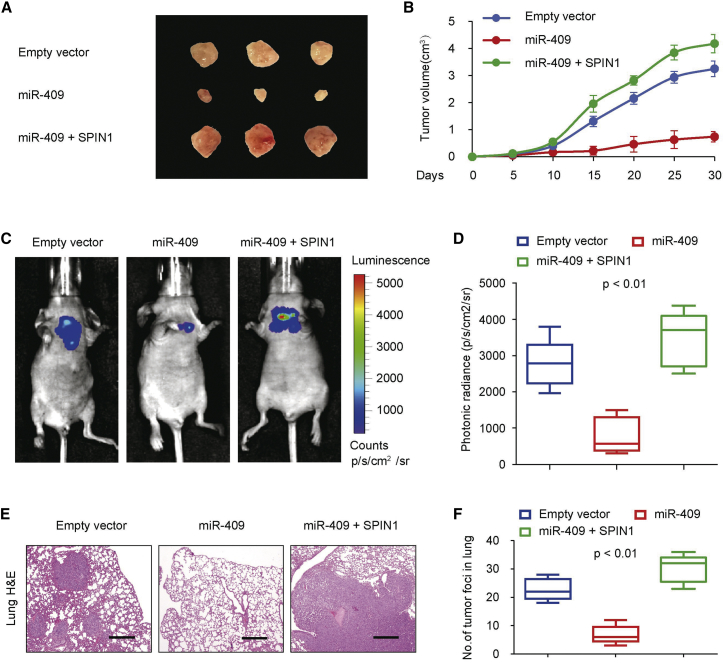
We further determined the effect of miR-409 on A549 cell growth in nude mice to investigate the miR-409 expression phenotype in vivo. We found that miR-409 overexpression markedly suppressed tumor growth in vivo (Figures 4A and 4B). Besides, tumors in mice inoculated with miR-409 plus SPIN1-overexpressing A549 cells revealed a reversal effect of miR-409 on NSCLC tumor growth (Figures 4A and 4B).
Figure 4.
miR-409 Inhibits the Initiation of NSCLC
(A and B) Stable A549 cells overexpressing miR-409 and miR-409 and SPIN1 were injected into nude mice (A). At the indicated times, tumors were measured with Vernier calipers (mean ± SD, n = 6) (B). (C and D) Bioluminescence imaging of metastasis of NSCLC cells in non-obese diabetic (NOD)-SCID mice 28 days after intravenous injection of cells infected with PCDH-control, PCDH-miR-409, or PCDH-miR-409 and SPIN1 via the lateral tail vein (C). The luminescence signal is represented by an overlaid false-color image, with the signal intensity indicated by the scale (D). (E and F) Representative foci of lungs were subjected to histological analyses (E). The data are shown as the mean ± SD (n = 6) (F).
Next we assessed the effect of miR-409 on lung burden. The data showed that the miR-409-expressing group had a more significant decrease in lung burden compared with the control group (Figure 4C). Similar results were also observed regarding the photonic radiance intensity of the lungs in the miR-409-expressing group (Figure 4D). The miR-409 plus SPIN1 group had impaired expression of miR-409 (Figure 4D). We next performed a histologic analysis of the mouse lungs. Indeed, the findings confirmed the tumor foci (Figures 4E and 4F). The miR-409-expressing group presented fewer tumor foci in the lungs than the control group, whereas the miR-409 plus SPIN1 group showed reversal effects of miR-409 on tumor foci. Together, these data strongly demonstrate that miR-409 and SPIN1 function as an important regulator in NSCLC dissemination.
miR-409 Expression in Human NSCLC Samples and the Correlation between miR-409 and SPIN1
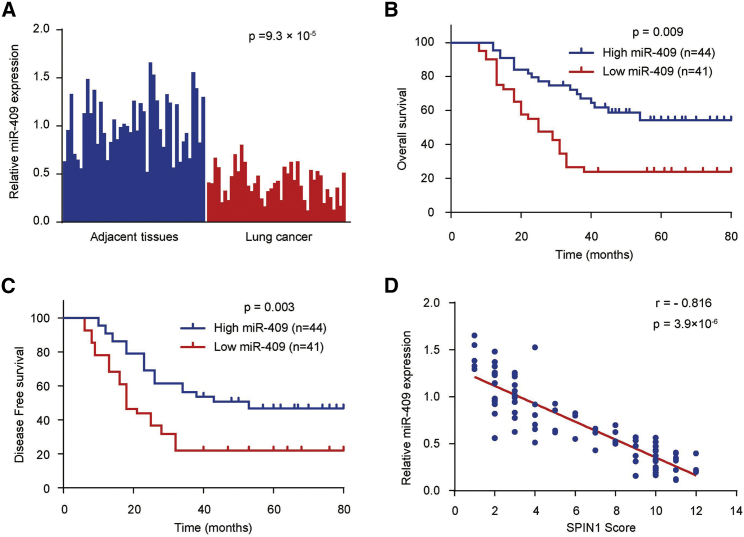
To investigate the clinical significance of miR-409 in NSCLC patients, we performed a qRT-PCR assay and examined the expression of miR-409 in 85 NSCLC samples and matched adjacent nontumor tissues. We found that the expression of miR-409 was significantly downregulated in NSCLC patients according to the qRT-PCR analysis (p = 9.3 × 10−5) (Figure 5A). We next investigated the association between miR-409 expression and clinicopathological characteristics to determine the clinical significance of miR-409. The data revealed that miR-409 expression was closely correlated with NSCLC histological stage, tumor size, pleural invasion, and metastasis (Table 2). A Kaplan-Meier survival analysis revealed that NSCLC patients with high expression levels of miR-409 had better OS (p = 0.009) and DFS (p = 0.003) than patients with low miR-409 expression levels, suggesting that miR-409 might be a predictor of better human NSCLC clinical outcomes (Figures 5B and 5C). Furthermore, miR-409 expression was negatively correlated with SPIN1 protein expression in NSCLC samples (p = 3.9 × 10−6, r = −0.816) (Figure 5D; Figure S4), which was in line with miR-409 inhibition of SPIN1 protein expression in cultured cells (Figure 2A; Figure S1). Collectively, these findings strongly indicate an important role of miR-409 and SPIN1 in the prognosis of human NSCLC.
Figure 5.
miR-409 Expression in Human NSCLC and the Correlation between miR-409 and SPIN1
(A) The expression of miR-409 in NSCLC tissues and matched adjacent tissues (n = 85) was compared using the Mann-Whitney U test. U6 small nuclear RNA was used as the internal control. (B) Kaplan-Meier survival curves and log rank tests were used to compare OS of NSCLC patients with high and low expression levels of miR-409. (C) Kaplan-Meier survival curves and log rank tests were used to compare the DFS of NSCLC patients with high and low expression levels of miR-409. (D) The relationship between miR-409 and SPIN1 expression based on western blot and immunohistochemistry assay was assessed by Spearman’s rank correlation analysis in NSCLC samples. The symbols represent individual samples.
Table 2.
Associations between miR-409 Expression and Clinicopathological Characteristics
| Characteristics | n | miR-409 Expression |
p Value | |
|---|---|---|---|---|
| High (n, %) | Low (n, %) | |||
| Ages | ||||
| ≥60 | 47 | 22 (46.8) | 25 (53.2) | 0.593 |
| <60 | 38 | 20 (52.6) | 18 (47.4) | |
| Gender | ||||
| Male | 62 | 33 (53.2) | 29 (46.8) | 0.658 |
| Female | 23 | 11 (47.8) | 12 (53.2) | |
| Tumor Size (cm) | ||||
| >3 | 34 | 11 (32.4) | 23 (67.6) | 5.148 × 10−5** |
| ≤3 | 51 | 39 (76.5) | 12 (23.5) | |
| TNM Stage | ||||
| I | 43 | 29 (67.4) | 14 (32.6) | 0.026** |
| II | 31 | 14 (45.2) | 17 (54.8) | |
| III | 11 | 3 (27.3) | 8 (72.7) | |
| Pleural invasion | ||||
| No | 68 | 47 (69.1) | 21 (30.9) | 0.010* |
| Yes | 17 | 6 (35.3) | 11 (64.7) | |
| Metastasis | ||||
| No | 51 | 38 (74.5) | 13 (25.5) | 1.165 × 10−4** |
| Yes | 34 | 11 (32.4) | 23 (67.6) | |
The p values were calculated by Pearson’s χ2 test. *p < 0.05, **p < 0.01. TNM, tumor, node, metastasis.
Discussion
Accumulating evidence has shown that miRNAs are aberrant in various cancers and closely correlated with progression and metastasis.30, 31, 32, 33, 34, 35, 36 Hence, identification of tumor-related miRNAs might be critical for current effective and specific therapy. miR-409 acts as a novel miRNA regulator in biological behavior in some tumors. miR-409 inhibits tumor cell invasion and metastasis by directly targeting radixin in gastric cancers.37 miR-409 also functions as a tumor suppressor in bladder cancer.38 Nevertheless, the exact function of miR-409 in human NSCLC remains largely unknown. In the present study, we found that miR-409 has a tumor-suppressive effect on human NSCLC migration, growth, and proliferation by directly targeting SPIN1. miR-409 regulated activation through the PI3K/AKT pathway in NSCLC, and miR-409 expression in human NSCLC was markedly downregulated compared with adjacent tissues. Furthermore, NSCLC patients with high levels of miR-409 displayed better OS and DFS, supporting that miR-409 functions as a novel predictive and prognostic regulator in human NSCLC.
SPIN1, a member of the SPIN/SSTY gene family, was first identified as a target highly expressed in ovarian cells.39 SPIN1 has been shown to regulate the process of spindle organization and chromosomal stability and plays an important role in the progression of cancer.15, 27, 29 Reducing SPIN1 expression levels in vitro and in xenograft mouse models strongly impairs proliferation and increases apoptosis of liposarcoma cells.40 SPIN1 has also been shown to function as a histone code reader to control skeletal muscle functional networks.41 In this work, we found that the miR-409/SPIN1 axis could regulate cell migration and proliferation in NSCLC cells through the PI3K/AKT pathway. We also identified that the SPIN1 overexpression directly promoted NSCLC progression. Importantly, SPIN1 was closely associated with NSCLC carcinogenesis-related cellular properties such as motility and migration, suggesting that SPIN1 might be a promising attractive therapeutic target for intervention in human NSCLC.
Currently, emerging data have revealed that SPIN1 inhibition affects tumor biological motility and activity. One recent study found that SPIN1 small-molecule inhibitors block transcriptional coactivator activity.42 SPIN1 was also upregulated in glioma compared with normal tissues, and SPIN1 has been identified to be a potential target of miR-489,17 which was also demonstrated in NSCLC cells in our study. However, the exact function of SPIN1 in NSCLC and the miRNAs in regulating SPIN1 expression in NSCLC still remains to be elucidated. Conceivably, SPIN1 inhibition might be a promising potential molecular therapeutic target for treating human NSCLC. In this study, we revealed that high expression of SPIN1 was more frequent in human NSCLC tissues and that NSCLC patients with high levels of SPIN1 presented with worse OS and DFS. In addition, miR-409 overexpression inhibited cell migration, growth, and proliferation in vitro and in vivo. Furthermore, we performed clinical analyses and revealed a negative correlation between miR-409 and SPIN1 expression in human NSCLC samples, suggesting that the miR-409/SPIN1 axis might be a novel promising candidate for the prevention human NSCLC.
Taken together, our data reveal that high expression of SPIN1 is more frequent in human NSCLC tissues and that NSCLC patients with high levels of SPIN1 presented with worse OS and DFS, suggesting that SPIN1 functions as an important independent biomarker that could predict the clinical outcome of human NSCLC. miR-409 could inhibit cell migration, growth, and proliferation in NSCLC by directly targeting SPIN1. The SPIN1 expression level was upregulated in NSCLC patients and negatively correlated with that of miR-409, suggesting that the miR-409/SPIN1 axis might be a potential predictive therapeutic candidate for human NSCLC clinical outcomes.
Materials and Methods
Patients and Specimens
This research was performed with informed consent from the patients and approved by the Institutional Review Committee of the General Hospital of the People’s Liberation Army (Beijing, China). 85 conventional NSCLC and adjacent noncancerous tissues were investigated on the basis of accepted radiological and pathological criteria. The follow-up information of the patients was updated every month. OS was defined as the time elapsed from surgery to death. Specimens were divided into two portions; one portion was used for histopathologic assessment, and the other portion was immediately snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction. The clinical and demographic characteristics are presented in Table S1. Clinical information was collected according to the records of the patients.
Histopathologic Assessment
For histopathologic assays, tissues were fixed in 4% buffered paraformaldehyde for 48 hr and subsequently decalcified with buffered EDTA (20% EDTA [pH 7.4]). Tissues were then embedded in paraffin, sectioned, and stained with H&E. For immunohistochemistry (IHC) assays, the sections were pretreated with trypsin (0.05%) for 10 min before treatment with 3% (v/v) H2O2 for 15 min. Then the sections were then blocked with 10% goat serum at room temperature for 1 hr. After washing with PBS, anti-SPIN1 antibody (1:50 dilution) was applied to the sections, and the sections were incubated at 4°C overnight. The sections were then washed with PBS for 15 min and incubated with biotinylated secondary antibody using a Histostain Plus kit (Invitrogen, Carlsbad, CA, USA). The sections were washed and incubated with 3,3′-diaminobenzidine (DAB) substrate for 2 min. The IHC staining was evaluated by two pathologists blinded to the origin of the specimen using light microscopy. The H-score method, which combines the values of immunoreaction intensity and the percentage of cells stained, was applied to investigate the total immunohistochemical scoring as described previously.28 Briefly, the H-score was achieved by multiplying the percentage of weakly stained cells (× 1), the percentage of moderately stained cells (× 2), and the percentage of strongly stained cells (× 3). A score of 2.1 or less was defined as low score, and a score between 2.1 and 3 was defined as a high score.
Cell Culture and Transfection
The A549 and H460 cell lines, which were tested for mycoplasma contamination, were bought from the American Type Culture Collection (Manassas, VA, USA). Cells were routinely cultured at 37°C in an atmosphere with 5% CO2 and in DMEM with high glucose supplemented with 100 μg/mL streptomycin and 10% fetal calf serum (FCS) and 100 IU/mL penicillin. For transfection, cells were seeded with the indicated plasmids in 6- or 24-well plates using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. The miRNA inhibitors (Ambion, Grand Island, NY, USA) were transfected at a concentration of 50 nM. The miRNA mimics were transfected into the cells using FuGENE HD (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
Plasmids and Reagents
Mutant or wild-type promoter-containing luciferase reporters were constructed by insertion of PCR-amplified promoter fragments from genomic DNA into the pGL4-Basic vector (Promega, Madison, WI, USA). The primer sequences are presented in Table S2. To introduce mutations into the seed sequences of the predicted miR-409 target sites within the 3′ UTR of SPIN1, recombinant PCR was performed using the primers mentioned (Table S2). Lentiviruses were produced by co-transfection of HEK293T cells with recombinant lentiviral vectors and pPACK Packaging Plasmid Mix (System Biosciences, Mountain View, CA, USA) using MegaTran reagent (OriGene, Rockville, MD, USA). After 48-hr transfection, the lentiviruses were collected and added to the medium of the target cells with 8 μg/mL Polybrene (Sigma-Aldrich, St. Louis, MO, USA). Stable cell lines were selected for approximately 2 months with 1 μg/mL puromycin. Individual clones or pooled clones were screened by standard immunoblotting protocols and produced similar results. Anti-SPIN1 (ab216498), anti-β-actin (ab8227), anti-CREB1 (ab32515), and anti-BCL2 (ab32124) were purchased from Abcam (Cambridge, MA, USA). Anti-p-AKT (4060), anti-AKT (9272), and anti-Cyclin D1 (2978) were purchased from Cell Signaling Technology (Beverly, MA, USA).
Cell Immunofluorescence
Cells were cultured on coverslips and exposed to treatment. Cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100 for 5 min. Blocking was performed with 3% normal serum for 20 min, and then the cells were stained with the indicated antibody. After washing, the nuclei were counterstained with DAPI. Immunofluorescence was visualized using an Olympus BX41 microscope, and images were captured using a DP70 Olympus digital camera at high resolution.
Western Blotting
Total protein extracts were prepared for western blot analysis as described previously.43 The immunocomplexes were visualized via chemiluminescence using an enhanced chemiluminescence (ECL) kit (Amersham Biosciences, Piscataway, NJ, USA).
Luciferase Assay
Cells at 60% confluence were seeded in 24-well plates. Using Lipofectamine 2000 reagent, reporter constructs containing the mutant or wild-type 3′ UTR of SPIN1 were co-transfected with miR-409 into cells according to the manufacturer’s protocol. The cells were collected after 48 hr and assessed for β-galactosidase and luciferase activities as described previously.28
Wound Healing Assays
Cells at 60% confluence were seeded in 6-well plates in culture medium for wound healing assays. After 24 hr, the confluent cellular monolayer was scratched with a fine pipette tip. For migration, using a microscope, the rate of wound closure was observed at the indicated times.
Anchorage-Dependent and Anchorage-Independent Growth Assays
Cell proliferation was examined using a CCK-8 Kit (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. To analyze anchorage-independent growth, transfected cells were seeded in 96-well plates and examined at 0, 24, 48, 72, and 96 hr as described previously.44
RNA Extraction and qRT-PCR
Total RNA was extracted and reverse-transcribed into cDNA using an RNeasy Mini kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. The CFX Connect Real-Time PCR Detection System (Life Science Research, Hercules, CA, USA) was used to detect and quantify miRNA expression. The relative expression level of the miRNA was calculated using the comparative cycle threshold (Ct) method. Universal small nuclear RNA U6 (RNU6B) was used as the endogenous control for the miRNAs. The sequences of the primers used for qRT-PCR analysis are listed in Table S2.
Animal Experiments
The animal studies were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at the General Hospital of the People’s Liberation Army. Approximately 1.3 × 107 A549 cells were injected into 6-week-old BALB/c mice. For the tumor growth model, cells labeled with firefly luciferase and stably transfected with the pCDH control vector, pCDH-miR-409, or PCDH-miR-409 and SPIN1 were injected subcutaneously. Tumor volume was examined according to the following formula: volume = (longest diameter × shortest diameter2)/2. Tumor growth was calculated by caliper measurements. Excised tumors were weighed, and portions were frozen in liquid nitrogen or fixed in 4% paraformaldehyde for further study. The animals were imaged on day 35 using the IVIS200 imaging system (Xenogen, Alameda, CA, USA). The mice were then killed, and the lungs were weighed and fixed with 4% paraformaldehyde for further study. For in vivo lung locus study, 1.1 × 107 A549 cells labeled with firefly luciferase carrying the indicated constructs were injected into the lateral tail vein of BALB/c female mice.
Statistical Analysis
To perform univariate and multivariate analyses, the Cox regression model was applied. The survival analysis was performed using the Kaplan-Meier method, and differences in survival curves were evaluated by log rank test. The qRT-PCR analysis was examined using one-way ANOVA with Tukey’s post hoc test. Correlation was determined through Pearson’s χ2 analysis using GraphPad Prism 6 (GraphPad, San Diego, CA, USA). All statistical tests were two-sided. Statistical calculations were applied using SPSS 17.0. The data are shown as the means ± SD. All in vitro experiments were carried out in triplicate and were repeated three times. p < 0.05 was considered statistically significant
Author Contributions
Q.S. and S.J. conceived the project and designed the experiments. Q.S., Q.J., J.X., F.L., and Y.C. performed the experiments. Q.S., Q.J., and Y.X. wrote the manuscript. All authors analyzed the data. Y.X. supervised the project.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This study was financially supported by grants from the Beijing Natural Science Foundation (7174349), the Chinese Geriatric Oncology Society Foundation (CGOS-05-2016-1-2-00300), and the National Natural Science Foundation (81672195 and 81371976).
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.08.020.
Contributor Information
Yameng Xu, Email: xuym0928@163.com.
Shunchang Jiao, Email: jiaosc@vip.sina.com.
Supplemental Information
References
- 1.Hirsch F.R., Scagliotti G.V., Mulshine J.L., Kwon R., Curran W.J., Jr., Wu Y.L., Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R., Karachaliou N. Lung cancer in 2014: optimizing lung cancer treatment approaches. Nat. Rev. Clin. Oncol. 2015;12:75–76. doi: 10.1038/nrclinonc.2014.225. [DOI] [PubMed] [Google Scholar]
- 3.Spiegel M.L., Goldman J.W., Wolf B.R., Nameth D.J., Grogan T.R., Lisberg A.E., Wong D.J.L., Ledezma B.A., Mendenhall M.A., Genshaft S.J. Non-small cell lung cancer clinical trials requiring biopsies with biomarker-specific results for enrollment provide unique challenges. Cancer. 2017;123:4800–4807. doi: 10.1002/cncr.31056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espana-Serrano L., Chougule M.B. Enhanced Anticancer Activity of PF-04691502, a Dual PI3K/mTOR Inhibitor, in Combination With VEGF siRNA Against Non-small-cell Lung Cancer. Mol. Ther. Nucleic Acids. 2016;5:e384. doi: 10.1038/mtna.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Q., Li A., Tian Y., Liu Y., Li T., Zhang C., Wu J.D., Han X., Wu K. The expression profile and clinic significance of the SIX family in non-small cell lung cancer. J. Hematol. Oncol. 2016;9:119. doi: 10.1186/s13045-016-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iaboni M., Russo V., Fontanella R., Roscigno G., Fiore D., Donnarumma E., Esposito C.L., Quintavalle C., Giangrande P.H., de Franciscis V., Condorelli G. Aptamer-miRNA-212 Conjugate Sensitizes NSCLC Cells to TRAIL. Mol. Ther. Nucleic Acids. 2016;5:e289. doi: 10.1038/mtna.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brule S.Y., Al-Baimani K., Jonker H., Zhang T., Nicholas G., Goss G., Laurie S.A., Wheatley-Price P. Palliative systemic therapy for advanced non-small cell lung cancer: Investigating disparities between patients who are treated versus those who are not. Lung Cancer. 2016;97:15–21. doi: 10.1016/j.lungcan.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Nagasaka M., Gadgeel S.M. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev. Anticancer Ther. 2018;18:63–70. doi: 10.1080/14737140.2018.1409624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bansal P., Rusthoven C., Boumber Y., Gan G.N. The role of local ablative therapy in oligometastatic non-small-cell lung cancer: hype or hope. Future Oncol. 2016;12:2713–2727. doi: 10.2217/fon-2016-0219. [DOI] [PubMed] [Google Scholar]
- 10.Couñago F., Rodríguez A., Calvo P., Luna J., Monroy J.L., Taboada B., Díaz V., Rodríguez de Dios N. Targeted therapy combined with radiotherapy in non-small-cell lung cancer: a review of the Oncologic Group for the Study of Lung Cancer (Spanish Radiation Oncology Society) Clin. Transl. Oncol. 2017;19:31–43. doi: 10.1007/s12094-016-1512-2. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Zhu T., Gao Y.F., Zheng W., Wang C.J., Xiao L., Huang M.S., Yin J.Y., Zhou H.H., Liu Z.Q. Targeting DNA Damage Response in the Radio(Chemo)therapy of Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2016;17:E839. doi: 10.3390/ijms17060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawe D.E., Christiansen D., Swaminath A., Ellis P.M., Rothney J., Rabbani R., Abou-Setta A.M., Zarychanski R., Mahmud S.M. Chemoradiotherapy versus radiotherapy alone in elderly patients with stage III non-small cell lung cancer: A systematic review and meta-analysis. Lung Cancer. 2016;99:180–185. doi: 10.1016/j.lungcan.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Ernani V., Steuer C.E., Jahanzeb M. The End of Nihilism: Systemic Therapy of Advanced Non-Small Cell Lung Cancer. Annu. Rev. Med. 2017;68:153–168. doi: 10.1146/annurev-med-042915-102442. [DOI] [PubMed] [Google Scholar]
- 14.Wang J.X., Zeng Q., Chen L., Du J.C., Yan X.L., Yuan H.F., Zhai C., Zhou J.N., Jia Y.L., Yue W., Pei X.T. SPINDLIN1 promotes cancer cell proliferation through activation of WNT/TCF-4 signaling. Mol. Cancer Res. 2012;10:326–335. doi: 10.1158/1541-7786.MCR-11-0440. [DOI] [PubMed] [Google Scholar]
- 15.Choi J.W., Zhao M.H., Liang S., Guo J., Lin Z.L., Li Y.H., Jo Y.J., Kim N.H., Cui X.S. Spindlin 1 is essential for metaphase II stage maintenance and chromosomal stability in porcine oocytes. Mol. Hum. Reprod. 2017;23:166–176. doi: 10.1093/molehr/gax005. [DOI] [PubMed] [Google Scholar]
- 16.Drago-Ferrante R., Pentimalli F., Carlisi D., De Blasio A., Saliba C., Baldacchino S., Degaetano J., Debono J., Caruana-Dingli G., Grech G. Suppressive role exerted by microRNA-29b-1-5p in triple negative breast cancer through SPIN1 regulation. Oncotarget. 2017;8:28939–28958. doi: 10.18632/oncotarget.15960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Ma X., Wang Y., Li G. miR-489 inhibits proliferation, cell cycle progression and induces apoptosis of glioma cells via targeting SPIN1-mediated PI3K/AKT pathway. Biomed. Pharmacother. 2017;93:435–443. doi: 10.1016/j.biopha.2017.06.058. [DOI] [PubMed] [Google Scholar]
- 18.Li Z., Rana T.M. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 19.Ivey K.N., Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Mendell J.T., Olson E.N. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 22.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang T.H., Tsai M.F., Gow C.H., Wu S.G., Liu Y.N., Chang Y.L., Yu S.L., Tsai H.C., Lin S.W., Chen Y.W. Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett. 2017;402:190–202. doi: 10.1016/j.canlet.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Wei C.H., Wu G., Cai Q., Gao X.C., Tong F., Zhou R., Zhang R.G., Dong J.H., Hu Y., Dong X.R. MicroRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway. J. Hematol. Oncol. 2017;10:125. doi: 10.1186/s13045-017-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Yang Y., Liu L., Zhang Y., Guan H., Wu J., Zhu X., Yuan J., Li M. MiR-503 targets PI3K p85 and IKK-β and suppresses progression of non-small cell lung cancer. Int. J. Cancer. 2014;135:1531–1542. doi: 10.1002/ijc.28799. [DOI] [PubMed] [Google Scholar]
- 26.Fang L., Cai J., Chen B., Wu S., Li R., Xu X., Yang Y., Guan H., Zhu X., Zhang L. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/β-catenin signalling. Nat. Commun. 2015;6:8640. doi: 10.1038/ncomms9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Wang Y.W., Xing A.Y., Xiang S., Shi D.B., Liu L., Li Y.X., Gao P. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J. Pathol. 2016;239:459–472. doi: 10.1002/path.4743. [DOI] [PubMed] [Google Scholar]
- 28.Ji Q., Xu X., Xu Y., Fan Z., Kang L., Li L., Liang Y., Guo J., Hong T., Li Z. miR-105/Runx2 axis mediates FGF2-induced ADAMTS expression in osteoarthritis cartilage. J. Mol. Med. (Berl.) 2016;94:681–694. doi: 10.1007/s00109-016-1380-9. [DOI] [PubMed] [Google Scholar]
- 29.Jiang F., Zhao Q., Qin L., Pang H., Pei X., Rao Z. Expression, purification, crystallization and preliminary X-ray analysis of human spindlin1, an ovarian cancer-related protein. Protein Pept. Lett. 2006;13:203–205. doi: 10.2174/092986606775101661. [DOI] [PubMed] [Google Scholar]
- 30.Ji Q., Xu X., Li L., Goodman S.B., Bi W., Xu M., Xu Y., Fan Z., Maloney W.J., Ye Q., Wang Y. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 2017;8:e3103. doi: 10.1038/cddis.2017.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Zhang Y., Wu H., Li Y., Zhang Y., Liu M., Li X., Tang H. miR-10a suppresses colorectal cancer metastasis by modulating the epithelial-to-mesenchymal transition and anoikis. Cell Death Dis. 2017;8:e2739. doi: 10.1038/cddis.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colden M., Dar A.A., Saini S., Dahiya P.V., Shahryari V., Yamamura S., Tanaka Y., Stein G., Dahiya R., Majid S. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis. 2017;8:e2572. doi: 10.1038/cddis.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan M.N., Yu X.T., Tang J., Zhou C.X., Wang C.L., Yin Q.Q., Gong X.F., He M., He J.R., Chen G.Q., Zhao Q. MicroRNA-494 inhibits breast cancer progression by directly targeting PAK1. Cell Death Dis. 2017;8:e2529. doi: 10.1038/cddis.2016.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang H., Liu S., Chen Y., Bai X., Liu L., Dong Y., Hu M., Su X., Chen Y., Huangfu L. miR-26a suppresses EMT by disrupting the Lin28B/let-7d axis: potential cross-talks among miRNAs in IPF. J. Mol. Med. (Berl.) 2016;94:655–665. doi: 10.1007/s00109-016-1381-8. [DOI] [PubMed] [Google Scholar]
- 35.Mutlu M., Raza U., Saatci Ö., Eyüpoğlu E., Yurdusev E., Şahin Ö. miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance. J. Mol. Med. (Berl.) 2016;94:629–644. doi: 10.1007/s00109-016-1420-5. [DOI] [PubMed] [Google Scholar]
- 36.Babu K.R., Muckenthaler M.U. miR-20a regulates expression of the iron exporter ferroportin in lung cancer. J. Mol. Med. (Berl.) 2016;94:347–359. doi: 10.1007/s00109-015-1362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng B., Liang L., Huang S., Zha R., Liu L., Jia D., Tian Q., Wang Q., Wang C., Long Z. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene. 2012;31:4509–4516. doi: 10.1038/onc.2011.581. [DOI] [PubMed] [Google Scholar]
- 38.Li J., Xu X., Meng S., Liang Z., Wang X., Xu M., Wang S., Li S., Zhu Y., Xie B. MET/SMAD3/SNAIL circuit mediated by miR-323a-3p is involved in regulating epithelial-mesenchymal transition progression in bladder cancer. Cell Death Dis. 2017;8:e3010. doi: 10.1038/cddis.2017.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chew T.G., Peaston A., Lim A.K., Lorthongpanich C., Knowles B.B., Solter D. A tudor domain protein SPINDLIN1 interacts with the mRNA-binding protein SERBP1 and is involved in mouse oocyte meiotic resumption. PLoS ONE. 2013;8:e69764. doi: 10.1371/journal.pone.0069764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franz H., Greschik H., Willmann D., Ozretić L., Jilg C.A., Wardelmann E., Jung M., Buettner R., Schüle R. The histone code reader SPIN1 controls RET signaling in liposarcoma. Oncotarget. 2015;6:4773–4789. doi: 10.18632/oncotarget.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greschik H., Duteil D., Messaddeq N., Willmann D., Arrigoni L., Sum M., Jung M., Metzger D., Manke T., Günther T., Schüle R. The histone code reader Spin1 controls skeletal muscle development. Cell Death Dis. 2017;8:e3173. doi: 10.1038/cddis.2017.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae N., Viviano M., Su X., Lv J., Cheng D., Sagum C., Castellano S., Bai X., Johnson C., Khalil M.I. Developing Spindlin1 small-molecule inhibitors by using protein microarrays. Nat. Chem. Biol. 2017;13:750–756. doi: 10.1038/nchembio.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji Q., Xu X., Zhang Q., Kang L., Xu Y., Zhang K., Li L., Liang Y., Hong T., Ye Q., Wang Y. The IL-1β/AP-1/miR-30a/ADAMTS-5 axis regulates cartilage matrix degradation in human osteoarthritis. J. Mol. Med. (Berl.) 2016;94:771–785. doi: 10.1007/s00109-016-1418-z. [DOI] [PubMed] [Google Scholar]
- 44.Chen X., Dong H., Liu S., Yu L., Yan D., Yao X., Sun W., Han D., Gao G. Long noncoding RNA MHENCR promotes melanoma progression via regulating miR-425/489-mediated PI3K-Akt pathway. Am. J. Transl. Res. 2017;9:90–102. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.