Abstract
Objective:
Association of multiple polymorphic variants with cervical cancer has been elucidated by several candidate gene based as well as genome-wide association studies. However, contradictory outcomes of those studies have failed to estimate the true effect of the polymorphic variants on cervical cancer.
Methods:
Literature mining of the PubMed database was done to gather all the publications related to genetic association with cervical cancer in India. Out of 98 PubMed hits only 29 genetic association studies were selected for meta-analysis based on specific inclusion criteria. A fixed-effect meta-analysis was performed to evaluate the overall association of the genetic polymorphisms with cervical cancer. Cochran’s Q test was performed to assess between study heterogeneity. Publication bias was also estimated by funnel plots and Egger’s regression test. Further, sub-group analysis was conducted by fixed-effect meta-regression to assess the impact of polymorphisms on cervical cancer in the presence of Human Papilloma Virus (HPV).
Result:
Following a fixed-effect model, meta-analysis was conducted that revealed 2 polymorphic variants viz. ‘deletion polymorphism (Del2) (OR=1.79, 95% CI= 1.08-2.95, P=0.023) in GSTM1’ and ‘rs1048943 (OR = 2.34, 95% CI=1.37-3.99, P=0.0018) in CYP1A1’ to be associated with cervical cancer. However, multiple testing correction showed only rs1048943 of CYP1A1 to be significantly associated (P-value=0.029) with cervical cancer with significant publication bias (P-value=0.0113) as estimated by Egger’s regression test. The polymorphic variants ‘rs1801131’, ‘rs1801133’, ‘rs2430561’, ‘rs1799782’, ‘rs25486’ and ‘rs25487’ showed significant (p<0.05) evidence of heterogeneity between studies by Cochran’s Q test and also by heterogeneity index (I2) calculation.
Conclusion:
Therefore, our study revealed significant association of rs1048943 in CYP1A1, but a nominal association of deletion polymorphism (Del2) in GSTM1 with cervical cancer, which provides a comprehensive insight on the true effect of the polymorphisms, reported in various case-control studies, on the risk of the development of cervical cancer in Indian women.
Keywords: Cervical cancer, meta-analysis, polymorphism, HPV, logistic regression
Introduction
Cervical cancer is the fourth most prevalent cancer in women worldwide with nearly 85% of reported incidences in the lesser socioeconomically developed regions of the world (Ferlay et al., 2015). Highest incidence and mortality rates of cervical cancer are reported from sub-Saharan Africa, Central and South America, South-eastern Asia, Central and Eastern Europe (Ferlay et al., 2015). A statistically significant decline in the incidence of cervical cancer in India is evident from long-term hospital based patient registries (Sreedevi et al., 2015; Yeole et al., 2004). However, India still shares 25% of global incidences with highest age standardized occurrence in South Asia. In India, nearly122,844 women are diagnosed with cervical cancer and 67,477 die from the disease every year (Sreedevi et al., 2015) ranking second in cancer related deaths in women (International Collaboration of Epidemiological Studies of Cervical, 2006). Aizawl district of Mizoram, a north-eastern state of India, accounts for the highest reported incidences of cervical cancer followed by Barshi and Bengaluru (Sreedevi et al., 2015).
Cancer of the cervix is generally classified into two broad histotypes based on their site of origin, viz. squamous cell carcinoma from ectocervix and adenocarcinoma from endocervix (Sreedevi et al., 2015). Several epidemiological and clinical reports states the development of cervical cancer to be multifactorial where human papillomavirus (HPV) infection is considered to be a major player in the etiology of cervical cancer (Bruni et al., 2017; Vaccarella et al., 2006) with evidences of also being a risk factor for other anogenital cancers, such as, anal cancer, vulval cancer, vaginal cancer, penile cancer and head and neck cancers (Bruni et al., 2017). Nearly 70% of all reported cervical cancer cases worldwide are caused mainly by HPV serotypes 16 and 18 (Bruni et al., 2017; Khaliq et al., 2012). Apart from HPV infection, other factors like smoking (Vaccarella et al., 2006; Ylitalo et al., 1999), immunosuppression (International Collaboration of Epidemiological Studies of Cervical, 2006; Vaccarella et al., 2006) high parity (Hildesheim et al., 2001; International Collaboration of Epidemiological Studies of Cervical, 2006), chronic use of oral contraceptives (Smith et al., 2003; Ylitalo et al., 1999), age of first pregnancy (International Collaboration of Epidemiological Studies of Cervical Cancer., 2006) also act as modifiers conferring risk to the development of cervical cancer. Interestingly, HPV negative incidences of cervical cancer (Barreto et al., 2013; de Sanjose et al., 2010; Guan et al., 2012; Igidbashian et al., 2014; Lai et al., 2007; Li et al., 2011) have been reported to be associated with adenocarcinoma of the uterine endocervix with poor disease free survival (DFS) (Rodriguez-Carunchio et al., 2015). HPV infection of the cervical epithelium are transient in nature that elicits immune responses to counter the infection (Hakama and Day, 1986). Interestingly, 80-90% of HPV infections are neutralized by the immune system within few years, but persistent infections of the cervical epithelium due to compromised immunity are associated with intraepithelial lesions that turn into invasive cervical carcinoma (Moscicki et al., 2012; Shvetsov et al., 2009). This differential nature of immune action is well attributed to the variations in the genes involved in generation and modification of immune responses to viral antigens.
Candidate gene based association studies have identified the impact of genetic polymorphisms in the genes belonging to xenobiotic metabolism (Abbas et al., 2014; Jain et al., 2017; Joseph et al., 2006; Satinder et al., 2017; Sobti et al., 2006), DNA repair (Konathala et al., 2017; Nagpal et al., 2002; Singhal et al., 2013), cell cycle (Katiyar et al., 2003; Pillai et al., 2002; Saranath et al., 2002; Satinder et al., 2008; Singhal et al., 2013; Thakur et al., 2009) and immune responses (Gangwar et al., 2009; Kordi Tamandani et al., 2008; Shekari et al., 2012; Singh et al., 2008; Singh et al., 2009; Singhal et al., 2015; Sobti et al., 2008), in modifying the risk of cervical cancer. Several genome-wide association studies (Chen et al., 2013; Chen et al., 2016; Leo et al., 2017; Lin et al., 2016; Miura et al., 2016) revealed polymorphisms in human leukocyte antigen (HLA)/MHC genes involved in immune response pathways, pertaining to cervical carcinogenesis. The HLA genes are very complex and are difficult to assess the influence of the SNPs on cervical cancer risk based on standard SNP gene effect model (Wang et al., 2015). However, most of these reports were based on Caucasian, Chinese and Japanese populations (Chen et al., 2013; Chen and Gyllensten, 2015; Chen et al., 2016; Leo et al., 2017; Lin et al., 2016; Miura et al., 2016), and most of the identified risk alleles have not been adequately evaluated in Indian population. Till date, the overall effects of the genes and the genetic variations on cervical cancer risk in Indian population have not been determined due to contradictory findings. In one study conducted on 147 patients and 165 controls in south Indian population at Thiruvananthapuram, the GSTT1 null allele shows no significant association with cervical cancer (Joseph et al., 2006), but in another study conducted on 150 patients and 150 controls in north Indian population of Chandigarh, the same null allele of GSTT1shows significant association with cervical cancer (Satinder et al., 2017).The conflicting results in two different reports on the same variant could be due to small sample sizes, racial and/or ethnic differences and/or clinical and genetic heterogeneity between populations.
Therefore, the combined effect of the variant(s) on the Indian population to get an accurate estimate of association between a genetic variant and cervical cancer risk needs to be assessed by meta-analysis (Lee, 2015), which is a powerful statistical tool to evaluate the true effect of polymorphisms on the disease status by pooling the individual study data. The study aims to assess the overall influence of genetic variants on cervical cancer risk in the context of Indian populations.
Materials and Methods
Data extraction
Data mining was done from the PubMed database (www.ncbi.nlm.nih.gov/pubmed) for the appropriate studies using the following search strings: (polymorphisms/single nucleotide polymorphisms /SNP/SNPs/SNVs/SNV/Mutation) AND (cervical cancer /cervical carcinoma) AND (India). Selection of the studies for inclusion in the meta-analysis was done in accordance with the following criteria: (a) the studies should be based samples from Indian population, (b) the studies should have case-control genotype data for the polymorphic variant reported, (c) the studies should be full research articles, (d) the studies should have reports on covariate risk factors, such as HPV status and/or smoking status and/or use of oral contraceptives and/or age of menarche, menopause and first pregnancy and/or parity, (e) all the candidate genes based association studies were considered that were published till November 2017, and (f) the polymorphisms with reports in at least two different studies were considered. The following data were extracted from the selected reports: (1) first author, (2) year of publication, (3) mean age with standard deviation, (4) sex, (5) smoking status, (6) HPV status, (7) age of menarche and menopause, (8) age of marriage and first pregnancy, (9) parity, (10) use of oral contraceptives, (11) genetic polymorphisms and (12) genotype specific case-control data. On the basis of the aforementioned criteria 29 papers involving 16 polymorphisms from 11 genes were selected for further meta-analysis (Figure 1).
Figure 1.
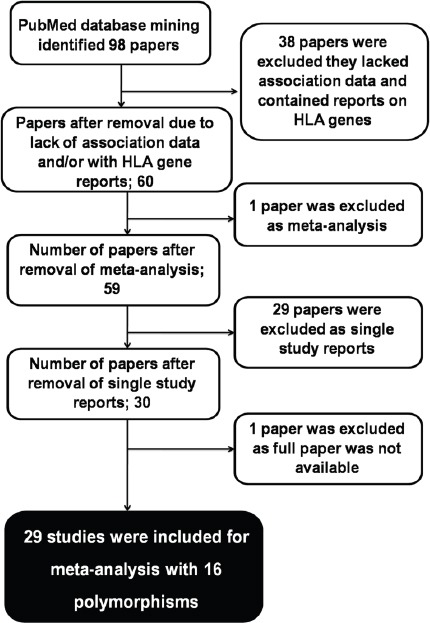
A Workflow for the Selection of Studies from PubMed Repository
Selection of genetic model
Apart from Del1, Del2, rs1695, rs1800871 and rs1801133, an additive (allelic) model was followed for all the 11 polymorphisms that assume the effect of the heterozygous genotype as intermediate between the two homozygotes. Del1, Del2, rs1695, rs1800871 and rs1801133 were analyzed in a recessive model.
Meta-analysis
Meta-analysis was performed in R, 3.4.2 (R Core Team, 2017) package ‘metafor’ (Viechtbauer, 2010) considering fixed-effect model (Borenstein et al., 2010), on genetic association reports pertaining to Indian population (Sengupta et al., 2017) ignoring ethnicity and geographical distribution stratification due to lack of sufficient data.
Evaluation of heterogeneity between selected studies
Cochran’s Q test (P<0.10) (Huedo-Medina et al., 2006) was done to evaluate the heterogeneity of inter-study variations. Further, heterogeneity index (I2) was calculated that measures the degree of inconsistency across studies using the formula: I2 = (Q-(n-1) n)/QX100 %, where ‘n’ is the number of studies. I2 value is expressed as percentage with grade cut-offs as 25%, 50% or 75%, which signifies the presence of low-, mid- or high-grade heterogeneity, respectively (Bedi et al., 2011; Jiang et al., 2012; Jin et al., 2009; Vyas et al., 2013). Hence, heterogeneity between studies for all SNPs were evaluated using H2 metrics at 10% level of significance and I2 metric, (Sengupta et al. 2017).
Evaluation of publication bias among the selected studies
Qualitative estimation of publication bias was done by visual inspection of funnel plots (Sterne and Egger, 2001). Further, quantitative estimation of publication bias was done by Egger’s regression test that evaluate the asymmetry of the funnel plots (P<0.05) using a weighted regression model with multiplicative dispersion, for only those polymorphic variants that are reported in 3 or more studies. Symmetry of the funnel plots shows absence of publication bias, whereas asymmetry depicts the presence of publication bias.
Evaluation of genetic association of reported polymorphisms with cervical cancer
Individual study level odds ratios and 95% confidence intervals (95% CI) for both additive (allelic) and recessive model along with their corresponding standard errors were first determined to evaluate the statistical association (p<0.05) between the reported polymorphisms and cervical cancer risk. Further a logistic regression of cancer status on variant genotype coded as 0 (homozygous wild type), 1 (heterozygous) and 2 (homozygous variant) was done. For, the four polymorphisms (Del1, Del2, rs1695, rs1800871 and rs1801133); 0 (homozygous wild type + heterozygous), 1 (homozygous variant) coding was used.
Results
Study characteristics
Extensive mining of PubMed (www.ncbi.nlm.nih.gov/pubmed) database generated 98 articles for the aforementioned search strings. Further textmining of the 98 articles identified 29 articles (Table 1) that actually fit all the inclusion criteria of the study proposed. The reported variables of the studies included in this meta-analysis are summarized in (Table A1). However, case-control genotype data of all the 16 polymorphisms were extracted from 29 selected articles (Table A2). The case-control data for covariates, particularly tobacco smoking, mean age at sampling, HPV status, age at menarche, age at menopause, age at first pregnancy, parity and use of oral contraceptives, were recorded from 29 selected articles for meta-analysis (Table A3).
Table 1.
The List of Candidate Gene-Based Association Studies Included in the Meta-Analysis along with the Study Numbers, First Author, Year of Publication, Covariate Counts and Reference Urls for PubMed Search
| First Author Year | Study Number | Number of Cases | Number of Controls | Smokers in Cases | Smokers in Controls | Non-Smokers in Cases | Non-Smokers in Controls | Mean Age of Cases | Mean Age of Controls | Number of HPV positives in Cases | Number of HPV positives in Controls | Number of HPV negative in Cases | Number of HPV negative in Controls | Geographical Location of the Study | Reference URLs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jain V et al.2017 | Study 20 | 100 | 100 | 0 | 0 | 0 | 0 | 50.9±8.2 | 44.05±10.4 | 0 | 0 | 0 | 0 | Raipur | https://www.ncbi.nlm.nih.gov/pubmed/28433806 |
| Satinder K et al.2017 | Study 25 | 150 | 150 | 0 | 0 | 0 | 0 | 48.5±9.4 | 46.1±11.2 | 0 | 0 | 0 | 0 | Chandigarh | https://www.ncbi.nlm.nih.gov/pubmed/28361858 |
| Bajpai D et al.2016 | Study 28 | 65 | 68 | 13 | 5 | 52 | 63 | 43.9±0.0 | 43.2±10.3 | 61 | 12 | 4 | 56 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/25812040 |
| Konathala G et al.2016 | Study 29 | 125 | 150 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Visakhapatnam | https://www.ncbi.nlm.nih.gov/pubmed/27942558 |
| Sharma A et al. 2015 | Study 3 | 135 | 457 | 22 | 39 | 113 | 418 | 42.1±11.7 | 41.1±8.9 | 89 | 91 | 46 | 366 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/26434855 |
| Abbas M et al. 2014 | Study 1 | 200 | 208 | 13 | 9 | 74 | 119 | 48.54±9.529 | 48.09±8.347 | 0 | 0 | 0 | 0 | Lucknow | https://www.ncbi.nlm.nih.gov/pubmed/24657182 |
| Singhal P et al.2014 | Study 5 | 256 | 250 | 0 | 0 | 0 | 0 | 48.6±11 | 46.07±6.6 | 229 | 12 | 27 | 238 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/25412954 |
| Singhal P et al.2013 | Study 12 | 182 | 182 | 0 | 0 | 0 | 0 | 49±0.0 | 49±9.64 | 166 | 8 | 16 | 174 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/23210739 |
| Shekari M et al.2012 | Study 7 | 200 | 200 | 90 | 62 | 110 | 138 | 48.55±9.43 | 48.81±9.64 | 0 | 0 | 0 | 0 | Chandigarh | https://www.ncbi.nlm.nih.gov/pubmed/22157213 |
| Prasad et al.2011 | Study 16 | 62 | 241 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Andhra Pradesh | https://www.ncbi.nlm.nih.gov/pubmed/21934341 |
| Kohar I et al. 2010 | Study 18 | 203 | 231 | 0 | 0 | 0 | 0 | 49.4±12.4 | 48.2±10.2 | 173 | 2 | 30 | 229 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/19793004 |
| Singh H et al.2009 | Study 6 | 150 | 162 | 0 | 0 | 0 | 0 | 47.2±8.8 | 48.3±8.3 | 0 | 0 | 0 | 0 | Lucknow | https://www.ncbi.nlm.nih.gov/pubmed/19823053 |
| Gangwar R et al.2009 | Study 8 | 200 | 230 | 62 | 32 | 138 | 198 | 44.8±9.3 | 46.7±9.9 | 0 | 0 | 0 | 0 | Lucknow | https://www.ncbi.nlm.nih.gov/pubmed/19681846 |
| Thakur N et al.2009 | Study 26 | 200 | 200 | 0 | 0 | 0 | 0 | 49±11.3 | 49±11.3 | 0 | 0 | 0 | 0 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/19489683 |
| Kordi TMK et al.2008 | Study 9 | 200 | 200 | 90 | 62 | 110 | 138 | 48.55±9.43 | 48.81±9.9 | 0 | 0 | 0 | 0 | Chandigarh | https://www.ncbi.nlm.nih.gov/pubmed/18806746 |
| Singh H et al.2008 | Study 10 | 150 | 162 | 0 | 0 | 0 | 0 | 47.2±8.8 | 48.3±9.64 | 0 | 0 | 0 | 0 | Lucknow | https://www.ncbi.nlm.nih.gov/pubmed/18333945 |
| Sobti RC et al.2008 | Study 11 | 150 | 200 | 65 | 57 | 83 | 74 | 48.55±9.43 | 48.81±8.3 | 0 | 0 | 0 | 0 | Chandigarh | https://www.ncbi.nlm.nih.gov/pubmed/18154955 |
| Nandan NK et al.2008 | Study 17 | 142 | 77 | 0 | 0 | 0 | 0 | 39±5 | 39±5 | 0 | 0 | 0 | 0 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/19356065 |
| Shekari M et al.2008 | Study 19 | 200 | 200 | 2 | 6 | 110 | 138 | 48.55±9.43 | 48.81±9.64 | 0 | 0 | 0 | 0 | Chandigarh | https://www.ncbi.nlm.nih.gov/pubmed/18351371 |
| Satinder K et al.2008 | Study 27 | 150 | 150 | 67 | 62 | 83 | 88 | 48.5±9.4 | 46.1±11.2 | 0 | 0 | 0 | 0 | Chandigarh | https://www.ncbi.nlm.nih.gov/pubmed/18548202 |
| Joseph T et al.2006 | Study 2 | 222 | 90 | 0 | 0 | 0 | 0 | 46±10.3 | 47±9.2 | 111 | 32 | 36 | 280 | Thiruvananthapuram | https://www.ncbi.nlm.nih.gov/pubmed/16360200 |
| Sobti RC et al.2006 | Study 4 | 103 | 103 | 45 | 13 | 58 | 90 | 48.6±9.9 | 48±11.3 | 0 | 0 | 0 | 0 | Chandigarh | https://www.ncbi.nlm.nih.gov/pubmed/16631467 |
| Bhattacharya P et al.2005 | Study 15 | 120 | 205 | 0 | 0 | 0 | 0 | 0 | 0 | 82 | 84 | 38 | 121 | Kolkata | https://www.ncbi.nlm.nih.gov/pubmed/16054204 |
| Mitra S et al. 2004 | Study 21 | 61 | 94 | 0 | 0 | 0 | 0 | 46.89±10.29 | 47.44±12.53 | 52 | 17 | 9 | 77 | Kolkata | https://www.ncbi.nlm.nih.gov/pubmed/15623478 |
| Katiyar S et al.2003 | Study 23 | 163 | 74 | 0 | 0 | 0 | 0 | 0 | 0 | 128 | 0 | 35 | 74 | New Delhi | https://www.ncbi.nlm.nih.gov/pubmed/14577584 |
| Pillai MR et al.2002 | Study 13 | 311 | 110 | 0 | 0 | 0 | 0 | 0 | 0 | 201 | 14 | 110 | 96 | Thiruvananthapuram | https://www.ncbi.nlm.nih.gov/pubmed/12458344 |
| Bhattacharya P et al.2002 | Study 14 | 55 | 201 | 0 | 0 | 0 | 0 | 0 | 0 | 46 | 84 | 9 | 117 | Kolkata | https://www.ncbi.nlm.nih.gov/pubmed/12406566 |
| Nagpal JK et al. 2002 | Study 22 | 111 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 77 | 11 | 34 | 18 | Cuttack | https://www.ncbi.nlm.nih.gov/pubmed/12534455 |
| Saranath D et al. 2002 | Study 24 | 337 | 164 | 0 | 0 | 0 | 0 | 0 | 0 | 258 | 37 | 79 | 127 | Mumbai | https://www.ncbi.nlm.nih.gov/pubmed/12144822 |
Meta-analysis of reported polymorphisms to determine the overall association with cervical cancer through
Meta-analysis was performed in R package ‘metafor’ using Fixed-effect model for 16 polymorphisms from 11 genes reported on Indian population. For 12 polymorphisms, study-specific crude Odds Ratio (OR) estimates and 95% Confidence Interval (CI) were determined using additive (allelic) model and for the remaining 4 polymorphisms recessive model was used. Due to few available study reports for the 16 polymorphisms in the context of Indian population, stratification based on the geographical distribution was not performed for the meta-analysis due to lack to consistent data. The polymorphic variants ‘rs1801131’, ‘rs1801133’, ‘rs2430561’, ‘rs1799782’, ‘rs25486’ and ‘rs25487’ were found to exhibit significant heterogeneity (P <0.10) between studies as revealed by Cochran’s Q test. The variants ‘rs1799782’ and ‘rs25486’ of XRCC1 gene showed high-grade heterogeneity according to I2 metric (I2>75%) while the variants ‘rs1801131’, ‘rs1801133’ and ‘rs25487’ showed mid-grade heterogeneity (I2>50%). The remaining 10 polymorphic variants showed low-grade or no heterogeneity (Table 2). After meta-analysis, 2 polymorphisms, viz. del2 (OR=1.79, 95% CI= 1.08-2.95, P=0.023) and rs1048943 (G versus A: crude OR = 2.34, 95% CI=1.37-3.99, P=0.0018) were found to be associated (p< 0.05) with cervical cancer. Further, multiple-testing adjustment by Benjamini-Höchberg FDR correction (p<0.05) for 16 variants revealed only rs1048943 of CYP1A1 to be associated (P=0.029) with cervical cancer based on additive (allelic) model in Indian population (Table 2). The association of rs1048943 of CYP1A1 and Del2 of GSTM1 with cervical cancer are shown in forest plots (Figure 2-a, Figure 3-a). Publication bias was qualitatively assessed by visual inspection of funnel plots that revealed evidence of publication bias for rs1048943 of CYP1A1gene but not for Del2 of GSTM1 (Figure 2-b, Figure 3-b). Further, Egger’s regression test for the quantitative assessment of funnel plot asymmetry (critical p<0.05) also showed evidence of significant publication bias (P=0.0113) for rs1048943 of CYP1A1gene (Table 3). However, the individual and overall study effects for each of the remaining fourteen variants (OR estimates, 95% CI) are also depicted in forest plots (Figures 1-14; Supplement and Supporting Data).
Table 2.
Combined Statistical Association of the Respective Variants with Cervical Cancer; with Crude Odds Ratio (OR), 95% CI and Heterogeneity Indices, I2, H2. P-values are adjusted by Benjamini-Höchberg FDR Correction.
| Variants | Number of Studies | Odds Ratio (OR) | 95% CI | P-value | I2 | H2 | Het.stat | Het.P.value | Padj |
|---|---|---|---|---|---|---|---|---|---|
| del 1 | 4 | 0.8347 | 0.468-1.487 | 0.5399 | 30.2584 | 1.4339 | 3.0935 | 0.3774 | 0.7853 |
| del 2 | 4 | 1.7881 | 1.081-2.955 | 0.0234 | 16.6965 | 1.2004 | 1.8402 | 0.6062 | 0.1873 |
| rs1695 | 2 | 1.2124 | 0.411-3.574 | 0.7270 | 0.0271 | 1.0003 | 0.0234 | 0.8784 | 0.8331 |
| rs1800871 | 2 | 0.6886 | 0.360-1.316 | 0.2592 | 0.0151 | 1.0002 | 0.0175 | 0.8949 | 0.5428 |
| rs1801133 | 4 | 1.0987 | 0.645-1.870 | 0.7290 | 67.7116 | 3.0971 | 9.6305 | 0.0220 | 0.8331 |
| rs1801131 | 2 | 0.5838 | 0.287-1.183 | 0.1357 | 69.8727 | 3.3192 | 3.6057 | 0.0576 | 0.4844 |
| rs4646903 | 4 | 1.3821 | 0.899-2.123 | 0.1400 | 41.2724 | 1.7028 | 3.8478 | 0.2784 | 0.4844 |
| rs1048943 | 3 | 2.3384 | 1.371-3.987 | 0.0018 | 11.2109 | 1.1263 | 1.0149 | 0.6020 | 0.0290 |
| rs1800872 | 2 | 1.3058 | 0.773-2.205 | 0.3185 | 40.0435 | 1.6679 | 1.5370 | 0.2151 | 0.5662 |
| rs2430561 | 2 | 1.0410 | 0.608-1.780 | 0.8835 | 67.7555 | 3.1013 | 3.3542 | 0.0670 | 0.8835 |
| Rs16944 | 2 | 1.4198 | 0.760-2.651 | 0.2714 | 15.9649 | 1.1900 | 0.7187 | 0.3966 | 0.5428 |
| rs1042522 | 8 | 0.8134 | 0.600-1.101 | 0.1816 | 23.3077 | 1.3039 | 5.4461 | 0.6057 | 0.4844 |
| rs603965 | 2 | 1.2190 | 0.708-2.096 | 0.4740 | 19.9871 | 1.2498 | 0.8427 | 0.3586 | 0.7584 |
| rs1799782 | 2 | 0.9418 | 0.488-1.814 | 0.8578 | 92.3336 | 13.0439 | 13.7906 | 0.0002 | 0.8835 |
| rs25486 | 2 | 1.5943 | 0.817-3.110 | 0.1713 | 85.5546 | 6.9226 | 7.5016 | 0.0062 | 0.4844 |
| rs25487 | 2 | 1.1416 | 0.614-2.121 | 0.6752 | 66.7560 | 3.0081 | 3.2455 | 0.0716 | 0.8331 |
P value<0.05*; Cochran’s Q test P-value<0.10
Figure 2.
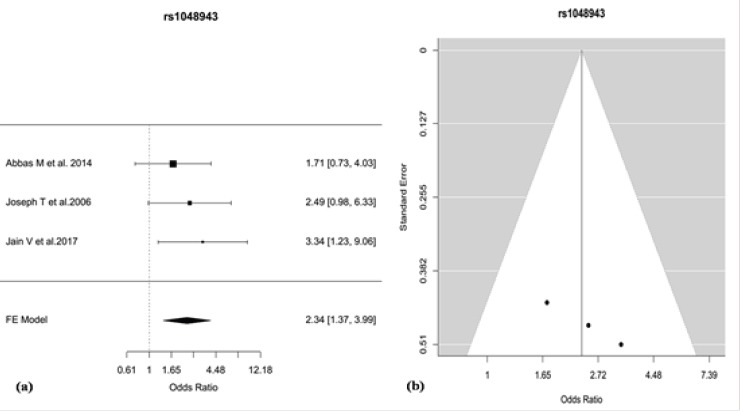
(a) Forest Plot for the Odds Ratios (ORs) of the SNP rs1048943 of CYP1A1 for the Association with Cervical Cancer, (b) Funnel Plot, Showing Evidence of Publication Bias between the Studies Reporting the SNP rs1048943.
Figure 3.
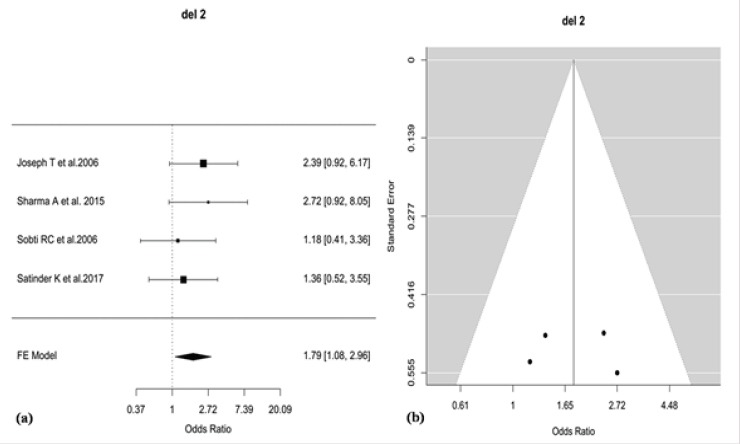
(a) Forest Plot for the Odds Ratios (ORs) of the Deletion Polymorphism (del2) of GSTM1 for the Association with Cervical Cancer, (b) Funnel Plot, Showing no Evidence of Publication bias between the Studies Reporting del2.
Table 3.
Egger’s Regression Test for Funnel Plot Asymmetry
| Gene name | Variant I.D | t | df | P-value* |
|---|---|---|---|---|
| GSTT1 | del 1 | 0.6324 | 2 | 0.5918 |
| GSTM1 | del 2 | 0.1516 | 2 | 0.8934 |
| MTHFR | rs1801133 | -0.5631 | 2 | 0.6301 |
| XRCC1 | rs25487 | 1.6167 | 4 | 0.1812 |
| TP53 | rs1042522 | -0.4649 | 6 | 0.6584 |
| CYP1A1 | rs4646903 | -0.6117 | 2 | 0.603 |
| CYP1A1 | rs1048943 | 56.5441 | 1 | 0.0113 |
P<0.05
Evaluation of covariate stratified genetic association of reported polymorphic variants with cervical cancer
The effect of the polymorphic variants on the development of cervical cancer influenced by the covariate risk factors, such as HPV status, was estimated. We collected HPV stratified summary data from the literature for only rs1042522 of TP53 due availability of sufficient data (Table A4). Study specific summary data viz. β-coefficient (log OR) and standard errors (SE) was obtained 7 studies for rs1042522 respectively. For some articles, stratified β-coefficient and SE were calculated using logistic regression from the covariate stratified genotype counts provided in the tables. The remaining 15 polymorphic variants were not stratified based on any of the covariate risk factors due to lack of consistent covariate stratified summary data in the selected studies.
Sub-group meta-analysis of rs1042522 of TP53 was done following the fixed-effect model (Borenstein et al., 2010). Using the covariate specific summary data separately within the “HPV positive” and “HPV negative” sub-groups. The sub-group meta-analysis stratified by HPV status revealed lack of significant associations with cervical cancer (Table 4) as depicted in forest plots (Figure 4).
Table 4.
Sub-Group Meta- Analysis of rs1042522 of TP53 with Cervical Cancer, Stratified by HPV Status, i.e. HPV Positive and HPV Negative. OR, Odds Ratio (OR); 95% CI and Heterogeneity indices, I2, H2
| Sub-Groups | Number of Studies | Odds Ratio (OR) | 95% CI | P-value | I2 | H2 | Het-stat | P-value (Het.) |
|---|---|---|---|---|---|---|---|---|
| For rs1042522 of TP53 | ||||||||
| HPV positive | 7 | 1.0743 | 0.7154-1.6132 | 0.7290 | 25.1340 | 1.3357 | 4.1915 | 0.6508 |
| HPV negative | 7 | 1.0177 | 0.6641-1.5594 | 0.9350 | 27.9676 | 1.3883 | 4.8911 | 0.5578 |
*P-value< 0.05
Figure 4.
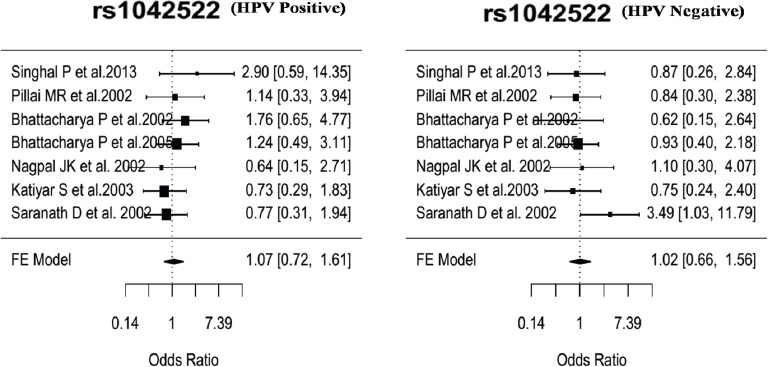
Forest Plot for the Odds Ratios (ORs) of the rs1042522 of TP53 for the Association with Cervical Cancer Stratified by HPV Status in Sub-Group Analysis between HPV Positive and HPV Negative.
Test for effect modification of rs1042522 of TP53 by HPV status using a fixed effect meta-regression model was done with all the sub-group specific summaries and specifying covariate status as a moderator variable in R package ‘metafor’ (Viechtbauer 2010). The difference in effect size among HPV positive individuals and HPV negative individuals as captured by the coefficient of the moderator term revealed that rs1042522 has no significant effect modification on cervical cancer risk by HPV status (Moderator Effect size (θ)= 0.05415944, θlow= -0.5352891, θhigh= 0.6435892; P= 0.85).
Discussion
Cervical cancer is a complex disease and is essentially an outcome of virus induced carcinogenesis. Human Papilloma virus (HPV) is the primal cause of cervical carcinogenesis but other environmental and social factors, like tobacco smoke, age of marriage and first pregnancy, use of oral contraceptive, number of sexual partners, modify the risk of the cancer (Fonseca-Moutinho, 2011; Hildesheim et al., 2001; Igidbashian et al., 2014; Vaccarella et al., 2006; Ylitalo et al., 1999). HPV infection acts as the primary etiological factor for the development of cervical cancer but all HPV infections do not result in cervical cancer. The immune system is well equipped to counter the viral attack and in most of the infected individuals the viral load is cleared out or become dormant by the immune responses. Persistent HPV infection along with other risk modifiers synergistically act to cause cellular transformations that leads to cervical cancer (Bosch et al., 2002; Hakama and Day, 1986; Moscicki et al., 2012). Recent candidate gene based investigations show genetic association of several polymorphisms with cervical cancer (Abbas et al., 2014; Bajpai et al., 2016; Bhattacharya et al., 2002; Bhattacharya and Sengupta, 2005; Gangwar et al., 2009; Jain et al., 2017; Joseph et al., 2006; Katiyar et al., 2003; Kohaar et al., 2010; Konathala et al., 2017; Kordi Tamandani et al., 2008; Mitra et al., 2005; Nagpal et al., 2002; Pillai et al., 2002; Prasad and Wilkhoo 2011; Saranath et al., 2002; Satinder et al., 2008; Satinder et al., 2017; Sharma et al., 2015; Shekari et al., 2008; Shekari et al., 2012; Singh et al., 2008; Singh et al., 2009; Singhal et al., 2013; Singhal et al., 2015; Sobti et al., 2006; Sobti et al., 2008; Thakur et al., 2009). A genome-wide association study (GWAS) revealed three haplotypes, HLA-DRB1*15/HLA-DQB1*0602/HLA-DQA1*0102, HLA-B*0702/HLA-C*0702, and HLA-DRB1*0401/HLA-DQA1*0301, to be associated with increased risk of both HPV16 and HPV18-driven development of both squamous cell carcinoma and adenocarcinoma of the cervix in populations of European descent (Leo et al., 2017). Another GWAS reported four new SNPs, viz. rs6812281, located at 4q34.3 (P < 5.0 × 10−8) rs4590782, located at 10q26.2 (P = 1.59 × 10−5), rs1742101 located at 14q32.11 (P = 7.11 × 10−6), and rs1364121 located at 16q23.3 (P = 3.15 × 10−6), that exhibits strong evidence of associations with response to neoadjuvant chemotherapy in cervical cancer in Chinese women (Li et al., 2017). More GWAS reports on Swedish population supported the association of previously identified loci at 6p21.3 (rs9271898, P = 1.2 × 10−24; rs2516448, 1.1 × 10−15; and rs3130196, 2.3 × 10−9, respectively) with cervical cancer. The study also confirmed associations of cervical cancer with reported classical HLA alleles including HLA-B*07:02, -B*15:01, -DRB1*13:01, -DRB1*15:01, -DQA1*01:03, -DQB1*06:03 and -DQB1*06:02. Further, an independent signal at rs73730372 at 6p21.3 (P = 3.0 × 10−19) was found to be an expression quantitative trait locus (eQTL) of both HLA-DQA1 and HLA-DQB1 conferring protection to cervical cancer (CIN3) (Chen et al., 2016).
This study is the first comprehensive meta-analysis from India on the available candidate gene based genetic association studies that gives us the most risk polymorphic variant responsible for the development of cervical cancer based on the available reports. Our meta-analysis revealed rs1048943 (exon 7, A>G) of CYP1A1 gene to be significantly associated with cervical cancer (Figure 2a) after Benjamini-Höchberg FDR correction (p=0.029). The absence of inter-study heterogeneity for the variant rs1048943 of CYP1A1 suggests that the association of the polymorphic variant with cervical cancer holds true. Moreover, the studies for the variant rs1048943 of CYP1A1 exhibits significant publication bias as confirmed by Egger’s regression test (Table 3) and visual inspection of funnel plots (Figure 2b), which indicates a probable overestimation under the influence of publication bias. For rs1048943 of CYP1A1, sufficient covariate-stratified summary data is absent to perform sub-group analysis that led to the failure of the determination of the true effect of the variant under the influence of covariate risk factors. Therefore, more studies are needed to gain a comprehensive insight on the overall and covariate-stratified effect of rs1048943 of CYP1A1 on cervical cancer biology.
Meta-analysis of rs1042522 of TP53 stratified by HPV status also revealed lack of significant association with cervical cancer in any of the sub-groups. However, No significant statistical interaction (effect modification) was found for rs1042522 of TP53 with HPV infection through our meta-regression analysis, which implies that the polymorphism has no modifier effect on cervical cancer by the status of HPV infection in Indian women. However, we surmise that there may be interaction in the biological mechanisms conferring susceptibility towards the development of cervical cancer. Due to wide variation in covariate-adjusted or covariate-stratified summary data in the various studies, moderate degree of heterogeneity could have led to loss of power in detecting effect modification. Due to inconsistency in reporting subgroup level summaries or covariate adjusted summaries across studies, we restricted our sub-group analysis with only rs1042522 of TP53. Further, we were unable to perform sub-group analysis based on covariates, such as age, sex, cancer subtypes, age of marriage, age at first pregnancy, use of oral contraceptives due to lack of sufficient reports on Indian population.
The SNP rs1048943 of CYP1A1 was found to be significantly associated with cervical cancer mostly in Caucasian and East Asian population (Wang et al., 2015) and lung cancer in Indian population (Sengupta et al., 2017), which shows colinearity of our finding on Indian population. The CYP1A1 (Cytochrome P4501A1; 15q22-24) gene encodes an essential phase I xenobiotic metabolism enzyme (512 aa length) found mainly in the endoplasmic reticulum and cytosol. The enzyme generates highly reactive electrophilic compounds from pro-carcinogenic xenobiotics, like polycyclic aromatic hydrocarbons (PAHs) and enhances the formation of genotoxic DNA adducts that results in mutagenesis and cellular transformation. CYP1A1 is also involved in the metabolism different drugs and endogenous steroid hormone molecules like oestrogen. The SNP rs1048943A>G in the exon 7 of CYP1A1 gene is a point mutation that results in the substitution of isoleucine by valine (Ile>Val) in the crucial heme-binding domain, which increases enzyme activity and confers risk of cervical carcinogenesis. Cervical carcinogenesis occurs due the synergistic effect of HPV infection and oestrogen metabolism (den Boon et al., 2015). In the development of cervical carcinogenesis the expression of oestrogen receptor alpha (ER-α) ablates in tumor cells but retained in tumor-associated stromal fibroblasts that alters several cross signaling pathways between stroma and tumor (den Boon et al., 2015). Cytochrome P4501A1 enzyme is involved in oxidation of oestrogen to catechols and oestrogen quinones (Spink et al., 1992; Zhang et al., 2007) that forms mutagenic stable and depurinating DNA adducts (Zhang et al., 2007; Zhu and Conney, 1998) facilitating HPV integration in the host genome (Joseph et al., 2006). Moreover, a polymorphism in CYP1A1 has been found to be associated with extended time for clearance of high risk HPV infection from cervical epithelia (Sudenga et al., 2014).
Our meta-analysis also revealed a marginal association of GSTM1 null genotypes with cervical cancer. Glutathione-S-transferases (GSTs) are a family of multifunctional enzymes that catalyzes the conjugation of glutathione to electrophilic substrates resulting in their enhanced renal clearance reducing carcinogenic load from the cell. The null genotype of GSTM1 has been reported to be associated with enhanced risk to cervical cancer (Hasan et al., 2015; Nunobiki et al., 2015) that justified our finding. Cervical cancer is an outcome of the combined effect of HPV infection with environmental and hormonal influence. Interestingly, our meta-analysis pooled out 11 genes that includes genes belonging to xenobiotic metabolism, DNA repair, cell cycle and immune response regulation. Probably these genes play as ‘modifiers’ that enhances the chance and severity of HPV infection in the cervical epithelia. Tobacco smoking has been reported as one of the major modifiers in cervical cancer risk (Fonseca-Moutinho, 2011; Ylitalo et al., 1999).
With the inclusion of more studies on genetic association with cervical cancer on different populations and sub-populations with contradictory outcomes; meta-analysis would become a very important tool to estimate the true effect of those variants on the cancer status considering the covariate stratifications of the studied population. Therefore, identification of the genetic variants for which there is evidence of influence on cervical cancer risk through meta-analysis, would provide new insights into the candidate biological pathways involved in the development of cervical cancer. This would further benefit in the assessment of population specific risk for accurate decision making, which could be of potential value in targeting primary prevention and population specific cervical cancer screening modalities and therapeutic interventions.
Our meta-analysis showed rs1048943A>G, in exon 7 of CYP1A1 to be associated with cervical cancer even after Benjamini-Höchberg FDR correction (P=0.029). However, the remaining variants failed to show significant association with cervical cancer after multiple-testing adjustment, which might be due to small sample size, ethnic differences or probably due to lack of sufficient data. Therefore, more studies on Indian population are needed along with covariate-adjusted summary data from individual studies, to gain more statistical power and accuracy for such a study.
Authors Contribution Statement
D. Sengupta and M. Sengupta conceptualized and designed the study. U. Guha, S. Mitra, S. Ghosh and D. Sengupta searched literature and extracted data. S. Bhattacharjee, M. Sengupta and D. Sengupta analyzed the data. D. Sengupta drafted the manuscript with important intellectual inputs from S. Bhattacharjee and M. Sengupta. S. Bhattacharjee and M. Sengupta critically revised the manuscript, figures, tables and supplementary data and supervised the entire work. All the authors gave approval for submission of the current version of the manuscript for publication and have full access to the study data.
Competing Financial Interests
The author(s) declare no competing financial interests.
Acknowledgements
This study has been supported by: The Department of Science and Technology (DST), Government of India - Promotion of University Research and Scientific Excellence (DST-PURSE) provided to Department of Genetics, University of Calcutta. D.Sengupta is supported by Senior Research Fellowship from University Grants Commission (UGC), Govt. of India.
References
- 1.Abbas M, Srivastava K, Imran M, Banerjee M. Association of cyp1a1 gene variants rs4646903 (t>c) and rs1048943 (a>g) with cervical cancer in a north indian population. Eur J Obstet Gynecol Reprod Biol. 2014;176:68–74. doi: 10.1016/j.ejogrb.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 2.Bajpai D, Banerjee A, Pathak S, et al. Single nucleotide polymorphisms in the DNA repair genes in hpv-positive cervical cancer. Eur J Cancer Prev. 2016;25:224–31. doi: 10.1097/CEJ.0000000000000159. [DOI] [PubMed] [Google Scholar]
- 3.Barreto CL, Martins DB, de Lima Filho JL, Magalhaes V. Detection of human papillomavirus in biopsies of patients with cervical cancer, and its association with prognosis. Arch Gynecol Obstet. 2013;288:643–8. doi: 10.1007/s00404-013-2803-2. [DOI] [PubMed] [Google Scholar]
- 4.Bedi U, Singh M, Singh P, et al. Effects of statins on progression of coronary artery disease as measured by intravascular ultrasound. J Clin Hypertens (Greenwich) 2011;13:492–6. doi: 10.1111/j.1751-7176.2011.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya P, Duttagupta C, Sengupta S. Proline homozygosity in codon 72 of p53: A risk genotype for human papillomavirus related cervical cancer in indian women. Cancer Lett. 2002;188:207–11. doi: 10.1016/s0304-3835(02)00430-5. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya P, Sengupta S. Lack of evidence that proline homozygosity at codon 72 of p53 and rare arginine allele at codon 31 of p21, jointly mediate cervical cancer susceptibility among indian women. Gynecol Oncol. 2005;99:176–82. doi: 10.1016/j.ygyno.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Borenstein M, Hedges LV, Higgins JP, Rothstein HR. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods. 2010;1:97–111. doi: 10.1002/jrsm.12. [DOI] [PubMed] [Google Scholar]
- 8.Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruni L B-RL, Albero G, Serrano B, et al. Ico information centre on hpv and cancer (hpv information centre) Human papillomavirus and related diseases in india. (Summary Report) 2017 [Google Scholar]
- 10.Chen D, Juko-Pecirep I, Hammer J, et al. Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst. 2013;105:624–33. doi: 10.1093/jnci/djt051. [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Gyllensten U. Lessons and implications from association studies and post-gwas analyses of cervical cancer. Trends Genet. 2015;31:41–54. doi: 10.1016/j.tig.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, Enroth S, Liu H, et al. Pooled analysis of genome-wide association studies of cervical intraepithelial neoplasia 3 (cin3) identifies a new susceptibility locus. Oncotarget. 2016;7:42216–24. doi: 10.18632/oncotarget.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–56. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 14.den Boon JA, Pyeon D, Wang SS, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112:3255–64. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int J Cancer. 2015;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 16.Fonseca-Moutinho JA. Smoking and cervical cancer. ISRN Obstet Gynecol. 2011:847684. doi: 10.5402/2011/847684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangwar R, Pandey S, Mittal RD. Association of interferon-gamma +874a polymorphism with the risk of developing cervical cancer in north-indian population. BJOG. 2009;116:1671–7. doi: 10.1111/j.1471-0528.2009.02307.x. [DOI] [PubMed] [Google Scholar]
- 18.Guan P, Howell-Jones R, Li N, et al. Human papillomavirus types in 115,789 hpv-positive women: A meta-analysis from cervical infection to cancer. Int J Cancer. 2012;131:2349–59. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 19.Hakama M MA, Day NE. Screening for cancer of the uterine cervix. From the iarc working group on cervical cancer screening and the uicc project group on the evaluation of screening programmes for cancer. IARC Sci Publ. 1986;76:1–315. [PubMed] [Google Scholar]
- 20.Hasan S, Hameed A, Saleem S, et al. The association of gstm1 and gstt1 polymorphisms with squamous cell carcinoma of cervix in pakistan. Tumour Biol. 2015;36:5195–9. doi: 10.1007/s13277-015-3175-y. [DOI] [PubMed] [Google Scholar]
- 21.Hildesheim A, Herrero R, Castle PE, et al. Hpv co-factors related to the development of cervical cancer: Results from a population-based study in costa rica. Br J Cancer. 2001;84:1219–26. doi: 10.1054/bjoc.2001.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or i2 index? Psychol Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 23.Igidbashian S, Schettino MT, Boveri S, et al. Tissue genotyping of 37 in situ and invasive cervical cancer with a concomitant negative hc2 hpv DNA test. J Low Genit Tract Dis. 2014;18:87–91. doi: 10.1097/LGT.0b013e3182909f86. [DOI] [PubMed] [Google Scholar]
- 24.International Collaboration of Epidemiological Studies of Cervical C. Cervical carcinoma and reproductive factors: Collaborative reanalysis of individual data on 16,563 women with cervical carcinoma and 33,542 women without cervical carcinoma from 25 epidemiological studies. Int J Cancer. 2006;119:1108–24. doi: 10.1002/ijc.21953. [DOI] [PubMed] [Google Scholar]
- 25.Jain V, Ratre YK, Amle D, Mishra PK, Patra PK. Polymorphism of cyp1a1 gene variants rs4646903 and rs1048943 relation to the incidence of cervical cancer in chhattisgarh. Environ Toxicol Pharmacol. 2017;52:188–92. doi: 10.1016/j.etap.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y, Zhang R, Zheng J, et al. Meta-analysis of 125 rheumatoid arthritis-related single nucleotide polymorphisms studied in the past two decades. PLoS One. 2012;7:e51571. doi: 10.1371/journal.pone.0051571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin DH, Lamberton GR, Broome DR, et al. Renal stone detection using unenhanced multidetector row computerized tomography--does section width matter? J Urol. 2009;181:2767–73. doi: 10.1016/j.juro.2009.01.092. [DOI] [PubMed] [Google Scholar]
- 28.Joseph T, Chacko P, Wesley R, et al. Germline genetic polymorphisms of cyp1a1, gstm1 and gstt1 genes in indian cervical cancer: Associations with tumor progression, age and human papillomavirus infection. Gynecol Oncol. 2006;101:411–7. doi: 10.1016/j.ygyno.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 29.Katiyar S, Thelma BK, Murthy NS, et al. Polymorphism of the p53 codon 72 arg/pro and the risk of hpv type 16/18-associated cervical and oral cancer in india. Mol Cell Biochem. 2003;252:117–24. doi: 10.1023/a:1025546610920. [DOI] [PubMed] [Google Scholar]
- 30.Khaliq SA, Shyum Naqvi SB, Fatima A. Human pappilomavirus (hpv) induced cancers and prevention by immunization. Pak J Pharm Sci. 2012;25:763–72. [PubMed] [Google Scholar]
- 31.Kohaar I, Kumar J, Thakur N, et al. Homocysteine levels are associated with cervical cancer independent of methylene tetrahydrofolate reductase gene (mthfr) polymorphisms in indian population. Biomarkers. 2010;15:61–8. doi: 10.3109/13547500903295881. [DOI] [PubMed] [Google Scholar]
- 32.Konathala G, Mandarapu R, Godi S. Data on polymorphism of xrcc1 and cervical cancer risk from south india. Data Brief. 2017;10:11–3. doi: 10.1016/j.dib.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kordi Tamandani MK, Sobti RC, Shekari M, Mukesh M, Suri V. Expression and polimorphism of ifn-gamma gene in patients with cervical cancer. Exp Oncol. 2008;30:224–9. [PubMed] [Google Scholar]
- 34.Lai CH, Huang HJ, Hsueh S, et al. Human papillomavirus genotype in cervical cancer: A population-based study. Int J Cancer. 2007;120:1999–6. doi: 10.1002/ijc.22538. [DOI] [PubMed] [Google Scholar]
- 35.Lee YH. Meta-analysis of genetic association studies. Ann Lab Med. 2015;35:283–7. doi: 10.3343/alm.2015.35.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leo PJ, Madeleine MM, Wang S, et al. Defining the genetic susceptibility to cervical neoplasia-a genome-wide association study. PLoS Genet. 2017;13:e1006866. doi: 10.1371/journal.pgen.1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer. 2011;128:927–35. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Huang K, Zhang Q, et al. Genome-wide association study identifies four snps associated with response to platinum-based neoadjuvant chemotherapy for cervical cancer. Sci Rep. 2017;7:41103. doi: 10.1038/srep41103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin H, Ma Y, Wei Y, Shang H. Genome-wide analysis of aberrant gene expression and methylation profiles reveals susceptibility genes and underlying mechanism of cervical cancer. Eur J Obstet Gynecol Reprod Biol. 2016;207:147–52. doi: 10.1016/j.ejogrb.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Mitra S, Misra C, Singh RK, Panda CK, Roychoudhury S. Association of specific genotype and haplotype of p53 gene with cervical cancer in india. J Clin Pathol. 2005;58:26–31. doi: 10.1136/jcp.2004.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miura K, Mishima H, Yasunami M, et al. A significant association between rs806 7378 at 17q12 and invasive cervical cancer originally identified by a genome-wide association study in han chinese is replicated in a japanese population. J Hum Genet. 2016;61:793–6. doi: 10.1038/jhg.2016.50. [DOI] [PubMed] [Google Scholar]
- 42.Moscicki AB, Schiffman M, Burchell A, et al. Updating the natural history of human papillomavirus and anogenital cancers. Vaccine. 2012;30:24–33. doi: 10.1016/j.vaccine.2012.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagpal JK, Sahni S, Das BR. P53 codon 72 polymorphism and susceptibility to development of human papilloma virus-associated cervical cancer in indian women. Eur J Clin Invest. 2002;32:943–8. doi: 10.1046/j.1365-2362.2002.01096.x. [DOI] [PubMed] [Google Scholar]
- 44.Nunobiki O, Ueda M, Akise H, et al. Gstm1, gstt1, and nqo1 polymorphisms in cervical carcinogenesis. Hum Cell. 2015;28:109–13. doi: 10.1007/s13577-015-0111-9. [DOI] [PubMed] [Google Scholar]
- 45.Pillai MR, Sreevidya S, Pollock BH, Jayaprakash PG, Herman B. Polymorphism at codon 72 of p53, human papillomavirus, and cervical cancer in south india. J Cancer Res Clin Oncol. 2002;128:627–31. doi: 10.1007/s00432-002-0383-9. [DOI] [PubMed] [Google Scholar]
- 46.Prasad VV, Wilkhoo H. Association of the functional polymorphism c677t in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Onkologie. 34:422–6. doi: 10.1159/000331131. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez-Carunchio L, Soveral I, Steenbergen RD, et al. Hpv-negative carcinoma of the uterine cervix: A distinct type of cervical cancer with poor prognosis. BJOG. 2015;122:119–27. doi: 10.1111/1471-0528.13071. [DOI] [PubMed] [Google Scholar]
- 48.Saranath D, Khan Z, Tandle AT, et al. Hpv16/18 prevalence in cervical lesions/cancers and p53 genotypes in cervical cancer patients from india. Gynecol Oncol. 2002;86:157–62. doi: 10.1006/gyno.2002.6735. [DOI] [PubMed] [Google Scholar]
- 49.Satinder K, Chander SR, Pushpinder K, Indu G, Veena J. Cyclin d1 (g870a) polymorphism and risk of cervix cancer: A case control study in north indian population. Mol Cell Biochem. 2008;315:151–7. doi: 10.1007/s11010-008-9799-0. [DOI] [PubMed] [Google Scholar]
- 50.Satinder K, Sobti RC, Pushpinder K. Impact of single nucleotide polymorphism in chemical metabolizing genes and exposure to wood smoke on risk of cervical cancer in north-indian women. Exp Oncol. 2017;39:69–74. [PubMed] [Google Scholar]
- 51.Sengupta D, Guha U, Bhattacharjee S, Sengupta M. Association of 12 polymorphic variants conferring genetic risk to lung cancer in indian population: An extensive meta-analysis. Environ Mol Mutagen. 2017;58:688–700. doi: 10.1002/em.22149. [DOI] [PubMed] [Google Scholar]
- 52.Sharma A, Gupta S, Sodhani P, et al. Glutathione s-transferase m1 and t1 polymorphisms, cigarette smoking and hpv infection in precancerous and cancerous lesions of the uterine cervix. Asian Pac J Cancer Prev. 2015;16:6429–38. doi: 10.7314/apjcp.2015.16.15.6429. [DOI] [PubMed] [Google Scholar]
- 53.Shekari M, Sobti RC, Kordi Tamandani DM, Suri V. Impact of methylenetetrahydrofolate reductase (mthfr) codon (677) and methionine synthase (ms) codon (2756) on risk of cervical carcinogenesis in north indian population. Arch Gynecol Obstet. 2008;278:517–24. doi: 10.1007/s00404-008-0623-6. [DOI] [PubMed] [Google Scholar]
- 54.Shekari M, Kordi-Tamandani DM, MalekZadeh K, et al. Effect of anti-inflammatory (il-4, il-10) cytokine genes in relation to risk of cervical carcinoma. Am J Clin Oncol. 2012;35:514–9. doi: 10.1097/COC.0b013e31822d9c12. [DOI] [PubMed] [Google Scholar]
- 55.Shvetsov YB, Hernandez BY, McDuffie K, et al. Duration and clearance of anal human papillomavirus (hpv) infection among women: The hawaii hpv cohort study. Clin Infect Dis. 2009;48:536–46. doi: 10.1086/596758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh H, Sachan R, Goel H, Mittal B. Genetic variants of interleukin-1rn and interleukin-1beta genes and risk of cervical cancer. BJOG. 2008;115:633–8. doi: 10.1111/j.1471-0528.2007.01655.x. [DOI] [PubMed] [Google Scholar]
- 57.Singh H, Jain M, Sachan R, Mittal B. Association of tnfa (-308g>a) and il-10 (-819c>t) promoter polymorphisms with risk of cervical cancer. Int J Gynecol Cancer. 2009;19:1190–4. doi: 10.1111/IGC.0b013e3181a3a3af. [DOI] [PubMed] [Google Scholar]
- 58.Singhal P, Hussain S, Thakur N, et al. Association of mdm2 and p53 polymorphisms with the advancement of cervical carcinoma. DNA Cell Biol. 2013;32:19–27. doi: 10.1089/dna.2012.1718. [DOI] [PubMed] [Google Scholar]
- 59.Singhal P, Kumar A, Bharadwaj S, Hussain S, Bharadwaj M. Association of il-10 gtc haplotype with serum level and hpv infection in the development of cervical carcinoma. Tumour Biol. 2015;36:2287–98. doi: 10.1007/s13277-014-2836-6. [DOI] [PubMed] [Google Scholar]
- 60.Smith JS, Green J, Berrington de Gonzalez A, et al. Cervical cancer and use of hormonal contraceptives: A systematic review. Lancet. 2003;361:1159–67. doi: 10.1016/s0140-6736(03)12949-2. [DOI] [PubMed] [Google Scholar]
- 61.Sobti RC, Kaur S, Kaur P, et al. Interaction of passive smoking with gst (gstm1, gstt1, and gstp1) genotypes in the risk of cervical cancer in india. Cancer Genet Cytogenet. 2006;166:117–23. doi: 10.1016/j.cancergencyto.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Sobti RC, Kordi Tamandani DM, Shekari M, et al. Interleukin 1 beta gene polymorphism and risk of cervical cancer. Int J Gynaecol Obstet. 2008;101:47–52. doi: 10.1016/j.ijgo.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 63.Spink DC, Eugster HP, Lincoln DW, et al. 17 beta-estradiol hydroxylation catalyzed by human cytochrome p450 1a1: A comparison of the activities induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in mcf-7 cells with those from heterologous expression of the cdna. Arch Biochem Biophys. 1992;293:342–8. doi: 10.1016/0003-9861(92)90404-k. [DOI] [PubMed] [Google Scholar]
- 64.Sreedevi A, Javed R, Dinesh A. Epidemiology of cervical cancer with special focus on india. Int J Womens Health. 2015;7:405–14. doi: 10.2147/IJWH.S50001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: Guidelines on choice of axis. J Clin Epidemiol. 2001;54:1046–55. doi: 10.1016/s0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- 66.Sudenga SL, Macaluso M, Partridge EE, Johanning GL, Piyathilake CJ. Functional variants in cyp1a1 and gstm1 are associated with clearance of cervical hpv infection. Gynecol Oncol. 2014;135:560–4. doi: 10.1016/j.ygyno.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Team RCR. A language and environment for statistical computing. vienna, austria: R foundation for statistical computing; 2017. URL https://www.R-project.org/ [Google Scholar]
- 68.Thakur N, Hussain S, Kohaar I, et al. Genetic variant of ccnd1: Association with hpv-mediated cervical cancer in indian population. Biomarkers. 2009;14:219–25. doi: 10.1080/13547500902825274. [DOI] [PubMed] [Google Scholar]
- 69.Vaccarella S, Herrero R, Dai M, et al. Reproductive factors, oral contraceptive use, and human papillomavirus infection: Pooled analysis of the iarc hpv prevalence surveys. Cancer Epidemiol Biomarkers Prev. 2006;15:2148–53. doi: 10.1158/1055-9965.EPI-06-0556. [DOI] [PubMed] [Google Scholar]
- 70.Viechtbauer W. Conducting meta-analyses in r with the metafor package. J Stat Soft. 2010;36:1–48. [Google Scholar]
- 71.Vyas A, Swaminathan PD, Zimmerman MB, Olshansky B. Are treatments for vasovagal syncope effective?A meta-analysis. Int J Cardiol. 2013;167:1906–11. doi: 10.1016/j.ijcard.2012.04.144. [DOI] [PubMed] [Google Scholar]
- 72.Wang S, Sun H, Jia Y, et al. Association of 42 snps with genetic risk for cervical cancer: An extensive meta-analysis. BMC Med Genet. 2015;16:25. doi: 10.1186/s12881-015-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeole BB, Kumar AV, Kurkure A, Sunny L. Population-based survival from cancers of breast, cervix and ovary in women in Mumbai, India. Asian Pac J Cancer Prev. 2004;5:308–15. [PubMed] [Google Scholar]
- 74.Ylitalo N, Sorensen P, Josefsson A, et al. Smoking and oral contraceptives as risk factors for cervical carcinoma in situ. Int J Cancer. 1999;81:357–65. doi: 10.1002/(sici)1097-0215(19990505)81:3<357::aid-ijc8>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, Gaikwad NW, Olson K, et al. Cytochrome p450 isoforms catalyze formation of catechol estrogen quinones that react with DNA. Metabolism. 2007;56:887–94. doi: 10.1016/j.metabol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: Review and perspectives. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]


