Abstract
Root hair patterning is best studied in Arabidopsis thaliana. A pattern of root hair and non-root hair files is governed by a gene-regulatory network of activators and inhibitors. Under phosphate starvation conditions, extra root hairs are formed in non-root hair positions. This raises the question, whether and how this environmental stimulus is mediated by the known root hair gene network. In this study, we provide genetic and molecular data on the role of ETC1 in the phosphate starvation induced ectopic root hair formation. We show that the expression in the epidermis is irregular and reduced and that a new expression domain is induced in the sub-epidermis. By expressing ETC1 in the sub-epidermis, we show that this is sufficient to induce extra root hair formation in N-files. This suggests that the phosphate induced expressional switch from epidermal to epidermal plus sub-epidermal expression of ETC1 is one environmental input to the underlying patterning network.
Keywords: root hair patterning, ETC1, phosphate starvation, expression domains, development
Introduction
In Arabidopsis thaliana, the root epidermis exhibits a regular pattern of root hair and non-root hair files. Root hairs develop from files of epidermal cells that are located over the cleft of two underlying cortical cells, whereas epidermal cells that are in contact with only one cortical cell become non-root hair cells (Dolan et al., 1993, 1994; Galway et al., 1994). This pattern is regulated by a complex genetic gene regulatory network that governs the determination of root hair and non-root hair files. The cortical-position dependent regulation of cell fate is mediated by the leucine-rich repeat receptor-like kinase SCRAMBLED (SCM) (Kwak et al., 2005). SCM in turn is regulated by the zinc finger protein JACKDAW (JKD) through a non-cell autonomous signal from the underlying cortical cells (Hassan et al., 2010). SCM negatively regulates the expression of the R2R3 MYB protein WEREWOLF (WER) (Kwak et al., 2005). WER (Lee and Schiefelbein, 1999) acts at an early stage in cell fate determination of the non-root hair cell files by forming an activator complex with the basic helix-loop-helix (bHLH) proteins GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) (Bernhardt et al., 2003) and the WD40 domain protein TRANSPARENT TESTA GLABRA1 (TTG1) (Walker et al., 1999).
In addition, five homologous R3MYB genes including CAPRICE (CPC), TRIPTYCHON (TRY), ENHANCER OF TRIPTYCHON AND CAPRICE 1, 2, and 3 (ETC 1, 2, and 3) regulate root hair formation in a redundant manner by suppressing non-root hair formation (Wada et al., 1997; Schellmann et al., 2002; Kirik et al., 2004a,b; Simon et al., 2007; Wester et al., 2009). The respective proteins are considered to compete with WER for binding to GL3 thereby suppressing the activator complex formation (Esch et al., 2003; Ryu et al., 2005; Tominaga et al., 2007). Intercellular interactions are mediated by movement of the R3MYB proteins and GL3. The R3MYB proteins are expressed in the non-root hair cells and move to the hair cells (Wada et al., 2002; Kang et al., 2013). GL3 is expressed in the hair cells and moves to the non-root hair cells (Bernhardt et al., 2005). As a result from these intercellular interactions, the activator complex activates the expression of the homeodomain leucine zipper protein GLABRA2 (GL2) in non-hair cells, where it represses root hair cell fate (Galway et al., 1994; Hung et al., 1998). Here, GL2 negatively regulates the expression of down stream genes. One of the down-stream genes is the basic helix loop helix gene ROOT HAIR DEFECTIVE 6 (RHD6) (Menand et al., 2007). RHD6 is essential for root hair initiation and also one of its direct targets ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4) is expressed in the root hair cells where it acts as a regulator of root hair growth (Yi et al., 2010).
Under Pi deficient growth conditions, A. thaliana plants produce higher density of root hairs (Bates and Lynch, 1996; Ma et al., 2001; Muller and Schmidt, 2004). This increase in root hair density can be explained by the decrease of the longitudinal length of epidermal cells though. However, the decrease in the epidermal cell length in wer, scm and cpc mutants under Pi starvation conditions did not account for the increase of root hair cells, pointing toward an additional mechanism triggering extra root hair formation (Savage et al., 2013). A role in the determination of root hair and non-root hair fate was suggested for BHLH32 (Chen et al., 2007). Mutations in BHLH32 lead to an increase of root hairs under Pi-starvation. It was suggested that BHLH32 regulates root hair patterning under Pi starvation through direct interaction with GL3 and TTG1 (Chen et al., 2007).
In the present study, we investigate the role of the root hair patterning machinery under phosphate-starvation. We show that in the absence of GL2, WER, and TTG1 phosphate starvation cannot trigger additional root hair formation. We further show that ETC1 plays a role in the increase of root hair number under Pi- conditions. We report that ETC1 expression changes such that it is irregular in the epidermis and turned on in the sub-epidermis. We provide evidence that ETC1 can move from the sub-epidermis and that this is sufficient to induce root hair formation.
Results and Discussion
Phosphate Starvation Promotes Root Hair Formation Through the Patterning Machinery
In order to study the possible role of genes controlling root hair pattering in the phosphate starvation response, we compared the formation of root hairs in root hair (H-position) and non-root hair (N-position) files between wild type and mutants. While previous studies used older plants, we aimed to capture in particular early events of pattern formation by analyzing root hair patterns on 7-day old seedlings grown either on Pi+ or Pi- conditions (Supplementary Table S1). We reasoned that this might reveal different results as in experiments done under low phosphate levels [e.g., cpc-1 (Ws), try-29760 (Col-0), and etc1-1 (Col-0) (Chen and Schmidt, 2015)] or phosphate shift experiments (Muller and Schmidt, 2004; Savage et al., 2013; Supplementary Table S1). In this study, we grew seedlings under long day conditions on agar plates. We analyzed root hair formation in H and N positions in gl2-1 (Ler), wer-1 (Col-0), ttg1-1 (Ler), cpc-2 (Col-0), try-JC (Col-0), etc1-1, and cpc-2 etc1-1 mutants. Due to high plasticity of quantitative root hair phenotypes we present the results of two independent experiments and consider only results that are statistically significant in both experiments.
As described previously, both wild-type ecotypes Ler and Col-0 produced extra root hairs in non-root hair cells when grown under Pi- conditions (Figure 1 and Supplementary Tables S2, S3) (Muller and Schmidt, 2004). As expected, we found ectopic root hairs in gl2-1, wer-1, and ttg1-1 mutants. In contrast to a previous study (Muller and Schmidt, 2004), we found no statistically significant difference of ectopic hairs in non-root hair files under Pi- conditions in these three mutants (Figure 1 and Supplementary Tables S2, S3). This suggests that they are required for Pi- induced extra root hair formation under our growth conditions. The analysis of the R3MYB mutants cpc-2, try-JC, and etc1-1 revealed different responses. In contrast to Chen and Schmidt (2015), root hair formation in try-JC mutants was indistinguishable from wild type under Pi+ and Pi-conditions (Figure 1 and Supplementary Tables S2, S4). cpc-2 mutants have a reduced number of root hairs in root hair positions that is strongly increased under Pi-conditions (Figure 1 and Supplementary Tables S2, S3) see also (Muller and Schmidt, 2004; Chen and Schmidt, 2015). Root hair number in etc1-1 mutants was similar to wild-type under Pi+ conditions (Kirik et al., 2004a). For the etc1-1 mutant, a slight decrease of root hair numbers in N-files under Pi- conditions was reported by others (Chen and Schmidt, 2015). We also observed a small decrease in one of our two sets of experiments (Figure 1 and Supplementary Tables S2–S4). Given that the reduction of root hairs is very low (in the 10% range) in the report by (Chen and Schmidt, 2015) as well as in our experiment it is conceivable that such a low deviation may be difficult to detect in different experiments.
FIGURE 1.
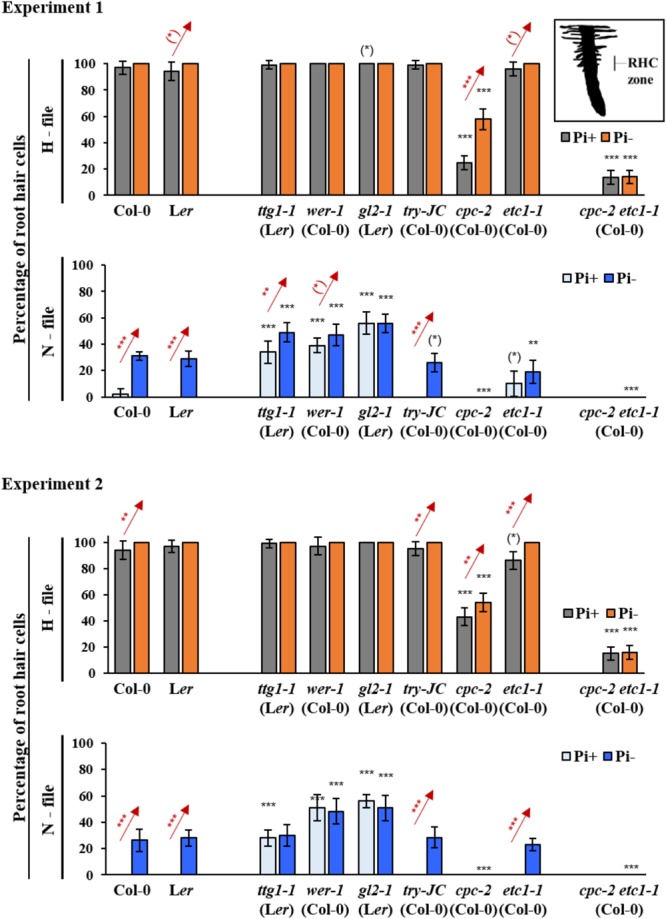
Quantitative analysis of root hair formation in response to phosphate starvation in wild type and mutant genotypes. Two independent experiments are shown in which root hair number is compared in N- and H files between at least 10 wild type and mutant seedlings under normal conditions and under phosphate starvation conditions. RHC zone indicates the region of root hair analysis. Red stars indicate a significant difference between Pi+ and Pi– conditions at ∗p < 0.5, ∗∗p < 0.01, ∗∗∗p < 0.001 (Wilcoxon test). Black stars indicate a significant difference between wild type and mutants at ∗p < 0.5, ∗∗p < 0.01, ∗∗∗p < 0.001 (Wilcoxon test).
We also analyzed the cpc-2 etc1-1 double mutant under Pi+ and Pi- conditions. Similar to previous studies (Savage et al., 2013; Chen and Schmidt, 2015), the cpc-2 etc1-1 double mutant did not produce ectopic root hairs under Pi- conditions suggesting no independent effect of ETC1 (Figure 1 and Supplementary Tables S2, S3). However, an ETC1 dependent response in root hair production in H files is evident when comparing the cpc-2 single mutant with the cpc-2 etc1-1 double mutant. While the cpc-2 mutant shows a higher number of root hairs in H files under phosphate starvation conditions, the double mutant does not respond. These results support the idea that ETC1 is involved in the regulation of root hair patterning under Pi- conditions.
Spatial Expression of Root Hair Patterning Genes Under Pi- Conditions
Several studies had shown that the expression levels of root hair patterning genes were altered under phosphate starvation (Wu et al., 2003; Misson et al., 2005; Lan et al., 2012; Chen and Schmidt, 2015). For ETC1 expression, conflicting results were obtained. Two studies reported an up-regulation of ETC1 expression (Misson et al., 2005; Lan et al., 2012) whereas no differences were detected in the study of Chen and Schmidt (2015). As for patterning processes expression levels are not necessarily relevant but rather the correct spatial expression matters, we decided to assess the spatial expression of CPC, GL2, TRY, and ETC1 under Pi+ and Pi- conditions using established promoter:GUS marker lines. To judge changes in the relative expression levels we monitored the GUS staining after 4 and 16 h. In agreement with other studies, the expression of pGL2:GUS was found in non-root hair files under normal conditions (Figures 2A – I, V) (Masucci and Schiefelbein, 1996). After 4 h of staining, we found almost no GUS staining in Pi- treated roots (Figure 2A – II) and after 16 h we observed a discontinuous staining in non-root hair files (Figure 2A – VI). This is consistent with the formation of extra root hairs in approximately 30% of the cells under Pi- conditions (Supplementary Table S2). As compared to normal growth conditions (Figure 2A – III; Lee and Schiefelbein, 2002) pCPC:GUS expression levels were clearly reduced and discontinuous in N-files in Pi- media grown seedlings after 4 h of GUS staining (Figure 2A – IV). pTRY:GUS was neither detected under normal conditions (Schellmann et al., 2002) nor in Pi- grown seedlings after 16 h GUS staining (Figures 2A – VII, VIII). Expression of pETC1:GUS was previously reported to be in N-files (Kirik et al., 2004a) which was confirmed in in situ hybridization experiments (Simon et al., 2007). Consistent with this we found pETC1:GUS expression in cell files under Pi+ conditions (Figures 2B – I, III, VII). Under Pi- conditions, the epidermal expression of ETC1 was irregular such that large regions including N-files showed almost no GUS staining and some files were normally stained (Figures 2B – II, IV, VIII). An additional expression domain was observed in sub-epidermal cells (Figures 2B – VI, VIII) which was never noticed under Pi+ conditions (Figure 2B – V).
FIGURE 2.
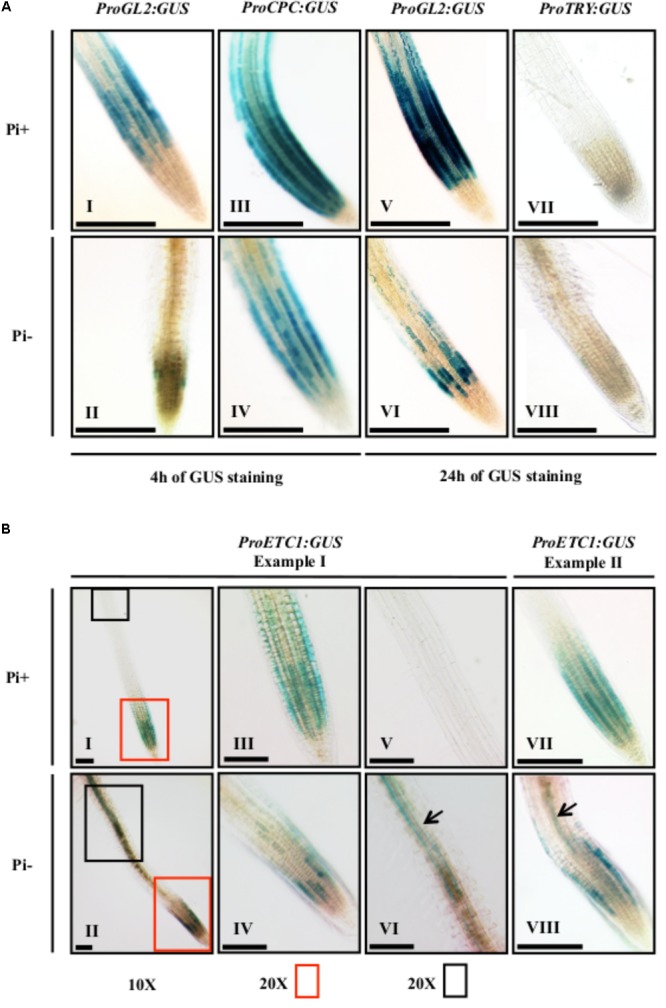
Histochemical GUS expression analysis of ProCPC, ProGL2, ProTRY, and ProETC1. GUS expression staining was done on 7-day old seedlings grown on Pi+ media and Pi– media. (A) GUS expression of ProCPC, ProGL2, and ProTRY. GUS staining was done for 4 h in seedlings (I–IV) and for 24 h in seedlings (V–VIII). A reduction in GUS expression was noticed for ProGL2 and ProCPC in seedlings grown under Pi– conditions (II, IV, VI) as compared to seedlings grown on Pi+ (I, III, V). No GUS expression was detected for ProTRY under both Pi+ and Pi– conditions (VII, VIII). (B) GUS expression of ProETC1. Two examples are shown to depict the variability of the expression pattern. Plant I (I, II) shows an overview and higher magnifications of the root tip (III, IV) and a more distal region (V, VI). A new expression domain of ProETC1 was found in the sub-epidermis under Pi– conditions (black arrows in VI and VIII). Scale bars: 200 μm.
Promoter Analysis of ETC1
The expression analysis of the ProETC1:GUS line indicates an expression switch from regular epidermal expression in N-files to subepidermal expression upon phosphate starvation. In an attempt to dissect these regulation events, we created a series of deletion constructs driving either the expression of the GUS-marker gene or the ETC1 CDS for rescue experiments (Figure 3A). As the etc1-1 single mutant root hair phenotype is fairly subtle and variable, we used the cpc-2 etc1-1 double mutant for rescue experiments. A 932 bp long promoter fragment was sufficient to mediate rescue of the cpc-2 etc1-1 mutant phenotype to a level found in the cpc-2 mutant and normal expression patterns (Figures 3A,B and Supplementary Tables S5–S9).
FIGURE 3.
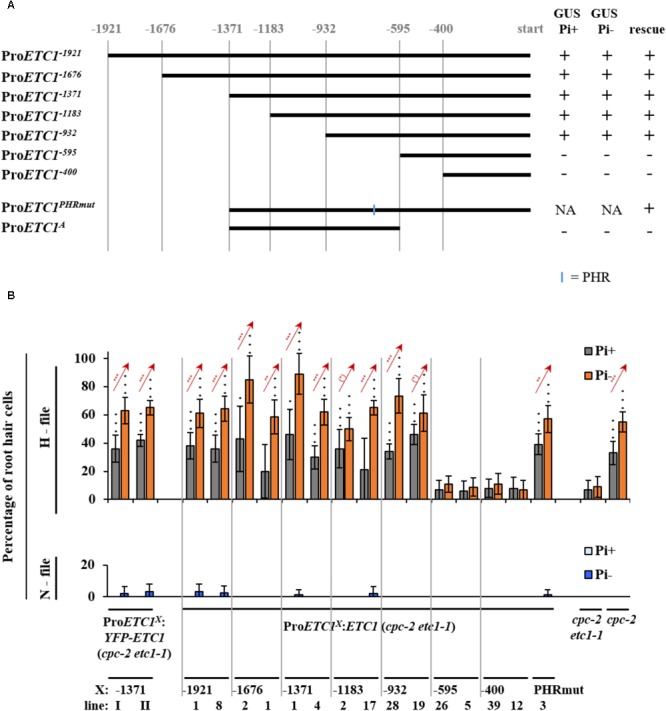
Promoter deletion analysis of ETC1. (A) Schematic diagram showing a serial deletion of the 5′ regulatory region of ETC1. Vertical lines show the relative position of the fragment with respect to the ETC1 start codon. The blue line indicates the location of the phosphate response binding (PHR-1) site. (B) Rescue analysis of the ETC1 promoter fragments in at least ten plants. Red stars indicate a significant difference between Pi+ and Pi– conditions at ∗p < 0.5, ∗∗p < 0.01, ∗∗∗p < 0.001 (Wilcoxon test). Black stars indicate a significant difference between wild type and mutants at ∗p < 0.5, ∗∗p < 0.01, ∗∗∗p < 0.001 (Wilcoxon test). + Indicates that the construct was able to rescue or was able to drive GUS expression, – indicates that the construct was not able to rescue or was not able to drive GUS expression.
As all ProETC1 fragments were able to rescue cpc-2 etc1-1 mutant phenotype except for ProETC1-595 and ProETC1-400, we expected the existence of regulatory sequence between -932 and -595 region to be essential for the expression of ETC1. At this time point we used the PLACE tool to search for MYB and phosphate response (PHR-1) binding sites https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace (Higo et al., 1999) (Supplementary Table S10). We found one PHR-1 binding site, however, changing its sequence from (gtatatcc) to (aaaaaaaa) did not alter the promoter rescuing ability to cpc-2 etc1-1 double mutant under phosphate starvation conditions (Figure 3B and Supplementary Tables S5–S9). This suggests the presence of other phosphate regulatory elements that control the ETC1 expression based on phosphate availability. We therefore analyzed the relevant promoter region using more recent tools and data sets. Toward this end we focused on 14 cis-regulatory elements from co-expression-derived modules that are involved in the root hair formation induced by phosphate deficiency (Salazar-Henao et al., 2016). Three of these cis-regulatory elements are present in the relevant ProETC1 fragment (Figure 4 and Supplementary Table S11): the AAAAG pattern, regulated by Dof (DNA-binding with one finger) transcription factors, the GATC pattern, regulated by GATA transcription factors and the CACGTG pattern, a G-box which occurs in phytochrome A-responsive promoters (Salazar-Henao et al., 2016). The CACGTG motif is of particular interest as both bHLH/MYB and other factors can potentially bind to this region. Thus this region might play a role to coordinate developmental regulation and the Pi starvation response.
FIGURE 4.
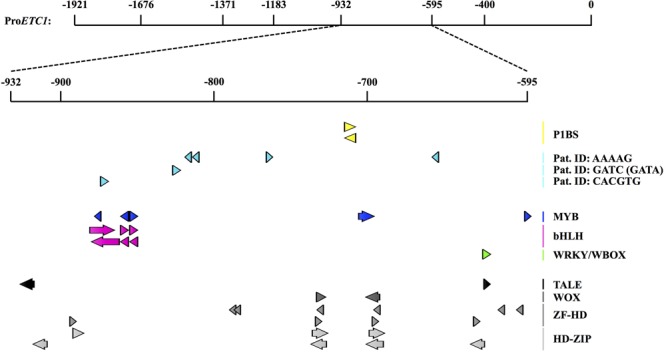
Cis-regulatory elements and motifs related to Pi responsiveness and patterning with the selected ProETC1 fragment. Detailed analysis of cis-regulatory motives in the ProETC1-932- -595 fragment using PlantPAN 2.0 (Chow et al., 2016) and manual analysis with motifs from a recent publication (Salazar-Henao et al., 2016). The figure was constructed using the CLC DNA Workbench (see text footnote 4). P1BS was previously identified by PLACE (Higo et al., 1999) mutated in this study and did not show to be causative. Recently, 14 root specific Pi-responsive motifs were identified (Salazar-Henao et al., 2016). Three of these were found in the selected ProETC1 fragment (labeled “Pat. ID”). Below, patterning related motifs extracted from a PlantPAN 2.0 analysis are shown. Orientation of motifs: +: arrows point to the right, – strand: arrow points to the left. P1BS, PHR1-binding site; Pat. ID, pattern identifier from Salazar-Henao et al. (2016); TALE, three amino acid loop extension (of homeodomains); WOX, WUS homeobox-containing; ZF-HD, zinc-finger-homeodomain; HD-ZIP, homeodomain-leucine zipper. All results and precise positions can be found in Supplementary Table S11.
Spatial Distribution of YFP-ETC1 Protein
The reduced expression of ETC1 in the epidermis is not consistent with a role of ETC1 in increased root hair formation under Pi- conditions. However, it is conceivable that ETC1 protein can move from the sub-epidermal cells and contributes to the regulation of epidermal cell fate. In this scenario, ETC1 might be expected to be transported equally well to N and H files. To explore this question, we established plants carrying the ProETC1:YFP-ETC1 construct. In a first step, we demonstrated that the promoter and the fusion proteins are fully functional by rescuing the cpc-2 etc1-1 double mutant (Figure 3B and Supplementary Table S6). ProETC1:YFP-ETC1 cpc-2 etc1-1 plants showed a root hair phenotype similar to cpc-2 mutant under Pi+ and Pi- conditions (Supplementary Table S6). Under Pi+ conditions, YFP-ETC1 protein was found in N- and H- files (Figure 5A). Thus, similar as shown for CPC (Wada et al., 2002), ETC1 is expressed in N-files and moves into the H files. In ProETC1:YFP-ETC1 plants grown under Pi-conditions, the YFP-ETC1 signal was observed in all root layers with elevated levels in the stele and epidermal cells (Figures 5A – II, IV, VI). When comparing the GUS expression with the YFP-ETC1 distribution we noted clear differences. In particular, the region more distal from the root tip displaying ETC1 expression in the sub-epidermis clearly exhibited YFP-ETC1 protein in the epidermis (Figures 2B – VI, 5A – II). This suggests that YFP-ETC1 RNA or protein can (Figure 5A – II) move between cell layers. We also noted that the pattern irregularities found for the GUS expression is much less pronounced for the YFP-ETC1 signal. This is likely to be due to YFP-ETC1 mobility that levels out expression differences between cells.
FIGURE 5.
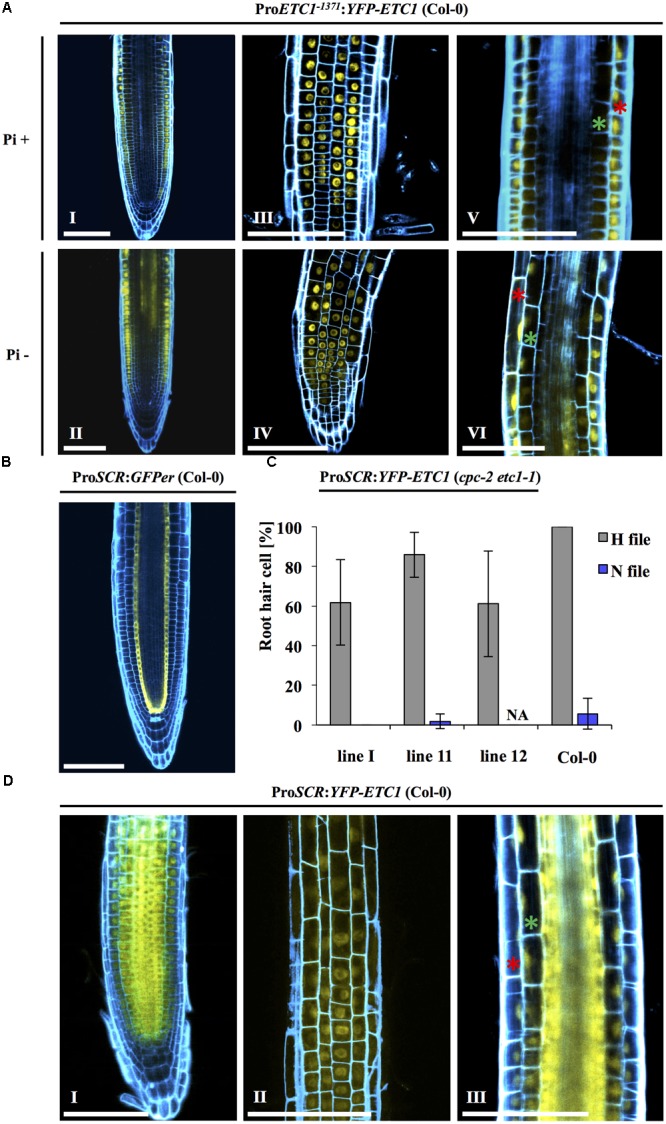
Intercellular motility and localization of YFP-ETC1 under the ETC1 and SCR promoters. (A) Root of a cpc-2 etc1-1 double mutant plant carrying ProETC1:YFP-ETC1 under Pi+ and Pi– conditions. Under Pi+ conditions, YFP-ETC1 is seen in the root hair and non-root hair file epidermal cells (I, III) but not in the ground tissue (V). Under Pi– conditions, YFP-ETC1 was observed in the root hair and non-root hair file epidermal cells (II, IV) and in the ground tissue (VI). (I, II) Pictures show overview, (III, IV) pictures show focal planes of the epidermis and (V, VI) pictures show the focal planes in the cortical cells and the stele. Red stars mark epidermal cells, green stars mark cortical cells. Scale bar = 100 μm. (B) CLSM pictures of roots from transgenic ProSCR:GFPer (Col-0) plants grown under Pi+ conditions. Blue: propidium iodide, yellow: YFP. (C) A diagram showing the rescuing ability of ProSCR:YFP-ETC1 in the cpc-2 etc1-1 double mutant under Pi+ conditions. (D) ProSCR:YFP-ETC1 (Pi+ conditions). (I) Overview shows YFP-ETC1 in all tissues. (II) Focal plane showing YFP-ETC1 in epidermal cells. (III) Focal plane showing YFP-ETC1 in the epidermal cells and the ground tissue. Red star labels an epidermal cell, green star labels a cortical cell. Scale bar = 100 μm.
To test whether movement of ETC1 from the subepidermis to the epidermis can regulate root hair patterning, we expressed the ETC1 CDS under the SCARECROW (SCR) promoter in the cpc-2 ect1-1 double mutant and analyzed the resulting plants for root hair rescue under Pi+ conditions. As expected, ProSCR:GFPer (Figure 5B) showed expression in the cortical/endodermis initial cells and in the endodermis (Helariutta et al., 2000). In ProSCR:YFP-ETC1 plants, YFP-ETC1 was detected in all cell layers indicating that YFP-ETC1 RNA or protein can move between several layers (Figure 5D). This is consistent with the report from Rim et al. (2011). In a systematic approach it was shown that transcription factors may be cell-autonomous, may move several cell layers or to all layers including the epidermis (Rim et al., 2011). Interestingly, CPC was shown to belong to the latter class supporting our finding that ETC1 can move through all cell layers.
The phenotypic analysis revealed, that the ProSCR:YFP-ETC1 cpc-2 etc1-1 plants exhibited more root hairs in H files than the cpc-2 single mutant (Figure 5C and Supplementary Table S12) indicating that expression of ETC1 in the stele is sufficient to rescue the mutant phenotype. It is therefore conceivable that the expression switch of ETC1 upon phosphate starvation from N-files to the sub-epidermis is functionally relevant and that it contributes to the ectopic root hair formation.
Conclusion and Outlook
Despite the functional redundancy of ETC1 with other R3 MYB genes such as CPC, ETC3, and TRY under normal growth conditions it is interesting that they appear to have divergent functions under Pi starvation conditions as noted before (Chen and Schmidt, 2015). The four paralogous genes strikingly differ with respect to changes in RNA abundance and root hair formation upon Pi- starvation (Chen and Schmidt, 2015). Among them, ETC1 appears to have a less prominent role as the number of root hairs is only weakly effected by Pi starvation (Chen and Schmidt, 2015). Only the comparison of cpc and the cpc etc1 double mutant unambiguously revealed a function of ETC1 in Pi starvation responses. However, the mechanism by which ETC1 contributes to Pi starvation response is novel and a potentially powerful principle to override other regulation events. The Pi starvation induced new expression domain in subepidermal cell layers in combination with the fact that ETC1 can travel through several cell layers represents a developmental switch by which a root hair promoting factor can be provided in all epidermal cells and thereby promoting root hair formation.
Materials and Methods
Plant Material, Culture Media, and Root Hair Analysis
The following A. thaliana lines were used in this study: Columbia (Col-0), Landsberg erecta (Ler), ttg1-1 (Ler) (Koornneef, 1981), gl2-1 (Ler) (Di Cristina et al., 1996), wer-1 (Col-0) (Lee and Schiefelbein, 1999), cpc-2 (Col-0) (Kurata et al., 2005), try-JC (Col-0) (Larkin et al., 1999) and etc1-1 (Kirik et al., 2004a), pGL2:GUS, pCPC:GUS (Col-0) (Pesch et al., 2013), pTRY:GUS (Col-0) (Pesch and Hulskamp, 2011) and pSCR:GFPer (Col-0) (Rishmawi et al., 2014). pETC1:GUS (Col-0), pETC1:YFP-ETC1 (Col-0), and pSCR:YFP-ETC1 (Col-0) were created in this study as described below. The cpc-2 etc1-1 double mutant was created by crossing cpc-2 with etc1-1.
Seeds were sterilized by shaking in 100% ethanol followed by adding 4% HCL, washing three times with sterile water. After 3 days of vernalization at 4°C seedlings were grown on vertically placed MS (Murashige and Skoog, 1962) agar plates with (Pi+) and without (Pi-) phosphate under long day conditions (16 h light/8 h dark) for 7 days. Pi+ and Pi- MS medium contained 2.06 mM NH4NO3, 1.88 mM KNO3, 0.31 mM MgSO4, 0.1 mM MnSO4, 0.03 mM ZnSO4, 0.1 mM CuSO4, 0.3 mM CaCl2, 5.0 mM KI, 0.1 mM CoCl2, 0.1 mM FeSO4, 0.1 mM EDTA, 0.1 mM H3BO3, and 1 mM Na2MoO4.2H2O supplemented with 1% (w/v) agar and 1% (w/v) sucrose. 1 mM KH2PO4 was added for MS Pi+ and replaced by 1 mM of KCL for MS Pi-. The pH was adjusted to 5.6–5.8 by adding NaOH.
For root hair analysis epidermal files were defined by their relative position to the cortical cells. At least 10 biological replicas were analyzed for each genotype. For each seedling, the number of root hairs was determined for 10 cells in the H-file and 10 cells in the N-position.
Plasmid Construction
For ProETC1:GUS constructs, serial deletions were created as presented in Figure 3. For all the constructs, genomic DNA of Col-0 and primers with attb sites were used: GW-ProETC1 F and GW-ProETC1 R primers for ProETC1-1921, GW-ProETC1-1676 F and GW-ProETC1 R primers for ProETC1-1676, GW-ProETC1-1371 F and GW-ProETC1 R primers for ProETC1-1371, GW-ProETC1-1183 F and GW-ProETC1 R primers for ProETC1-1183, GW-ProETC1-932 F and GW-ProETC1 R primers for ProETC1-932, GW-ProETC1-595 F and GW-ProETC1 R primers for ProETC1-595, GW-ProETC1-400 F and GW-ProETC1 R primers for ProETC1-400. Each of the produced fragments was introduced in pDONR201 entry vector. The entry vectors were used to transfer the ETC1 fragments to the destination vector (ProBat-TL-B-C-GUS-FUS-w/oPromoter) through LR reactions (Clontech).
ProBat-B-C-GUS-FUS-w/oPromotor was generated as follows. ProBat-B-sYFPN-w/oPromoter (provided by Cordula Jörgens) contains unique restriction sites replacing Pro35S-TL upstream of a GatewayTM (Invitrogen1) cassette in ProBat-TL-B-sYFPN (J.F. Uhrig, unpublished data). sYFPN was removed (SpeI) to generate ProBat-B-w/oPromotor. GUS-FUS with attached SpeI sites was amplified with ANS89SpeI-HA-GUS-s and ANS90SpeI-GUS-as (Supplementary Table S12) from an entry vector (Rudolph et al., 2003) and inserted into the SpeI site following the GatewayTM cassette of ProBat-B-w/oPromotor to obtain ProBat-B-C-GUS-FUS-w/oPromotor.
For the rescuing experiments with ProETC1 fragments ETC1 CDS was amplified from cDNA using the GW-ETC1 cDNA F and GW-ETC1 CDS R primers (Supplementary Table S13) and introduced in pDONR201. The ETC1 CDS was introduced in the ProBat-B-w/oPromotor destination vector by LR reaction (Clontech). Each fragment of ProETC1 was introduced in ProBat-B-w/oPromotor-ETC1 CDS by blunt ending cloning using the Smi1 restriction enzyme.
For pENSG-GW-ProETC1:YFP-ETC1: The ETC1 CDS was introduced in pENSG-YFP destination vector (Wester et al., 2009) by LR reaction (Clontech). The ETC1 promoter (1371 bp upstream of start codon) was amplified using Asc1-pETC1 F and Xho1-pETC1 R primers (Supplementary Table S13) and introduced in pJET1.2 (Fermentas, United States). Finally ProETC1 was cloned in the pENSG-YFP-ETC1 destination vector as an Asc1 and Xho1 fragment.
For pENSG-GW-ProSCR:YFP-ETC1 was created analogous as described for pENSG-GW-ProETC1:YFP-ETC1 using a ProSCR CDS amplified using Asc1-pSCR F and Xho1-pSCR R primers (Supplementary Table S13).
pENSG-GW-ProETC1PHR1mut:YFP-ETC1 the ETC1 CDS was introduced by LR reaction as previously described. ProETC1PHR1mut promoter was amplified in three steps. The first fragment was amplified using Asc1-ProETC1 F and phr1mut- R primers. The second fragment was amplified using Xho1-ProETC1 R and phr1mut- F primers. These two fragments were used as a template for the third PCR using Asc1-pETC1 F and Xho1-pETC1 R primers (Supplementary Table S13). The phr1mut-F and phr1mut-R primers contain the substituted nucleotides that will convert the PHR-1 binding sequence from (gtatatcc) to (aaaaaaaa). The ProETC1PHR1mut was cloned in the ProENSG-YFP-ETC1 destination vector as an Asc1 and Xho1 fragment.
GUS Staining
GUS staining was done with 7-day old seedlings as described previously (Vroemen et al., 1996). Seedlings were fixed (50% methanol; 10% acetic acid) at 4°C for 24 h, transferred to 80% ethanol for 2 h, washed three times with water and analyzed by transmission light microscopy.
Microscopy and Image Acquisition
Pictures of GUS stained roots were acquired using a Leica DMRA microscope (Leica Microsystems) equipped with DISKUS software (Carl H. Hilgers-Technisches Büro).
Confocal laser-scanning microscopy (CLSM) was performed using a Leica TCS-SP8 confocal microscope equipped with the Leica LAS AF software. For imaging a Leica 1.2 NA, 63× water objective was used. The propidium iodide staining was excited using a DPSS laser at 561 nm, the GFP/YFP signal was excited using an argon laser at 488/514 nm. For the protein localization, seedlings were stained with 100 μg/ml of propidium iodide for 1 min followed by washing the samples with water. The analyses were done by sequential scanning starting with the respective higher wavelength. CLSM pictures (focal planes) of each channel were merged in Fiji (Schindelin et al., 2012) and the colors were set as follows: “Cyan Hot” for propidium iodide and “Yellow” for YFP. For each channel, only brightness and contrast were additionally adjusted.
Statistical Analysis and Motif Analysis
Statistical analyses were done with R using RStudio (R Core Team, 2017). Normality was tested for with the Kolmogorov–Smirnov test. As data for almost all genotypes and conditions were not normally distributed, an unpaired two-samples Wilcoxon test was conducted.
The motif analysis was conducted using PlantPAN 2.02 (Chow et al., 2016) and manual analysis with 14 cis-regulatory elements related to the formation of phosphate deficiency-induced root hairs before (Salazar-Henao et al., 2016). The figure was constructed using the CLC DNA Workbench3.
Author Contributions
LR did the most experiments. HW did the confocal image acquisition and root hair phenotyping analysis of selected lines. LR, MH, and AS conceived the study and wrote the manuscript. AS and LR analyzed the data. AS did the statistics and motif analysis. All authors have approved the final article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Cordula Jörgens for providing the pBat-B-sYFPN-w/oPromotor vector.
Funding. This work was supported by the Cluster of Excellence on Plant Sciences (Grant No. EXC 1028).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01411/full#supplementary-material
References
- Bates T. R., Lynch J. P. (1996). Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 19 529–538. 10.1111/j.1365-3040.1996.tb00386.x [DOI] [Google Scholar]
- Bernhardt C., Lee M. M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130 6431–6439. 10.1242/dev.00880 [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M. Z., Gonzalez A., Lloyd A., Schiefelbein J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132 291–298. 10.1242/dev.01565 [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Schmidt W. (2015). The paralogous R3 MYB proteins CAPRICE, TRIPTYCHON and ENHANCER OF TRY AND CPC1 play pleiotropic and partly non-redundant roles in the phosphate starvation response of Arabidopsis roots. J. Exp. Bot. 66 4821–4834. 10.1093/jxb/erv259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. H., Nimmo G. A., Jenkins G. I., Nimmo H. G. (2007). BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem. J. 405 191–198. 10.1042/BJ20070102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. N., Zheng H. Q., Wu N. Y., Chien C. H., Huang H. D., Lee T. Y., et al. (2016). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44 D1154–D1160. 10.1093/nar/gkv1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., et al. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10 393–402. 10.1046/j.1365-313X.1996.10030393.x [DOI] [PubMed] [Google Scholar]
- Dolan L., Duckett C. M., Grierson C., Linstead P., Schneider K., Lawson E., et al. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120 2465–2474. 9501025 [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., et al. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119 71–84. [DOI] [PubMed] [Google Scholar]
- Esch J. J., Chen M., Sanders M., Hillestad M., Ndkium S., Idelkope B., et al. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130 5885–5894. 10.1242/dev.00812 [DOI] [PubMed] [Google Scholar]
- Galway M. E., Masucci J. D., Lloyd A. M., Walbot V., Davis R. W., Schiefelbein J. W. (1994). The Ttg gene is required to specify epidermal-cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166 740–754. 10.1006/dbio.1994.1352 [DOI] [PubMed] [Google Scholar]
- Hassan H., Scheres B., Blilou I. (2010). JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137 1523–1529. 10.1242/dev.048777 [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 555–567. 10.1016/S0092-8674(00)80865-X [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 297–300. 10.1093/nar/27.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. Y., Lin Y., Zhang M., Pollock S., Marks M. D., Schiefelbein J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117 73–84. 10.1104/pp.117.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. H., Song S. K., Schiefelbein J., Lee M. M. (2013). Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol. 163 193–204. 10.1104/pp.113.221028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Simon M., Huelskamp M., Schiefelbein J. (2004a). The enhancer of try and CPC1 gene acts redundantly with triptychon and caprice in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268 506–513. 10.1016/j.ydbio.2003.12.037 [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Wester K., Schiefelbein J., Hulskamp M. (2004b). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55 389–398. [DOI] [PubMed] [Google Scholar]
- Koornneef M. (1981). The complex syndrome of ttg mutants. Arabidopsis Inform. Serv. 18 45–51. [Google Scholar]
- Kurata T., Ishida T., Kawabata-Awai C., Noguchi M., Hattori S., Sano R., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132 5387–5398. 10.1242/dev.02139 [DOI] [PubMed] [Google Scholar]
- Kwak S. H., Shen R., Schiefelbein J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307 1111–1113. 10.1126/science.1105373 [DOI] [PubMed] [Google Scholar]
- Lan P., Li W., Schmidt W. (2012). Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics 11 1156–1166. 10.1074/mcp.M112.020461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J. C., Walker J. D., Bolognesi-Winfield A. C., Gray J. C., Walker A. R. (1999). Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 151 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 473–483. [DOI] [PubMed] [Google Scholar]
- Lee M. M., Schiefelbein J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14 611–618. 10.1105/tpc.010434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Bielenberg D. G., Brown K. M., Lynch J. P. (2001). Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 24 459–467. 10.1046/j.1365-3040.2001.00695.x [DOI] [Google Scholar]
- Masucci J. D., Schiefelbein J. W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8 1505–1517. 10.1105/tpc.8.9.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., et al. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316 1477–1480. 10.1126/science.1142618 [DOI] [PubMed] [Google Scholar]
- Misson J., Raghothama K. G., Jain A., Jouhet J., Block M. A., Bligny R., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U.S.A. 102 11934–11939. 10.1073/pnas.0505266102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Schmidt W. (2004). Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 134 409–419. 10.1104/pp.103.029066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Pesch M., Hulskamp M. (2011). Role of TRIPTYCHON in trichome patterning in Arabidopsis. BMC Plant Biol. 11:130. 10.1186/1471-2229-11-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M., Schultheiss I., Digiuni S., Uhrig J. F., Hulskamp M. (2013). Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140 3456–3467. 10.1242/dev.094698 [DOI] [PubMed] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rim Y., Huang L., Chu H., Han X., Cho W. K., Jeon C. O., et al. (2011). Analysis of Arabidopsis transcription factor families revealed extensive capacity for cell-to-cell movement as well as discrete trafficking patterns. Mol. Cells 32 519–526. 10.1007/s10059-011-0135-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L., Pesch M., Juengst C., Schauss A. C., Schrader A., Hulskamp M. (2014). Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol. 165 186–195. 10.1104/pp.113.233775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph C., Schreier P. H., Uhrig J. F. (2003). Peptide-mediated broad-spectrum plant resistance to tospoviruses. Proc. Natl. Acad. Sci. U.S.A. 100 4429–4434. 10.1073/pnas.0730832100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K. H., Kang Y. H., Park Y. H., Hwang I., Schiefelbein J., Lee M. M. (2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132 4765–4775. 10.1242/dev.02055 [DOI] [PubMed] [Google Scholar]
- Salazar-Henao J. E., Lin W. D., Schmidt W. (2016). Discriminative gene co-expression network analysis uncovers novel modules involved in the formation of phosphate deficiency-induced root hairs in Arabidopsis. Sci. Rep. 6:26820. 10.1038/srep26820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage N., Yang T. J., Chen C. Y., Lin K. L., Monk N. A., Schmidt W. (2013). Positional signaling and expression of ENHANCER OF TRY AND CPC1 are tuned to increase root hair density in response to phosphate deficiency in Arabidopsis thaliana. PLoS One 8:e75452. 10.1371/journal.pone.0075452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21 5036–5046. 10.1093/emboj/cdf524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Lee M. M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311 566–578. 10.1016/j.ydbio.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Okada K., Wada T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19 2264–2277. 10.1105/tpc.106.045732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen C. W., Langeveld S., Mayer U., Ripper G., Jurgens G., VanKammen A., et al. (1996). Pattern formation in the arabidopsis embryo revealed by position-specific lipid transfer protein gene expression. Plant Cell 8 783–791. 10.1105/tpc.8.5.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Kurata T., Tominaga R., Koshino-Kimura Y., Tachibana T., Goto K., et al. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129 5409–5419. 10.1242/dev.00111 [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277 1113–1116. 10.1126/science.277.5329.1113 [DOI] [PubMed] [Google Scholar]
- Walker A. R., Davison P. A., Bolognesi-Winfield A. C., James C. M., Srinivasan N., Blundell T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1349. 10.1105/tpc.11.7.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester K., Digiuni S., Geier F., Timmer J., Fleck C., Hulskamp M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136 1487–1496. 10.1242/dev.021733 [DOI] [PubMed] [Google Scholar]
- Wu P., Ma L., Hou X., Wang M., Wu Y., Liu F., et al. (2003). Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 132 1260–1271. 10.1104/pp.103.021022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L. (2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42 264–267. 10.1038/ng.529 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


