Abstract
RNA-Seq is a widely used method for studying the behavior of genes under different biological conditions. An essential step in an RNA-Seq study is normalization, in which raw data are adjusted to account for factors that prevent direct comparison of expression measures. Errors in normalization can have a significant impact on downstream analysis, such as inflated false positives in differential expression analysis. An underemphasized feature of normalization is the assumptions on which the methods rely and how the validity of these assumptions can have a substantial impact on the performance of the methods. In this article, we explain how assumptions provide the link between raw RNA-Seq read counts and meaningful measures of gene expression. We examine normalization methods from the perspective of their assumptions, as an understanding of methodological assumptions is necessary for choosing methods appropriate for the data at hand. Furthermore, we discuss why normalization methods perform poorly when their assumptions are violated and how this causes problems in subsequent analysis. To analyze a biological experiment, researchers must select a normalization method with assumptions that are met and that produces a meaningful measure of expression for the given experiment.
Keywords: RNA-Seq, normalization, assumptions, differential expression, spike-in control, transcriptome size
Introduction
The introduction of microarrays provided the ability to study many genes in an organism under different biological conditions, with a dramatic reduction in expense and time from previous methods [1]. More recently, high-throughput sequencing has become an affordable and effective way of examining gene behavior and has been applied to a wide range of biological studies. For example, specific questions about transcriptomes and splicing can now be addressed [2], and the study of techniques for the analysis of high-throughput sequencing data continues to be a hot topic, involving researchers from biology, statistics and computer science.
High-throughput sequencing with RNA, commonly referred to as RNA-Seq, involves mapping sequenced fragments of cDNA. In RNA-Seq, the RNA is fragmented and then reverse transcribed to cDNA (or reverse transcribed then fragmented). These fragments are then sequenced, producing reads that are aligned back to a pre-sequenced reference genome or transcriptome [2–4], or in some cases assembled without the reference [3]. The number of reads mapped to a gene is used to quantify its expression.
To convert raw read counts into informative measures of gene expression, normalization is needed to account for factors that affect the number of reads mapped to a gene, like length [5], GC-content [6] and sequencing depth [7]. Length and GC-content are within-sample effects, meaning that they affect the comparison of read counts between different genes in a sample. Sequencing depth, on the other hand, is a between-sample effect that alters the comparison of read counts between the same gene in different samples. Here we focus on between-sample normalization, which is needed to account for technical effects (differences not because of the biological conditions of interest) that prevent read count data from accurately reflecting differences in expression [7]. In RNA-Seq, a cDNA library is constructed and then a portion of the molecules is sequenced to produce reads [8]. Experimental variability, such as variability in the total number of molecules sequenced, can lead to different total read counts in different samples; this is referred to as differences in sequencing depth, and the total number of reads in a sample is the library size of that sample [9]. When one sample has more reads than another, non-differentially expressed genes will tend to have higher read counts in that sample [7] and so a correction is necessary. For applications requiring both between-sample and within-sample normalization, performing both types of normalization may be necessary; for example, Risso et al. recommend using within-sample GC-content normalization in combination with between-sample normalization [6]. An additional issue when applying normalization methods is the total number of reads; in this article, we assume the samples are sequenced sufficiently deeply for all normalization methods considered.
Many normalization schemes have been proposed to account for between-sample effects in RNA-Seq data [9], and several attempts have been made to determine the best strategy [9–16]. However, little attention has been paid to the assumptions on which the different normalization methods rely. Several authors have identified situations in which a few highly expressed genes make up a large proportion of the total reads [9, 10, 13], which could result in differences in distribution of read counts among genes. Others have found cases in which most or all genes are up-regulated in one condition [17–20]. These situations, especially a global shift in expression, violate assumptions of many commonly used methods and so result in errors in downstream analysis. Furthermore, biological experiments in which assumptions are unwittingly violated may mean that there are flaws in comparisons of normalization methods and in the conclusions drawn from these experiments. As we have evidence of violated assumptions in some biological experiments, but not the extent to which assumptions are violated in others, it has been suggested that many prior conclusions are incorrect and a reanalysis of published results is necessary [21].
The goal of this article is to present normalization methods in the context of their assumptions, and to evaluate the effect and importance of assumptions on the performance of different normalization methods. We believe that a focus on assumptions can aid in evaluating different methods, and in choosing an appropriate method given knowledge of which assumptions are reasonable to make for the experiment at hand. With this in mind, we group between-sample methods by the assumptions they rely on and their strategy for normalization. We explain the reason the assumptions are necessary and the result of using a method when the assumptions do not hold. Finally, we examine previous research that aims to determine which normalization method is better from the perspective of why some methods perform better than the other in specific situations.
Gene expression and normalization
The goal of normalization is for differences in normalized read counts to represent differences in true expression. Normalization is correct when the relationship between normalized read counts is correct. Given that the actual product of gene expression is never measured, we consider the true expression of a gene to be the amount of mRNA/cell it produces.
This appears to be the definition commonly used in previous work, as prior research considers a gene to be differentially expressed (DE) across different biological conditions if there is a difference in the amount of mRNA/cell it produces under these conditions. For example, authors discussing a global shift in expression talk about a global change in the absolute amount of RNA from a fixed number of cells [22]. In this article, we view expression and differential expression in terms of absolute quantities of mRNA/cell, and keeping this perspective in mind is important for understanding our discussion of normalization methods and their assumptions. However, it is important to note that other definitions of expression and differential expression are possible [23], and beginning with a different definition may change which methods are appropriate for a given RNA-Seq experiment. For example, for certain biological experiments, one might be interested in detecting differences in mRNA/transcriptome (that is, a gene’s proportion of mRNA of all mRNA transcribed) rather than mRNA/cell [23].
Considering DE genes is helpful for understanding normalization. As stated above, correct normalization will result in correct relationships between normalized read counts. In terms of differential expression, this means that non-DE genes should on average have the same normalized read counts across conditions, while DE genes should have normalized read counts whose differences (ratios) across conditions represent the true differences (ratios) in mRNA/cell. As with microarrays, a common use of RNA-Seq is to investigate the differential expression of an organism’s genes under different biological conditions [2], but normalization is needed in any RNA-Seq study where the relationship between normalized read counts must be correct, not just in differential expression analyses. In this article, for simplicity we restrict our examples to the most basic case of two biological conditions, which will generally be referred to as A and B. Our results, however, hold for any number of conditions.
Gene expression is measured with RNA-Seq using the number of reads aligned to each gene under each biological condition [3]. However, a naive comparison of read counts for a given gene under the different conditions is problematic for two reasons. First, the number of reads aligned to a given gene in a given sample is generally considered a random variable [24] (though non-random events, such as inconsistent fragment amplification or poor amplification of certain sequences, can impact the final read count), and so read count comparisons must take into account the variability of these random variables. Second, the total number of reads can vary across samples [2], and so a large difference in a gene’s read count between different conditions may simply be the result of differential coverage, rather than of differential expression. It is the second problem that necessitates normalization of read counts before differential expression analysis can be performed [2, 4].
Normalization is an essential step in an RNA-Seq analysis, in which the read count matrix is transformed to allow for meaningful comparisons of counts across samples. With the advent of RNA-Seq technology, it was initially believed that normalization would not be necessary [3], but normalization has been found to be indispensable for correct analysis of RNA-Seq data. Indeed, Bullard et al. [10] found that the normalization procedure used in a differential expression pipeline had the largest impact on the results of the analysis, even more than the choice of test statistic used in hypothesis tests for differential expression.
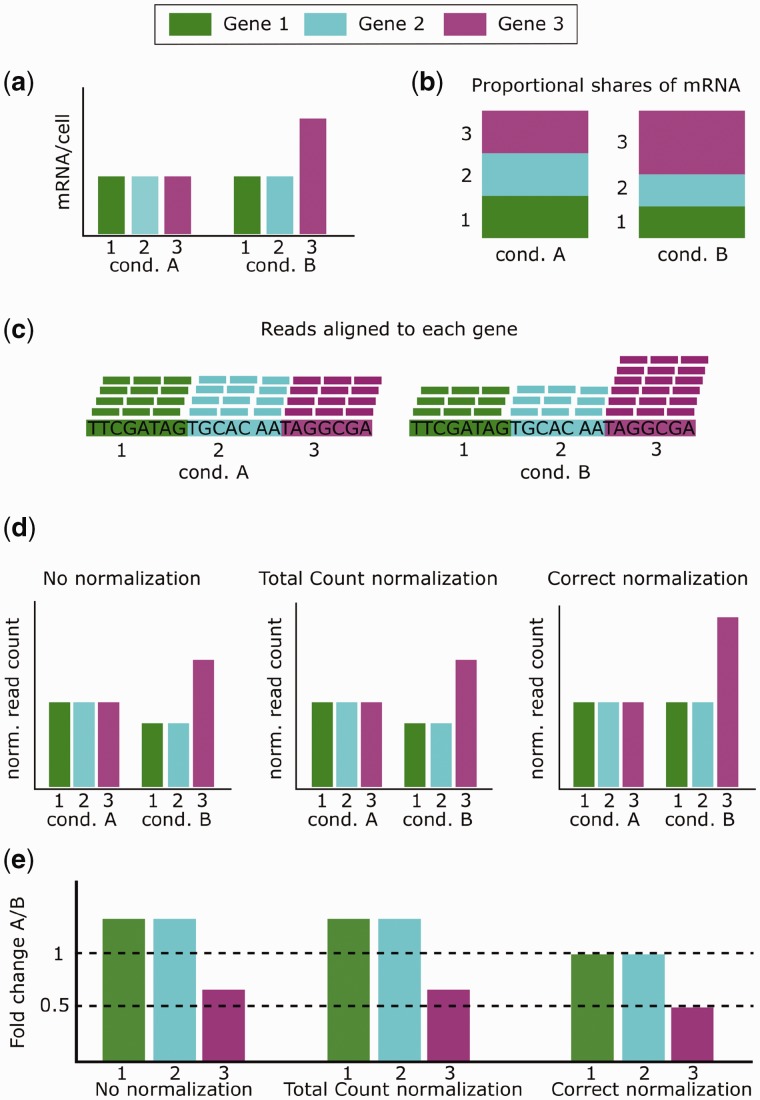
Another reason normalization is required is that the proportion of mRNA corresponding to a given gene may change across biological conditions. In the sample of molecules sequenced, the number of molecules (and so by extension the number of reads) corresponding to a given gene is tied to that gene’s share of the population of molecules available for sequencing. Hence, when there are a few genes that are highly expressed in only one of the conditions, the few genes will make up a greater share of the total molecules and so a smaller fraction of the reads will be left for the other genes [7]. This can cause the false appearance of differential expression for the non-DE genes, and normalization is needed to account for this difference. A visualization of such a situation is presented in Figure 1. Of the three genes, one is up-regulated while the other two are non-DE (Figure 1A). The one highly expressed gene leads to differences in shares of the proportion of mRNA for each gene (Figure 1B), which in turn causes differences in the share of reads aligned to each gene, even if the total number of reads is the same in each condition (Figure 1C). If the differences in read share are not corrected by normalization (Figure 1D), then the apparent fold change for every gene will be wrong (Figure 1E). Correct normalization, on the other hand, equilibrates the read counts for the two non-DE genes (Figure 1D) and thereby leads to accurate observed fold changes (Figure 1E).
Figure 1.
One highly expressed gene. An experiment is performed with conditions A and B to compare expression for the three genes (1, 2 and 3). (A) Gene 3 is 2-fold up-regulated under condition B, while the other genes are not DE; the quantity of mRNA/cell (in bp) is the same for genes 1 and 2, but is twice as high for gene 3 under condition B. (B) Because of the change in expression of gene 3, the shares of mRNA in the cell are different between conditions. Under condition A, each gene gets one-third, whereas under condition B, gene 3 gets half while the other two get one-fourth. (C) Differences in shares of mRNA are reflected in the shares of reads. Each sample has the same total number of reads, but the distribution is different between the conditions, matching the distribution of mRNA in (B). (D) When no normalization is performed, there are apparent differences in read counts for all three genes. Total count normalization produces the exact same result as no normalization at all, as the total read count for each sample is the same. In truth, there is no difference in expression for genes 1 and 2, and the relative count for gene 3 should be higher than found by no normalization or total count normalization. Correct normalization, therefore, makes the read counts of the non-DE genes equivalent, which also makes the relative expression of gene 3 correct. (E) No normalization and total count normalization fail to equilibrate the read counts of the non-DE genes, resulting in each gene appearing DE, and the truly DE gene (gene 3) having the wrong fold change. Correct normalization reveals no difference in expression for the non-DE genes and the correct fold change for gene 3.
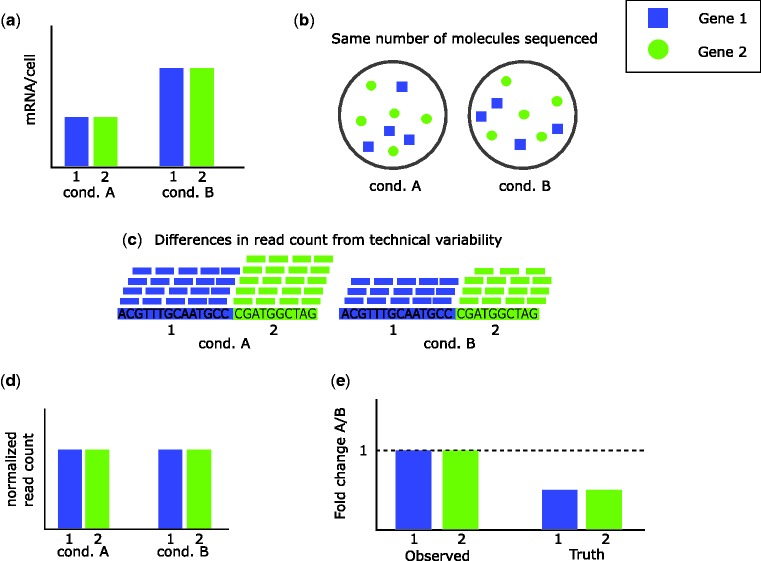
As normalization methods have developed, it has become clear that initial approaches fail in cases of a shift in expression for many or all genes [22]. In cases like Figure 1, a small number of highly expressed genes creates the appearance that non-DE are DE, but the false DE calls may be corrected by normalizing read counts so that the expression levels of non-DE genes are equivalent. In contrast, in the case of a global shift in expression, it may appear that DE genes are non-DE or that up-regulated genes are down-regulated [22]. An example is presented in Figure 2. All genes are up-regulated 2-fold under condition B (Figure 2A), but roughly the same number of molecules are sequenced (Figure 2B). This conceals the fact that one condition results in twice as much total expression, and the only differences in read counts between the two conditions is because of technical variability (e.g. sequencing depth; Figure 2C). Conventional normalization approaches account for the technical differences, resulting in the same normalized read counts under each condition (Figure 2D). Conventional normalization fails to reflect the 2-fold up-regulation under condition B, and examining the observed fold changes (Figure 2E), it appears that neither gene is DE when in truth both are. A further need for normalization is therefore in cases of global shifts in expression, in which it is necessary to take into account the differences in overall expression between conditions.
Figure 2.
Global shift in expression. There are two genes, and an experiment is performed to compare expression between condition A and condition B. (A) There is global up-regulation under condition B versus condition A, with both genes having twice the expression under condition B. Within each condition, the two genes produce the same amount of mRNA/cell (measured in bp). (B) In the RNA-Seq experiment, the same number of molecules are sequenced from each of the two samples. Proportionally, the mRNA composition is the same under each condition, and so the composition of molecules sequenced is also the same. Within each condition, the two genes produce the same amount of mRNA (in bp) but gene 2 is four-fifth the length of gene 1, so must produce five-fourth the number of molecules that gene 1 does. (C) Sequenced reads are aligned to the reference genome and mapped to each gene. The distribution of reads is the same in each sample, but by chance the sample for condition A happens to have more reads in total. (D) Normalization is performed, which removes the differences in read count from technical variability, so the read count for each gene is the same across conditions. (E) Because the normalized read counts are the same, the observed fold change for each gene is 1, indicating no differential expression. However, genes are really twice as expressed under condition B and so in truth we should see half the expression when comparing A with B.
To address the variety of needs for normalization, a corresponding variety of normalization methods has been developed. To correctly normalize, each method requires one or more assumptions about the experiment and gene expression. Assumptions are necessary for converting read counts into meaningful measures of expression. In the following sections, we organize normalization methods into groups of methods that rely on similar assumptions.
Normalization methods and assumptions
Here we group normalization methods that have similar assumptions and approaches to normalization. Short descriptions of the methods are provided; more detailed information on the method specifics is available in the Supplementary Information.
Recall that for our purposes, a gene is DE across a set of conditions if that gene produces different levels of mRNA/cell under the different conditions. For a normalization method to work, the normalized read counts must be representative of the true mRNA/cell values. That is, if a gene produces twice as much mRNA/cell under condition A as under condition B, then the normalized read count for that gene should on average be twice as big under condition A as under condition B. However, RNA-Seq, on the other hand, initially produces relative measures of expression [25]. As shown in Figure 2, the number of reads aligned to a given gene reflects the sequencing depth and that gene’s share of the population of mRNA molecules. We should not expect a gene with twice as much mRNA/cell to have twice the number of reads. To correctly normalize, then, we must make some assumptions so that initial raw read counts can be converted into a measure comparable across samples. Different groups of normalization methods discussed here take different approaches, and so require different assumptions to produce correctly normalized values. These assumptions often deal with the total amount of mRNA/cell or the amount of ‘symmetry’ in the differential expression.
We say that differential expression is symmetric between two conditions when the number of genes up-regulated in each condition is equal. Figure 3 demonstrates the four possible combinations of symmetry/asymmetry and same/different total mRNA/cell. Figure 3 will be referenced to illustrate situations in which assumptions are and are not met.
Figure 3.
Differential expression and (a)symmetry. There are six genes, and two experimental conditions. (A) Differential expression is asymmetric (three up-regulated genes under condition A, one under condition B). The total mRNA/cell (summed over the six genes) is the same under both conditions. (B) Differential expression is asymmetric. The total mRNA/cell is different (less total mRNA/cell under condition B). (C) Differential expression is symmetric (two up-regulated genes under each condition). The total mRNA/cell is the same under both conditions. (D) Differential expression is symmetric. The total mRNA/cell is different (more total mRNA/cell under condition B).
Normalization by library size
The normalization by library size aims to remove differences in sequencing depth simply by dividing by the total number of reads in each sample [9].
Assumptions
Same total expression: The amount of total expression is the same under the different experimental conditions. That is, each condition has the same amount of mRNA/cell. Figure 3A and C show examples in which this assumption holds.
Methods
Total Count normalization [9] divides each read count by the number of reads in its sample. The reads per kilobase per million mapped reads (RPKM) [26] method is essentially the same as Total Count normalization, but with the added component of accounting for gene length as well. FPKM [27] and ERPKM [12] are variants of RPKM.
Motivation
After dividing by library size, the normalized counts reflect the proportion of total mRNA/cell taken up by each gene. If the total mRNA/cell is the same across conditions, this proportion reflects absolute mRNA/cell for each gene.
Normalization by distribution/testing
If technical effects are the same for DE and non-DE genes, then normalization could be done by equilibrating expression levels for non-DE genes. This set of methods attempts to capture information from non-DE genes. Normalization by distribution compares distributions (either of read counts or some function of read counts) across samples; normalization by testing attempts to detect a set of non-DE genes through hypothesis testing.
Assumptions
DE and non-DE genes behave the same: Technical effects are the same for DE and non-DE genes.
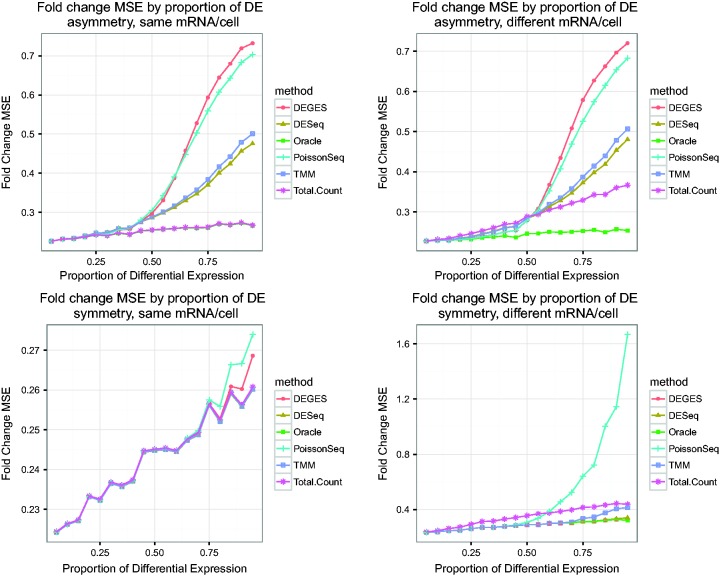
Balanced expression: There is roughly symmetric differential expression across conditions (same number of up-regulated and down-regulated genes). This assumption holds in Figure 3C and D. Normalization by testing can tolerate a larger difference in number of up- and down-regulated genes for higher proportions of DE than can normalization by distribution (see Figure 6).
Figure 6.
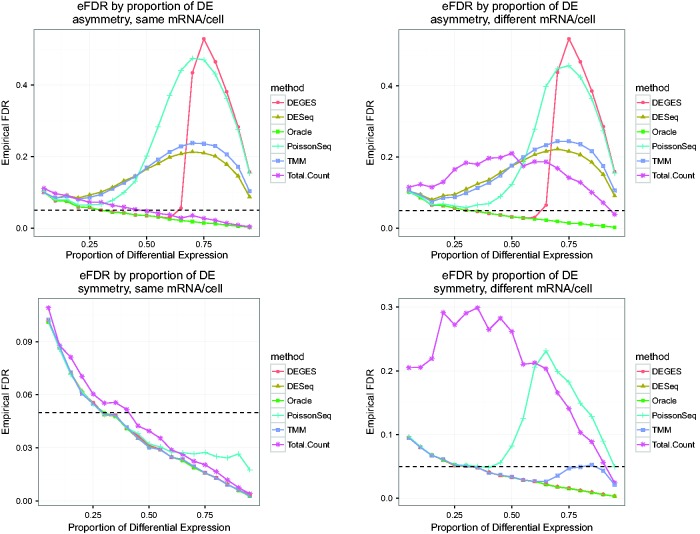
Impact of amount of asymmetry and amount of mRNA/cell on error control, 10 000 genes and four samples. These plots show the average empirical FDR of several methods on simulated data with varying proportions of differential expression (5–95%). The simulations are performed with two conditions, with 10 000 genes and two replicates per condition. Asymmetric differential expression was simulated as 75% of the set of DE genes up-regulated in one condition and 25% up-regulated in the other. Under symmetric differential expression, 50% of DE genes are up-regulated in each condition. For simulations with the same mRNA/cell, non-DE genes had the same proportion of reads in each condition; simulations with different mRNA/cell resulted in non-DE genes having different shares of the reads in the different conditions. The black dashed line is at 0.05, the nominal FDR using the Benjamini–Hochberg adjustment. Deviations of the oracle value from the nominal value (starting above 0.05 and falling below as the proportion of DE increases) are a result of DESeq2 hypothesis testing and the conservativeness of Benjamini–Hochberg.
Methods
Normalization by distribution. Quantile normalization [28] forces the distribution of the normalized data to be the same for each sample by replacing each quantile with the average (or median) of that quantile across all samples. Other methods do not force all quantiles to be the same, but instead focus on a specific quantile. Upper Quartile normalization [10] divides each read count by the 75th percentile of the read counts in its sample. Median normalization [9] is essentially the same, but uses the median rather than the 75th percentile. The DESeq normalization method [24] finds the ratio of each read count to the geometric mean of all read counts for that gene across all samples (the denominator serving as a pseudo-reference sample [24]). The median of these ratios for a sample, called the ‘size factor’, is used to scale that sample. This idea was expanded in the CuffDiff 2 software; CuffDiff normalization calculates ‘internal’ and ‘external’ size factors using the DESeq approach. The internal size factors are found for each sample by only considering other samples performed under the same biological condition when taking the geometric mean, while the external size factors are calculated after normalization by the internal size factors. The Trimmed Mean of the M-values (TMM) [7] approach is to choose a sample as a reference sample, then calculate fold changes and absolute expression levels relative to that sample. The genes are trimmed twice by these two values, to remove DE genes, then the trimmed mean of the fold changes is found for each sample. Read counts are scaled by this trimmed mean and the total count of their sample. Note: The edgeR package [29] uses TMM normalization, and so TMM could reasonably be called edgeR normalization instead. However, the name TMM seems to be more commonly used in the literature, and so we use it here. Median Ratio normalization (MRN) [14] is a method similar to TMM, with the goal of being more robust. In MRN, read counts are divided by the total count of their sample, then averaged across all samples in a condition for a given gene. This produces an average count-normalized value for each gene and each condition, and the median of the ratios of these values between conditions is taken. The original counts are then normalized by this median and their library size.
Normalization by testing. ‘PoissonSeq’ [30] uses an iterative process that alternates between estimating a set of non-DE genes, and estimating the scaling factor for each sample using that set. Given estimates of the scaling factor, expected values for the read counts can be determined and non-DE genes are identified using a goodness-of-fit test. A similar iterative strategy is implemented by Differentially Expressed Gene Elimination Strategy (DEGES) [11], which alternates between calculating scaling factors from a set of genes identified as non-DE and estimating which genes are non-DE using differential expression hypothesis testing.
Motivation
Non-DE genes should have, on average, the same normalized counts across conditions. Clearly, we want to normalize to equilibrate the non-DE genes. If technical effects impact non-DE genes and DE genes alike, then we can normalize all genes with the same normalization factor as the non-DE genes. So, we need to compare the non-DE genes; assuming balanced expression means we can estimate the differences in read counts between non-DE genes across samples.
Normalization by controls
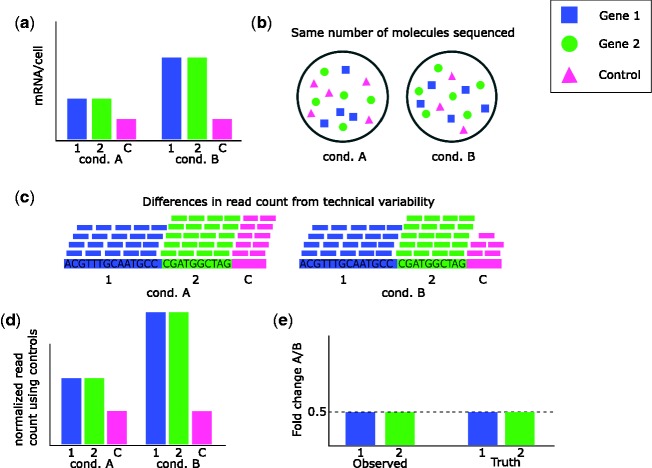
Controls are needed for normalization when the assumptions of other methods are violated. For example, Figure 2 demonstrates how a global shift in expression can go undetected. When controls are used, such as the negative controls illustrated in Figure 4, then it is possible to correctly normalize by performing normalization on the controls. Because the controls are not affected by the biological conditions but the same amount of controls/cell are present in each condition (Figure 4A), then different numbers of control molecules are sequenced (Figure 4B). This leads to a share of the reads reflective of the share of mRNA for the control (Figure 4C). By normalizing on the control, the correct levels of expression are seen (Figure 4D) and so accurate fold changes are observed (Figure 4E).
Figure 4.
Use of negative controls with shift in expression. Two genes are investigated for differential expression between condition A and condition B. A negative control is used for normalization (could be a known non-DE gene or spike-in control). (A) Both non-control genes are up-regulated under condition B versus condition A, having twice the expression under condition B. As a negative control, the control has the same expression under both conditions. (B) In the RNA-Seq experiment, the same number of molecules is sequenced from each sample. As the control has a smaller share of the mRNA in condition B, there are fewer control molecules in the sample for condition B. (C) Variability leads to differences in the total read count for the two samples. The share of the reads aligned to the control is the share of mRNA from the control. (D) The control should have the same expression in both conditions, so normalization is performed to equalize the normalized read count for the control, resulting in normalized read counts that reflect the correct mRNA/cell levels. (E) Because normalized counts correctly reflect mRNA/cell, the observed fold change agrees with the truth.
Assumptions
Existence of controls: The controls needed for the experiment do in fact exist, and their expression behaves as expected (e.g. for negative controls they are non-DE under the conditions of the experiment).
Controls behave like non-control genes: The technical effects for the controls in some way reflect the technical effects for all the genes, so that the controls can be used for normalization.
Methods
Housekeeping genes. Housekeeping genes (HG) are genes that play a role in the basic functions of a cell [31], and so are believed to be non-DE under the biological conditions of interest [10, 31]. HG normalization assumes that these genes are truly not DE, and furthermore that they are affected by technical effects the same way as DE genes. These HG must be identified a priori, and the appropriate choice of HG likely changes across different conditions and cell/tissue types. Normalization using HG can either equalize the read count of the gene (if one housekeeping gene) [10] or perform a conventional normalization procedure on a set of HG [9]. It is generally recommended to use a set of HG, as the use of one housekeeping gene is not robust.
Conventional normalization with spike-ins. A set of synthetic spike-in controls is available through the External RNA Controls Consortium (ERCC) [32], and these can be used instead of HG. The use of spike-ins with conventional methods assumes that the spike-ins are not affected by the biological conditions under investigation, and that they have the same technical effects as the real genes [33]. Conventional normalization methods, such as Upper Quartile, may be applied to the spike-ins [33], as with HG controls. The conventional normalization methods are applied only to the spike-ins, and then used to calculate normalization factors for all genes. One approach is proposed by Lovén et al. [22], which uses cyclic loess normalization on the spike-ins (CLS). Spike-ins are added to RNA in proportion to the number of cells from which RNA is extracted. Then, cyclic loess normalization is performed on the RPKM values (more details can be found in the Supplementary Information). The loess curve is fit using only the spike-ins, but used to adjust all RPKM values so that the other genes are normalized with the spike-in information, which is not affected by differential expression.
Factor analysis of controls. To address perceived problems with the use of spike-ins, Remove Unwanted Variation (RUV) [33] uses factor analysis to remove factors of unwanted variation in RNA-Seq data. Using a set of negative control genes or samples, singular value decomposition is used to estimate a matrix for the factors of unwanted variation. Normalization to remove the factors of unwanted variation is then performed. It is divided into three sub-methods: RUVg, RUVs and RUVr. The two assumptions listed above indicate slightly different things for the different sub-methods, and RUVr does not require controls (it is an adaptation of the RUV method to be used when controls are not available) [33]. Here we list the meaning of the assumptions for each of the three sub-methods:
RUVg. Existence of controls: negative controls exist (non-DE across conditions). Controls behave like non-control genes: the factors of unwanted variation for the controls span the same space as the factors for the entire set of genes.
RUVs. Existence of controls: negative controls exist (non-DE across conditions) and there are also negative control samples (expression not related to biological condition). Controls behave like non-control genes: the factors of unwanted variation for the controls span the same space as the factors for the entire set of genes, and the factors of unwanted variation are not correlated with experimental condition.
RUVr. Does not require existence of controls. Assumes that factors of ‘wanted’ variation are known (i.e. the design matrix) and the factors of unwanted variation are not correlated with experimental condition.
Motivation
Controls should be non-DE across conditions, and hence, on average, normalized counts for the controls should be the same across conditions. If technical effects impact controls like they impact genes, then we can apply the adjustment for the controls to all genes. The reasoning for normalization by controls is similar to normalization by distribution/testing, but in the former it is assumed that an explicit set of controls is known, while in the latter we aim to capture the information from non-DE genes without knowing beforehand which genes are non-DE.
Importance of the assumptions
At first glance it makes sense that correcting for differences in sequencing depth can be done simply by library size normalization, which works if the total amount of mRNA in each cell is the same across experimental conditions. Then, a gene that produces the same amount of mRNA under each condition will produce the same proportion of total mRNA in each condition. We thus expect the same proportion of reads to be aligned to that gene under each condition, and Total Count normalization gives us exactly the proportion of reads aligned to each gene. Likewise, differences in expression correspond to differences in proportion of reads in the sample. However, differences in total mRNA/cell can lead to both failing to detect DE genes (Figure 2) and incorrectly calling non-DE genes DE (Figure 1) when normalization by library size is performed in situations where total mRNA/cell is not constant.
On the other hand, normalization by distribution and by testing are impacted by differences in the number of up-regulated versus down-regulated genes, but not by the relative amounts of mRNA/cell. The greater the disparity between the number of up-regulated genes and the number of down-regulated genes under a given condition, the higher the asymmetry of the differential expression under that condition. Both Figures 1 and 2 show differences in total expression (mRNA/cell) between the two conditions, but there is much more asymmetry in Figure 2 (that is, 100% of the genes are up-regulated). Accordingly, normalization by distribution and by testing can handle differences in mRNA/cell in the case of a few highly expressed genes (small asymmetry), but not a global shift in expression (large asymmetry). If there are only a few DE genes, these DE genes will not do much to change the estimated normalization factor. For example, the Upper Quartile normalization strategy compares the 75th percentile of read counts between samples. If the 75th percentile of all the read counts is similar to the 75th percentile of the non-DE read counts, this is a reasonable approach. The normalization statistic for all genes will be similar to the normalization statistic for non-DE genes if there are only a few DE genes. The two statistics will also be similar when there is a small proportion of asymmetry. When differential expression is mostly symmetric, the values for DE genes should more or less balance out on either side of the statistic for non-DE genes, so that the statistic for all genes is close to the statistic for non-DE genes. A small proportion of asymmetry can allow distribution/testing methods to tolerate higher proportions of differential expression.
Knowledge of the assumptions made by each normalization method allows for good predictions of which biological experiments are suitable for each method. Normalization by library size should work well when total mRNA/cell is equivalent across conditions, regardless of the amount of asymmetry (Figure 3A and C). On the other hand, normalization by distribution/testing should generally work well when there is symmetry, regardless of differences in mRNA/cell (Figure 3C and D). When there is both asymmetry and different levels of total mRNA/cell (Figure 3B), we expect both sets of methods to perform poorly.
Simulations
To demonstrate the effects of asymmetry and different mRNA/cell in a controlled scenario, we examined the performance of several normalization methods on simulated data (Figures 5–8). Simulations were performed with two combinations of number of genes and number of samples: 10 000 genes and 4 samples (two replicates per condition), and 1000 genes and 10 samples (five replicates per condition). Figures 5 and 7 show the error in fold change estimates for the different normalization methods, while Figures 6 and 8 show empirical error rates in detecting differential expression. The use of simulation allows us to isolate the effects of asymmetry and mRNA/cell, and to vary the amount of differential expression to see the effect of each combination of (a)symmetry and same/different mRNA/cell at each level of differential expression. We recognize that real experiments contain additional sources of bias not present in our simulations. Such biases are beyond the scope of this article; as we are focused on the effects of normalization, we control for other sources of error.
Figure 5.
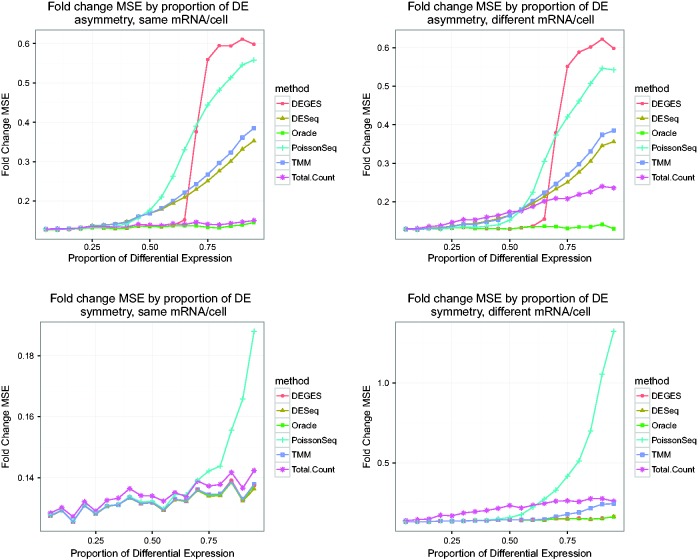
Impact of amount of asymmetry and amount of mRNA/cell on fold change estimates, 10 000 genes and four samples. These plots show the average log fold-change MSE for non-DE genes of several methods. Simulated data are used, with 10 000 genes and two replicates per condition, and varying proportions of differential expression (5–95%). Genes simulated to be non-DE should have an observed log fold-change close to 0; the MSE is thus calculated by averaging the squared observed log fold-changes for each non-DE gene (treating the true log fold-change as 0). Because of variability in the generation of read count data, the observed log fold-change will in general not be exactly 0, so the Oracle normalization method (normalizing the data with the correct normalization factors given the simulation) serves as a baseline. Methods with MSEs that closely follow those of Oracle normalization are doing well. Asymmetric differential expression was simulated as 75% of the set of DE genes up-regulated in one condition and 25% up-regulated in the other. Under symmetric differential expression, 50% of DE genes are up-regulated in each condition. For simulations with the same mRNA/cell, non-DE genes had the same proportion of reads in each condition; simulations with different mRNA/cell resulted in non-DE genes having different shares of the reads in the different conditions.
Figure 7.
Impact of amount of asymmetry and amount of mRNA/cell on fold change estimates, 1000 genes and 10 samples. These plots show the average log fold-change MSE for non-DE genes of several methods. Simulated data are used, with 1000 genes and 5 replicates per condition, and varying proportions of differential expression (5–95%). Genes simulated to be non-DE should have an observed log fold-change close to 0; the MSE is thus calculated by averaging the squared observed log fold-changes for each non-DE gene (treating the true log fold-change as 0). Because of variability in the generation of read count data, the observed log fold-change will in general not be exactly 0, so the Oracle normalization method (normalizing the data with the correct normalization factors given the simulation) serves as a baseline. Methods with MSEs that closely follow those of Oracle normalization are doing well. Asymmetric differential expression was simulated as 75% of the set of DE genes up-regulated in one condition and 25% up-regulated in the other. Under symmetric differential expression, 50% of DE genes are up-regulated in each condition. For simulations with the same mRNA/cell, non-DE genes had the same proportion of reads in each condition; simulations with different mRNA/cell resulted in non-DE genes having different shares of the reads in the different conditions.
Figure 8.
Impact of amount of asymmetry and amount of mRNA/cell on error control, 1000 genes and 10 samples. These plots show the average empirical FDR of several methods on simulated data with varying proportions of differential expression (5–95%). The simulations are performed with two conditions, with 1000 genes and five replicates per condition. Asymmetric differential expression was simulated as 75% of the set of DE genes up-regulated in one condition and 25% up-regulated in the other. Under symmetric differential expression, 50% of DE genes are up-regulated in each condition. For simulations with the same mRNA/cell, non-DE genes had the same proportion of reads in each condition; simulations with different mRNA/cell resulted in non-DE genes having different shares of the reads in the different conditions. The black dashed line is at 0.05, the nominal FDR using the Benjamini–Hochberg adjustment. Deviations of the oracle value from the nominal value (starting above 0.05 and falling below as the proportion of DE increases) are a result of DESeq2 hypothesis testing and the conservativeness of Benjamini–Hochberg.
For the simulations, we chose methods that were representative and generally perform well in the literature, as summarized in Table 2 (except for Total Count normalization, as all normalization by library size methods perform poorly in the literature). We used Total Count, DESeq, TMM, PoissonSeq, DEGES and finally Oracle normalization that uses the true normalization factor known from the simulation parameters. To measure how well the methods performed normalization, we used a method similar to Maza et al. [14] and calculated the empirical mean squared error (MSE) of the log fold change (LFC) for non-DE genes (Figures 5 and 7), comparing each observed LFC to 0. As these genes are not DE, if normalization is performed correctly, then the LFC between samples of different conditions should be close to 0. Oracle normalization provides the baseline for the MSE under perfect normalization; methods that track closely with the Oracle are performing well.
Table 2.
Literature comparing normalization methods
| Paper goal | Evaluation criteria | Approximate ranking |
|---|---|---|
| Global compare | Equiv. normalized count distribution between replicates (real data); variance of normalized counts within condition (real data); equiv. expression of HG (real data); agreement on DE calls (real data); false positives and power (simulation) [9]. | DESeq & TMM |
| UQ & Med | ||
| Q | ||
| RPKM & TC | ||
| Introduces UQ | DE detection compared with qRT-PCR (ROC curves) (real data); variability between replicates after normalization (real data); bias in fold-change estimation compared with qRT-PCR (real data) [10]. | UQ |
| Q | ||
| TC | ||
| Introduces MRN | False positives, false negatives and power (simulation); MSE of expression fold-change estimates (simulation); number of DE calls and agreement on DE calls (real data) [14]. | MRN |
| DESeq & TMM | ||
| TC | ||
| UQ & Med | ||
| FPKM | ||
| Global compare | Equiv. normalized count distribution between replicates (real data); variance of normalized counts within condition (real data); agreement on DE calls (real data); variability of results under different filtering techniques (real data) [13]. | DESeq |
| TMM | ||
| UQ, Med, & Q | ||
| RPKM & TC | ||
| (RUVg considered, but assumptions not met) | ||
| Global compare | Correlation between normalized counts and qRT-PCR data (real and simulated data) [12]. | All were equivalent (DESeq, Med, Q, RPKM and ERPKM, TMM, UQ) |
| Global compare | Bias and variance in fold change estimation (compared with HG) (real data); sensitivity and specificity in DE calls (using genes believed to be DE and non-DE) (real data); prediction of DE genes (real data); agreement on DE calls (real data) [16]. | DESeq |
| PS | ||
| Q | ||
| UQ | ||
| TMM | ||
| Global compare | Clustering of normalized counts agrees with condition (real data); correlation between fold change estimates and qRT-PCR fold changes (real data) [15]. | All were equivalent (DESeq, PS, UQ, TMM, Q, CuffDiff) |
| Introduces DEGES | ROC curves and AUC (real and simulated data) [11]. | DEGES strategy using a normalization method generally performed better than that method by itself |
| Introduces CLS | Observed fold change for normalized data (real data) [22]. | CLS |
| RPKM | ||
| Introduces RUV | PCA (real data); variance and distribution of normalized data (real data); distribution of P-values (real data); clustering and proportion of reads mapping to spike-ins (real data); MA plots (real data); ROC curves (real data); comparison with qRT-PCR (real data) [33]. | RUV (UQ, CLS, RPKM, TMM, DESeq and Q) |
Several papers that include comparisons of DE assumption normalization methods are summarized here. Short descriptions of the criteria used to evaluate the normalization methods are provided, and the final results of the paper are condensed into an approximate ranking of the methods considered (best performing methods at the top). These rankings are not explicit in all papers and for some have been inferred from the paper’s discussion of the strengths and weaknesses of the different methods. Abbreviations: UQ, Upper Quartile; Med, Median; Q, Quantile; TC, Total Count; MRN, Median Ratio; PS, PoissonSeq; CLS, Cyclic Loess on Spike-ins.
The results are the same regardless of the number of genes or samples, demonstrating that the results do not depend on the number of genes or samples. Figures 5 and 7 show the MSE results of the simulations, confirming that the methods perform as expected. Total Count normalization follows the Oracle closely when there is the same total mRNA/cell, but diverges quickly when there is different mRNA/cell. DESeq, TMM and DEGES perform well when there is symmetry, for all proportions of differential expression. PoissonSeq does well under symmetry until too high a proportion of differential expression is reached, at which point it diverges. This is likely because PoissonSeq normalization uses a set of genes of a fixed size for normalization; when the proportion of differential expression is too high, the set necessarily contains DE genes that skew the normalization estimate. When there is asymmetry, the normalization by distribution/testing methods can tolerate a small proportion of differential expression but eventually reach a break-down point.
The effects on downstream analysis of applying the different normalization methods are shown in Figures 6 and 8, which show empirical false discovery rate (eFDR) measures for each method after testing for differential expression (note: the downward trend in the Oracle eFDR is because of the use of the Benjamini–Hochberg (BH) procedure to control FDR, which is conservative and controls at a level directly related to the proportion of true null hypotheses, i.e. non-DE genes). When methods normalize correctly, as shown in Figures 5 and 7, the subsequent tests for differential expression are able to control the false discovery rate in the absence of additional sources of error. However, when normalization fails and the observed fold changes depart sufficiently from the truth, the result is inflated false positives. Our work illustrates how heavily analysis relies on correct normalization, which in turn relies on assumptions. When the assumptions are violated, normalization fails (Figures 5 and 7) and as a result so does the downstream analysis (Figures 6 and 8). As the figures demonstrate, the optimal normalization methods heavily depend on the biological circumstances, and so we can give no clear guideline for which normalization method to use without knowing the conditions at hand. Additionally, we emphasize that the simulations are designed to isolate the effects of incorrect normalization; analysis of real RNA-Seq data will likely include additional biases that, if not accounted for, can lead to spurious results even if normalization is correct. The eFDR numbers given in the simulations should not be treated as predictions of what the true FDR will be in an experimental setting, but rather provide a way to compare different methods with all other factors being equal. As the methods generally perform similarly for a small proportion of differential expression, under these conditions the choice of method is less important. However, as studies have demonstrated the existence of global shifts in expression [17–20], we believe that the assumption of a small proportion of differential expression can be dangerous. Hence, it is important to consider the performance of the different methods for a wide range of differential expression.
Simulation details
To assess the downstream results of violating the assumptions of different normalization methods, simulations were run in which the average MSE, on non-DE LFCs, and average eFDR were computed for different proportions of differential expression (proportion of genes which are truly DE), amounts of asymmetry and relative amounts of mRNA/cell. The code for the simulations and the plots of the results can be found at https://github.com/ciaranlevans/rnaSeqAssumptions, and was adapted from the R code used in the simulations of Law et al. [34].
For each of the two combinations of number of genes and number of samples, four sets of simulations were performed, one for each combination of asymmetry versus symmetry and same mRNA/cell versus different mRNA/cell. In each simulation, read count data were generated, then normalized according to one of six different methods: DEGES, DESeq, Oracle (normalization with the true scaling factors, used for benchmarking other normalization methods), PoissonSeq, TMM and Total Count. The normalization methods were selected to represent different types of normalization: by library size (Total Count), by distribution (DESeq and TMM) and by testing (PoissonSeq and DEGES). DESeq and TMM were chosen to represent normalization by distribution methods, as they are widely studied and generally perform well relative to other methods (Table 2). Simulated RNA-Seq data were generated, then each normalization method was performed. After normalization, two normalized columns of the read count matrix (one from each condition) were compared to produce LFCs for the non-DE genes. These observed LFCs should be close to 0, so the MSE was calculated by averaging the squared LFCs for the non-DE genes. Differential expression hypothesis testing was performed on the data for each normalization method. Testing was done separately from normalization, and was performed with the DESeq2 [35] package after normalization with each method (the data were not re-normalized with DESeq2). As in DESeq2, and as is common in differential expression studies, P-values were adjusted using the Benjamini–Hochberg procedure for FDR control [36]. Using the adjusted P-values, and knowledge of which genes were simulated to be DE, the average eFDR (observed proportion of false discoveries out of all discoveries) was calculated across 50 repetitions.
Simulations begin by creating initial proportions of expression, representing the proportion of the total expression for each gene and each sample, with 10 000 genes and 4 samples (two samples per condition), or 1000 genes and 10 samples (five samples per condition). A random subset of genes is chosen to be DE, with the number determined by the specified proportion of differential expression.
Asymmetry, same mRNA/cell: Differential expression is asymmetric (more genes up-regulated under one condition than the other), but the absolute expression is the same for each condition. In all, 75% of DE genes were 2-fold up-regulated under condition A, and 25% were 4-fold up-regulated under condition B.
Asymmetry, different mRNA/cell: Differential expression is asymmetric, and the absolute expression is different under the different conditions. In all, 75% of DE genes were 2-fold up-regulated under condition A, and 25% were 2-fold up-regulated under condition B.
Symmetry, same mRNA/cell: Differential expression is symmetric (same number up-regulated under each condition), and the absolute expression is the same for each condition. In all, 50% of DE genes were 2-fold up-regulated under condition A, and 50% were 2-fold up-regulated under condition B.
Symmetry, different mRNA/cell: Differential expression is symmetric, but the absolute expression is different under the different conditions. In all, 50% of genes are 4-fold up-regulated under condition A, and 50% are 6-fold up-regulated under condition B.
Experimental data
In addition to a simulation study, we examined the performance of normalization methods on RNA-Seq data from the SEQC project [37], in which the SEQC/MAQC-III consortium studied RNA-Seq technology across different platforms and alignment methods. We used sequencing data from the Australian Genome Research Facility, performed on the Illumina HiSeq 2000 platform and mapped with the AceView annotations. The SEQC project collected data for four different samples A, B, C and D, with many replicates per sample [37]. Following [33] and [10], for tests of differential expression we performed comparisons between samples A and B (from Agilent’s UHRR cells and Life Technologies’ HBRR cells, respectively). Data were obtained from the seqc R package that is available through Bioconductor [37]. Our code for analysis of the SEQC data can be found at https://github.com/ciaranlevans/rnaSeqAssumptions.
Additionally, the SEQC data include TaqMan qRT-PCR measurements on about 1000 genes [37]. PCR data are often used to determine ‘true’ differential expression and assess false positives and false negatives in an RNA-Seq analysis; for example, [10] and [33] both use SEQC PCR data to evaluate performance of differential expression testing. For the purposes of this section we will use PCR data as a benchmark for assessing differential expression calls. However, we note that the practice of treating PCR as a ‘gold standard’ may not always be justified: there has been concern over possible errors in PCR data [38], and PCR data may not detect global shifts in expression in the absence of reliable controls [40].
The full SEQC qRT-PCR data contain 1044 genes. We matched the PCR data with SEQC RNA-Seq data, selecting genes that were represented in both data sets with enough information, and removed duplicated genes. This results in 733 unique genes with both RNA-Seq and PCR measurements. Following the examples of [33] and [10], we divide the PCR-validated genes into groups of ‘non-DE’, ‘no-call’ and ‘DE’ based on their absolute average LFC (respective ranges are < 0.2, and > 1).
After selecting genes to use in our analysis, we compared the PCR expression measures between samples A and B by computing LFCs of the average expression. The distribution of the mean LFCs is symmetric about 0 (Figure 9), with 401 genes expressed more in sample A (LFC > 0) and 332 expressed more in sample B (LFC < 0). The PCR data identified 268 DE genes with higher expression in sample A (LFC > 1), and 203 DE genes with higher expression in sample B (LFC ). Approximately the same number of up-regulated genes are observed in each condition, indicating that differential expression is symmetric. Additionally, the distribution has a similar shape on each side of 0 (Figure 9). There is no reason to suspect that there are systematic differences between the amounts of mRNA/cell produced by genes with higher expression in sample A versus higher expression in sample B, and so having the same distributional shape suggests that each sample produces approximately the same mRNA/cell.
Figure 9.

Distribution of qRT-PCR mean LFC. The histogram shows the distribution of the LFC comparing the average PCR measures of expression between SEQC samples A and B in each gene. The distribution is symmetric around 0, indicating that each sample has the same number of up- and down-regulated genes. Additionally, the shape of the distribution is similar on both sides of 0, suggesting that there are similar amounts of mRNA/cell for each sample.
Symmetric expression and the same mRNA/cell indicate that all normalization methods should perform approximately equivalently, as illustrated by the simulations. Using each normalization method from the simulations (DEGES, DESeq, PoissonSeq, TMM and Total Count), we performed normalization and differential expression testing, using the DESeq2 package for the hypothesis testing. To compare the different normalization methods, we compared the results of differential expression testing with the calls from the PCR analysis.
By varying the significance cutoff for the DESeq2 P-values, we can change which genes are called DE from the RNA-Seq analysis. That is, if the level of significance is set at 0.05, there will be fewer false positives (and more false negatives) than if the level of significance is set at 0.1. We then compared the RNA-Seq calls with the ‘true’ PCR calls to create Receiver Operating Characteristic (ROC) curves for each method (Figure 10), with the no-call genes ignored when making the ROC curves. A steep initial slope of the ROC curve indicates a large gain in sensitivity (ability to correctly determine DE genes, i.e. true positives) for a small loss in specificity (ability to correctly determine non-DE genes, i.e. true negatives). Methods perform better when, for a given level of specificity, they have higher sensitivity. Graphically, this corresponds to the top left in the ROC graph. In the case analyzed here we expect each method to perform similarly; DESeq, Total Count, TMM and DEGES do so, but PoissonSeq performs somewhat worse than the others, with an ROC curve noticeably below the rest.
Figure 10.
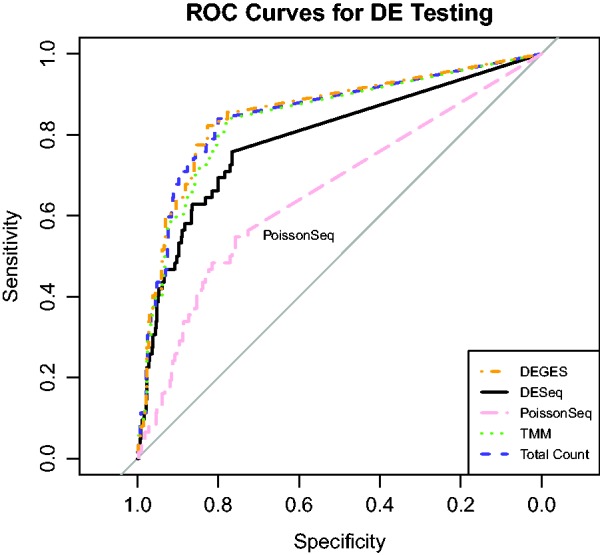
ROC curves for each normalization method using SEQC data. This figure displays the ROC performance of each method using RNA-Seq data for 733 PCR-validated genes. False positives and false negatives are determined by the PCR validation, and no-call genes are ignored in the construction of the ROC curves.
For further comparison with our simulations, we calculated eFDRs for each method. In our simulations, we had two or five replicates for each condition, but in the SEQC data there are 64 replicates for sample A and sample B. To make eFDR calculations comparable with our simulations, we randomly selected two replicates from sample A and two from sample B, then tested for differential expression (using the BH procedure at level 0.05) and calculated an eFDR using the PCR data as a benchmark. Selecting two replicates and testing for DE was repeated 100 times to get an average eFDR for each normalization method. We then performed the same procedure with random selections of five replicates from sample A and five from sample B. The results are displayed in Table 1. Note that the eFDR calculations treat no-call genes as DE, so the exact values in Table 1 are likely not a true representation of the FDR. Rather, we are interested in the relationship between the values for each method. We note that, consistent with our simulations, all methods are approximately equal with a slightly higher empirical FDR for PoissonSeq, and furthermore that empirical FDR increases when more samples are used in the differential expression testing.
Table 1.
Empirical FDR for SEQC RNA-seq data with two and five replicates per condition
| DESeq | Total Count | TMM | DEGES | PoissonSeq | |
|---|---|---|---|---|---|
| 2 replicates | 0.0552 (0.0042) | 0.0489 (0.0048) | 0.0521 (0.0043) | 0.0532 (0.0052) | 0.0730 (0.0049) |
| 5 replicates | 0.0714 (0.0024) | 0.0687 (0.0028) | 0.0700 (0.0030) | 0.0699 (0.0027) | 0.0822 (0.0024) |
Two replicates from sample A and two from sample B were randomly chosen and used to test for differential expression. The empirical FDR was then calculated, and the process repeated 100 times. The procedure was also performed using five replicates from sample A and five from sample B. The average eFDR results are displayed in this table, with the standard deviation of the eFDRs across 100 repetitions given in parentheses. Note: The empirical FDR is calculated as the ratio of the number of non-DE genes (as determined by PCR), which are called DE by RNA-Seq testing to the total number of genes called DE by RNA-Seq testing, effectively treating ‘no call’ PCR genes as DE.
Initial ROC analysis was performed using a full set of 733 genes, for which differential expression is approximately symmetric. To evaluate the different normalization methods under asymmetric differential expression, we took a subset of 619 PCR-validated genes such that 75% of DE genes were up-regulated (according to PCR) in sample A, and the remaining 25% were up-regulated in sample B (the DE genes made up about 57% of the 619 genes in the subset). As illustrated by the simulations, we expect DEGES, DESeq, PoissonSeq and TMM to perform worse if the proportion of differential expression is high enough. The performance of Total Count normalization depends on the relative levels of mRNA/cell, which we are unable to definitively measure with the RNA-Seq or PCR data. However, under our previous assumption that production of mRNA/cell is unrelated to whether a gene is up-regulated or down-regulated in sample A, we would expect some difference in mRNA/cell when there is asymmetric differential expression.
Using the subset of genes with asymmetric differential expression, we again performed an ROC curve analysis (Figure 11). As expected, DEGES, DESeq, PoissonSeq and TMM each perform worse with asymmetric differential expression than with symmetric differential expression (each has a lower ROC curve in Figure 11 than in Figure 10). Total Count normalization also performs worse with the asymmetric differential expression, but does noticeably better than the other methods.
Figure 11.
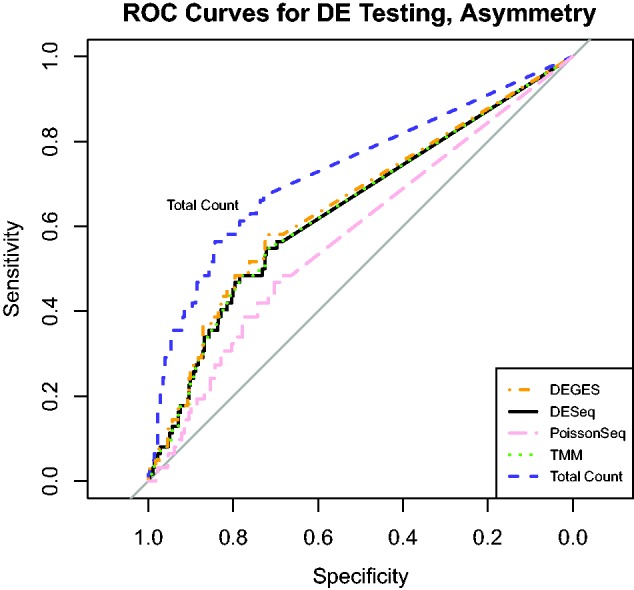
ROC curves for each normalization method using SEQC data. This figure displays the ROC performance of each method using RNA-Seq data for 619 PCR-validated genes. False positives and false negatives are determined by the PCR validation, and no-call genes are ignored in the construction of the ROC curves. The genes are a subset chosen for asymmetric differential expression, so that 75% of DE genes are up-regulated in sample A and 25% are up-regulated in sample B.
Our simulations help illustrate that with symmetric differential expression and similar mRNA/cell, performance of each normalization method should be approximately equivalent. This is indeed what we observe with the SEQC PCR-validated data, which appear to occur under those conditions. Based on our analysis of the assumptions of each method, backed by our simulation data, we expect differences in performance under different conditions. For example, many methods should perform worse under asymmetric differential expression, which was observed in the SEQC data by taking a subset of genes to force asymmetric expression. We are not aware of any large PCR-validated data set that has strongly asymmetric expression and/or a global shift in expression in the full data set, without taking subsets (though as discussed above, it is not clear that PCR data could detect a global shift).
Evaluation of methods and assumptions
Several papers have investigated the different normalization methods described in the previous section. Table 2 summarizes these comparisons by giving approximate rankings of the methods evaluated in each comparison. Here we expand on these rankings to remark on several key ideas.
Differences in mRNA/cell result in poor performance of library size normalization
As shown in Table 2, in many comparisons, Total Count and RPKM/FPKM perform worse than all other methods, and several authors expressly recommend against its use [9]. A likely cause of this is that in these evaluations, the assumption required for library size normalization (same amount of mRNA/cell) is violated. For example, Dillies et al. [9] observed that a few highly expressed genes had a large share of the read counts in Mus musculus data they compared. Bullard et al. [10] and Lin et al. [13] reported similar findings. Bullard saw 50% of the reads concentrated in 5% of the genes, and Lin found 50% of the reads in 45 genes for male flies and 186 genes for female flies. With such a large proportion of the reads aligned to a small fraction of the genes, if these genes are DE, it is likely that there will be different amounts of mRNA/cell across the conditions, and Bullard et al. [10] did observe that the highly expressed genes were DE.
DESeq and TMM generally perform well, but validity is not certain
Dillies et al. [9], for example, note that DESeq and TMM are the only methods that perform well both with the ability to detect DE genes and with controlling false positives. This supports the conclusion of Bullard et al. [10], who concluded that normalization has the biggest impact on detection of DE genes.
Given that several authors have found that a few highly expressed genes have a large share of total expression [9, 10, 13] and these genes may be DE, it is clear that assuming the same amount of mRNA/cell is not always reasonable. The good performance of DESeq and TMM in these studies suggests that perhaps their assumptions (DE and non-DE genes behave the same, balanced expression) are fairly reasonable, or at least not too violated, for the data analyzed in the comparisons. However, it is possible that for the real data analyzed in these comparisons there is a global shift in expression that is not picked up by these normalization methods. For example, a global shift has been observed in DE analysis with low and high c-Myc conditions [19, 20], and this shift was undetected without the use of spike-in controls [22]. Other researchers [17, 18] have found similar global up-regulation when using spike-ins, and it has been suggested that such shifts were not detected by previous research owing to lack of proper normalization [18]. Even qRT-PCR, often treated as a ‘gold standard’ for evaluating the performance of DE analysis methods, might not be able to detect a global shift without controls. Normalization for qRT-PCR often relies on HG [10, 39, 40]. In the absence of non-DE genes, as occurs with a global shift in expression, qRT-PCR results might not be accurate. Furthermore, the use of PCR as a gold standard for evaluation of normalization methods has been called into question, as despite being highly accurate, PCR can contain errors [38]. Hence, for methods which normalize by distribution or by testing, it is difficult or impossible to know whether their assumptions have been met without additional information.
Potential lack of HG
The possible absence of HG poses a problem for HG normalization of RNA-Seq data as well as PCR data. While Bullard et al. [10] found that HG normalization performed equivalently to UQ, the housekeeping gene they used (POLR2A) was selected based on previous studies and they caution that such information may not always be available. Dillies et al. [9] also selected HG from previous research, and state that one cannot be certain HG will always be non-DE. As mentioned above, several authors have found global shifts in expression, which would leave few, if any, non-DE HG for use in normalization [17–20].
External controls may be needed
In the case of a global shift in expression, the assumptions are violated for normalization methods that do not rely on external controls. Global up-regulation necessarily leads to different amounts of mRNA/cell (library size normalization), highly asymmetric expression (distribution/testing normalization) and an absence of non-DE genes (HG normalization). Without the use of external controls, it is possible that many experiments have resulted in incorrect conclusions [21]. Normalization with spike-in controls attempts to rectify the problems of asymmetry, by relying on genes/spike-ins that should have the same expression under the different conditions.
Mixed performance of spike-ins
As we have seen, these methods come with their own set of assumptions, and it is not clear that these assumptions can always be trusted. In an assessment of ERCC spike-in controls, Jiang et al. found that only small fractions (0.5% and 0.01%) of spike-in reads were incorrectly aligned to the actual genome of the organisms in their experiment (Drosophila and humans) [32]. This indicates that as desired, there will be little error introduced into the read counts by the controls. Furthermore, Jiang et al. found a linear relationship between the amount of spike-in and read count [32], which is evidence that the spike-in read counts are representative of expression level. However, Risso et al. [33] found violations of both assumptions necessary for basic spike-in normalization (the assumptions that spike-ins are non-DE across conditions and have the same technical effects as genes), and Qing et al. [41] found that read counts for the spike-ins depended in part on the mRNA enrichment protocol used in the experiment.
Recommendations: appropriate method depends on DE definition and assumptions
Different circumstances call for different normalization methods. Correct normalization should cause non-DE genes to have the same (expected) normalized read count across conditions. This requires a definition of differential expression. In this article, we defined differential expression in terms of differences in mRNA/cell across conditions, and it appears that this is the definition used in previous research evaluating normalization methods. Consequently, the majority of the commentary and recommendations presented here is in the context of mRNA/cell differential expression. However, other definitions of differential expression are possible and may be appropriate/necessary in certain conditions [23]. One alternative is to define a gene as DE if its share of mRNA in the transcriptome is different across conditions; this bases differential expression on relative, rather than absolute, measures of expression. The mRNA/transcriptome definition may be appropriate in some circumstances: Ignatov et al. [42] performed an experiment which found down-regulation of every gene when using the mRNA/cell definition, so they chose instead to look for differences in per transcriptome expression.
Choosing a normalization method depends on the definition of differential expression. For example, library size normalization generally performs poorly when defining DE in terms of mRNA/cell, but should produce exactly the desired measure when defining DE in terms of mRNA/transcriptome. Hence, choosing a normalization method for an RNA-Seq experiment must begin with choosing a definition of differential expression. While one definition may be less often used than the other, it is necessary to make a choice between the two definitions, and the choice is particularly important if there is a possibility of a global shift in expression.
Once differential expression is defined, the next step is to determine which assumptions are appropriate for the experiment at hand, and then choose a method that follows those assumptions. Assumptions of each method depend on the definition of differential expression; in this article, we consider the assumptions necessary for each method under mRNA/cell differential expression. However, these assumptions will not be the same for mRNA/transcriptome differential expression. For example, the assumption for library size normalization discussed above is that the total mRNA/cell is the same under each condition. This assumption is necessary for the relative measures of expression obtained via library size normalization to be valid measures of absolute expression. If a relative definition of DE is used instead, such as mRNA/transcriptome, then it is not necessary to assume equivalent total mRNA/cell across conditions.
If spike-ins can be trusted, they are important to use in normalization because there may be previously unknown shifts in expression that cannot be detected without controls, and HG do not seem a reliable choice for controls. RUV aims to address the shortcomings of spike-ins, so may be a good method to use when spike-ins are available.
However, there are situations in which spike-in methods are not an option. Coate and Doyle [23] note that application of spike-in methods requires the ability to count the number of cells used in RNA extraction, and cell counting is not possible in some tissue types. In these cases, normalization by distribution/testing appears to be the best option, and DESeq especially has generally been shown to perform well.
Conclusion
The use of RNA-Seq experiments to study organisms’ genomes is becoming ubiquitous, and the explosion in the use of sequencing technology has led to a related explosion in the development of statistical methods for processing and analyzing RNA-Seq data. As previous research has demonstrated [10], proper normalization is an essential step in the analysis pipeline. We have seen that incorrect normalization can result in downstream errors such as inflated false positives. The need for normalization arises from the inherent variability in the collection of RNA-Seq data, and a variety of normalization methods have been devised to combat this variability. As we have seen, the literature has not reached a consensus on which normalization method to use.
Both the simulations and the real data allow us to understand the effects of symmetric versus asymmetric differential expression and the effects of differing amounts of mRNA/cell. The simulations isolated all other conditions and allowed for a direct comparison between methods. The real data told the same story as the simulated data with respect to the (a)symmetry of the differential expression, validating the more complete simulation results. In particular, it is worth noting that the performance of Total Count normalization depends on the amount of mRNA/cell and not differential expression symmetry. Indeed, Total Count normalization outperforms the other normalization methods when the data are asymmetric with same mRNA/cell, though we do not know how often such conditions occur in real, full data.
Each normalization procedure relies on assumptions, and when violated, the procedures lead to incorrect results. For each assumption, there is evidence that it may not hold in some experiments. Part of an analysis of RNA-Seq data requires choosing a normalization procedure, and keeping the assumptions of each method in mind can help to make the appropriate choice for the experiment at hand. However, there may be many situations in which the validity of any assumption is unknown for the given experiment. In such cases, normalization with external controls would be the appropriate choice if the external controls can be trusted. Unfortunately, several authors have found problems with spike-ins and so propose additional methods to handle these issues. It is clear that spike-ins are necessary in some circumstances, and we hope that as research progresses their performance will improve.
To the best of our knowledge, there does not exist an extensive analysis of published data, which evaluates the assumptions of normalization methods. Given the potential violations to each normalization assumption, knowledge of the extent to which each assumption holds in a given experiment would be instrumental in helping to choose a normalization method for RNA-Seq analysis. There is no clear way to perform such an evaluation, however, considering that violations of assumptions (such as a global shift) may go undetected without additional information, and the requisite information may not be present in the original experiment.
Key Points
Assumptions allow normalization to translate raw read counts into meaningful measures of expression.
The correct normalization method to use depends on which assumptions are valid for the biological experiment.
Incorrect normalization leads to problems in downstream analysis, such as inflated false positives, that mean results cannot be trusted.
No normalization method is perfect, and for every method there exists cases for which the assumptions are violated. There are examples of global shifts in expression that violate assumptions of conventional normalization methods, requiring controls.
An understanding of assumptions can help pick the most suitable normalization method for a given experiment.
Supplementary Material
Funding
This research was supported in part by grants to Pomona College [52007555] and Harvey Mudd College [52007544] from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program.
Ciaran Evans is a PhD student in statistics at Carnegie Mellon University. He is interested in applications of statistics to high-throughput genetic data.
Johanna Hardin is a Professor of Mathematics at Pomona College. She works on different types of analyses of high-throughput genetic data that do not conform to the usual assumptions needed for statistical analyses.
Daniel M. Stoebel is an Associate Professor of Biology at Harvey Mudd College. He works on the molecular biology and evolution of bacterial regulatory networks, particularly the Escherichia coli global transcriptional regulator RpoS.
References
- 1. Shendure J. The beginning of the end for microarrays? Nat Methods 2008;5(7):585–7. [DOI] [PubMed] [Google Scholar]
- 2. Oshlack A, Robinson MD, Young MD.. From RNA-seq reads to differential expression results. Genome Biol 2010;11(12):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Z, Gerstein M, Snyder M.. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10(1):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Auer PL, Srivastava S, Doerge R.. Differential expression - the next generation and beyond. Brief Funct Genomics 2012;11(1):57–62. [DOI] [PubMed] [Google Scholar]
- 5. Oshlack A, Wakefield MJ.. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 2009;4(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Risso D, Schwartz K, Sherlock G, et al.GC-content normalization for RNA-Seq data. BMC Bioinformatics 2011;12(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robinson MD, Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McIntyre L, Lopiano K, Morse A, et al.RNA-seq: technical variability and sampling. BMC Genomics 2011;12(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dillies MA, Rau A, Aubert J, et al.A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform 2013;14(6):671–83. [DOI] [PubMed] [Google Scholar]
- 10. Bullard JH, Purdom E, Hansen KD, et al.Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 2010;11(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kadota K, Nishiyama T, Shimizu K.. A normalization strategy for comparing tag count data. Algorithms Mol Biol 2012;7(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li P, Piao Y, Shon H, et al.Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-Seq data. BMC Bioinformatics 2015;16(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin Y, Golovnina K, Chen Z, et al.Comparison of normalization and differential expression analyses using RNA-Seq data from 726 individual Drosophila melanogaster. BMC Genomics 2016;17(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maza E, Frasse P, Senin P, et al.Comparison of normalization methods for differential gene expression analysis in RNA-Seq experiments: a matter of relative size of studied transcriptomes. Commun Integr Biol 2013;6(6):e25849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rapaport F, Khanin R, Liang Y, et al.Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol 2013;14(9):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zyprych-Walczak J, Szabelska A, Handschuh L, et al.The impact of normalization methods on RNA-seq data analysis. BioMed Res Int 2015;2015:621690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Athanasiadou N, Neymotin B, Brandt N, et al. Growth rate-dependent global amplification of gene expression. bioRxiv2016. doi: 10.1101/044735
- 18. Hu Z, Chen K, Xia Z, et al.Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 2014;28(4):396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin C, Lovén J, Rahl P, et al.Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012;151(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nie Z, Hu G, Cui K, et al.c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012;151(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen K, Hu Z, Xia Z, et al.The overlooked fact: fundamental need for spike-in controls for virtually all genome-wide analyses. Mol Cell Biol 2015;36(5):662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lovén J, Orlando D, Sigova A, et al.Revisiting global gene expression analysis. Cell 2012;151(3):476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coate JE, Doyle JJ.. Variation in transcriptome size: are we getting the message? Chromosoma 2015;124(1):27–43. [DOI] [PubMed] [Google Scholar]
- 24. Anders S, Huber W.. Differential expression analysis for sequence count data. Genome Biol 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pachter L. Models for transcript quantification from RNA-Seq. arXiv2011. arXiv:1104.3889v2.
- 26. Mortazavi A, Williams BA, McCue K, et al.Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5(7):621–8. [DOI] [PubMed] [Google Scholar]
- 27. Trapnell C, Williams BA, Pertea G, et al.Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isofrom switching during cell differentiation. Nat Biotechnol 2010;28(5):511–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bolstad B, Irizarry R, Åstrand M, et al.A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19(2):185–93. [DOI] [PubMed] [Google Scholar]
- 29. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Witten D, Johnstone I, et al.Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics 2012;13(3):523–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eisenberg E, Levanon E.. Human housekeeping genes, revisted. Hum Genet 2013;29(10):569–74. [DOI] [PubMed] [Google Scholar]
- 32. Jiang L, Schlesinger F, Davis C, et al.Synthetic spike-in standards for RNA-seq experiments. Genome Res 2011;21(9):1543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Risso D, Ngai J, Speed T, et al.Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014;32(9):896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Law CW, Chen Y, Shi W, et al.voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15(2):R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57(1):289–300. [Google Scholar]
- 37. Su Z, Labaj PP, Li S, et al.A comprehensive assessment of RNA-Seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat Biotechnol 2014;32:903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun Z, Zhu Y.. Systematic comparison of RNA-Seq normalization methods using measurement error models. Bioinformatics 2012;28(20):2584–91. [DOI] [PubMed] [Google Scholar]
- 39. Lee P, Sladek R, Greenwood C, et al.Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res 2002;12(2):292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kanno J, Aisaki K, Igarashi K, et al.“Per cell” normalization method for mRNA measurement by quantitative PCR and microarrays. BMC Genomics 2006;7(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qing T, Yu Y, Du T, et al.mRNA enrichment protocols determine the quantification characteristics of external RNA spike-in controls in RNA-Seq studies. Sci China Life Sci 2013;56(2):134–42. [DOI] [PubMed] [Google Scholar]
- 42. Ignatov DV, Salina EG, Fursov MV, et al.Dormant non-culturable Mycobacterium tuberculosis retains stable low-abundant mRNA. BMC Genomics 2015;16(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trapnell C, Hendrickson G, Sauvageau M, et al.Differential analysis of gene regulation at transcript resolution with RNA-Seq. Nat Biotechnol 2013;31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.