Abstract
Objective:
To develop and evaluate an efficient Trie structure for large-scale, rule-based clinical natural language processing (NLP), which we call n-trie.
Background:
Despite the popularity of machine learning techniques in natural language processing, rule-based systems boast important advantages: distinctive transparency, ease of incorporating external knowledge, and less demanding annotation requirements. However, processing efficiency remains a major obstacle for adopting standard rule-base NLP solutions in big data analyses.
Methods:
We developed n-trie to specifically address the token-based nature of context detection, an important facet of clinical NLP that is known to slow down NLP pipelines. N-trie, a new rule processing engine using a revised Trie structure, allows fast execution of lexicon-based NLP rules. To determine its applicability and evaluate its performance, we applied the n-trie engine in an implementation (called FastContext) of the ConText algorithm and compared its processing speed and accuracy with JavaConText and GeneralConText, two widely used Java ConText implementations, as well as with a standalone machine learning NegEx implementation, NegScope.
Results:
The n-trie engine ran two orders of magnitude faster and was far less sensitive to rule set size than the comparison implementations, and it proved faster than the best machine learning negation detector. Additionally, the engine consistently gained accuracy improvement as the rule set increased (the desired outcome of adding new rules), while the other implementations did not.
Conclusions:
The n-trie engine is an efficient, scalable engine to support NLP rule processing and shows the potential for application in other NLP tasks beyond context detection.
Keywords: Natural Language Processing, Medical Informatics Applications, Algorithms, Data Accuracy
Graphical abstract:
2. Introduction1
Algorithmic processing efficiency becomes increasingly important as the size of clinical datasets grows, especially in the era of “Big Data” [1]. In the specific domain of clinical natural language processing (NLP), even small increases in processing throughput are important when handling very large note corpora. An extreme example is the U.S. Veterans Administration VINCI national data warehouse, which contains > 2 billion clinical notes. Divita et al. demonstrated the effect of note processing efficiency in VINCI studies [2]. In their work, 6 million records is considered a representative national sample for many applications. Shaving off 100 milliseconds of processing time per note in their benchmark system (401 milliseconds was their nominal per-note-processing time) would save nearly a week of clock time for a corpus of that size.
With the growing interest in Data Science and Big Data, information extraction and retrieval will continue to be a trend in clinical research and practice [3]. Warehoused clinical data growth, spurred in the U.S. by the HITECH Act [4], has escalated the need for faster and more accurate information processing. To improve clinical NLP processing efficiency, we investigated the bottlenecks in typical processing pipelines. Context detection, a rule-based NLP component (defined below), consumed ~70% of processing time in Divita’s report.
In this paper, we show that Trie-based rule processing performs well in an important area of clinical NLP. We also present the n-trie (as in “trie for NLP”) engine, an optimized Trie rule-processing engine for NLP and demonstrate its efficacy and processing performance with context detection as the use case, specifically the ConText algorithm [5]. Finally, we discuss the engine’s potential in other clinical NLP use cases.
3. Background
3.1. Rule-based systems are not dead
Traditionally there are three approaches to information extraction (IE) from clinical text: rule-based, machine learning-based, or a hybrid of the two. Although machine learning exhibits promising performance when provided with enough labeled data and a well-defined target structure, its disadvantages in real-world applications are significant: the inner workings of all but a few models are hard to extract (especially true of the currently popular deep neural network models), they are difficult to maintain (e.g., updating or adapting them requires re-training), they are hard to debug (i.e., one can only reduce errors by tuning parameters or by adopting heuristics), and they are hard to enhance with new domain knowledge [6]. In many IE tasks rule-based systems show comparable or superior extraction performance compared to state-of-the-art machine learning solutions [7–10]. Whether one approach or the other has an advantage regarding human labor cost is debatable. Rule-based approaches are generally labor intensive when developing rules. On the other hand, creating a labeled training set and choosing, in a principled and disciplined way, a mathematical model optimized for a specific NLP task requires significant effort and expertise with the machine learning approach.
3.2. Previous research related to optimizing rule processing execution time
Most rule-based systems use regular expressions to define rules or part of rules. In the rule execution experiment conducted by Reiss et al. [11], regular expression execution time accounts for 90% of the overall running time. Chiticariu et al. described SystemT, a rule processing engine that utilizes a set of optimization strategies to speed up rule execution [9]. The most effective strategy involves prioritizing matching rare elements in rules to reduce the matching workload when the rules contain the logical conjunction of multiple elements. However, this strategy only works when prior statistical knowledge about the IE search space is available.
A related optimization that may potentially improve rule processing time uses character trigram indices [12]. After building the character trigram index, regular expressions can be converted to logic-joined trigram search queries. In this way, rule matching speed can be improved by reducing the search space through limiting regular expression execution within trigram-matched query results. In a sense, this approach resembles a very shallow version of the hash tree algorithm we adopted in the work described below. However, trigram indexing requires extra space and substantial preprocessing.
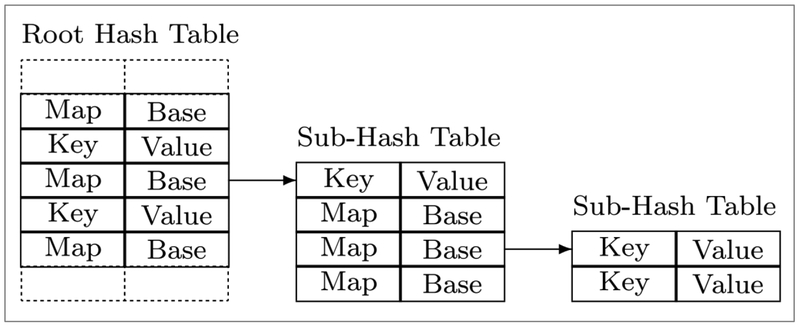
The third related approach is to utilize specifically designed data structures to optimize the string matching process. Since the fundamental goal of rule processing is matching the rules to target text strings, any string matching algorithm potentially could be adopted. For instance, the Trie structure has been studied to host rule dictionaries in hopes of speeding up processing time. A variety of Tries have been demonstrated to be highly effective at string searching, such as the ternary Trie [13], the Patricia Tries[14], the Cache-Efficient Trie [15], and the Hash Array Mapped Trie (HAMT) [16]. HAMP is the ancestor of our n-trie (see section 4.1), which has a speed-optimized design. It consists of a root hash table with entries of key/value pairs or pointers to sub-hash tables (see Figure 1). Each sub-hash table has the same structure as the root hash table.
Figure 1.
Hash Array Mapped Trie (HAMT) Structure [16]
These earlier Trie data structures, based as they are on character strings, frequently prove less flexible and more inconvenient to use for defining clinical NLP rules. Often an NLP rule needs a component that can represent a specific token (e.g., “ruled out” or “denies”). It is difficult to build an efficient character-based Trie for large sets of such rules.
3.3. Contextual Information
The following subsections briefly review the background of context detection --- the use case to demonstrate the n-trie engine. One can think of the term “contextual information” as a set of modifiers associated with a specific mention of a concept. In the domain of clinical information extraction, contextual information typically includes three types of modifiers: negation (whether the target concept exists, does not exist, or is possible/uncertain), experiencer (whether the target concept refers to the current patient or to someone else), and temporality (whether the target concept is currently true, historically true, or hypothetical) [7]. In some studies, the “certainty” (possible/uncertain) is separated as an independent modifier [17,18].
Negation is the best-studied modifier and refers to an assertion about what a patient does not have, such as medical conditions or symptoms (e.g., “Patient denies exertional dyspnea”). Previous studies discovered that clinical notes contain a significant number of negative statements [19]. In the corpus used in this study, SemEval-2015 Task 14: Analysis of Clinical Text [20], more than one-fifth of the identified disorder concepts were negated. These negations are often clinically meaningful. They serve critical roles in supporting differential diagnosis generation and treatment planning. For instance, in an ultrasound report, the context of “No atrial septal defect was found” versus the context of “Atrial septal defect is present” would lead to completely different diagnostic and treatment strategies.
3.4. Context Detectors
Several rule-based context detectors have been developed and evaluated, such as NegExpander [21], NegFinder [19], and NegEx [22] plus its descendent ConText [5,23]. Rule-based detectors typically define a set of regular expressions that look for trigger terms across a given scope of tokens, usually augmented with additional regular expressions designed to suppress false positives (e.g., double negatives). NegExpander identifies negation words and conjunctions, and asserts the conjunctive noun phrases as negated. However, it cannot adjust the negation scope based on relevant semantic clues, e.g., “although.” NegFinder introduces “negation terminators” to overcome this limitation. Notwithstanding, it cannot handle pseudo-negation words, such as double negations. NegEx and ConText use “pseudo” triggers to deal with these situations. Goryachev et al. [8] compared four different negation detection methods to process discharge summaries, including NegEx, NegExpander, and two other simple machine learning (ML) approaches. The rule-based NegEx (F1-score: 0.89) and NegExpander (F1-score: 0.91) outperformed the two ML approaches (F1-scores: 0.78 and 0.86, respectively). Several ML-based or hybrid systems have been described [17,24–28]. Only NegScope [27], a negation detector evaluated on radiology reports and biomedical abstracts in the Bioscope Corpus [18] and Cogley et al.’s system [28] (a temporality and experiencer detector applied to 120 history and physical notes), outperformed the rule-based systems (Cogley’s system only outperformed ConText in temporality assertions). None of the reports noted above provided any information on processing efficiency.
Although these reported results suggest that the context detection task has been “solved,” there is evidence that ML and rule-based context algorithms show a drop in performance when applied to clinical corpora they have not been trained on [29]. This suggests applying these solutions in real-world applications will require additional tuning. Patterson and Hurdle showed that clinical note authors in different clinical settings use distinct sub-languages [30] and Doing-Harris et al. showed that those sub-languages change across institutions [31]. These studies suggest that, when applied to a new corpus, ML-based approaches require new annotations in order to be retrained. Rule-based systems will need rule modifications to cover patterns previously unseen. In our experience the latter is less labor intensive but, as we noted, rule-based systems can be computationally expensive. Thus, we choose this ConText detection challenge as the demonstration use case for the n-trie engine.
4. Material and Methods
4.1. N-trie structure
The n-trie structure is a variation of HAMT. Observing that token-based formats are dominant in clinical rule-based NLP and that most clinical NLP pipelines require tokenization as a preprocessing step, we implemented n-trie in a token-based fashion to take advantage of tokenized text. Compared with the HAMT, we further simplified the structure by unifying the two types of entries (key/value pairs and sub-hash table pointers). First, n-trie constructs a nested-map structure from the rules. The top level is a single map, where the keys are the first word in each rule. The value associated with a top-level key is a child map that consists of the second words of all the rules starting with that first word, and an “END” key if the first word itself can be matched to a rule alone. The “END” key’s value is a list of matched rule IDs. The actual rules are maintained in an array, which can be quickly located and allow storing multiple rule properties. In the case of ConText rules, these properties include the trigger names, the trigger types, the “affect directions”, and the window sizes. Similarly, the rest of the words in the rules will be included in the subsequent descendent maps. With this data structure the n-trie engine can process all the rules without looping through them, leading to rapid processing.
To support wildcard-like functionality, we added a special key “\w+” to allow matching any word during the rule processing. For instance, “given \w+ history of” can match “given his history of,” “given the history of,” etc. Additionally, we added another two keys “>” and “<”, followed by a number to match a number that is greater or less than a number defined in a rule. For instance, “> 3 months ago” will match “4 months ago”, “5 months ago”, etc. This type of rule can be used to define a specific time window before an encounter that can be considered to include or exclude related mentions. The three special keys are summarized based on previous context rules and our current project needs, which cover most of our clinical use cases.
4.2. FastContext Implementation
To demonstrate the n-trie engine we use it to implement FastContext, a fast and efficient rule-based ConText detector. FastContext supports all three types of clinical context lexical cues (trigger terms, pseudo-trigger terms, and termination terms) and three directional properties (forward, backward, and bidirectional) for each modifier. Also, FastContext supports rule window size configuration (i.e., the maximum scope where a context cue can take effect). This last feature helps restrict the context scope in corpora where sentences are difficult to segment, which is common in clinical documents [32].
A few examples below in Table 1 illustrate how the FastConText rules can be configured.
Table 1.
FastConText rule examples.
| Text Snippet | Trigger Direction |
Context and Its Window |
Rule Interpretation |
|---|---|---|---|
| “can rule out” | Forward Trigger | Negated 10 | Means if the phrase “can rule out” is found, this context cue will trigger a “negated” affect in the forward direction: the words that come after it (within 10 token windows size of the same sentence) will be negated. |
| “although” | Forward Termination | Negated 30 | This is a termination rule; it will stop the negation cue on its left side (if any) from affecting negation to the right side. Window size is not applicable in termination rules. |
| “false negative” | Both | Pseudonegated 30 | This is a pseudo-trigger rule. The phrase “false negative” is a double negation, which has negation cues but is not meant to negate. Whenever this rule is matched, it will ignore the negation trigger cues “false” or “negative.” Since its effect is to overwrite other rules, its own direction and window size do not have any specific effect, but it keeps rule formats consistent. |
The Java implementation of FastConText is available on GitHub [33], along with a comprehensive set of rules refined from the original ConText rule set, additional rules contributed from Divita’s U.S. Veterans Administration VINCI group, and rules of our own design.
4.3. Evaluation
We evaluated the n-trie engine by comparing FastContext with two other popular ConText implementations: JavaConText [34] and GeneralConText [35] (the second generation of NegEx), measuring both speed and accuracy. Since NegEx is directly incorporated in GeneralConText, comparing FastContext to GeneralConText directly measures n-trie’s negation utility against that of NegEx.
All the experiments above were conducted within the same hardware configuration environment: eight dedicated compute nodes, each with eight Intel Xeon E5530 processors; 24Gb total RAM per node. A run of an implementation over the target corpora was threaded in parallel and repeated 200 times. The evaluation was run under CentOS 6.7 Linux running the Java 1.8 virtual machine.
Additionally, we compared the processing speed between FastContext and the machine learning-based NegScope. However, we could not make a fair comparison between FastContext and NegScope regarding the accuracy of negation detection. Because, as of this writing, the clinical text dataset of the Bioscope corpus[18] is no longer available, we are not able to evaluate FastContext’s performance on that dataset. On the other hand, we do not have the NegScope-annotated format of the SemEval dataset (see the next subsection below), so we are constrained to only measuring processing speed differences. This experiment was conducted on a single compute node with 28 Intel Xeon E5-2680 v4 processors, 128Gb RAM, under CentOS 7.4 running Java 1.8.
4.3.1. Dataset
For this evaluation, we used the SemEval 2015 “Analysis of Clinical Text” task dataset [20]. This dataset has 431 narrative clinical notes with 19,512 manually annotated disorder concepts and corresponding modifiers. SemEval annotations differ slightly from ConText annotations. To minimize the modification of ConText implementations, we treated a SemEval annotation that has “Negation: yes” + “Uncertainty: true” as equivalent to ConText’s “Negation: possible.” Table 2 shows the distribution of different context modifiers. Because the SemEval dataset does not contain temporality labels, we could not evaluate the accuracy performance of temporality detection, although FastContext is capable of detecting it.
Table 2.
Data composition of the SemEval 2015 test dataset
| Context Type | Modifiers | Number of Test Cases |
Percent |
|---|---|---|---|
| Negation | Negated | 3,621 | 22% |
| Affirmed | 11,796 | 70% | |
| Possible | 1,333 | 8% | |
| Experiencer | Patient | 16,536 | 99% |
| Other | 214 | 1% |
Note: Each concept has both negation context and experiencer context.
To prepare the evaluation dataset, the notes were pre-processed by our tokenizer and sentence segmenter [36]. Tokenized words were rejoined using whitespaces to provide fair input for the three implementations. JavaConText and GeneralConText do not take tokenized words as input, instead they only take input in string format and split words on whitespaces themselves. We excluded 1,743 concepts that were comprised of non-contiguous words and 1,019 concepts segmented into sentences without context information as annotated by SemEval, leaving 16,750 concepts.
4.3.2. Processing Speed Evaluation
To assess the baseline, the original rules of JavaConText and GeneralConText were used. To evaluate processing speed performance as we increased rule set size, we started with the initial 409 ConText rules directly derived from JavaConText and kept them in the same order. Then we added an additional 440 of our own rules plus those from Divita’s VINCI rule set. Rules were added incrementally to the three implementations at a pace of 50 rules per step (40 rules in the last step and 849 final rules in total). At each step, the added rules were randomly selected from the new 440 rules. Two hundred runs were repeated at every step. The average processing times across different implementations were compared at each step.
To compare the speed between FastContext and NegScope, the full rule set was used to configure FastContext and the default pre-trained clinical models were used to configure NegScope (recall that we are only evaluating processing speed in this comparison, not accuracy). Each of them processed the SemEval dataset 200 times and resulting times were averaged.
4.3.3. Context Assertion Performance Evaluation (Accuracy)
To determine how well each of the three implementations accurately interpret context assertions as the rule set grows, the same rule-adding steps were used as above. The assertion performance, which we call accuracy here, was measured as F1-scores using the standard formula:
Because 200 runs at each step were repeated, the average F1-score of each step was calculated by taking the mean of each set of 200 scores.
Although the Negation modifier has three values: “Affirm”, “Negated” and “Possible”, the ConText algorithm is designed to detect the latter two, because “Affirm” is considered as the default value when no negation cue is found. Thus, we only calculated the F1-scores for the “Negated” and “Possible” detection. Similarly, because “Patient” is the default value of Experiencer, only the F1-scores for “Other” detection were calculated. GeneralConText was not evaluated for “Possible” detection because it does not support the “Possible” modifier.
5. Results
5.1. Speed Comparison Against the Other Two ConText Implementations and NegScope
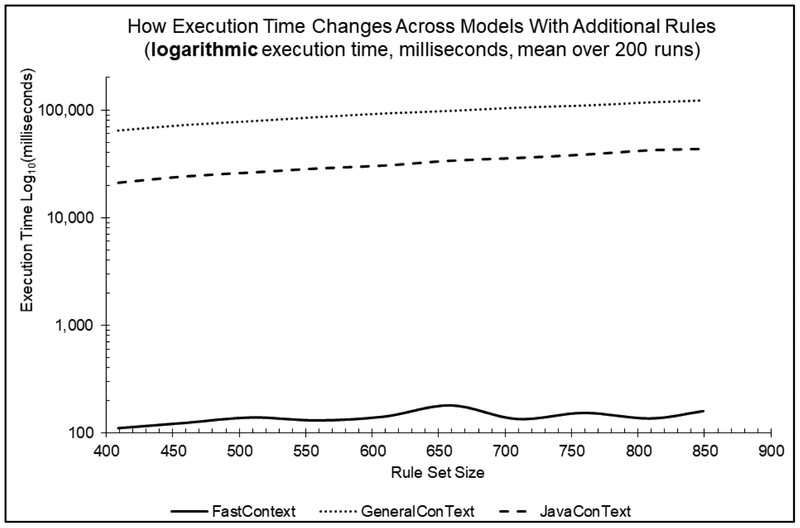
Table 3 shows the average processing time of the three implementations as the size of the rule set grows. Figure 2 shows the same data graphically (note the use of a logarithmic scale).
Table 3.
The three implementations’ average corpus processing time (in milliseconds) over 200 runs
| Number of Rules |
GeneralConText (ms) |
JavaConText (ms) |
FastContext (ms) |
Multiples of FastContext Avg. Run Time (i.e., how many times faster FastContext runs) |
|
|---|---|---|---|---|---|
| GeneralConText | JavaConText | ||||
| 409 | 64,440 | 21,091 | 110 | 580 | 190 |
| 459 | 72,559 | 24,142 | 123 | 590 | 200 |
| 509 | 78,740 | 26,244 | 139 | 570 | 190 |
| 559 | 85,838 | 28,606 | 130 | 660 | 220 |
| 609 | 92,848 | 30,443 | 141 | 660 | 220 |
| 659 | 98,195 | 33,782 | 180 | 550 | 190 |
| 709 | 105,200 | 35,795 | 134 | 790 | 270 |
| 759 | 110,010 | 38,430 | 153 | 720 | 250 |
| 809 | 117,707 | 42,229 | 136 | 870 | 310 |
| 849 | 122,761 | 43,491 | 159 | 770 | 270 |
Figure 2.
Processing time for the three implementations as the rule set grow (log-linear scale).
FastContext exhibits a compelling speed improvement over JavaConText and GeneralConText. In every run it is at least two orders of magnitude faster than the other two approaches. Additionally, the processing times of both GeneralContext and JavaConText clearly increase as the rule set grows, while FastContext’s processing time remains stable.
Table 4 shows the speed advantage of FastContext over NegScope (~4,000 times faster).
Table 4.
The average corpus processing time (in milliseconds) over 200 runs between FastContext and NegScope
| FastContext (full rule set) | NegScope (default model) | |
|---|---|---|
| Average processing time (ms) | 79* | 367,106 |
This is about twice as fast as the corresponding run reported in Table 3. We attribute this to the upgraded hardware compute node use for this comparison.
5.2. Accuracy Comparison of Three ConText Algorithm Implementations
5.2.1. Comparing The Accuracy Of Negation Detection
The accuracy, how well the models compared to the annotated reference standard from SemEval 2015, of negation detection is reflected by the average F1-scores of “Negated” detection and “Possible” detection. Figure 3 compares the average F1-scores of “Negated” detection among the three implementations. At every rule amount level, FastContext outperforms the other two.
Figure 3.
The average F1-scores for "Negation" detection of the three implementations with respect to increasing rule set size.
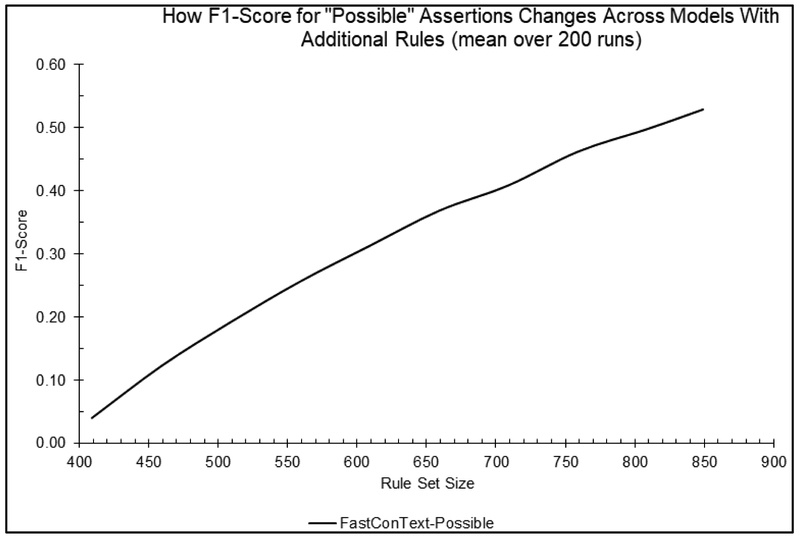
Figure 4 compares the F1-scores of the “Possible” detection for the FastContext model (JavaContext’s performance curve remained flat and well below 0.05; and GeneralContext does not report Possible matches). FastContext’s accuracy of “Possible” detection effectively improves as the rule amount grows, while JavaConText is barely effective (F1-scores well below 0.05).
Figure 4.
The Average accuracy performance for FastContext as the rule set grows (JavaContext’s performance curve remained flat and well below 0.0005; and GeneralContext does not report Possible matches).
5.2.2. Comparing the accuracy of experiencer detection
In “Other” detection (i.e., distinguishing whether an extracted concept applies to the current patient or not), FastContext exhibits superior performance compared to JavaConText and GeneralConText. In Figure 5, FastContext shows a steeper slope than the other two, indicating that FastContext uses the new added rules more effectively. JavaConText shows a slight accuracy drop as the rule set increases.
Figure 5.
The average accuracy performance for the three model implementations assessing “Other” experiencer as the rule set grows.
6. Discussion
There are many ways to improve processing efficiency [4], for example, the use of parallel processing augmented with multiple pipeline stages for slow components. This approach is usually implemented using a complex framework like UIMA-AS [37]. The approach we advocate here is far simpler, requires no additional hardware, and is well within the reach of any NLP group. We note that Divita’s principled approach [4], exploiting hardware resources using multithreads, did not achieve performance approaching the two-order of magnitude improvement we showed here. This speed improvement is crucial when processing large corpora. It saves both time and expense. With the accumulation of rules over time, the speed advantage of FastContext will be even more evident.
Importantly, the n-trie engine achieves accelerated performance without sacrificing accuracy. In our experiment, FastContext improves accuracy by avoiding the shortcomings of JavaConText and GeneralConText, such as non-exhaustive search for the most suitable rules (both models), non-discriminative to the context clues’ directions (GeneralConText), and insufficient support for all types of triggers (GeneralConText).
The speed comparison results demonstrate the scalability of Trie-based approaches. Based on this study of a context assertion clinical NLP use case, the utility of the Trie-based n-trie engine seems clear. One particularly important advantage accruing to Trie approaches is their resilience in the face of increasing rule set size (see Figures 2). JavaConText and GeneralConText show processing times that increase with the size of the rule set, while FastContext does not show a significant increase. There is a minor non-linear processing time increase, but that could be due to the unpredictability of CPU frequency fluctuation. Because FastContext runs so quickly, its runtime measurement may be more sensitive to hardware performance fluctuations, especially when compared with the runtime of the other two implementations (each takes two-orders of magnitude longer to execute). There can be some processing time increase when dealing with larger rule sets, for example, caused by more collisions or switching to larger capacity hash functions. Although this type of increase is hard to predict analytically, it is known to be less than linear.
Beyond context detection, n-trie engine can be used in other NLP tasks. A simplified version of FastContext (without tracking the context window and using “termination” rules to adjust the context window) can be directly used as a fast named entity recognition tool [38]. In addition to the simplicity and clarity of its human-readable rule grammar (e.g., as compared with the regular expressions), “pseudo” rule support can be used to easily configure rule overwrite. For instance, if we want to match “IUD” but not “IUD appointment”, a rule of “IUD” and a “pseudo” rule “IUD appointment” will work together gracefully.
We only discussed a token-based n-trie engine in this paper, mainly because context-oriented rules, in particular, are token oriented. If a few rules may be easier to write in a character-based format, these easily can be converted to tokened based versions by spelling out the tokens that represented using wildcards. Note also that the token-based engine has far better speed in this n-trie structure compared with character-based engines. Last but by no means least, as anyone who has struggled using regular expressions can attest, token-based rules are more human readable. Nevertheless, this n-trie engine does have a character-based variant with the capability of supporting character-based wildcards. This variant was implemented to detect clinical sentence boundaries, outperforming three state-of-the-art clinical text sentence segmenters [36].
Limitations
This study is limited by the evaluation set which is highly imbalanced for experiencer: only 214 out of 16,750 test cases are annotated as “Other,” which reflects the nature of clinical records in the real world, where the focus is primarily on the current patient. We avoided improper evaluation by computing only the F1-score on “Other” detection. This non-patient detection is critical for other clinical NLP scenarios, for instance, to identify the family cancer history of a patient.
We could not compare accuracy for temporality (described in section 3.3), because temporality labels are not available in the dataset. Finally, we could not make a fair accuracy comparison between FastContext and NegScope owing to dataset access limitations, but we were able to compare processing efficiency.
7. Conclusion
N-trie engine, a Trie-based rule processor designed for clinical NLP, is a fast, efficient, and scalable rule processing engine that supports a wide range of rule-based NLP tasks. Using the n-trie engine to implement the ConText algorithm is demonstrated to be effective and efficient, essentially removing the performance bottleneck of the context assertion stage in a clinical NLP pipeline (found to approach 70% of pipeline processing time by Divita et al.). In our evaluation, FastContext outperforms JavaConText and GeneralConText in terms of both speed and accuracy. In addition, FastContext's speed performance is only slightly affected by increasing rule set size. Its accuracy performance does not depend on the order of rules, which simplifies rule set customization.
Highlights.
N-trie, a new hash trie IE rule engine designed for clinical NLP, is introduced
The engine was designed to increase the efficiency/scalability of rule-base NLP
Using the ConText algorithm as a use case, N-trie exhibits superior execution time
N-trie gracefully accommodates the addition of new rules to improve accuracy
Trie-based hashing has significant potential in other rule-based NLP tasks
Acknowledgments:
We would like to acknowledge the critical technical insights and manuscript review provided by Dr. Kristina Doing-Harris, Dr. Ellen Riloff, and Sean Igo. We are grateful to Dr. Olga Patterson, Thomas Ginter, and Guy Divita for sharing the ConText rules they refined at the Salt Lake City Veterans Administration Medical Center.
Funding: This work was supported by a grant from the National Library of Medicine (NIH) grant number 5R01LM010981, as well as by operational funds from the University of Utah Hospital. Neither sponsor played a role in the design of the study nor in data collection, analysis, or interpretation.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used in this article: “F1-score” is the harmonic mean of precision and recall and is defined in section 3.2.3; “NLP” is “Natural Language Processing; “IE” is “information extraction”.
References
- [1].Ross MK, Wei W, Ohno-Machado L, “Big Data” and the Electronic Health Record, Yearb. Med. Inform 9 (2014) 97–104. doi: 10.15265/IY-2014-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Divita G, Carter M, Redd A, Zeng Q, Gupta K, Trautner B, Samore M, Gundlapalli A, Scaling-up NLP Pipelines to Process Large Corpora of Clinical Notes, Methods Inf. Med 54 (2015) 548–552. doi: 10.3414/ME14-02-0018. [DOI] [PubMed] [Google Scholar]
- [3].Jensen PB, Jensen LJ, Brunak S, Mining electronic health records: towards better research applications and clinical care, Nat. Rev. Genet 13 (2012) 395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- [4].Blumenthal D, Launching HITECH, N. Engl. J. Med 362 (2010) 382–385. doi: 10.1056/NEJMp0912825. [DOI] [PubMed] [Google Scholar]
- [5].Harkema H, Dowling JN, Thornblade T, Chapman WW, ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports, J. Biomed. Inform 42 (2009) 839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chiticariu L, Li Y, Reiss FR, Rule-Based Information Extraction is Dead! Long Live Rule-Based Information Extraction Systems!, in: EMNLP, 2013: pp. 827–832. http://www.aclweb.Org/website/old_anthology/D/D13/D13-1079.pdf (accessed September 4, 2015). [Google Scholar]
- [7].Goryachev Sergey, Sordo M, Zeng QT, Ngo L, Implementation and evaluation of four different methods of negation detection, Boston MA: DSG; (2006). http://www.researchgate.net/profile/Qing_Zeng-Treitler/publication/267552783_Implementation_and_Evaluation_of_Four_Different_Methods_of_Negation_Detection/links/54ac0cf20cf2bce6aa1dee7f.pdf (accessed July 18, 2015). [Google Scholar]
- [8].Chiticariu L, Krishnamurthy R, Li Y, Raghavan S, Reiss FR, Vaithyanathan S, SystemT: an algebraic approach to declarative information extraction, in: Proc. 48th Annu. Meet. Assoc. Comput. Linguist, Association for Computational Linguistics, 2010: pp. 128–137. http://dl.acm.org/citation.cfm?id=1858695 (accessed April 1, 2016). [Google Scholar]
- [9].Chiticariu L, Krishnamurthy R, Li Y, Reiss F, Vaithyanathan S, Domain Adaptation of Rule-based Annotators for Named-entity Recognition Tasks, in: Proc. 2010 Conf. Empir. Methods Nat. Lang. Process, Association for Computational Linguistics, Stroudsburg, PA, USA, 2010: pp. 1002–1012. http://dl.acm.org/citation.cfm?id=1870658.1870756 (accessed April 1, 2016). [Google Scholar]
- [10].Uzuner Ö, Solti I, Cadag E, Extracting medication information from clinical text, J. Am. Med. Inform. Assoc 17 (2010) 514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Reiss F, Raghavan S, Krishnamurthy R, Zhu H, Vaithyanathan S, An Algebraic Approach to Rule-Based Information Extraction, in: IEEE 24th Int. Conf. Data Eng. 2008 ICDE 2008, 2008: pp. 933–942. doi: 10.1109/ICDE.2008.4497502. [DOI] [Google Scholar]
- [12].Regular Expression Matching with a Trigram Index, (n.d.). https://swtch.com/~rsc/regexp/regexp4.html (accessed April 1, 2016). [Google Scholar]
- [13].Bentley JL, Sedgewick R, Fast Algorithms for Sorting and Searching Strings, in: Proc. Eighth Annu. ACM-SIAM Symp. Discrete Algorithms, Society for Industrial and Applied Mathematics, Philadelphia, PA, USA, 1997: pp. 360–369. http://dl.acm.org/citation.cfm?id=314161.314321 (accessed May 16, 2018). [Google Scholar]
- [14].Morrison DR, PATRICIA—Practical Algorithm To Retrieve Information Coded in Alphanumeric, J ACM. 15 (1968) 514–534. doi: 10.1145/321479.321481. [DOI] [Google Scholar]
- [15].Acharya A, Zhu H, Shen K, Adaptive Algorithms for Cache-Efficient Trie Search, in: Sel. Pap. Int. Workshop Algorithm Eng. Exp, Springer-Verlag, Berlin, Heidelberg, 1999: pp. 296–311. http://dl.acm.org/citation.cfm?id=646678.702159 (accessed May 16, 2018). [Google Scholar]
- [16].Bagwell P, Ideal Hash Trees, in: 2001. http://citeseerx.ist.psu.edu/viewdoc/citations;jsessionid=BB8EFE2C60C3CB5D01AF6F3FB329849E?doi=10.1.1.21.6279 (accessed May 16, 2018). [Google Scholar]
- [17].Cruz Díaz NP, Maña López MJ, Vázquez JM, Álvarez VP, A machine-learning approach to negation and speculation detection in clinical texts, J. Am. Soc. Inf. Sci. Technol 63 (2012) 1398–1410. doi: 10.1002/asi.22679. [DOI] [Google Scholar]
- [18].Szarvas G, Vincze V, Farkas R, Csirik J, The BioScope Corpus: Annotation for Negation, Uncertainty and Their Scope in Biomedical Texts, in: Proc. Workshop Curr. Trends Biomed. Nat. Lang. Process, Association for Computational Linguistics, Stroudsburg, PA, USA, 2008: pp. 38–45. http://dl.acm.org/citation.cfm?id=1572306.1572314 (accessed July 18, 2015). [Google Scholar]
- [19].Mutalik PG, Deshpande A, Nadkarni PM, Use of General-purpose Negation Detection to Augment Concept Indexing of Medical Documents, J. Am. Med. Inform. Assoc. JAMIA 8 (2001) 598–609. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC130070/ (accessed July 19, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].SemEval-2015 Task 14: Analysis of Clinical Text, (n.d.). http://alt.qcri.org/semeval2015/task14/ (accessed July 25, 2015).
- [21].Aronow DB, Fangfang F, Croft WB, Ad Hoc Classification of Radiology Reports, J. Am. Med. Inform. Assoc 6 (1999) 393–411. doi: 10.1136/jamia.1999.0060393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG, A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries, J. Biomed. Inform 34 (2001) 301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- [23].Chapman WW, Hilert D, Velupillai S, Kvist M, Skeppstedt M, Chapman BE, Conway M, Tharp M, Mowery DL, Deleger L, Extending the NegEx Lexicon for Multiple Languages, Stud. Health Technol. Inform 192 (2013) 677–681. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3923890/ (accessed June 22, 2015). [PMC free article] [PubMed] [Google Scholar]
- [24].Huang Y, Lowe HJ, A Novel Hybrid Approach to Automated Negation Detection in Clinical Radiology Reports, J. Am. Med. Inform. Assoc 14 (2007) 304–311. doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Morante R, Daelemans W, A Metalearning Approach to Processing the Scope of Negation, in: Proc. Thirteen. Conf. Comput. Nat. Lang. Learn, Association for Computational Linguistics, Stroudsburg, PA, USA, 2009: pp. 21–29. http://dl.acm.org/citation.cfm?id=1596374.1596381 (accessed April 10, 2016). [Google Scholar]
- [26].Fujikawa K, Seki K, Uehara K, A Hybrid Approach to Finding Negated and Uncertain Expressions in Biomedical Documents, in: Proc. 2Nd Int. Workshop Manag. Interoperability Complex. Health Syst, ACM, New York, NY, USA, 2012: pp. 67–74. doi: 10.1145/2389672.2389685. [DOI] [Google Scholar]
- [27].Agarwal S, Yu H, Biomedical negation scope detection with conditional random fields, J. Am. Med. Inform. Assoc 17 (2010) 696–701. doi: 10.1136/jamia.2010.003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cogley J, Stokes N, Carthy J, Dunnion J, Analyzing Patient Records to Establish if and when a Patient Suffered from a Medical Condition, in: Proc. 2012 Workshop Biomed. Nat. Lang. Process, Association for Computational Linguistics, Stroudsburg, PA, USA, 2012: pp. 38–46. http://dl.acm.org/citation.cfm?id=2391123.2391129 (accessed November 26, 2016). [Google Scholar]
- [29].Wu S, Masanz J, Coarr M, Halgrim S, Carrell D, Clark C, Miller T, Negation’s Not Solved: Reconsidering Negation Annotation and Evaluation, MITRE Corp. (n.d.). http://www.mitre.org/publications/technical-papers/negations-not-solved-reconsidering-negation-annotation-and-evaluation (accessed April 10, 2016). [Google Scholar]
- [30].Patterson O, Hurdle JF, Document clustering of clinical narratives: a systematic study of clinical sublanguages, AMIA Annu. Symp. Proc. AMIA Symp 2011 (2011) 1099–1107. [PMC free article] [PubMed] [Google Scholar]
- [31].Doing-Harris K, Patterson O, Igo S, Hurdle J, Document Sublanguage Clustering to Detect Medical Specialty in Cross-institutional Clinical Texts, Proc. ACM Int. Workshop Data Text Min. Biomed. Inform. ACM Int. Workshop Data Text Min. Biomed. Inform 2013 (2013) 9–12. doi: 10.1145/2512089.2512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF, others, Extracting information from textual documents in the electronic health record: a review of recent research, Yearb Med Inf. 35 (2008) 128–44. [PubMed] [Google Scholar]
- [33].Shi Jianlin, FastContext, GitHub. (n.d.). https://github.com/jianlins/FastContext (accessed December 20, 2017). [Google Scholar]
- [34].JavaConText.zip - negex - JavaConText - a Java implementation of the ConText algorithm. - negation identification for clinical conditions - Google Project Hosting, (n.d.). https://storage.googleapis.com/google-code-archive-downloads/v2/code.google.com/negex/JavaConText.zip (accessed January 5, 2018).
- [35].GeneralConText. Java.v. 1.0_10272010.zip - negex - General ConText Java Implementation v.1.0 (Imre Solti & Junebae Kye) & De-Identified Annotations for Negation (Chapman) Input: Sentence to be analyzed. Output: NEGATION scope as token offset or flag if nega, (n.d.). https://storage.googleapis.com/google-code-archive-downloads/v2/code.google.com/negex/GeneralConText.Java.v.1.0_10272010.zip (accessed January 5, 2018).
- [36].Shi Jianlin, Mowery Danielle, Doing-Harris Kristina M., Hurdle John F., RuSH: a Rule-based Segmentation Tool Using Hashing for Extremely Accurate Sentence Segmentation of Clinical Text, in: AMIA Annu Symp Proc, Chicago, Ill, 2016: p. 1587 https://knowledge.amia.org/amia-63300-1.3360278/t005-1.3362920/f005-1.3362921/2495498-1.3363244/2495498-1.3363247. [Google Scholar]
- [37].UIMA Asynchronous Scaleout, (n.d.). https://uima.apache.org/d/uima-as-2.9.0/uima_async_scaleout.html (accessed December 20, 2017). [Google Scholar]
- [38].Shi J, Mowery D, Zhang M, Sanders J, Chapman W, Gawron L, Extracting Intrauterine Device Usage from Clinical Texts Using Natural Language Processing, in: 2017 IEEE Int. Conf. Healthc. Inform. ICHI, 2017: pp. 568–571. doi: 10.1109/ICHI.2017.21. [DOI] [Google Scholar]