Abstract
The order Mucorales, the largest in number of species within the Mucoromycotina, comprises typically fast-growing saprotrophic fungi. During a study of the fungal diversity of undiscovered taxa in Korea, two novel mucoralean strains, CNUFC-GWD3-9 and CNUFC-EGF1-4, were isolated from specific habitats including freshwater and fecal samples, respectively. On the basis of their morphological characteristics and sequence analyses of internal transcribed spacer and large subunit ribosomal DNA, the CNUFC-GWD3-9 and CNUFC-EGF1-4 isolates were confirmed to be Gilbertella persicaria and Pilobolus crystallinus, respectively. It is ecologically, pathologically, and mycologically significant to find such rare zygomycetous fungi in such specific habitats.
KEYWORDS: Mucorales, phylogeny, rare fungi, undiscovered taxa
1. Introduction
Previously, taxa of the former phylum Zygomycota were distributed among the phylum Glomeromycota and four subphyla incertae sedis, namely Mucoromycotina, Kickxellomycotina, Zoopagomycotina, and Entomophthoromycotina [1]. Recently, Spatafora et al. [2] proposed two new phyla, Mucoromycota and Zoopagomycota, on the basis of phylogenetic analyses of a genome-scale dataset for 46 taxa, including 25 zygomycetes and 192 proteins. According to these results, Mucoromycota and Zoopagomycota were newly formalized phyla of fungi and comprised six subphyla. The phylum Mucoromycota comprises the subphyla Mucoromycotina, Mortierellomycotina, and Glomeromycotina, whereas the phylum Zoopagomycota comprises the subphyla Entomophthoromycotina, Zoopagomycotina, and Kickxellomycotina.
Mucorales is the largest order within the Mucoromycotina and comprises 15 families, 57 genera, and ∼334 species [3]. Most mucoralean species are saprotrophic and grow on different organic substrates, such as fruits, soil, dung, and plants [4,5]. Several species are parasites or pathogens of animals, plants, and fungi [4,5]. A few species of which cause human and animal diseases called mucormycosis, as well as allergic reactions [6]. The traditional classification of Mucorales has been determined on the basis of morphological characteristics, such as the size and shape of the sporangium, sporangiophore, sporangiospore (asexual reproduction), and zygospore (sexual reproduction) [4,5]. Recently, several molecular studies evaluating mucoralean species indicated that some of the genera may be polyphyletic [4,5].
The genus Gilbertella belongs to the subphylum Mucoromycotina, order Mucorales, family Choanephoraceae. It was first named Choanephora persicaria by Eddy in 1925 [7] and then renamed as the genus Gilbertella by Hesseltine in 1960 [8].
Species of this genus are characterized as having sporangia with a persistent wall dehiscing through a longitudinal suture; sporangiospores with apical, hyaline appendages; and Mucor-type zygospores [9]. Previously, the genus Gilbertella was assigned within the Choanephoraceae because it had not been seen since its original description [7]. Hesseltine [8] placed the genus within the Mucoraceae because the zygospores are of Mucor-type. Later, Gilbertella was confirmed through studies of DNA sequence data as in fact belonging to the family Choanephoraceae [10].
G. persicaria, which is heterothallic, has a sporangial wall that splits into hemispheres at maturity, and sporangiospores that bear long filamentous appendages on the ends. This species has been reported as a plant pathogen of peach, pear, tomato, and some tropical fruits [7,11,12]. In Index Fungorum (2018; http://www.indexfungorum.org), the genus Gilbertella contains only one species, named G. persicaria (E.D. Eddy) Hesselt.
Genus Pilobolus Tode (Pilobolaceae, Mucorales) is characterized by positive phototropism and its method of spore dispersal; that is, through the ballistic discharge caused by the elevated pressure generated by subsporangial swelling of the sporangiophore [13,14]. Pilobolus species are attached to the substrate by an absorptive structure, the swollen trophocyst, which is semi-immersed in the substrate [14]. The trophocysts are generally ovoid to globose, whereas the rhizoidal extension is long and cylindrical [14]. The sporangiophores are straight, unbranched, and positively phototropic, with two rings of orange pigment at the base and near the subsporangial vesicle [14]. The sporangia are hemispherical and contain the spores, which are globose or ellipsoidal depending on the species [14]. Zygospores are formed in the substrate and have apposed suspensors [15].
Pilobolus species are coprophilous and have typically been detected on herbivore dung and are frequently observed sporulating on this substrate [16–18]. Coprophilous fungi play an important role in the recycling of nutrients in animal dung [19]. In Index Fungorum 2018, the genus Pilobolus contains 15 species.
In Korea, within the Choanephoraceae, only three species have been described, whereas species belonging to the Pilobolaceae have not yet been described.
The aim of the present study was to perform molecular and morphological analyses to characterize two novel mucoralean species from specific habitats such as freshwater and animal feces in Korea: G. persicaria and P. crystallinus.
2. Materials and methods
2.1. Sampling and isolation of fungal strain
Fecal samples of water deer were collected on Eulsukdo Island (35°6′17.92″ N, 128°56′24.52″ E; located in Busan, Korea) in June 2017. The samples were transferred to sterile 50-mL conical tubes (SPL Life Sciences Co., Pocheon, Korea), and stored at 4 °C until examination. The fecal samples were placed onto sterile moist Whatman’s filter paper in a Petri dish using sterile forceps and incubated in a moist chamber at 25 °C for 6–9 days.
Freshwater samples were collected from the Geum River (36°27′47.32″ N, 127°6′3.24″ E; located in Gongju, Korea) in August 2017. These samples were transported in sterile 50 mL conical tubes and stored at 4 °C until examination. Fungi were isolated by the direct plating method. In brief, plant debris in the freshwater samples was placed onto synthetic mucor agar (SMA; 40 g of dextrose, 2 g of asparagine, 0.5 g of KH2PO4, 0.25 g of MgSO4·7H2O, 0.5 g of thiamine chloride, and 15 g of agar in 1 L of deionized water) using sterile forceps and incubated at 25 °C for 1–3 days. To isolate pure cultures, individual colonies of varied morphologies were picked up, transferred to potato dextrose agar (39 g of PDA in 1 L of deionized water; Becton, Dickinson and Co., Sparks, MD) plates, and subcultured until pure mycelia were obtained. All pure isolates, including those of G. persicaria and P. crystallinus, were stored in 20% glycerol at −80 °C at the Environmental Microbiology Laboratory Fungarium (Chonnam National University, Gwangju, Korea), as CNUFC-GWD3-9 and CNUFC-EGF1-4, respectively. Strain CNUFC-EGF1-4 was also deposited at the Culture Collection of the National Institute of Biological Resources (NIBR, Incheon, Korea), whereas strain CNUFC-GWD3-9 was also deposited at the Culture Collection of the Nakdonggang National Institute of Biological Resources (NNIBR, Sangju, Korea).
2.2. Morphological studies
For detailed morphological studies, strain CNUFC-GWD3-9 was cultured on SMA, PDA, and malt extract agar (33.6 g of MEA in 1 L of deionized water; Becton, Dickinson and Co.). The plates were incubated at 5 °C, 15 °C, 25 °C, 35 °C, and 40 °C in the dark for 2–3 days. Samples were mounted in distilled water and observed using an Olympus BX51 microscope with differential interference contrast (DIC) optics (Olympus, Tokyo, Japan). CNUFC-EGF1-4 strain was cultured on dung agar medium (2 g of water deer dung and 2 g of agar in 100 mL of deionized water) and the plates were incubated at 20 °C, 25 °C, and 35 °C in the dark for 7–14 days. In addition, fungal spores of strain CNUFC-EGF1-4 were inoculated on surface-sterilized pieces of water deer dung by touching with a sterile needle, and the plates were then incubated at 25 °C in the dark for 7–14 days. Samples were observed under an Olympus BX51 microscope with DIC optics.
2.3. DNA extraction, PCR, and sequencing
Genomic DNA was extracted directly from mycelia and spores of the fungal isolates, using the Solg Genomic DNA Prep Kit for fungi (SolGent Co. Ltd., Daejeon, Korea). The internal transcribed spacer (ITS) and large subunit (LSU) regions were amplified with the primer pairs ITS1 and ITS4 [20], and LROR and LR5F [21,22], respectively (Table 3). The PCR amplification mixture (total volume, 20 µL) contained fungal DNA template, 5 pmol/µL of each primer, and Accupower PCR Premix (Taq DNA polymerase, dNTPs, buffer, and a tracking dye; Bioneer Corp., Daejeon, Korea). The PCR products were purified using the Accuprep PCR Purification Kit (Bioneer Corp.) according to the manufacturer’s instructions. DNA sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA).
Table 3.
Morphological characteristics of CNUFC-EGF1-4 and the reference species Pilobolus crystallinus.
| Characteristic | CNUFC-EGF1-4 | Pilobolus crystallinusa |
|---|---|---|
| Trophocysts | Subglobose to ellipsoidal, 199.0–409.8 × 147.5–186.9 µm | Oblong, 500–575 × 200–230 µm |
| Sporangiophores | Variable in length, 57.0–122.7 µm wide | 5–15 mm long, 115–160 µm wide |
| Subsporangial vesicles | Ovoid, orange ring at the base, 298.0–677.9 × 175.0–548.7 µm | Oviform, colorless except for an orange ring at the base, 400–920 × 350–720 µm |
| Sporangia | Hemispherical, umbonate, first yellow or brown and then black when mature, 169.5–371.5 µm × 151.5–295.5 µm | Semiglobose, black, 237–529 µm wide near the base, 138–345 µm high |
| Columellae | Ellipsoidal to mammiform, 110.3–186.7 µm × 122.1–230.5 µm | Broadly conical, 92–287 µm high, 172–345 µm wide below |
| Sporangiospores | Elliptical, hyaline, yellowish, 6.0–8.5 × 4.0–5.5 μm | Elliptical, hyaline, dark yellow in mass, 7–10 × 4–6 µm |
| Zygospores | Not observed | Unknown |
From the description by Boedijin [18].
2.4. Phylogenetic analysis
The sequences were aligned with Clustal_X v.2.0 [23] and edited using Bioedit v.7.2.5 software [24]. Phylogenetic trees based on the ITS and D1/D2 rDNA sequences were constructed using the neighbor-joining method in MEGA 6 [25]. The reliability of the internal branches was assessed using the p-distance substitution model, with 1,000 bootstrap replications. The CNUFC-GWD3-9, CNUFC-GWD3-10, CNUFC-EGF1-4, and CNUFC-EGF1-5 sequences were deposited in the NCBI database under the accession numbers shown in Table 1.
Table 1.
Taxa, collection numbers, sequences, and GenBank accession numbers used in this study.
| Taxon name | Collection No. (Isolate No.) | GenBank accession No. |
|
|---|---|---|---|
| ITS | LSU | ||
| Backusella circina | CBS 128.70 (T) | – | JN206529 |
| B. circina | KH10 | – | JX644493 |
| B. indica | CBS 786.70 | – | JN206526 |
| B. lamprospora | CBS 195.28 | – | JN206530 |
| B. lamprospora | CBS 118.08 (T) | – | JN206531 |
| B. recurva | CBS 318.52 | – | JN206522 |
| B. tuberculispora | CBS 562.66 | – | JN206525 |
| Benjaminiella poitrasii | CBS 158.68 (T) | – | JN206411 |
| Blakeslea trispora | CBS 130.59 | JN206227 | – |
| Bl. trispora | EML-PUKI88 | KY047144 | – |
| Bl. trispora | CBS 564.91 | JN206230 | JN206515 |
| Choanephora cucurbitarum | CBS 120.25 | JN206231 | – |
| C. cucurbitarum | CBS 674.93 | JN206233 | JN206514 |
| C. infundibulifera | CBS 153.51 | JN206236 | JN206513 |
| C. infundibulifera | CBS 155.51 | JN206237 | – |
| Cokeromyces recurvatus | CBS 158.50 (T) | – | HM849699 |
| Gilbertella persicaria | CBS 785.97 | JN206218 | – |
| G. persicaria | CBS 190.32 (T) | HM999958 | HM849691 |
| G. persicaria | CBS 246.59 | JN206222 | – |
| G. persicaria | CBS 442.64 | JN206219 | – |
| G. persicaria | CBS 532.77 | JN206224 | JN206517 |
| G. persicaria | CBS 565.91 | JN206226 | – |
| G. persicaria | CNUFC-GWD3-9 | MG906872 | MG906876 |
| G. persicaria | CNUFC-GWD3-10 | MG906873 | MG906877 |
| Hyphomucor assamensis | CBS 415.77 | JN206211 | – |
| Pilobolus crystallinus | ATCC 11505 | FJ160947 | – |
| P. crystallinus | ATCC 36186 | FJ160949 | – |
| P. crystallinus | ATCC 46942 | FJ160958 | – |
| P. crystallinus | KH25 | JX644569 | – |
| P. crystallinus | TZS | JN942691 | JN982943 |
| P. crystallinus | TZS990207 | JN942689 | JN982939 |
| P. crystallinus | CNUFC-EGF1-4 | MG906874 | MG906878 |
| P. crystallinus | CNUFC-EGF1-5 | MG906875 | MG906879 |
| P. heterosporus | IUE 120 | HM049566 | – |
| P. heterosporus | IUE 706 | HM049604 | – |
| P. heterosporus | IUE 906 | HM049615 | – |
| P. kleinii | ATCC 36185 | FJ160957 | – |
| P. kleinii | IUE 205 | HM049567 | – |
| P. kleinii | IUE 305 | HM049574 | – |
| P. longipes | IUE 340 | FJ160950 | – |
| P. longipes | IUE 409 | FJ160951 | – |
| P. longipes | IUE 563 | FJ160952 | – |
| P. pullus | IUE 0014 | HQ877876 | – |
| P. pullus | IUE 0017 | HQ877877 | – |
| P. roridus | CHC | JN942692 | JN982944 |
| P. roridus | IUE 319 | HM049579 | – |
| P. roridus | IUE 415 | FJ160948 | – |
| P. roridus | IUE 918 | HM049619 | – |
| P. sphaerosporus | ATCC 14499 | FJ160954 | – |
| P. sphaerosporus | ATCC 22499 | DQ059382 | – |
| P. sphaerosporus | IUE 916 | HM049616 | – |
| P. sphaerosporus | UAMH 1312 | FJ160953 | – |
| P. umbonatus | CBS 302.83 | JN206274 | HM849665 |
| P. umbonatus | CBS 425.50 | JN206275 | – |
| P. umbonatus | KH24 | JX644571 | JX644519 |
| P. umbonatus | NRRL 6349 | FJ160955 | – |
| P. umbonatus | UAMH 7297 | FJ160956 | – |
| P. umbonatus | UAMH 7298 | DQ058412 | – |
| Poitrasia circinans | CBS 153.58 (T) | JN206239 | JN206516 |
| Pt. circinans | CBS 647.70 | JN206240 | – |
| Rhizopus americanus | CBS 340.62 | HM999967 | – |
| R. koreanus | EML-HO95-1 | – | KU058196 |
| R. sexualis | CBS 336.39 (T) | – | HM849673 |
| Syzygites megalocarpus | CBS 372.39 | – | JN206401 |
| Umbelopsis isabellina | CBS 560.63 | – | JN206573 |
| Utharomyces epallocaulus | CBS 329.73 | – | HM849660 |
Bold letters indicate the isolates and accession numbers determined in our study.
ITS: internal transcribed spacer; ATCC: American Type Culture Collection (Manassas, VA, USA); CBS: Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands); CNUFC: Chonnam National University Fungal Collection (Gwangju, South Korea); EML: Environmental Microbiology Laboratory (Fungarium, Chonnam National University, Gwangju, South Korea); NRRL (Agricultural Research Service Culture Collection, Peoria, IL, USA); T: ex-type strain.
2.5. Pectinase activity assay
To detect pectinase activity, we used the medium as described by Hankin and Anagnostakis [26]. The medium contained 1 g of yeast extract, 20 g of agar, 10 g of pectin (citrus), NaNO3, 2 g; KCl, 0.5 g; MgSO4.7H2O, 0.5 g; K2HPO4, 1.0 g; FeSO4.7H2O, 0.01 g and 1 L of distilled water; pH 6.8 or 7.0. Strain CNUFC-GWD3-9 was cultured on potato dextrose agar (PDA; Becton, Dickinson and Co.) at 25°С for 7 days. 7 agar pieces 6.5 mm of fungal mycelia were put in to 50 ml potato dextrose broth (PDB) in a 100 ml Erlenmeyer flask, previously sterilized at 121°С for 15 min. Strain CNUFC-GWD3-9 was incubated at 25°С for 5 days at 130 rpm in a horizontal shaker incubator. The broth culture was centrifuged at 13,000 × g for 20 min at 4 °C. A 50-µL aliquot of the supernatant was transferred to a paper disc (diameter, 8 mm), and then the disc was placed on the surface of a potato-dextrose-agar (PDA) plate (90 mm × 15 mm). After 3 days of incubation at 25 °C, plates were flooded with a 1% aqueous solution of hexadecyltrimethylammonium bromide (Fisher Chemical Co., Fairlawn, NJ). Clear zones around a colony indicated degradation of the pectin.
3. Results
3.1. Molecular phylogenetic status
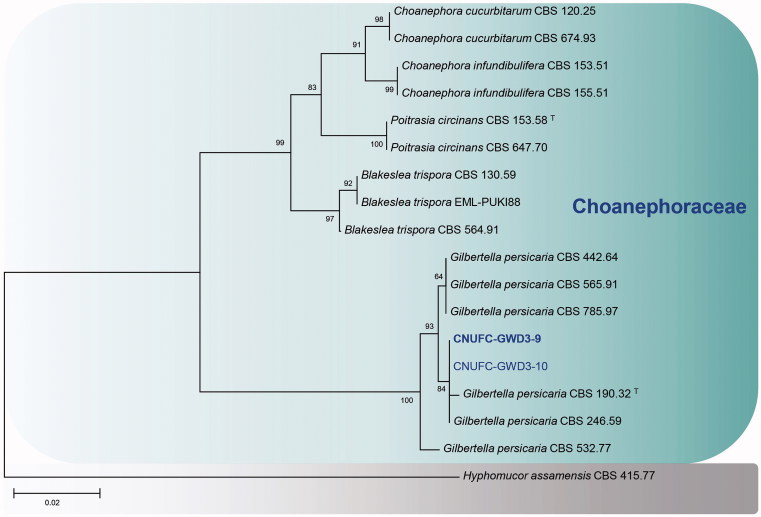
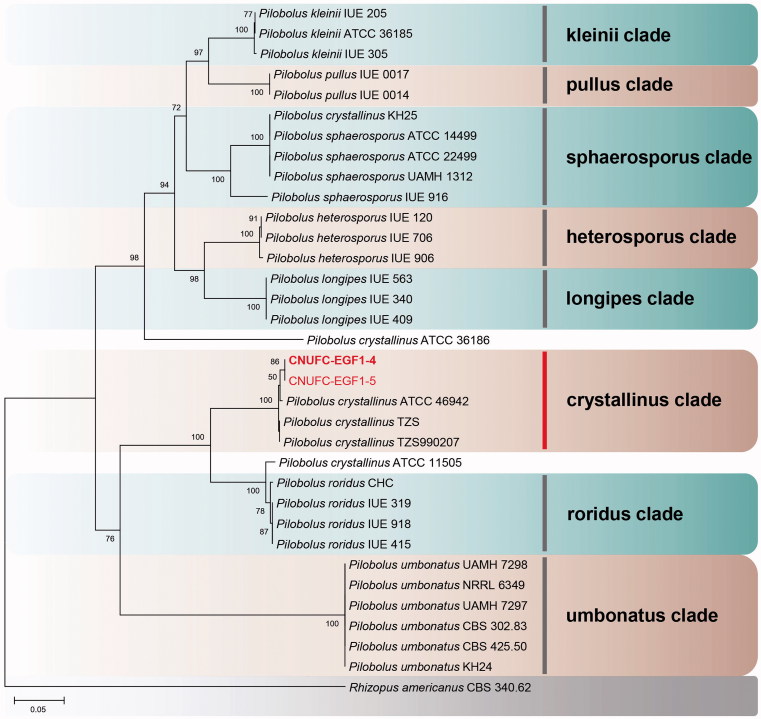
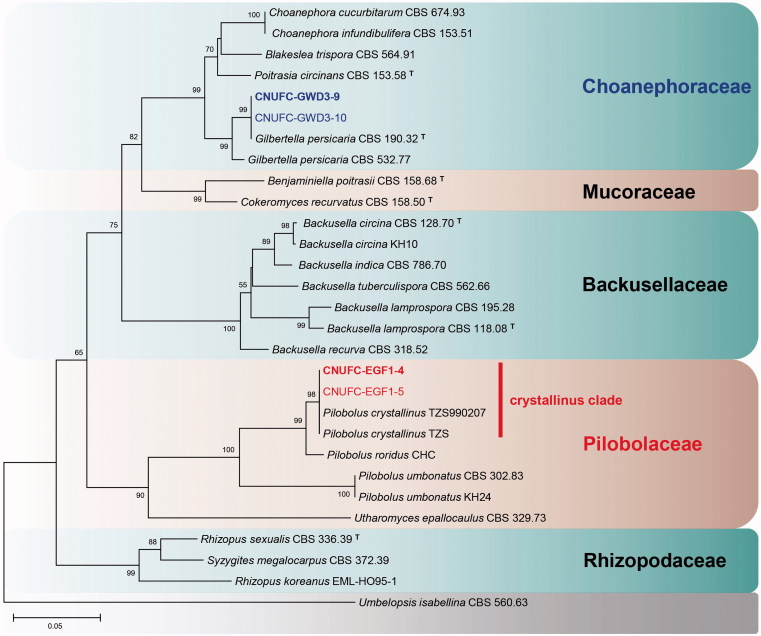
A BLASTn search showed that the ITS rDNA sequences of CNUFC-GWD3-9 and CNUFC-EGF1-4 have high sequence similarities of 99.7% (490/491 bp) and 99.3% (572/576 bp) with G. persicaria (NR111692) and P. crystallinus (FJ160958), respectively. In the BLASTn analysis of the 28S rDNA sequences, CNUFC-GWD3-9 and CNUFC-EGF1-4 strains revealed 100% (653/653 bp) and 100% (538/538 bp) identity values with G. persicaria (JN939197) and P. crystallinus (JN982939), respectively. In the trees, they were grouped separately but placed into the same clade with the reference of Gilbertella and Pilobolus (Figures 1–3).
Figure 1.
Phylogenetic tree based on neighbor-joining analysis of internal transcribed spacer rDNA sequences for Gilbertella persicaria CNUFC-GWD3-9 and G. persicaria CNUFC-GWD3-10. Hyphomucor assamensis was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
Figure 2.
Phylogenetic tree based on neighbor-joining analysis of internal transcribed spacer rDNA sequences for Pilobolus crystallinus CNUFC-EGF1-4 and P. crystallinus CNUFC-EGF1-5. Rhizopus americanus was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
Figure 3.
Phylogenetic tree based on neighbor-joining analysis of 28S rDNA sequences for Gilbertella persicaria CNUFC-GWD3-9, G. persicaria CNUFC-GWD3-10, Pilobolus crystallinus CNUFC-EGF1-4, and P. crystallinus CNUFC-EGF1-5. Umbelopsis isabellina was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
3.2. Morphological characterization
Morphological structures for G. persicaria CNUFC-GWD3-9 and P. crystallinus CNUFC-EGF1-4 are described in details below.
3.2.1. Taxonomy of CNUFC-GWD3-9
Gilbertella persicaria (E.D. Eddy) Hesselt., Bulletin of the Torrey Botanical Club 87 (1): 24 (1960) (Table 2 and Figure 4)
Table 2.
Morphological characteristics of CNUFC-GWD3-9 and the reference species Gilbertella persicaria.
| Characteristic | CNUFC-GWD3-9 | Gilbertella persicariaa |
|---|---|---|
| Colony color | Rapid-growing, first white and then grayish yellow | Rapid-growing, first white and then grayish olive |
| Sporangiophores | 10.5–50.0 µm in width, variable in length | Up to 40–50 µm in width, up to 3–4 mm in height |
| Sporangia | Many-spored, globose to subglobose, first white-yellowish and then brown or black when mature, 36.5–250.5 × 37.2–253.5 µm | Many-spored, globose to irregularly globose, first white and then yellow and then black and glistening when mature, 40–260 µm in diameter |
| Columellae | Variable in shape, ovoid to pyriform, subglobose, 20.5–110.7 × 25.2–139.0 µm | Variable in shape depending on size, 40–119 × 20–170 µm |
| Sporangiospores | Irregular in shape, mainly ellipsoidal, 5.9–15.5 × 4.5–8.9 μm | Short oval and rather irregular in shape, 5–13 × 4.5–11 μm, up to 8.6 × 17 μm |
| Chlamydospores | Present | Present |
| Zygospores | Not observed | Present |
From the description by Hesseltine [8].
Figure 4.
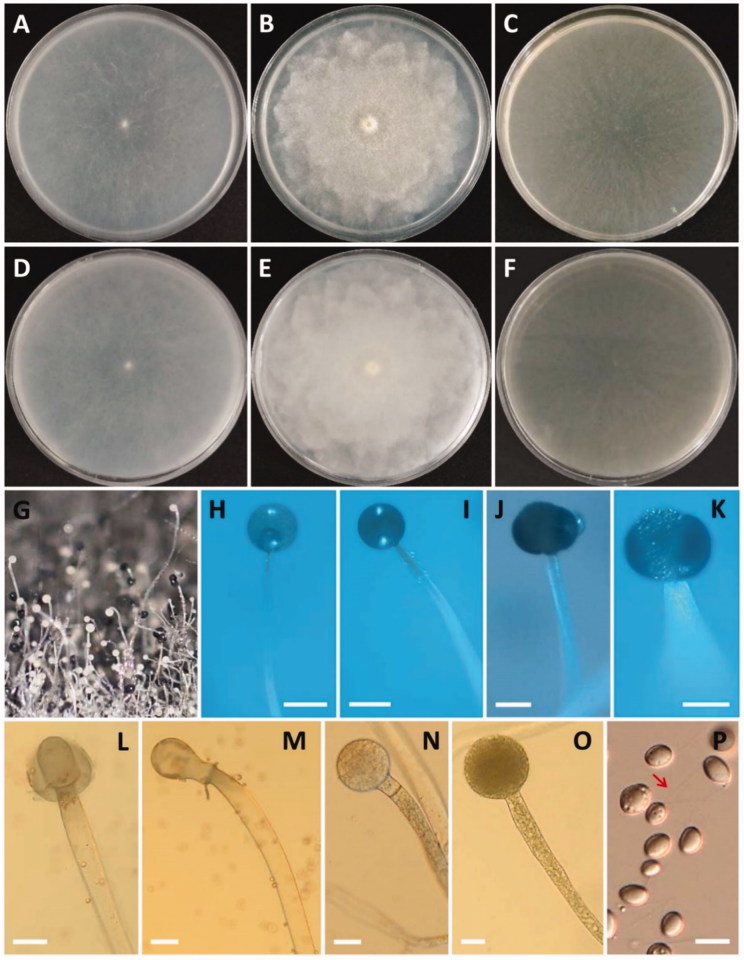
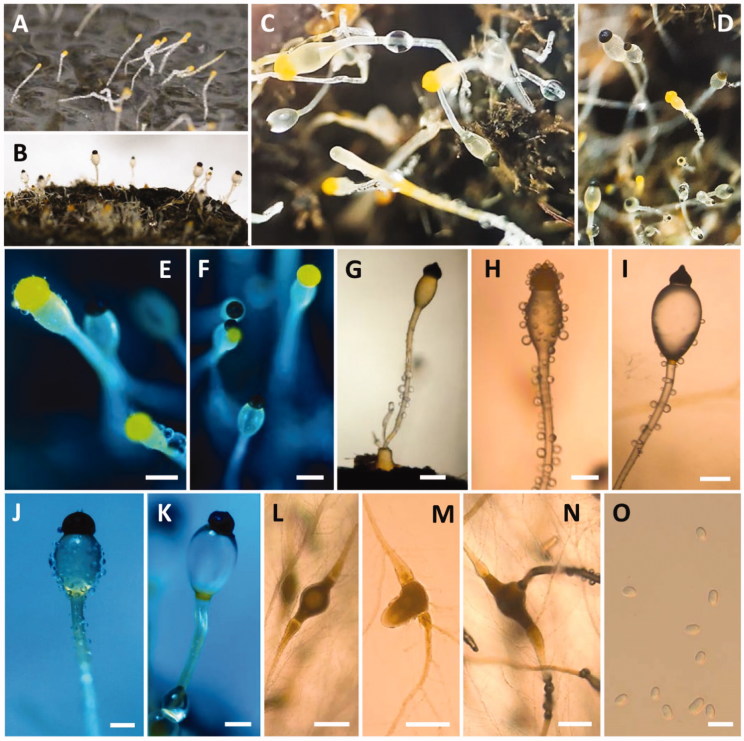
Morphology of Gilbertella persicaria CNUFC-GWD3-9. (A, D) Colony on synthetic mucor agar; (B, E) Colony on potato dextrose agar; (C, F) Colony on malt extract agar (A–C, top view; D–F, reverse view); (G–I, N, O) Immature and mature sporangia and sporangiophores; (J, K) Wall suturing in two equal halves; (L, M) Columellae with collarette; (P) Sporangiospore with appendage (red arrow) (scale bars H, I = 200 μm; J–M = 50 μm; N, O = 20 μm; P = 10 μm).
≡Choanephora persicaria E.D. Eddy, Phytopathology 15: 610 (1925)
Description: Colonies grew rapidly at 25 °C on SMA, filling the Petri dish after 2 days of incubation. The colony color was initially white and later grayish yellow. The colony reverse side was white and later pale yellow. Sporangiophores were 10.5–50.0 µm wide, variable in length, hyaline, light brown to grayish, sometimes branched and uncommonly had a septum under the sporangia. The sporangia separated longitudinally into two halves, were globose to subglobose, many-spored, initially white-yellowish and then turning brown or black at maturity, and measured 36.5–250.5 × 37.2–253.5 µm. Columellae were variable in shape, ovoid to pyriform, subglobose, and measured 20.5–110.7 × 25.2–139.0 µm. Sporangiospores were irregular in shape, mainly ellipsoidal, and measured 5.9–15.5 × 4.5–8.9 µm. Chlamydospore formations were well defined on the medium. Zygospores were not observed. Subsidiarily, the colonies grew slowly on SMA, PDA, and MEA at 5 °C. Among these, the best mycelial growth and sporulation were on PDA at 25 °C.
3.2.2. Taxonomy of CNUFC-EGF1-4
Pilobolus crystallinus (F.H. Wigg.) Tode, Schr. Berlin. Ges. naturf. Freunde: 46 (1784) (Table 3, Figure 5)
Figure 5.
Morphology of Pilobolus crystallinus CNUFC-EGF1-4. (A) Young sporangia and sporangiophores on dung agar medium; (B–K) Yellow and black sporangia, subsporangial vesicles, and sporangiophores (B–G, J, K, on water deer dung); (L–N) Substrate mycelia with trophocysts and rhizoidal extensions; (O) Sporangiospores (scale bars E–N = 200 μm; O = 10 μm).
≡Hydrogera crystallina F.H. Wigg., Primitiae Florae Holsaticae: 110 (1780)
≡Mucor urceolatus Dicks., Fasciculus plantarum cryptogamicarum Britanniae 1: 25, t.3:6 (1785)
Description: Colonies grew slowly at 25 °C on dung containing a medium. Trophocysts were subglobose to ellipsoidal and measured 199.0–409.8 × 147.5–186.9 µm. Sporangiophores were 57.0–122.7 µm wide, variable in length, erect, nonseptate, and unbranched. Subsporangial vesicles were ovoid, with an orange ring at the base, and measured 298.0–677.9 × 175.0–548.7 µm. Sporangia were hemispherical, umbonate, yellow or brown when young and turning black at maturity, and measured 169.5–371.5 × 151.5–295.5 µm. Columellae were ellipsoidal to mammiform and measured 110.3–186.7 × 122.1–230.5 µm. Sporangiospores were elliptical, hyaline, yellowish, and measured 6.0–8.5 × 4.0–5.5 µm.
4. Discussion
Until now, the distribution and occurrence of mucoralean species from dung and freshwater sources is poorly studied. As there have been no reports related to Gilbertella and Pilobolus species in Korea, the purpose of this paper was to describe and illustrate two rare species: Gilbertella and Pilobolus from specific sources such as freshwater and water deer dung in Korea, respectively.
In our phylogenetic analyses, the isolates of CNUFC-GWD3-9 and CNUFC-GWD3-10 were grouped with strains of G. persicaria CBS 190.32 (ex-type strain) (Figures 1 and 3). The morphological characteristics of an isolate of G. persicaria were almost identical with those previously described by Hesseltine [8], except for some narrower sporangiospores.
Our G. persicaria isolate presented sporangiophores that were sometimes branched, which was not recognized by Hesseltine [8].
Species of G. persicaria have been reported to produce extracellular enzymes such as endoglucanase, β-glucosidase, lipase, and pectinase [27–29]. Similarly, our strain, CNUFC-GWD3-9, showed pectinase activity, suggesting its potential as a source of novel enzyme (data not shown).
G. persicaria were often isolated from peach, pear, tomato, and dragon fruit by other researchers [7, 11, 12, 30–32]. However, this is the first isolation of G. persicaria from a freshwater source. Based on a recent literature, members of Ascomycota are dominant in freshwater environment with ∼622 species (170 genera), including more than 531 species of Hyphomycetes (55 genera), and 183 species of Trichomycetes (3 orders, no longer regarded as fungi); whereas the information about freshwater-derived fungi belonging to Basidiomycetes and Zygomycetes was rare [33,34]. Hence, further understanding about the biodiversity of Zygomycetes in freshwater is needed.
In contrast, isolates CNUFC-EGF1-4 and CNUFC-EGF1-5 were clustered with P. crystallinus species in a well-supported clade (Figures 2 and 3). Although most of the morphological features of our isolate were similar to those of P. crystallinus described by Boedijin [21], there were several differences in the diameter of subsporangial vesicles and sporangia. Subsporangial vesicles sizes reported in the literature range from 400–920 × 350–720 µm [21], which are larger than our maximum measurement. According to Foos et al. [35], although the size and shape of the sporangiospores have been detected to be stable within species [36], species descriptions typically give a large range of sporangiospore sizes. Moreover, the sizes of many structures used for species identification vary greatly depending on changes in the environmental conditions [35,37]. In this study, rDNA ITS gene provided sufficient phylogenetic information for the separation of Pilobolus species (Figure 2). However, isolate KH25 named as P. crystallinus was clustered with the other P. sphaerosporus species. Besides, isolates ATCC 36186 and ATCC 11505 named as P. crystallinus were not clustered with the other P. crystallinus species. Our results revealed that the isolate P. crystallinus KH25 should be changed to P. sphaerosporus. In addition, based on the sequences of ITS rDNA, we showed that the group containing species P. crystallinus is polyphyletic.
Despite the wide intraspecific variation found among some taxa, the ITS and D1/D2 regions have been used as appropriate barcode markers for identifying mucoralean fungi at the species level [4,5]. Currently, the traditional method of fungal identification is still mainly in use, as further studies are required to reconcile the molecular and morphological conceptions of families and genera. In the present study, we also used the molecular strategy for fungal identification at the level of species, specifically utilizing of ITS rDNA gene sequence and phylogenetic analysis. In 2011, Foos et al. [35] conducted sequence analysis of the ITS region of rRNA, small subunit of 18S rRNA, and LSU (23S) of mitochondrial rRNA, and showed that the genus Pilobolus is polyphyletic. The results revealed that molecular phylogenetic identification of Pilobolus species based on sequence analysis of pure culture isolates was more reliable than the traditional method of identification [35,38]. Our phylogenetic trees also agree with those reported by Foos et al. [35]. Therefore, these results confirmed that the isolate CNUFC-EGF1-4 belongs to the species P. crystallinus. Although a large number of species of fungi have been reported from the dung of different animal taxa, few have been reported from water deer dung. Thus, diversity of rare dung fungi or dung-derived fungi is to be investigated consistently.
Our findings contribute to the current knowledge of the diversity of the order Mucorales in Korea. However, data regarding the diversity of the order Mucorales in Korea are still lacking, further studies on the classification of different orders and families within the Mucoromycotina are required to expand our knowledge of rare undiscovered taxa with specific habitats in Korea.
Funding Statement
This work was supported by the Graduate Program for the Undiscovered Taxa of Korea, and by the Project on Survey and Discovery of Indigenous Fungal Species of Korea funded by the NIBR, and in part by the Project on Discovery of Fungi from Freshwater and Collection of Fungarium funded by the NNIBR of the Ministry of Environment (MOE). This work was in part supported by the BK21 plus program through the National Research Foundation (NRF) funded by the Ministry of Education of Korea.
Acknowledgment
We are grateful to Dr. Paul M. Kirk for his kindness in reviewing the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Hibbett DS, Binder M, Bischoff JF, et al. . A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. [DOI] [PubMed] [Google Scholar]
- 2.Spatafora JW, Chang Y, Benny GL, et al. . A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108:1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benny GL, Humber RA, Voigt K. The zygomycetous fungi: the Phylum Entomophthoromycota and subphyla Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina In McLaughlin DJ, Blackwell M, Spatafora JW, editors. Systematics of fungi, 2nd ed Vol. VII, The Mycota., part A. Springer Verlag: New York, United States; 2014; p. 209–250. [Google Scholar]
- 4.Walther G, Pawłowska J, Alastruey-Izquierdo A, et al. . DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia. 2013;30:11–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann K, Pawłowska J, Walther G, et al. . The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 2013;30:57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen TT, Lee SH, Bae S, et al. . Characterization of two new records of Zygomycete species belonging to undiscovered taxa in Korea. Mycobiology. 2016;44:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy ED. A storage rot of peaches caused by a new species of Choanephora. Phytopathology. 1925;15:607–610. [Google Scholar]
- 8.Hesseltine CW. Gilbertella gen. nov. (Mucorales). Bull Torrey Bot Club. 1960;87:21–30. [Google Scholar]
- 9.Benny GL. Gilbertellaceae, a new family of the Mucorales (Zygomycetes). Mycologia. 1991;83:150–157. [Google Scholar]
- 10.Voigt K, Olsson L. Molecular phylogenetic and scanning electron microscopical analyses places the Choanephoraceae and Gilbertellaceae in a monophyletic group within the Mucorales (Zygomycetes, Fungi). Acta Biol Hung. 2008;59:365–383. [DOI] [PubMed] [Google Scholar]
- 11.Mehrotra MD. Fruit rot of tomato caused by Gilbertella persicaria. Sydowia. 1964;17:17–19. [Google Scholar]
- 12.Pinho DB, Pereira OL, Soares DJ. First report of Gilbertella persicaria as the cause of soft rot of fruit of Syzygium cumini. Australasian Plant Dis Notes. 2014;9:143–146. [Google Scholar]
- 13.Page RM. Light and the asexual reproduction of Pilobolus. Science. 1962;138:1238–1245. [DOI] [PubMed] [Google Scholar]
- 14.Viriato A. Pilobolus species found on herbivore dung from the São Paulo Zoological Park, Brazil. Acta Bot Bras. 2008;22:614–620. [Google Scholar]
- 15.Zygomycota; [cited 2018 Mar 19]. Available from: http://www.zygomycetes.org.
- 16.Krug JC, Benny GL, Keller HW. Coprophilous fungi In Mueller GM, Bills GF, Foster MS, editors. Biodiversity of fungi. 1st ed Elsevier Academic Press: Burlington, United States; 2004; p. 468–499, 9780125095518. [Google Scholar]
- 17.Souza CAF, Lima DX, Gurgel LMS, et al. . Coprophilous Mucorales (ex Zygomycota) from three areas in the semi-arid of Pernambuco, Brazil. Braz J Microbiol. 2017;48:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boedijin KB. Notes on the Mucorales of Indonesia. Sydowia Ann Mycol Ser II. 1958;12:321–362. [Google Scholar]
- 19.Richardson MJ. Records of coprophilous fungi from the Lesser Antilles and Puerto Rico. Caribb J Sci. 2008;44:206–214. [Google Scholar]
- 20.White TJ, Bruns T, Lee S, et al. . Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In Innis M.A., Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. Academic Press: New York, United States; 1990; p. 315–322. [Google Scholar]
- 21.Vilgalys R, Hester M. . Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HB. Molecular phylogenetic status of Korean strain of Podosphaera xanthii, a causal pathogen of powdery mildew on japanese thistle (Cirsium japonicum) in Korea. J Microbiol. 2012;50:1075–1080. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Gibson TJ, Plewniak F, et al. . The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hankin L, Anagnostakis SL. The use of solid media for detection of enzyme production by fungi. Mycologia. 1975;67:597–607. [Google Scholar]
- 27.Takó M, Farkas E, Lung S, et al. . Identification of acid-and thermotolerant extracellular beta-glucosidase activities in Zygomycetes fungi. Acta Biol Hung. 2010;61:101–110. [DOI] [PubMed] [Google Scholar]
- 28.Takó M, Kotogán A, Krisch J, et al. . Enhanced production of industrial enzymes in Mucoromycotina fungi during solid-state fermentation of agricultural wastes/by-products. Acta Biol Hung. 2015;66:348–360. [DOI] [PubMed] [Google Scholar]
- 29.William S.Akiko A. history of soy flour, grits and flakes (510 CE to 2013): extensively annotated bibliography and sourcebook (illustrated ed.). Soyinfo Center. 2013; p. 371. [Google Scholar]
- 30.Mehrotra MD. Fruit rot of pear caused by Gilbertella persicaria var. indica. Sydowia. 1964;17:124–125. [Google Scholar]
- 31.Mehrotra MD. Fruit rot of peach by Gilbertella persicaria var. indica from India. Mycopathol Mycol Appl. 1966;29:151–154. [Google Scholar]
- 32.Guo LW, Wu YX, Mao ZC, et al. . Storage rot of dragon fruit caused by Gilbertella persicaria. Plant Dis. 2012;96:1826. [DOI] [PubMed] [Google Scholar]
- 33.Jones EBG, Hyde KD, Pang KL. Freshwater fungi and fungal-like organisms. Boston: De Gruyter; 2014. [Google Scholar]
- 34.Nguyen TT, Pangging M, Lee HB. Three unrecorded fungal species from fecal and freshwater sample in Korea. Kor J Mycol. 2017;45:304–318. [Google Scholar]
- 35.Foos KM, May NL, Beach DL, et al. . Phylogeny of Pilobolaceae. Mycologia. 2011;103:36–44. [DOI] [PubMed] [Google Scholar]
- 36.Foos KM, Jeffries BS. Sporangiospore variability in Pilobolus. Proc Indiana Acad Sci. 1988;98:105–108. [Google Scholar]
- 37.Hu FM, Zheng RY, Chen GQ. A redelimitation of the species of Pilobolus. Mycosystema. 1989;2:111–133. [Google Scholar]
- 38.Foos KM, Sheehan KB. Molecular identification of Pilobolus species from Yellowstone National Park. Mycologia. 2011;103:1208–1215. [DOI] [PubMed] [Google Scholar]