Abstract
Immunologically based clinical trials performed thus far have failed to cure type 1 diabetes (T1D), in part because these approaches were nonspecific. Because the disease is driven by autoreactive CD4 T cells, which destroy β cells, transplantation of hematopoietic stem and progenitor cells (HSPCs) has been recently offered as a therapy for T1D. Our transcriptomic profiling of HSPCs revealed that these cells are deficient in programmed death ligand 1 (PD-L1), an important immune checkpoint, in the T1D nonobese diabetic (NOD) mouse model. Notably, the immunoregulatory molecule PD-L1 plays a determinant role in controlling/inhibiting activated T cells and thus maintains immune tolerance. Furthermore, our genome-wide and bioinformatic analysis revealed the existence of a network of microRNAs (miRNAs) controlling PD-L1 expression, and silencing one of key altered miRNAs restored PD-L1 expression in HSPCs. We therefore sought to determine whether restoration of this defect would cure T1D as an alternative to immunosuppression. Genetically engineered or pharmacologically modulated HSPCs overexpressing PD-L1 inhibited the autoimmune response in vitro, reverted diabetes in newly hyperglycemic NOD mice in vivo, and homed to the pancreas of hyperglycemic NOD mice. The PD-L1 expression defect was confirmed in human HSPCs in T1D patients as well, and pharmacologically modulated human HSPCs also inhibited the autoimmune response in vitro. Targeting a specific immune checkpoint defect in HSPCs thus may contribute to establishing a cure for T1D.
INTRODUCTION
Since the search for feasible and safe immunological approaches to reestablish tolerance toward islet autoantigens and preserve β cell function in type 1 diabetes (T1D) began, little progress has been made clinically (1–4). However, most immunotherapies tested thus far are simply broadly immunosuppressive and are not linked to any immunological abnormalities detected in T1D (5). Couri et al. (6) evaluated the safety and efficacy of autologous hematopoietic stem and progenitor cell (HSPC) transplantation in combination with thymoglobulin plus cyclophosphamide as induction in newly diagnosed T1D patients. The latest multicenter analysis on 65 newly diagnosed T1D patients treated with autologous HSPC transplantation achieved insulin independence in nearly 60% of treated patients (7), suggesting that HSPCs may be a therapeutic option for selected T1D patients. HSPCs are endowed with immunoregulatory properties, which have been shown to be linked to the expression of the immune checkpoint PD-L1 (also known as CD274) (8). PD-L1 is the ligand for the inhibitory programmed death 1 (PD-1) receptor, expressed primarily on activated T cells (9). Crosslinking of PD-L1 and PD-1 inhibits T cell activation and favors their exhaustion/apoptosis (10); mice deficient in PD-L1/PD-1 develop accelerated diabetes (9). On the basis of these data, we hypothesized that a defect in expression of the immune checkpoint PD-L1 in HSPCs plays a role in T1D and that genetic or pharmacological restoration of this defect would cure T1D. PD-L1+ HSPCs may play an important endogenous immunoregulatory role, capable of eliminating autoreactive T cells but eventually becoming defective in T1D. This deficiency may serve as a novel mechanism of disease for T1D and allow for establishment of a therapeutic approach based on the restoration of the PD-L1 defect in HSPCs.
RESULTS
A defect in PD-L1 is evident in HSPCs from nonobese diabetic mice
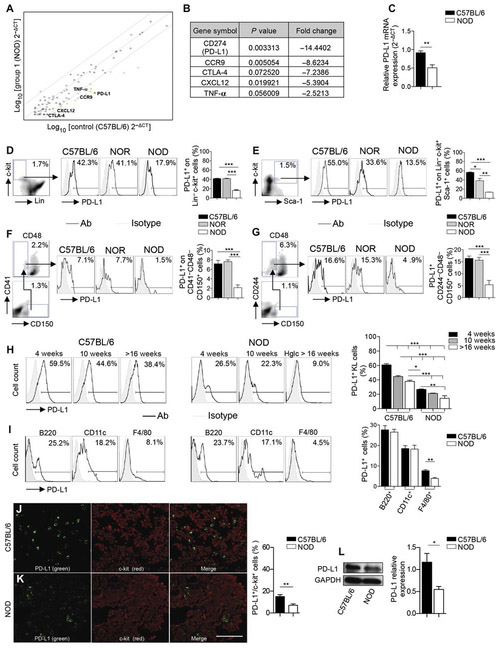
To identify any defects in immunoregulatory molecules in HSPCs derived from nonobese diabetic (NOD) mice, we performed broad transcriptomic profiling of immune-related molecules in murine HSPCs. Sca-1+Lineage−c-kit+ (KLS) cells (or a subset of murine HSPCs) obtained from 10-week-old normoglycemic NOD mice had decreased PD-L1 transcripts as compared to HSPCs obtained from C57BL/6 mice (Fig. 1, A and B, and table S1). Measurement of PD-L1 mRNA expression by reverse transcription polymerase chain reaction (RT-PCR) confirmed reduction in NOD HSPCs as well (Fig. 1C). We next used a range of techniques to demonstrate the defect in PD-L1 expression in a variety of bone marrow HSPCs, including KLS cells, Lineage−c-kit+ (KL) cells, and long-term repopulating HSPCs (CD41−CD48−CD150+ and CD244−CD48−CD150+ cells), and compared it to the expression observed in NOR (NOD-related diabetes-resistant) and C57BL/6 mice (Fig. 1, D to G). The overall PD-L1 defect is primarily confined to NOD mice (Fig. 1, D to G). We sought then to explore any association of the PD-L1 defect in HSPCs with age or disease status. We noticed a slight decline in the number of KL–PD-L1+ cells in both strains with progressive age but again with a clear defect in NOD mice (Fig. 1H). Other costimulatory molecules were evaluated as well, and no major significant differences were observed in HSPCs (fig. S1, A to D), suggesting a uniqueness of the PD-L1 defect. The PD-L1 defect was primarily confined to HSPCs in NOD mice, although other bone marrow–derived myeloid immune cells were slightly deficient in PD-L1 expression (that is, F4/80+ and CD11b+ cells; Fig. 1I and fig. S1, E to M). A subset of CD11c+ cells in NOD mice were PD-L1 high, whereas all CD11c+ cells in C57BL/6 mice expressed a low level of PD-L1; this could be a compensatory effect in myeloid cells (Fig. 1I). To understand the extent of the PD-L1 defect within the HSPC niche, we analyzed bone marrow tissues using confocal imaging. Fewer c-kit+PD-L1+ cells were observed in samples obtained from NOD as compared to C57BL/6 control mice (Fig. 1, J and K). Western blotting confirmed reduced PD-L1 protein expression on KL cells obtained from NOD bone marrow compared to C57BL/6 bone marrow (Fig. 1L). Our data confirmed the existence of a defect in PD-L1 expression in HSPCs in NOD mice.
Fig. 1. PD-L1 is down-regulated In HSPCs from NOD mice.
(A and B) Transcriptomic profiling of KLS cells obtained from bone marrow of NOD and C57BL/6 mice; n = 3 samples per group were evaluated. Statistical analysis was performed also by using the software available (RT2 profiler PCR Array Data Analysis, Qiagen). TNF-α, tumor necrosis factor–α. (C) Bar graph representing mRNA expression of PD-L1 as measured by quantitative RT-PCR (qRT-PCR) in KL cells, collected from bone marrow of C57BL/6 and NOD mice. All samples were run in triplicate and normalized to expression of the housekeeping gene GAPDH. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D to G) Representative flow cytometric analysis and quantitative bar graphs assessing PD-L1 expression in four populations of HSPCs in C57BL/6 and NOD mice. (H) Representative flow cytometry and quantitative bar graphs of PD-L1 expression in KL cells obtained from the bone marrow of C57BL/6 and NOD mice at different ages; n = 3 mice per group were evaluated and for statistical analysis, one-way analysis of variance (ANOVA) followed by Bonferroni multiple comparison test for group comparisons between C57BL/6 and NOD mice. Lin, Lineage; Ab, antibody; Hglc, hyperglycemic. (I) Representative flow cytometric analysis and quantitative bar graphs of PD-L1 expression in KL cells and in other nonprogenitor cells from bone marrow or spleen respectively obtained from C57BL/6 and NOD mice; n = 3 mice per group were evaluated. (J and K) Confocal imaging and quantification of bone marrow sections of C57BL/6 and NOD mice for c-kit (shown in red) and PD-L1 (shown in green) staining (n = 5 sections per strain); the quantification of the orange-stained bone marrow element was performed by ImageJ, and statistical significance was performed using two-tailed unpaired t test with Welch’s correction. Histology magnification, ×63. Scale bar, 40 μm. (L) Western blotting and quantitative bar graphs confirming reduced PD-L1 protein expression in KL cells obtained from bone marrow of C57BL/6 and NOD mice (n = 3 samples per group), with GAPDH used as an internal control. Data are expressed as means ± SEM. Data are representative of at least n = 3 mice. *P < 0.05; **P <0.01; ***P < 0.001.
The PD-L1 defect is associated with an altered network of PD-L1–related microRNAs
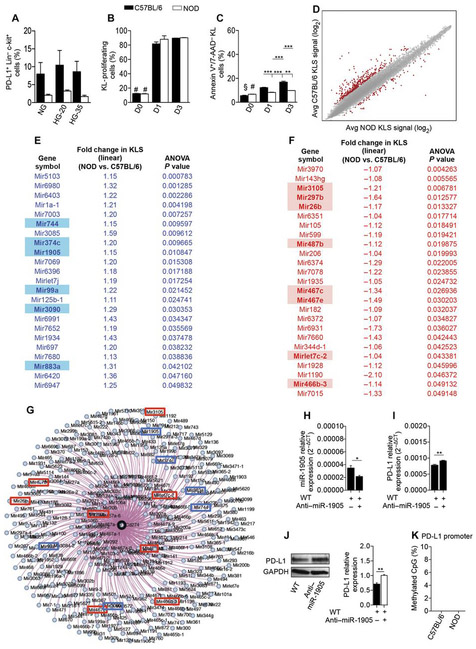
To better understand the mechanism behind the PD-L1 defect in HSPCs of NOD mice, we performed a series of in vitro experiments. We first tested the effect of high glucose on PD-L1 expression and then evaluated the existence of any HSPC survival defect that could explain the deficiency in PD-L1. Isolated KL cells from NOD and C57BL/6 mice were cultured for 3 days in high glucose, and there was no evidence of a high glucose–associated effect on PD-L1 expression (Fig. 2A), although we cannot exclude the fact that the observed PD-L1 defect maybe caused by a metabolic derivative of high glucose. No differences in the proliferation rate, and a slight difference in the percentage of apoptotic cells, were detected among KL cells from NOD or C57BL/6 mice (Fig. 2, B and C).
Fig. 2. Mechanism of PD-L1 down-regulation in NOD HSPCs.
(A) Bar graphs depicting percentage of PD-L1+ cells within KL cells isolated from bone marrow of C57BL/6 and NOD mice and cultured for 3 days in normal glucose, in 20 mM, or in 35 mM high glucose. Experiments were run in triplicate. NG, normal glucose; HG-20, 20 mM high glucose; HG-35, 35 mM high glucose. (B) Proliferation rates of carboxy-fluorescein succinimidyl ester (CFSE)–labeled KL cells obtained from C57BL/6 and NOD bone marrow at baseline and after 1 and 3 days of culture. (C) Frequency of apoptosis of KL cells obtained from bone marrow of C57BL/6 and NOD mice at baseline and after 1 and 3 days of culture. For statistical analysis in (A) to (C), one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons between C57BL/6 and NOD mice; in (A): P = not significant (ns); in (B): #P < 0.0001 versus all except NOD-day 0 (D0) (P = ns); in (C): §P < 0.0001 versus all except NOD-D0 (P = ns) and NOD-D1 (P < 0.001); #P < 0.0001 versus all except NOD–D1 (P =ns). 7-AAD, 7-aminoactinomycin D. (D) MA plot for GWAS performed in KLS cells obtained from bone marrow of C57BL/6 compared to NOD mice; MA-plot, log2 normalized expression levels of expression for KLS cells from C57BL/6 (y axis) in comparison with KLS cells from NOD (x axis). Avg, average. (E and F) List of miRNAs significantly up-regulated (E) and down-regulated (F) in KLS cells obtained from the bone marrow of NOD as compared to C57BL/6 mice. (G) miRNA network controlling PD-L1 gene expression generated with MGI. (H and I) KL cells obtained from bone marrow of NOD mice were cultured in the presence of miR-1905 inhibitor, and wild-type (WT) (untreated KL) were used as controls. qRT-PCR of miR-1905 (H) and qRT-PCR of PD-L1 (I) are shown. Experiments were run in triplicate, and statistical significance was determined using two-tailed unpaired Student’s t test. (J) Western blotting and quantitative bar graphs of PD-L1 protein expression in KL cells obtained from bone marrow of NOD mice cultured in the presence of miR-1905 inhibitor, and WT (untreated KL) were used as controls, with GAPDH used as an internal control. Experiments were run in triplicate, and statistical significance was determined using two-tailed unpaired Student’s t test (P= 0.0005). (K) Methylation status of the PD-L1 locus in KLS cells obtained from the bone marrow of C57BL/6 and NOD mice. Experiments were run in triplicate, and statistical significance comparing methylated CpG% of C57BL/6 to NOD was determined using two-tailed unpaired Student’s t test. Data are expressed as means ± SEM. Data are representative of at least n = 3 mice. *P < 0.05; **P <0.01; ***P < 0.0001.
When extending our investigation into analysis of gene expression profiles, Affymetrix microarray analysis revealed 48 microRNAs (miRNAs) differentially expressed in KLS cells of NOD and C57BL/6 mice (Fig. 2, D to F, and table S2). Data related to up-regulated and down-regulated miRNAs observed in the genome-wide analysis study (GWAS) performed on KLS cells obtained from NOD and C57BL/6 mice are described in table S2, shown as an MA plot in Fig. 2D and listed in Fig. 2 (E and F). Multiple databases and bioinformatics tools [Mouse Genome Informatics (MGI), DIANA-microT-CDS (http://microrna.gr/microT-CDS), and MouseMine (www.mousemine.org)] enabled us to identify ~330 miRNAs predicted to target the PD-L1 gene and revealed a comprehensive miRNA network associated with PD-L1 (Fig. 2G). Fourteen miRNAs appeared as both key players in controlling PD-L1 expression and altered in KLS cells obtained from NOD mice (Fig. 2, E to G). Next, to test the proof of concept that an altered miRNA network may have influenced PD-L1 expression on HSPCs, we silenced miR-1905, one of the miRNAs found altered in NOD HSPCs, in isolated KL cells extracted from bone marrow of NOD mice (Fig. 2, H and I). Although silencing one miRNA may influence other target genes, we chose to target miR-1905 because of the fact that it was the only mature miRNA among the six identified as relevant to PD-L1. Furthermore, miR-1905 was confirmed as having the higher miRNA target gene score or prediction score by the online software “DIANA-microT-CDS (www.microrna.gr/microT),” thus indicating a higher probability of affecting PD-L1 expression as compared to other miRNAs on the list. miR-1905 antagomir increased the expression of PD-L1 transcripts and protein in HSPCs (Fig. 2, I and J). An altered network of miRNAs may be responsible for PD-L1 reduced expression in HSPCs. Finally, we demonstrated the absence of a methylation of the PD-L1 promoter in KLS cells from NOD mice, which could have accounted for the PD-L1 defect (Fig. 2K).
Genetically engineered NOD HSPCs abrogate the autoimmune response in vitro and in vivo and revert hyperglycemia
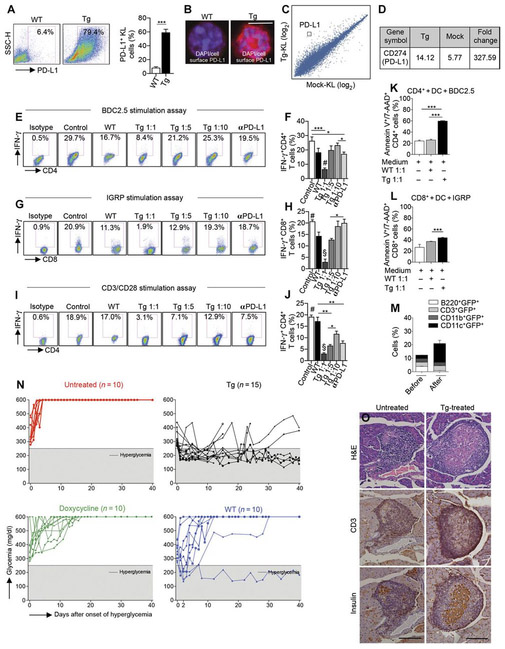
We next tested the effect of a genetic engineering approach to overcome the PD-L1 defect in NOD HSPCs. We genetically engineered murine KL cells ex vivo to generate PD-L1.Tg (transgenic) KL cells (Fig. 3A) from normoglycemic NOD mice by using third-generation self-inactivating lentiviral vectors (Lvs), a technique that has potential use in vivo because of its high efficiency and low risk of genotoxicity (fig. S2A) (11). Immunofluorescence and GWAS of these PD-L1.Tg KL cells confirmed the increase in PD-L1 expression compared to mock–Lv-transduced KL cells (Fig. 3, B to D, and table S3). We then explored the immunoregulatory properties of PD-L1.Tg KL cells in an autoimmune setting in vitro. PD-L1.Tg KL cells generated from normoglycemic NOD mice were cocultured at different ratios with T cells (1:1, 1:5, and 1:10) with CD11c+ dendritic cells (DCs) and BDC2.5 Tg CD4+CD25− T cells in the presence of the CD4-restricted islet mimotope peptide BDC2.5. A significant decrease (P < 0.05) in the percentage of interferon γ-positive (IFN-γ+)CD4+ T cells, as quantified by flow cytometry, was evident when naïve T cells were cocultured with PD-L1.Tg KL cells as compared to those cultured alone or cocultured with untransduced KL cells (Fig. 3, E and F). The gating strategy was determined using nonreactive isotype-matched control monoclonal antibodies (mAbs) in each culture condition, in which 99% of nonreactive cells were excluded. When PD-L1.Tg KL cells were precultured at a ratio of 1:1 to T cells with an anti–PD-L1–blocking mAb, the aforementioned immunoregulatory effect was severely hampered (Fig. 3, E and F). The robust and PD-L1-dependent immunoregulatory properties of PD-L1.Tg KL cells were confirmed using a CD8-restricted islet peptide–based assay [islet-specific glucose-6-phosphatase catalytic subunit–related protein (IGRP)] (Fig. 3, G and H) and an assay not specific to the autoimmune setting [anti-CD3/anti-CD28 stimulation (Fig. 3, I and J)], thus confirming that PD-L1. Tg HSPCs abrogate the autoimmune response in vitro. BDC2.5 Tg CD4+CD25− T cells appear to be more susceptible to the effect of PD-L1 up-regulation as compared to IGRP Tg CD8+ T cells. To understand the mechanism by which PD-L1.Tg KL cells exert their immunosuppressive effects on autoreactive T cells, we performed an apoptosis assay in vitro during a diabetogenic autoimmune response by coculturing IGRP Tg CD8+ T cells from NOD 8.3 (stimulated with the islet peptide IGRP) or CD4+CD25− T cells from NOD BDC2.5 (stimulated with the islet peptide BDC2.5) in the presence of PD-L1.Tg KL or WT KL cells. PD-L1.Tg KL cells induced late cell death of autoreactive CD4+ and CD8+ T cells, whereas only a minor effect was observed, primarily on autoreactive CD8+ T cells, when WT KL cells were added (Fig. 3, K and L). We explored whether a possible conversion into myeloid-suppressive cells may partially explain the immunoregulatory effects of PD-L1. Tg KL cells. Whereas after transduction the presence of T/B cell markers was very scant, myeloid markers were strongly expressed (Fig. 3M and fig. S2B).
Fig. 3. Genetically engineered PD-Ll.Tg KL cells abrogate the autoimmune response in vitro and revert diabetes in hyperglycemic NOD mice in vivo.
(A) Freshly isolated murine KL cells were transduced with PD-L1 lentiviral particles, and 24 hours after transduction, cells were collected for fluorescence-activated cell sorting (FACS) analysis; representative flow cytometric analysis and quantitative bar graph of KL cells obtained from bone marrow of NOD mice before and after transduction with PD-L1 lentivirus. Experiments were run in triplicate, and statistical significance comparing WT to Tg postlentiviral PD-L1 transduction was determined by using two-tailed unpaired t test. (B) Confocal imaging of KL cells obtained from bone marrow of NOD mice pre- and postlentiviral PD-L1 transduction confirmed PD-L1 up-regulation. Histology magnification, ×63. Scale bar, 50 μm. DAPI, 4’,6- diamidino-2-phenylindole. (C and D) MA plot and PD-L1 fold change for gene expression level in KL cells obtained from bone marrow of NOD mice transduced with PD-L1 lentivirus as compared to mock-transduced KL cells, demonstrating PD-L1 up-regulation. Experiments were run in triplicate, and statistical analysis was performed using pairwise ANOVA test. (E and F) Representative flow cytometric analysis and quantitative bar graph of IFN-γ+CD4+ T cells isolated from NOD-BDC2.5 T cell receptor (TCR) Tg mice stimulated with BDC2.5 peptide in the presence of DCs (control) or upon coculture with untransduced KL cells (WT), with PD-L1.Tg KL cells (at different ratios), or with PD-L1.Tg KL cells pretreated with anti–PD-L1–blocking mAb, with the isotype control also shown. αPD-L1, anti–PD-L1–blocking mAb. (G and H) Representative flow cytometric analysis and quantitative bar graph of IFN-γ+CD8+ T cells isolated from NOD-8.3 TCR Tg mice stimulated with IGRP peptide in the presence of DCs (control), or upon coculture with WTKL cells, with PD-L1.Tg KL cells (at different ratios), or with PD-L1.Tg KL cells pretreated with PD-L1–blocking mAb. Experiments were run in triplicate, and statistical significance was determined by using two-tailed unpaired t test; #P < 0.05 versus all except Tg 1:10 and anti–PD-L1–blocking mAb (P = ns); §P < 0.05 versus all. (I and J) Representative flow cytometric analysis and quantitative bar graph of IFN-γ+CD4+ T cells isolated from normoglycemic NOD mice stimulated with soluble anti-CD3/anti-CD28 (control), or upon coculture with WT KL cells, with PD-L1.Tg KL cells (at different ratios), or with PD-L1.Tg KL cells pretreated with PD-L1–blocking mAb. PD-L1.Tg KL cells strongly abrogate the CD4- and CD8-restricted autoimmune response and anti–CD3/CD28-dependent T cell stimulation in vitro. All experiments were run at least in triplicate, and statistical significance was determined using two-tailed unpaired t test; #P < 0.05 versus all except WT (P = ns); §P < 0.05 versus all. (K and L) Naive CD4+CD25− T cells isolated from BDC2.5 TCR Tg NOD mice or CD8+ T cells from 8.3 TCR Tg NOD mice and stimulated with BDC2.5 or IGRP islet peptides and CD11c+ DCs were cocultured with KL cells (WT) or PD-L1.Tg KL cells, and the rate of apoptosis of CD4 or CD8T cells was assessed by flow cytometry. PD-L1.Tg KL cells’ effect on cell death in autoreactive CD4+ and CD8+T cells as compared to WT KL cells. Experiments were run in triplicate, and statistical significance was determined by using two-tailed unpaired t test. (M) Quantitative bar graphs for lymphoid and myeloid markers of isolated KL cells before and after lentiviral transduction. GFP+, green fluorescent protein. (N) Newly hyperglycemic NOD mice were treated with WT KL cells, with PD-L1.Tg KL cells, and with doxycycline or were left untreated. (O) Representative immunohistochemical hematoxylin and eosin (H&E) analysis and CD3/insulin staining in serial pancreatic islet tissue sections from PD-L1.Tg KL cell–treated or untreated newly hyperglycemic NOD mice. Histology magnification, ×20. Scale bars, 200 mm. *P < 0.05; **P <0.01; ***P < 0.0001.
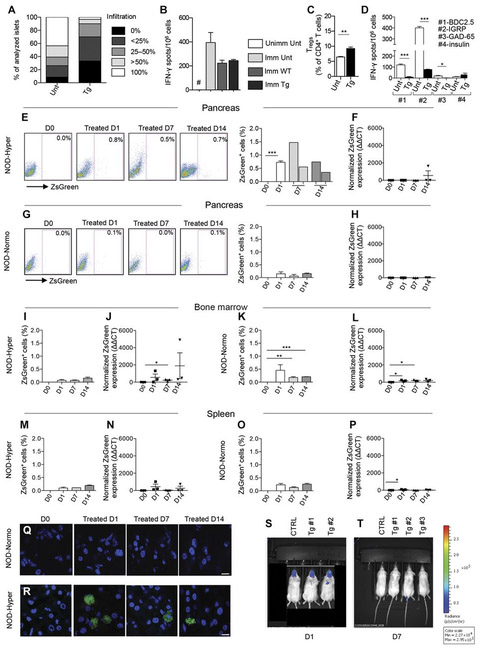
To evaluate the immunoregulatory properties of PD-L1.Tg KL cells in vivo, we adoptively transferred newly hyperglycemic NOD mice intravenously with 3 × 106 PD-L1.Tg KL cells (Fig. 3N), and they received doxycycline in the water (2 mg/ml) to induce PD-L1 expression, until the completion of the study, or with 3 × 106 untransduced KL cells (Fig. 3N), respectively. PD-L1.Tg KL cells successfully reverted hyperglycemia in 100% of treated hyperglycemic NOD mice with nearly 30% of treated mice remaining normoglycemic in the long term (Fig. 3N and fig. S2J), whereas none of the untreated hyperglycemic NOD mice (Fig. 3N) or those treated with doxycycline (Fig. 3N) reverted to normoglycemia. When untransduced KL cells were used, one hyperglycemic NOD mouse reverted to normoglycemia, and another showed a mild transient improvement of glycemic levels (Fig. 3N). Pathology of the pancreas of PD-L1.Tg KL cell–treated hyperglycemic NOD mice revealed reduced islet infiltration, with fewer CD3+ cells, preserved insulin staining, and improved insulitis score as compared to hyperglycemic untreated NOD mice (Figs. 3O and 4A). We then evaluated whether immunocompetence was preserved in NOD mice during treatment with PD-L1.Tg KL cells. PD-L1.Tg KL cell–treated NOD mice, untransduced KL cell–treated NOD mice, and untreated NOD mice were immunized at day 14 after the onset of hyperglycemia with ovalbumin (OVA), and after 3 days, splenocytes were harvested and rechallenged in vitro with OVA. In a 24-hour enzyme-linked immunospot (ELISpot) assay, the T cell response against the OVA peptide was measured as number of IFN-γ-producing cells. Treated NOD mice were capable of mounting a regular immune response to OVA similar to other groups tested and were thus immunocompetent (Fig. 4B). Immunophenotyping of PD-L1.Tg KL cell–treated hyperglycemic NOD mice showed at day 14 after treatment a twofold increase in the percentage of FoxP3+ regulatory CD4+ T cells as compared to untreated mice (Fig. 4C). Furthermore, a reduction in IFN-γ-producing cells was evident in PD-L1.Tg KL cell–treated as compared to untreated hyperglycemic NOD mice in an ex vivo assay, when splenocytes were challenged with islet peptides at day 40 after treatment (Fig. 4D).
Fig. 4. Genetically engineered PD-L1.Tg KL cells traffic to the pancreas in hyperglycemic NOD mice.
(A) Insulitis score: n = 9 sections per group were analyzed. Unt, untreated. (B) OVA rechallenge test. Data are representative of one experiment performed in three mice per group, and statistical significance was determined by using two-tailed unpaired t test. Unimm Unt, unimmunized untreated; Imm Unt, immunized untreated; Imm WT, immunized treated with KL; Imm Tg, immunized treated with PD-L1.Tg KL cells. (C) Immunophenotypic analysis of lymphocytes isolated from spleens by flow cytometry of FoxP3+ regulatory T cells (Tregs) in PD-L1.Tg KL cell–treated NOD mice as compared to untreated NOD mice. Data are representative of one experiment performed in three mice per group, and statistical significance was determined by using two-tailed unpaired t test. (D) Quantification of IFN-γ-producing cells (with number of spots normalized for background) in an ex vivo assay, in which splenocytes were challenged with islet peptides [BDC2.5, IGRP, glutamic acid decarboxylase 65 (GAD-65), and insulin] 40 days after treatment in newly hyperglycemic PD-L1.Tg KL cell–treated NOD mice or in untreated hyperglycemic NOD mice. Data are expressed as means ± SEM, and statistical significance was determined by using two-tailed unpaired t test. Data are representative of at least n = 3 mice. *P < 005; **P < 0.01; ***P < 0.001. #P < 0.05 versus all; §P < 0.05 versus all. (E and G) Representative flow cytometric analysis and quantitative bar graphs of ZsGreen+PD-L1. Tg KL cells in the pancreas of hyperglycemic (Hyper) and normoglycemic (Normo) NOD mice at 1,7, and 14 days after treatment with PD-L1.Tg KL cells. Experiments were run in triplicate [in (F): in duplicate], and statistical significance was determined by using two-tailed unpaired t test. (F and H) Quantification of ZsGreen mRNA in the pancreas of hyperglycemic and normoglycemic NOD mice by qRT-PCR after treatment with PD-L1.Tg KL cells. (I and K) Bar graphs depicting flow cytometric quantification of ZsGreen+PD-L1.Tg KL cells and (J and L) quantification of ZsGreen mRNA by qRT-PCR in the bone marrow of hyperglycemic and normoglycemic NOD mice, after treatment with PD-L1.Tg KL cells. Experiments were run in triplicate [in (M): in duplicate], and statistical significance was determined by using two-tailed unpaired t test. (M and O) Bar graphs for flow cytometric quantification of ZsGreen+PD-L1.Tg KL cells and (N and P) quantification of ZsGreen mRNA by qRT-PCR in the spleen of hyperglycemic and normoglycemic NOD mice. (Q and R) Confocal imaging of pancreatic sections obtained from normoglycemic or hyperglycemic NOD mice after 1,7, and 14 days after treatment with ZsGreen+PD-L1.Tg KL cells. Histology magnification, ×63 in all confocal images. Scale bars, 5 μm. (S and T) Luminescent images of NOD mice adoptively transferred with luciferase+PD-L1.Tg KL cells after 1 and 7 days of treatment. Data are expressed as means ± SEM. Data are representative of at least n = 2 mice. Statistical significance was determined using two-tailed unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001. CTRL, control.
Genetically engineered HSPCs traffic to the pancreas in hyperglycemic NOD mice
To explore the fate of infused PD-L1.Tg KL cells in NOD mice, we performed a set of tracking experiments in the pancreas, spleen, pancreatic draining lymph nodes (PLNs), and bone marrow using the tracer ZsGreen, present on the vector used to transduce PD-L1. Tg KL cells. PD-L1.Tg KL cells were adoptively transferred into normoglycemic and hyperglycemic NOD mice, and tissues were harvested at days 1, 7, and 14 after infusion. ZsGreen+ cells and ZsGreen mRNA expression were quantified in all tissues by flow cytometry and qRT-PCR, respectively. PD-L1.Tg KL cells preferentially trafficked to the pancreas once infused into hyperglycemic NOD (Fig. 4, E and F), although they homed to a lesser extent to the PLN, bone marrow, and spleen (Fig. 4, I, J, M, and N, and fig. S2C). Conversely, in normoglycemic NOD mice, PD-L1.Tg KL cells rarely trafficked to the pancreas, to the spleen, or to the PLN (Fig. 4, G, H, O, and P, and fig. S2D) but instead preferentially homed to the bone marrow (Fig. 4, K and L). Confocal imaging confirmed that ZsGreen+ cells were absent in the pancreas of normoglycemic NOD mice (Fig. 4Q), whereas they were detectable in the pancreata of hyperglycemic NOD mice, particularly at day 1 after PD-L1.Tg KL cell infusion (Fig. 4R). Although preferential homing of PD-L1.Tg KL cells to the bone marrow and to the spleen in hyperglycemic NOD mice was observed based on ZsGreen transcript quantification by qRT-PCR, flow cytometry and confocal imaging consistently showed trafficking of PD-L1.Tg KL cells to the pancreas. We also performed a tracking experiment using GFP+ KL cells (WT, untransduced KL cells) infused into hyperglycemic NOD mice and examined these mice by flow cytometry at days 1, 7, and 14; results showed no migration into the pancreas (fig. S2E). Moreover, we explored the chemokine receptor profile of PD-L1.Tg KL cells to determine which chemokine receptors are more likely involved in the homing of Tg HSPCs. Results showed that CXCR4 is expressed by the largest proportion of cells and thus highly involved in PD-L1.Tg KL cell trafficking (table S4). Bioluminescence imaging of NOD mice adoptively transferred with luciferase+PD-L1.Tg KL cells showed a rapid disappearance of luciferase+PD-L1.Tg KL cells from the peripheral blood (Fig. 4, S and T). We also examined whether PD-L1.Tg KL cells differentiated after their infusion into NOD mice. Tracking studies revealed a predominant transformation into myeloid cells (for example, CD11b) into the PLN of treated mice (fig. S2, F to I). These PD-L1–expressing myeloid cells may interact with autoreactive CD4 and CD8 T cells and may be contributing to their demise.
Pharmacologically modulated HSPCs abrogate the autoimmune response in vitro
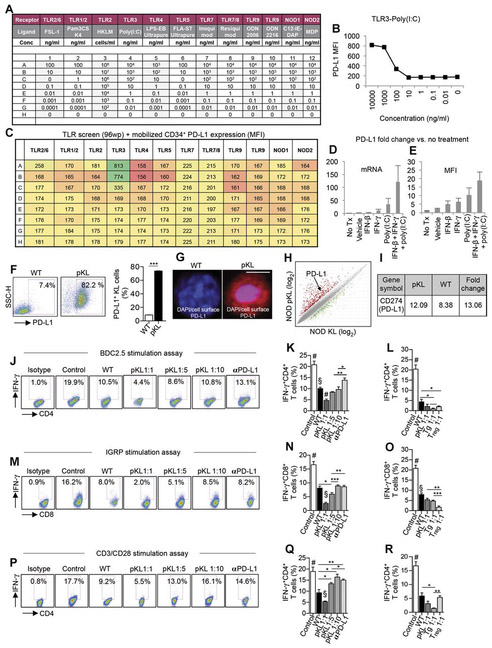
To offer an alternative approach to gene therapy, we explored the feasibility of pharmacological modulation of PD-L1. We first tested the ability of single agents potentially capable of up-regulating PD-L1 in human CD34+ cells (Fig. 5, A to C) and then established a cocktail of three agents {IFN-β, IFN-γ, and polyinosinic-polycytidylic acid [poly(I:C)]}, which robustly up-regulates PD-L1 (Fig. 5, D and E). We then confirmed the ability of the cocktail to up-regulate PD-L1 in murine KL cells isolated from normoglycemic NOD mice, thus generating pharmacologically modulated KL (pKL) cells (Fig. 5, F to I, and tables S5 and S6). We then analyzed the expression of costimulatory molecules and proinflammatory and anti-inflammatory cytokines by flow cytometry, which demonstrated up-regulation of interleukin-4 (IL-4), PD-1, CD80, CD86, and ICOSL in pKL cells as compared to unmodulated-KL (Vehicle-KL) cells, with CD40 comparatively down-regulated in pKL cells (fig. S3, A and B). We explored the immunoregulatory properties of pKL cells in an autoimmune setting in vitro. pKL cells generated from normoglycemic NOD mice were cocultured at ratios of 1:1, 1:5, and 1:10 to T cells with CD11c+ DCs and BDC2.5 Tg CD4+CD25− T cells in the presence of BDC2.5 peptide. Quantification by flow cytometry revealed a pronounced and significant decrease (P < 0.01) in the percentage of IFN-γ+CD4+ T cells when pKL cells were added to the assay as compared to when unmodulated KL cells were used (Fig. 5, J and K). Immunoregulation was determined to be PD-L1–dependent by preculturing pKL cells at a ratio of 1:1 to T cells with an anti–PD-L1–blocking mAb, which resulted in a marked reduction in the immunoregulatory effect (Fig. 5, J and K). These effects and PD-L1 dependency were confirmed in CD8-dependent (Fig. 5, M and N) and in non–autoimmune-specific anti-CD3/anti-CD28 (Fig. 5, P and Q) assays. Finally, we compared the immunosuppressive effects of PD-L1.Tg KL cells, pKL cells, and unmodulated KL cells (WT) with that of CD4+CD25+ regulatory T cells similarly obtained from normoglycemic NOD mice. In the three aforementioned CD4- and CD8-restricted autoimmune and anti-CD3/anti-CD28 assays, PD-L1.Tg KL and pKL cells exerted robust immunoregulatory properties almost comparable or higher to those obtained with freshly isolated CD4+CD25+ regulatory T cells from normoglycemic NOD (10 weeks old); this was particularly evident for PD-L1.Tg KL cells (Fig. 5, L, O, and R, and fig. S4, A to C). A less pronounced effect (but still significantly suppressive; P < 0.05) was observed when unmodulated KL cells (WT) were added to the assays. Pharmacologically modulated HSPCs are thus endowed with immunoregulatory properties and abrogate the autoimmune response in vitro.
Fig. 5. pKL cells abrogate the autoimmune response in vitro.
(A to C) Results of screening of small molecules tested for their ability to up-regulate PD-L1 [mean fluorescence intensity (MFI)] on mobilized CD34+ cells obtained from healthy controls, the three-color coding shown in (C) represents lowest PD-L1 MFI values (orange), median PD-L1 MFI values (yellow), and highest PD-L1 MFI values (green). TLR, toll-like receptor; wp, well plate. (D and E) PD-L1 expression (mRNA and MFI) fold change was quantified for each component of the small-molecule cocktail tested singularly or in combination. Tx, treatment. (F) Representative flow cytometric analysis and quantitative bar graph of PD-L1 expression on KL cells from NOD mice pre- and postpharmacological modulation with a cocktail of small molecules (n = 3 from two independent experiments), and statistical significance was performed by using twotailed unpaired t test. (G) Confocal imaging of KL cells pre- and postmodulation with cocktail of small molecules, showing DAPI (in blue) and PD-L1 (in red) staining. Histology magnification, ×63. Scale bar, 50 μm. (H and I) MA plot and fold change for gene expression in KL cells obtained from bone marrow of NOD mice and pKL as compared to unmodulated KL cells (Vehicle-KL cells, WT) n = 3 samples per condition, and statistical significance was performed using pairwise ANOVA test. (J and K) Representative flow cytometric analysis and quantitative bar graph for IFN-γ+CD4+ T cells isolated from NOD-BDC2.5 TCRTg mice and stimulated with BDC2.5 peptide in the presence of DCs (Controls) or upon coculture with unmodulated KL (WT), with pKL cells (at different ratios), or with pKL cells pretreated with anti–PD-L1–blocking mAb with the isotype control also shown; n = 3 samples per condition, and statistical significance was performed using two-tailed unpaired t test. #P < 0.05 versus all; §P < 0.05 versus all except Tg. 1:10. (L) Bar graph for flow cytometric quantification of IFN-γ+CD4+T cells after coculture of CD4+ T cells isolated from NOD-BDC2.5 TCR Tg mice stimulated with BDC2.5 peptide in the presence of DCs (control) or upon coculture with unmodulated KL cells (WT), with pKL cells, with PD-L1.Tg KL cells, or with CD4+CD25+ T regulatory cells; n = 4 samples per condition were used, and statistical significance was performed using two-tailed unpaired t test; #P < 0.05 versus all. (M and N) Representative flow cytometric analysis and quantitative bar graph for IFN-γ+CD8+ T cells isolated from NOD-8.3 TCR Tg mice and stimulated with IGRP peptide in the presence of DCs (control) or upon coculture with unmodulated KL cells (WT), with pKL cells (at different ratios), or with pKL cells pretreated with anti–PD-L1–blocking mAb. Experiments were run in triplicate (n = 3 for controls, all the rest n ≥ 5), and statistical analysis was performed using two-tailed unpaired t test. (O) Bar graph for flow cytometric quantification of IFN-γ+CD8+T cells after coculture of CD8+T cells isolated from NOD-8.35 TCR Tg mice stimulated by IGRP peptide in the presence of DCs (control) or upon coculture with unmodulated KL cells(WT), with pKL cells, with PD-L1.Tg KL cells, or with CD4+CD25+T regulatory cells. Experiments were run in triplicate (n = 4 for controls and WT, all the rest n = 3), and statistical analysis was performed using two-tailed unpaired t test (P and Q) Representative flow cytometric analysis and quantitative bar graph for IFN-γ+CD4+ T cells isolated from NOD mice and stimulated with soluble anti-CD3/anti-CD28 (control) or upon coculture with unmodulated KL cells (WT), with pKL cells (at different ratios), or with pKL cells pretreated with PD-L1 -blocking/-neutralizing mAb. Experiments were run in triplicate [n = 3 for all conditions except for anti–PD-Ll (n = 4)], and statistical analysis was performed using two-tailed unpaired t test; in (R): #P < 0.05 versus all except anti–PD-Ll–blocking mAb and pKL 1:10 (P = ns); §P < 0.05 versus all. (R) Bar graph depicting flow cytometric quantification of IFN-γ+CD4+ Tcells within CD4+CD25− T cells isolated from NOD mice and stimulated with soluble anti-CD3/anti-CD28 (control) or upon coculture with WT, with pKL, with PD-L1.Tg KL cells, or with CD4+CD25+ T regulatory cells. Experiments were run in triplicate [n = 3 for all conditions, except for WT (n = 4)], and statistical analysis was performed using two-tailed unpaired t test. Data are expressed as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.0001. #P < 0.05 versus all; §P < 0.05 versus all.
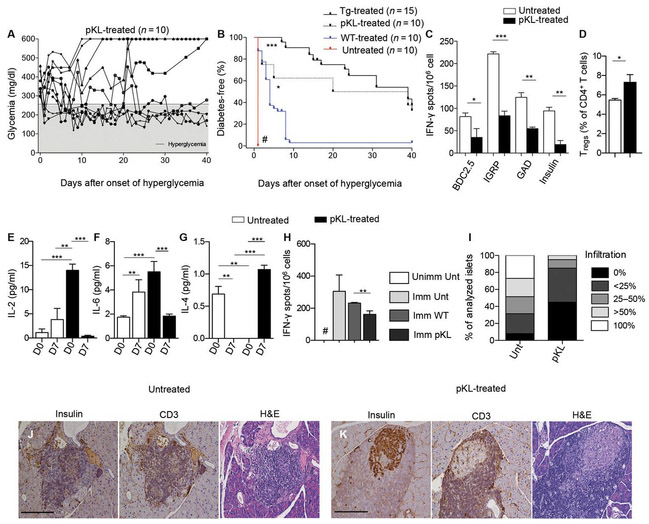
Pharmacologically modulated HSPCs revert hyperglycemia in vivo
To evaluate the effect of pKL cells in vivo, newly hyperglycemic NOD mice were adoptively transferred with 3 × 106 pKL cells (Fig. 6A). pKL cells successfully reverted diabetes in 40% of treated newly hyperglycemic NOD mice, with 30% remaining normoglycemic until the completion of the study at day 40. Kaplan-Meier analysis showed a greater effect of PD-L1.Tg KL cells in reverting hyperglycemia in NOD mice, as compared to pKL cells (Fig. 6B). Quantification of IFN-γ-producing cells in an ex vivo assay where splenocytes were challenged with islet peptides at day 40 (BDC2.5, IGRP, GAD-65, and insulin) revealed a reduction in IFN-γ-producing cells in pKL cell–treated hyperglycemic NOD mice (Fig. 6C). Immunophenotyping of treated NOD mice at day 40 after treatment showed a marked increase in FoxP3+ regulatory CD4+ T cells (Fig. 6D). A reduction in the peripheral levels of proinflammatory (IL-2/IL-6; Fig. 6, E and F) and an increase in anti-inflammatory (IL-4; Fig. 6G) cytokines were evident in pKL cell–treated NOD mice. We then evaluated whether immunocompetence was maintained during treatment of NOD mice with pKL cells, as we did when using PD-L1.Tg KL cells. pKL cell–treated NOD mice were capable of mounting a regular immune response to OVA once immunized and rechallenged in vitro with OVA similar to untreated NOD mice and were thus immunocompetent (Fig. 6H). Pathology of the pancreas of pKL cell–treated NOD mice revealed mild infiltration of the islets, with preserved insulin staining and reduced insulitis score as compared to untreated hyperglycemic NOD mice (Fig. 6, I to K). Finally, we tested the effects of a PD-1 signaling mAb capable of mimicking PD-L1/PD-1 cross-linking. PD-1 mAb treatment did not delay diabetes onset in prediabetic NOD mice nor did it revert diabetes in newly hyperglycemic NOD mice (fig. S4, D and E). It is possible that the mAb used was unable to reach the inflamed islets within the pancreas, which conversely could be accessed by modified HSPCs because of their CXCR4 expression, which allows the trafficking of HSPCs into inflamed areas that release high levels of CXCL12.
Fig. 6. pKL cells revert hyperglycemia in NOD mice in vivo.
(A and B) Newly hyperglycemic NOD mice were treated with WT cells, pKL cells, or Tg cells. Data are representative of 10 untreated mice, 10 mice treated with WT cells, 10 mice treated with pKL cells, and 15 mice treated with Tg cells. Incidence of diabetes in all groups of NOD mice (untreated, WT-treated, pKL-treated, and Tg-treated mice) was compared using the log-rank (Mantel-Cox) test;#P <0.0001 versus all, P < 0.0001 for Tg versus WT, and P <0.05 for pKL versus WT. (C) Quantification of IFN-γ–producing cells in an in vitro assay, in which splenocytes isolated from newly hyperglycemic pKL-treated NOD mice or in untreated hyperglycemic NOD mice 40 days after treatment were challenged with islet peptides after treatment with pKL, normalized for background. Experiments were run in triplicate, and statistical analysis was performed using two-tailed unpaired t test. (D) Immunophenotype of lymphocytes isolated from spleen of pKL-treated newly hyperglycemic NOD mice showed an increase in the percentage of FoxP3+ regulatory T cells. Experiments were run in triplicate (n = 3 pKL-treatedand n = 5 untreated), and statistical analysis was performed using two-tailed unpaired t test. (E to G) Bar graph showing the levels of pro- (IL-2/IL-6) and anti-inflammatory (IL-4) cytokines as measured by Luminex in the serum of untreated or pKL-treated newly hyperglycemic NOD mice at baseline and at 7 days after treatment. Experiments were run in triplicate per group, and condition (D0 and D7) and statistical analysis were performed using two-tailed unpaired t test. (H) OVA rechallenge. Experiments were run in triplicate per group, and statistical analysis were performed using two-tailed unpaired t test, #P < 0.05 versus all. (I) Insulitis score in untreated and pKL-treated newly hyperglycemic NOD mice. (J and K) Representative immunohistochemical H&E analysis and CD3/insulin staining in serial pancreatic islet tissue sections from pKL-treated or untreated NOD mice; n = 9 sections per group were analyzed. Histology magnification, ×20. Scale bars, 200 μm. Data are expressed as means ± SEM. Data are representative of at least n = 3 mice. *P < 0.05; **P < 0.01; ***P < 0.0001; #P < 0.05 versus all.
The PD-L1 defect is evident in human HSPCs from T1D patients and is associated with an altered miRNA network, and pharmacologically modulated HSPCs reinstate PD-L1 expression and immunoregulatory properties
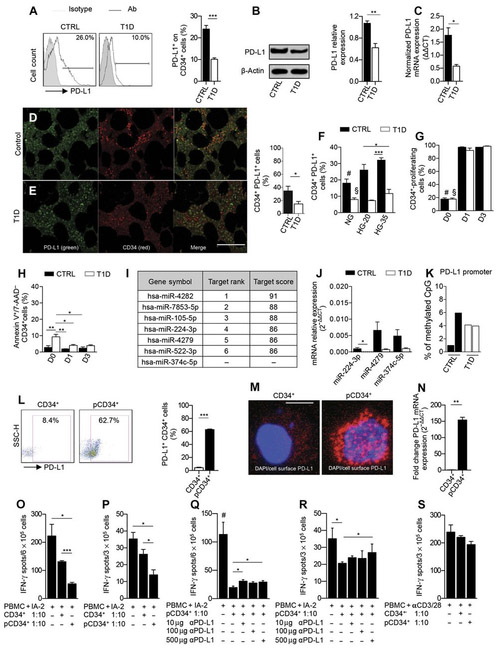
To assess whether patients with T1D displayed defects in HSPCs similar to those observed in our preclinical models, PD-L1 expression was analyzed on CD34+ cells isolated from peripheral blood (table S7). In line with our findings in NOD mice, fewer PD-L1+CD34+ cells were detectable in T1D patients as compared to healthy controls (Fig. 7A). The defect was evident in newly diagnosed T1D patients as well, when it is unlikely there is any effect of high glucose (fig. S5A). Western blot and PCR analysis confirmed reduced PD-L1 protein and mRNA expression in CD34+ cells obtained from T1D patients as compared to those obtained from healthy controls (Fig. 7, B and C). Other relevant immune cells were not deficient in PD-L1 (fig. S5, B to D), thus confirming that the PD-L1 defect is mainly restrained to HSPCs in T1D patients. PD-L2 was expressed on CD34+ cells from T1D patients at higher levels as compared to controls; however, PD-L2 has been described to be less relevant for T1D onset as shown by the lack of effect when knocking down PD-L2 in NOD mice (fig. S5, E to G) (9). PD-1 expression was significantly slightly reduced (P < 0.05) by MFI in T1D patients compared to healthy controls (fig. S5, H to J). Finally, we confirmed by confocal imaging a reduced number ofPD-L1+CD34+ cells in the bone marrow of T1D patients as compared to healthy controls (Fig. 7, D and E). Our data confirmed the existence of a defect in PD-L1 expression in HSPCs of T1D patients. We analyzed the effect of plerixafor-mediated mobilization, used in clinic (8, 12), on PD-L1 expression in CD34+ cells obtained from five T1D patients and eight healthy controls (table S8). Whereas the percentage and absolute number of CD34+ cells significantly increased (P < 0.05), the percentage of PD-L1+CD34+ cells remained stable in T1D patients but decreased after mobilization in healthy controls, highlighting that CD34+ cell mobilization may have failed in previous clinical trials because of PD-L1 down-regulation (fig. S8, A to C).
Fig. 7. The PD-L1 defect is evident in HSPCs from T1D patients.
(A) Representative flow cytometric and quantitative bar graph of PD-L1+CD34+ cells from patients with T1D as compared to healthy controls (n = 10 from each group), and statistical significance was performed by using two-tailed unpaired t test. (B) Western blot analysis and (C) qRT-PCR confirmed the PD-L1 defect in CD34+ cells from T1D patients (n = 3 in each group), and statistical significance was performed by using two-tailed unpaired t test. (D and E) Confocal imaging and quantitative bar graph of bone marrow sections obtained from T1D patients and healthy controls showing PD-L1 (green) and CD34 (red) staining; the quantification of the orange-stained bone marrow element was performed by ImageJ. Data are representative of n = 5 sections per group, and statistical significance was performed using two-tailed unpaired t test with Welch’s correction. Histology magnification, ×63. Scale bar, 40 μm. (F) Bar graph depicting the percentage of PD-L1 on CD34+ cells obtained from peripheral blood ofT1D patients or healthy controls at baseline or cultured for 3 days in normal glucose, in 20 mM, or in 35 mM high glucose (n = 3 samples from each group), and statistical significance was performed using two-tailed unpaired t test; #P < 0.05 versus all except T1D HG-35 and CTRL HG-20; §P < 0.05 versus all except T1D HG-35 (ns) and T1D HG-20 (ns). (G) CFSE-based proliferation assay of peripheral CD34+ cells obtained from T1D and healthy control patients at baseline and after 1 and 3 days of culture (n = 3 samples from each group), and statistical significance was performed using two-tailed unpaired t test; #P < 0.0001 versus all except T1D-D0; §P < 0.0001 versus all except CTRL-D0 (ns). (H) Frequency of apoptosis of CD34+ cells obtained from T1D and healthy control patients at baseline and after 1 and 3 days of culture. Experiments were run in triplicate, and statistical significance was performed using two-tailed unpaired t test. (I) Table of human miRNAs, discovered by bioinformatic approach, involved in the regulation of PD-L1 expression. (J) qRT-PCR showed differentially expressed miRNA in human CD34+ cells obtained from T1D patients as compared to controls (at least n = 5 samples from each group), and statistical significance was performed using two-tailed unpaired t test with Welch’s correction. (K) DNA methylation status of the PD-L1 gene promoter in peripheral CD34+ cells obtained from T1D patients as compared to healthy controls (n = 2 samples from each group). Statistical significance was performed using two-tailed unpaired t test with Welch’s correction. (L) Representative flow cytometric and quantitative bar graph of PD-L1 expression on peripheral CD34+ cells from T1D patients pre- and postpharmacological modulation with a cocktail of small molecules. Experiments were run in triplicate, and statistical significance was performed using two-tailed unpaired t test. (M) Confocal imaging of PD-L1 expression on CD34+ cells from T1D patients pre- and postpharmacological modulation. Histology magnification, ×63. Scale bar, 50 μm. (N) PD-L1 expression fold change in pCD34+ after 24 hours and in vehicle-treated CD34+ cells as assessed by RT-PCR. Experiments were run in duplicate, and statistical significance was performed using two-tailed unpaired t test. (O to R) Quantification of IFN-γ–producing cells where human PBMCs from T1D were challenged with IA-2 in the presence of unmodulated CD34+ or pCD34+ cells with or without an anti–PD-L1–blocking mAb. Experiments were run in triplicate, and statistical significance was performed using two-tailed unpaired t test. Experiments were performed at least in triplicate; in (R): Data related to anti–PD-L1 treatment were performed in duplicate, and statistical significance was performed using two-tailed unpaired t test. (S) Quantification of IFN-γ–producing cells where human PBMCs from T1D already stimulated with anti-CD3/anti-CD28 were cocultured in the presence of unmodulated CD34+ or pCD34+ cells with or without an anti–PD-L1–blocking mAb. Experiments were run in duplicate, and statistical significance was performed using two-tailed unpaired t test. Data are expressed as means ± SEM. *P < 0.05; **P <0.01; ***P < 0.001. #P < 0.05 versus all.
To understand the immunological basis of the PD-L1 defect in human HSPCs, we performed in vitro experiments similar to those we performed in mice. We did not find any potential high glucose–associated effect on PD-L1 expression on CD34+ cells or only small differences, if any, in the proliferation and apoptosis rate in CD34+ cells obtained from T1D patients and controls (Fig. 7, F to H). Bioinformatic analysis of miRNAs showed a number of miRNA species involved in controlling PD-L1 expression (Fig. 7I). qRT-PCR analysis of several relevant miRNAs confirmed that a number of miRNA species were differentially expressed in human CD34+ cells obtained from T1D patients as compared to controls (Fig. 7J), with no differences observed in the methylation status of the promoter of the PD-L1 gene (Fig. 7K). In line with preclinical findings, an altered miRNA network is evident in HSPCs of T1D patients.
We tested the effect of overcoming PD-L1 deficiency in human HSPCs by using the same cocktail of small molecules tested in NOD mice. As shown by flow cytometric analysis, confocal imaging, and qRT-PCR, modulation of CD34+ cells with a cocktail of three agents [IFN-β, IFN-γ, and poly(I:C)], up-regulated PD-L1 expression in human CD34+ cells obtained from T1D patients [pharmacologically modulated CD34+ (pCD34+)] as compared to unmodulated CD34+ cells (Fig. 7, L to N). Immunophenotyping and transcriptome profiling of pCD34+ cells (figs. S6, A and B, and S7, A and B, and table S9) confirmed specific PD-L1 up-regulation in pCD34+ cells as compared to unmodulated CD34+ cells. To study the ex vivo immunoregulatory functions of pCD34+, CD34-depleted peripheral blood mononuclear cells (PBMCs) were cocultured with unmodulated CD34+ or pCD34+ cells in the presence of insulin-associated autoantigen 2 (I-A2), and the number of IFN-γ-producing cells was quantified in an ELISPot assay. The addition of unmodulated CD34+ or pCD34+ resulted in a significant decrease (P < 0.05) in the number of IFN-γ-producing cells as compared to those obtained when PBMCs were cultured without unmodulated CD34+/pCD34+ cells in the presence of the islet peptide IA-2 (Fig. 7, O and P). The suppression was more pronounced when pCD34+ cells were added, suggesting that pCD34+ cells are endowed with greater immunoregulatory activity than unmodulated CD34+ cells (Fig. 7, O and P). To further confirm that the immunosuppressive effect exerted by pCD34+ cells was primarily due to PD-L1 expression, we precultured pCD34+ cells in the presence of an anti–PD-L1–blocking mAb and then tested them in the autoimmune assay. Ab-mediated PD-L1 blockade hampered the immunoregulatory effect exerted by pCD34+ cells (Fig. 7, Q and R). Surprisingly, the effects of pCD34+ were not confirmed in a non–autoimmune-specific anti-CD3/anti-CD28 assay (Fig. 7S). Overall, our data confirmed that pCD34+ cells are endowed with PD-L1–dependent immunoregulatory properties.
DISCUSSION
T1D is regarded as one of the most aggressive autoimmune diseases and requires lifelong exogenous insulin administration. Efforts to halt β cell decline or stall chronic complications are ongoing (13–15); however, the immunotherapies tested thus far have failed, mostly because of their lack of specificity as well as the fact that they are usually simply adopted from other settings (for example, kidney transplantation) (16–18). The need for more T1D-tailored therapies led us to explore the existence of immune checkpoint abnormalities that may be relevant to the disease. Various pieces of evidence led us to hypothesize that an HSPC-specific PD-L1 defect may be involved in the onset of T1D and that the resolution of this defect may provide a cure for the disease. First of all, the expansion and reinfusion of autologous HSPCs were the most potent therapy in reverting hyperglycemia in T1D patients (7); second, there is a strong link between the PD-L1 defect and T1D (9, 19); and finally, PD-L1 is a key player in HSPC immunobiology, such that the lack of PD-L1 reduces the ability of HSPCs to abrogate the immune response (8). Whereas immunosuppressant treatment alone [that is, antithymocyte globulin (ATG)] failed to preserve β cell function in recent onset T1D, HSPCs plus immunosuppressant were successful in the Voltarelli trial (20). This result suggests either that there is a synergistic effect between HSPCs and immunosuppression or that a defect prevents HSPCs from being fully effective in their suppression. Transcriptomic profiling, flow cytometric analysis, RT-PCR, and direct analysis of bone marrow showed a reduction in PD-L1 expression in HSPCs in both NOD mice and T1D patients. Whereas high glucose, altered HSPC survival, or epigenetic abnormalities cannot account for the impaired PD-L1 expression, gene expression profiling unveiled abnormalities in the HSPC miRNA network in T1D that may be responsible for the PD-L1 defect. Therefore, we developed a genetic approach to overcome the PD-L1 defect and generated PD-L1.Tg HSPCs, which successfully abrogated the autoimmune response in vitro in a PD-L1–dependent manner. Notably, our PD-L1.Tg HSPCs successfully converted all treated hyperglycemic NOD mice to normoglycemia, with suppression of the autoimmune response. Tracking studies suggested that PD-L1.Tg HSPCs preferentially homed to the inflamed pancreas, most likely because of substantial CXCR4 expression, which is in line with the CXCL12 shown to be released by inflamed pancreatic islets (21). The persistence of a regular immune response against OVA, but not against islet peptides, suggests both the maintenance of immunocompetence and the idea that autoreactive T cells only are strictly dependent from the PD-L1/PD-1 pathway for tolerance/anergy induction.
Once in the pancreas, PD-L1.Tg HSPCs may induce cell death of autoreactive T cells or may just render them unresponsive. The recent progress in the field of gene therapy (22) provides a basis for the potential use of the aforementioned genetic approach in T1D as well. Clinically relevant, our pharmacologically modulated HSPCs also exhibited immunoregulatory effects because they markedly abrogated CD4- and CD8-restricted autoimmune responses in vitro and reverted diabetes in nearly 40% of newly hyperglycemic NOD mice. The human data parallel the preclinical findings, confirming the presence of the PD-L1 defect in human CD34+ cells. Our results have two major implications. First, we may have identified a path involved in the onset of T1D, and the PD-L1 defect in HSPCs may have a permissive role on the generation of autoreactive T cells (23). We can now propose a working hypothesis, in which an altered network of miRNAs is responsible for the PD-L1 defect in HSPCs in T1D. The genetic and pharmacological restoration of this PD-L1 defect in HSPCs in T1D generates a new pool of PD-L1+ HSPCs, which, once adoptively transferred, traffic to the pancreas, and they may be able to either delete or to render unresponsive autoreactive T cells via a PD-L1/PD-1–dependent mechanism, thus reverting hyperglycemia (fig. S8D). However, it is also possible that other HSPC-derived cells may abrogate the autoimmune response in the pancreas.
Our study thus provides key insight into the potential role of miRNAs in the regulation of PD-L1 expression of HSPCs and potentially of T1D pathogenesis. Second, expression of PD-L1 in HSPCs may be used as a tool for targeted immunotherapy in T1D, which appears more efficacious than mAbs in animal models and also appears to be safe (24, 25).
MATERIALS AND METHODS
Study design
The objective of our study was to demonstrate the existence of a defect in PD-L1 expression on HSPCs in NOD mice and T1D individuals. We then sought to use genetic engineering/pharmacological modulation to restore/up-regulate PD-L1 defect in HSPC and study their effect on autoimmune diabetes. NOD mice were treated and followed until day 40 or earlier to avoid the wasting syndrome. Between 10 and 15 NOD mice were allocated in the different treatment groups randomly with our NOD colony exhibiting a consistent diabetes penetrance (that is, 80% at 25 weeks) (2, 3). Sample analysis and pathology were blinded. Primary data were reported in table S10.
Human studies
T1D patients and healthy patients matched for age and gender were enrolled (table S7). This study was conducted in accordance with Institutional Review Board approval (BCH 3851).
In vitro human studies
Isolated human CD34+ hematopoietic stem cells were stimulated for 24 hours with human IFN-β (hIFN-β), hIFN-γ, and poly(I:C). PD-L1 expression was evaluated before and after culture by different techniques (qRT-PCR, FACS, and confocal imaging) (26). PBMCs isolated from T1D patients were cultured for 2 days in the presence of I-A2 peptide. Cells were plated with or without CD34+ or pCD34+ cells. hIFN-γ spots were counted using an ELISpot reader.
Animal studies
Animal studies were conducted in NOD and C57BL/6 mice; all the mice were used according to institutional guidelines, and animal protocol was approved by the Boston Children’s Hospital Institutional Animal Care and Use Committee. A complete description of the different murine strains used is provided in the Supplementary Materials.
In vitro murine studies
Murine bone marrow KL cells were transduced with PD-L1 lentivirus, and 24 hours after transduction PD-L1 expression was evaluated by multiple techniques (qRT-PCR, FACS, and confocal imaging). In vitro assays were performed by coculturing KL–PD-L1.Tg KL cells, unmodulated KL cells, or pKL with CD4+CD25−/CD8+ T cells extracted from splenocytes of NOD-BDC2.5 TCR Tg mice or 8.3 TCR Tg NOD mice in the presence of islet mimotope peptides.
In vivo interventional murine studies
Newly diabetic NOD mice were treated with PD-L1.Tg KL cells, unmodulated KL cells, or pKL, and glycemia was monitored daily (27). Mechanistic studies were conducted on different groups of treated NOD mice and compared to untreated NOD mice (ELISPot, flow cytometry, Luminex).
Statistical analysis
Unless otherwise indicated, all data, including human data, were shown as means ± SEM. Statistical analysis was performed using unpaired Student’s t test. A two-sided value of P ≤ 0.05 was considered statistically significant. Kaplan-Meier curve analysis with Wilcoxon test was used to analyze the development of diabetes in mice. For multiple comparisons, one-way ANOVA followed by Bonferroni posttest between the group of interests and all other groups was used. For GWAS, data were presented as Robust Multi-array Average normalized intensities, and statistical analysis was performed using pairwise ANOVA test. Two-tailed unpaired t test or two-tailed unpaired t test with Welch’s correction (if applicable) was used for comparison between C57BL/6 and NOD mice. One-way ANOVA followed by Bonferroni multiple comparison test was used for group comparison between C57BL/6 mice (used as control) and NOD or NOR mice or DBA/1J or BALB/C and for comparison between healthy control, long-standing T1D, and new-onset T1D. For transcriptomic analysis, statistical analysis was performed using the software available (RT2 profiler PCR Array Data Analysis, Qiagen). For GWAS, statistical analysis was performed using pairwise ANOVA test. Diabetes incidence among different groups of NOD mice (untreated, WT-treated, pKL-treated, and Tg-treated mice) was analyzed with the log-rank (Mantel-Cox) test. All mechanistic and in vitro studies were performed in triplicate (unless otherwise indicated), and the statistical analyses used are reported in the figure legends. All graphs and statistical tests were generated using GraphPad Prism software version 5.0b (GraphPad Software Inc.) and were performed at the 5% significance level. More detailed Materials and Methods are available in the Supplementary Materials.
Supplementary Material
PD-L1 expression in non-HSPCs.
Generation of PD-L1.Tg KL cells and their tracking.
Immunophenotype of pKL cells.
Anti–PD-1 studies.
CD34+ cells characterization in T1D patients and in healthy controls.
Immunophenotype of pCD34+.
Transcriptome of pCD34+.
CD34+ cell mobilization with plerixafor and working hypothesis.
Transcriptomic profiling of murine KLS cells (provided as an Excel file).
Genome-wide expression analysis of murine KLS cells (provided as an Excel file).
Genome-wide expression analysis of Tg KL cells.
Chemokine receptors expression in different groups of KL cells.
Genome-wide expression analysis of pKL cells: up-regulated genes (provided as an Excel file).
Genome-wide expression analysis of pKL cells: down-regulated genes (provided as an Excel file).
Characteristics of patients enrolled in the study.
Characteristics of patients enrolled in the plerixafor mobilization study.
Transcriptome of pCD34+ cells (provided as an Excel file).
Primary data (provided as an Excel file).
Acknowledgments:
We thank M.Jurewicz for editing. We thank the Fondazione Romeo ed Enrica Invernizzi for its support.
Funding: P.F. is supported by European Foundation for the Study of Diabetes/Sanofi European Research Programme, an American Heart Association Grant-in-Aid, and Fate Therapeutic research grant.
Footnotes
Competing interests: P.F. is the inventor on patent application (PCT/US2016/043053) submitted by Boston Children’s Hospital that covers the immunoregulatory properties of HSPCs. All other authors declare that they have no competing interests.
Data and materials availability: The data for this study have been deposited in the database dbGaP (database of Genotypes and Phenotypes).
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/9/416/eaam7543/DC1
Materials and Methods
REFERENCES AND NOTES
- 1.Bluestone JA, Herold K, Eisenbarth G, Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 464, 1293–1300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiorina P, Jurewicz M, Augello A, Vergani A, Dada S, La Rosa S, Selig M, Godwin J, Law K, Placidi C, Smith RN, Capella C, Rodig S, Adra CN, Atkinson M, Sayegh MH, Abdi R, Immunomodulatory function of bone marrow-derived mesenchymal stem cells in experimental autoimmune type 1 diabetes. J. Immunol 183, 993–1004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiorina P, Vergani A, Dada S, Jurewicz M, Wong M, Law K, Wu E, Tian Z, Abdi R, Guleria I, Rodig S, Dunussi-Joannopoulos K, Bluestone J, Sayegh MH, Targeting CD22 reprograms B-cells and reverses autoimmune diabetes. Diabetes 57, 3013–3024 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vergani A, D’Addio F, Jurewicz M, Petrelli A, Watanabe T, Liu K, Law K, Schuetz C, Carvello M, Orsenigo E, Deng S, Rodig SJ, Ansari JM, Staudacher C, Abdi R, Williams J, Markmann J, Atkinson M, Sayegh MH, Fiorina P, A novel clinically relevant strategy to abrogate autoimmunity and regulate alloimmunity in NOD mice. Diabetes 59, 2253–2264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Nasr M, D’Addio F, Usuelli V, Tezza S, Abdi R, Fiorina P, The rise, fall, and resurgence of immunotherapy in type 1 diabetes. Pharmacol. Res 98, 31–38 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Couri CE, Oliveira MCB, Stracieri ABPL, Moraes DA, Pieroni F, Barros GMN, Madeira MIA, Malmegrim KCR, Foss-Freitas MC, Simoes BP, Martinez EZ, Foss MC, Burt RK, Voltarelli JC, C-peptide levels and insulin independence following autologous nonmyeloablative hematopoietic stem cell transplantation in newly diagnosed type 1 diabetes mellitus. JAMA 301, 1573–1579 (2009). [DOI] [PubMed] [Google Scholar]
- 7.D’Addio F, Valderrama Vasquez A, Ben Nasr M, Franek E, Zhu D, Li L, Ning G, Snarski E, Fiorina P, Autologous nonmyeloablative hematopoietic stem cell transplantation in new-onset type 1 diabetes: A multicenter analysis. Diabetes 63, 3041–3046 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Fiorina P, Jurewicz M, Vergani A, Petrelli A, Carvello M, D’Addio F, Godwin JG, Law K, Wu E, Tian Z, Thoma G, Kovarik J, La Rosa S, Capella C, Rodig S, Zerwes Hans-Guenter, Sayegh MH, Abdi R, Targeting the CXCR4-CXCL12 axis mobilizes autologous hematopoietic stem cells and prolongs islet allograft survival via programmed death ligand 1. J. Immunol 186, 121–131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansari MJ, Salama AD, Chitnis T, Smith RN, Yagita H, Akiba H, Yamazaki T, Azuma M, Iwai H, Khoury SJ, Auchincloss H Jr., Sayegh MH, The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J. Exp. Med 198, 63–69 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, Saito T, Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med 209, 1201–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunting KD, Qu C-K, The hematopoietic stem cell landscape. Methods Mol. Biol 1185, 3–6 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Scandling JD, Busque S, Shizuru JA, Lowsky R, Hoppe R, Dejbakhsh-Jones S, Jensen K, Shori A, Strober JA, Lavori P, Turnbull BB, Engleman EG, Strober S, Chimerism, graft survival, and withdrawal of immunosuppressive drugs in HLA matched and mismatched patients after living donor kidney and hematopoietic cell transplantation. Am. J. Transplant 15, 695–704 (2015). [DOI] [PubMed] [Google Scholar]
- 13.von Herrath MG, Korsgren O, Atkinson MA, Factors impeding the discovery of an intervention-based treatment for type 1 diabetes. Clin. Exp. Immunol 183, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Control Diabetes and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group, Lachin JM, Genuth S, Cleary P, Davis MD, Nathan DM, Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. N. Engl. J. Med 342, 381–389 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson MA, von Herrath M, Powers AC, Clare-Salzler M, Current concepts on the pathogenesis of type 1 diabetes—Considerations for attempts to prevent and reverse the disease. Diabetes Care 38, 979–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitelman SE, Gottlieb PA, Rigby MR, Felner EI, Willi SM, Fisher LK, Moran A, Gottschalk M, Moore WV, Pinckney A, Keyes-Elstein L, Aggarwal S, Phippard D, Sayre PH, Ding L, Bluestone JA, Ehlers MR; START Study Team, Antithymocyte globulin treatment for patients with recent-onset type 1 diabetes: 12-month results of a randomised, placebo-controlled, phase 2 trial. Lancet Diabetes Endocrinol 1, 306–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold KC, Gitelman SE, Ehlers MR, Gottlieb PA, Greenbaum CJ, Hagopian W, Boyle KD, Keyes-Elstein L, Aggarwal S, Phippard D, Sayre PH, McNamara J, Bluestone JA; AbATE Study Team, Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: Metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes 62, 3766–3774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, Becker DJ, Gitelman SE, Goland R, Gottlieb PA, Marks JB, McGee PF, Moran AM, Raskin P, Rodriguez H, Schatz A, Wherrett D, Wilson DM, Lachin JM, Skyler JS; Type 1 Diabetes TrialNet Anti-CD20 Study Group, Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N. Engl. J. Med 361, 2143–2152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guleria I, Gubbels Bupp M, Dada S, Fife B, Tang Q, Ansari MJ, Trikudanathan S, Vadivel N, Fiorina P, Yagita H, Azuma M, Atkinson M, Bluestone JA, Sayegh MH, Mechanisms of PDL1-mediated regulation of autoimmune diabetes. Clin. Immunol 125, 16–25 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Voltarelli JC, Couri CEB, Stracieri ABPL, Oliveira MC, Moraes DA, Pieroni F, Coutinho M, Malmegrim KCR, Foss-Freitas MC, Simões BP, Foss MC, Squiers E, Burt RK, Autologous nonmyeloablative hematopoietic stem cell transplantation in newly diagnosed type 1 diabetes mellitus. JAMA 297, 1568–1576 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Cowley MJ, Weinberg A, Zammit NW, Walters SN, Hawthorne WJ, Loudovaris T, Thomas H, Kay T, Gunton JE, Alexander SI, Kaplan W, Chapman J, O’Connell PJ, Grey ST, Human islets express a marked proinflammatory molecular signature prior to transplantation. Cell Transplant 21, 2063–2078 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Sessa M, Lorioli L, Fumagalli F, Acquati S, Redaelli D, Baldoli C, Canale S, Lopez D, Morena F, Calabria A, Fiori R, Silvani P, Rancoita PMV, Gabaldo M, Benedicenti F, Antonioli G, Assanelli A, Cicalese MP, Del Carro U, Sora MG, Martino S, Quattrini A, Montini E, Di Serio C, Ciceri F, Roncarolo MG, Aiuti A, Naldini L, Biffi A, Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: An ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet 388, 476–487 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Riella LV, Chock S, Liu T, Zhao X, Yuan X, Paterson AM, Watanabe T, Vanguri V, Yagita H, Azuma M, Blazar BR, Freeman GJ, Rodig SJ, Sharpe AH, Chandraker A, Sayegh MH, The novel costimulatory programmed death ligand 1/B7.1 pathway is functional in inhibiting alloimmune responses in vivo. J. Immunol 187,1113–1119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haller MJ, Wasserfall CH, McGrail KM, Cintron M, Brusko TM, Wingard JR, Kelly SS, Shuster JJ, Atkinson MA, Schatz DA, Autologous umbilical cord blood transfusion in very young children with type 1 diabetes. Diabetes Care 32, 2041–2046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiorina P, Voltarelli J, Zavazava N, Immunological applications of stem cells in type 1 diabetes. Endocr. Rev 32, 725–754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Addio F, La Rosa S, Maestroni A, Jung P, Orsenigo E, Ben Nasr M, Tezza S, Bassi R, Finzi G, Marando A, Vergani A, Frego R, Albarello L, Andolfo A, Manuguerra R, Viale E, Staudacher C, Corradi D, Batlle E, Breault D, Secchi A, Folli F, Fiorina P, Circulating IGF-I and IGFBP3 levels control human colonic stem cell function and are disrupted in diabetic enteropathy. Cell Stem Cell 17, 486–498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill RG, Pagni PP, Kupfer T, Wasserfall CH, Deng S, Posgai A, Manenkova Y, Bel Hani A, Straub L, Bernstein P, Atkinson MA, Herold KC, von Herrath M, Staeva T, Ehlers MR, Nepom GT, A preclinical consortium approach for assessing the efficacy of combined anti-CD3 plus IL-1 Blockade in reversing new-onset autoimmune diabetes in NOD mice. Diabetes 65, 1310–1316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PD-L1 expression in non-HSPCs.
Generation of PD-L1.Tg KL cells and their tracking.
Immunophenotype of pKL cells.
Anti–PD-1 studies.
CD34+ cells characterization in T1D patients and in healthy controls.
Immunophenotype of pCD34+.
Transcriptome of pCD34+.
CD34+ cell mobilization with plerixafor and working hypothesis.
Transcriptomic profiling of murine KLS cells (provided as an Excel file).
Genome-wide expression analysis of murine KLS cells (provided as an Excel file).
Genome-wide expression analysis of Tg KL cells.
Chemokine receptors expression in different groups of KL cells.
Genome-wide expression analysis of pKL cells: up-regulated genes (provided as an Excel file).
Genome-wide expression analysis of pKL cells: down-regulated genes (provided as an Excel file).
Characteristics of patients enrolled in the study.
Characteristics of patients enrolled in the plerixafor mobilization study.
Transcriptome of pCD34+ cells (provided as an Excel file).
Primary data (provided as an Excel file).