Abstract
Background
Fibroblast growth factor 23 (FGF23), a bone-derived hormone that regulates phosphorus and vitamin D metabolism, contributes to the pathogenesis of mineral and bone disorders in CKD and is an emerging cardiovascular risk factor. Central elements of FGF23 regulation remain incompletely understood; genetic variation may help explain interindividual differences.
Methods
We performed a meta-analysis of genome-wide association studies of circulating FGF23 concentrations among 16,624 participants of European ancestry from seven cohort studies, excluding participants with eGFR<30 ml/min per 1.73 m2 to focus on FGF23 under normal conditions. We evaluated the association of single-nucleotide polymorphisms (SNPs) with natural log–transformed FGF23 concentration, adjusted for age, sex, study site, and principal components of ancestry. A second model additionally adjusted for BMI and eGFR.
Results
We discovered 154 SNPs from five independent regions associated with FGF23 concentration. The SNP with the strongest association, rs17216707 (P=3.0×10−24), lies upstream of CYP24A1, which encodes the primary catabolic enzyme for 1,25-dihydroxyvitamin D and 25-hydroxyvitamin D. Each additional copy of the T allele at this locus is associated with 5% higher FGF23 concentration. Another locus strongly associated with variations in FGF23 concentration is rs11741640, within RGS14 and upstream of SLC34A1 (a gene involved in renal phosphate transport). Additional adjustment for BMI and eGFR did not materially alter the magnitude of these associations. Another top locus (within ABO, the ABO blood group transferase gene) was no longer statistically significant at the genome-wide level.
Conclusions
Common genetic variants located near genes involved in vitamin D metabolism and renal phosphate transport are associated with differences in circulating FGF23 concentrations.
Keywords: human genetics, fibroblast growth factor 23, mineral metabolism
Visual Abstract
Fibroblast growth factor 23 (FGF23) is secreted from bone and plays a role in both phosphate homeostasis and the vitamin D endocrine system. FGF23 induces urinary phosphate excretion by suppressing the expression of the sodium phosphate cotransporter in kidney proximal tubular cells. FGF23 also suppresses the synthesis of calcitriol [1,25(OH)2D] by inhibiting its production via 1,25α-hydroxylase (CYP27B1) and stimulating its degradation via 1,24,25-hydroxylase (CYP24A1).1,2 In turn, calcitriol directly enhances the transcription of FGF23 to complete a feedback loop. The effects of FGF23 on phosphate and vitamin D homeostasis require klotho as an obligatory coligand, which activates fibroblast growth factor receptors and their downstream signaling molecules.3 Optimal phosphate balance is important for many physiologic functions, from cell signaling to energy metabolism and skeletal mineralization. In humans, high FGF23 concentrations contribute to the pathogenesis of mineral and bone disorders in CKD and are associated with ventricular hypertrophy, cardiovascular events, and premature death.4–12
Despite intense investigation, central elements of FGF23 regulation remain unknown. The primary role of FGF23 in phosphate homeostasis suggests regulation by serum phosphate concentrations; however, experimental studies have not detected direct actions of serum phosphate on FGF23. Rare heritable disorders are characterized by dysregulated FGF23 metabolism, yet only two (autosomal dominant hypophosphatemic rickets and familial hyperphosphatemic tumoral calcinosis) involve direct mutation of the FGF23 gene.13–15 Mutations in other bone metabolism genes cause familial tumoral calcinosis (GALNT3, klotho), X-linked phosphatemic rickets (PHEX), and autosomal recessive hypophosphatemic rickets (DMP1, FAM20C, ENPP1), that subsequently alter FGF23 metabolism through mechanisms that are not completely understood.16–23 We performed a meta-analysis of genome-wide association studies of circulating FGF23 concentrations among 16,624 individuals of European ancestry from seven cohort studies, and replicated in 4443 individuals of African ancestry from three cohorts, to investigate the role of common genetic variants on this hormone.
Methods
Study Populations
Seven cohorts contributed to the meta-analysis, by providing study-specific genome-wide analyses of FGF23 concentrations, for a total of 16,624 individuals of European ancestry (Table 1). Contributing studies included the Atherosclerosis Risk in Communities Study (ARIC; number of individuals of European ancestry, n=8594),24 the Indiana Sisters Study (n=1128),25 Osteoporotic Fractures in Men Study–Goteborg (MrOS GBG, n=937),26 the Multi-Ethnic Study of Atherosclerosis (MESA; number of individuals of European ancestry, n=2163),27 MrOS Malmo26 (n=894), the Osteoporosis Prospective Risk Assessment Study (OPRA, n=920),28 and the Cardiovascular Health Study (CHS; number of individuals of European ancestry, n=1988).29 Detailed information on the study cohorts and methods is provided in Supplemental Table 1 and Table 1.
Table 1.
Cohort and study participant (European descent) characteristics
| Characteristic | ARIC | CHS | Indiana | MESA | MrOS GBG | MrOS Malmo | OPRA |
|---|---|---|---|---|---|---|---|
| Individuals of European descent | 8594 | 1988 | 1128 | 2163 | 937 | 894 | 920 |
| Age, yr | 57.1 (5.7) | 78.0 (4.4) | 36.4 (8.5) | 62.6 (10.3) | 72.3 (5.7) | 73.2 (5.8) | 75.2 (0.1) |
| Men, n (%) | 3962 (46.1) | 754 (38.3) | 0 (0) | 1025 (47.3) | 937 (100) | 894 (100) | 0 (0) |
| BMI, kg/m2 | 27.3 (5.0) | 26.6 (4.5) | 26.3 (6.0) | 27.7 (5.1) | 27.5 (3.7) | 27.5 (3.7) | 26.2 (4.1) |
| eGFR (CKD-EPI), ml/min per 1.73 m2 | 87.8 (13.2) | 72.1 (17.7) | 103.5 (7.4) | 76.0 (17.1) | 71.6 (19.3) | 68.8 (19.3) | 68.0 (12.6) |
| FGF23, pg/ml (intact) | 45.5 (17.3) | — | 39.4 (17.4) | 41.9 (17.6) | 46.2 (22.5) | 55.9 (27.7) | — |
| FGF23, pg/ml (C-terminal) | — | 12.9 (21.3) | — | — | — | — | 8.3 (9.1) |
Data are mean (SD) or number (%), as appropriate. Indiana, Indiana Sisters Cohort; GBG, Goteburg; CKD-EPI, CKD Epidemiology Collaboration equation; —, not available.
We calculated the eGFR using the creatinine-based CKD Epidemiology Collaboration equation equation,30 and excluded participants with eGFR<30 ml/min per 1.73 m2. This exclusion was chosen because our intent was to study FGF23 under normal conditions, because the strong influence of kidney disease may overwhelm potentially more subtle influences of individual SNPs on circulating FGF23. The individual studies were approved by the local research ethics committees, and informed consent was obtained from all participants.
Measurement of FGF23
Circulating FGF23 concentrations were measured by ELISA (Kainos Laboratories, Inc., Tokyo, Japan), which detects the full-length, biologically intact FGF23 molecule via midmolecule and distal epitopes, in the ARIC, Indiana Sisters, MrOS GBG and Malmo, and MESA cohorts. In the CHS and OPRA cohorts, FGF23 was measured using a C-terminal ELISA kit (Immutopics, San Clemente, CA).
Genotyping and Quality Control
Genome-wide genotyping and imputation with reference to the 1000 Genomes Phase 3 genotypes were performed independently in each cohort. Each cohort applied sample and SNP-based quality control measures for missingness, minor allele frequency, and Hardy–Weinberg equilibrium (Supplemental Table 1). Poorly imputed SNPs were excluded if R2<0.3 or proper-info was <0.4. Population stratification and relatedness were assessed using the ancestry principal components as previously described.31 All cohorts used EIGENSTRAT for principal components of ancestry computation.
Statistical Analyses
Each SNP was tested for association with natural log–transformed FGF23 using linear regression in two additive genetic models.32 Model 1 was adjusted for age, sex, and the first ten principal components of ancestry.
FGF23 concentrations are strongly associated with eGFR and body mass index (BMI). To detect FGF23 loci independent of these pathways and to diminish associations with effects modulated through these factors, model 2 added adjustment for BMI, and both linear and quadratic terms for the eGFR.30
Genomic control parameters were estimated for each cohort and appropriate genomic control correction was applied to input statistics before performing meta-analysis to correct for residual cryptic relatedness or population stratification.33 There was little evidence for population stratification at study level (median genomic inflation factor, λ=1.03) or meta-analysis level (λ=1.007). A fixed-effects inverse-variance weighted meta-analysis using METAL was performed across cohorts on the β coefficient/SEM from each cohort.34 Secondarily, random-effects DerSimonian and Laird models were performed using Stata 15.1 (Statacorp, College Station, TX).35
Genetic differentiation was estimated using the Weir unbiased estimator of the fixation index, calculated using the variance in allele frequencies among European and African ancestry samples from the 1000 Genomes and standardized according to the mean allele frequency in the combined sample.36
We examined the consistency of the magnitude and direction of associations across individual studies using forest plots. The Locuszoom tool was used to make regional association plots,37 and Manhattan and QQ-plots were plotted with the qqman R package.38
Proportion of Phenotypic Variance Explained
The proportion of variance (PVE) in circulating FGF23 levels explained by each top novel locus, jointly across all cohorts, was estimated as:
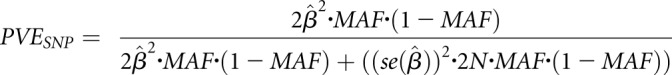 |
where 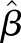 ,
, 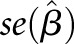 , N, and MAF are the effect size estimate of each minor allele on the relative concentration of FGF23, SEM of the effect size, sample size, and MAF for the SNP, respectively.39
, N, and MAF are the effect size estimate of each minor allele on the relative concentration of FGF23, SEM of the effect size, sample size, and MAF for the SNP, respectively.39
Follow-Up in African Ancestry Cohorts
SNPs found to be associated with FGF23 concentration in individuals of European ancestry were evaluated for replication among individuals of African ancestry from three cohorts: ARIC (n=2464), MESA (n=1510), and CHS (n=469). These analyses were performed using the same quality control and imputation exclusions, and meta-analysis was performed using METAL.34
Sensitivity Analyses
A sensitivity analysis was conducted in which we excluded the CHS and OPRA GWAS results, which were on the basis of C-terminal FGF23 measurements, in order to evaluate whether their inclusion had altered the findings.
eQTL and Functionality Prediction
Functional annotations were assigned to all SNPs from the meta-analysis with P<5×10−6 using ANNOVAR software.40 We used the MetaXcan software to identify genes whose expression levels were significantly associated with circulating FGF23.41,42 MetaXcan uses the FGF23 GWAS summary results and gene expression levels predicted by genetic variants in a library of tissues from the Genotype-Tissue Expression (GTEx) project to impute the genetic component of gene expression in different tissues (thereby eliminating the need to directly measure gene expression levels) and to correlate the imputed gene expressions with the phenotype of interest.43,44 For the purposes of this study, we used covariance matrices built for ten relevant tissues from GTEx (i.e., whole blood cells, transformed fibroblast cells, subcutaneous adipose, skeletal muscle, liver, tibial nerve, left ventricle of the heart, coronary artery, aortic artery, and atrial appendage). Of note, reliable data from kidney and bone tissue are not currently available in GTEx.
MetaXcan is an extension of PredixCan,39 a gene-based approach that uses GWAS summary results to impute the genetic component of gene expression in different tissues (thus eliminating the need to directly measure gene expression levels), and correlates the imputed gene expressions with phenotypes of interest.
Associations with Other Traits
We performed evaluations of SNP associations with publicly available results generated from consortia investigating other traits. Specifically, we evaluated SNPs associated with eGFR from CKD-GEN,45 coronary artery disease in CARDIOGRAM,46 parathyroid hormone,47 and bone mineral density from the GEFOS.48 In total, we performed 20 tests, corresponding to four traits tested for association against the five SNPs. Significance was evaluated at the Bonferroni-corrected level of 0.0025.
Results
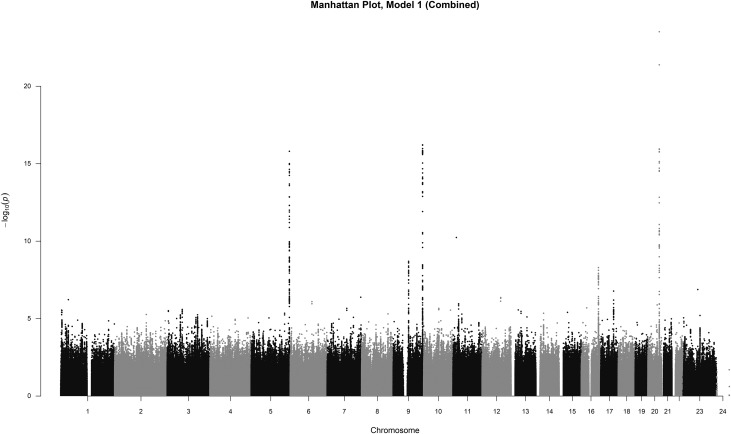
The SNP-based meta-analysis identified 192 SNPs associated with circulating FGF23 at genome-wide significance level (P<5×10−8). These SNPs were located in five genomic regions, 5q35.3, 9q21.11, 9q34.2, 16q23.2, and 20q13.2 (Figure 1). The top SNP in each region and genes contained in the region were 20q13.2, rs17216707 (P=3.0×10−24; CYP24A1); 9q34.2, rs2769071 (P=6.1×10−17; ABO); 5q35.3, rs11741640 (P=1.6×10−16; RGS14); 9q21.11, rs17479566 (P=2.0×10−9; LINC01506); and 16q23.2, rs9925837 (P=5.1×10−9; LINC01229). In aggregate, the top five loci explained 3% of the variability in circulating FGF23. Inspection of the quantile-quantile plot suggested an excess of association signals beyond those expected (Supplemental Figure 1). All 341 SNPs from the meta-analysis with a P value less than a suggestive significance threshold of P<5×10−6 are detailed in Supplemental Table 2.
Figure 1.
Manhattan plot of SNPs for FGF23. SNPs are plotted on the x axis according to their position on each chromosome against association with ln(FGF23) on the y axis (shown as –log10 P value).
The top SNP in the 20q13.2 region, having the strongest association with FGF23 concentration, was rs17216707 (P=3.0×10−24; Supplemental Figure 2A, Table 2). This intergenic SNP lies 38 kb upstream of CYP24A1 (cytochrome P450, family 24, subfamily A, polypeptide 1). Each additional copy of the rs17216707 T allele was associated with 5.4% higher FGF23 concentration, after adjustment for age, sex, and the first ten principal components of ancestry (model 1). The CYP24A1 gene encodes the major enzyme responsible for catabolizing calcitriol to water-soluble 1,24,25-trihydroxyvitamin D (1,24,25(OH)3D) and 25(OH)D to 25-hydroxyvitamin D (25(OH)D) to 24,25-dihydroxyvitamin D (24,25(OH)2D) for excretion.49
Table 2.
Associations of top single nucleotide polymorphisms with ln-transformed FGF23 concentrations
| SNP | Nearest Genea | Chr | Position | FGF23 Increasing Allele | Other Allele | FGF23 Increasing Allele Frequencyb | Model 1 | Model 2c | ||
|---|---|---|---|---|---|---|---|---|---|---|
| (n=16,624) | (n=15,496) | |||||||||
| βd (SEM) | P Value | β (SEM) | P Value | |||||||
| rs17216707 | CYP24A1 | 20 | 52732362 | T | C | 0.80 | 0.054 (0.005) | 3.0×10−24 | 0.044 (0.005) | 1.5×10−21 |
| rs2769071 | ABO | 9 | 136145974 | G | A | 0.37 | 0.037 (0.005) | 6.1×10−17 | 0.031 (0.008) | 3.0×10−5 |
| rs11741640 | RGS14 | 5 | 176792743 | G | A | 0.73 | 0.039 (0.005) | 1.6×10−16 | 0.038 (0.005) | 2.8×10−16 |
| rs17479566 | LINC01506 | 9 | 71198013 | T | C | 0.22 | 0.031 (0.005) | 2.0×10−9 | 0.039 (0.009) | 6.8×10−6 |
| rs9925837 | LINC01229 | 16 | 79927303 | G | A | 0.13 | 0.035 (0.006) | 5.1×10−9 | 0.035 (0.006) | 2.1×10−9 |
Only the top SNP from each region is shown. Model 1 includes age, sex, and first ten principal components of ancestry. SNP, single nucleotide polymorphism; Chr, chromosome.
Nearest gene by physical distance to the lead SNP.
Allele frequency data from 1000 Genomes Phase 1 genotype data.
Results for model 2 were not available for the Indiana Sisters Study.
β-estimates are interpreted as the relative difference in FGF23 concentration per minor allele; e.g., 0.051 is a 5.1% higher FGF23 concentration per additional allele.
rs2769071, a variant on chromosome 9q34.2, intronic in the ABO (ABO blood group transferase gene), was also associated with FGF23 concentrations (P=6.1×10−17; Supplemental Figure 2B, Table 2). Every additional minor allele at the locus was associated with 3.7% higher circulating FGF23 concentrations. The association did not remain statistically significant after adjustment for BMI, eGFR, and eGFR squared (P=3.0×10−5).
Within the 5q35.3 region, the variant most strongly associated with circulating FGF23 concentrations was rs11741640 (P=1.6×10−16; Table 2). This SNP is intronic within RGS14 (regulator of G-protein signaling 14) and is in linkage disequilibrium (LD) with SNPs in adjacent genes, SLC34A1 (solute carrier family 34 [type 2 sodium/phosphate cotransporter], member 1, responsible for phosphate reabsorption in the proximal tubule) (e.g., rs12659266, R2=0.78), and FGFR4 (fibroblast growth factor receptor 4) (e.g., rs244707, R2=0.43) (Supplemental Figure 2C).11,50,51
Other top SNPs significantly associated with FGF23 included rs17479566, an SNP in the 9q21.11 region, residing intergenically between LINC01506 (long intergenic non–protein coding RNA 1506) and PIP5K1B (phosphatidylinositol-4-phosphate 5-kinase type 1 β; Supplemental Figure 2D), and rs9925837, an intergenic SNP positioned between LINC01229 (long intergenic non–protein coding RNA 1229) and LOC102724084 (uncharacterized noncoding RNA) in the 16q23.2 region (P=5.1×10−9; Supplemental Figure 2E, Table 2).
The primary regression coefficients and interpretation of our results were not affected by further adjustment for BMI, eGFR, and eGFR squared (model 2) for rs17216707, rs11741640, or rs9925837 (Supplemental Figure 3). However, the P values for SNPs rs2769071 and rs17479566 were attenuated by factors of 1012 and 103, respectively (Table 2).
Replication in Individuals of African Ancestry
In populations of African ancestry, the effect estimates for the five top SNPs were in the same direction as in individuals of European ancestry and one SNP (rs9925837) was nominally associated (P<0.05) with FGF23 concentrations (Table 3). Rs17216707, rs11741640, and rs9925837 showed the most allelic differentiation between individuals of African and European ancestry.
Table 3.
Associations of top single nucleotide polymorphisms with ln-transformed FGF23 concentrations among individuals of African ancestry (n=4443)
| SNP | Nearest Gene | Chr | Position | FGF23 Increasing Allele | Other Allele | FGF23 Increasing Allele Frequencya | βb (SEM) | P Value | Fstc |
|---|---|---|---|---|---|---|---|---|---|
| rs17216707 | CYP24A1 | 20 | 52732362 | T | C | 0.97 | 0.044 (0.037) | 0.24 | 0.162 |
| rs2769071 | ABO | 9 | 136145974 | G | A | 0.48 | 0.010 (0.012) | 0.41 | 0.009 |
| rs11741640 | RGS14 | 5 | 176792743 | G | A | 0.96 | 0.008 (0.027) | 0.78 | 0.195 |
| rs17479566 | LINC01506 | 9 | 71198013 | T | C | 0.93 | 0.022 (0.020) | 0.29 | 0.087 |
| rs9925837 | LINC01229 | 16 | 79927303 | G | A | 0.28 | 0.029 (0.015) | 0.05 | 0.232 |
Only the top SNP from each region is shown. SNP, single nucleotide polymorphism; Chr, chromosome.
Allele frequency data from 1000 Genome Phase 1 genotype data among African samples.
Effect estimate adjusted for age, sex, and first ten principal components of ancestry. β-estimates are interpreted as the relative difference in FGF23 concentration per minor allele; e.g., 0.038 is a 3.8% higher FGF23 concentration per additional allele.
Fst is an estimate of genetic differentiation calculated using the variance in allele frequencies among European- and African-ancestry samples from the 1000 Genomes and standardized according to the mean allele frequency in the combined sample.36
Sensitivity Analysis
We repeated our meta-analysis after excluding the CHS and OPRA, in which circulating concentrations of C-terminal FGF23 were measured. This meta-analysis identified 161 SNPs associated with circulating intact FGF23 at genome-wide significance level (P<5×10−8), located in four genomic regions, 5q35.3, 9q21.11, 9q34.2, and 20q13.2. The top SNP in each region and genes contained in the region were 20q13.2, rs17216707 (β [SEM]=0.057 [0.006]; P=1.0×10−25; CYP24A1); 9q34.2, rs2769071 (β [SEM]=0.037 [0.005]; P=7.1×10−16; ABO); 5q35.3, rs11741640 (β [SEM]=0.038 [0.005]; P=1.6×10−14; RGS14); and 9q21.11, rs17479566 (β [SEM]=0.032 [0.005]; P=3.8×10−9; LINC01506) (Supplemental Table 3). Although the association at rs9925837 did not meet genome-wide significance, the magnitude of the effect was not meaningfully different after exclusion of CHS and OPRA (β [SEM]=0.032 [0.006], P=2.2×10−7).
Functional Evaluation
Of the 192 SNPs that met genome-wide significance (P<5×10−8) in the primary analysis, 98 were in intergenic regions, 84 overlapped introns, one was exonic (rs5030873, for SLC34A1), one overlapped a 3′ untranslated region (rs3812035, for SLC34A1), four overlapped the 1-kb region upstream of a transcription start site (all for RGS14), and four overlapped the 1-kb region downstream of a transcription end site (three for RGS14 and one for LMAN2) (Supplemental Table 2). Analysis with MetaXcan to evaluate the association with genetically predicted gene expression implicated RGS14 as the most significant genetic result and revealed a significant association of FGF23 with increased expression at that gene in multiple tissues (Supplemental Table 4).
Assessment for Association of SNPs with Related Traits
Each of the top SNPs was associated with parathyroid hormone concentration; four of the five were significantly associated at the Bonferroni-corrected P value threshold of 0.003 (Table 4). We also observed associations of four of the five SNPs with eGFR, and of rs2769071 with coronary artery disease and bone mineral density. At this locus, the FGF23 increasing allele was associated with 4.5% greater odds of coronary artery disease (P=3.3×10−6) and lower BMD (β=−0.0197, P=2.7×10−8).
Table 4.
Association between replicated SNPs and FGF23-related phenotypes
| SNP | Nearest Gene | Allele | Coronary Artery Diseasea | Bone Mineral Densityb (standardized mean difference) | Parathyroid Hormonec (% difference) | eGFRd (ml/min per 1.73 m2) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (SEM) | P Value | β (SEM) | P Value | β (SEM) | P Value | β (SEM) | P Value | |||
| rs17216707 | CYP24A1 | T | 0.98 (0.012) | 0.13 | −0.0036 (0.0044) | 0.26 | −5.4% (0.053) | 4.7×10−25 | −0.97% (0.13) | 5.4×10−15 |
| rs2769071 | ABO | G | 1.05 (0.01) | 3.3×10−6 | −0.0197 (0.0036) | 2.7 ×10−8 | +1.2% (0.044) | 6.3×10−4 | +0.01% (0.10) | 0.91 |
| rs11741640 | RGS14 | G | 0.98 (0.010) | 0.04 | −0.0049 (0.0038) | 0.10 | −3.1% (0.031) | 3.4×10−24 | +0.75% (0.11) | 4.5×10−12 |
| rs17479566 | LINC01506 | T | 0.98 (0.011) | 0.10 | +0.0066 (0.0041) | 0.18 | –1.7% (0.047) | 5.1×10−5 | +0.56% (0.12) | 1.5×10−6 |
| rs9925837 | LINC01229 | G | 1.00 (0.013) | 0.87 | −0.0035 (0.0047) | 0.62 | +1.0% (0.055) | 0.02 | +0.51% (0.14) | 1.7×10−4 |
In total, we performed 20 tests, corresponding to four traits tested for association against the five SNPs. Significance was evaluated at the Bonferroni-corrected level of 0.0025. SNP, single nucleotide polymorphism; OR, odds ratio; GWAS, Genome-wide association study.
Association estimates from GWAS meta-analysis of coronary artery disease among 60,801 cases and 123,504 controls from 48 studies within the Coronary Artery Disease Genome Wide Replication and Meta-Analysis Consortium.46
Estimates from meta-analysis of bone mineral density of the femoral neck GWAS among 32,961 individuals from 17 studies within the Genetic Factors for Osteoporosis Consortium.48
Estimates from meta-analysis of parathyroid hormone GWAS among 29,155 individuals from 13 cohort studies.47
Estimates from meta-analysis of eGFR in 110,517 individuals from 33 studies within the Chronic Kidney Disease Genetics (CKD-Gen) CKDGen Consortium.45
Discussion
In this first reported GWAS of FGF23, we identified five loci, located on chromosomes 5, 9, 16, and 20, that were associated with variation in FGF23 concentrations. The strongest association was observed for rs17216707, located 37 kb upstream of CYP24A1, encoding the primary catabolic enzyme for calcitriol [1,25(OH)2D]. Every additional T allele at this locus was associated with a 5.1% difference in circulating FGF23. Other significant associations included SNPs located near the gene encoding proteins related to the first discovered blood group system, ABO, and genes encoding the regulator of G-protein signaling 14 and the Na/Pi cotransporter solute carrier family 34 member 1. In replication analyses conducted in blacks, the effect estimates for the top five SNPs were in the same direction as in the individuals of European ancestry; however, confidence intervals were wider, and the results were not statistically significant.
Calcitriol, the activated form of vitamin D, directly promotes FGF23 gene transcription via enhancement of the promotor region. The cytochrome p450 enzyme CYP24A1 is critically important for maintaining 1,25(OH)2D concentrations within a tight physiologic range and preventing vitamin D toxicity by catalyzing the conversion of 1,25(OH)2D to 1,24,25(OH)3D for subsequent excretion.49 We found the T allele of rs17216707 to be associated with greater circulating FGF23 concentrations, suggesting that the associated haplotype may confer suppressed CYP24A1 activity, with a resultant increase in 1,25(OH)2D and stimulation of FGF23 production. The T allele was also strongly associated with lower eGFR in previous GWASs,52,53 and yet adjustment for eGFR in the association of the SNP with FGF23 did not alter the magnitude of the association. The association of the T allele of rs17216707 with lower serum concentrations of PTH is also consistent with suppressed CYP24A1 activity, decelerated 1,25(OH)2D catabolism, and increased 1,25(OH)2D levels. In murine homolog models of autosomal dominant phosphatemic rickets (ADHR) and X-linked hypophosphatemic rickets (XLH), high FGF23 states, CYP24A1 expression in the kidney is elevated.54,55 Interestingly, when these mouse strains were crossed with CYP24A1-null mice, the bone abnormalities improved, without improvements in the serum biochemical profile.56
rs2769071 is located within the ABO blood group locus on chromosome 9, which encodes alpha 1–3-N-acetylgalactosaminyltransferase, a major determinant of ABO blood type. The C allele at rs2769071 is highly correlated (r2=0.86) with a single base pair insertion/deletion (indel) variant in exon 6 of ABO (rs8176719) which characterizes the human O blood group.57 This SNP is associated with ABO gene expression in most tissues in GTEx. Using available GWAS data of other phenotypes, we observed a significant association of variation at this locus to be associated with higher odds of coronary artery disease, lower bone mineral density, and higher parathyroid hormone levels. In previous published GWASs, this SNP has been found to be associated with plasma vWf and Factor VII,58 coagulation factors levels,59 and myocardial infarction in patients with angiographic coronary artery disease.60 Recent studies have suggested that serum iron may be a regulator of FGF23 homeostasis, and ferritin levels have been shown to differ between blood groups.61–63 The attenuation of the statistical significance of the association with adjustment for eGFR and BMI suggests that the contribution of genetic risk to FGF23 within the ABO loci may be mediated by the SNP’s effects on these covariates. However, variation at rs2769071 was not associated with circulating eGFR45 in the CKD-GEN GWAS or BMI in the GWAS of 700,000 individuals from the GIANT Consortium.64 The functional relevance of this polymorphism to mineral metabolism and FGF23 regulation awaits further elucidation.
rs11741640 is located within RGS14, which encodes a G-protein signal–regulating protein and was the only replicated SNP significantly associated with both intact and C-terminal FGF23. Previous GWASs of serum phosphate and parathyroid hormone concentrations reported strong associations with rs4074995, which is in near-perfect LD with rs11741640 (R2=0.93).47,65 In our gene-level expression-based analyses with MetaXcan, RGS14 was the gene with the strongest association of genetically determined expression with FGF23 levels. We found that increased expression of RGS14 was associated with higher levels of FGF23 across many tissues, including in heart and muscle tissue, lending biologic support to its implication in mineral metabolism. rs11741640 is also in strong LD with multiple SNPs within the gene coding for type 2a sodium phosphate cotransporter (Npt2a, SLC34A1) and fibroblast growth factor receptor 4 (FGFR4). FGF23 acts on the kidney to induce phosphaturia by reducing the expression of Npt2a in the brush border of the proximal tubular cells, and FGFR4 has been proposed as a mediator in the associations of FGF23 with left ventricular hypertrophy.11
Strengths of our study include the large and diverse sample, the population-based settings, and the comprehensive set of common genetic variants examined. Potential limitations include a restriction to common variants only, discovery efforts in an exclusively European ancestry sample, limited African ancestry and cFGF23 samples, and a lack of kidney or bone tissue in the gene expression–based association methods.
Although the observed variants were associated with small differences in FGF23 concentrations, a difference in circulating FGF23 of 5% is a risk equivalent for heart failure in the MESA study of 15 ml/min per 1.73 m2 lower eGFR or 5 mm Hg higher systolic BP.4 Further, it is possible that the effects may be more pronounced among individuals with mineral metabolism disturbances, such as those with CKD mineral and bone disorder. It will be crucial to test these results among individuals with CKD, in whom the biologic implications and potential treatment options are most relevant. The clinical relevance of the GWAS findings may not be specifically the magnitude of the difference in FGF23 concentrations, but the underlying biology that is revealed.
In summary, we demonstrate that common genetic variants are associated with circulating FGF23 in adults from the general population. Several of the genetic variants are closely linked with enzymes, transporters, and receptors known to be critical to vitamin D and phosphate homeostasis. Further studies are needed to confirm the SNPs and target loci and elucidate the biologic role of the implicated genes. Candidate genes may be explored in more comprehensive metabolic studies of FGF23 metabolism and in translational animal models that will shed new light on the mechanisms and clinical implications of FGF23 in vitamin D and phosphate homeostasis and its treatment.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors wish to thank the investigators, staff, and participants of the individual participating studies for their valuable contributions.
C.R.-C.’s work is supported by K01DK09019 from the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK). The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, R01HL086694, National Human Genome Research Institute grant U01HG004402, and National Institutes of Health (NIH) contract HHSN268200625226C. FGF23 measurements were supported by R01HL103706. Infrastructure was partly supported by grant number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. The work within the Indiana Sisters cohort was supported by NIH grants R01AG041517, P30AR072581, P01AG018397, R21AR061078, K23AR057096, UL1RR025761, and M01RR00750 and by a donation from the Scottish Rite of Indianapolis Foundation. Genotyping services were provided by the Center for Inherited Disease Research, which is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract HHSN268200782096C). This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. The Osteoporotic Fractures in Men (MrOS) Study is supported by the following NIH institutes: the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Center for Advancing Translational Sciences, and the NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. MrOS Sweden is supported by the Swedish Research Council, Läkarutbildningsavtalet/Avtal om läkarutbildning och forskning research grants in Gothenburg, and the King Gustav V and Queen Victoria Frimurarestiftelse Research Foundation. The work was also supported by the UK Medical Research Council Lifecourse Epidemiology Unit (4050502589). The Osteoporosis Prospective Risk Assessment study was supported by Swedish Research Council grants (K2009-53X-14691-07-3 through K2015-52X-14691-13-4). This Cardiovascular Health Study (CHS) research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268200960009C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, and N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL085251, R01HL105756, R01HL103612, R01HL120393, and U01HL130114, with additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support was provided through R01AG023629 from the NIA. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, Clinical and Translational Science Institute grant UL1TR001881, and the NIDDK Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Support for FGF23 measurements was provided by American Heart Association grant 0575021N to J.H.I. Infrastructure for the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium is supported in part by the NHLBI grant R01HL105756. The Multi-Ethnic Study of Atherosclerosis (MESA) and the MESA Single Nucleotide Polymorphism Health Association Resource (SHARe) project are conducted and supported by the NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420. Funding for FGF23 measurements was provided by R01HL096875. Support is also provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071258, and R01HL071259; by the National Center for Research Resources, grant UL1RR033176; the National Center for Advancing Translational Sciences, grant UL1TR001881; and the NIDDK Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Funding for SHARe genotyping was provided by NHLBI contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and MIT (Boston, MA) using the Affymetrix Genome-Wide Human SNP Array 6.0.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018020192/-/DCSupplemental.
References
- 1.Hu MC, Shi M, Zhang J, Pastor J, Nakatani T, Lanske B, et al.: Klotho: A novel phosphaturic substance acting as an autocrine enzyme in the renal proximal tubule. FASEB J 24: 3438–3450, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimada T, Hasegawa H, Yamazaki Y, Muto T, Hino R, Takeuchi Y, et al.: FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J Bone Miner Res 19: 429–435, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, et al.: Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem 281: 6120–6123, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kestenbaum B, Sachs MC, Hoofnagle AN, Siscovick DS, Ix JH, Robinson-Cohen C, et al.: Fibroblast growth factor-23 and cardiovascular disease in the general Population: The multi-ethnic study of atherosclerosis. Circ Heart Fail 7: 409–417, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ix JH, Katz R, Kestenbaum BR, de Boer IH, Chonchol M, Mukamal KJ, et al.: Fibroblast growth factor-23 and death, heart failure, and cardiovascular events in community-living individuals: CHS (Cardiovascular Health Study). J Am Coll Cardiol 60: 200–207, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isakova T, Wahl P, Vargas GS, Gutiérrez OM, Scialla J, Xie H, et al.: Fibroblast growth factor 23 is elevated before parathyroid hormone and phosphate in chronic kidney disease. Kidney Int 79: 1370–1378, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutiérrez OM, Mannstadt M, Isakova T, Rauh-Hain JA, Tamez H, Shah A, et al.: Fibroblast growth factor 23 and mortality among patients undergoing hemodialysis. N Engl J Med 359: 584–592, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kendrick J, Cheung AK, Kaufman JS, Greene T, Roberts WL, Smits G, et al.: HOST Investigators : FGF-23 associates with death, cardiovascular events, and initiation of chronic dialysis. J Am Soc Nephrol 22: 1913–1922, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scialla JJ, Xie H, Rahman M, Anderson AH, Isakova T, Ojo A, et al.: Chronic Renal Insufficiency Cohort (CRIC) Study Investigators : Fibroblast growth factor-23 and cardiovascular events in CKD. J Am Soc Nephrol 25: 349–360, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ärnlöv J, Carlsson AC, Sundström J, Ingelsson E, Larsson A, Lind L, et al.: Higher fibroblast growth factor-23 increases the risk of all-cause and cardiovascular mortality in the community. Kidney Int 83: 160–166, 2013 [DOI] [PubMed] [Google Scholar]
- 11.Grabner A, Schramm K, Silswal N, Hendrix M, Yanucil C, Czaya B, et al.: FGF23/FGFR4-mediated left ventricular hypertrophy is reversible. Sci Rep 7: 1993, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutsey PL, Alonso A, Selvin E, Pankow JS, Michos ED, Agarwal SK, et al.: Fibroblast growth factor-23 and incident coronary heart disease, heart failure, and cardiovascular mortality: The Atherosclerosis Risk in Communities study. J Am Heart Assoc 3: e000936, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ADHR Consortium : Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet 26: 345–348, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Econs MJ, McEnery PT, Lennon F, Speer MC: Autosomal dominant hypophosphatemic rickets is linked to chromosome 12p13. J Clin Invest 100: 2653–2657, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson T, Yu X, Davis SI, Draman MS, Mooney SD, Cullen MJ, et al.: A novel recessive mutation in fibroblast growth factor-23 causes familial tumoral calcinosis. J Clin Endocrinol Metab 90: 2424–2427, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al.: Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet 38: 1310–1315, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenz-Depiereux B, Bastepe M, Benet-Pagès A, Amyere M, Wagenstaller J, Müller-Barth U, et al.: DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet 38: 1248–1250, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Econs MJ, Fain PR, Norman M, Speer MC, Pericak-Vance MA, Becker PA, et al.: Flanking markers define the X-linked hypophosphatemic rickets gene locus. J Bone Miner Res 8: 1149–1152, 1993 [DOI] [PubMed] [Google Scholar]
- 19.Econs MJ, Francis F, Rowe PS, Speer MC, O’Riordan JL, Lehrach H, et al.: Dinucleotide repeat polymorphism at the DXS1683 locus. Hum Mol Genet 3: 680, 1994 [DOI] [PubMed] [Google Scholar]
- 20.Francis F, Rowe PS, Econs MJ, See CG, Benham F, O’Riordan JL, et al.: A YAC contig spanning the hypophosphatemic rickets disease gene (HYP) candidate region. Genomics 21: 229–237, 1994 [DOI] [PubMed] [Google Scholar]
- 21.Rowe PS, Goulding JN, Francis F, Oudet C, Econs MJ, Hanauer A, et al.: The gene for X-linked hypophosphataemic rickets maps to a 200-300kb region in Xp22.1, and is located on a single YAC containing a putative vitamin D response element (VDRE). Hum Genet 97: 345–352, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Francis F, Hennig S, Korn B, Reinhardt R, de Jong P, Poustka A, et al.: A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. The HYP Consortium. Nat Genet 11: 130–136, 1995 [DOI] [PubMed] [Google Scholar]
- 23.Econs MJ, Rowe PS, Francis F, Barker DF, Speer MC, Norman M, et al.: Fine structure mapping of the human X-linked hypophosphatemic rickets gene locus. J Clin Endocrinol Metab 79: 1351–1354, 1994 [DOI] [PubMed] [Google Scholar]
- 24.The ARIC Investigators: The Atherosclerosis Risk in Communities (ARIC) Study: Design and objectives. The ARIC investigators. Am J Epidemiol 129: 687–702, 1989 [PubMed] [Google Scholar]
- 25.Peacock M, Koller DL, Hui S, Johnston CC, Foroud T, Econs MJ: Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporos Int 15: 489–496, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Mellström D, Johnell O, Ljunggren O, Eriksson A-L, Lorentzon M, Mallmin H, et al.: Free testosterone is an independent predictor of BMD and prevalent fractures in elderly men: MrOS Sweden. J Bone Miner Res 21: 529–535, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, et al.: Multi-Ethnic Study of Atherosclerosis: Objectives and design. Am J Epidemiol 156: 871–881, 2002 [DOI] [PubMed] [Google Scholar]
- 28.Gerdhem P, Brändström H, Stiger F, Obrant K, Melhus H, Ljunggren O, et al.: Association of the collagen type 1 (COL1A 1) Sp1 binding site polymorphism to femoral neck bone mineral density and wrist fracture in 1044 elderly Swedish women. Calcif Tissue Int 74: 264–269, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al.: The Cardiovascular Health Study: Design and rationale. Ann Epidemiol 1: 263–276, 1991 [DOI] [PubMed] [Google Scholar]
- 30.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, et al.: CKD-EPI Investigators : Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 367: 20–29, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Zheng J, Li Y, Abecasis GR, Scheet P: A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet Epidemiol 35: 102–110, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devlin B, Roeder K: Genomic control for association studies. Biometrics 55: 997–1004, 1999 [DOI] [PubMed] [Google Scholar]
- 34.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials 7: 177–188, 1986 [DOI] [PubMed] [Google Scholar]
- 36.Weir BS, Cockerham CC: Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370, 1984 [DOI] [PubMed] [Google Scholar]
- 37.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al.: LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner SD: qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. Journal of Open Source Software 3: 731, 2018 [Google Scholar]
- 39.Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA, et al.: A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS One 10: e0120758, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Wang K: Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10: 1556–1566, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbeira A, Shah KP, Torres JM, Wheeler HE, Torstenson ES, Edwards T, et al.: MetaXcan: Summary statistics based gene-level association method infers accurate PrediXcan results [Internet]. bioRxiv 2016. Available at: https://www.biorxiv.org/content/early/2016/03/23/045260. Accessed November 10, 2017
- 42.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al.: GTEx Consortium : A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47: 1091–1098, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, et al.: GTEx Consortium : A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv Biobank 13: 311–319, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al.: GTEx Consortium : Human genomics. The human transcriptome across tissues and individuals. Science 348: 660–665, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, et al.: 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep 7: 45040, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikpay M, Goel A, Won H-H, Hall LM, Willenborg C, Kanoni S, et al.: A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47: 1121–1130, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson-Cohen C, Lutsey PL, Kleber ME, Nielson CM, Mitchell BD, Bis JC, et al. : Genetic variants associated with circulating parathyroid hormone. J Am Soc Nephrol 28: 1553–1565, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y-H, Duncan EL, Ntzani EE, et al.: Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44: 491–501, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones G, Prosser DE, Kaufmann M: 25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): Its important role in the degradation of vitamin D. Arch Biochem Biophys 523: 9–18, 2012 [DOI] [PubMed] [Google Scholar]
- 50.Tenenhouse HS: Regulation of phosphorus homeostasis by the type iia na/phosphate cotransporter. Annu Rev Nutr 25: 197–214, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Branch MR, Hepler JR: Endogenous RGS14 is a cytoplasmic-nuclear shuttling protein that localizes to juxtanuclear membranes and chromatin-rich regions of the nucleus. PLoS One 12: e0184497, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, et al.: ICBP Consortium; AGEN Consortium; CARDIOGRAM; CHARGe-Heart Failure Group; ECHOGen Consortium : Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7: 10023, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahajan A, Rodan AR, Le TH, Gaulton KJ, Haessler J, Stilp AM, et al.: SUMMIT Consortium; BioBank Japan Project : Trans-ethnic fine mapping highlights kidney-function genes linked to salt sensitivity. Am J Hum Genet 99: 636–646, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai X-Y, Miao D, Goltzman D, Karaplis AC: The autosomal dominant hypophosphatemic rickets R176Q mutation in fibroblast growth factor 23 resists proteolytic cleavage and enhances in vivo biological potency. J Biol Chem 278: 9843–9849, 2003 [DOI] [PubMed] [Google Scholar]
- 55.Roy S, Martel J, Ma S, Tenenhouse HS: Increased renal 25-hydroxyvitamin D3-24-hydroxylase messenger ribonucleic acid and immunoreactive protein in phosphate-deprived Hyp mice: A mechanism for accelerated 1,25-dihydroxyvitamin D3 catabolism in X-linked hypophosphatemic rickets. Endocrinology 134: 1761–1767, 1994 [DOI] [PubMed] [Google Scholar]
- 56.Bai X, Miao D, Xiao S, Qiu D, St-Arnaud R, Petkovich M, et al.: CYP24 inhibition as a therapeutic target in FGF23-mediated renal phosphate wasting disorders. J Clin Invest 126: 667–680, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto F, Clausen H, White T, Marken J, Hakomori S: Molecular genetic basis of the histo-blood group ABO system. Nature 345: 229–233, 1990 [DOI] [PubMed] [Google Scholar]
- 58.Smith NL, Chen M-H, Dehghan A, Strachan DP, Basu S, Soranzo N, et al.: Wellcome Trust Case Control Consortium : Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation 121: 1382–1392, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desch KC, Ozel AB, Siemieniak D, Kalish Y, Shavit JA, Thornburg CD, et al.: Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proc Natl Acad Sci U S A 110: 588–593, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, et al.: Myocardial Infarction Genetics Consortium; Wellcome Trust Case Control Consortium : Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. Lancet 377: 383–392, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewerin C, Ljunggren Ö, Nilsson-Ehle H, Karlsson MK, Herlitz H, Lorentzon M, et al.: Low serum iron is associated with high serum intact FGF23 in elderly men: The Swedish MrOS study. Bone 98: 1–8, 2017 [DOI] [PubMed] [Google Scholar]
- 62.Imel EA, Liu Z, McQueen AK, Acton D, Acton A, Padgett LR, et al.: Serum fibroblast growth factor 23, serum iron and bone mineral density in premenopausal women. Bone 86: 98–105, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rigas AS, Berkfors AA, Pedersen OB, Sørensen E, Nielsen KR, Larsen MH, et al.: Reduced ferritin levels in individuals with non-O blood group: Results from the Danish Blood Donor Study. Transfusion 57: 2914–2919, 2017 [DOI] [PubMed] [Google Scholar]
- 64.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al.: GIANT Consortium : Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry [published online ahead of print August 16, 2018]. Hum Mol Genet doi: 10.1093/hmg/ddy271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kestenbaum B, Glazer NL, Köttgen A, Felix JF, Hwang SJ, Liu Y, et al. : Common genetic variants associate with serum phosphorus concentration. J Am Soc Nephrol 7: 1223–1232, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.