Abstract
miR-214 plays a major role in the self-renewal of skin tissue. However, whether miR-214 regulates the proliferation and differentiation of human hair follicle stem cells (HFSCs) is unknown. Primary HFSCs were isolated from human scalp skin tissue, cultured, and identified using flow cytometry. An miR-214 mimic and inhibitor were constructed for transfection into HFSCs. The MTS and colony formation assays examined cell proliferation. Immunofluorescence detected the localization and expression levels of TCF4, β-catenin, and differentiation markers. Luciferase reporter and TOP/FOP Flash assays investigated whether miR-214 targeted EZH2 and regulated the Wnt/β-catenin signaling pathway. Western blot determined the expression levels of enhancer of zeste homolog 2 (EZH2), Wnt/β-catenin signaling-related proteins, and HFSC differentiation markers in cells subjected to miR-214 transfection. miR-214 expression was remarkably decreased during the proliferation and differentiation of HFSCs into transit-amplifying (TA) cells. Downregulation of miR-214 promotes the proliferation and differentiation of HFSCs. Overexpression of miR-214 led to decreased expression of EZH2, β-catenin, and TCF-4, whereas downregulation of miR-214 resulted in increased expression of EZH2, β-catenin, and TCF-4 as well as TA differentiation markers. Immunofluorescence assay revealed that inhibiting miR-214 triggered the entry of β-catenin and TCF-4 into the nucleus. The luciferase reporter and TOP/FOP Flash assays demonstrated that miR-214 directly targets EZH2 and affects Wnt/β-catenin signaling. The miR-214/EZH2/β-catenin axis could be considered a candidate target in tissue engineering and regenerative medicine for HFSCs.
Keywords: miR-214, Hair follicles stem cells, Transit-amplifying cells, EZH2, Wnt/β-catenin signal
Introduction
Hair follicle stem cells (HFSCs), which reside in the basal region of the follicles, are essential in maintaining homeostasis and self-renewal of skin tissue [1]. Previous studies demonstrated that HFSCs are a potential source of seed cells for skin repair and re-epithelialization [2]. Transit-amplifying (TA) cells are committed progenitors among adult stem cells (ASCs), and their terminally differentiated daughter cells serve as the main intermediate cells of HFSC differentiation into epidermal cells [3, 4]. Hitherto, the signaling pathway and the molecular biological mechanism underlying the regulation of HFS proliferation and differentiation into TA cells has not been elucidated.
Enhancer of zeste homolog 2 (EZH2), a catalytic component of polycomb repressive complex 2 (PRC2), epigenetically regulates chromatin structure and gene expression via tri-methylation on histone H3K27 and the recruitment of DNA methyltransferases for gene silencing [5]. Accumulating evidence has revealed that EZH2 is critical in stem cell maintenance and lineage specification in multiple tissues, including osteogenesis, neurogenesis, hematopoiesis, epidermal differentiation, and hepatogenesis [6]. The PcG proteins form chromatin-remodeling complexes known as polycomb repressor complexes (PRCs). Comprised of EZH2, Eed, and Suz12, PRC2 is recruited to chromatin, where the methyltransferase EZH2 catalyzes H3 trimethylation on lysine 27 (triMeK27-H3). This histone marker then provides a platform to recruit PRC1 (which aids in PcG-mediated repression either by compacting chromatin or by interfering with the transcription machinery). Without EZH2 activity, PRC1 cannot be recruited to chromatin, and PcG-mediated repression is not established [7]. A previous study demonstrated that EZH2 was involved in the regulation of epidermal differentiation. In epidermal progenitor cells, elevated expression of EZH2 restricts proliferation by repressing the cyclin-dependent kinase inhibitors INK4A and INK4B, which in turn, suppresses cell cycle progression. During epidermal differentiation, the diminshed levels of EZH2 activate the expression of INK4A and INK4B, which slows cell proliferation and removes histone H3K27me3. This phenomenon recruits the transcriptional factor AP1 to structural genes that are required for epidermal differentiation [7].
The Wnt/β-catenin signaling pathway plays a major role in regulating epidermal stem cell renewal and lineage selection of HFSCs [8]. Previous studies have shown that β-catenin regulates stem cell differentiation by activating downstream genes. After its accumulation into the nucleus, β-catenin binds to TCF/LEF and stimulates related downstream genes, including c-myc, cyclinD1, and keratin 14/15, all of which play critical roles in promoting HFSC differentiation into TA cells [9, 10].
MicroRNAs (miRNAs) represent a subset of small, noncoding RNAs that can directly bind to the 3′-untranslated region (3′-UTR) of targeting mRNAs to posttranscriptionally regulate gene expression, resulting in either translational repression or mRNA degradation [11]. miRNAs participate in a variety of biological processes, including cell apoptosis, proliferation, differentiation, and survival [12]. In addition to signaling/transcription factor-mediated and epigenetic regulatory mechanisms, miRNAs also control HFSC development [13, 14]. miR-214 is one of the most studied miRNAs and is located within the sequence of the long noncoding Dmn3os transcript [15]. miR-214 is highly conserved across mulitple species and is involved in numerous physiological and pathological processes. It also coordinates the cell fate, differentiation, and morphogenesis of muscle, skeleton, skin, the cardiovascular system, and the nervous system [16–19].
Collectively, the Wnt/β-catenin pathway and EZH2 have emerged as two main signaling/protein nodes that regulate HFSC proliferation and differentiation. However, the type of epigenetic factors or upstream factors that regulate or interact with Wnt/β-catenin signaling or EZH2 into modify HFSC activity remains unknown. Although previous studies have explored the role of Wnt/β-catenin and EZH2 on regulating the activity of HFSCs, the detailed molecular mechanisms underlying this process necessitate further investigation. Therefore, the present study was carried out to examine the negative effect of miR-214 on HFSC proliferation and differentiation and whether EZH2 and the Wnt/β-catenin signaling pathway are involved.
Materials and methods
Isolation and culture of HFSCs
Scalp tissues were obtained from six patients with a scalp laceration and contusion at our hospital. The study was approved by the Ethics Committee of the hospital, and all the participants provided written informed consent. The skin was cut into 2 mm × 2 mm blocks using ophthalmic scissors; the blocks were rinsed three times with phosphate-buffered saline (PBS). They were then treated with 1% dispase and 1% collagenase IV at 37 °C for 90 min followed by three PBS washes. Under a stereomicroscope, the hair follicle was separated from the connective tissue sheath using a syringe needle. The two ends were cut off, leaving the bulge that was cultured for 2 days in Matrigel Basement Membrane Matrix (BD, NY, USA) with 1 mL of complete medium (869 μL/mL DMEM/F12, 100 μL/mL KSR, 10 μL/mL penicillin–streptomycin, 10 μL/mL l-glutamine, 10 μL/mL MEM nonessential amino acids, 20 ng/mL EGF, 10 ng/mL bFGF, 1 μL/mL 2-mercaptoethanol, and 10 ng/mL hydrocortisone) at 37 °C with 5% CO2. After tissue adherence, 5 mL of complete medium was slowly added. To induce HFSC differentiation, lentiviral vectors overexpressing human β-catenin and c-myc were transfected into HFSCs as described previously (GeneChem, Shanghai, China) [10]. The medium was changed every 3 days. Cell growth, proliferation and differentiation were all observed through an inverted microscope (Ti-E, Nikon, Tokyo, Japan). The images of 10 × 10 or 10 × 20 areas were photographed as appropriate.
HFSC sorting and verification
Flow cytometry was performed for sorting and verification of HFSCs. The identification of HFSCs was confirmed by the following expression pattern: LGR5 (+), CD200 (++), CD271 (−), and CK15 (−) [20]. Briefly, third-generation HFSCs were collected to prepare suspensions at a density of 1 × 105 cells/mL. A total of 20 µL of fluorescein isothiocyanate (FITC)-labeled mouse anti-human LGR5, CD200, CD271, and K15 monoclonal antibodies (Abcam, Cambridge, UK) were added to the cell suspension, followed by an incubation on ice for 20 min in the dark; tubes without antibodies were used as a negative control. Next, 2 mL of PBS was added to each tube followed by centrifugation at 1000 rpm for 6 min. The cells were washed two times with 0.5 mL of PBS. Flow cytometry was performed using a BD FACS Calibur™ (BD Biosciences, NJ, USA). The experiments were repeated three times.
RNA extraction and real-time quantitative RT-PCR
Total RNA was extracted from the HFSC cultures using TRIzol (Invitrogen, CA, USA). The quality and quantity of the isolated total RNA were assessed using an Agilent 2100 Bioanalyzer and NanoDrop ND-1000 Spectrophotometer (Agilent, Santa Clara, CA, USA), respectively. For miR-214 detection, total RNA samples were polyadenylated and reverse transcribed using a two-step quantitative RT-PCR with an NCode™ VILO™ miRNA cDNA Synthesis Kit and EXPRESS SYBR® GreenER™ miRNA qRT-PCR Kits (Invitrogen) according to the manufacturer’s instructions. The sequence-specific forward and reverse primers for hsa-miR-214 and U6 were 5′-ACACTCCAGCTGGGACAGCAGGCACAGACAGGCA-3′, 5′-CTCGCTTCGGCAGCACA-3′ and 5′-CTCGCTTCGGCAGCACA-3′, 5′-AACGCTTCACGAATTTGCGT-3′, respectively. U6 was used as an internal control. The expression levels of miR-214 were calculated using the ratio of miR-214/U6 expression.
Cell transfection
HFSCs were divided into six groups: normal (untransfected HFSCs), negative control (Mock), miR-214 mimic, miR-214 inhibitor, EZH2 siRNA, and XAV-939 (Wnt/β-catenin inhibitor). The sequences in the vectors transfected in the mock, miR-214 mimic, and miR-214 inhibitor groups were 5′-CCUGACAAUUAGUAUUU-3′ GenePharma, Shanghai, China), 5′-ACAGGUAGCUGAACACUGGGUU-3′ (Synbio Technologies), and 5′-UCACAGUGCUCAUCAUGAAUAA-3′ (Shanghai Bioleaf, Shanghai, China), respectively. The sequence for EZH2 siRNA was 5′-GAGGTTCAGACGAGCTGAT-3′ (Ribobio, Guangzhou, China). HFSCs (200 μL/well) in the logarithmic growth phase were seeded in a 6-well plate in antibiotic-free complete medium. When the cells reached 30–50% confluence, they were transfected with miR-214 NC vector, miR-214 mimic, and miR-214 inhibitor (100 pmol; final concentration, 50 nM) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific Inc.). The transfected cells were placed in a 5% CO2 incubator at 37 °C. The medium was replaced with complete medium after a 6–8 h incubation at 37 °C, and the transfected cells were further incubated at 37 °C for 24–48 h for subsequent experiments.
Cell viability and proliferation examined by the MTS assay
Cell viability and proliferation were determined by the MTS [(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl) 2-(4-sulfophenyl)-2H-tetrazolium] assay (Promega, Madison, WI, USA). Briefly, cells were seeded in 96-well plates and cultured at 37 °C for 24 h before transfection. Next, the cells were transfected with the corresponding oligonucleotides. At different time points, the MTS solution (20 μL) was added to each well of the plate, and the plates were incubated at 37 °C for 1 h. Subsequently, the absorbance was measured at 450 nm.
Colony formation assay
Cells from the six groups were cultured for five days and then transferred to 6-well plates at a density of 500 cells/well. The cells were fixed with 4% paraformaldehyde and stained with crystal violet for 20 min. Colonies comprising ≥ 50 cells were detected.
Cell cycle assay
Cells from the six groups were harvested and suspended in 70% cold ethanol overnight. Then, the cells were stained with propidium iodide (PI) (Vazyme, Nanjing, China) for 30 min and analyzed. The proportion of cells in different phases of the cell cycle was calculated and compared.
Luciferase reporter assay
For the luciferase reporter assays, 3 × 104 HSCF cells were plated in collagen-coated wells of a 12-well plate in DMEM. After 72 h, the cells were transfected with either EZH2 3′-UTR Lenti-reporter-Luc vector (ABM, EZH2-UTR-wt) or EZH2 3′-UTR Lenti-reporter-Luc vector with a mutated miR-214 binding site (EZH2-UTR-mt) and the indicated miRNAs or controls. Briefly, 0.33 μg/well of the reporter plasmids were co-transfected with the miR-214 mimics, miR-214 inhibitor, or respective controls (2 μL/well) using jetPRIME® (Polyplus-transfection SA) according to the manufacturer’s protocol. pRenilla (0.1 μg/well) was included in all the transfection assays as an internal control. Luciferase and Renilla (Promega) activity assays were conducted 48 h posttransfection. The relative luciferase unit (RLU) was defined as the ratio of luciferase activity to Renilla activity with the control group set as 1.0.
TOP/FOP Flash assay
The TOP/FOP Flash assay is a luciferase reporter assay that specifically assesses the activity of β-catenin and T cell factor (TCF) signaling. Cells (3 × 104 cells/well in 24-well plates) were transfected with 0.1 g of either TOP Flash or FOP Flash (Upstate Biotechnology, Lake Placid, NY, USA) and 5 ng pRL-SV40 (Promega) using Lipofectamine 2000. After 24 h, the medium was replaced with basic complete medium containing either miR-214 mimic or inhibitor, and the cells were incubated for an additional 24 h. After the cells were lysed, the luciferase assay was subsequently carried out according to the manufacturer’s instructions. The luciferase activity of each sample was normalized with the respective Renilla luciferase activity.
Western blot analysis of EZH2, Wnt/β-catenin signaling proteins, and TA differentiation markers
Cells were lysed on ice using 1X SDS lysis buffer [100 mM 2-ME, 50 mM Tris–HCl (pH 6.8), 2% w/v SDS, 10% glycerol]. The total protein concentration was determined using the bicinchoninic acid (BCA) method. An equivalent of 40 μg of protein was separated by 10–12% SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare Life Sciences). Subsequently, the membranes were probed with anti-GAPDH (Santa Cruz Biotechnology Inc.), anti-β-catenin (BD Bioscience, USA), anti-TCF-4 (BD Bioscience), anti-LEF-1 (Abcam, Cambridge, UK), anti-cyclin D1 (Abcam), anti-EZH2 (R&D, Minneapolis, USA), anti-K15 (Abcam), anti-K19 (Abcam), and anti-Integrin-α6 (Abcam) antibodies overnight at 4 °C followed by an incubation with horseradish peroxidase-conjugated secondary antibodies (Bio-Rad Laboratories Inc., Hercules, CA, USA) for 1 h. Then, the immunoreactive bands were visualized using a ChemiDoc XRS system (Bio-Rad Laboratories Inc.).
Immunofluorescence assay and DAPI staining
For the immunofluorescence studies, cells were grown on poly-d-lysine-coated glass coverslips, fixed with 4% paraformaldehyde for 20 min at 37 °C, and permeabilized with 0.1% Triton X-100/PBS for 3 min at room temperature before incubation with the specific or control antibodies (anti-β-catenin and anti-TCF-4 (Abcam), respectively). Then, the cells were fixed with methanol:acetic acid (3:1, v/v) prior to PBS washes and counterstained with DAPI (4,6-diamidino-2-phenylindole, dihydrochloride) (Vector Laboratories Inc., Burlingame, CA, USA) in the dark. The images of the stained cells were visualized and captured using an Olympus confocal microscope.
Statistical analysis
The results are expressed as the mean ± SD. Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad, San Diego, CA, USA) by t-test and/or one-way ANOVA. The least-significant-difference (LSD) method of multiple comparisons was used when ANOVA showed a statistical significance. All the data represent the mean value from three independent experiments with 6 samples each. p < 0.05 was regarded as statistically significant.
Results
miR-214 expression is attenuated during HFSC proliferation and differentiation
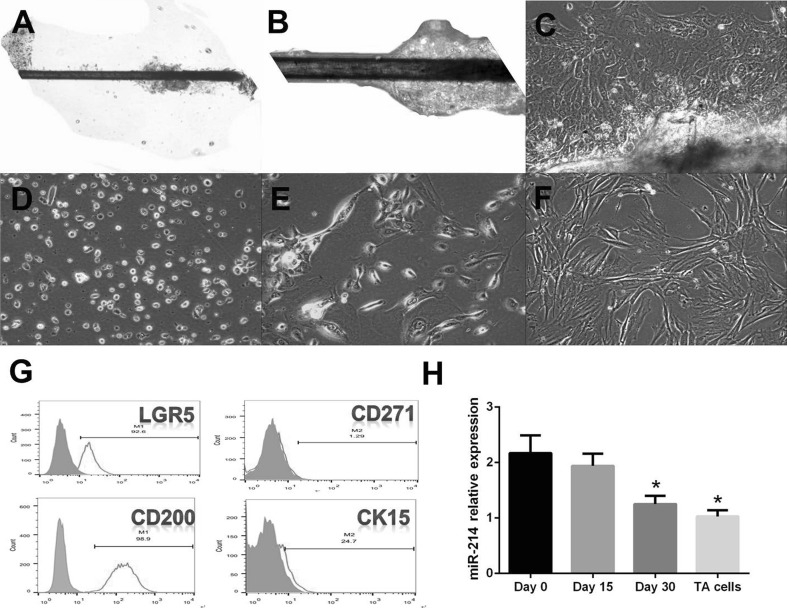
Primary HFSCs were successfully cultured (Fig. 1A–F). HFSCs were sorted and identified as LGR5 (+), CD200 (++), CD271 (−), and CK15 (−) (Fig. 1G). To investigate the putative role of miR-214 in the proliferation and differentiation processes, we examined its expression during differentiation. The levels of miR-214 were estimated by RT-PCR at different time points during HFSC differentiation into TA cells. The changes in the expression of miR-214 during HFSC differentiation into TA cells were further confirmed when HFSCs were induced for 15 or 30 days to promote differentiation into TA cells. The results showed that miR-214 was slightly decreased after the 15-day stimulation and was significantly attenuated after the 30-day induction; however, it was still higher than the levels in TA cells (Fig. 1H) (p < 0.05). Therefore, miR-214 expression gradually decreased during the differentiation of HFSCs into TA cells compared to the levels observed on day 0 (*p < 0.05).
Fig. 1.
Primary HFSC culture and identification of HFSCs and miR-214 expression during proliferation and differentiation stages. A–F Primary HFSCs were successfully cultured and subcultured. A, B Day 15, HFSCs emerge from the tissue (10 × 10 and 10 × 20); C Day 21, HFSCs emerge from the tissue (10 × 10); D Day 7 after primary culture (scattered HFSCs, 10 × 10); E: 1st generation; F 2nd generation; G Identification of HFSCs by flow cytometry using LGR5 (+), CD200 (++), CD271 (−), and CK15 (−) as markers; H miR-214 expression during differentiation to TA cells
miR-214 inhibits cell proliferation and alters the cell cycle progression of human HFSCs
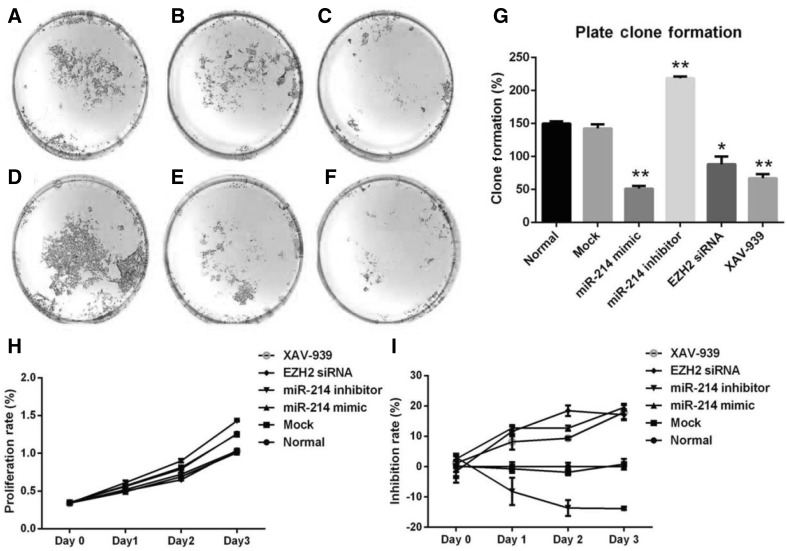
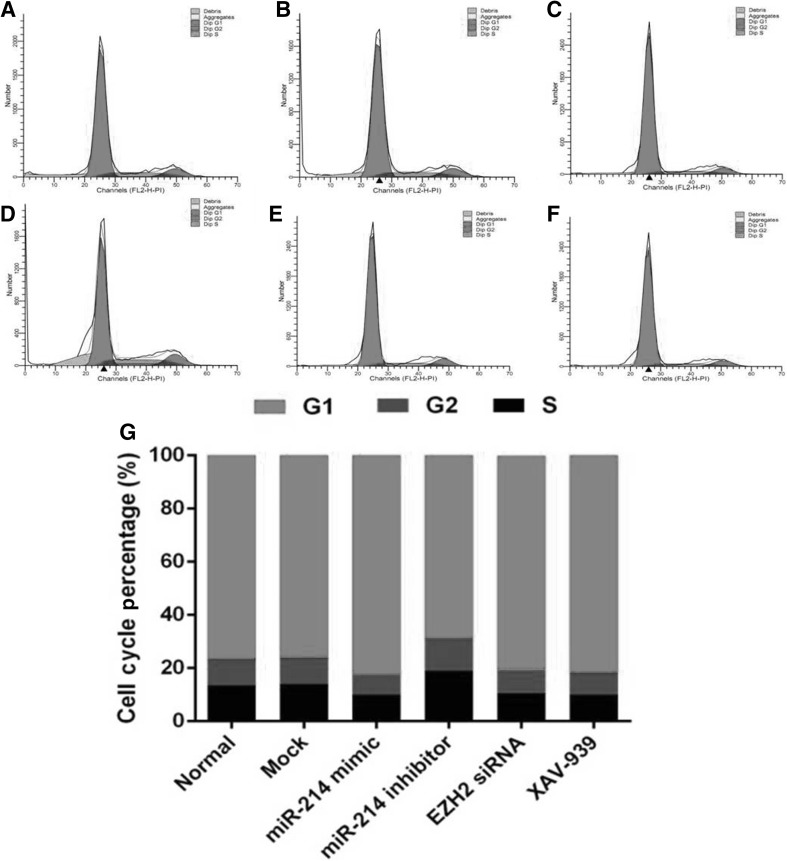
To investigate the biological function of miR-214 expression in HFSCs, cells were transfected with miR-214 mimic or miR-214 inhibitor (Fig. 2A–I). The MTS assay demonstrated ameliorated cell proliferation after upregulation of miR-214 following transfection of HFSCs with miR-214 mimic (Fig. 2H, I). Conversely, cell proliferation was enhanced after downregulation of miR-214 following transfection of HFSCs with miR-214 inhibitor (Fig. 2H, I). Furthermore, the colony formation assays demonstrated that the relative number and size of the colonies were fewer and smaller, respectively, after upregulation of miR-214 in HFSCs (Fig. 2C); however, they were greater and larger, respectively after downregulation of miR-214 in HFSCs (Fig. 2D). To further explore whether EZH2 and Wnt/β-catenin signaling are involved, EZH2 siRNA and the Wnt inhibitor XAV-939 were added to cells. The results showed that both EZH2 siRNA (Fig. 2E) and XAV-939 (Fig. 2F) decreased the proliferation of HFSCs compared to that of the control cells. As shown in Fig. 3, miR-214 mimic promoted cell cycle progression and exerted an increase in the G1/S fraction in HFSCs. Conversely, inhibition of miR-214 resulted in a decreasing trend in the cell population in G1/S phase. These findings suggested that downregulation of miR-214 accelerates cell cycle progression in HFSCs. In addition, EZH2 siRNA and XAV-939 treatment increased the ratio of cells in G1/S phase similar to the effect of miR-214 mimic.
Fig. 2.
Clone formation and MTS assays following miR-214 transfection. A Normal group; B Mock; C miR-214 mimic; D miR-214 inhibitor; E EZH2 siRNA; F XAV-939; G Plate clone formation statistics (*p < 0.05 vs. control, **p < 0.01 vs. control); H Proliferation rate; I Inhibition rate
Fig. 3.
FACS analysis of the effect of altering miR-214 expression on cell cycle progression. A Normal group; B Mock; C miR-214 mimic; D miR-214 inhibitor; E EZH2 siRNA; F XAV-939; G Statistics of the cell cycle distribution
miR-214 directly targets EZH2 during HFSC proliferation and their differentiation into TA cells
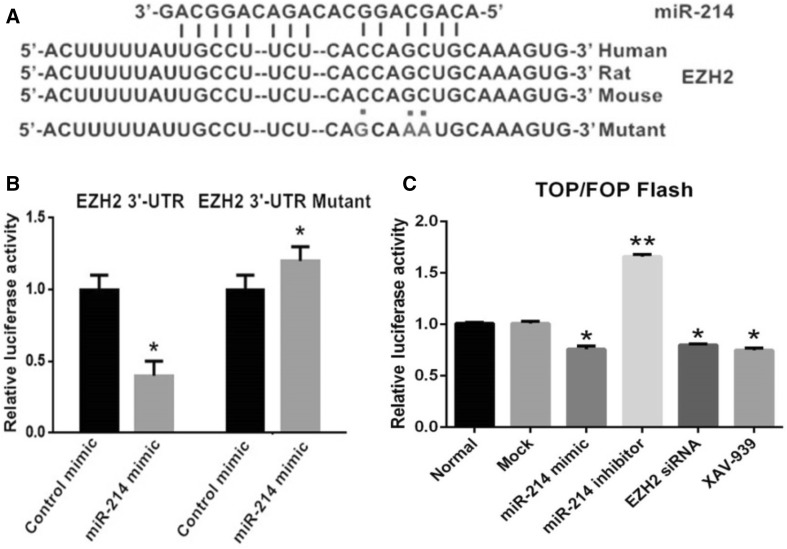
To determine the target genes and signaling pathway of miR-214, candidates were searched using DIANA TOOLS v7.0 (http://diana.imis.athena-innovation.gr/DianaTools). The results showed that miR-214 targeted the EZH2 gene at the designated positions (Fig. 4A). To analyze the relationship between miR-214 and EZH2, a luciferase reporter assay containing wild-type (wt) or mutant (mut) miR-214 target sites in the EZH2 3′-UTR was carried out. HFSCs were co-transfected with miR-214 mimic and either wild-type recombinant plasmid (Wt-miR-214/EZH2) or mutated recombinant plasmid (Mut-miR-214/EZH2). The luciferase activity assay indicated that transfection with miR-214 mimic did not significantly affect the luciferase activity of the cells in the Mut-miR-214/EZH2 plasmid group compared to cells in the negative control group. By contrast, the luciferase activity of cells in the Wt-miR-214/EZH2 plasmid group decreased by 60% compared to the activity in the negative control group (p < 0.05) (Fig. 4B). These results supported the bioinformatics predictions and indicated that EZH2 might be a direct target of miR-214.
Fig. 4.
Analysis of the miR-214 binding to the EZH2 3′-UTR by the luciferase activity assay. A miR-214 sequence with the 3′-UTRs of EZH2 in different species. The position of the miR-214 binding site on human EZH2 mRNA and the EZH2 3′-UTR mutant is depicted. B Addition of miR-214 mimic attenuated the EZH2 3′-UTR reporter gene activity. Mutation of the miR-214 putative target site blocked the suppressive effect of miR-214 on the target, suggesting EZH2 as a miR-214 target gene. p < 0.05 versus control mimic. C TOP Flash and FOP Flash assays assessed the effect of miR-214 and EZH2 interference on the Wnt/β-catenin signaling pathway (*p < 0.05, **p < 0.01)
miR-214 directly regulates the Wnt/β-catenin signaling pathway during HFSC proliferation and their differentiation into TA cells
miR-214 might exhibit a critical role in regulating the activity of the Wnt/β-catenin signaling pathway. In an effort to determine the relationship between the expression of miR-214 and the activity of the Wnt/β-catenin signaling pathway, we employed TOP Flash and FOP Flash reporters, which are widely used to evaluate β-catenin-dependent signaling; thus, the effects of miR-214 on Wnt/β-catenin signaling were assessed. The luciferase activity of the cells altered as predicted; Wnt/β-catenin signaling was inhibited when the levels of miR-214 were upregulated. On the other hand, silencing miR-214 significantly enhanced Wnt/β-catenin signaling. In addition, application of EZH2 siRNA also attenuated Wnt/β-catenin signaling in a similar manner as XAV-939 treatment (Fig. 4C).
miR-214 influences the differentiation of HFSCs into TA cells
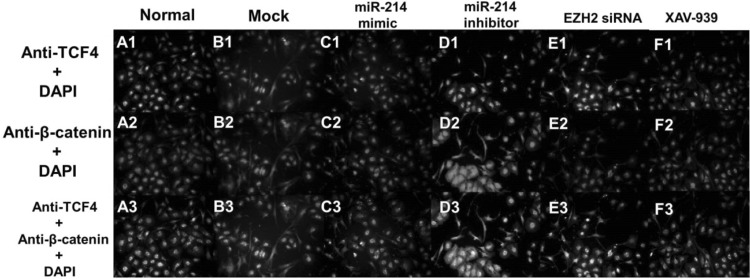
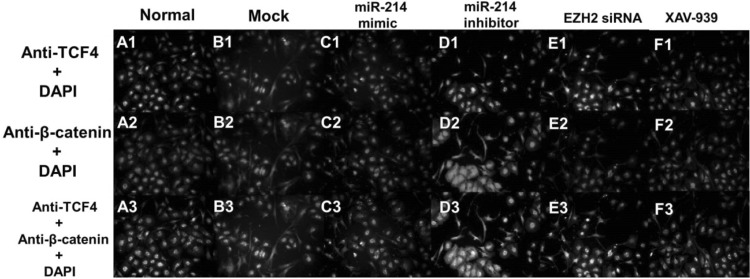
β-Catenin and TCF-4 are the two key proteins involved in the proliferation of HFSCs and their differentiation to TA cells. Next, we used Western blotting to investigate the protein expression levels of β-catenin and TCF-4 (Fig. 5) and found that downregulation of miR-214 enhances the expression levels of β-catenin and TCF-4. On the other hand, overexpression of miR-214 leads to the significantly decreased expression of β-catenin and TCF-4. Fluorescence microscopy revealed that the localization of β-catenin in the cells shifted from nuclear to cytoplasmic when miR-214 expression increased; simultaneously, the TCF-4 levels in the nucleus decreased (Fig. 6). Conversely, downregulation of miR-214 promoted the entry of β-catenin and TCF-4 into the nucleus from the cytoplasm (Fig. 6). Next, we explored the expression levels of TA differentiation markers, including Integrin-α6, K15, and K19, and found that inhibiting miR-214 promoted the expression of Integrin-α6, K15, and K19 compared to the levels in the controls. In contrast, upregulation of miR-214 decreased the expression of Integrin-α6, K15, and K19. Moreover, we also found that EZH2 siRNA and XAV-939 attenuated the expressions of all three markers (Fig. 5).
Fig. 5.
Western blot analysis of the expression levels of β-catenin, Cyclin D1, EZH2, TCF4, and LEF1 and the TA cell differentiation markers Integrin α6, CK15, and CK19 after transfection. The data represent the mean value from three independent experiments
Fig. 6.
Immunofluorescence localization of TCF4 and β-catenin following miR-214 transfection. Green color: anti-TCF4; red color: anti-β-catenin; blue color: DAPI staining; yellow: overlap of green and red color. (Color figure online)
Discussion
Epidermal stem cells (ESCs) represent one of the earliest seed cells used in skin tissue engineering. However, their low adherence rate, slow proliferation, brittle cell membrane, and poor adhesion to the wound surface has restricted their clinical application [21]. Recently, HFSCs have been used as seed cells in skin tissue engineering. HFSCs exhibit stronger proliferative abilities and differentiation orientation than ESCs [22]. Recently, epigenetic factors have been identified to play important roles in regulating HFSC differentiation into TA cells.
MicroRNAs, one family of the most important epigenetic factors, are short regulatory RNAs that are able to posttranscriptionally modulate gene expression and that play crucial roles in the control of physiological and pathological processes, including the regulation of stem cell proliferation and differentiation. There is increasing evidence regarding the role of microRNAs (miRNAs) in a variety of pathological conditions, such as preeclampsia, endometriosis and cancer [23–26]. miR-214 has been previously implicated in skin renewal [19]. Previous studies also demonstrated that miR-214 transgenic mice exhibited decreased epidermal proliferation, a number of all types of HFs, variable sizes of hair bulbs, and the formation of thinner hair compared to wild-type controls via regulation of Wnt [19]. Moreover, activation of Wnt signaling rescues these skin phenotype changes in miR-214 transgenic mice [19]. The current study demonstrated that EZH2 is a target of miR-214. Several studies have shown that β-catenin is also a direct target of miR-214 [27–29].
Our present study highlights two different mechanisms by which miR-214 regulates the biological activity of HFSC proliferation and differentiation to TA cells. First, we showed that miR-214 could inhibit the activity of the Wnt/β-catenin signaling pathway. The Wnt signaling pathway is a potent regulator of cell proliferation and differentiation. Many effects of miR-214 on skin and HF development are most likely associated with interference of cell cycle regulation. In our study, activation of miR-214 alters the cell cycle through downregulation of cyclin D1 expression. Next, we found that miR-214 negatively regulates the expression of EZH2 via direct interaction with the 3′-UTR of EZH2 mRNA. EZH2, which plays an important biological role, has been reported to be a target gene of miR-214 in various cell types. For example, miR-214 regulates EZH2 in skeletal muscle and in embryonic stem cells [18]. In addition, miR-214 also protects erythroid cells against oxidative stress by directly targeting EZH2 [30].
To further investigate the effect of miR-214 on the phenotypic changes in HFSCs, we transfected HFSCs with miR-214 mimic and miR-214 inhibitor and found that overexpression of miR-214 significantly inhibited HFSC growth and differentiation to TA cells, while reduced expression of miR-214 promoted HFSC growth and differentiation to TA cells. Western blot analysis of Wnt/β-catenin-related proteins and the TA cell differentiation markers K15, K19, and Integrin-α6 showed that miR-214 could influence HFSC differentiation into TA cells. In addition, miR-214 interference altered the expression levels of the β-catenin and TCF-4 and influenced the entry of both proteins into the nucleus. Previous studies have found that β-catenin and its downstream factors c-myc, cyclin D, and K15 are key factors for TA cell formation [10]. Together, these findings suggested that miR-214 could regulate the human HFSC proliferation and differentiation by targeting EZH2 and the Wnt/β-catenin signaling pathway.
The Wnt/β-catenin signaling pathway has emerged as the dominant pathway regulating the patterning of skin and influencing the decisions of embryonic and adult stem cells to adopt the various cell lineages of the skin and its appendages. Subsequently, this pathway controls the function of differentiated skin cells [8]. Although the Wnt/β-catenin signaling pathway regulates the activity of HFSCs, the underlying molecular mechanisms are yet poorly understood. Therefore, finding a new modulator that can regulate this pathway is imperative. Wnt/β-catenin signaling plays a critical role in HFSC proliferation and differentiation. In addition, the TOP/FOP Flash assay showed that the EZH2 siRNA affected Wnt/β-catenin signaling, implicating that miR-214 might also influence Wnt/β-catenin signaling by targeting EZH2. A previous study showed that β-catenin is a downstream target of EZH2; hence, it can be directly or indirectly regulated via EZH2 to modulate the β-catenin signaling pathway [31]. Ectopic EZH2 expression increases the active β-catenin levels in immortalized human hepatocytes [28]. Furthermore, EZH2 also binds to the promoters of a panel of Wnt signal antagonists, wherein the H3K27me3-repressive marker was also enriched to activate the Wnt signal [28]. In the mechanism of the EZH2-mediated activation of Wnt/β-catenin signaling, EZH2 serves as a transcriptional repressor and represses Wnt antagonists, including RAF1, CXXC4, AXIN2/beta TrCP, and p53 [32–34].
Nevertheless, the present study has several limitations. First, this was an in vitro experimental study. Further in vitro or in vivo studies are essential to substantiate these findings. Second, the targets of miRNAs may not be unique as we could not deduce whether other signaling pathways might also be involved in miR-214 interference.
Taken together, these data showed that miR-214 is downregulated during HFSC proliferation and differentiation into TA cells. The reduction in the expression level of miR-214 resulted in the upregulation of EZH2 and β-catenin. In addition, the downregulation of miR-214 resulted in increased proliferation and differentiation into TA cells. On the other hand, the ectopic expression of miR-214 in HFSCs suppressed cell growth and differentiation to TA cells in vitro via inhibition of EZH2 and Wnt/β-catenin signaling.
Acknowledgement
Funding was provided by the National Natural Science Foundation of China (81372114).
Conflicts of interest
The authors have no financial conflicts of interest.
Ethical approval
The study was approved by the Ethical Committee of ChenZhou No.1 People’s Hospital (CZSDYRMYY-2017-18). All the donators signed the informed consent. There are no animal experiments carried out for this article.
Contributor Information
Cheng Peng, Email: pcheng83@sina.com.
Ming-Sheng Zhang, Email: mszrch3@163.com.
References
- 1.Yang L, Peng R. Unveiling hair follicle stem cells. Stem Cell Rev Rep. 2010;6:658–664. doi: 10.1007/s12015-010-9172-z. [DOI] [PubMed] [Google Scholar]
- 2.Ojeh N, Pastar I, Tomic-Canic M, Stojadinovic O. Stem cells in skin regeneration, wound healing, and their clinical applications. Int J Mol Sci. 2015;16:25476–25501. doi: 10.3390/ijms161025476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Díaz-Flores L, Jr, Madrid JF, Gutiérrez R, Varela H, Valladares F, Alvarez-Arguelles H, et al. Adult stem and transit-amplifying cell location. Histol Histopathol. 2006;21:995–1027. doi: 10.14670/HH-21.995. [DOI] [PubMed] [Google Scholar]
- 4.Hsu YC, Li L, Fuchs E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell. 2014;157:935–949. doi: 10.1016/j.cell.2014.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou RH, Chiu L, Yu YL, Shyu WC. The potential roles of EZH2 in regenerative medicine. Cell Transplant. 2015;24:313–317. doi: 10.3727/096368915X686823. [DOI] [PubMed] [Google Scholar]
- 6.Chou RH, Yu YL, Hung MC. The roles of EZH2 in cell lineage commitment. Am J Transl Res. 2011;3:243–250. [PMC free article] [PubMed] [Google Scholar]
- 7.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lien WH, Polak L, Lin M, Lay K, Zheng D, Fuchs E. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat Cell Biol. 2014;16:179–190. doi: 10.1038/ncb2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong SY, Reiter JF. Wounding mobilizes hair follicle stem cells to form tumors. Proc Natl Acad Sci U S A. 2011;108:4093–4098. doi: 10.1073/pnas.1013098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Q, Yu W, Fang Y, Yao M, Yang P. Beta-catenin can induce hair follicle stem cell differentiation into transit-amplifying cells through c-myc activation. Tissue Cell. 2017;49:28–34. doi: 10.1016/j.tice.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Cech TR, Steitz JA. The noncoding RNA revolution—trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ning MS, Andl T. Control by a hair’s breadth: the role of microRNAs in the skin. Cell Mol Life Sci. 2013;70:1149–1169. doi: 10.1007/s00018-012-1117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi R, Fuchs E. MicroRNAs and their roles in mammalian stem cells. J Cell Sci. 2011;124:1775–1783. doi: 10.1242/jcs.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penna E, Orso F, Taverna D. miR-214 as a key hub that controls cancer networks: small player, multiple functions. J Investig Dermatol. 2015;135:960–969. doi: 10.1038/jid.2014.479. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y, Ponnusamy M, Zhang L, Zhang Y, Liu C, Yu W, et al. The role of miR-214 in cardiovascular diseases. Eur J Pharmacol. 2017;816:138–145. doi: 10.1016/j.ejphar.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Zhao C, Sun W, Zhang P, Ling S, Li Y, Zhao D. miR-214 promotes osteoclastogenesis by targeting Pten/PI3 k/Akt pathway. RNA Biol. 2015;12:343–353. doi: 10.1080/15476286.2015.1017205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed MI, Alam M, Emelianov VU, Poterlowicz K, Patel A, Sharov AA, et al. MicroRNA-214 controls skin and hair follicle development by modulating the activity of the Wnt pathway. J Cell Biol. 2014;207:549–567. doi: 10.1083/jcb.201404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue K, Aoi N, Sato T, Yamauchi Y, Suga H, Eto H, et al. Differential expression of stem-cell-associated markers in human hair follicle epithelial cells. Lab Invest. 2009;89:844–856. doi: 10.1038/labinvest.2009.48. [DOI] [PubMed] [Google Scholar]
- 21.Eckert RL, Adhikary G, Balasubramanian S, Rorke EA, Vemuri MC, Boucher SE, et al. Biochemistry of epidermal stem cells. Biochim Biophys Acta. 2013;1830:2427–2434. doi: 10.1016/j.bbagen.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mokos ZB, Mosler EL. Advances in a rapidly emerging field of hair follicle stem cell research. Coll Antropol. 2014;38:373–378. [PubMed] [Google Scholar]
- 23.Vitale S, Laganà A, Capriglione S, Angioli R, La Rosa V, Lopez S, et al. Target therapies for uterine carcinosarcomas: current evidence and future perspectives. Int J Mol Sci. 2017;18:E1100. doi: 10.3390/ijms18051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laganà AS, Salmeri FM, Vitale SG, Triolo O, Götte M. Stem cell trafficking during endometriosis. Reprod Sci. 2017 doi: 10.1177/1933719116687661. [DOI] [PubMed] [Google Scholar]
- 25.Chiofalo B, Laganà AS, Vaiarelli A, La Rosa VL, Rossetti D, Palmara V, et al. Do miRNAs play a role in fetal growth restriction? A fresh look to a busy corner. Biomed Res Int. 2017;2017:6073167. doi: 10.1155/2017/6073167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laganà AS, Vitale SG, Sapia F, Valenti G, Corrado F, Padula F, et al. miRNA expression for early diagnosis of preeclampsia onset: hope or hype? J Matern Fetal Neonatal Med. 2018;31:817–821. doi: 10.1080/14767058.2017.1296426. [DOI] [PubMed] [Google Scholar]
- 27.Yi SJ, Li LL, Tu WB. MiR-214 negatively regulates proliferation and WNT/beta-catenin signaling in breast cancer. Eur Rev Med Pharmacol Sci. 2016;20:5148–5154. [PubMed] [Google Scholar]
- 28.Cao F, Zhan J, Chen X, Zhang K, Lai R, Feng Z. miR-214 promotes periodontal ligament stem cell osteoblastic differentiation by modulating Wnt/β-catenin signaling. Mol Med Rep. 2017;16:9301–9308. doi: 10.3892/mmr.2017.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JP, Zhuang HT, Xin MY, Zhou YL. MiR-214 inhibits human mesenchymal stem cells differentiating into osteoblasts through targeting beta-catenin. Eur Rev Med Pharmacol Sci. 2017;21:4777–4783. [PubMed] [Google Scholar]
- 30.Gao M, Liu Y, Chen Y, Yin C, Chen JJ, Liu S. miR-214 protects erythroid cells against oxidative stress by targeting ATF4 and EZH2. Free Radic Biol Med. 2016;92:39–49. doi: 10.1016/j.freeradbiomed.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Cheng AS, Lau SS, Chen Y, Kondo Y, Li MS, Feng H, et al. EZH2-Mediated concordant repression of Wnt antagonists promotes-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71:4028–4039. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- 32.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu H, Sun J, Wang F, Feng L, Ma Y, Shen Q, et al. Enhancer of zeste homolog 2 activates wnt signaling through downregulating CXXC finger protein 4. Cell Death Dis. 2013;4:e776. doi: 10.1038/cddis.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tseng RC, Lin RK, Wen CK, Tseng C, Hsu HS, Hsu WH, et al. Epigenetic silencing of AXIN2/betaTrCP and deregulation of p53-mediated control lead to wild-type β-catenin nuclear accumulation in lung tumorigenesis. Oncogene. 2008;27:4488–4496. doi: 10.1038/onc.2008.83. [DOI] [PubMed] [Google Scholar]