Abstract
Adult mammalian hearts have a very limited regeneration capacity, due largely to a lack of cardiomyocyte (CM) proliferation. It was recently reported that epicardial, but not myocardial, follistatin-like 1 (Fstl1) activates CM proliferation and cardiac regeneration after myocardial infarction (MI). Furthermore, bacterially synthesized human FSTL 1 (hFSTL1) was found to induce CM proliferation, whereas hFSTL1 synthesized in mammals did not, suggesting that post-translational modifications (e.g., glycosylation) of the hFSTL1 protein affect its regenerative activity. We used modified mRNA (modRNA) technology to investigate the possible role of specific hFSTL1 N-glycosylation sites in the induction, by hFSTL1, of CM proliferation and cardiac regeneration. We found that the mutation of a single site (N180Q) was sufficient and necessary to increase the proliferation of rat neonatal and mouse adult CMs in vitro and after MI in vivo, respectively. A single administration of the modRNA construct encoding the N180Q mutant significantly increased cardiac function, decreased scar size, and increased capillary density 28 days post-MI. Overall, our data suggest that the delivery of N180Q hFSTL1 modRNA to the myocardium can mimic the beneficial effect of epicardial hFSTL1, triggering marked CM proliferation and cardiac regeneration in a mouse MI model.
Keywords: cardiomyocytes proilferation, cardiac regeneration, gene therapy
Introduction
Most patients now survive their first myocardial infarction (MI), largely due to the use of coronary artery reperfusion technologies within hours of infarction. The high mortality traditionally associated with heart attacks has been replaced by an increase in the prevalence of chronic heart failure (HF) and its medical complications, disability and soaring costs.1 Thus, success in the treatment of heart attacks (or acute MI), coupled with the rising prevalence of obesity, diabetes, and hypertension, has dramatically increased the burden of HF, which now accounts for over 13% of all deaths worldwide and is projected to cost over $70 billion in the US alone by 2030.2
The pathophysiology of HF due to MI includes the injury and death of cardiomyocytes (CMs) and blood vessels, followed by their replacement with non-contractile scar tissue, in a process called cardiac remodeling. No current therapy, not even revascularization by coronary artery reperfusion, can promote the regeneration of lost CMs or blood vessels or prevent progression to HF.
The reactivation of CM proliferation is a key element in cardiac regeneration strategies. In zebrafish and newt, cardiac regeneration is mostly mediated by CM proliferation.3, 4, 5, 6 In mammals, CM proliferation is a distinct pathway for heart growth and regeneration during fetal development.7, 8 It has been shown that a subset of fetal genes is upregulated in adult CMs after injury, which suggests that adult CMs are not terminally differentiated and retain some degree of plasticity.7, 9 Several studies have shown that adult CMs can be induced to re-enter the cell cycle, by proteins,10, 11, 12, 13, 14, 15 viruses,12, 13, 16, 17, 18 microRNAs,19, 20, 21, 22, 23 long non-coding RNAs,24 small molecules,25 or transgenesis with pro-proliferation genes in mouse models.26, 27, 28, 29, 30, 31
Using adenovirus-mediated transformation with a dominant-negative (DN) p38 MAP kinase, Engel et al.12 showed that inhibiting p38 MAP kinase and adding fibroblast growth factor 1 (FGF1), either alone or together with neuregulin 1 (NRG1) or interleukin-1β (IL-1β), reactivated the proliferation of neonatal and adult mammalian CMs. Similarly, the addition of periostin13 or NRG111 induces cardiac repair via CM proliferation, but this effect is restricted to mononuclear cells in rodents. Viruses, such adenovirus or adeno-associated virus (AAV), have been used to inhibit proteins, such as p73, through a DN approach,16 or to deliver proteins, such as YAP,17 to increase CM proliferation. A recent screen of cell-cycle regulators showed that cyclin-dependent kinase 1 (CDK1), CDK4, cyclin B1, and cyclin D1 efficiently induced cell division in post-mitotic mouse, rat, and human CMs.18
Non-coding RNAs, such as microRNAs (miRs) and long non-coding RNAs (lincRNAs) also induce CM proliferation. A systematic screen with miR mimics showed that both miR-199a and miR-590 induce CM proliferation in neonatal and adult CMs when delivered by AAV or as a single-stranded RNA.19, 20, 21 Non-coding RNAs of the miR-15 family have been shown to inhibit the proliferation of neonatal CMs in vitro.22 In addition, miR-99/100 and Let-7a/c have been shown to regulate CM proliferation and cardiac regeneration in both zebrafish and mouse hearts.23 See et al.24 showed that lincRNA regulates the dedifferentiation of CMs and their entry into the cell cycle in vivo. A screening of chemical-compound libraries with a Fucci-based CM system to assess passage through the cell cycle in CMs identified carbacyclin as an inducer of neonatal and post-natal CM proliferation via PPARδ activation.25
Several transgenic mouse models overexpressing pro-proliferative genes or with conditional knockouts have been used to investigate the roles of various genes in CM proliferation after injury. In transgenic mouse models of MI and HF, constitutively active ERBB2 (caERBB2) was found to promote cardiac regeneration through the induction of CM proliferation.26 In addition, studies of transgenic mice have shown that Meis1 loss of function in adult CMs induces adult CM proliferation, whereas Meis1 overexpression in these cells limits mouse neonatal heart regeneration following MI.29 Similarly, Martin and colleagues27, 30 identified Hippo signaling as an endogenous repressor of adult CM renewal and regeneration. Transgenic mouse models with conditional knockouts of cell-cycle-inducer genes, such as the c-myc and cyclin D2 genes, display significant proliferation of adult CMs after activation;28, 31 however, these studies also clearly showed that the long-term expression of pro-proliferative genes resulted in cardiac hypertrophy.31
In 2015, Wei et al.14 showed that follistatin-like 1 protein (Fstl1) levels decline in the epicardium but increase in the myocardium after MI. Epicardial Fstl1 promotes CM proliferation and cardiac regeneration, whereas myocardial Fstl1 does not. In addition, bacterially synthesized human FSTL 1 (hFSTL1) promotes regeneration after MI in rodent and pig models, improving animal survival, decreasing scar size, and leading to a recovery of cardiac function and the formation of new CMs and blood vessels.14 Bacterially synthesized hFSTL1 induces CM proliferation in vitro and in vivo, whereas the same protein produced in mammals does not.14
FSTL1 (also known as FRP1, FSL1, OCC1, and Tsc36) is only distantly related to follistatin. FSTL1 belongs to the FS calcium-binding domain protein family. All members of this family contain a signal peptide, FS domains (one such domain in FSTL1) and consensus N-glycosylation sites (three for FSTL1, N180, N175, and N144).32
Several studies have shown that the delivery of specific genes can increase CM proliferation, but several obstacles to this approach remain: the choice of delivery method,33, 34, 35 the persistence of expression (too short for protein, or too long for AAV), and difficulties relating to local administration or dosing. These challenges highlight the need for an efficient gene delivery approach capable of delivering genes safely and locally to the heart in a transient, efficient, and controlled manner. We and others have shown that gene transfer with modRNA technology is a highly efficient approach to the rapid transient transfection of heart cells, without the induction of innate immunity.33, 36, 37, 38, 39, 40 We recently showed that modRNA in which all the uridine residues were replaced with N1-methylpseudouridine-5′-triphosphate (1-mψU) was less immunogenic, more resistant to RNase, and more efficiently translated than unmodified mRNA in cardiac cells and tissue.40 The delivery of modRNA to cardiac cells and tissue leads to the immediate transient production of large amounts of protein, with pulse-like kinetics (duration of ∼5–7 days in vitro and ∼10 days in vivo) in several cell types in the heart, including CMs.40
In this study, we used modRNA technology to investigate the impact of N-glycosylation on the regenerative activity of hFSTL1 in vitro and in vivo. We found that single replacement of asparagine (N) with glutamine (Q) in the N-glycosylation site at position 180 of hFSTL1 were sufficient and necessary to activate CM proliferation and limit cardiac remodeling post-MI, following the delivery of the modRNA to the myocardium.
Results
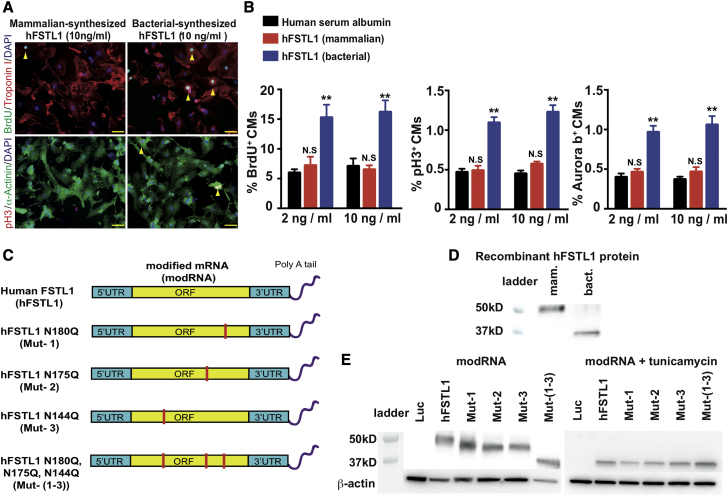
It has recently been shown14 that bacterially synthesized hFSTL1 or epicardial Fstl1 increases CM proliferation and promotes cardiac regeneration, whereas hFSTL1 synthesized in mammals or myocardial Fstl1 does not. These findings suggested a possible role for glycosylation in the function of hFSTL1 in cardiac regeneration. We first confirmed these observations by culturing neonatal rat CMs in the presence of mammalian or bacterially synthesized hFSTL1 (Figures 1A and 1B). Our results were consistent with the previous report,14 showing that only bacterially synthesized hFSTL1 significantly increased the expression of classical markers of CM proliferation (bromodeoxyuridine [BrdU], phosphohistone H3 [pH3], Aurora B).
Figure 1.
Bacterially Synthesized hFSTL1 Induces the Proliferation of Rat Neonatal CMs In Vitro, Whereas Mammalian-Synthesized hFSTL1 Does Not
(A) Representative images of proliferation marker (BrdU or PH3) levels in neonatal rat CMs cultured for 3 days in the presence of bacterially synthesized or mammalian-synthesized hFSTL1. (B) Quantification of classical markers of proliferation (BrdU, PH3, and Aurora B) in neonatal rat CMs cultured for 3 days in the presence of bacterially synthesized or mammalian-synthesized hFSTL1. (C) Design of different modRNAs with mutations of different N-glycosylation sites. (D and E) Western blot analysis of bacterially synthesized hFSTL1, mammalian-synthesized hFSTL1 (D) or the various hFSTL1 N-glycosylation mutants with or without tunicamycin (E). Representative results from three independent experiments are shown; **p < 0.01; N.S., not significant, n = 3, one-way ANOVA with Bonferroni post-hoc tests to correct for multiple testing. Scale bars, 50 μm.
We investigated whether a lack of glycosylation of the bacterially synthesized hFSTL1 was responsible for its pro-proliferative effect on CMs. We used an established modRNA system, developed in our laboratory, to generate two control modRNAs: the luciferase modRNA, which has no biological activity (modRNA control genes37, 38), and the hFSTL1 modRNA, which has all three of the identified N-glycosylation sites.32 We compared the induction of proliferation between the control modRNAs and (1) hFSTL1 modRNAs with single point mutations converting the asparagine resiudues of the individual N-glycosylation sites to glutamines (N180Q [Mut-1], N175Q [Mut-2], or N144Q [Mut-3]), (2) a mutated hFSTL1 modRNA in which all three N-glycosylation sites were mutated (N180Q, N175Q, and N144Q [Mut-(1–3)]) (Figures 1C and S1). We checked that the mutation of the N-glycosylation sites in the various mutant modRNAs did indeed impair glycosylation in the resulting proteins, by comparing the migration of the proteins generated with those of the mammalian and bacterially generated hFSTL1 proteins on SDS-polyacrylamide gels.
Consistent with reported findings,14 the glycosylated mammalian-synthesized hFSTL1 (∼50 kD) migrated significantly more slowly than the non-glycosylated bacterially synthesized hFSTL1 (∼35 kD) (Figure 1D). We showed, with mutated hFSTL1 modRNAs translated in 3T3 mammalian cells, that the hFSTL1 modRNA protein obtained had a molecular weight of ∼50 kD and that each of the modRNAs with a single N-glycosylation site mutation (N180Q, N175Q, or N144Q) generated a smaller hFSTL1 protein. Furthermore, the triple-mutant variant migrated to a position corresponding to ∼35 kD, a molecular weight similar to that for the bacterially synthesized hFSTL1. Finally, we confirmed that the differences in protein size were due to a lack of N-glycosylation by transfecting cells with the various hFSTL1 modRNAs in the presence of the N-glycosylation inhibitor tunicamycin. Tunicamycin treatment eliminated the size differences between the mutated and non-mutated hFSTL1 proteins generated from the different hFSTL1 modRNAs (all were ∼35 kD in size; Figure 1E).
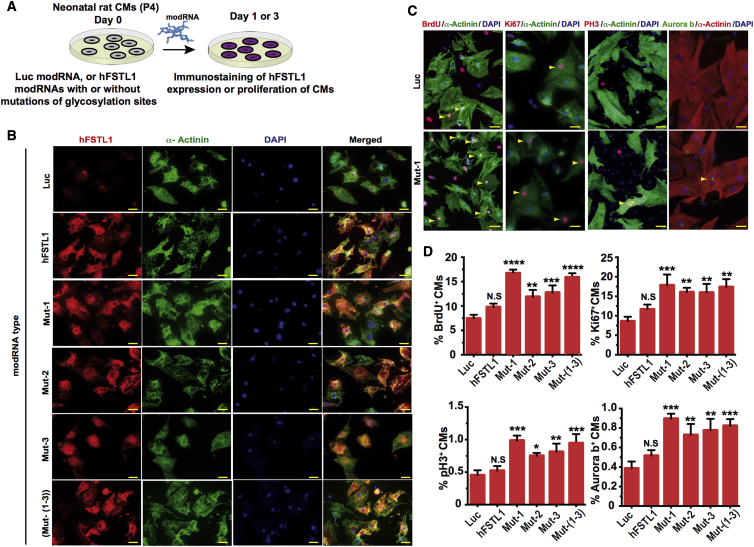
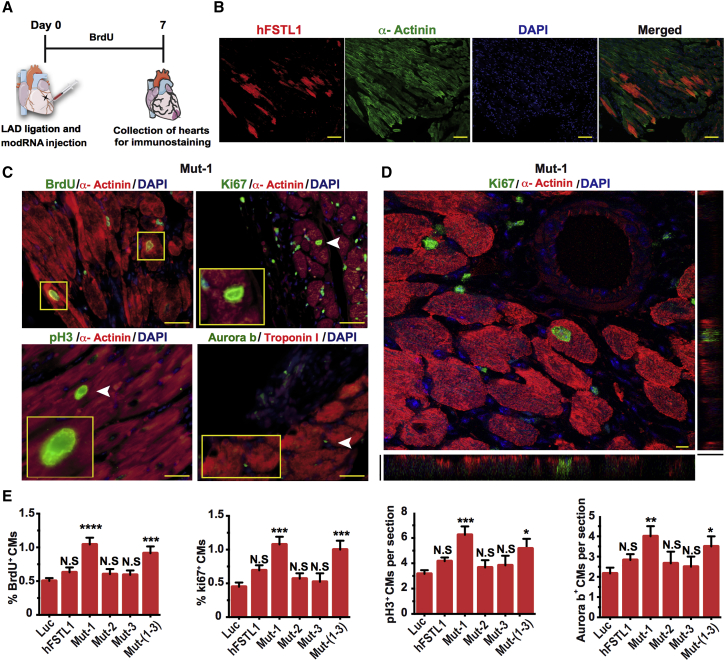
We assessed the ability of the various mutated hFSTL1 modRNAs to induce the proliferation of neonatal rat CMs in vitro. We first confirmed the successful translation of these modRNAs in neonatal rat CMs 1 day post-transfection (Figures 2A and 2B). We then assessed neonatal rat CM proliferation 3 days post-transfection. We found that only the mutated hFSTL1 modRNAs (with mutations at a single N-glycosylation site or at all three sites) significantly increased the proliferation of rat neonatal CMs 3 days post-modRNA transfection (Figures 2C and 2D). We then assessed the ability of mutated hFSTL1 modRNAs to increase the proliferation of adult mouse CMs in vivo. We used our mouse model of short-term MI, in which we ligate the mouse left anterior descending (LAD) coronary artery in the heart and immediate delivery of the control (Luc) or mutated hFSTL1 modRNAs. 7 days post-MI and modRNA delivery hearts were collected and evaluated (Figure 3A). Seven days post-transfection, hFSTL1 expression was detected in the heart (Figure 3B) and was associated with a significant enhancement of CM proliferation relative to control constructs for a single mutant hFSTL1 (N180Q, hereafter, Mut-1) or the triple mutant hFSTL1 (N180Q and N175Q and N144Q, hereafter, Mut-(1–3)) modRNAs delivered in vivo. The other mutated hFSTL1 (N175Q [Mut-2] or N144Q [Mut-3]) modRNAs and the hFSTL1 modRNA had no significant increment of CM proliferation in vivo (Figures 3C–3E). CM proliferation in vivo was validated by confocal microscopy, which showed that Ki67+ CM nuclei were surrounded on all sides by a single CM (α-actinin+; Figure 3D). The CM proliferation induced by mutated hFSTL1 (Mut-1 or Mut-(1–3)) modRNAs was not associated with an increase in CM size or cardiac hypertrophy (Figure S2). Nevertheless, all hFSTL1 modRNAs, whether or not they carried mutations, induced a significant increase in leukocyte (CD45+) infiltration (Figures S3A–S3C) and non-CM proliferation (Figures S3D–S3F) when delivered to the heart 7 days post-MI.
Figure 2.
N-Glycosylation-Deficient hFSTL1 modRNA Induces Rat Neonatal CM Proliferation In Vitro
(A) Experimental timeline for evaluation of the ability of different hFSTL1 modRNAs with mutations of different N-glycosylation sites to increase neonatal rat CM proliferation in vitro 3 days post-transfection. (B) Immunostaining of neonatal rat CMs 1 day post-transfection with the Luc control, hFSTL1, or various hFSTL1 mutant modRNAs. (C) Representative images of the expression of classical proliferation markers (BrdU, Ki67, PH3, and Aurora B) in neonatal rat CMs 3 days post-transfection with Luc control or Mut-1 modRNA. Yellow arrowheads indicate proliferating CMs. (D) Quantification of classical markers of proliferation in rat CMs 3 days post-treatment with various modRNAs. Representative results from three independent experiments are shown; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05; N.S., not significant, n = 3, one-way ANOVA with Bonferroni post-hoc correction for multiple testing. Scale bar, 20 μm.
Figure 3.
A Point Mutation Affecting a Single hFSTL1 N-Glycosylation Site (N180Q) Increases CM Proliferation Post-MI
(A) Experimental timeline for evaluation of the ability of hFSTL1 modRNAs with mutations of different N-glycosylation sites to increase CM proliferation in vivo 7 days post-MI. (B) Representative in vivo immunostaining of hFSTL1 post-modRNA delivery and MI. (C) Representative images of the expression of classical markers of proliferation (BrdU, Ki67, PH3, and Aurora B) in vivo 7 days post-MI. (D) Representative confocal microscopy images of Ki 67 and α-actinin expression in vivo 7 days post-MI. (E) Quantification of classical proliferation markers in CMs 7 days post-MI and treatment with various modRNAs. Representative results from two independent experiments are shown; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05; N.S., not significant, n = 5, one-way ANOVA with Bonferroni post-hoc correction for multiple testing. Scale bars, 50 μm (B and C) or 5 μm (D).
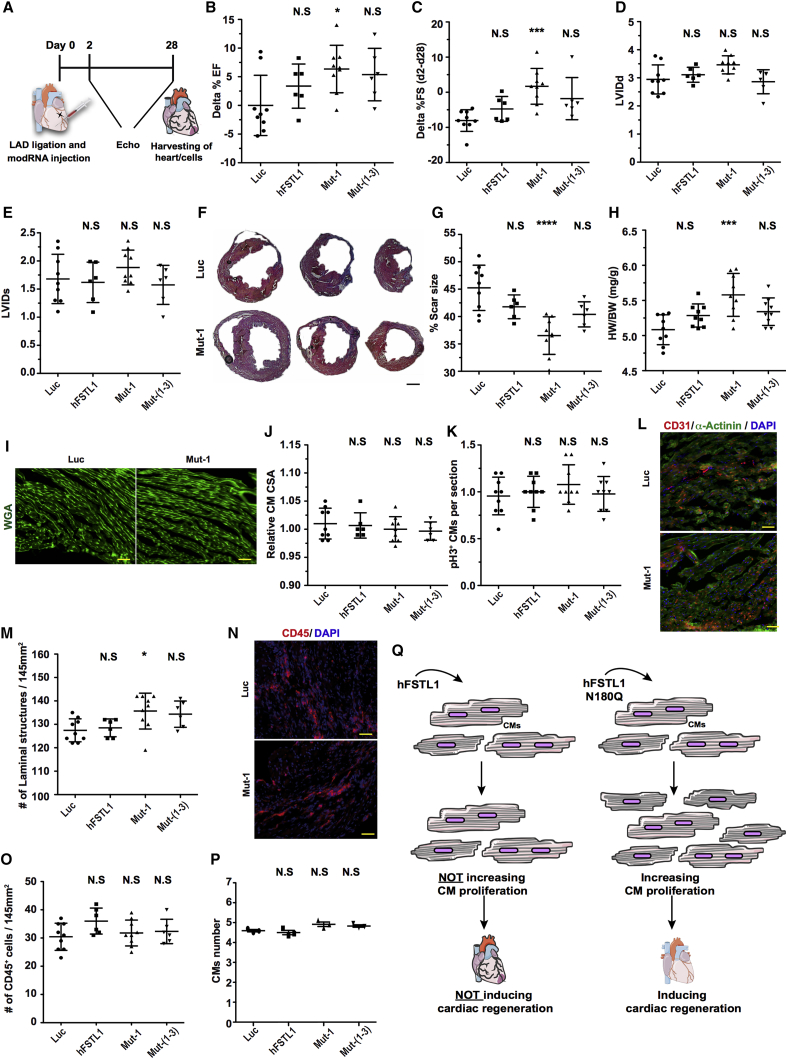
After confirming that the Mut-1 and Mut-(1–3) modRNAs induced CM proliferation in vivo post-injury, we evaluated their ability to prevent cardiac remodeling post-MI. We used our model of long-term MI in mice, in which mice were kept for 28 days post-MI and modRNA delivery. Echography was used to evaluate cardiac function (delta % ejection fraction [EF] and fractional shortening [FS]). Cardiac function was found to have improved significantly 28 days post-MI and -Mut-1 modRNA treatment, relative to treatment with the luciferase control, as shown by the significant increase in delta of %EF (from Luc control to Mut-1 at day 28 post-MI) or delta of %FS (from baseline to day 2 post-MI and day 28 post-MI) (Figures 4A–4C). Left ventricular internal diameter at the end of diastole and systole was not significantly modified by the various modRNAs (Figures 4D and 4E). Masson trichrome staining was used to evaluate the scar size in hearts on day 28 (Figures 4F and 4G). Scars were significantly smaller after treatment with Mut-1 than after treatment with the control Luc modRNA (Figure 4G). Hearts expressing the Mut-1 modRNA post-MI had a significantly higher heart weight-to-body weight ratio (Figure 4H). The increase in heart weight was not due to changes in CM size (hypertrophy), as no such change in size was detected in Mut-1 modRNA-expressing CMs 28 days post-MI (Figures 4I and 4J). CM proliferation 28 days post-MI did not differ between Mut-1 and control hearts (Figure 4K), but capillary density in the left ventricle was significantly higher in Mut-1 hearts than in control hearts (Figures 4L and 4M). No significant increase in the number of leukocytes (CD45+; Figures 4N and 4O) or CMs (Figure 4P) was detected 28 days post-MI for any of the treatments.
Figure 4.
A Point Mutation Affecting a Single hFSTL1 N-Glycosylation Site (N180Q) Improves Cardiac Function and Outcome Post-MI
(A) Experimental timeline for evaluation of cardiac function and outcome post-MI and the delivery of Luc (modRNA control), hFSTL1, or mutated hFSTL1 modRNAs (affecting a single N-glycosylation site [N180Q, Mut-1] or all three N-glycosylation sites [N180Q, N175Q, N144Q; Mut-(1–3)]). (B) Echographic evaluation of % ejection fraction 28 days post-MI and modRNA delivery. (C) Echographic evaluation of the change in the percentage fractional shortening between day 2 (baseline) and day 28 post-MI. (D and E) Echography was used to evaluate left ventricular internal diameter at end diastole (D), LVIDd, and end systole (E), LVIDs, 28 days after LAD ligation. (F) Representative image of Masson trichrome staining to evaluate scar size 28 days post-MI. (G and H) Quantification of scar size (G) and heart weight-to-body weight ratio (H). (I) Representative image of wheatgerm agglutinin (WGA) staining to evaluate CM size. (J and K) Quantification of CM size (J) and PH3 expression in CMs (K). (L) Representative image of CD31 staining to evaluate capillary density. (M) Quantification of capillary density. (N) Representative image of CD45 staining to evaluate leukocyte infiltration in the left ventricle. (O and P) Quantification of CD45 cells in the left ventricle (O) and of the number of CMs per heart (P) for the different treatments 28 days post-MI. (Q) Summary of the effect of the loss of a single N-glycosylation site (N180Q) of hFSTL1 on the induction of CM proliferation and cardiac regeneration following myocardial transfection post-MI. Representative results from two independent experiments are shown; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05; N.S., not significant. n = 9 for Luc and Mut-1 and n = 6 for hFSTL1 and Mut-(1–3). One-way ANOVA, with Bonferroni correction for multiple testing. Scale bars, 1 mm (F) or 50 μm (I, L, and N).
Overall, our functional data suggest that a single injection of Mut-1 modRNA significantly improves cardiac function, increases angiogenesis, and decreases scar size 28 days post-MI. We then investigated whether a second administration of Mut-1, immediately, or 10 days post-MI, further increased the regenerative effect. We observed no significant difference in the change in cardiac function, scar size, CM proliferation, capillary density, or CM size relative control. The double administration Mut-1 therefore had no additive or synergistic effect on the heart post-MI (Figure S4).
Discussion
Regenerative processes can occur in the mammalian heart immediately after birth and are mostly mediated by the proliferation of CMs.41 Over the last few decades, several groups have attempted to enhance CM proliferation with secreted proteins as a means of inducing cardiac regeneration and repair.11, 12, 13 This approach has several disadvantages, including a nonspecific organ distribution of the proteins used, due to their systemic delivery via intravenous injection. Protein delivery specifically to the heart was recently achieved by direct injection into the myocardium, and epicardial delivery has been achieved with a patch.14, 15 These approaches enhance the biological effects of the proteins, as secreted proteins usually have a very short half-life, and their local delivery accelerates the activation of CM proliferation and renders this process more efficient. However, it is clear that methods for longer-term expression and for controllable cardiac delivery are required. We recently optimized the delivery of modRNA into the heart40 and showed that it led to about 10 days of protein expression in various cell types, including CMs, endothelial cells, smooth muscle cells, and cardiac fibroblasts.37, 38, 40 The effect of non-glycosylated bacterially synthesized hFSTL1 on CM proliferation observed here (Figures 1A and 1B) is consistent with previous reports14 and justifies our investigation of the role of hFSTL1 glycosylation in inducing CM proliferation. This work provides insight into the structural or functional determinants of CM proliferation present in FTSL1. Using a modRNA delivery method (Figure 1C), we show here that the mutation of all three N-glycosylation sites in hFSTL1 results in faster migration on SDS-polyacrylamide gels at a molecular weight similar to that for non-glycosylated bacterially synthesized hFSTL1 (Figures 1D and 1E), suggesting that these mutations abolish the glycosylation of the protein. The mutation of just one of these sites in the hFSTL1 protein also increased migration on SDS-polyacrylamide gels, albeit to a lesser extent, suggesting that the glycosylation state of hFSTL1 may determine its cardiac regeneration capacity. We also show that the pro-proliferative effects of hFSTL1 are dependent on a lack of N-glycosylation at a particular hFSTL1 glycosylation site (N180Q, Mut-1 modRNA). Mut-1 induced higher levels of proliferation than the other two modRNAs with single mutations of N-glycosylation sites (N175Q and N144Q; Mut-2 and Mut-3; Figures 2C and 2D). Moreover, in adult mouse CMs, only Mut-1 and Mut-(1–3) (which includes the N180Q mutation) modRNAs significantly enhanced CM proliferation post-MI without inducing CM hypertrophy (Figures 3C–3E and S2), highlighting the importance of the N180 glycosylation site for the pro-proliferative effect observed in vivo. In the mouse model of long-term MI, only the Mut-1 modRNA significantly increased heart function and reduced scar size 28 days post-MI (Figure 4). We suggest that the lack of a functional N180 glycosylation site modifies the glycosylation pattern of hFSTL1 and, possibly, its secondary structure, allowing it to activate unknown receptors inducing CM proliferation and cardiac regeneration. Our data also suggest that double mutants of N180 and another N-glycosylation site on hFSTL1 (N175 or N144) reduce the proliferative and regenerative activity of this protein. Further studies are required to investigate the interplay between structure and function. It will also be interesting to investigate the role of the N180 glycosylation site of hFSTL1 in other species with and without cardiac regeneration capacity. The impact of Mut-1 remained significant even in the presence of an undesirable increase in non-CM proliferation and an inflammatory response observed with hFSTL1 or any of the mutated hFSTL1 modRNAs post-MI (Figure S3). These findings are consistent with those of Chaly et al.42 demonstrating a pro-inflammatory effect of Fstl1 in mice. They raise the possibility that we may be able to increase the beneficial effect of Mut-1 modRNA on cardiac regeneration by attenuating the inflammatory environment in the heart (e.g., by administering anti-inflammatory drugs or by expressing anti-inflammatory genes via the same modRNA delivery system). No additive effect on cardiac function was observed with two administrations of mut-1 modRNA, possibly due to the compromising effects of the inflammatory environment or other parameters, such as the opening of the chest for a second time and physiological injury to the heart due to the injection (Figure S4). This work extends previous findings14 by showing that Mut-1 modRNA induces cardiac regeneration when delivered directly into the myocardium by the modRNA delivery method, rather than only after its delivery to the epicardium. In summary, hFSTL1 has a limited but significant effect on CM proliferation and cardiac regeneration post-MI, which is dependent on its N180 glycosylation site. A single mutation of this glycosylation site, converting the asparagine residue to a glutamine, increases the ability of hFSTL1 to enhance CM proliferation and to improve cardiac regeneration post-MI (Figure 4Q). This study has several important implications for basic research, methodology research, and treatment. The identification of a specific hFSTL1 N-glycosylation site responsible for CM proliferation will pave the way for further investigations of the role of N-glycosylation in proliferation or in other cell functions related to regeneration. Such studies could be performed in CMs or other cell types or organs with, or even without, regenerative capacity. Moreover, as only the Mut-1 modRNA-induced CM proliferation, it should be possible to identify the cognate receptor of hFSTL1. In terms of methodology, this study describes faster and easier methods for evaluating N-glycosylation sites in proteins than those based on DNA plasmids, viruses, or purified proteins. Finally, to the best of our knowledge, this is the first report of the induction of transient but sustained CM proliferation by a modRNA. Our data suggest that Mut-1 modRNA is a strong candidate therapy for the induction of CM proliferation and cardiac regeneration post-MI. Mut-1 thus has potential clinical applications, as modRNA is a safe, transient, and clinically applicable gene-delivery method. Nevertheless, several questions remain unanswered. The mechanism of action of hFSTL1 in CMs and the identity of its cognate receptor remain unknown. In addition, the ability of Mut-1 to enhance CM proliferation or cardiac regeneration in large animal models (e.g., sheep or pigs) has not yet been assessed. As both modRNA and myocardial delivery are clinically relevant, we plan to investigate these issues further.
Materials and Methods
Animals
We used age-matched male and female Swiss Webster mice (CFW). Mice were anesthetized with a mixture of 1%–2% isoflurane in air. Various modRNAs (100 μg/heart) in sucrose citrate buffer were injected directly into the myocardium during open chest surgery (please see “modRNA Transfection In Vitro and In Vivo” below for additional technical information). Three to eight animals were used for each experiment. Mice were reared in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Icahn School of Medicine at Mount Sinai.
modRNA Synthesis
ModRNAs were synthesized in house. All DNA plasmid templates used for modRNA production were purchased from GeneArt and Invitrogen and were verified by sequencing. PCR-tailed DNA plasmids (120 poly(A) tail) were used for in vitro transcription to generate modRNAs (see Table S1 for complete list of open reading frame sequences used for modRNA synthesis in this study) with a customized ribonucleotide blend of anti-reverse cap analog, 3′-O-Me-m7G(5′)ppp(5′)G (6 mM, TriLink Biotechnologies), guanosine triphosphate (1.5 mM, Life Technologies), adenosine triphosphate (7.5 mM, Life Technologies), cytidine triphosphate (7.5 mM, Life Technologies), and 1-mψU (7.5 mM, TriLink Biotechnologies), as previously described.37, 38, 39, 40 We purified the mRNA with the Megaclear kit (Life Technologies) and treated it with Antarctic phosphatase (New England Biolabs) for 1 hr, before repurification with the Megaclear kit. The mRNA was precipitated by incubation overnight with ammonium acetate and ethanol at 4°C and resuspended in 10 mM TrisHCl, 1 mM EDTA. We quantified the mRNA with a Nanodrop spectrometer (Thermo Scientific) and checked its quality with a bioanalyzer (Agilent Technologies). A more detailed description of the protocol for modRNA synthesis is provided elsewhere.39
Transfection with modRNA In Vitro and In Vivo
Neonatal CMs were transfected in vitro in the presence of Lipofectamine RNAiMAX transfection reagent (Life Technologies), used according to the manufacturer’s instructions. Cells were transfected with modRNA in vitro in sucrose citrate buffer consisting of 20 μL of a 0.3 g/mL solution of sucrose (15% sucrose) in nuclease-free water, 20 μL of 0.1 M citrate (pH 7; Sigma), and 20 μL of modRNA solution (various concentrations in saline), with the final volume made up to 60 μL with nuclease-free water (5% sucrose and 0.03 M citrate final concentration). The transfection mixture was injected directly (three individual injections, 20 μL each) into the myocardium at three different sites (two injections in the border zone and one at the heart apex).
Echocardiography
We evaluated the dimensions and function of the left ventricle by performing transthoracic two-dimensional echocardiography. In a double-blind study (neither the surgeon nor the echography technician was aware of the treatment), various modRNAs were injected into CFW mice (8 to 12 weeks old). Animals underwent echography onsite with a GE Cares machine (V7R5049) equipped with a 40-MHz mouse ultrasound probe. Mice were anesthetized with a mixture of 1%–2% isoflurane in air, and imaging was performed on days 2 and 28 post-LAD-ligation.43 The EF and FS were calculated as percentages from the diastolic volume (EDV) and end systolic volume (ESV) dimensions on an M-mode ultrasound scan. The following formulas were used: % EF = (EDV − ESV)/EDV*100, and % FS = (left ventricular internal dimension at end-diastole (LVIDd)) − (left ventricular internal dimension at end-systolic (LVIDs))/LVIDd*100. Echocardiograms were performed on six or nine hearts/treatment group.
Isolation of CMs from Adult Mice and Neonatal Rats
Adult mouse CMs were isolated by Langendorff’s method from CFW mice 28 days after MI and modRNA injection, as previously described.37 The number of CMs was calculated as the mean of three to four different counts/sample of three hearts/group obtained with a hemocytometer. CMs were isolated from the hearts of 4-day-old (neonatal) rats as previously described.17 Ventricular CMs were isolated from the harvested ventricles of 4-day-old Sprague-Dawley rats (Jackson). The cells were isolated by multiple rounds of digestion with 0.14 mg/mL collagenase II (Invitrogen). After each round of digestion, the supernatant was collected in a tube containing horse serum (Invitrogen). The total cell suspension was centrifuged at 300 g for 4–5 min. Supernatants were discarded and cells were resuspended in DMEM (Gibco) supplemented with 0.1 mM ascorbic acid (Sigma), 0.5% insulin-transferrin-selenium (100×), penicillin (100 U/mL), and streptomycin (100 μg/mL). CMs were separated from the other cell types by plating on plastic culture dishes for 90 min. Most of the non-myocytes adhered to the dish, whereas the CMs remained in suspension. The CMs were then collected and used to seed 24-well plates at a density of 1 × 105 cells/well. The isolated CMs were incubated for 48 hr in DMEM supplemented with 5% horse serum plus ara-C. The medium was then replaced with DMEM without antibiotics, and the cells were treated as described.
Confocal Microscopy
Confocal images were acquired on a TCS SP8 STED3× microscope (Leica Microsystems) equipped with a tunable white-light laser. Z stack images were acquired with a 63× 1.40-NA OIL HC PL APO objective and gated HyD detectors (0.3–6 ns). In total, 18 stacks were obtained; x-y = 184.7 × 184.7 μm and z = 5.37 μm. Confocal microscopy was performed at the Microscopy CoRE of the Icahn School of Medicine at Mount Sinai.
Western Blot Analysis
3T3 (fibroblast) cells were transfected with 2 mg/mL luciferase, hFSTL1, Mut-1, Mut-2, Mut-3, or Mut-(1–3) modRNA, in the presence of RNAiMAX transfection reagent (Life Technologies), with or without tunicamycin. The medium was replaced with fresh DMEM (Gibco) medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL). We isolated total protein from the cells 18 hr after transfection and used it for western blots. In brief, equal amounts of protein were resolved by electrophoresis in 4%–15% Mini-PROTEAN TGX stain-free gels (Bio-Rad) and blotted onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked (5% non-fat dry milk [DM] in Tris-buffered saline (TBS; 50 mM Tris-HCl [pH 7.4], 150 mM NaCl), for 1 hr at room temperature and were then incubated with primary antibodies diluted in 5% milk in TBS (overnight, 4°C). We used goat anti-hFSTL1 (1:1,000, AF1694) and mouse monoclonal anti-B-actin (horseradish peroxidase [HRP] conjugate 1:3,000, #12262; Cell Signaling) antibodies. Antigen or antibody complexes were visualized with ChemiDoc Touch imaging system (Bio-Rad).
Mouse Model of MI and Histology
We anesthetized 8- to 12-week-old CFW mice with isoflurane. MI was induced by permanent ligation of the LAD artery, as previously described.43 In brief, the left thoracic region was shaved and sterilized. After intubation, the heart was exposed by left thoracotomy. The LAD was ligated with a suture, and the thoracotomy and skin were sutured closed in layers. Excess air was removed from the thoracic cavity, and the mouse was removed from ventilation once normal breathing had been re-established. We investigated the effect of modRNA on cardiovascular outcome after MI by injecting modRNAs (100 μg/heart) into the infarct border zone immediately after LAD ligation. The peri-infarct zone near the apex was fixed in 4% paraformaldehyde (PFA) for cryosectioning and immunostaining. In all experiments, the surgeon and the person analyzing the data were blind to treatment group. For heart histology analyses, mice were weighed and their hearts were collected at the time points indicated. The hearts were removed, briefly washed in PBS, perfused with perfusion buffer, weighed, and fixed by incubation in 4% PFA with PBS overnight on a shaker. They were then washed with PBS for 1 hr and incubated overnight in 30% sucrose in PBS at 4°C. The hearts were fixed in optimal cutting temperature (OCT) compound and frozen at −80°C. Transverse sections of the heart (8- to 9-μm thick) were cut on a cryostat. The slides were processed for immunostaining (see below) or histological scar staining with Masson’s trichrome staining kit (Sigma), according to standard procedures. We used MIQuant software for Masson’s trichrome-stained heart scar analysis. The ratio of the weight of the heart to total body weight, in grams, was calculated at the end of the experiment (day 28 post-MI). Weight was determined on a balance.
Immunostaining of Heart Sections
Frozen sections of mouse heart were rehydrated in PBS for 5–10 min and permeabilized by incubation in 0.1% Triton X-100 in PBS (PBST) for 7 min. The sections were treated with 3% H2O2 for 5 min and washed three times with PBST for 5 min each. The samples were blocked with PBS + 5% normal donkey serum + 0.1% Triton X-100 (PBSST) for 1–2 hr at room temperature, and primary antibodies (see complete list of primary antibodies used for this study in Table S1) diluted in PBSST were then added. The sections were incubated overnight at 4°C. They were then washed with PBST (five times, for 4 min each) and incubated with the secondary antibody (Invitrogen, 1:200) diluted in PBST for 2 hr at room temperature. The samples were washed with PBST (three times, for 5 min each) and stained with Hoechst 33342 (1 μg/mL) diluted in PBST for 7 min. After five washes with PBST and one wash in tap water (for 4 min each), the sections were mounted in mounting medium (VECTASHIELD) for imaging. Stained slides were stored at 4°C. All staining reactions were performed on two to three sections/heart and six or nine hearts/group. For BrdU analysis by immunostaining, BrdU (1 mg/mL, Sigma) was added to the drinking water of adult mice (2–3 months old) for 7 days before the hearts were harvested. BrdU-containing water bottles were shielded from the light to prevent BrdU degradation, and the water was replaced twice weekly. Quantitative analyses of CMs positive for Ki 67, phosphohistone H3 (PH3), BrdU, and Aurora B were performed on multiple fields for three independent samples per group and three sections/heart. We counted a total of ∼1–2 × 103 CMs per section. For the immunostaining of neonatal CMs in vitro, modRNA-transfected neonatal CMs were fixed on coverslips by incubation with 3.7% PFA for 15 min at room temperature and washed three times with PBS for 5 minutes each. The cells were permeabilized by incubation with 0.5% Triton X-100 in PBS for 10 min at room temperature and blocked by incubation with 5% normal donkey serum + 0.5% Tween 20 for 30 min. The coverslips bearing the cells were incubated with primary antibodies (see Table S1) in a humid chamber for 1 hr at room temperature. The cells were washed three times in PBS and incubated with the corresponding secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 555, and Alexa Fluor 647, and Hoechst 33342 staining was performed to visualize the nuclei (all from Invitrogen). Fluorescence was assessed on images obtained with 10×, 20×, and 40× objectives on a Zeiss fluorescence microscope.
Capillary Density Measurement
We analyzed angiogenesis in the left ventricle 28 days post-MI by immunostaining with an antibody directed against CD31 (also known as platelet endothelial cell adhesion molecule 1 [PECAM-1]; R&D Biosystems). Frozen heart sections were stained as described above. Images were obtained from the stained slides with a 20× objective, and counts of CD31-positive cells per 145 mm2 area in the left ventricle were obtained. Quantitative analyses were performed with the counting of multiple fields from six or nine independent hearts per group, and three sections/heart (∼50–100 cells per field assessed, with up to ∼300–600 cells per sample in total).
Wheatgerm Agglutinin
The cross-sectional area of the CMs was determined by washing frozen heart sections twice in PBS for 5 min each and then incubating them for 1 hr at room temperature with an Alexa Fluor 488-conjugated primary antibody against wheatgerm agglutinin (WGA) (50 μg/mL, Invitrogen). The sections were washed in PBS and mounted on slides in DAPI-supplemented Vectashield (Vector Labs, CA). The images were captured with a 20× objective and ImageJ software was used to determine the area of each cell. Quantitative analyses were performed by counting cells on multiple fields from six or nine independent hearts per group, and three sections/heart (∼50 cells per field assessed, with a total of up to ∼250 cells per sample).
Statistical Analysis
Statistical significance was determined by one-way ANOVA, with Bonferroni post-hoc tests. We considered p values < 0.05 to be significant. All graphs show mean values, and values are reported as the mean ± SEM. We quantified CD31 luminal structures, WGA, CD45, BrdU+, Ki67+, pH3+ or Aurora B+ CMs, on at least three heart sections per heart, for three to nine mice per group. For in vitro immunofluorescence analyses, we used 50 CMs in five to eight random fields, for two different subpopulations per experiment (total, 500 CMs).
Author Contributions
A.M. designed and carried out most of the experiments, analyzed most of the data, and wrote the manuscript. N.S. and A.A.K. performed experiments and obtained and analyzed immunostaining data. M.T.K.S. prepared modRNAs for this study. E.C. carried out all the surgical interventions on mice. L.Z. designed the experiments, analyzed the data, and wrote the manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We thank Pilar Ruiz-Lozano, Mark Mercola, Irsa Munir, Talha Mehmood, Nishat Sultana, Yoav Hadas, and Jason Kondrat for their help with this manuscript. This work was funded by a cardiology start-up grant awarded to the Zangi laboratory and also by NIH grant R01 HL142768-01. Confocal microscopy was performed at the Microscopy CoRE of the Icahn School of Medicine at Mount Sinai, with funding from an NIH Shared Instrumentation Grant (FAIN, S10OD021838).
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.08.021.
Supplemental Information
References
- 1.Warraich H.J., Hernandez A.F., Allen L.A. How medicine has changed the end of life for patients with cardiovascular disease. J. Am. Coll. Cardiol. 2017;70:1276–1289. doi: 10.1016/j.jacc.2017.07.735. [DOI] [PubMed] [Google Scholar]
- 2.Heidenreich P.A., Albert N.M., Allen L.A., Bluemke D.A., Butler J., Fonarow G.C., Ikonomidis J.S., Khavjou O., Konstam M.A., Maddox T.M., American Heart Association Advocacy Coordinating Committee. Council on Arteriosclerosis, Thrombosis and Vascular Biology. Council on Cardiovascular Radiology and Intervention. Council on Clinical Cardiology. Council on Epidemiology and Prevention. Stroke Council Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ Heart Fail. 2013;6:606–619. doi: 10.1161/HHF.0b013e318291329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader D., Oberpriller J.O. Repair and reorganization of minced cardiac muscle in the adult newt (Notophthalmus viridescens) J. Morphol. 1978;155:349–357. doi: 10.1002/jmor.1051550307. [DOI] [PubMed] [Google Scholar]
- 4.Major R.J., Poss K.D. Zebrafish heart regeneration as a model for cardiac tissue repair. Drug Discov. Today Dis. Models. 2007;4:219–225. doi: 10.1016/j.ddmod.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poss K.D. Getting to the heart of regeneration in zebrafish. Semin. Cell Dev. Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Singh B.N., Koyano-Nakagawa N., Garry J.P., Weaver C.V. Heart of newt: a recipe for regeneration. J. Cardiovasc. Transl. Res. 2010;3:397–409. doi: 10.1007/s12265-010-9191-9. [DOI] [PubMed] [Google Scholar]
- 7.Leone M., Magadum A., Engel F.B. Cardiomyocyte proliferation in cardiac development and regeneration: a guide to methodologies and interpretations. Am. J. Physiol. Heart Circ. Physiol. 2015;309:H1237–H1250. doi: 10.1152/ajpheart.00559.2015. [DOI] [PubMed] [Google Scholar]
- 8.Heo J.S., Lee J.C. β-Catenin mediates cyclic strain-stimulated cardiomyogenesis in mouse embryonic stem cells through ROS-dependent and integrin-mediated PI3K/Akt pathways. J. Cell. Biochem. 2011;112:1880–1889. doi: 10.1002/jcb.23108. [DOI] [PubMed] [Google Scholar]
- 9.Engel F.B. Cardiomyocyte proliferation: a platform for mammalian cardiac repair. Cell Cycle. 2005;4:1360–1363. doi: 10.4161/cc.4.10.2081. [DOI] [PubMed] [Google Scholar]
- 10.Beigi F., Schmeckpeper J., Pow-Anpongkul P., Payne J.A., Zhang L., Zhang Z., Huang J., Mirotsou M., Dzau V.J. C3orf58, a novel paracrine protein, stimulates cardiomyocyte cell-cycle progression through the PI3K-AKT-CDK7 pathway. Circ. Res. 2013;113:372–380. doi: 10.1161/CIRCRESAHA.113.301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bersell K., Arab S., Haring B., Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 12.Engel F.B., Schebesta M., Duong M.T., Lu G., Ren S., Madwed J.B., Jiang H., Wang Y., Keating M.T. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kühn B., del Monte F., Hajjar R.J., Chang Y.S., Lebeche D., Arab S., Keating M.T. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat. Med. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 14.Wei K., Serpooshan V., Hurtado C., Diez-Cuñado M., Zhao M., Maruyama S., Zhu W., Fajardo G., Noseda M., Nakamura K. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassat E., Mutlak Y.E., Genzelinakh A., Shadrin I.Y., Baruch Umansky K., Yifa O., Kain D., Rajchman D., Leach J., Riabov Bassat D. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547:179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebelt H., Zhang Y., Köhler K., Xu J., Gajawada P., Boettger T., Hollemann T., Müller-Werdan U., Werdan K., Braun T. Directed expression of dominant-negative p73 enables proliferation of cardiomyocytes in mice. J. Mol. Cell. Cardiol. 2008;45:411–419. doi: 10.1016/j.yjmcc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Lin Z., von Gise A., Zhou P., Gu F., Ma Q., Jiang J., Yau A.L., Buck J.N., Gouin K.A., van Gorp P.R. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 2014;115:354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura W., Xiao F., Canseco D.C., Muralidhar S., Thet S., Zhang H.M., Abderrahman Y., Chen R., Garcia J.A., Shelton J.M. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature. 2015;523:226–230. doi: 10.1038/nature14582. [DOI] [PubMed] [Google Scholar]
- 19.Chen J., Huang Z.P., Seok H.Y., Ding J., Kataoka M., Zhang Z., Hu X., Wang G., Lin Z., Wang S. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S., Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 21.Lesizza P., Prosdocimo G., Martinelli V., Sinagra G., Zacchigna S., Giacca M. Single-dose intracardiac injection of pro-regenerative MicroRNAs improves cardiac function after myocardial infarction. Circ. Res. 2017;120:1298–1304. doi: 10.1161/CIRCRESAHA.116.309589. [DOI] [PubMed] [Google Scholar]
- 22.Porrello E.R., Johnson B.A., Aurora A.B., Simpson E., Nam Y.J., Matkovich S.J., Dorn G.W., 2nd, van Rooij E., Olson E.N. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguirre A., Montserrat N., Zacchigna S., Nivet E., Hishida T., Krause M.N., Kurian L., Ocampo A., Vazquez-Ferrer E., Rodriguez-Esteban C. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15:589–604. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.See K., Tan W.L.W., Lim E.H., Tiang Z., Lee L.T., Li P.Y.Q., Luu T.D.A., Ackers-Johnson M., Foo R.S. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat. Commun. 2017;8:225. doi: 10.1038/s41467-017-00319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magadum A., Ding Y., He L., Kim T., Vasudevarao M.D., Long Q., Yang K., Wickramasinghe N., Renikunta H.V., Dubois N. Live cell screening platform identifies PPARδ as a regulator of cardiomyocyte proliferation and cardiac repair. Cell Res. 2017;27:1002–1019. doi: 10.1038/cr.2017.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Uva G., Aharonov A., Lauriola M., Kain D., Yahalom-Ronen Y., Carvalho S., Weisinger K., Bassat E., Rajchman D., Yifa O. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015;17:627–638. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 27.Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R.L., Martin J.F. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao H.S., Kang P.M., Nagashima H., Yamasaki N., Usheva A., Ding B., Lorell B.H., Izumo S. Cardiac-specific overexpression of cyclin-dependent kinase 2 increases smaller mononuclear cardiomyocytes. Circ. Res. 2001;88:443–450. doi: 10.1161/01.res.88.4.443. [DOI] [PubMed] [Google Scholar]
- 29.Mahmoud A.I., Kocabas F., Muralidhar S.A., Kimura W., Koura A.S., Thet S., Porrello E.R., Sadek H.A. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heallen T., Morikawa Y., Leach J., Tao G., Willerson J.T., Johnson R.L., Martin J.F. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H.G., Chen Q., Wolfram J.A., Richardson S.L., Liner A., Siedlak S.L., Zhu X., Ziats N.P., Fujioka H., Felsher D.W. Cell cycle re-entry and mitochondrial defects in myc-mediated hypertrophic cardiomyopathy and heart failure. PLoS ONE. 2009;4:e7172. doi: 10.1371/journal.pone.0007172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zwijsen A., Blockx H., Van Arnhem W., Willems J., Fransen L., Devos K., Raymackers J., Van de Voorde A., Slegers H. Characterization of a rat C6 glioma-secreted follistatin-related protein (FRP). Cloning and sequence of the human homologue. Eur. J. Biochem. 1994;225:937–946. doi: 10.1111/j.1432-1033.1994.0937b.x. [DOI] [PubMed] [Google Scholar]
- 33.Turnbull I.C., Eltoukhy A.A., Fish K.M., Nonnenmacher M., Ishikawa K., Chen J., Hajjar R.J., Anderson D.G., Costa K.D. Myocardial delivery of lipidoid nanoparticle carrying modRNA induces rapid and transient expression. Mol. Ther. 2016;24:66–75. doi: 10.1038/mt.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chien K.R., Zangi L., Lui K.O. Synthetic chemically modified mRNA (modRNA): toward a new technology platform for cardiovascular biology and medicine. Cold Spring Harb. Perspect. Med. 2014;5:a014035. doi: 10.1101/cshperspect.a014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Z., Pu W.T. Strategies for cardiac regeneration and repair. Sci. Transl. Med. 2014;6:239rv1. doi: 10.1126/scitranslmed.3006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C.L., Leblond A.L., Turner E.C., Kumar A.H., Martin K., Whelan D., O’Sullivan D.M., Caplice N.M. Synthetic chemically modified mrna-based delivery of cytoprotective factor promotes early cardiomyocyte survival post-acute myocardial infarction. Mol. Pharm. 2015;12:991–996. doi: 10.1021/mp5006239. [DOI] [PubMed] [Google Scholar]
- 37.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zangi L., Oliveira M.S., Ye L.Y., Ma Q., Sultana N., Hadas Y., Chepurko E., Später D., Zhou B., Chew W.L. Insulin-like growth factor 1 receptor-dependent pathway drives epicardial adipose tissue formation after myocardial injury. Circulation. 2017;135:59–72. doi: 10.1161/CIRCULATIONAHA.116.022064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondrat J., Sultana N., Zangi L. Synthesis of modified mRNA for myocardial delivery. Methods Mol. Biol. 2017;1521:127–138. doi: 10.1007/978-1-4939-6588-5_8. [DOI] [PubMed] [Google Scholar]
- 40.Sultana N., Magadum A., Hadas Y., Kondrat J., Singh N., Youssef E., Calderon D., Chepurko E., Dubois N., Hajjar R.J., Zangi L. Optimizing cardiac delivery of modified mRNA. Mol. Ther. 2017;25:1306–1315. doi: 10.1016/j.ymthe.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaly Y., Fu Y., Marinov A., Hostager B., Yan W., Campfield B., Kellum J.A., Bushnell D., Wang Y., Vockley J., Hirsch R. Follistatin-like protein 1 enhances NLRP3 inflammasome-mediated IL-1β secretion from monocytes and macrophages. Eur. J. Immunol. 2014;44:1467–1479. doi: 10.1002/eji.201344063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarnavski O., McMullen J.R., Schinke M., Nie Q., Kong S., Izumo S. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol. Genomics. 2004;16:349–360. doi: 10.1152/physiolgenomics.00041.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.