ABSTRACT
This study was performed to elucidate the effects of linoleic acid (LA), oleic acid (OA) and their combination (LA + OA) on cell proliferation, apoptosis, necrosis, and the lipid metabolism related gene expression in bovine satellite cells (BSCs), isolated from bovine muscles. Cell viability was significantly increased with the OA and LA treatment. Furthermore, LA + OA enhanced cell proliferation in a dose-dependent manner (10 to 100 µM), whereas it lowered at 250 µM. In addition, a cell-cycle analysis showed that 100 µM of LA and OA markedly decreased the G0/G1 phase proportion (62.58% and 61.33%, respectively), compared to controls (68.02%), whereas the S-phase cells’ proportion was increased. The ratio of G2/M phase cells was not significantly different among the groups. Moreover, analyses with AO/EtBr staining showed that no apoptosis occurred. Necrosis were determined by flow cytometry using Annexin V-FITC/PI staining which revealed no early apoptosis in the cells pretreated with LA or OA, but occurred in the LA + OA group. We also analyzed the mRNA expression of lipid metabolizing genes such as peroxisome proliferator receptor alfa (PPARα), peroxisome proliferator receptor gamma (PPARγ), acyl-CoA oxidase (ACOX), lipoprotein lipase (LPL), carnitine palmitoyl transferase (CPT-1), and fatty-acid binding protein4 (FABP4), which were upregulated in LA or OA treated cells compared to the control group. In essence, LA and OA alone promote the cell proliferation without any apoptosis and necrosis, which might upregulate the lipid metabolism related gene expressions, and increase fatty-acid oxidation in the BSCs’ lipid metabolism.
KEYWORDS: Bovine satellite cells, cell proliferation, gene expression, linoleic acid, oleic acid
Introduction
Fats are essential for living organisms to provide energy to cells and stop pathogens from entering the body. Many studies have reported the effect of fatty acids on muscle functions (Lee et al. 2009). For example, polyunsaturated fatty acids (PUFAs) could prevent cardiovascular disease and possible benefits of using PUFA supplements were shown in several clinical evaluations (Ander et al. 2003). Similarly, monounsaturated fatty acids (MUFAs) are able to reduce bad blood cholesterol levels which can lower the risk of heart disease and stroke. They also provide nutrients for the development and maintenance of body cells. A diet rich in saturated fats or linoleic acid (LA) and n-6 PUFAs leads to insulin resistance, mostly through their effects on oxidative skeletal muscles (Storlien et al. 1986; Holness et al. 2000). Previous studies have also confirmed that LA and OA exert a proliferative effect in primary cultured vascular smooth muscle cells (Rao et al. 1995; Lu et al. 1998).
Cell division, also called mitosis, is extremely important for all living organisms because without it, reproduction cannot occur. Mitosis includes cell growth and cell proliferation based on the segregation of replicated DNA and chromosomes into two separate cells. Primarily, cellular proliferation is arranged by the cell cycle’s regulation, which consists of four distinct chronological phases (G0/G1, S, G2, and M). This coordination mainly takes place at G1/S and G2/M transitions. The cell division is tightly regulated by the activation and inactivation of proteins that are variable in expression depending on their stage in the cell cycle (Elledge 1996; Stiller 2004; van den Heuvel 2005). Generally, long-chain PUFAs can inhibit cell proliferation and promote necrosis via apoptosis or their concentrations which is different than in saturated fatty acids (SFAs). However, some studies have shown a positive effect of PUFAs on cell proliferation (Finstad et al. 1994; Terano et al. 1999; Maurin et al. 2002; Yonezawa et al. 2008).
The peroxisome proliferators-activated receptor (PPAR) family has three subtypes called PPARα, PPARγ, and PPARβ. PPARα and PPARγ are hormone receptors located in the nuclear membrane and they play an important role in the lipid and glucose metabolisms. As a transcription factor, they regulate the fatty-acid metabolism and transportation by controlling mitochondrial β-oxidation, the peroxisomal β-oxidation expression, and upstream target genes, such as acyl-CoA oxidase (ACOX), fatty-acid binding protein (FABP), and carnitine palmitoyl transferase (CPT) genes (Schoonjans et al. 1995; Niot et al. 1997; Feige et al. 2006). This interaction enhances the expression of genes that code for proteins such as fatty acid synthase (FAS) and fatty-acid binding proteins, leading to the growth of adipocytes and the accumulation of fatty acids. Therefore, this experiment investigated the impact of OA, LA, and OA + LA on the proliferation, apoptotic morphological changes, and necrosis, as well as whether both LA and OA can impact the mRNA expression pattern of lipid metabolism genes in bovine satellite cells.
Materials and methods
Chemicals and reagents
Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were obtained from GIBCO (NY, USA). Fatty acids were acquired from Sigma-Aldrich (St. Louis, USA) as well as bovine serum albumin (BSA), 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT), dimethyl sulfoxide (DMSO), propidium iodide (#P4170), and acridine orange/ethidium bromide stain. A FITC/PI Annexin V apoptosis detection kit (Cat # 640914) was purchased from BioLegend, San Diego, California, USA and oligo-primers were synthesized by Bioneer (Daejeon, South Korea). All chemicals were of analytical grade.
Preparation of linoleic and oleic acid
Long-chain fatty acids such as linoleic acid (n-6 PUFA, C18:2) and oleic acid (n-9 MUFA, C18:1) were used in this experiment. They were prepared according to Cousin et al. (2001). Briefly, the stock solution of 60 mM LCFA was pre-arranged in an equimolar solution of NaOH by saponification at 70°C and a 10% (w/v) fatty-acid-free BSA solution in ultrapure water at 55°C. Various concentrations of sodium linoleate or oleate were complexed to BSA and then mixed by shaking. The mixture was incubated at 55°C for 10 min, cooled down to room temperature, and then sterile-filtered. The resulting solutions were added to the cell culture medium for reaching the desired final concentrations (10, 50, 100, and 250 µM) and 0.1% BSA before the experiment. The stock solutions were kept at −20°C until use.
Isolation of satellite cells from the bovine muscle
The satellite cells were isolated according to the method of Dodson et al. (1987) with minor modifications: Using sterile techniques, the longissimus dorsi muscle (500 g) was dissected from 30 month old Hanwoo steer (Korean native cattle) immediately after slaughter, transported to the laboratory, and subsequent procedures were conducted in a tissue culture hood. After removal of the epimysium and most of the fat, the muscle strips were ground by a sterile meat grinder and incubated with 1% pronase solution (Sigma-Aldrich) at 37°C for 60 min. Followed by enzymatic digestion with 1% pronase, single cells were separated by repeated centrifugation at 1,500 × g for 4 min at room temperature. The primary muscle cells were cultured in DMEM (GIBCO) supplemented with 15% FBS (GIBCO), 100 μg/mL streptomycin, and 100 IU/mL penicillin (Sigma-Aldrich) in a humidified incubator at 37°C with 5% CO2.
Magnetic assorted cell sorting (MACS) of satellite cells
Satellite cells were isolated from the muscle using a magnetic cell-sorting system (AutoMACS, Milteny Biotech, Bergisch Gladbach, Germany). At 80% confluence, the cells were collected and re-suspended in 1 × PBS (GIBCO), supplemented with 0.5% BSA and 2 mM EDTA. After centrifugation at 1,500 × g for 5 min, the cell pellet was re-suspended in 1 × PBS (100 μL) with 10 μg anti-Mcadherin antibodies (BD BioScience, San Diego, CA) and incubated with 20 μL of anti-mouse IgG1micro beads at 4°C for 30 min. Finally, the cell suspension (107 cells/2 mL PBS) was loaded into a magnetic cell-sorting system to isolate satellite cells and afterwards, the positive cells were counted by using a hemocytometer as well as the percentage of satellite cells determined. The satellite cells were cultivated in a growth medium and were subcultured to obtain 80% confluence and finally cells from fifth passage were used for the current study.
Cell viability assay
Cell proliferation was determined by MTT assay. The cells were seeded at 2 × 105 cells/mL in 96 well plates and maintained for 48 h in a complete growth medium. They were then exposed to LA and OA (both 0, 10, 50, 100, and 250 µM) in a growth medium for 24 h and 48 h. The cells were then incubated with 5 mg/mL MTT for 4 h at 37oC after which the formazan crystals were dissolved in DMSO. The absorbance of each well was measured at 490 nm by using a microplate reader (Multiskan GO, ThermoFisher Scientific, USA). The results are displayed as a percentage of untreated controls. Cell viability was calculated by the following formula: cell viability = (ODtreated – ODblank)/(ODcontrol – ODblank) wells × 100.
Cell-cycle analysis by flow cytometry
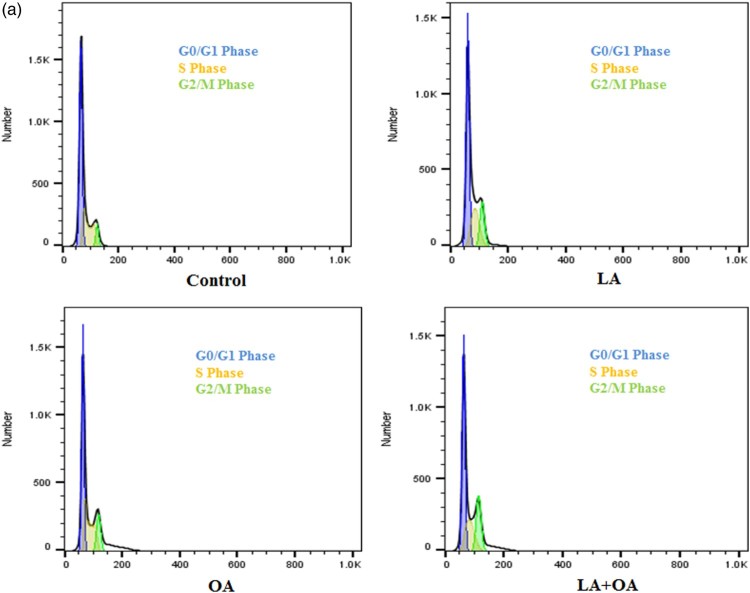
The cells were seeded into six-well plates at a density of 2 × 105 cells per well and incubated for 48 h. They were cultured in DEME supplemented with 10% of FBS and incubated at 37°C as well as 5% CO2. The medium was removed and replaced with another medium (final DMSO concentration 0.05% v/v) containing LA and OA (100 µM). After incubation for 24 h, the cell layer was trypsinized, washed with cold PBS, and fixed with 70% ethanol. RNAse (0.2 mg/mL) and propidium iodide (0.02 mg/mL) in the amount of 20 µL each were added to the cell suspensions following which the mixtures were incubated at 37°C for 30 min. The samples were then analyzed with FACS Calibur flow cytometry. Differences in DNA mass detected by fluorescence channel 2 allowed allocation of the cells to the G1, S, and G2 phases (Figure 2) of the cell cycle using the FlowJo 10.0.7 software (Treestar Inc., Ashland, USA).
Apoptosis assay by AO/EtBr staining methods
The cells were seeded onto chamber slides in six-well plates at a density of 2 × 105 cells per well. After the LA and OA treatments, they were incubated in LA and OA (100 µM) for 24 h. The cells were cultured in DEME supplemented with 10% of FBS and incubated at 37°C in 5% CO2. After removal of the medium, they were fixed with methanol:acetic acid (3:1). Following incubation for 1 h at room temperature, the methanol and acetic acid were removed and the cells were washed with ice-cold PBS. The cell nuclei were counterstained with AO/EtBr (100 µg/mL AO, 100 µg/mL EtBr) for 10–20 min and then examined under a fluorescence microscope (# LSM 510 META, Carl Zeiss, Jena, Germany).
Determination of apoptosis and necrosis by annexin V-FITC/PI-Staining
The FITC/PI Annexin V apoptosis detection kit (BioLegend, Cat # 640914, San Diego, California, USA) was used according to the manufacturer’s instructions for the apoptosis detection of LA- and OA-treated cells. The cells (2 × 105) were treated with LA and OA for 24 h. After incubation, the cells were harvested, washed with PBS, suspended in Annexin V binding buffer, and incubated with FITC-labeled Annexin V and PI for 15 min at room temperature in the dark. The Annexin V-FITC/PI-stained cells were analyzed using a BD FACS Calibur flow cytometer (BD) and the data analyzed using the FlowJo 10.0.7 software (Treestar Inc., Ashland, USA).
RNA extraction and the quantitative real-time polymerase chain reaction (qPCR)
After treatment, total cellular RNA was extracted from the cells by using the TRIzol Reagent (Invitrogen, NY, USA), following the manufacturer's instructions. Total RNA was quantified by Nano Drop (Thermo Fisher Scientific) at 260 nm/280 nm absorbance. The quality of total RNA was assessed by using an ExperionTM Automated Electrophoresis System (BIO-RAD) with RNA chip kits (ExperionTM RNA StdSens Reagents, #700–7259, BIO-RAD) and RNA samples of good quality were selected for further reverse transcription. Total RNA (1 μg) was reverse-transcribed into cDNA using the iScriptTM cDNA Synthesis kit (BIO-RAD, Hercules, CA, USA) according to the manufacturer's instructions. The reverse transcription was processed at 25°C for 18 sec followed by 11 cycles, 48°C for 4 min, and the enzyme reaction was inhibited at 55°C for 18 sec. Real-Time PCR was performed using the SsoFastTM EvaGreen® Supermix (BIO-RAD) and CFX96TM Real-Time PCR Detection System (BIO-RAD). cDNA was amplified for each gene and the reaction was carried out according to the manufacturer’s instructions (BIO-RAD). The thermal cycling parameters were as follows: 95°C for 2 min, followed by 40 cycles at 95°C for 5 s, 59°C for 5 s, and 65°C for 5 s. The ΔΔCT method was used to determine the relative fold-changes and all data were normalized with the housekeeping gene GAPDH. The qPCR results were calculated by using the ΔCt value (Ct gene of interest – Ct reference gene). The relative gene expression was obtained by ΔΔCt methods (ΔCt sample – ΔCt calibrator) with the control group as a calibrator for comparing all unknown sample gene expression levels. The conversion between ΔΔCt and relative gene expression levels is as follows: Fold induction = 2−ΔΔCt, where 2−ΔΔCt is the relative gene expression (Livak and Schmittgen 2001).
Statistical analyses
The effect of the treatments was analyzed by a one-way analysis of variance using the SPSS program (version 18). Tukey’s test was used for multiple comparisons. Statistically significance was determined as p < 0.05.
Results
Effect of LA and OA on cell proliferation
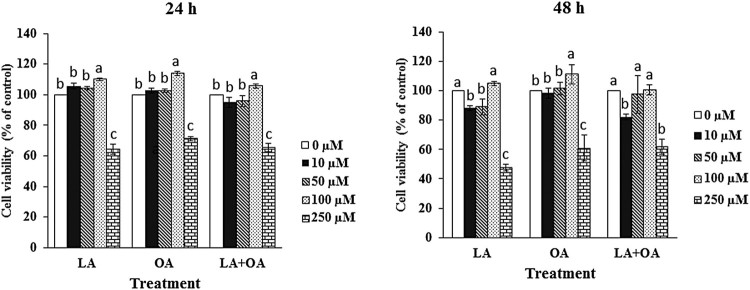
The different concentrations of LA, OA, and LA + OA altered the cells viability at 24 and 48 h as compared to the control group. At 24 h treatment with LA and OA, cell proliferation increased in the dose of 10 µM to 100 µM, while it decreased at 250 µM compared to the untreated control group (Figure 1). Cell proliferation (p < 0.05) increased significantly in response to 100 µM of LA, OA, and LA + OA at 24 h. Besides, there was no significant effect in response to 100 µM of LA, LA + OA at 48 h compared to control group, however OA treated group increased (p < 0.05) among the groups. These results suggest that a treatment with low concentrations (10 µM to 100 µM) of LA and OA alone could markedly promote the BSCs’ cells rather than a high concentration (250 µM) Table 1.
Figure 1.
Different long-chain fatty acids induce cell death. Cells were incubated with 10–250 μM of oleic acid (C18:1), linoleic acid (C18:2), and a combination of both for 24 h, 48 h and then, cell viability was estimated by MTT assay. The values are the mean ± SE. Mean values with differing letters differ significantly (p < 0.05).
Table 1. Primer information for qPCR.
| Gene | Accession number | Sequence (5’– 3’) | Size (bp) | Annealing (°C) |
|---|---|---|---|---|
| PPARα | EF534215 | F- CCTGGCTTCTCCAATCTTGAC R- GCAAATGATGGCAGCGACA |
252 | 59 |
| PPARγ | NM_181024 | F- GTGAAGCCCATTGAGGACAT R- AGCTGCACGTGTTCTGTCAC |
148 | 58 |
| LPL | NM_001075120 | F- ACTTGCCACCTCATTCCTG R- ACCCAACTCTCATACATTCCTG |
119 | 56 |
| ACOX | NM_001035289 | F- GAGTGAGCTGCCTGAGCTTC R- TTGTCCAGGACGTGAAAGC |
62 | 59 |
| CPT-1 | GW342984 | F- GTCTCCAAGGCTCCGACAA R-AAGACCCGAATGAAAGTA |
193 | 58 |
| FABP4 | BT10868 | F- GCTGCACTTCTTTCTCACCT R- TTCCTGGTAGCAAAGCCCAC |
140 | 58 |
| GAPDH | NM_001034034 | F- CACCCTCAAGATTGTCAGC R- TAAGTCCCTCCACGATGC |
98 | 57 |
PPARα, Peroxisome proliferator receptor alfa; PPARγ, Peroxisome proliferator receptor gamma; LPL, Lipoprotein lipase; ACOX, Acyl-CoA oxidase; CPT-1, Carnitine palmitoyl transferase; FABP4, Fatty-acid-binding protein4; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Effect of LA and OA on the cell-cycle analysis
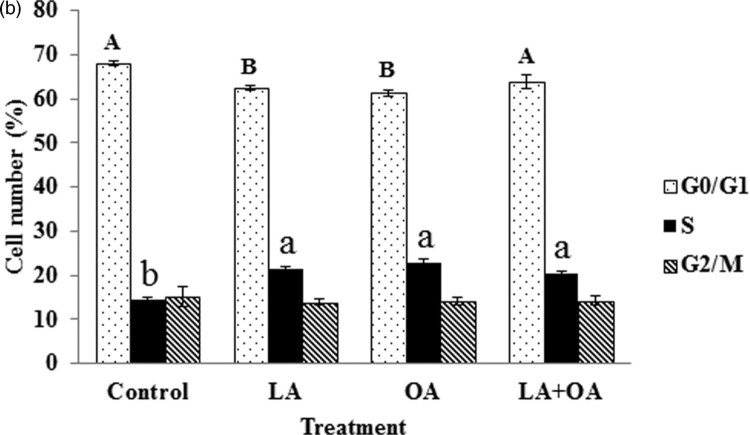
The status of the cell cycle for cells treated with LA, OA, and LA + OA was analyzed (Figures 2 and 3). In the controls, the percentage of the cells at the G0/G1 phase was 68.02% while it was 62.58%, 61.33% and 63.92% for the LA, OA, and LA + OA treated groups, respectively. Decreases of 5.44% for LA, 6.69% for OA, and 4.10% for LA + OA at the G0/G1 phase were observed, accompanied by a parallel increase in the S phase (Figure 3). The ratio of G2/M phase cells was not significant among the treatment groups (p > 0.05). The results indicate that PUFA and MUFA promote cell proliferation by accelerating at the G0/G1 phase to the S phase. The effect on the cell cycle was consistent with the cell viability assay findings.
Figure 2.
Analysis by flow cytometry of cell cycle profiles. Effect of LA, OA, and LA + OA at 100 µM on a cell cycle for 24 h.
Figure 3.
Cell-cycle analysis after fatty-acid treatment. Cell-cycle analysis (G0/G1, S, and G2/M) of BSCs’ exposure to 100 µM of LA and OA for 24 h. Mean values with differing letters differ significantly (p < 0.05). Large character for G0/G1 and small character for S phase. Lack of letter is not significant.
Apoptosis assay with AO/EB staining
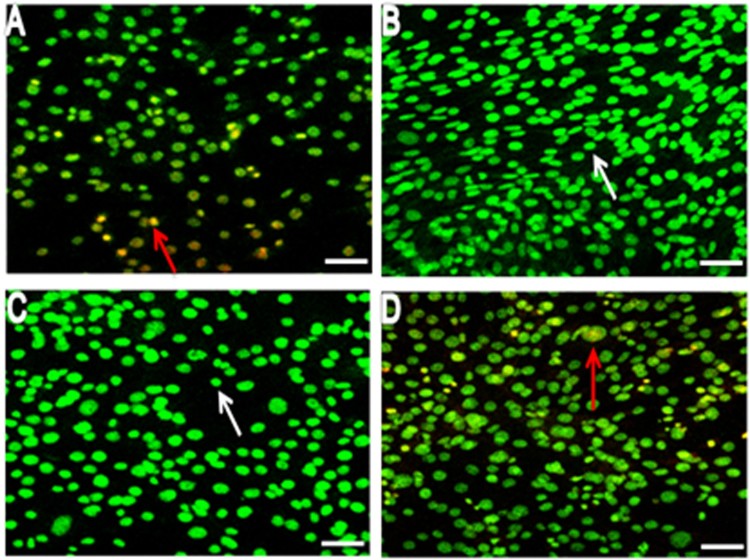
Acridine orange (AO) and ethidium bromide (EtBr) were used to differentiate the morphological changes in apoptotic and/or viable cells (Figure 4A, B, C and D). AO is a crucial dye that stains nuclear DNA across an intact cell membrane while EB only stains cells that have lost membrane integrity. To observe the morphological changes, BSCs were treated with 100 µM of LA and OA each for 24 h and the cells were examined under a fluorescent microscope. In the LA and OA treated groups, living cells were stained bright green spots (Figure 4B and C), while 5% of apoptotic BSCs were obtained in the control cells. In contrast, the treatment of 100 µM of LA + OA showed condensed nuclei, membrane blebbing and apoptotic bodies (EtBr stained cells) in BSCs.
Figure 4.
Morphological observation at 24 h in exposure to 100 µM fatty acids A (Control), B (OA), C (LA), and D (LA + OA) using AO/EB double-staining with confocal microscopy; Arrows, white Normal cells, redViable apoptotic cells. Bar scale 100 µm.
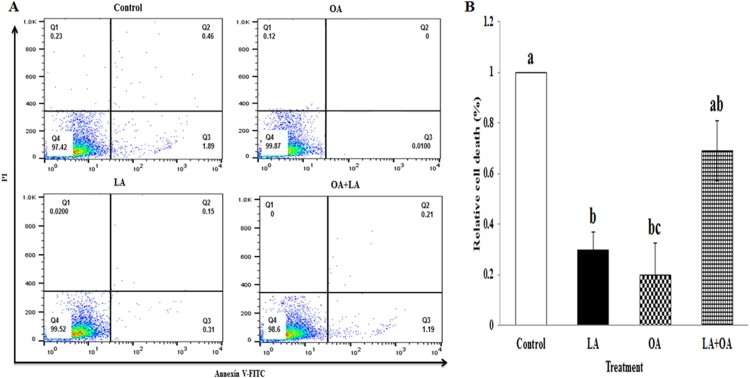
Annexin V-FITC flow cytometric analysis for determination of apoptosis and necrosis
Furthermore, (early and late) apoptosis assessed by the induction of FFAs was confirmed by the Annexin V–PI double-labeling assay. Figure 5A displays represent Annexin V-FITC/PI results, which indicate that OA, LA, and LA + OA treatments induced the increase of early-stage cell death in BSCs which was 0.01%, 0.31%, and 1.19%, while late cell deaths accounted for 0%, 0.15%, and 0.21%, respectively. Total cell death was significantly lower in the LA and OA treatment groups than in the controls, while there was a marked elevation in the LA + OA group compared to the OA-treated one (Figure 5B). Hence the treatment of LA and OA alone could promote cell growth and inhibit apoptosis compared to the combined treatment (LA + OA).
Figure 5.
FACS analysis of cell death. Effect of LA, OA, and the combination of both on cell death in exposure to 100 µM fatty acids for 24 h A (Control, LA, OA, LA + OA) and B, graphical representation. Each treatment was performed in triplicate. Mean values with differing letters differ significantly (p < 0.05).
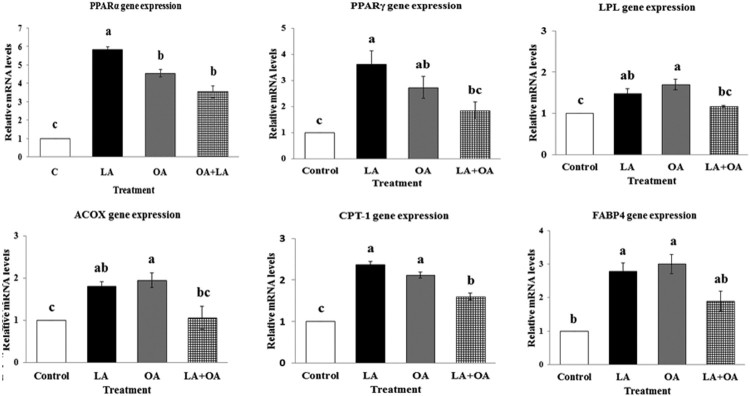
Effect of LA and OA on mRNA expression levels of selected genes in the lipid metabolism
To determine the effect of USFAs, we measured the mRNA levels of genes involved in the lipid metabolism of bovine primary satellite cells after treatment with LA, OA, and LA + OA for 24 h. The data presented in Figure 6 indicated effects of linoleate and oleate on the expression of PPARα, PPARγ, LPL, ACOX, CPT-1, and FABP4 by quantitative real-time PCR analysis. OA and LA showed marked effects on the lipid metabolism gene expressions. Incubation of BSCs with 100 µM LA and OA each resulted in a significant increment of PPARα, PPARγ, ACOX, LPL, CPT-1, and FABP4 genes’ mRNA levels compared to the control group (p < 0.05).
Figure 6.
Expression levels of peroxisome proliferator receptor alfa (PPARα), peroxisome proliferator receptor gamma (PPARγ), lipoprotein lipase (LPL), acyl-CoA oxidase (ACOX) genes, carnitine palmitoyl transferase (CPT-1), and fatty-acid-binding protein4 (FABP4) genes. The mRNA levels were determined by qPCR and the results were normalized with the housekeeping gene GAPDH. Values are the mean ± SE. Mean values with differing letters differ significantly (p < 0.05).
Discussion
Long-chain fatty acids (LCFAs) are important sources of energy and affect various tissues in numerous aspects (Chawla et al. 2001). In this study, the effect of unsaturated fatty acids on cell viability was determined by MTT assay results which showed that the cell viability of BSCs significantly increased in the treatment of 100 µM LA and OA when incubated alone or together. It was reported that USFAs (LA and OA) can promote cell proliferation and survival via the phosphatidylinositol 3-kinase (PI3K) pathway (Hardy et al. 2000). Further, incubation with linoleate for 24 h significantly promoted cell proliferation and cell viability (Yonezawa et al. 2008; Marei et al. 2010). The n−6 PUFAs, linoleic acid (LA), γ-linoleic acid (GLA), mono-unsaturated fatty acids (MUFAs), and oleic acid (OA) increased the proliferation of C2C12 cells (Lee et al. 2009). However, other studies with humans and rats showed that higher concentrations of dietary PUFAs may inhibit cell proliferation (Maurin et al. 2002; van Beelen et al. 2007; Yonezawa et al. 2008), which was similarly observed in this investigation. Also, many researchers have reported that MUFAs such as OA are either cytoprotective or non-toxic in several types of cells (Mu et al. 2001; Eitel et al. 2002; Akazawa et al. 2010; Suzuki et al. 2011).
Cyclin-dependent kinases (CDKs) belong to a group of protein kinases (serine/threonine kinases) activated via formation of a complex with cyclin molecules, involved in cell-cycle regulation. The level of CDK remains constant in a cell, while the cyclin level fluctuates depending on the cell-cycle stage. CDKs play a vital role in gene expressions and the cell-cycle regulation (Saeed et al. 2012). However, unsaturated fatty acids, namely OA, GLA, AA, DHA, and LA stimulate the proliferation of C2C12 cells (Lee et al. 2009). A prior study asserted that the presence of double bonds may be important for fatty acids to enhance the proliferation of skeletal muscle cells. Hence, our results also suggest that LA and OA which have double bonds, might increase the cell proliferation in BSCs. In another investigation, linoleate promoted primary cultured hepatocytes of geese (Pan et al. 2011). OA also induced the proliferation of rat aortic smooth muscle cells by activating protein kinase C (Lu et al. 1996). Such findings corroborate that fatty acids play an important role in regulating skeletal muscle growth.
To the best of our knowledge, studies to detect gene expressions in primary cultured bovine satellite cells exposed to LA and OA singly or in combination are very rare. PPARα gene expression was significantly upregulate (p < 0.50) in LA, OA and LA + OA treated groups than the control group. However, there were no significant effect between OA and LA + OA groups. This is in agreement with the observation that n-3 PUFAs and n-6 PUFAs are more potent activators of PPAR-α in vivo (Clarke 2001). It was shown that LA and OA upregulate the PPARγ gene in BSCs as compared to a combination of the two (LA + OA). In this study, the examined lipid metabolism genes, such as ACOX and LPL, also increased in the LA and OA treated groups. These results are similar to some previous studies of mammalian and fish cells (Swagell et al. 2007; Bionaz et al. 2008; Kjaer et al. 2008). The mRNA level of PPARγ increased more in the LA treated group than in the OA and LA + OA groups. In goose primary hepatocytes Pan et al. (2011) observed that linoleate elevated the PPARγ mRNA level at a certain concentration but was reduced with increasing concentrations of linoleate. In mammals, PUFAs modulate gene expressions in different systems by regulating transcription factors such as peroxisome proliferator receptors (Clarke et al. 2002; Ringseis et al. 2007; Sato et al. 2007). LPL is a rate-limiting triacylglycerol (TG) enzyme. Several investigations have reported an inverse relationship between LPL content and plasma TG levels (Inoue et al. 1999, Wong et al. 2002, Augustus et al. 2003). In this study, the cells incubated with LA or OA exhibited an increased LPL mRNA level, which may affect the TG level in bovine satellite cells. In vivo and in vitro studies demonstrated that LA lowers the LPL mRNA level in chicken and fish adipocytes (Liang et al. 2002; Montalto and Bensadoun, 1993), which indicates that LPL regulation by fatty acids might be tissue-specific.
We also measured CPT-1, which plays a key role in fatty-acid β-oxidation: Compared with the control group, linoleate and oleate increased the mRNA expression of CPT-1, although LA + OA significantly decreased it more than the LA or OA group which denotes that fatty acid β-oxidation increased, consistent with a PPAR agonist improving hepatic steatosis via improvement in fatty-acid beta-oxidation and the direct prevention of inflammations (Nagasawa et al. 2006; Litvinov et al. 2010). Notably, FABP4 was regulated by LA or OA. In the present results, LA or OA promote the expression of lipid catabolism genes and then increase the β-oxidation. Similarly oleic acid tended to promote fatty acid de novo synthesis and fatty acid oxidation in HepG2 cell (Kohjima et al. 2009). PUFAs suppress the expression of the genes responsible for fatty acid and triglyceride synthesis while also stimulating the expression of the genes involved in fatty-acid oxidation (Xu et al. 2002, 2001). LA or OA play a significant role in the lipid metabolism, which suggests that diets containing appropriate amounts of LA and OA could influence the lipid catabolism, and the research results may offer help for further animal lipid metabolism studies.
Conclusion
LA and OA stimulated cell proliferation and the cell-cycle conversion from the G0/G1 phase to the S phase. The transition from one cell-cycle phase to another phase occurs in an orderly fashion, it is regulated by different cellular proteins, and demonstrates that LA and OA play a positive role in the proliferation of BSCs. Both LA and OA upregulated the PPARγ gene expression and transformed the expression of some lipid metabolism related genes. This study confirmed that the unsaturated fatty acid could influence cell proliferation and the lipid metabolism in BSCs.
Funding Statement
This work was supported by Cooperative Research Program for Agriculture Science and Technology Development, Rural Development Administration, Republic of Korea: [Project No. PJ01197803].
Acknowledgements
This work was carried out with the support of the “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01197803), Rural Development Administration, Republic of Korea. Annexin V-FITC/PI-stained cells were analyzed using a BD FACS Calibur flow cytometer (USA, BD) installed at the Center for University-Wide Research Facilities (CURF) at Chonbuk National University. We thank Ms. Eun-Jin Choi at the CURF of Chonbuk National University.
Disclosure statement
The authors declare no potential conflict of interest.
ORCID
Shah Ahmed Belalhttp://orcid.org/0000-0001-5353-2439
References
- Akazawa Y, Cazanave S, Mott JL, Elmi N, Bronk SF, Kohno S, Charlton MR, Gores GJ.. 2010. Palmitoleate attenuates palmitate induced Bim and PUMA up-regulation and hepatocyte lipoapoptosis. J Hepatol. 52:586–593. doi: 10.1016/j.jhep.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ander BP, Dupasquier CMC, Prociuk MA, Pierce GN.. 2003. Polyunsaturated fatty acids and their effects on cardiovascular disease. Exp Clin Cardiol. 8(4):164–172. [PMC free article] [PubMed] [Google Scholar]
- Augustus AS, Kako Y, Yagyu H, Goldberg IJ.. 2003. Routes of FA delivery to cardiac muscle: modulation of lipoprotein lipolysis alters uptake of TG-derived FA. Am J Physiol Endocrinol Metab. 284:E331–E339. doi: 10.1152/ajpendo.00298.2002 [DOI] [PubMed] [Google Scholar]
- Bionaz M, Baumrucker CR, Shirk E, Van Den Heuve JP, Block E, Varga GA.. 2008. Short communication: characterization of Madin-Darby bovine kidney cell line for peroxisome proliferator activated receptors: temporal response and sensitivity to fatty acids. J Dairy Sci. 91:2808–2813. doi: 10.3168/jds.2007-0789 [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ.. 2001. Nuclear receptors and lipid physiology: opening the X-files. Sci. 294:1866–1870. doi: 10.1126/science.294.5548.1866 [DOI] [PubMed] [Google Scholar]
- Clarke SD.2001. Polyunsaturated fatty acid regulation of gene transcription: a molecular mechanism to improve the metabolic syndrome. J Nutr. 131:1129–1132. doi: 10.1093/jn/131.4.1129 [DOI] [PubMed] [Google Scholar]
- Clarke SD, Gasperikova D, Nelson C, Lapillonne A, Heird WC.. 2002. Fatty acid regulation of gene expression: a genomic explanation for the benefits of the Mediterranean diet. Ann NY Acad Sci. 967:283–298. doi: 10.1111/j.1749-6632.2002.tb04284.x [DOI] [PubMed] [Google Scholar]
- Cousin SP, Hügl SR, Wrede CE, Kajio H, Myers MG Jr., Rhodes CJ.. 2001. Free fatty acid-induced inhibition of glucose and insulin-like growth factor I-induced deoxyribonucleic acid synthesis in the pancreatic b-cell line INS-1. Endocr. 142:229–240. doi: 10.1210/endo.142.1.7863 [DOI] [PubMed] [Google Scholar]
- Dodson MV, Martin EL, Brannon MA, Mathison BA, Mcfarland DC.. 1987. Optimization of bovine satellite cell derived myotube formation in vitro. Tissue Cell. 19(2):159–166. doi: 10.1016/0040-8166(87)90001-2 [DOI] [PubMed] [Google Scholar]
- Eitel K, Staiger H, Brendel MD, Brandhorst D, Bretzel RG, Haring HU, Kellerer M.. 2002. Different role of saturated and unsaturated fatty acids in beta-cell apoptosis. Biochem Biophys Res Commun. 299:853–856. doi: 10.1016/S0006-291X(02)02752-3 [DOI] [PubMed] [Google Scholar]
- Elledge SJ.1996. Cell cycle checkpoints: preventing an identity crisis. Sci. 274:1664–1672. doi: 10.1126/science.274.5293.1664 [DOI] [PubMed] [Google Scholar]
- Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W.. 2006. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog Lipid Res. 45:120–159. doi: 10.1016/j.plipres.2005.12.002 [DOI] [PubMed] [Google Scholar]
- Finstad HS, Kolset SO, Holme JA, Wiger R, Farrants AK, Blomhoff R, Drevon CA.. 1994. Effect of n-3 and n-6 fatty acids on proliferation and differentiation of promyelocytic leukemic HL-60 cells. Blood. 84:3799–3809. [PubMed] [Google Scholar]
- Hardy S, Langelier Y, Prentki M.. 2000. Oleate activates phosphatidylinositol 3-kinase and promotes proliferation and reduces apoptosis of MDA-MB-231 breast cancer cells, whereas palmitate has opposite effects. Cancer Res. 60:6353–6358. [PubMed] [Google Scholar]
- Holness MJ, Kraus A, Harris RA, Sugden MC.. 2000. Targeted upregulation of pyruvate dehydrogenase kinase (PDK)-4 in slow-twitch skeletal muscle underlies the stable modification of the regulatory characteristics of PDK induced by high-fat feeding. Diabetes. 49:775–781. doi: 10.2337/diabetes.49.5.775 [DOI] [PubMed] [Google Scholar]
- Inoue M, Wu CZ, Dou DQ, Chen YJ, Ogihara Y.. 1999. Lipoprotein lipase activation by red ginseng saponins in hyperlipidemia model animals. Phytomedicine. 6:257–265. doi: 10.1016/S0944-7113(99)80018-X [DOI] [PubMed] [Google Scholar]
- Kjaer MA, Vegusdal A, Gjøen T, Rustan AC, Todorčević M, Ruyte B.. 2008. Effect of rapeseed oil and dietary n-3 fatty acids on triacylglycerol synthesis and secretion in Atlantic salmon hepatocytes. Biochim Biophys Acta. 1781:112–122. doi: 10.1016/j.bbalip.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Kohjima M, Enjoji M, Higuchi N, Kato M, Kotoh K, Nakashima M, Nakamuta M.. 2009. The effects of unsaturated fatty acids on lipid metabolism in HepG2 cells. In Vitro Cell Dev Biol Anim. 45:6–9. doi: 10.1007/s11626-008-9144-7 [DOI] [PubMed] [Google Scholar]
- Lee JH, Tachibana H, Morinaga Y, Fujimura Y, Yamada K.. 2009. Modulation of proliferation and differentiation of C2C12 skeletal muscle cells by fatty acids. Life Sci. 84:415–420. doi: 10.1016/j.lfs.2009.01.004 [DOI] [PubMed] [Google Scholar]
- Liang XF, Ogata HY, Oku H.. 2002. Lipoprotein lipase gene expression in the liver and visceral adipose tissue of fed and starved red sea bream Pagrus major. Comp Biochem Physiol A. 132:913–919. doi: 10.1016/S1095-6433(02)00118-6 [DOI] [PubMed] [Google Scholar]
- Litvinov D, Selvarajan K, Garelnabi M, Brophy L, Parthasarathy S.. 2010. Antiatherosclerotic actions of azelaic acid, an end product of linoleic acid peroxidation, in mice. Atherosclerosis. 209:449–454. doi: 10.1016/j.atherosclerosis.2009.09.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real time quantitative PCR and the 2− ΔΔCT method. Methods. 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu G, Meier KE, Jaffa AA, Rosenzweing SA, Egan BM.. 1998. Oleic acid and angiotensin II induce a synergistic mitogenic response in vascular smooth muscle cells. Hypertens. 31:978–985. doi: 10.1161/01.HYP.31.4.978 [DOI] [PubMed] [Google Scholar]
- Lu G, Morinelli TA, Meier KE, Rosenzweing SA, Egan BM.. 1996. Oleic acid-induced mitogenic signaling in vascular smooth muscle cells. A role for protein kinase C. Circ Res. 79:611–619. doi: 10.1161/01.RES.79.3.611 [DOI] [PubMed] [Google Scholar]
- Marei WF, Wathes DC, Fouladi-Nashta AA.. 2010. Impact of linoleic acid on bovine oocyte maturation and embryo development. Reprod. 139:979–988. doi: 10.1530/REP-09-0503 [DOI] [PubMed] [Google Scholar]
- Maurin AC, Chavassieux PM, Vericel E, Meunier PJ.. 2002. Role of polyunsaturated fatty acids in the inhibitory effect of human adipocytes on osteoblastic proliferation. Bone. 31:260–266. doi: 10.1016/S8756-3282(02)00805-0 [DOI] [PubMed] [Google Scholar]
- Montalto MB, Bensadoun A.. 1993. Lipoprotein lipase synthesis and secretion: effects of concentration and type of fatty acids in adipocyte cell culture. J Lipid Res. 34:397–407. [PubMed] [Google Scholar]
- Mu YM, Yanase T, Nishi Y, Tanaka A, Saito M, Jin CH, Mukasa C, Okabe T, Nomura M, Goto K, Nawata H.. 2001. Saturated FFAs, palmitic acid and stearic acid, induce apoptosis in human granulosa cells. Endocrinol. 142:3590–3597. doi: 10.1210/endo.142.8.8293 [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Inada Y, Nakano S, Tamura T, Takahashi T, Maruyama K, Yamazaki Y, Kuroda J, Shibata N.. 2006. Effects of bezafibrate, PPAR pan-agonist, and GW501516, PPARdelta agonist, on development of steatohepatitis in mice fed a methionine- and choline-deficient diet. Eur J Pharmacol. 536:182–191. doi: 10.1016/j.ejphar.2006.02.028 [DOI] [PubMed] [Google Scholar]
- Niot I, Poirier H, Besnard P.. 1997. Regulation of gene expression by fatty acids: special reference to fatty acid-binding protein (FABP). Biochimie. 79:129–133. doi: 10.1016/S0300-9084(97)81504-0 [DOI] [PubMed] [Google Scholar]
- Pan Z, Wang J, Tang H, Li L, Lv J, Han C, Xia L, Xu F.. 2011. Effects of linoleate on cell viability and lipid metabolic homeostasis in goose primary hepatocytes. Comp Biochem Physiol A. 159:113–118. doi: 10.1016/j.cbpa.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Rao GN, Alexander RW, Runge MS.. 1995. Linoleic acid and its metabolites, hydroperoxyoctadecadienoic acids, stimulate c-fos, c-jun and c-myc mRNA expression, MAP kinase activation and growth in rat aortic smooth muscle cells. J Clin Invest. 96:842–847. doi: 10.1172/JCI118130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringseis R, Muschick A, Eder K.. 2007. Dietary oxidized fat prevents ethanol-induced triacylglycerol accumulation and increases expression of PPARα target genes in rat liver. J Nutr. 137:77–83. doi: 10.1093/jn/137.1.77 [DOI] [PubMed] [Google Scholar]
- Saeed U, Jalal N, Ashraf M.. 2012. Roles of cyclin dependent kinase and Cdk- activating kinase in cell cycle regulation: contemplation of intracellular interactions and functional characterization. Global J Med Res. 12(11):47–52. Version 1. [Google Scholar]
- Sato K, Arai H, Mizuno A, Fukaya M.. 2007. Dietary palatinose and oleic acid ameliorate disorders of glucose and lipid metabolism in zucker fatty rats. J Nutr. 137:1908–1915. doi: 10.1093/jn/137.8.1908 [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Watanabe M, Suzuki H, Mahfoudi A, Krey G, Wahli W, Staels B, Yamamoto T, Auwerx J.. 1995. Induction of the acyl-coenzyme a synthetase gene by fibrates and fatty acids is mediated by a peroxisome proliferator response element in the C promoter. J Biol Chem. 270:19269–19276. doi: 10.1074/jbc.270.33.19269 [DOI] [PubMed] [Google Scholar]
- Stiller CA.2004. Epidemiology and genetics of childhood cancer. Oncogene. 23:6429–6444. doi: 10.1038/sj.onc.1207717 [DOI] [PubMed] [Google Scholar]
- Storlien LH, James DE, Burleigh KM, Chisholm DJ, Kraegen EW.. 1986. Fat feeding causes widespread in vivo insulin resistance, decreased energy expenditure, and obesity in rats. American J Physiol. 251:E576–E583. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Akahane K, Nakamura J, Naruse K, Kamiya H, Himeno T, Nakamura N, Shibata T, Kondo M, Nagasaki H, et al. 2011. Palmitate induces apoptosis in Schwann cells via both ceramide-dependent and independent pathways. Neurosci. 176:188–198. doi: 10.1016/j.neuroscience.2010.11.035 [DOI] [PubMed] [Google Scholar]
- Swagell CD, Henly DC, Morris CP.. 2007. Regulation of human hepatocyte gene expression by fatty acids. Biochem Biophys Res Commun. 362:374–380. doi: 10.1016/j.bbrc.2007.07.191 [DOI] [PubMed] [Google Scholar]
- Terano T, Tanaka T, Tamura Y, Kitagawa M, Higashi H, Saito Y, Hirai A.. 1999. Eicosapentaenoic acid and docosahexaenoic acid inhibit vascularsmooth muscle cell proliferation by inhibiting phosphorylation of Cdk2–cyclinE complex. Biochem Biophys Res Commun. 254:502–506. doi: 10.1006/bbrc.1998.9976 [DOI] [PubMed] [Google Scholar]
- van Beelen VA, Roeleveld J, Mooibroek H, Sijtsma L, Bino RJ, Bosch D, Rietjens IM, Alink GM.. 2007. A comparative study on the effect of algal and fish oil on viability and cell proliferation of Caco-2 cells. Food Chem Toxicol. 45(5):716–724. doi: 10.1016/j.fct.2006.10.017 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S.2005. Cell cycle regulation. Wormbook. 2:1–16. doi: 10.1895/wormbook.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H, Schotz MC.. 2002. The lipase gene family. J Lipid Res. 43:993–999. doi: 10.1194/jlr.R200007-JLR200 [DOI] [PubMed] [Google Scholar]
- Xu J, Cho H, O'Malley S, Park JH, Clarke SD.. 2002. Dietary polyunsaturated fats regulate rat liver sterol regulatory element binding proteins-1 and -2 in three distinct stages and by different mechanisms. J Nutr. 132:3333–3339. doi: 10.1093/jn/132.11.3333 [DOI] [PubMed] [Google Scholar]
- Xu J, Teran-Garcia M, Park JH, Nakamura MT, Clarke SD.. 2001. Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. J Biol Chem. 276:9800–9807. doi: 10.1074/jbc.M008973200 [DOI] [PubMed] [Google Scholar]
- Yonezawa T, Haga S, Kobayashi Y, Katoh K, Obara Y.. 2008. Unsaturated fatty acids promote proliferation via ERK1/2 and Akt pathway in bovine mammary epithelial cells. Biochem Biophys Res Commun. 367:729–735. doi: 10.1016/j.bbrc.2007.12.190 [DOI] [PubMed] [Google Scholar]