Abstract
Chrysanthemum coronarium is an economically important plant in Asia, and used medicinally, ornamentally and as a vegetable. In April 2017, leaf spot disease on C. coronarium was observed in Shiyan, Hubei, China. A single-spore isolate was obtained and identified based on morphology and sequence analysis using four regions (rDNA ITS, GAPDH, EF-1α, and RPB2). The results indicated that the fungus is Alternaria argyranthemi. The pathogenicity tests revealed that the species could cause severe leaf spot and blight disease on the host. This is the first report of leaf spot disease on C. coronarium caused by A. argyranthemi in the world, which is also a new record of Alternaria species in China.
Keywords: Asteraceae, leaf spot disease, morphology, multigene phylogenetic analysis
1. Introduction
Chrysanthemum coronarium L. (=Glebionis coronaria), commonly called garland chrysanthemum, is annual herbaceous plant belonging to the family Asteraceae and widely distributed in the Mediterranean region and several Asian countries [1]. The green leaves and shoots are appreciated in China, Korea, and Japan as a popular vegetable due to their fragrance, flavor, and nutritional quality [2]. The species is also used as an ornamental and in folk medicine. Furthermore, the antioxidative, antimicrobial, anticancerous, nematicidal, phytotoxic and insecticidal biological properties have been found in the numerous components extracted from C. coronarium flowerheads [3–5].
The plant can be infested by various fungi, and there have been reports of Cercospora spp., Entyloma spp., Peronospora spp., Puccinia spp. and so on (https://fungaldatabases/). Uematsu et al. [6] found that the Colletotrichum carthami could induce severe anthracnose on C. coronarium var. spatiosum. A new species, Gibellulopsis chrysanthemi was isolated from rotted leaves of C. coronarium and demonstrated to reproduce the natural symptoms in pathogencity test [7, 8]. Though Alternaria leaf spots and blight diseases are commonly found on asteraceous plants, and over 30 Alternaria species have been reported [9], there is no report about Alternaria spp. causing diseases on C. coronarium.
During a survey of large-spored Alternaria species in China, a unique species of the genus Alternaria with solitary conidia was observed on leaf spots of C. coronarium from Shiyan, Hubei, China, in April 2017. The study aimed to identify the Alternaria species associated with a leaf spot disease of C. coronarium based on morphological and molecular characteristics and to reveal their pathogenicity on C. coronarium.
2. Symptoms and isolation
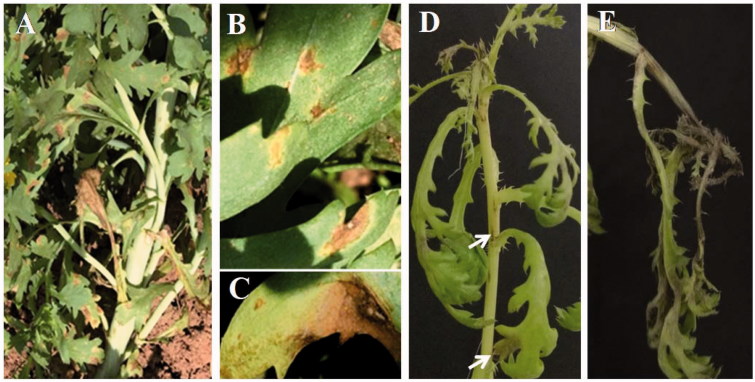
The leaf spot or blight symptoms were most severe on the lower part of the plants (Figure 1(A)). The initial symptoms appeared as irregular small brown spots with faint yellowish halos (Figure 1(B)). The typical character was round or irregular necrotic spots surrounded by yellow halo. There was a tiny white spot in the center of each larger necrotic spot. Leaf spots enlarged and their colors darkened as infection progressed, and developed into full blight symptoms (Figure 1(C)). Severely infected leaves fell off.
Figure 1.
Symptoms on Chrysanthemum coronarium caused by Alternaria argyranthemi in field. (A) Leaf spots in field. (B, C) Different stages of natural leaf spots. (D) Necrotic spots after 3-day-inoculation of spore suspension for the pathogenecity tests. (E) Leaf blight symptoms observed after 6-day-inoculation.
Specimens with leaf spot symptoms were randomly sampled. Infected tissues were cut into small pieces and incubated on sterile wet paper at 25 °C in the dark. After 1 d, a type of Alternaria conidia was observed. A single conidium was picked up under a stereomicroscope and placed on the surface of potato dextrose agar (PDA; Difco, Montreal, Canada). The plates were transferred into an incubator and kept at 25 °C without light. Six pure cultures were placed on PDA slants and in glycerol stock solutions and deposited in the Culture Collection of Yangtze University (YZU). One of deposited strains, YZU 171067 was selected for morphological characterization and DNA sequencing.
3. Morphological observation
Isolates were grown on PDA and PCA (containing 20 g potato, 20 g carrot, 20 g agar per liter) at 25 °C for 3–5 d. Mycelial plugs (6 mm diameter) were collected from the edge of colonies and placed onto the center of PDA and PCA plates (90 mm diameter). The cultures were maintained at 25 °C for 7 d in darkness to determine the cultural characteristics. To assess conidial morphology, mycelia of the isolate YZU 171067 were transferred to PCA and incubated at 22 °C for 7 d [9]. However, conidia were rarely found on PCA, but chlamydospores (microsclerotia) were abundant. Then, the mycelia were placed on healthy leaves of C. coronarium to induce disease symptoms. Diseased leaves were cut into 1 cm length, and placed onto wet filter paper in plates and incubated at 22 °C with light/dark periods of 8 h/16 h for conidial production. After 1 d, the conidiophores were examined using a Leica M205A stereomicroscope (Germany, L 161). Conidia were collected and mounted in lactophenol picric acid solution for further measurement. Conidia (n = 50) were randomly chosen and measured under an Olympus BX50 light microscope (Japan, L 165).
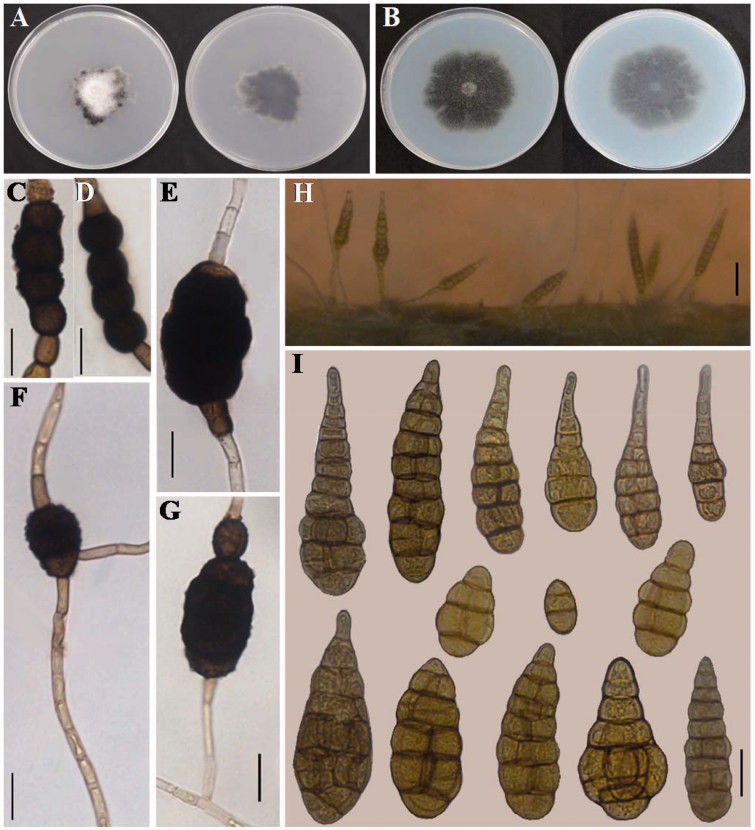
Colonies of the isolate YZU 171067 on PDA and PCA medium were strikingly different from each other after 7 d. Colonies on PDA (Figure 2(A)) were 23–30 mm across, irregular effuse, white to dark olive brown, and velvety with thick hyphae covering the center and dark olive brown on the reverse side. Colonies on PCA (Figure 2(B)) were 33–38 mm across, densely radial, pale gray to dark olive brown with a similar color on the reverse side. Chlamydospores were golden brown to dark brown with punctate ornamentation, toruloid or oval, 30–100 × 18–30 μm in the middle of hyaline hyphae (Figure (2C–G)). However, there were few conidia found on PCA colonies. To measure the conidia, the strain YZU 171067 was stimulated to produce conidia by inoculation onto the host C. coronarium and incubation at 22 °C (Figure 2(H)). Conidia produced after 1 d were predominantly solitary, medium yellowish tan to brown, smooth, narrowly ellipsoid to obclavate, 50–120 × 19–30 μm, with 4–9 transversal septa and 0–3 longitudinal septa in some of the transverse divisions, normally with blunt tapered beaks, 10–25 × 4–8 μm (Figure 2(I)). The conidial morphology is in agreement with Alternaria argyranthemi described by Simmons [9]. The species produces enormous numbers of chlamydospores (microsclerotia) on PCA and V8 media [9], which was also observed during this study. However, few conidia were observed on PCA, which is in contrast to the results of Simmons [9]. Chlamydospores are readily identifiable characteristics among the morphologically similar species (A. sonchi, A. brassicae, and A. panax) [9]. This is a particularly key character to identify this species.
Figure 2.
Morphology of Alternaria argyranthemi from Chrysanthemum coronarium. (A) Colony on PDA. (B) Colony on PCA. (C–G) Chlamydospores (microsclerotia) formed on PCA at 22 °C for 7 days (bar = 20 μm). (H) Sporulation patterns on the host plant (bar = 50 μm). (I) Conidia produced on diseased leaves inoculated at 22 °C (bar = 20 μm).
4. Molecular analyses
Genomic DNA of the fungus was extracted using the method described by Cenis [10]. The DNA pellet was dissolved in 40 µL of ddH2O and stored at –20 °C until required for further use. Four target regions were selected for PCR amplification: rDNA ITS (ITS4/ITS5) [11], GAPDH (gpd1/gpd2) [12], EF-1α (EF1-728F/EF1-986R) [13] and RPB2 (RPB2-6F/RPB2-7cR) [14]. PCR amplification was performed in 20 µL (10 µL 2 × Taq PCR StarMix, 7 µL ddH2O, 2 µL DNA, 0.5 µL of each primer) reactions using an ABI Veriti 96-well Thermal Cycler. Successfully amplified DNA was purified and sequenced with the same primers by BGI (Beijing Genomics Institute). The sequences were compared to GenBank (https://www.ncbi.nlm.nih.gov/) using BLASTn, and most highly matching sequences were downloaded. The sequences were adjusted and compared using PHYDIT 3.2 software (Korea, L 281). Four regions were combined, aligned and constructed a neighbor-joining tree by MEGA 7.0 [15], the evolutionary distances were computed using Kimura 2-parameter method and with 1000 bootstrap replicates.
The resulting sequences of the rDNA ITS, GAPDH, EF-1α and RPB2 region have been deposited in GenBank (Table 1). Sequences of the rDNA ITS, GAPDH and RPB2 showed 100% identity to the type strain of A. argyranthemi CBS 116530, with two nucleotides different in the EF-1α sequence. Their phylogenetic relationship was exhibited in the neighbor-joining phylogram generated from the four gene sequences. The isolates YZU 171067 and CBS 116530 were clustered together supported by 100% bootstrap value (Figure 3). In a taxonomic analysis of the genus Alternaria, A. argyranthemi fell into a monotypic lineage (containing one species), and was not assigned to any of the 24 sections [16]. The gene sequences of Alt a1 (Alt-a1-for/Alta1-rev) [17] and ATPase (ATPDF1/ATPDR1) [18] were also amplified, sequenced and submitted to GenBank with accession No. MG647616 and MG647615, respectively. The results also supported the identification of YZU 171067 as A. argyranthemi.
Table 1.
Isolates used in this study for the phylogenetic analysis.
| GenBank accession No. |
|||||||
|---|---|---|---|---|---|---|---|
| Species | Strain | Host | Location | rDNA ITS | GAPDH | EF-1α | RPB2 |
| A. argyranthemi | CBS 116530 | Argyranthemum frutescens | New Zealand | KC584181 | KC584098 | KC584637 | KC584378 |
| YZU 171067 | Chrysanthemum coronarium | China | MG647618 | MG674139 | MG647619 | MG647617 | |
| A. brassicae | CBS 116528 | Brassica oleracea | USA | KC584185 | KC584102 | KC584641 | KC584382 |
| A. botrytis | CBS 197.67 | Contaminant | USA | KC584243 | KC584168 | KC584736 | KC584476 |
| A. capsici-annui | CBS 504.74 | Capsicum annuum | Unkonw | KC584187 | KC584105 | KC584644 | KC584385 |
| A. panax | CBS 482.81 | Aralia racemosa | USA | KC584209 | KC584128 | KC584675 | KC584417 |
| A. penicillata | CBS 116607 | Papaver rhoeas | Austria | KC584229 | KC584153 | KC584706 | KC584447 |
| A. sonchi | CBS 119675 | Sonchus asper | Canada | KC584220 | KC584142 | KC584691 | KC584433 |
Figure 3.
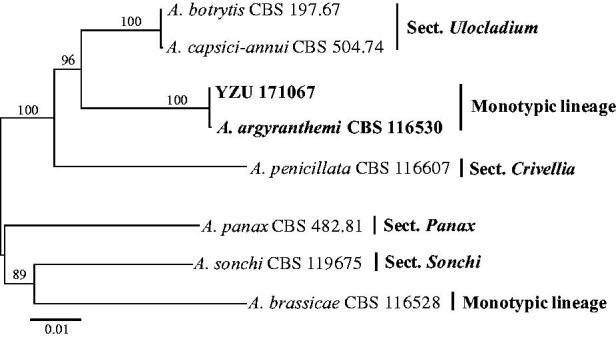
Neighbor-joining tree generated from Alternaria argyranthemi YZU 171067 from Chrysanthemum coronarium and its related species based on a combined datasets of rDNA ITS, GAPDH, EF-1α and RPB2 gene sequences. Bootstrap values were determined using 1000 replicates.
5. Pathogenicity tests
The chlamydospore of YZU 171067 grown on PCA after 7 d was flushed by ddH2O, using two layers of sterile gauze to filter out hyphae to obtain a chlamydospore suspension, the suspension was adjusted to 1 × 105 spores/mL. Intact potted C. coronarium plants were sprayed with the spore suspension until the leaves were uniformly wet. As a control, plants were inoculated with sterile distilled water. All the plants were incubated at 80–90% relative humidity in separate containers around 25 °C. Disease progress was monitored daily for 7 d. The fungus was re-isolated from symptomatic tissues to address Koch’s postulates. The tests were repeated three times.
After inoculation with the chlamydospores suspension, the symptoms on leaves appeared as little brown to black, water-soaked spots after 24 h. It enlarged on leaves with large irregular spots after 3 d. Furthermore, stems were also infected with small long fusiform necrotic symptoms, and the top shoots were severely infected and became blighted (Figure 1(D)). Later, foliar lesions enlarged and coalesced to form large areas of necrosis on leaves and stem, finally resulting in defoliation (Figure 1(E)). No symptoms were observed in the control leaves inoculated with sterile distilled water. Also, the pathogenicity tests revealed that the fungus is the causal agent causing leaf spot and blight disease on the host. Nishikawa and Nakashima [19] had been tested the pathogenicity of A. cinerariae on C. coronarium (Glebionis coronaria) and found it to be non-pathogenic on this plant.
6. Conclusions
In this study, the fungus isolated from C. coronarium was identified as Alternaria argyranthemi. E.G. Simmons & C.F. Hill based on the morphology and molecular analysis. To our knowledge, this is the first report of leaf spot disease on C. coronarium caused by A. argyranthemi in the world, which is also a new record of Alternaria species in China.
Funding Statement
The work was supported by the National Natural Science Foundation of China [31400014] and under the Young Scientist Foundation of Yangtze University [2016cqr08].
Acknowledgements
We sincerely thank T. Hsiang for his assistance during the preparation of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1. Basta A, Pavlovi M, Couladis M, et al. Essential oil composition of the flower heads of Chrysanthemum coronarium L from Greece. Flavour Fragr J. 2007;22:197–200. [Google Scholar]
- 2. Chung K, Park Y. Development of an agrobacterium-mediated transformation system for regenerating garland chrysanthemum (Chrysanthemum coronarium L.). J Plant Biol. 2005;48:136–141. [Google Scholar]
- 3. Bar-Eyal M, Sharon E, Spiegel Y. Nematicidal activity of Chrysanthemum coronarium . Eur J Plant Pathol. 2006;114:427–433. [Google Scholar]
- 4. Haouas D, Cioni PL, Halima-Kamel MB, et al. Chemical composition and bioactivities of three Chrysanthemum essential oils against Tribolium confusum, (du Val) (Coleoptera: Tenebrionidae). J Pest Sci. 2012;85:367–379. [Google Scholar]
- 5. Hosni K, Hassen I, Sebei H, et al. Secondary metabolites from Chrysanthemum coronarium (Garland) flowerheads: chemical composition and biological activities. Ind Crop Prod. 2013;44:263–271. [Google Scholar]
- 6. Uematsu S, Kageyama K, Moriwaki J, et al. Colletotrichum carthami comb. nov., an anthracnose pathogen of safflower, garland chrysanthemum and pot marigold, revived by molecular phylogeny with authentic herbarium specimens. J Gen Plant Pathol. 2012;78:316–330. [Google Scholar]
- 7. Kawaradani M, Taguchi K, Okada K, et al. Seedling rot of garland chrysanthemum caused by Gibellulopsis chrysanthemi and ecological characters of the causal fungus. J Gen Plant Pathol. 2013;79:346–349. [Google Scholar]
- 8. Hirooka Y, Kawaradani M, Sato T. Description of Gibellulopsis chrysanthemi sp. nov. from leaves of garland chrysanthemum. Mycol Progress. 2014;13:13–19. [Google Scholar]
- 9. Simmons EG. Alternaria: an identification manual. CBS Biodiversity Series 6. Utrecht: CBS Fungal Biodiversity Centre; 2007. [Google Scholar]
- 10. Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20:2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. White TJ, Bruns TD, Lee SB, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (Eds.). PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322. [Google Scholar]
- 12. Berbee ML, Pirseyedi M, Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from its and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. [Google Scholar]
- 13. Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- 14. Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA Polymerse II Subunit. Mol Biol Evol. 1999;16:1799–1808. [DOI] [PubMed] [Google Scholar]
- 15. Kumar S, Stecher G, Tamura K. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woudenberg JHC, Groenewald JZ, Binder M, et al. Alternaria redefined. Stud Mycol. 2013;75:171–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong SG, Cramer RA, Lawrence CB, et al. Alt a 1 allergen homologs from Alternaria, and related taxa: analysis of phylogenetic content and secondary structure. Fungal Genet Biol. 2005;42:119–129. [DOI] [PubMed] [Google Scholar]
- 18. Lawrence DP, Gannibal PB, Dugan FM, et al. Characterization of Alternaria isolates from the infectoria species-group and a new taxon from Arrhenatherum, Pseudoalternaria arrhenatheria sp. nov. Mycol Progress. 2014;13:257–276. [Google Scholar]
- 19. Nishikawa J, Nakashima C. Morphological variation and experimental host range of Alternaria cinerariae . Mycoscience. 2015;56:141–149. [Google Scholar]