Abstract
Persistent high-risk HPV infection is the main cause of cervical cancer. The HPV oncogene E7 plays an important role in HPV carcinogenesis. Currently, HPV vaccines do not offer an effective treatment for women who already present with cervical disease, and recommended periodical cervical screenings are difficult to perform in countries and areas lacking medical resources. Our aim was to develop nanoparticles (NPs) based on poly (β-amino ester) (PBAE) and HPV16 E7-targeting CRISPR/short hairpin RNA (shRNA) to reduce the levels of HPV16 E7 as a preliminary form of a drug to treat HPV infection and its related cervical malignancy. Our NPs showed low toxicity in cells and mouse organs. By reducing the expression of HPV16 E7, our NPs could inhibit the growth of cervical cancer cells and xenograft tumors in nude mice, and they could reverse the malignant cervical epithelium phenotype in HPV16 transgenic mice. The performance of NPs containing shRNA is better than that of NPs containing CRISPR. HPV-targeting NPs consisting of PBAE and CRISPR/shRNA could potentially be developed as drugs to treat HPV infection and HPV-related cervical malignancy.
Keywords: cervical cancer, HPV, PBAE
Wang and colleagues report that nanoparticles based on PBAE and CRISPR/shRNA could reduce the expression of HPV16 E7, inhibit the growth of cervical cancer cells, and reverse the malignant cervical epithelium phenotype. These nanoparticles could potentially be developed as drugs to treat HPV infection and HPV-related cervical malignancy.
Introduction
Cervical cancer is a serious threat to women’s health and is the third most common malignancy in women worldwide.1 Over 85% of new cervical cancer cases and cervical cancer-related deaths occur in developing countries. High-risk human papillomavirus (HR-HPV) infection is the main cause of cervical cancer;2 type 16 (HPV16) is the most common type of HPV and is found in more than 60% of cervical cancer cases.3 The E6 and E7 oncoproteins are the primary cause of HPV-related carcinogenesis, and they have been shown to target the tumor protein p53 (P53) and retinoblastoma (RB) proteins, promoting cell proliferation and leading to the development of carcinoma.4 The E7 oncogene is a stronger carcinogen than E6, and reports have shown that cervical cancer could be induced in HPV16 E7 transgenic mice by estrogen treatment.5, 6 Another possible cause of cervical cancer is HPV integration, which could result in E7 being retained in the human genome.7, 8 Since the progression from HPV infection to cervical intraepithelial neoplasia (CIN) and eventually to cervical cancer occurs over a long period of time, HPV, especially E7, is a critical target for the prevention and treatment of cervical cancer.
Several currently available methods for the prevention and treatment of HPV infection have disadvantages. Interferon is often used to treat viral infections by improving the local immune response. However, this is not an HPV-specific treatment. Physical therapy, such as microwave treatment, loop electrosurgical excision procedure (LEEP) treatment, and cervical conization, cannot clear HPV infection and may even cause cervical insufficiency,9 which likely results in higher premature birth and abortion.10 HPV vaccines are widely used to prevent HPV infection.11 Currently, HPV vaccine can prevent most nine types of HPV infections (9-valent HPV vaccine), and it could prevent infection and disease related to HPV31, 33, 45, 52, and 58 in a susceptible population and generated an antibody response to HPV6, 11, 16, and 18 that was noninferior to that generated by the quadrivalent human papillomavirus (qHPV) vaccine.12 However, the 9-valent HPV vaccine did not prevent infection and disease related to HPV types beyond the nine types covered by the vaccine.12 Furthermore, prophylactic vaccines cannot offer an effective treatment for women who already present with cervical diseases.13
An increasing number of gene-targeting technologies based on recombinant plasmids have been developed in recent years, such as targeted gene silencing by short hairpin RNA (shRNA) or RNAi14, 15 and targeted gene knockout by CRISPR/Cas.16 The stability of these plasmids is promising for their prospective use in pharmaceutical industrial production. However, as these plasmids have to cross organ, tissue, and cell barriers to enter the cytoplasm, where they can induce sequence-specific mRNA degradation or DNA double-strand breaks, their application in therapy is limited by the lack of safe and efficient carriers. Poly (β-amino ester) (PBAE) is a biocompatible polymer with the advantages of pH sensitivity, low toxicity, high water solubility, fast drug delivery at acidic pH values (suitable for the acidic environment in the vagina), and high transfection efficiency.17, 18, 19, 20, 21 Thus, we explored whether nanoparticles (NPs) consisting of PBAE and recombinant plasmids could reverse the HPV16-related cervical malignancy, providing novel ideas for the development of HPV-targeting drugs.
Results
Synthesis and Characterization of NPs
The PBAE polymer for delivering plasmids was synthesized using three raw materials: 1,4-butanediol diacrylate, 4-amino-1-butanol, and 1-(3-aminopropyl)-4-methylpiperazine. The synthetic scheme is shown in Figure S1. We first generated NPs using PBAE and GFP plasmid at different weight ratios (PBAE/GFP). Agarose gel electrophoresis showed that the plasmid could be released from the NPs and migrated into the gel until the weight ratio reached 60:1 (Figure S2A). Dynamic light scattering indicated that the NPs were 110.2–174.7 nm in size, and their zeta potentials ranged from 13.82 to 22.28 mV (Figures S2B and S2C; Table S1). The transmission electron microscopy (TEM) micrograph showed the NPs were about 90 nm in size, and it demonstrated a uniform distribution (Figure S2D).
NPs Showed Low Toxicity in Cervical Cancer Cells and Mice
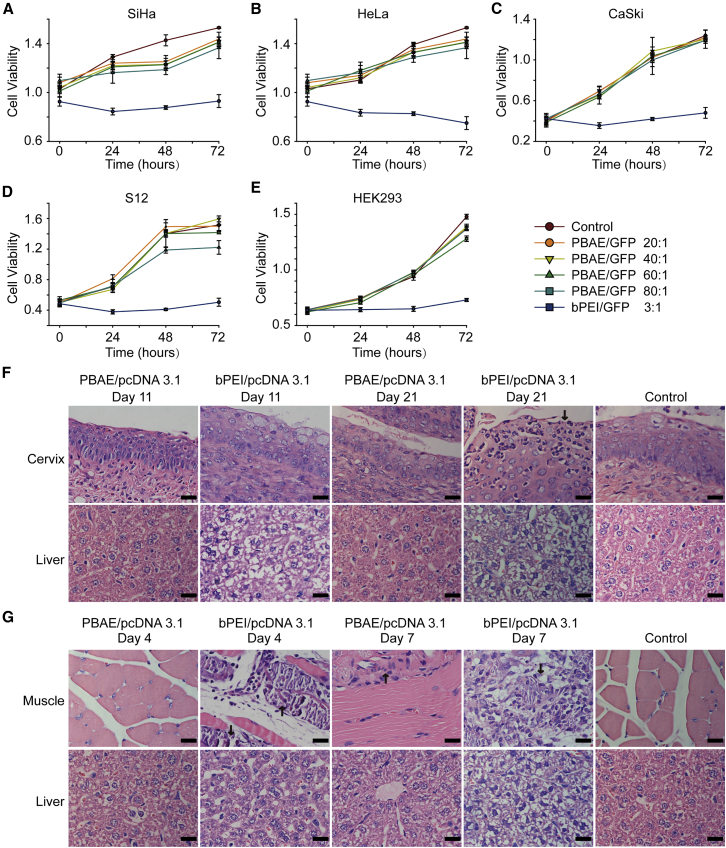
Toxicity is a primary concern in the development of NP pharmaceuticals. To investigate the cytotoxicity of our NPs, SiHa, HeLa, CaSki, S12, and HEK293 cells were treated with NPs consisting of PBAE and GFP plasmid for 6 hr and further cultured for 72 hr. Cell viability was tested at different time points by CCK-8 assays (Figures 1A–1E). No significant alteration in cell viability in response to our NPs was observed compared with control groups. The cell monolayers were also co-incubated with branched poly (ethyleneimine) (bPEI)/GFP (weight ratio 3:1) complexes, which inhibited cell growth significantly (Figures 1A–1E). These data indicated that our NPs did not show obvious cytotoxicity or deleterious effects on cell monolayers. To further test the toxicity in vivo, NPs consisting of PBAE or bPEI carrying pcDNA3.1 (a non-fluorescent and non-toxic empty vector) were injected into the thigh muscles of mice. H&E staining and TUNEL staining showed that cell apoptosis was low in the mouse thigh muscles injected with PBAE/pcDNA3.1 but increased sharply in those injected with bPEI/pcDNA3.1 (Figure S3).
Figure 1.
Analysis of NP Toxicity in Cell Lines and Mouse Organs
(A–E) Cytotoxicity of PBAE/GFP (weight ratio 20:1, 40:1, 60:1, and 80:1, GFP 100 ng/well) in (A) SiHa, (B) HeLa, (C) CaSki, (D) S12, and (E) HEK293 cells. CCK-8 assays were used to determine the cytotoxicity of NPs, which was compared with that of bPEI/GFP (weight ratio 3:1). Toxicity is given as the viability of the cells remaining after treatment for 0, 24, 48, and 72 hr. Each point represents the mean ± SD (n = 4). (F and G) Representative images of H&E staining of (F) uterine cervixes and livers of mice that were vaginally treated with NPs and (G) thigh muscles and livers of mice whose thigh muscles were injected with NPs. The NPs were generated using PBAE/pcDNA3.1 (weight ratio 60:1) and bPEI/pcDNA3.1 (weight ratio 3:1). Mouse vaginas were treated by pipetting with NPs carrying 10 μg plasmid once per day for 20 days, and the uterine cervixes and livers were harvested on the 11th and 21st days after the initial pipetting. Mouse thigh muscles were injected with NPs carrying 100 μg plasmid once per day for 3 days, and the muscles and livers were harvested on the fourth and seventh days after initial injection. Arrows indicate the necrotic areas. Scale bars, 20 μm.
We further examined the toxicity of a maximum dose of NPs upon vaginal and intramuscular administration. Considering that a total of 200–300 μg plasmid was used in the subsequent HPV treatment experiments, we injected NPs carrying 10 μg pcDNA3.1 into the vaginas of C57BL/6 mice once a day for 20 days, and we examined the toxicity on the 11th and 21st days after the initial injection. Administration of bPEI/pcDNA3.1 resulted in an increased number of vaginal localized inflammatory necrosis and pycnosis of hepatocyte nuclei, whereas similar changes were not observed after PBAE/pcDNA3.1 treatment (Figure 1F). We also administered NPs carrying 100 μg pcDNA3.1 to the thigh muscles of C57BL/6 mice once a day for 3 days, and we examined the toxicity on the fourth and seventh days. Injection of bPEI/pcDNA3.1 resulted in massive necrosis of the muscle cells and pycnosis of hepatocyte nuclei, indicating muscle and liver damage, while injection of PBAE/pcDNA3.1 did not cause significant changes (Figure 1G). Significant toxicity of heart, spleen, lung, and kidney organs was not observed after muscular or vaginal administration (Figures S4 and S5).The hepatotoxicity results suggest that attention should be paid to side effects when developing future vaginal medications, since these medications may be absorbed through the vagina into the circulation and may subsequently damage the liver. These data indicated that our NPs exhibited low toxicity and good biocompatibility, which are necessary features of potential drugs.
NPs Were Internalized by Cervical Cancer Cells and Cervical Tissues
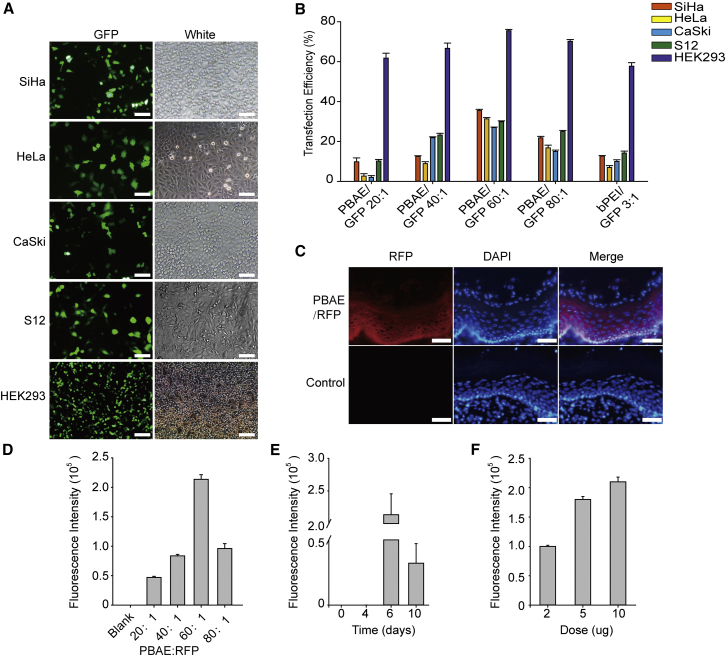
Efficient cellular uptake and expression are major requirements for the therapeutic application of plasmids. To investigate the expression efficiency of our NPs, we treated cells with NPs consisting of PBAE and GFP plasmid. Our NPs with a weight ratio of 60:1 exhibited the highest transfection efficiency (35.5% in SiHa cells, 31.3% in HeLa cells, 27.0% in CaSki cells, 29.9% in S12 cells, and 75.5% in HEK293 cells) compared with NPs consisting of bPEI and GFP with a weight ratio of 3:1 (Figures 2A and 2B). TEM imaging of 293 cells transfected with PBAE/GFP showed that NPs were in the vesicles of the cytoplasm, indicating that our NPs were uptaken into the cells by endocytosis (Figure S6). Meanwhile, co-localization of green and red fluorescence confirmed the expression of GFP protein in the cytoplasm (Figure S7). These data demonstrated that our NPs were taken up by cells and the plasmids were released into the cytoplasm and expressed proteins.
Figure 2.
Uptake of NPs by Cell Lines and Mouse Uterine Cervixes
(A) Representative images of SiHa, HeLa, CaSki, S12, and HEK293 cells 72 hr after treatment with NPs (PBAE/GFP, weight ratio 60:1, GFP 1 ng/μL). (B) Statistical uptake efficiency of PBAE/GFP NPs with different weight ratios (20:1, 40:1, 60:1, and 80:1) compared with that of bPEI/GFP (weight ratio 3:1) by flow cytometry. (C) Representative images of fluorescence in the uterine cervixes of C57BL/6 mice treated with NPs (PBAE/RFP, weight ratio 60:1, 10 μg RFP once per day for 3 days) compared with cervixes treated with 25 mM sodium acetate. Scale bars, 20 μm. Control means the vagina of C57BL/6 with no NP treatment. (D–F) Comparison of the uptake of NPs (D) with different weight ratios (PBAE/RFP, 10 μg RFP once a day for 3 days), (E) at different time points after treatment (PBAE/RFP, weight ratio 60:1, 10 μg RFP once a day for 3 days), and (F) with different doses of RFP (PBAE/RFP, weight ratio 60:1, once a day for 3 days) in the cervixes of C57BL/6 female mice, as determined by fluorescence intensity.
We next evaluated the uptake of PBAE/red fluorescent protein (RFP) NPs by vaginal administration in C57BL/6 mice (Figure 2C). Considering that the best treatment effect may result from the best transfection efficiency, we tested different in vivo transfection parameters, including PBAE/RFP weight ratios, detection time points after treatment, and the dosage of the transfected plasmid. We observed that a weight ratio of 60:1, a plasmid quantity of 10 μg once a day for 3 days, and detection on the sixth day after initial administration resulted in the best transfection efficiency (Figures 2D–2F). The results provided guidance for the application of NPs generated with PBAE and HPV-targeting plasmids in the next step.
HPV16 E7 Expression Was Downregulated by NPs in Cervical Cancer Cells and Samples
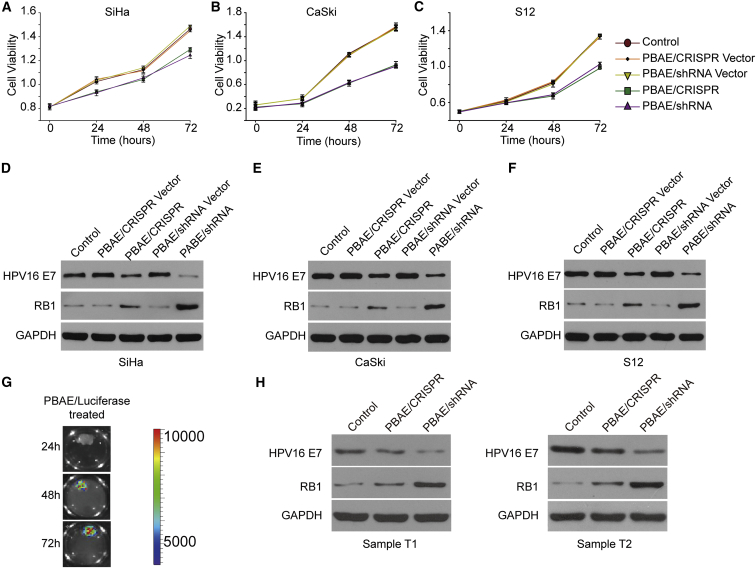
We constructed HPV16 E7-targeting CRISPR and shRNA plasmids (Figure S8; Table S2), and we fabricated NPs from PBAE and these plasmids. HPV16-positive cells (SiHa, CaSki, and S12) and HPV16-negative cells (HeLa) were treated with our NPs, and the cell viability was detected at 0, 24, 48, and 72 hr after treatment by CCK-8 assay. Three shRNAs and three CRISPRs were selected to form NPs, and these NPs all could reduce E7 expression with different levels in SiHa cells (Figures S9A and S9B). Then we selected one shRNA and one CRISPR with the best performance as the representative for further study. The viability of cells treated with our NPs was significantly decreased compared with that of the untreated cells (Figures 3A–3C), whereas no differences were observed in HeLa cells (Figure S10).
Figure 3.
HPV16 E7-Targeting Ability of NPs in HPV16-Positive Cervical Cell Lines and Cervical Cancer Samples
(A–C) The cell viabilities of (A) SiHa, (B) CaSki, and (C) S12 cells were determined at 0, 24, 48, and 72 hr after HPV16 E7-targeting NP treatment by CCK-8 assay (PBAE/CRISPR and PBAE/shRNA, weight ratio 60:1, 1 ng/μL plasmid). The data represent the mean ± SD (n = 3). (D–F) The HPV16 E7 and RB1 protein expression levels in (D) SiHa, (E) CaSki, and (F) S12 cells 72 hr after NP treatment were determined by western blot analysis (PBAE/plasmid, weight ratio 60:1, 1 ng/μL plasmid). (G) Fluorescence intensity of cervical cancer samples 24–72 hr after treatment with NPs (PBAE/luciferase, weight ratio 60:1, 100 ng/μL plasmid). Cervical cancer samples were cultured in KSFM after surgery. (H) HPV16 E7 and RB1 protein expression in HPV16-positive cervical cancer samples 72 hr after treatment with NPs was determined by western blot analysis (PBAE/CRISPR and PBAE/shRNA, weight ratio 60:1, 100 ng/μL plasmid). Cervical cancer samples were cultured in KSFM after surgery (Table S4). T1 and T2, cervical cancer samples 1 and 2.
We further examined the protein expression of HPV16 E7 and RB1 by western blot analysis, as protein expression is more solid evidence of an effect, and we found that our NPs caused decreased HPV16 E7 and restored RB1 protein expression in the three HPV16-positive cell lines. It is worth noting that the effect of the PBAE/shRNA NPs was stronger than that of the PBAE/CRISPR NPs (Figures 3D–3F). Similar results were also obtained from the combination use of PBAE/CRISPR and PBAE/shRNA NPs (Figure S9C). Although the therapeutic effect did not increase synergistically, the combination treatment of CRISPR and shRNA may clear HPV episome and integrated HPV simultaneously, indicating a bright prospect in clinical application. These data demonstrated that our NPs inhibited the growth of cervical cancer cells by downregulating HPV16 E7 and subsequently restoring RB1 expression.
To further demonstrate the application potential of NP drugs in the treatment of cervical cancer, cervical cancer samples were randomly collected in Tongji Hospital (Table S3), cultured in 12-well plates with 1 mL karatinocyte serum-free medium (KSFM), and treated with NPs consisting of PBAE and luciferase. We observed that 72 hr of treatment resulted in the strongest luciferase signals (Figure 3G). We randomly collected two HPV16-positive cervical cancer samples, divided them into three equal parts, and treated them with PBAE/CRISPR and PBAE/shRNA NPs. As expected, the protein expression of HPV16 E7 was decreased, while that of RB1 was restored after treatment with HPV-targeting NPs for 72 hr (Figure 3H). These data further indicated the potential value of our NPs in the treatment of cervical cancer.
NPs Inhibited the Growth of Xenografts in Nude Mice
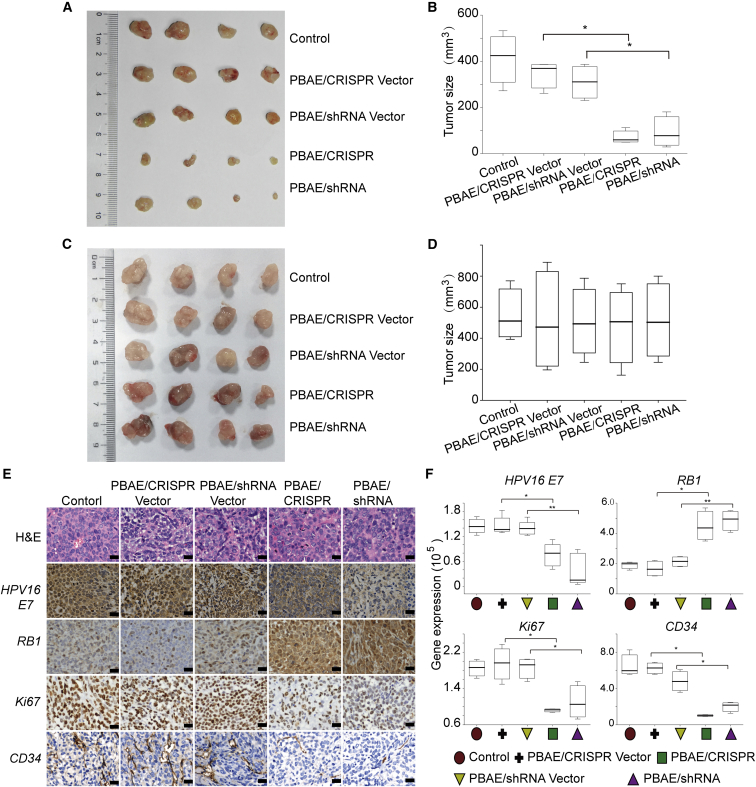
To further explore the impact of our NPs on tumor growth in vivo, nude mice were inoculated subcutaneously with SiHa (HPV16 positive) and HeLa cells (HPV16 negative, HPV18 positive), and xenograft tumors were allowed to grow to approximately 35 mm3. NPs were then administered intratumorally every 4 days. Tumor growth was observed continuously for 20 days. The tumors injected with our HPV16-targeting NPs (PBAE/CRISPR and PBAE/shRNA) showed slower growth rates, and the average xenograft volumes were 52 and 60 mm3, respectively, significantly smaller than those of tumors injected with control and plasmid only (Figures 4A and 4B). However, there was no significant change in the volumes of HeLa cell xenograft tumors (Figures 4C and 4D). We also examined the protein expression of HPV16 E7, Ki67, RB1, and CD34 in SiHa cell xenografts by immunohistochemistry (IHC) staining. The expression of the tumor suppressor gene RB1 was restored, while the expression of HPV16 E7, the proliferation-associated gene Ki67, and vascular endothelial growth-related gene CD34 was decreased after treatment with our HPV16-targeting NPs (Figures 4E and 4F). These data indicated that our HPV16-targeting NPs significantly inhibit the growth of cervical cancer cells.
Figure 4.
Tumorigenicity of Xenografts in Nude Mice Treated with HPV16 E7-Targeting NPs
SiHa and HeLa cells were injected subcutaneously into the right flanks of BALB/c-nu mice. HPV16 E7-targeting NPs (PBAE/CRISPR and PBAE/shRNA, weight ratio 60:1, 60 μg plasmid every 4 days) were injected intratumorally when the xenografts reached approximately 35 mm3. (A–D) The subcutaneously formed tumors of (A) SiHa and (C) HeLa were photographed, and the estimated sizes of (B) SiHa and (D) HeLa were measured after treatment with NPs. (E and F) Shown are (E) representative images of H&E and IHC staining and (F) a comparison of the protein expression of HPV16 E7, RB1, Ki67, and CD34 in xenografts with or without NP treatment. Scale bars, 20 μm. The protein expression was measured using the optical staining intensities by ImagePro Plus (version 6.0). The data represent the mean ± SD (n = 4). *p < 0.05, **p < 0.01.
NPs Reversed the Malignant Cervical Epithelium Phenotype of HPV16 Transgenic Mice
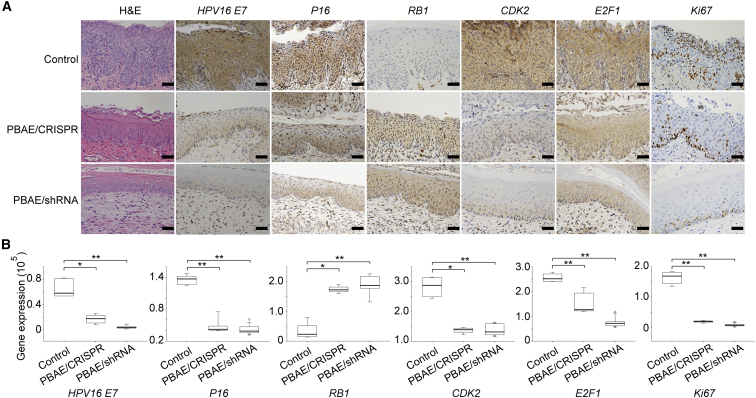
In the control group of HPV16 transgenic mice, we observed thickened cervical epithelia, the disappearance of epithelial stratification, prominent papillary growth of the basal layer, and abnormal nucleus enlargement. After 20 days of treatment with our NPs (especially PBAE/shRNA), the malignant phenotype of the epithelium was reversed, as evidenced by the reappearance of epithelial stratification, smoothing of the basal layer, and reduced nuclear size (Figure 5A). The better performance of the PBAE/shRNA NPs was consistent with the data obtained in cervical cancer cell experiments (Figure 3). We further examined the changes in protein expression by IHC. After treatment with our NPs, the expression of HPV16 E7, the cell cycle-related protein CDK2, the transcription factor E2F1, and the proliferation-related protein Ki67 decreased significantly, while the expression of the tumor suppressor gene RB1 was restored (Figures 5A and 5B). The expression of P16, a surrogate marker of HR-HPV, also decreased significantly (Figures 5A and 5B). These data suggest that our NPs can effectively reduce the expression of the HPV16 E7 oncoprotein and regulate the related signaling pathways, thereby inhibiting cell proliferation, promoting apoptosis, and reversing the cervical epithelial malignant phenotype of HPV16 transgenic mice.
Figure 5.
Protein Expression Changes in Cervical Tissues from HPV16 Transgenic Mice Treated with HPV16 E7-Targeting NPs
(A and B) Shown are (A) representative images of H&E and IHC staining and (B) a comparison of the protein expression of HPV16 E7, P16, RB1, CDK2, E2F1, and Ki67 in cervical tissues from HPV16 transgenic mice treated with or without NPs (PBAE/CRISPR and PBAE/shRNA, weight ratio 60:1, 10 μg plasmid DNA per day for 20 days). Scale bars, 20 μm. The protein expression was measured using the optical staining intensities by ImagePro Plus (version 6.0). The data represent the mean ± SD (n = 4). *p < 0.05, **p < 0.01.
Discussion
HPV infection is an established cause of cervical cancer.22 Cervical cancer prevention mainly depends on periodical cervical screening23 and the use of HPV vaccines.11, 24, 25 A randomized, double-blind trial suggests the 9-valent HPV (HPV6, 11, 16, 18, 31, 33, 45, 52, and 58) vaccine prevents HPV infection, cytological abnormalities, and high-grade lesions,26 but the vaccinated women are still threatened by other HR-HPV types (HPV39, 56, 59, 68, etc.). HPV vaccines did not prevent infection and disease related to HPV types beyond the types covered by vaccines.12 HPV vaccines can stimulate the body to produce antibodies to HPV and prevent it from infecting cells,12, 27 but it does not offer an effective treatment for women who already present with cervical disease.13
An individual-based mathematical model suggests cervical screening can be modified to start at later ages, occurring at decreased frequency in HPV-vaccinated women,28 but vaccination does not completely prevent HPV infection. Therefore, a drug for the treatment of HPV as a supplement to the vaccine is needed. Periodical cervical screening is a passive measure and is difficult to perform in countries and areas lacking medical resources.29, 30 To meet the requirement of patients threatened by HPV, we combined PBAE and CRISPR/shRNA to design a novel drug for targeted HPV therapy.
The E6 and E7 oncoproteins expressed by HPV are primarily responsible for HPV-related malignancies. The E6 and E7 oncogenes inhibit apoptosis and promote proliferation by affecting the p53- and RB1-signaling pathways, and eventually they cause cervical cancer.4, 31, 32, 33 At present, two types of molecular biology techniques have been developed to target specific genes. One is the silencing technique, with shRNA as the representative, which specifically degrades genes at the mRNA level.14, 34, 35 The other is the knockout technique, with CRISPR/Cas as the representative, which takes advantage of double-strand DNA cleavage and error-prone repair.16, 36, 37, 38 Currently, researchers have reported the application of the two techniques in the HPV-related cancer treatment.39, 40, 41, 42 They silenced the HPV E6 and E7 expression in cervical carcinoma cells by RNAi;40 established lentiviral vector-mediated shRNA targeting HPV16 E6/E7 transcripts, transducing the lentiviral construct into cervical HPV16-positive cell lines Siha and Caski;43 and used retroviruses that expressed shRNA targeting the HPV16 E6 and E7 genes to infect human oropharyngeal squamous cancer cell.44 A dual-expression plasmid that co-expressed E6-specific small interfering RNA (siRNA) and wild-type p53 was developed, and it could cause a robust suppression of tumor growth in vitro and in vivo.45 The CRISPR/Cas system disrupting HPV16 E7 could induce apoptosis and growth inhibition in HPV16-positive human cervical cancer cells in vitro.41 CRISPR/Cas9 targeting HPV16 E6/E7 was also suggested could be used as a chemosensitizer of cisplatin chemotherapy in cervical cancer cells.46
Our data indicated that the performance of silencing was better than that of knockout in inhibiting E7 oncogene expression (Figures 3D–3F and 4). This may be because the silencing technique functions directly at the mRNA level after plasmid transcription, while the efficiency of the knockout technique is relatively low, as it requires protein translation after plasmid transcription. Moreover, silencing is transient, while the DNA damage caused by knockout is permanent. Considering that both techniques have potential off-target effects that could cause unintended gene expression changes, the silencing technique may be safer and more suitable for drug development.
These techniques or functional plasmids alone are not enough for the development of appropriate drugs to treat HPV infection and cervical cancer. They are unable to pass through the barriers of organs, tissues, and cells, and, therefore, they require a suitable carrier. A variety of nanomaterials has been shown to deliver nucleic acids (plasmid or RNAi) for gene therapy, including vaginal application.47 Liposome-templated hydrogel NPs (LHNPs) were suggested as a versatile CRISPR/Cas9 delivery tool for experimentally studying the biology of cancer as well as for clinically translating cancer gene therapy.48 Folate receptor-targeted liposome (F-LP) was used to deliver a CRISPR plasmid DNA co-expressing Cas9 and single-guide RNA targeting the DNA methyltransferase 1 (DNMT1) gene to treat ovarian cancer.49 Cell-penetrating peptide (CPP)-modified nano-micelles, comprising polyethylene glycol-polycaprolactone (PEG-PCL) copolymers, were synthesized to improve the efficiency of siRNA delivery to the brain.50 Urocanic acid-modified chitosan/siCD98 NPs could reduce colitis in mice.51 siRNA-loaded biodegradable poly(lactic-co-glycolic acid) (PLGA) NPs targeted against EGFP successfully silenced the expression of EGFP in transgenic GFP mice by vaginal instillation.52 These previous studies indicated prospect in clinic application of nanomaterials in gene therapy.
In our study, we used the non-viral nanomaterial PBAE for plasmid delivery, which has many advantages, such as low toxicity, biodegradability, high transfection efficiency, and stable polyplex formulation.17, 18, 19, 20, 21 PBAE has been used in the treatment of small cell lung cancer, malignant brain glioma, breast cancer, melanoma, and pulmonary fibrosis, and it has shown promising results.18, 21, 53, 54, 55 However, the stability of PBAE in blood is a main limitation for the application, and PBAE can also cause hemolysis at large doses; thus, some investigators have made attempts to modifiy PBAEs with mannose and poly(ethylene glycol) to improve stability and reduce toxicity.21, 56 Here we suggest that PBAE is particularly suitable for vaginal administration; the acidic environment of the vagina is good for the stability of PBAE NPs, and less PBAE in blood will reduce the possibility of side effects such as hemolysis. Optimization of PBAE materials and CRISPR/shRNA plasmids is worthy of further study.
NPs have a diameter ranging from 10 to 1,000 nm and may or may not contain drug substance.57 The size of NPs provide that they have the capacity of encapsulating drugs.58 The particle size of NPs synthesized by PBAE ranged from 100 to 400 nm in previous reports.18, 21, 59, 60 In this study, our NPs were 110.2–174.7 nm in size and were uniformly distributed. They also had low toxicity and high transfection efficiency. We observed that our NPs could effectively inhibit the growth of xenografts in nude mice (Figure 4) and significantly reduce the expression of E7 in HPV16-positive cervical cancer samples (Figure 3). In the HPV16 transgenic mouse model in particular, our NPs successfully reversed the malignant phenotype of the cervical epithelium by inhibiting the expression of the HPV oncoprotein (Figure 5). Our NPs are already very close to a drug form that could be used in clinical vaginal medication.
We studied NPs targeting HPV16 E7 oncogene as a representative. Other HPV16 elements and other types of HPV could also contribute to cervical tumorigenesis. HPV E6 can cause carcinogenesis by degrading P53,61 and the integration of the HPV origin into the host genome can increase the genomic instability.62 Multiple HPV infections (HPV16, 18, 31, and 59) often exist in lots of patients. HPV-targeting nanodrugs for HPV16 E6, origin, and other HPV types could be fabricated using a similar method in the future. In the clinical settings, we may first detect the HPV type and also the integrated HPV elements using high-throughput sequencing,8 then apply specific HPV-targeting nanodrugs for individual therapy.
In conclusion, we established NPs based on PBAE and silencing or knockout plasmids for the treatment of HPV infection, and we systematically evaluated the toxicity and efficacy of these NPs. Our work provides novel ideas for the translational application of nanomaterials and plasmids to the clinical treatment of HPV, and it sheds new light on the prevention and treatment of cervical cancer.
Materials and Methods
Material Preparation
1,4-Butanediol diacrylate and 4-amino-1-butanol were purchased from TCI (B2935, A1013, Japan). 1-(3-aminopropyl)-4-methylpiperazine was purchased from Alfa Aesar (L04876, USA). Branched poly (ethyleneimine) (water-free, 25 kDa, bPEI) was purchased from Sigma-Aldrich (408727, USA). The plasmids pDest-EGFP-N1 (plasmid 31796, GFP), pAAV-CAG-RFP (plasmid 22910, RFP), and M69 pCMV pGL3 luciferase (plasmid 17186, luciferase) and the HPV16 E7-targeting CRISPR vector pSpCas9 (BB)-2A-GFP (plasmid 48138) were purchased from Addgene. The HPV16 E7-targeting shRNA vector pSIREN_U6-shRNA_FF6-CMV-iRFP was purchased from BioVector NTCC, China. pcDNA3.1/Hygro (+) (pcDNA3.1) was purchased from Invitrogen (USA). The HPV16 E7-targeting shRNA was constructed in our laboratory, and it was chosen from three shRNAs according to the effective knockdown of HPV16 E7 (Figures S8A and S8B; Table S2). The HPV16 E7-targeting CRISPR plasmid was synthesized by Genscript Biotechnology (Shanghai, China), and the sequence was chosen as the best of three guide RNAs (gRNAs) designed from the ATUM website (https://www.atum.bio) (Figures S8C and S8D; Table S2). Plasmid DNA was prepared using an endotoxin-free plasmid extraction kit (Omega, USA) and stored at −80°C.
C57BL/6 female mice and specific-pathogen-free BALB/c-nu nude mice were purchased from BEIJING HFK BIOSCIENCE and housed at the Experimental Animal Center, Tongji Medical College, Huazhong University of Science and Technology (HUST, Wuhan, China).
K14-HPV16 transgenic mice were provided by the National Cancer Institute (NCI) Mouse Repository (Frederick, MD, USA) (strain nomenclature: FVB.Cg-Tg (KRT14-HPV16)wt1Dh) and housed at the Experimental Animal Center, Tongji Medical College, Huazhong University of Science and Technology (HUST, Wuhan, China). Breeding and genotyping of the mice were performed as described previously.63
PBAE Polymer Synthesis
Considering that PBAE may be synthesized by a number of different acrylate-terminated backbone monomers and amine-terminated side-chain monomers and end-cap monomers, after literature review18, 20, 64 and with the experimental verification, 1,4-butanediol diacrylate, 4-amino-1-butanol, and 1- (3-aminopropyl)-4-methylpiperazine were selected to form PBAE polymer as follows: briefly, 1, 4-butanediol diacrylate was mixed with 4-amino-1-butanol and stirred on a magnetic stir-plate at 90°C for 24 hr with 2 mL DMSO at a 1.2:1 molar ratio. After stirring, 1-(3-aminopropyl)-4-methylpiperazine was added to the mixture (10-fold), and the mixture was stirred for 30 s, incubated at room temperature for 1 hr, and precipitated in anhydrous diethyl ether (Figure S1). The polymer was washed three times with ether and kept under a vacuum with desiccant for 48 hr to remove the final traces of ether. The purified polymer was then stored at 4°C. For further use, the polymer was dissolved in DMSO at 100 mg/mL and stored at −20°C.
Cell Culture
The cervical cancer cell lines SiHa (HPV16 positive), CaSki (HPV16 positive), HeLa (HPV16 negative, HPV18 positive) and the HEK cell line HEK293 (HPV negative) were purchased from ATCC and passaged in our laboratory. The S12 cell line (HPV16 positive) is an immortalized human cervical keratinocyte cell line, and it was a generous gift from professor Kenneth Raj (Health Protection Agency) with permission from the original owner professor Margaret Stanley.65, 66 SiHa, HeLa, CaSki, and HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco) and 100 U/mL penicillin and streptomycin (Invitrogen) at 37°C in a humidified incubator with 5% CO2. S12 cells were maintained in a 1:1 mixture of DMEM/F12 (Gibco) and Ham’s F12 (Gibco) medium supplemented with 5% FBS, 24.3 mg/mL adenine, 0.5 mg/mL hydrocortisone, 8.4 ng/mL cholera toxin, 5 mg/mL insulin, and 10 ng/mL epidermal growth factor (EGF).
NP Fabrication and Characterization
PBAE and plasmids were separately diluted in a 25 mM sodium acetate solution (pH 5). The PBAE solution was added dropwise to the DNA solution and mixed gently for 30 s, and the mixture was incubated at room temperature for 15 min for complete NP formation. The particle size and zeta potential of the NPs were measured by laser light scattering (DB-525 Zeta PALS; Brookhaven Instruments, Holtsville, NY, USA).
Biocompatibility of NPs
SiHa, HeLa, CaSki, S12, and HEK293 cells were seeded in 96-well plates and incubated overnight. PBAE was diluted in 25 mM sodium acetate (pH 5) with a GFP plasmid at different weight ratios (20:1, 40:1, 60:1, and 80:1, PBAE to GFP). bPEI/GFP (weight ratio 3:1) diluted to 1 μg/μL in PBS was used as a control. Each well was transfected with 100 ng GFP. The culture medium was replaced after 6 hr of treatment with our NPs and the controls. Cell viability was defined as the metabolic activity retained in each well after transfection, and it was measured at different time points using a Cell Counting Kit-8 (CCK-8, Dojindo) according to the manufacturer’s instructions.
A total of 100 μL NPs (PBAE/pcDNA3.1, 10 μg pcDNA3.1 for the low dose and 100 μg pcDNA3.1 for the maximum dose) was injected into the thigh muscles of C57BL/6 mice once a day for 3 days, and bPEI/pcDNA3.1 was injected as a positive control. On the fourth and seventh days after the initial injection, the thigh muscles and other organs were harvested. PBAE/pcDNA3.1 and bPEI/pcDNA3.1 (20 μL NPs containing 10 μg pcDNA3.1) were pipetted into the vaginas of mice once a day for 20 days; the cervix and other organs were then harvested. Toxicity was evaluated by H&E staining and TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling) staining.
In Vitro, Ex Vivo, and In Vivo Uptake of NPs
All cells were seeded in sterile 12-well plates and incubated overnight at 37°C. The culture medium was replaced with a solution containing 125 μL NPs carrying 1 μg plasmid. bPEI/plasmid polyplexes (weight ratio 3:1, 1 μg plasmid) were formed by mixing in PBS buffer for 15 min and then added to cells. The medium was replaced after 6 hr of incubation.
Fresh cervical cancer samples (HPV types were confirmed by Sanger PCR; Tables S4 and S5) were randomly collected from patients in Tongji Hospital, immediately washed with Dulbecco’s PBS (DPBS) supplemented with 100 U/mL penicillin and streptomycin, and cultured in 12-well plates with 1 mL KSFM (10725018, Gibco). Then, 150 μL PBAE/plasmid NPs carrying 100 μg plasmid DNA was added to the tissues. The KSFM was replaced after 6 hr of incubation.
The vaginas of C57BL/6 mice were rinsed 3 times by PBS to clear the vaginal mucus. PBAE/plasmid NPs containing 10 μg plasmid were injected into the vaginas of C57BL/6 mice (4 weeks old) once a day for 3 days. Another 3 days later, uterine cervixes and vaginas were isolated after the mice were euthanized, and they were cryosectioned at 7 μm using a frozen section machine (Thermo Fisher Scientific).
TEM Imaging
PBAE/GFP NPs were formed and deposited on carbon-coated copper grids with porous carbon films, and they were characterized using scanning electron microscopy with a Hitachi model microscope (Hitachi HT7700, Japan).
293 cells were transfected with PBAE/GFP NPs carrying 1 mg/mL GFP, and they were cultured with DMEM. The cells were collected, washed with PBS, fixed with 2% glutaraldehyde buffer, dried by critical point drying with liquid CO2 dryer (HCP-2 type; Hitachi), coated with osmium with a model osmium plasma coater (Nippon Laser & Electronic Laboratory), and examined by scanning electron microscopy with a Hitachi model microscope (Hitachi HT7700, Japan).
qRT-PCR Analysis
Total RNA was extracted from cells using Trizol reagent (Takara, Dalian, China) 72 hr after treatment with NPs. cDNA (2 μg) was generated using SMART MMLV Reverse Transcriptase (639522, Takara, Dalian, China). qRT-PCR was performed using All-in-One qPCR Mix on a CFX96 Real-Time system (Bio-Rad, USA). Gene expression in each sample was normalized to GAPDH expression. For qRT-PCR primers, please see Table S3. The experiments were performed in triplicates, and the relative RNA expression was calculated using the comparative Ct method.
Western Blot Analysis
Cells and tissues were lysed on ice for 30 min in lysis buffer containing 150 mmol/L NaCl, 1% sodium deoxycholate, 50 mmol/L Tris, 1% Triton X-100, 0.1% SDS, and a protease inhibitor cocktail. The primary antibodies used were rabbit anti-GAPDH (1:1,000, AM1020a, Abgent), rabbit anti-HPV16 E7 (1:200, orb10837, Biorbyt), and rabbit anti-RB1 (1:1,000, 10048-2-Ig, Proteintech Group).
IHC and Immunofluorescence Staining
After the mice were euthanized, the uterine cervixes, vaginas, and xenografts were isolated and fixed (4% paraformaldehyde). Paraffin-embedded sections (5 μm) were subjected to IHC staining according to the Proteintech protocol (http://www.ptgcn.com/support/protocols). The slides were incubated overnight at 4°C with rabbit anti-HPV16 E7 (1:50, orb10573, Biorbyt), rabbit anti-P16 (1:100, A11337, Abclonal), rabbit anti-RB1 (1:100, 10048-2-Ig, Proteintech), rabbit anti-Ki67 (1:100, ab16667, Abcam), rabbit anti-CD34 (1:100, BA0532, Wuhan Boster Biological Engineering), rabbit anti-CDK2 (1:400, ab6538, Abcam), and rabbit anti-E2F1 (1:200, 12171-1-Ap, Proteintech) primary antibodies. Antibody detection was performed using diaminobezidin (DAB). Photographs were taken of three randomly chosen fields using cellSens Dimension (version 1.8.1, Olympus), and the staining intensity was measured using ImagePro Plus (version 6.0, Media Cybernetics).
SiHa cells were transfected with PBAE/GFP carrying 1 ng/μL GFP plasmids, fixed, and subjected to immunofluorescence staining (red) according to the Proteintech protocol. The cells were incubated overnight at 4°C with rabbit anti-GFP (1:200, 10048-2-Ig, Proteintech).
Xenografts and Transgenic Mouse Experiments
4-week-old BALB/c-nu nude mice were injected subcutaneously in the right flank with 5 × 106 SiHa and HeLa cells. The mice were randomly divided into groups. After the xenografts grew to approximately 35 mm3, we intratumorally injected 100 μL NPs carrying 60 μg plasmid DNA every 4 days. The tumor growth was measured every 4 days until 20 days using a digital caliper, and the tumor size was calculated using the following formula: L × W2 × 0.5. The subcutaneous tumors were collected after the mice were euthanized. All experimental protocols were approved by the Institutional Animal Care and Use Committee of HUST, and the study was carried out in strict accordance with the Guidelines for the Welfare of Animals in Experimental Neoplasia.
HPV16-positive female mice (6–8 weeks old) were randomly assigned to three groups for drug treatment. The mice were anesthetized with 1% pentobarbital, the vaginas were washed with 20 μL saline, and 20 μL NPs consisting of PBAE and HPV-targeting CRISPR/shRNA containing 10 μg plasmid DNA were pipetted into the vaginas every day for 20 days. The mice were kept anesthetized in the dorsal position for at least 30 min to keep the NPs in the vagina. After 20 days of treatment, the mice were euthanized, and the vaginas were dissected, fixed in 4% paraformaldehyde, and analyzed by IHC.
Author Contributions
D.M. and H.W. designed and supervised the research together with D.Z. and H.S. S.T. and Z.Z. provided technical support. D.Z., H.S., L.W., Z.H., and L.Y. performed the experiments. X.T., W.D., C.R., and C.G. obtained specimens. J.C., M.D., R.L., J.H., L.X., and P.W. provided analysis and interpretation of data. The manuscript was drafted by D.Z. and H.S. All authors critically reviewed the article and approved the final manuscript.
Conflicts of Interest
The authors disclose no potential conflicts of interest.
Acknowledgments
This work was supported by funds from the National Development Program (973) for the Key Basic Research of China (2013CB911304 and 2015CB553903), the National Key Research & Development Program of China (2016YFC0902900), the National Science-technology Supporting Plan Projects (2015BAI13B05), the National Natural Science Foundation of China (81372805, 81472783, 81502253, 81402158, 81502252, 81403166, 81230038, 81630060, 81372806, and 81373360), and the Natural Science Foundation of Hubei Province (2015CFB492). We thank professor Kenneth Raj (Health Protection Agency, Didcot, UK) for providing the S12 cell line, which was permitted by the primary owner professor Margaret Stanley (Division of Virology, National Institute for Medical Research, London, UK).
Footnotes
Supplemental Information includes ten figures and five tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.07.019.
Contributor Information
Ding Ma, Email: dingma424@yahoo.com.
Hui Wang, Email: huit71@sohu.com.
Supplemental Information
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Francis S.A., Nelson J., Liverpool J., Soogun S., Mofammere N., Thorpe R.J., Jr. Examining attitudes and knowledge about HPV and cervical cancer risk among female clinic attendees in Johannesburg, South Africa. Vaccine. 2010;28:8026–8032. doi: 10.1016/j.vaccine.2010.08.090. [DOI] [PubMed] [Google Scholar]
- 3.Phelps W.C., Yee C.L., Münger K., Howley P.M. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 4.Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 5.Arbeit J.M., Howley P.M., Hanahan D. Chronic estrogen-induced cervical and vaginal squamous carcinogenesis in human papillomavirus type 16 transgenic mice. Proc. Natl. Acad. Sci. USA. 1996;93:2930–2935. doi: 10.1073/pnas.93.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elson D.A., Riley R.R., Lacey A., Thordarson G., Talamantes F.J., Arbeit J.M. Sensitivity of the cervical transformation zone to estrogen-induced squamous carcinogenesis. Cancer Res. 2000;60:1267–1275. [PubMed] [Google Scholar]
- 7.Pett M., Coleman N. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J. Pathol. 2007;212:356–367. doi: 10.1002/path.2192. [DOI] [PubMed] [Google Scholar]
- 8.Hu Z., Zhu D., Wang W., Li W., Jia W., Zeng X., Ding W., Yu L., Wang X., Wang L. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 2015;47:158–163. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 9.Ciavattini A., Clemente N., Delli Carpini G., Gentili C., Di Giuseppe J., Barbadoro P., Prospero E., Liverani C.A. Loop electrosurgical excision procedure and risk of miscarriage. Fertil. Steril. 2015;103:1043–1048. doi: 10.1016/j.fertnstert.2014.12.112. [DOI] [PubMed] [Google Scholar]
- 10.Guo H.J., Guo R.X., Liu Y.L. Effects of loop electrosurgical excision procedure or cold knife conization on pregnancy outcomes. Eur. J. Gynaecol. Oncol. 2013;34:79–82. [PubMed] [Google Scholar]
- 11.Saslow D., Castle P.E., Cox J.T., Davey D.D., Einstein M.H., Ferris D.G., Goldie S.J., Harper D.M., Kinney W., Moscicki A.B., Gynecologic Cancer Advisory Group American Cancer Society Guideline for human papillomavirus (HPV) vaccine use to prevent cervical cancer and its precursors. CA Cancer J. Clin. 2007;57:7–28. doi: 10.3322/canjclin.57.1.7. [DOI] [PubMed] [Google Scholar]
- 12.Joura E.A., Giuliano A.R., Iversen O.E., Bouchard C., Mao C., Mehlsen J., Moreira E.D., Jr., Ngan Y., Petersen L.K., Lazcano-Ponce E., Broad Spectrum HPV Vaccine Study A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N. Engl. J. Med. 2015;372:711–723. doi: 10.1056/NEJMoa1405044. [DOI] [PubMed] [Google Scholar]
- 13.Rangel-Colmenero B.R., Gomez-Gutierrez J.G., Villatoro-Hernández J., Zavala-Flores L.M., Quistián-Martínez D., Rojas-Martínez A., Arce-Mendoza A.Y., Guzmán-López S., Montes-de-Oca-Luna R., Saucedo-Cárdenas O. Enhancement of Ad-CRT/E7-mediated antitumor effect by preimmunization with L. lactis expressing HPV-16 E7. Viral Immunol. 2014;27:463–467. doi: 10.1089/vim.2014.0055. [DOI] [PubMed] [Google Scholar]
- 14.Pahle J., Walther W. Vectors and strategies for nonviral cancer gene therapy. Expert Opin. Biol. Ther. 2016;16:443–461. doi: 10.1517/14712598.2016.1134480. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Ding M., Xu K., Mao L., Zheng J. shRNA-armed conditionally replicative adenoviruses: a promising approach for cancer therapy. Oncotarget. 2016;7:29824–29834. doi: 10.18632/oncotarget.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida T., Lai T.C., Kwon G.S., Sako K. pH- and ion-sensitive polymers for drug delivery. Expert Opin. Drug Deliv. 2013;10:1497–1513. doi: 10.1517/17425247.2013.821978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerrero-Cázares H., Tzeng S.Y., Young N.P., Abutaleb A.O., Quiñones-Hinojosa A., Green J.J. Biodegradable polymeric nanoparticles show high efficacy and specificity at DNA delivery to human glioblastoma in vitro and in vivo. ACS Nano. 2014;8:5141–5153. doi: 10.1021/nn501197v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F., Cho S.W., Son S.M., Bogatyrev S.R., Singh D., Green J.J., Mei Y., Park S., Bhang S.H., Kim B.S. Genetic engineering of human stem cells for enhanced angiogenesis using biodegradable polymeric nanoparticles. Proc. Natl. Acad. Sci. USA. 2010;107:3317–3322. doi: 10.1073/pnas.0905432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzeng S.Y., Higgins L.J., Pomper M.G., Green J.J. Student award winner in the Ph.D. category for the 2013 society for biomaterials annual meeting and exposition, april 10-13, 2013, Boston, Massachusetts : biomaterial-mediated cancer-specific DNA delivery to liver cell cultures using synthetic poly(beta-amino ester)s. J. Biomed. Mater. Res. A. 2013;101:1837–1845. doi: 10.1002/jbm.a.34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J., Kang Y., Tzeng S.Y., Green J.J. Synthesis and application of poly(ethylene glycol)-co-poly(β-amino ester) copolymers for small cell lung cancer gene therapy. Acta Biomater. 2016;41:293–301. doi: 10.1016/j.actbio.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zandberg D.P., Bhargava R., Badin S., Cullen K.J. The role of human papillomavirus in nongenital cancers. CA Cancer J. Clin. 2013;63:57–81. doi: 10.3322/caac.21167. [DOI] [PubMed] [Google Scholar]
- 23.Naucler P., Ryd W., Törnberg S., Strand A., Wadell G., Elfgren K., Rådberg T., Strander B., Johansson B., Forslund O. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N. Engl. J. Med. 2007;357:1589–1597. doi: 10.1056/NEJMoa073204. [DOI] [PubMed] [Google Scholar]
- 24.(2006). HPV vaccine fights cervical cancer. CA Cancer J. Clin. 56, 249–250. [DOI] [PubMed]
- 25.Kim J.J., Goldie S.J. Health and economic implications of HPV vaccination in the United States. N. Engl. J. Med. 2008;359:821–832. doi: 10.1056/NEJMsa0707052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huh W.K., Joura E.A., Giuliano A.R., Iversen O.E., de Andrade R.P., Ault K.A., Bartholomew D., Cestero R.M., Fedrizzi E.N., Hirschberg A.L. Final efficacy, immunogenicity, and safety analyses of a nine-valent human papillomavirus vaccine in women aged 16-26 years: a randomised, double-blind trial. Lancet. 2017;390:2143–2159. doi: 10.1016/S0140-6736(17)31821-4. [DOI] [PubMed] [Google Scholar]
- 27.Bogani G., Leone Roberti Maggiore U., Signorelli M., Martinelli F., Ditto A., Sabatucci I., Mosca L., Lorusso D., Raspagliesi F. The role of human papillomavirus vaccines in cervical cancer: Prevention and treatment. Crit. Rev. Oncol. Hematol. 2018;122:92–97. doi: 10.1016/j.critrevonc.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Kim J.J., Burger E.A., Sy S., Campos N.G. Optimal Cervical Cancer Screening in Women Vaccinated Against Human Papillomavirus. J. Natl. Cancer Inst. 2016;109:djw216. doi: 10.1093/jnci/djw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campos N.G., Sharma M., Clark A., Kim J.J., Resch S.C. Resources Required for Cervical Cancer Prevention in Low- and Middle-Income Countries. PLoS ONE. 2016;11:e0164000. doi: 10.1371/journal.pone.0164000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldhaber-Fiebert J.D., Goldie S.J. Estimating the cost of cervical cancer screening in five developing countries. Cost Eff. Resour. Alloc. 2006;4:13. doi: 10.1186/1478-7547-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 32.Werness B.A., Levine A.J., Howley P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 33.Münger K., Basile J.R., Duensing S., Eichten A., Gonzalez S.L., Grace M., Zacny V.L. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001;20:7888–7898. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- 34.Cullen B.R. Enhancing and confirming the specificity of RNAi experiments. Nat. Methods. 2006;3:677–681. doi: 10.1038/nmeth913. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Grainger D.W. RNA therapeutics targeting osteoclast-mediated excessive bone resorption. Adv. Drug Deliv. Rev. 2012;64:1341–1357. doi: 10.1016/j.addr.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 38.Ul Ain Q., Chung J.Y., Kim Y.H. Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. J. Control. Release. 2015;205:120–127. doi: 10.1016/j.jconrel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 39.Bousarghin L., Touze A., Gaud G., Iochmann S., Alvarez E., Reverdiau P., Gaitan J., Jourdan M.L., Sizaret P.Y., Coursaget P.L. Inhibition of cervical cancer cell growth by human papillomavirus virus-like particles packaged with human papillomavirus oncoprotein short hairpin RNAs. Mol. Cancer Ther. 2009;8:357–365. doi: 10.1158/1535-7163.MCT-08-0626. [DOI] [PubMed] [Google Scholar]
- 40.Sima N., Wang W., Kong D., Deng D., Xu Q., Zhou J., Xu G., Meng L., Lu Y., Wang S., Ma D. RNA interference against HPV16 E7 oncogene leads to viral E6 and E7 suppression in cervical cancer cells and apoptosis via upregulation of Rb and p53. Apoptosis. 2008;13:273–281. doi: 10.1007/s10495-007-0163-8. [DOI] [PubMed] [Google Scholar]
- 41.Hu Z., Yu L., Zhu D., Ding W., Wang X., Zhang C., Wang L., Jiang X., Shen H., He D. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. BioMed Res. Int. 2014;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J., Li B., Peng C., Wang F., Fu Z., Zhou C., Hong D., Ye F., Lü W., Xie X. Inhibition of cervical cancer cell growth in vitro and in vivo by lentiviral-vector mediated shRNA targeting the common promoter of HPV16 E6 and E7 oncogenes. Antiviral Res. 2013;98:305–313. doi: 10.1016/j.antiviral.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Rampias T., Sasaki C., Weinberger P., Psyrri A. E6 and e7 gene silencing and transformed phenotype of human papillomavirus 16-positive oropharyngeal cancer cells. J. Natl. Cancer Inst. 2009;101:412–423. doi: 10.1093/jnci/djp017. [DOI] [PubMed] [Google Scholar]
- 45.Li X., Li Y., Hu J., Wang B., Zhao L., Ji K., Guo B., Yin D., Du Y., Kopecko D.J. Plasmid-based E6-specific siRNA and co-expression of wild-type p53 suppresses the growth of cervical cancer in vitro and in vivo. Cancer Lett. 2013;335:242–250. doi: 10.1016/j.canlet.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhen S., Lu J.J., Wang L.J., Sun X.M., Zhang J.Q., Li X., Luo W.J., Zhao L. In Vitro and In Vivo Synergistic Therapeutic Effect of Cisplatin with Human Papillomavirus16 E6/E7 CRISPR/Cas9 on Cervical Cancer Cell Line. Transl. Oncol. 2016;9:498–504. doi: 10.1016/j.tranon.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung H.S., Rajasekaran N., Ju W., Shin Y.K. Human Papillomavirus: Current and Future RNAi Therapeutic Strategies for Cervical Cancer. J. Clin. Med. 2015;4:1126–1155. doi: 10.3390/jcm4051126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Z., Liu F., Chen Y., Liu J., Wang X., Chen A.T., Deng G., Zhang H., Liu J., Hong Z., Zhou J. Targeted Delivery of CRISPR/Cas9-Mediated Cancer Gene Therapy via Liposome-Templated Hydrogel Nanoparticles. Adv. Funct. Mater. 2017;27:1703036. doi: 10.1002/adfm.201703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Liang H., Liu J., Wang Z. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int. J. Pharm. 2018;546:215–225. doi: 10.1016/j.ijpharm.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 50.Kanazawa T., Akiyama F., Kakizaki S., Takashima Y., Seta Y. Delivery of siRNA to the brain using a combination of nose-to-brain delivery and cell-penetrating peptide-modified nano-micelles. Biomaterials. 2013;34:9220–9226. doi: 10.1016/j.biomaterials.2013.08.036. [DOI] [PubMed] [Google Scholar]
- 51.Xiao B., Laroui H., Viennois E., Ayyadurai S., Charania M.A., Zhang Y., Zhang Z., Baker M.T., Zhang B., Gewirtz A.T., Merlin D. Nanoparticles with surface antibody against CD98 and carrying CD98 small interfering RNA reduce colitis in mice. Gastroenterology. 2014;146 doi: 10.1053/j.gastro.2014.01.056. 1289–300.e1, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodrow K.A., Cu Y., Booth C.J., Saucier-Sawyer J.K., Wood M.J., Saltzman W.M. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat. Mater. 2009;8:526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J., Sunshine J.C., Green J.J. Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(β-amino ester) polyplexes in human breast cancer cells. Bioconjug. Chem. 2014;25:43–51. doi: 10.1021/bc4002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastorakos P., da Silva A.L., Chisholm J., Song E., Choi W.K., Boyle M.P., Morales M.M., Hanes J., Suk J.S. Highly compacted biodegradable DNA nanoparticles capable of overcoming the mucus barrier for inhaled lung gene therapy. Proc. Natl. Acad. Sci. USA. 2015;112:8720–8725. doi: 10.1073/pnas.1502281112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Min S., Jin Y., Hou C.Y., Kim J., Green J.J., Kang T.J., Cho S.W. Bacterial tRNase-Based Gene Therapy with Poly(β-Amino Ester) Nanoparticles for Suppressing Melanoma Tumor Growth and Relapse. Adv. Healthc. Mater. 2018 doi: 10.1002/adhm.201800052. Published online June 10, 2018. [DOI] [PubMed] [Google Scholar]
- 56.Jones C.H., Chen M., Ravikrishnan A., Reddinger R., Zhang G., Hakansson A.P., Pfeifer B.A. Mannosylated poly(beta-amino esters) for targeted antigen presenting cell immune modulation. Biomaterials. 2015;37:333–344. doi: 10.1016/j.biomaterials.2014.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saravanan M., Asmalash T., Gebrekidan A., Gebreegziabiher D., Araya T., Hilekiros H., Barabadi H., Ramanathan K. Nano-Medicine as a Newly Emerging Approach to Combat Human Immunodeficiency Virus (HIV) Pharm. Nanotechnol. 2018;6:17–27. doi: 10.2174/2211738506666180209095710. [DOI] [PubMed] [Google Scholar]
- 58.Medina-Alarcón K.P., Voltan A.R., Fonseca-Santos B., Moro I.J., de Oliveira Souza F., Chorilli M., Soares C.P., Dos Santos A.G., Mendes-Giannini M.J.S., Fusco-Almeida A.M. Highlights in nanocarriers for the treatment against cervical cancer. Mater. Sci. Eng. C. 2017;80:748–759. doi: 10.1016/j.msec.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 59.Kozielski K.L., Tzeng S.Y., De Mendoza B.A., Green J.J. Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells. ACS Nano. 2014;8:3232–3241. doi: 10.1021/nn500704t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segovia N., Dosta P., Cascante A., Ramos V., Borrós S. Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization. Acta Biomater. 2014;10:2147–2158. doi: 10.1016/j.actbio.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 61.Ferenczy A., Franco E. Persistent human papillomavirus infection and cervical neoplasia. Lancet Oncol. 2002;3:11–16. doi: 10.1016/s1470-2045(01)00617-9. [DOI] [PubMed] [Google Scholar]
- 62.Kadaja M., Isok-Paas H., Laos T., Ustav E., Ustav M. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 2009;5:e1000397. doi: 10.1371/journal.ppat.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sepkovic D.W., Stein J., Carlisle A.D., Ksieski H.B., Auborn K., Bradlow H.L. Diindolylmethane inhibits cervical dysplasia, alters estrogen metabolism, and enhances immune response in the K14-HPV16 transgenic mouse model. Cancer Epidemiol. Biomarkers Prev. 2009;18:2957–2964. doi: 10.1158/1055-9965.EPI-09-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamat C.D., Shmueli R.B., Connis N., Rudin C.M., Green J.J., Hann C.L. Poly(β-amino ester) nanoparticle delivery of TP53 has activity against small cell lung cancer in vitro and in vivo. Mol. Cancer Ther. 2013;12:405–415. doi: 10.1158/1535-7163.MCT-12-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bechtold V., Beard P., Raj K. Human papillomavirus type 16 E2 protein has no effect on transcription from episomal viral DNA. J. Virol. 2003;77:2021–2028. doi: 10.1128/JVI.77.3.2021-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhakta M.S., Segal D.J. The generation of zinc finger proteins by modular assembly. Methods Mol. Biol. 2010;649:3–30. doi: 10.1007/978-1-60761-753-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.