Abstract
The disabling degenerative disease osteoarthritis (OA) is prevalent among the global population. Articular cartilage degeneration is a central feature of OA; therefore, a better understanding of the mechanisms that maintain cartilage homeostasis is vital for developing effective therapeutic interventions. MicroRNAs (miRs) modulate cell signaling pathways and various processes in articular cartilage via posttranscriptional repression of target genes. As dysregulated miRs frequently alter the homeostasis of articular cartilage, modulating select miRs presents a potential therapeutic opportunity for OA. Here, we review key miRs that have been shown to modulate cartilage-protective or -destructive mechanisms and signaling pathways. Additionally, we use an integrative computational biology approach to provide insight into predicted miR gene targets that may contribute to OA pathogenesis, and highlight the complexity of miR signaling in OA by generating both unique and overlapping gene targets of miRs that mediate protective or destructive effects. Early OA detection would enable effective prevention; thus, miRs are being explored as diagnostic biomarkers. We discuss these ongoing efforts and the applicability of miR mimics and antisense inhibitors as potential OA therapeutics.
Introduction
MicroRNAs (miRs) are a class of small, noncoding RNAs that regulate cellular processes through RNA silencing and posttranscriptional regulation of gene expression. Primary miR (pri-miR) transcripts may originate from intergenic, intronic, or exonic regions of host genes (1). In the nucleus, pri-miRs are transcribed and then cleaved by the ribonuclease Drosha to produce a precursor-miR (pre-miR) (2). Exportin 5 transports pre-miRs to the cytoplasm where they undergo a final cleavage by the endonuclease Dicer, giving rise to mature miRs (3). Mature miRs bind to complementary messenger RNA (mRNA) sequences of target genes via the RNA-induced silencing complex (RISC). Interactions between miRs and targets with a high degree of complementarity result in mRNA degradation, while imperfect interactions between miR and target transcripts usually result in translational repression (4). Until recently, the main focus of miR research has been to identify downstream target genes and biological functions. Researchers are now exploring upstream signaling pathways that regulate miR expression. miRs are mostly regulated by the promoters of their host genes, which are transcriptionally induced in cis or trans by transcription factors (5, 6). For example, let-7e and miR-98 are estradiol-regulated miRs that reduce c-Myc and E2F2 expression in breast cancer cells (7). Estrogen can also act indirectly by inducing c-Myc to bind to the miR-17–92 promoter (8). miR-146a, which exhibits cartilage-destructive effects, is induced by LPS in an NF-κB–dependent manner in human monocytes (9). Despite substantial progress in understanding miR expression patterns in osteoarthritis (OA), their regulatory mechanisms are still widely unknown. Bioinformatic algorithms predict that each miR can regulate hundreds of mRNA targets and individual genes are regulated by multiple miRs, thereby mediating a diverse array of biological functions across most signaling cascades. Some miRs target multiple genes in the same pathway (intrapathway), while others target genes across diverse pathways (universal), emphasizing the importance of miRs in regulating whole signaling cascades (10). Not surprisingly, dysregulation of miR expression contributes to pathologies of various diseases, including cancers, neurodegenerative diseases, and metabolic disorders (11–13).
In the past few years, the OA research community has been considerably interested in miRs, especially as potential OA biomarkers and therapeutic targets. In this review, we provide a comprehensive summary of miRs known to be involved in cartilage-protective or -destructive mechanisms. Using computational approaches, we further highlight the complexity of miR regulation in articular cartilage by identifying unique and overlapping gene targets and signaling pathways of miRs that mediate protective or destructive effects to expand on their biological relevance and therapeutic potential. Finally, we discuss the current understanding of miRs as OA biomarkers and future strategies that may facilitate translation of miR-targeted therapies from bench to bedside.
Cartilage homeostasis and OA
Healthy articular cartilage is paramount to normal and pain-free joint function. Articular cartilage is a 2- to 4-mm-thick tissue of hyaline cartilage comprising chondrocytes surrounded by an extracellular matrix (ECM). The ECM primarily contains type II collagen, proteoglycans, and non-collagenous proteins, which are structurally and spatially organized for optimum tensile strength and resistance to compressive forces (14). Risk factors such as age, sex, genetics, and obesity adversely affect matrix quality, resulting in cartilage function decline (15–18). Articular cartilage degeneration, particularly in appendicular joints, is central to the clinical syndrome of OA. OA pathologies also include increased chondrocyte proliferation and fibrocartilage formation resulting from the synthesis of abnormal matrix components, predominantly type I collagen (19). Fibrocartilage lacks the appropriate biomechanical properties of articular cartilage and, consequently, undergoes degeneration (20). Intact articular cartilage that borders fibrocartilage subsequently degenerates, resulting in OA progression.
Articular cartilage cannot self-heal and, as this tissue lacks vessels and innervation, is unable to take advantage of vascular system–invoked reparative processes. As Hunter observed in 1743, cartilage “once destroyed, is not repaired” (21). Over time, catabolic activity outcompetes anabolic attempts, disrupting cartilage homeostasis. Other joint components, including the subchondral bone and synovium, contribute to cartilage destruction and OA progression through various mechanisms, such as catabolic enzyme expression, paracrine modulation of neighboring cells via inflammatory cytokines, and release of other molecular regulators, including miRs (22–24).
miRs and articular cartilage
miRs that are differentially expressed in cartilage, synovial fluid, and blood of patients with OA compared with those from healthy individuals likely contribute to OA pathophysiology (25–31). For instance, a miR signature comprising 9 increased and 7 decreased miRs was identified in human OA cartilage compared with normal cartilage (26). Proteomic analysis of predicted gene targets of OA-associated miRs identified SRY-box 11 (SOX11), CCR3, and WW domain–containing oxidoreductase (WWOX), all of which are differentially expressed in OA chondrocytes and may alter cartilage homeostasis. Thus, microarray screening can identify candidate miRs that can be used to predict mRNA targets, proteins, and pathways that may contribute to OA pathogenesis. This approach identifies potential therapeutic targets for further research.
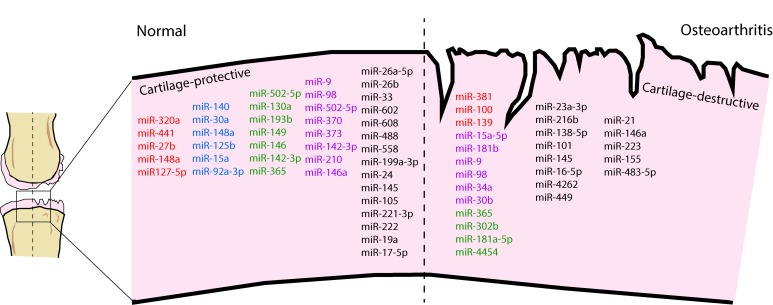
Cellular and tissue maintenance mechanisms are critical for preservation of cartilage integrity and function. Autophagy is fundamental in maintaining chondrocyte homeostasis and adjusts cell metabolism in response to various stresses by removing damaged and unnecessary intracellular organelles and proteins (32). During OA, autophagy-related proteins are markedly reduced in articular cartilage and contribute to cartilage degeneration (33). miRs regulate autophagy machinery in cartilage by directly targeting 3′-UTRs of autophagy genes, including beclin1 (BECN1) and autophagy-associated gene 5 (ATG5) (34, 35). miRs can also be packaged in 30- to 100-nm-sized extracellular vesicles called exosomes (36). Exosomes present another level of communication between joint tissues and the systemic circulation, as they are released into the synovial fluid and influx into joints from circulating blood via the vascularized synovial membrane (37, 38). Thus, miRs not only function as cell-autonomous regulatory molecules but also mediate cell-to-cell and tissue-to-tissue communication in joints. Overall, some miRs are involved in mediating protective mechanisms within the joint, while others have destructive consequences contributing to OA-associated molecular changes. We summarize miRs that have been shown to modulate cartilage-protective or -destructive mechanisms in OA in Figure 1, and describe their functional roles in Supplemental Tables 1 and 2 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121630DS1).
Figure 1. miRNAs involved in cartilage-protective and cartilage-destructive mechanisms.
miRNAs highlighted in red directly target MMPs. miRNAs highlighted in blue directly target ADAMTSs. miRNAs highlighted in green regulate inflammatory pathways. miRNAs highlighted in purple regulate cellular apoptosis mechanisms. The remaining miRNAs either maintain or disrupt the articular cartilage via alternate mechanisms. See Supplemental Tables 1 and 2.
miRs involved in cartilage-protective mechanisms
Inflammation and catabolic enzyme–regulating miRs.
In OA, articular cartilage degradation triggers inflammatory responses and cytokine production in surrounding tissues of the joint. These inflammatory molecules stimulate further ECM catabolism via increased protease production in chondrocytes (22). miR expression is affected by proinflammatory cytokines, which contribute to differential expression of target genes that promote OA progression. Furthermore, miRs can directly modulate proinflammatory cytokine expression. For instance, inhibition of miR-203 in LPS-treated mouse chondrocytes reduces cell viability, increases apoptosis, and further stimulates proinflammatory cytokine production (39). Maternally expressed gene 3 (MEG3) is a long noncoding RNA (lncRNA) that acts as a competing endogenous RNA, or sponge, for miR-203. MEG3 knockdown modulates LPS-induced inflammation by increasing miR-203 and decreasing proinflammatory cytokines (39). Overall, these studies suggest that miR-203 may have an antiinflammatory role in OA. miR-92a-3p, which is involved in late chondrogenesis of human mesenchymal stem cells (hMSCs), is downregulated in human OA cartilage and in response to IL-1β in vitro (40, 41). A disintegrin-like metalloproteinase with thrombospondin type 1 motifs (ADAMTS-4 and -5, also known as aggrecanase-1 and -2) and histone deacetylase 2 (HDAC2) are validated miR-92a-3p targets; thus, IL-1β–mediated decreases of miR-92a-3p likely promote aggrecan degradation in OA (40, 41).
Several other miRs also protect cartilage from proteolytic ECM destruction by modulating ADAMTS-5 expression. miR-140 is one of the first miRs shown to contribute to articular cartilage development and homeostasis, and normal endochondral bone development (42, 43). miR-140 is markedly downregulated in human OA cartilage compared with normal cartilage (26, 44). Moreover, miR-140 knockout in mice accelerates proteoglycan loss and fibrillation of articular cartilage by dysregulating ADAMTS-5 expression (43). In rats, intra-articular injection of miR-140 restores ECM homeostasis and prevents OA progression (45). Similarly to miR-140, miR-30a, which directly targets Adamts5, is decreased by IL-1β–induced activator protein (AP1) expression in human chondrocytes in vitro and in OA tissues in vivo (46). Thus, decreased expression of miR-140 and miR-30a in chondrocytes, in part through inflammatory signaling, may contribute to increased aggrecanase activity and the catabolic shift in OA.
Some miRs protect against OA by modulating the expression of transcription factors induced by inflammatory signaling pathways. For instance, miR-210, which is downregulated in OA tissue (26), promotes survival of LPS-treated rat chondrocytes by inhibiting NF-κB signaling and apoptosis (47). In OA chondrocytes miR-210 overexpression inhibits HIF-3α expression, further promoting chondrocyte proliferation and ECM deposition (48). Similarly, injection of miR-210–expressing lentivirus into rats with surgically induced OA reduces select cytokine levels in synovial fluid (47).
Other miRs target components of inflammatory signaling pathway receptor complexes to modulate inflammatory signaling–induced joint degeneration. For example, miR-502-5p, which is downregulated in OA articular tissues and IL-1β–induced chondrocytes (49), targets the 3′-UTR of the gene encoding TNF receptor–associated factor 2 (TRAF2), inhibiting NF-κB signaling and protecting chondrocytes from IL-1β–induced apoptosis (49). miR-145, a Sox9-mediated chondrogenic inhibitor, is downregulated in both TNF-α–induced chondrocytes and OA cartilage, resulting in induction of downstream matrix-degrading enzymes (50, 51). miR-145, regulated by p65, inhibits cartilage degradation by suppressing mitogen-activated protein kinase 4 (MKK4), which decreases matrix-degrading enzyme production and inactivates c-JUN N-terminal kinase (JNK) and p38 (50).
As nitric oxide (NO) production promotes cartilage degeneration, miRs that inhibit inducible NO synthase (iNOS) expression may impart protective effects. For instance, while miR-26a-5p directly targets IL-1β–induced iNOS in human OA chondrocytes, it is also downregulated by IL-1β signaling in chondrocytes (52), limiting its protective function. In addition to MMP-19, miR-193b-3p inhibits iNOS in human chondrocytes. miR-193b-3p promotes chondrogenic differentiation of hMSCs but is reduced in OA and in IL-1β–treated chondrocytes (53, 54). Increasing miR-193b-3p expression may inhibit degradation of ECM components and moderate inflammation through regulation of iNOS (54).
Thus, selective miRs are capable of imparting protective effects in cartilage and maintaining tissue homeostasis by targeting inflammatory signaling and cartilage catabolic components. However, reductions in expression of cartilage-protective miRs as part of OA pathogenesis may contribute towards altered cartilage homeostasis and eventual degradation.
Senescence-regulating miRs.
Both intrinsic and extrinsic mechanisms contribute to chondrocyte senescence as articular cartilage ages. Premature cell cycle arrest can occur due to stress-induced premature senescence (SIPS), such as in response to extrinsic stresses including oxidative stress and inflammation (55). Senescent chondrocytes exhibit a senescent secretory phenotype (SSP) characterized by increased IL-6, IL-1, MMPs, and growth factors. Accumulation of SSP chondrocytes alters articular cartilage homeostasis and drives aging-related cartilage destruction (56). p16INK4a, a marker of cellular senescence, is highly expressed in human OA cartilage (57), and chondrocytes with p16INK4a overexpression produce significantly higher MMP1 and MMP13 levels compared with control chondrocytes (58). Bioinformatic analysis identified miR-24, which is significantly downregulated in OA cartilage, as a negative regulator of p16INK4a. Transfection of a miR-24 antagonist in vitro markedly increases p16INK4a and MMP1 secretion, indicating that miR-24 is protective against p16INK4a-induced senescence and cartilage catabolism (58). In addition to preventing senescence, miR-24 promotes cell proliferation and inhibits chondrocyte apoptosis in rats, possibly via regulating the proto-oncogene c-Myc and downregulating MAPK signaling (59), activities that likely help in reducing OA pathogenic processes.
miRs involved in cartilage-destructive mechanisms
Chondrocyte maturation transcription factor–regulating miRs.
miRs can promote cell differentiation and increase catabolic effector gene expression by targeting various transcription-regulating factors. For example, miR-139 is increased in OA articular cartilage and inhibits cell proliferation and viability by suppressing expression of eukaryotic translation initiation factor 4 G2 (EIF4G2) and insulin-like growth factor 1 receptor (IGF1R), which is involved in cell proliferation (60, 61). miR-381, involved in late chondrogenesis and endochondral ossification, is increased in various models of cartilage degeneration and directly inhibits the expression of histone deacetylase 4 (HDAC4), a key regulator of runt-related transcription factor 2 (RUNX2) and MMP13, promoting a catabolic chondrocyte phenotype (62, 63). Mechanical stress, which contributes to articular cartilage catabolic processes and disrupts cartilage homeostasis (64), regulates expression of miR-365 via cyclical loading in vitro and in vivo (65). miR-365 is highly expressed in cartilage from primary OA patients and those with trauma-induced OA (65) and, like miR-381, directly inhibits HDAC4, promoting chondrocyte hypertrophy and catabolic enzyme expression (65, 66). A separate study showed that miR-365 is downregulated in OA cartilage, thus sparing cartilage from increased catabolic gene expression, in part, by sustaining expression of HIF-2α, another target gene of miR-365 (67). These discrepancies in miR-365 detection in OA could result from OA cartilage donor age, disease stage, or control-tissue criteria, all of which must be considered when interpreting results and selecting miR targets for therapeutic development.
Similarly to OA cartilage, miRs present in synovial fluid can help distinguish between early- and late-stage radiographic knee OA. miR-23a-3p is significantly increased in synovial fluid late-stage OA compared with early-stage OA, consistent with expression patterns in arthritic cartilage compared with normal tissue, and in synovium explants treated with IL-1β compared with untreated explants (30, 68). miR-23a-3p overexpression in OA cartilage suppresses ECM synthesis by targeting SMAD3 (68). In contrast, IL-1β signaling inhibits miR-23a-3p expression in cartilage explants (30). Collectively, these studies indicate that a negative-feedback regulatory mechanism involving inflammatory signals could manage expression of miR-23a-3p in OA.
miR-101 is increased in IL-1β–stimulated chondrocytes and mono-iodoacetate–induced (MIA-induced) OA models, and promotes OA progression, in part, by regulating expression of type II collagen (Col2a1) and aggrecan by decreasing Sox9, the essential transcription factor for chondrocyte differentiation (69, 70). In vivo, miR-101 also induces cartilage degradation–related genes including Il6, Adamts1, Adamts5, and periostin (Postn) (70). Antisense-mediated inhibition of miR-101 protects cartilage from MIA-induced degradation in rats, highlighting the feasibility of miR inhibitors as therapeutic options for OA (discussed further below). miR-145, which also directly targets SOX9 (71), is increased in human OA chondrocytes and is further enhanced by IL-1β stimulation (72). Like miR-23a-3p, Smad3 is another direct target of miR-145 (72, 73). Thus, miRs target a number of key transcription factors expressed as part of OA molecular pathology that modulate the chondrocyte phenotype and cartilage homeostasis towards a catabolic phenotype.
Apoptosis-regulating miRs.
An accumulating body of evidence suggests that chondrocyte apoptosis reduces cellularity and compromises ECM maintenance, leading to articular cartilage degradation and OA progression (74). Selected miRs are vital regulators of this cell death process. miR-98 promotes chondrocyte apoptosis, as miR-98 silencing reduces chondrocyte apoptosis and inhibits cartilage degradation in rat models of OA (75). Apoptosis-promoting effects of miR-98 are, in part, mediated through regulation of the antiapoptotic target gene B cell lymphoma 2 (Bcl-2) (76). Moreover, in vitro expression of miR-139 and miR-9 in chondrocytes reduces BCL2 and B cell lymphoma–extra large (BCLXL), promoting caspase 3/7 activity–induced apoptosis (61). miR-9 expression is significantly downregulated in both human and rat knee OA cartilage compared with normal tissues, inversely correlates with its direct target NF-κB1, and indirectly targets IL-6 and MMP13 (77, 78). Thus, modulation of miR-9 expression in OA may enhance chondrocyte proliferation and suppress apoptosis. miR-181a regulates OA pathogenesis by inducing chondrocyte apoptosis via downregulating glycerol-3-phosphate dehydrogenase 1–like protein (GPD1L) (79) and tumor suppressor PTEN (80). GDP1L regulates the hydroxylation of HIF-1α, which is vital for chondrocyte homeostasis (81). In LPS-treated chondrocytes, miR-146a expression is increased and targets CXCR4, deactivating the PI3K/AKT and Wnt/β-catenin pathways, subsequently reducing chondrocyte viability, promoting apoptosis, and increasing expression of inflammatory cytokines (82). Additional roles of miR-146a in OA pathophysiology are discussed below.
miR-34a is a p53-targeted miR with apoptotic and antiproliferative effects in some human cancers (83, 84). This miR is induced by IL-1β in rat chondrocytes and promotes apoptosis, induces iNOS expression, and decreases Col2a1 expression (85). miR-4262, which targets SIRT1, is increased in TNF-α–treated rat chondrocytes and promotes chondrocyte apoptosis and inhibits autophagy, contributing to OA pathogenesis (86). Loss of matrix synthesis proteins and elevation of matrix-degrading enzymes, such as MMP-13 and ADAMTS-5, was also observed downstream of miR-4262 (86).
In facet joint (FJ) cartilage, both miR-181a-5p and miR-4454 are upregulated. Moreover, expression of these miRs positively correlated with FJ OA severity (87). This study showed that both miRs promote chondrocyte apoptosis, inflammation, and catabolic activity in FJ cartilage.
Autophagy-regulating miRs.
In cartilage, age-related loss of autophagy has been associated with cell death and cartilage degeneration, while adequate autophagy signaling is essential for maintaining cartilage homeostasis (88–90). A number of miRs increased in OA pathology target autophagy-related genes, shifting cartilage homeostasis towards catabolism. For instance, miR-155 is upregulated in human OA knee cartilage compared with normal cartilage, based on next-generation sequencing. Analysis revealed multiple putative autophagy-related miR-155 gene targets, including ATG3, GABA type A receptor–associated protein-like 1 (GABARAPL1), ATG5, ATG2B, lysosome-associated membrane protein 2 (LAMP2), and forkhead box O3 (FOXO3) (35). In human chondrocytes, miR-155 mimic reduced mRNA and protein levels of autophagy-related target genes, as well as other nonpredicted targets, including unc-51–like autophagy-activating kinase 1 (ULK1), ATG14, and microtubule-associated protein 1 light chain 3 (MAP1LC3) (35), possibly through indirect mechanisms. miR-146a is also upregulated in OA cartilage and contributes to cartilage degeneration; however, in later disease stages, miR-146a is substantially downregulated compared with normal tissue (91–93). Aberrant miR-146a expression also contributes to pathogenesis of systemic lupus erythematosus (SLE), psoriasis, and Sjögren’s syndrome, demonstrating its role in various rheumatic diseases (94–96). Besides regulating chondrocyte apoptosis in response to mechanical injury, miR-146a promotes autophagy under hypoxic conditions (92, 97). HIF-1α induces miR-146a, leading to decreased expression of the autophagy inhibitor BCL-2. Therefore, miR-146a likely promotes both cartilage destruction and autophagy-mediated chondrocyte homeostasis, depending on disease stage. mir-17-5p belongs to the oncogenic miR-17–92 cluster, which is essential for normal skeletal growth and embryonic development (98). miR-17-5p decreases autophagy in OA chondrocytes by targeting the crucial autophagy regulator p62 (also known as SQSTM-1) (99). Compared with normal tissue, miR-20 expression is elevated in human OA chondrocytes and suppresses autophagy partly through inhibition of target gene ATG10 (100). miR-30b targets autophagy pathway regulators BECN1 and ATG5 (34), and overexpression of this miR in TNF-α–treated chondrocytes suppresses autophagy and ECM gene expression while upregulating proapoptotic genes. Inhibition of one or combinations of these miRs could protect against OA pathogenesis by promoting autophagy and associated homeostatic mechanisms in chondrocytes.
Synovial membrane contribution to joint pathology via miRs
Fibroblast-like synoviocytes (FLSs) respond to inflammatory mediators, including IL-1β, TNF-α, and ligands of TLR2, TLR3, and TLR4, by modulating miR expression profiles (101). For example, miR-155 increases in response to proinflammatory stimuli and inhibits MMP3 and MMP1 expression, thereby suppressing FLS proliferation and invasion (102, 103). TNF-α–mediated NF-κB activation induces the miR-17–92 cluster, which includes miR-18a and miR-19b, in FLSs. miR-18a increases secretion of MMP1 and inflammatory cytokines (104), whereas miR-19b increases basal cytokine production to exacerbate inflammatory activation of FLSs (105). Elevated miR-203 levels also facilitate FLS activation by increasing NF-κB–mediated expression and secretion of MMP1 and IL-6 (106). Thus, miR-18a, miR-19b, and miR-203 may contribute to FLS-mediated cartilage destruction and immune cell infiltration by aggravating the activated FLS phenotype.
The miR-29 family is highly expressed in human cartilage during end-stage OA and immediately after destabilization of the medial meniscus (DMM) surgery in mice, highlighting its involvement in early disease and maintaining articular cartilage homeostasis (107). However, miR-29 expression is diminished in end-stage knee OA synovium and associates with synovial lining hypertrophy, inflammation, and fibrosis (108). Intra-articular miR-29a injection maintains synovial homeostasis by targeting VEGF and inhibiting OA-related angiogenesis. Interestingly, reduced miR-29 is also implicated in fibrogenesis of systemic sclerosis, an autoimmune rheumatic disease (109). miR-125b-5p expression increases with OA severity (110), and miR-125b-5p overexpression inhibits OA synovial cell proliferation and promotes apoptosis by targeting synoviolin (SYVN1), suggesting miR-125b-5p upregulation is an attempt to attenuate synovial hyperplasia and fibrosis (110). Thus, miR-29 family miRs and miR-125b-5p expression may be part of an effort to maintain normal synovial function rather than contribute to pathologic OA disease progression. miRs in FLSs also target other noninflammatory, noncatabolic pathways that contribute to homeostasis or promote pathological states (see Supplemental Tables 1 and 2).
Unique and overlapping targets of cartilage-protective and -destructive miRs
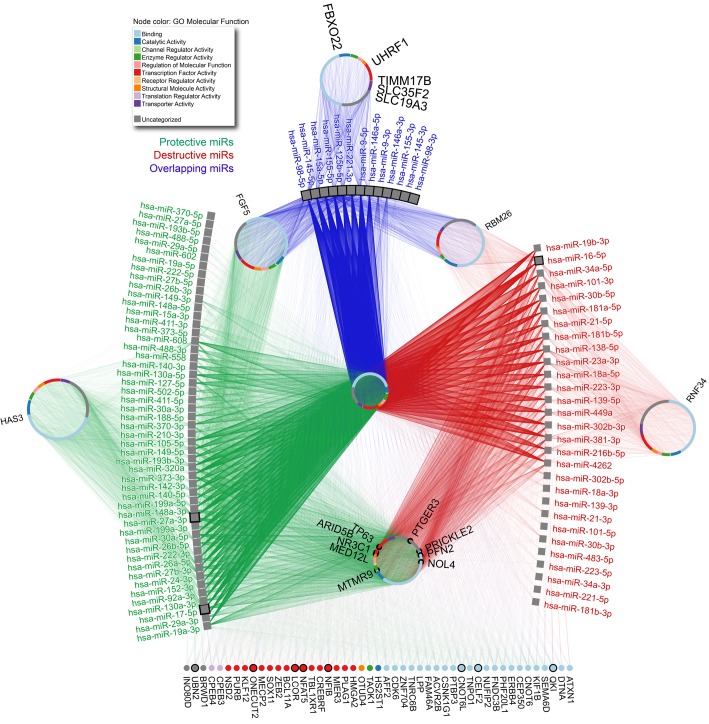
As detailed above, miRs can regulate both cartilage-protective and -destructive mechanisms, resulting in a complex signaling network. To allow investigation of a broader biological relevance and therapeutic potential of miRs in the maintenance of articular cartilage homeostasis, we used an integrative computational biology approach to identify unique and overlapping gene targets, and signaling pathways of miRs that have been reported to mediate cartilage-protective and -destructive mechanisms (Supplemental Tables 1 and 2).
First, we used mirDIP (http://ophid.utoronto.ca/mirDIP) ver. 4.1.6.6 to identify gene targets for selected miRs (111). To increase the quality of predicted targets, we focused only on predictions with very high support (top 1%), resulting in some miRs not having any targets. Second, NAViGaTOR (http://ophid.utoronto.ca/navigator) ver. 3.0.3 was used to further annotate, visualize, and prioritize identified gene targets (Figure 2) (112). This analysis identified both unique and overlapping targets, as well as genes regulated by many miRs exhibiting cartilage-protective or -destructive mechanisms. Interestingly, most of these prioritized targets (39 out of 47, see Figure 2) exhibit binding activity or transcription factor functions. Link protein N-terminal peptide (LPP) is predicted to be regulated by a high number of both protective and destructive miRs. This glycoprotein strengthens the binding between aggrecan and hyaluronan (HA) and can function as a growth factor to stimulate synthesis of type II collagen and proteoglycans in human articular cartilage and intervertebral disc (113–115). Given the number of LPP-targeting miRs, miR-mediated LPP regulation may substantially contribute to maintaining ECM integrity in OA. HA is synthesized by chondrocytes and is integral for articular cartilage structure and function; thus, modifications in size and concentration of HA can affect cartilage homeostasis (116). Our prediction indicates that HA synthase 3 (HAS3) is exclusively regulated by miRs exhibiting cartilage-protective mechanisms, suggesting that these miRs could play a role in modulating HA polymers in cartilage (117). Ring finger protein 34 (RNF34), also known as human ring finger homologous to inhibitor of apoptosis protein type (hRFI), is among the predicted genes targeted by only those miRs that exhibit cartilage-destructive mechanisms. hRFI has not been investigated in OA; however, it has antiapoptotic functions in various cancers (118, 119). Thus, select cartilage-destructive miRs may promote apoptosis by targeting RNF34. Most gene targets predicted by mirDIP have not been implicated or investigated in OA, apart from a select few. Owing to the vast number of connected miRs, the putative targets highlighted in Figure 2 are potentially significant contributors to the pathogenesis of OA. Third, we performed a comprehensive pathway enrichment analysis of identified miR targets using pathDIP (http://ophid.utoronto.ca/pathDIP) ver. 3.0.27.2 (120). We used extended pathway associations that integrate core pathways with gene-pathway associations predicted using physical protein interactions from IID (http://ophid.utoronto.ca/iid) ver. 04-2018 (121).
Figure 2. Unique and overlapping gene targets regulated by miRNAs involved in cartilage-protective and -destructive mechanisms.
Considering protective and destructive miRNAs, we used mirDIP ver. 4.1.6.6 portal to identify high-confidence mRNA targets. The resulting network was further annotated with gene ontology (GO) molecular function in NAViGaTOR version 3.0.3. Edge color corresponds to specific or overlapping miRNA-gene relationships.
A total of 2,224 pathways were significantly (P < 0.05; Bonferroni corrected for multiple testing) enriched for protective miR gene targets (including WNT pathways [n = 35; P < 2.36 × 10–6], TGF pathways [n = 28; P < 5.05 × 10–8], epidermal growth factor receptor [ERBB] pathways [n = 24; P < 3.17 × 10–4], and TNF pathways [n = 18; P < 1.78 × 10–3]). Active EGFR signaling maintains structural, functional, and mechanical properties of articular cartilage (122). Reduced EGFR signaling dramatically accelerates cartilage degeneration and OA progression in mouse models (122, 123). Term enrichment analysis of significantly enriched OA-associated pathways identified cancer, MAPK, and WNT as the most frequent terms. Extracellular stimuli, including proinflammatory cytokines, transduce signals into the nucleus, in part through MAPK signaling, activating genes that promote cellular development, proliferation, and apoptosis (124). MAPK signaling is required for chondrogenic regulation and cartilage development (125, 126) but can also negatively regulate articular chondrocyte stability by mediating catabolic responses to inflammatory cytokines. In fact, p38 MAPK signaling promotes OA chondrocyte apoptosis in response to proinflammatory stimuli (127).
A total of 2,169 pathways were significantly enriched for destructive miR targets (including WNT pathways [n = 36; P < 1.39 × 10–2], TGF pathways [n = 28; P < 5.55 × 10–10], and ERBB pathways [n = 24; P < 1.96 × 10–2]). Term enrichment analysis of OA-destructive miR targets revealed a high frequency of the terms MAPK, cancer, WNT, and regulation. The TGF-β pathway is a key signaling pathway in OA, is necessary for normal cartilage development (128), and serves both protective and destructive roles in the synovial joint (129–132). Similarly, pathway enrichment analysis predicts TGF-β signaling to be influenced by both protective and destructive miRs. WNT signaling is also among the pathways enriched in both protective and destructive miR targets, supporting previous findings that propose cartilage homeostasis depends on fine-tuning of WNT signaling and not binary activation or suppression (133).
miRs as biomarkers
As they are stable, highly sensitive, and easy to detect, miRs can be valuable OA biomarkers. Extracellular miRs in plasma, serum, or urine could be used to noninvasively diagnose or prognosticate OA severity. miRs in OA synovial fluid are similar to those secreted by synovial tissues, suggesting that these miRs potentially originate from synovial membrane tissue; however, there is no correlation between plasma and synovial fluid miRs (134). An initial screen of 752 miRs in synovial fluid from patients with early- and late-stage radiographic knee OA identified a panel of miRs capable of differentiating knee OA stage (30). miR-378-5p was detectable in late-stage OA synovial fluid but largely undetectable in early-stage OA synovial fluid, thereby providing a distinct synovial fluid miR signature with potential to predict early- from late-stage radiographic knee OA. Interestingly, increased serum miR-378 is being investigated as a biomarker for renal cell carcinoma and gastric cancer, demonstrating multiple diagnostic potentials of miR-378 in different biofluids (135, 136).
While miR-132 has diagnostic potential to differentiate healthy controls from patients with OA or rheumatoid arthritis (RA), it does not differentiate RA and OA patients. In contrast, miR-16, miR-146a, miR-155, and miR-223 are enriched in RA synovial fluid compared with OA synovial fluid, and can differentiate between patient cohorts, supporting miRs in synovial fluid as biomarkers of specific arthropathies compared with those in plasma (134). However, plasma miR expression profiles in patients with early-to-intermediate radiologic knee OA compared with healthy controls identified 12 of 380 analyzed miRs as highly expressed in OA that could clearly differentiate OA individuals from healthy controls (31). As discussed above, miR-181a-5p and miR-4454 positively correlate with the degree of facet cartilage destruction (87); however, detection of these miRs in blood and a correlation with cartilage tissue levels is required for future use as noninvasive biomarkers.
In a large prospective population-based study, 12 (miR-122, -25, - 28-3p, -93, -140, -191, -342-3p, -146b, -454, -885-5p, let-7b, and let-7e) of 377 analyzed miRs were differentially detected in serum of OA patients (29). Further validation revealed that circulating let-7e levels inversely associate with the severity of knee or hip OA. Serum levels of miR-454 and miR-885-5p also differentiate between individuals receiving joint arthroplasty and healthy controls; however, these associations are not consistently significant. Thus, specific miRs, such as let-7e, and to a lesser extent miR-454 and miR-885-5p, have potential to predict severe knee and hip OA (29). These miRs have not been investigated for early OA detection.
A recent screen of 2,549 miRs revealed that miR-140-3p, miR-671-3p, and miR-33b-3p are downregulated in the serum of OA patients compared with healthy individuals, with miR-140-3p and miR-671-3p also downregulated in OA articular cartilage compared with healthy cartilage (137). These results complement previous studies showing reduced miR-140 in OA cartilage and knee synovial fluid (26, 43, 44, 138). Target gene analysis revealed that these miRs are involved in regulating metabolic processes, such as lipid and cholesterol metabolism, that could affect OA progression (139). Larger, independent cohorts with diverse demographic and anthropometric characteristics are needed to validate these miRs as reliable biomarkers.
There is a great need for reliable biomarkers to detect early OA, as symptoms only begin to surface after cartilage is degraded past the point of intrinsic repair. While differences in miR expression profiles are evident between mouse models of posttraumatic OA and inflammatory arthritis, dysregulation of serum miRs between mouse models of arthritis and controls at early stages of disease are not detectable, suggesting circulating miRs may not be useful predictors of early cartilage degeneration. However, miRs in cartilage correlate with early disease. For example, miR-146 is highly expressed in low-grade OA cartilage and decreases with increasing cartilage degeneration, suggesting miR-146 could be an early OA indicator (140). miR-146 is also markedly downregulated in cartilage obtained from patients undergoing total knee replacements (end-stage OA) compared with normal cartilage (27). Furthermore, miR-29–family expression increases immediately upon surgical induction in a murine cartilage injury model (107). Thus, changes in miR-29–family expression appear to correlate with OA onset, while miR-146 may indicate severity. Overall, serum miRs in OA are not yet a reliable tool for detecting early stages of cartilage damage but can be predictive of OA progression.
Kung et al. recently profiled early OA mouse cartilage and subchondral bone miR expression to gain further insight into potential regulators of OA initiation (141) and discovered 139 mouse cartilage–specific miRs dysregulated early (1 and 6 weeks) after DMM surgery. Bioinformatic analysis revealed that OA pathology–associated miR-mRNA target interactions overlap with previously identified dysregulated human miRs, suggesting that these miRs (miR-15/16-5p, miR-26b-5p, miR-30c-5p, miR-98-5p, miR-149-5p, miR-210-3p, and miR-342-3p) associate with both OA initiation and progression. A similar study was conducted in an attempt to identify dysregulated synovial tissue miRs in early OA; however, no differential miR expression was observed between DMM and control mice (142).
miR mimics and antisense inhibitors as therapeutic agents
Early therapeutic intervention is crucial to improve OA patient outcomes. Reliable and accurate miR biomarkers for OA will be valuable for early diagnosis; however, specific miRs for disease progression monitoring, determination of treatment responses, or therapeutic targeting are also of interest. Several miR antagonists and replacement therapies are being studied preclinically and in clinical trials (143). The miR-122–specific inhibitor miravirsen showed promising results in a completed phase IIa clinical trial of patients with chronic hepatitis C (ClinicalTrials.gov Identifier NCT01200420). The miR-34 mimic MRX34 reduces cell proliferation in multiple cancers and inhibits the formation of cancer stem cells in preclinical studies (144–146). In 2013, MRX34 was the first miR mimic to enter clinical trials; however, the phase 1 study was halted in September of 2016 due to multiple immune-related severe adverse events (ClinicalTrials.gov Identifier NCT01829971). Miragen Therapeutics has miR antagonist antimiR-155 and miR mimic promiR-29b in clinical trials to treat cutaneous T cell lymphoma and fibrosis, respectively (ClinicalTrials.gov Identifier NCT02580552 and NCT02603224). Interestingly, miR-155 is upregulated in OA chondrocytes and contributes to autophagy dysfunction and OA pathogenesis; however, in vitro inhibition of miR-155 is chondroprotective by enhancing autophagy (35). One advantage of miR therapeutics for treating OA is the ability to locally deliver treatment via intra-articular injection. The encapsulated and isolated structure of a synovial joint allows for fewer off-target effects and adverse events that can result from systemic exposure (as seen with MRX34), where miR expression and function may differ from pathological or homeostatic roles in the joint. miR-targeted therapy appears to be a promising therapeutic avenue; however, off-target effects are possible due to multiple gene targets of miRs (direct or indirect).
In preclinical and clinical studies, mesenchymal stem/stromal cells (MSCs) can protect and repair articular cartilage (147–161), and the MSC secretome, by way of exosomes, has been shown to possess paracrine factors required to mediate tissue repair (162–165). In accordance with these findings, injection of MSC-derived exosomes on a weekly basis repairs osteochondral defects in a rat model (166). The cartilage regenerative effects of embryonic stem cell–derived (ESC-derived) MSC exosomes were also investigated in a DMM-induced mouse OA model (167). Intra-articular injection of ESC-derived MSC exosomes twice a week for 4 weeks prevented cartilage destruction, increased type II collagen, and reduced ADAMTS5 expression. This approach represents a cell-free, lipid-based, safer therapeutic approach for administration of disease-modifying factors and eliminates challenges of cell-based MSC therapies. Of particular relevance, Chen et al. showed that besides proteins and lipids, biologically functional pre-miRs are enriched in secreted exosomes and can exert their functions after being readily taken up by cells (168). Experiments in which argonaut-2 (Ago2), a regulator of the biological function of miRs, is knocked down showed that miR composition mediates the neuroprotective effect of exosomes for treatment of degenerative ocular disease (169). Tao et al. showed that overexpressed miR-140-5p in synovial MSC exosomes promotes chondrocyte proliferation and migration, thereby delaying progression in an OA rat model (170). Thus, it is possible to manipulate the miR content in exosomes to modulate miR expression in the joint and restore joint homeostasis. While further characterization of MSC-exosome miRs is required, these studies show a potential application of MSC-exosome miRs — with or without miR modification — for OA treatment. Additionally, synthetic lipid–based vesicles resembling exosomes could be generated with a subset of miR mimics and antisense inhibitors to mediate OA disease modification (171). Thus, exosomes could serve as a delivery tool for OA-modulating miRs.
Conclusions
Research of OA-associated miRs is still in its infancy and further work is required before translation to clinical application. Research advances in various fields highlight the potential of miRs as indicators of disease activity and therapeutic targets, with preclinical animal models of OA producing encouraging results. miRs are not purely tissue specific and many remain to be identified. This further emphasizes the need for unbiased, comprehensive, sequencing-based assays combined with systematic computational analysis to identify OA miRs for diagnosis, prognosis, and treatment. Of note, some miRs that actively maintain articular cartilage homeostasis (protection or destruction) may operate through common gene targets and signaling pathways, as identified by our integrated computational analysis. Such comprehensive analysis may open up new therapeutic avenues for targeting multiple miRs by modulating common downstream gene targets or signaling pathways. These data also suggest that targeting miRs for therapeutic benefit may pose significant challenges, as miRs operate through multiple gene targets and signaling pathways.
Author contributions
HE and MK were responsible for the conception and design of the review. HE was responsible for data analysis and interpretation and was the major contributor in drafting the review. JR contributed to the design, critical revision, and editing of the review. IJ analyzed data, performed computational analyses for miR gene target and pathway predictions, and interpreted the data. All authors read and approved the final manuscript to be published.
Supplementary Material
Acknowledgments
This review was prepared by searching PubMed and ClinicalTrials.gov databases. The PubMed search was restricted to English-language, full-length articles. Main key search terms were osteoarthritis, microRNA, articular cartilage, homeostasis, biomarker, chondrogenesis, synovial fibroblasts, facet cartilage, and clinical trial. The ClinicalTrials.gov database search was limited to relevant therapies. M. Kapoor is supported in part by the Krembil Foundation, Canadian Institute of Health Research (grant 285893), Canadian Foundation for Innovation, Stem Cell Network and The Arthritis Program, University Health Network. I. Jurisica is supported in part by the Canada Research Chair Program (CRC grant 225404), Krembil Foundation, Ontario Research Fund (grants GL2-01-030 and 34876), Natural Sciences Research Council (NSERC grant 203475), Canada Foundation for Innovation (CFI grants 225404 and 30865), and IBM.
Version 1. 09/06/2018
Electronic publication
Footnotes
Conflict of interest: A US provisional patent application (62/299,305, filed February 24, 2016) and a PCT international patent application (PCT/CA2017/000019, filed Jan 31, 2017) have been filed with respect to therapeutic and diagnostic uses of miRNA-181a-5p and -4454.
Published: September 6, 2018
Reference information: JCI Insight. 2018;3(17):e121630. https://doi.org/10.1172/jci.insight.121630.
Contributor Information
Helal Endisha, Email: helal.endisha@mail.utoronto.ca.
Igor Jurisica, Email: juris@ai.utoronto.ca.
Mohit Kapoor, Email: mkapoor@uhnresearch.ca.
References
- 1.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1(1):31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22(22):3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat-Nakshatri P, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37(14):4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellano L, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106(37):15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome--using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One. 2011;6(2):e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Femminella GD, Ferrara N, Rengo G. The emerging role of microRNAs in Alzheimer’s disease. Front Physiol. 2015;6:40. doi: 10.3389/fphys.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erggelet C, Mandelbaum BR. Articular cartilage biology. In: Principles of Cartilage Repair. Heidelberg, Germany: Steinkopff-Verlag Heidelberg; 2008:3–5. [Google Scholar]
- 15.Temple MM, et al. Age- and site-associated biomechanical weakening of human articular cartilage of the femoral condyle. Osteoarthr Cartil. 2007;15(9):1042–1052. doi: 10.1016/j.joca.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Hudelmaier M, et al. Age-related changes in the morphology and deformational behavior of knee joint cartilage. Arthritis Rheum. 2001;44(11):2556–2561. doi: 10.1002/1529-0131(200111)44:11<2556::AID-ART436>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Kizawa H, et al. An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat Genet. 2005;37(2):138–144. doi: 10.1038/ng1496. [DOI] [PubMed] [Google Scholar]
- 18.Mouritzen U, Christgau S, Lehmann HJ, Tankó LB, Christiansen C. Cartilage turnover assessed with a newly developed assay measuring collagen type II degradation products: influence of age, sex, menopause, hormone replacement therapy, and body mass index. Ann Rheum Dis. 2003;62(4):332–336. doi: 10.1136/ard.62.4.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maldonado M, Nam J. The role of changes in extracellular matrix of cartilage in the presence of inflammation on the pathology of osteoarthritis. Biomed Res Int. 2013;2013:284873. doi: 10.1155/2013/284873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae DK, Yoon KH, Song SJ. Cartilage healing after microfracture in osteoarthritic knees. Arthroscopy. 2006;22(4):367–374. doi: 10.1016/j.arthro.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Hunter W. Of the structure and diseases of articulating cartilages, by William Hunter, surgeon. Philosophical Transactions. 1742;42(462–471):514–521. [Google Scholar]
- 22.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7(1):33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 23.Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011;23(5):471–478. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato T, et al. Exosomes from IL-1β stimulated synovial fibroblasts induce osteoarthritic changes in articular chondrocytes. Arthritis Res Ther. 2014;16(4):R163. doi: 10.1186/ar4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012;18(2):109–118. doi: 10.1016/j.molmed.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3(11):e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones SW, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthr Cartil. 2009;17(4):464–472. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Díaz-Prado S, et al. Characterization of microRNA expression profiles in normal and osteoarthritic human chondrocytes. BMC Musculoskelet Disord. 2012;13:144. doi: 10.1186/1471-2474-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyer C, et al. Signature of circulating microRNAs in osteoarthritis. Ann Rheum Dis. 2015;74(3):e18. doi: 10.1136/annrheumdis-2013-204698. [DOI] [PubMed] [Google Scholar]
- 30.Li YH, et al. Identification of synovial fluid microRNA signature in knee osteoarthritis: differentiating early- and late-stage knee osteoarthritis. Osteoarthr Cartil. 2016;24(9):1577–1586. doi: 10.1016/j.joca.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 31.Borgonio Cuadra VM, González-Huerta NC, Romero-Córdoba S, Hidalgo-Miranda A, Miranda-Duarte A. Altered expression of circulating microRNA in plasma of patients with primary osteoarthritis and in silico analysis of their pathways. PLoS One. 2014;9(6):e97690. doi: 10.1371/journal.pone.0097690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lotz MK, Caramés B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nat Rev Rheumatol. 2011;7(10):579–587. doi: 10.1038/nrrheum.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockel JS, Kapoor M. Autophagy: controlling cell fate in rheumatic diseases. Nat Rev Rheumatol. 2016;12(9):517–531. doi: 10.1038/nrrheum.2016.92. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Jin T, Lu Y. AntimiR-30b inhibits TNF-α mediated apoptosis and attenuated cartilage degradation through enhancing autophagy. Cell Physiol Biochem. 2016;40(5):883–894. doi: 10.1159/000453147. [DOI] [PubMed] [Google Scholar]
- 35.D’Adamo S, Alvarez-Garcia O, Muramatsu Y, Flamigni F, Lotz MK. MicroRNA-155 suppresses autophagy in chondrocytes by modulating expression of autophagy proteins. Osteoarthr Cartil. 2016;24(6):1082–1091. doi: 10.1016/j.joca.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosenza S, Ruiz M, Maumus M, Jorgensen C, Noël D. Pathogenic or Therapeutic Extracellular Vesicles in Rheumatic Diseases: Role of Mesenchymal Stem Cell-Derived Vesicles. Int J Mol Sci. 2017;18(4):889. doi: 10.3390/ijms18040889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cosenza S, Ruiz M, Toupet K, Jorgensen C, Noël D. Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci Rep. 2017;7(1):16214. doi: 10.1038/s41598-017-15376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, et al. Long noncoding RNA maternally expressed gene 3 knockdown alleviates lipopolysaccharide-induced inflammatory injury by up-regulation of miR-203 in ATDC5 cells. Biomed Pharmacother. 2018;100:240–249. doi: 10.1016/j.biopha.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Mao G, et al. MicroRNA-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthr Cartil. 2017;25(4):521–532. doi: 10.1016/j.joca.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Mao G, et al. MicroRNA-92a-3p regulates aggrecanase-1 and aggrecanase-2 expression in chondrogenesis and IL-1β-induced catabolism in human articular chondrocytes. Cell Physiol Biochem. 2017;44(1):38–52. doi: 10.1159/000484579. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31(14):3019–3028. doi: 10.1128/MCB.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyaki S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24(11):1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyaki S, et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60(9):2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Si HB, et al. Intra-articular injection of microRNA-140 (miRNA-140) alleviates osteoarthritis (OA) progression by modulating extracellular matrix (ECM) homeostasis in rats. Osteoarthr Cartil. 2017;25(10):1698–1707. doi: 10.1016/j.joca.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Ji Q, et al. The IL-1β/AP-1/miR-30a/ADAMTS-5 axis regulates cartilage matrix degradation in human osteoarthritis. J Mol Med. 2016;94(7):771–785. doi: 10.1007/s00109-016-1418-z. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D, Cao X, Li J, Zhao G. MiR-210 inhibits NF-κB signaling pathway by targeting DR6 in osteoarthritis. Sci Rep. 2015;5:12775. doi: 10.1038/srep12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Meng D, Li G, Xu J, Tian K, Li Y. Overexpression of microRNA-210 promotes chondrocyte proliferation and extracellular matrix deposition by targeting HIF-3α in osteoarthritis. Mol Med Rep. 2016;13(3):2769–2776. doi: 10.3892/mmr.2016.4878. [DOI] [PubMed] [Google Scholar]
- 49.Zhang G, Sun Y, Wang Y, Liu R, Bao Y, Li Q. MiR-502-5p inhibits IL-1β-induced chondrocyte injury by targeting TRAF2. Cell Immunol. 2016;302:50–57. doi: 10.1016/j.cellimm.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 50.Hu G, et al. MicroRNA-145 attenuates TNF-α-driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis. 2017;8(10):e3140. doi: 10.1038/cddis.2017.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 2011;6(7):e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rasheed Z, Al-Shobaili HA, Rasheed N, Mahmood A, Khan MI. MicroRNA-26a-5p regulates the expression of inducible nitric oxide synthase via activation of NF-κB pathway in human osteoarthritis chondrocytes. Arch Biochem Biophys. 2016;594:61–67. doi: 10.1016/j.abb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Meng F, et al. MicroRNA-193b-3p regulates chondrogenesis and chondrocyte metabolism by targeting HDAC3. Theranostics. 2018;8(10):2862–2883. doi: 10.7150/thno.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang ZK, et al. MicroRNA-193b-3p regulates matrix metalloproteinase 19 expression in interleukin-1β-induced human chondrocytes. J Cell Biochem. 2018;119(6):4775–4782. doi: 10.1002/jcb.26669. [DOI] [PubMed] [Google Scholar]
- 55.Mobasheri A, Matta C, Zákány R, Musumeci G. Chondrosenescence: definition, hallmarks and potential role in the pathogenesis of osteoarthritis. Maturitas. 2015;80(3):237–244. doi: 10.1016/j.maturitas.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Loeser RF. Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthr Cartil. 2009;17(8):971–979. doi: 10.1016/j.joca.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou HW, Lou SQ, Zhang K. Recovery of function in osteoarthritic chondrocytes induced by p16INK4a-specific siRNA in vitro. Rheumatology (Oxford) 2004;43(5):555–568. doi: 10.1093/rheumatology/keh127. [DOI] [PubMed] [Google Scholar]
- 58.Philipot D, et al. p16INK4a and its regulator miR-24 link senescence and chondrocyte terminal differentiation-associated matrix remodeling in osteoarthritis. Arthritis Res Ther. 2014;16(1):R58. doi: 10.1186/ar4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu YH, et al. Effects of microRNA-24 targeting C-myc on apoptosis, proliferation and cytokine expressions in chondrocytes of rats with osteoarthritis via MAPK signaling pathway. J Cell Biochem. doi: 10.1002/jcb.26514. doi: 10.1002/jcb.26514. [published online ahead of print November 16, 2017]. [DOI] [PubMed] [Google Scholar]
- 60.Hu W, Zhang W, Li F, Guo F, Chen A. miR-139 is up-regulated in osteoarthritis and inhibits chondrocyte proliferation and migration possibly via suppressing EIF4G2 and IGF1R. Biochem Biophys Res Commun. 2016;474(2):296–302. doi: 10.1016/j.bbrc.2016.03.164. [DOI] [PubMed] [Google Scholar]
- 61.Makki MS, Haqqi TM. miR-139 modulates MCPIP1/IL-6 expression and induces apoptosis in human OA chondrocytes. Exp Mol Med. 2015;47:e189. doi: 10.1038/emm.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen W, et al. MicroRNA-381 Regulates Chondrocyte Hypertrophy by Inhibiting Histone Deacetylase 4 Expression. Int J Mol Sci. 2016;17(9):1377. doi: 10.3390/ijms17091377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hou C, et al. The role of microRNA-381 in chondrogenesis and interleukin-1-β induced chondrocyte responses. Cell Physiol Biochem. 2015;36(5):1753–1766. doi: 10.1159/000430148. [DOI] [PubMed] [Google Scholar]
- 64.Buckwalter JA, Anderson DD, Brown TD, Tochigi Y, Martin JA. The roles of mechanical stresses in the pathogenesis of osteoarthritis: implications for treatment of joint injuries. Cartilage. 2013;4(4):286–294. doi: 10.1177/1947603513495889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X, Guan Y, Tian S, Wang Y, Sun K, Chen Q. Mechanical and IL-1β responsive miR-365 contributes to osteoarthritis development by targeting histone deacetylase 4. Int J Mol Sci. 2016;17(4):436. doi: 10.3390/ijms17040436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Usui T, et al. HDAC4 mediates development of hypertension via vascular inflammation in spontaneous hypertensive rats. Am J Physiol Heart Circ Physiol. 2012;302(9):H1894–H1904. doi: 10.1152/ajpheart.01039.2011. [DOI] [PubMed] [Google Scholar]
- 67.Hwang HS, Park SJ, Lee MH, Kim HA. MicroRNA-365 regulates IL-1β-induced catabolic factor expression by targeting HIF-2α in primary chondrocytes. Sci Rep. 2017;7(1):17889. doi: 10.1038/s41598-017-18059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang L, et al. MicroRNA-23a-3p promotes the development of osteoarthritis by directly targeting SMAD3 in chondrocytes. Biochem Biophys Res Commun. 2016;478(1):467–473. doi: 10.1016/j.bbrc.2016.06.071. [DOI] [PubMed] [Google Scholar]
- 69.Dai L, Zhang X, Hu X, Zhou C, Ao Y. Silencing of microRNA-101 prevents IL-1β-induced extracellular matrix degradation in chondrocytes. Arthritis Res Ther. 2012;14(6):R268. doi: 10.1186/ar4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai L, et al. Silencing of miR-101 prevents cartilage degradation by regulating extracellular matrix-related genes in a rat model of osteoarthritis. Mol Ther. 2015;23(8):1331–1340. doi: 10.1038/mt.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287(2):916–924. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang B, et al. Effect of microRNA-145 on IL-1β-induced cartilage degradation in human chondrocytes. FEBS Lett. 2014;588(14):2344–2352. doi: 10.1016/j.febslet.2014.05.033. [DOI] [PubMed] [Google Scholar]
- 73.Chen CG, Thuillier D, Chin EN, Alliston T. Chondrocyte-intrinsic Smad3 represses Runx2-inducible matrix metalloproteinase 13 expression to maintain articular cartilage and prevent osteoarthritis. Arthritis Rheum. 2012;64(10):3278–3289. doi: 10.1002/art.34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aigner T, Söder S, Gebhard PM, McAlinden A, Haag J. Mechanisms of disease: role of chondrocytes in the pathogenesis of osteoarthritis--structure, chaos and senescence. Nat Clin Pract Rheumatol. 2007;3(7):391–399. doi: 10.1038/ncprheum0534. [DOI] [PubMed] [Google Scholar]
- 75.Wang J, et al. MiR-98 promotes chondrocyte apoptosis by decreasing Bcl-2 expression in a rat model of osteoarthritis. Acta Biochim Biophys Sin (Shanghai) 2016;48(10):923–929. doi: 10.1093/abbs/gmw084. [DOI] [PubMed] [Google Scholar]
- 76.Boise LH, Gottschalk AR, Quintáns J, Thompson CB. Bcl-2 and Bcl-2-related proteins in apoptosis regulation. Curr Top Microbiol Immunol. 1995;200:107–121. doi: 10.1007/978-3-642-79437-7_8. [DOI] [PubMed] [Google Scholar]
- 77.Gu R, Liu N, Luo S, Huang W, Zha Z, Yang J. MicroRNA-9 regulates the development of knee osteoarthritis through the NF-kappaB1 pathway in chondrocytes. Medicine (Baltimore) 2016;95(36):e4315. doi: 10.1097/MD.0000000000004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang H, Song B, Pan Z. Downregulation of microRNA-9 increases matrix metalloproteinase-13 expression levels and facilitates osteoarthritis onset. Mol Med Rep. 2018;17(3):3708–3714. doi: 10.3892/mmr.2017.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhai X, et al. miR-181a modulates chondrocyte apoptosis by targeting glycerol-3-phosphate dehydrogenase 1-like protein (GPD1L) in osteoarthritis. Med Sci Monit. 2017;23:1224–1231. doi: 10.12659/MSM.899228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu XF, Zhou ZH, Zou J. MicroRNA-181 inhibits proliferation and promotes apoptosis of chondrocytes in osteoarthritis by targeting PTEN. Biochem Cell Biol. 2017;95(3):437–444. doi: 10.1139/bcb-2016-0078. [DOI] [PubMed] [Google Scholar]
- 81.Yudoh K, Nakamura H, Masuko-Hongo K, Kato T, Nishioka K. Catabolic stress induces expression of hypoxia-inducible factor (HIF)-1 alpha in articular chondrocytes: involvement of HIF-1 alpha in the pathogenesis of osteoarthritis. Arthritis Res Ther. 2005;7(4):R904–R914. doi: 10.1186/ar1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun T, et al. MiR-146a aggravates LPS-induced inflammatory injury by targeting CXCR4 in the articular chondrocytes. Cell Physiol Biochem. 2017;44(4):1282–1294. doi: 10.1159/000485488. [DOI] [PubMed] [Google Scholar]
- 83.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 85.Abouheif MM, Nakasa T, Shibuya H, Niimoto T, Kongcharoensombat W, Ochi M. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 2010;49(11):2054–2060. doi: 10.1093/rheumatology/keq247. [DOI] [PubMed] [Google Scholar]
- 86.Sun W, Li Y, Wei S. miR-4262 regulates chondrocyte viability, apoptosis, autophagy by targeting SIRT1 and activating PI3K/AKT/mTOR signaling pathway in rats with osteoarthritis. Exp Ther Med. 2018;15(1):1119–1128. doi: 10.3892/etm.2017.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakamura A, et al. Identification of microRNA-181a-5p and microRNA-4454 as mediators of facet cartilage degeneration. JCI Insight. 2016;1(12):e86820. doi: 10.1172/jci.insight.86820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carames B, et al. The relationship of autophagy defects to cartilage damage during joint aging in a mouse model. Arthritis Rheumatol. 2015;67(6):1568–1576. doi: 10.1002/art.39073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caramés B, Hasegawa A, Taniguchi N, Miyaki S, Blanco FJ, Lotz M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann Rheum Dis. 2012;71(4):575–581. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. 2015;74(7):1432–1440. doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 91.Li J, et al. miR-146a, an IL-1β responsive miRNA, induces vascular endothelial growth factor and chondrocyte apoptosis by targeting Smad4. Arthritis Res Ther. 2012;14(2):R75. doi: 10.1186/ar3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jin L, et al. Role of miR-146a in human chondrocyte apoptosis in response to mechanical pressure injury in vitro. Int J Mol Med. 2014;34(2):451–463. doi: 10.3892/ijmm.2014.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jones SW, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthr Cartil. 2009;17(4):464–472. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 94.Tang Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 95.Pauley KM, et al. Altered miR-146a expression in Sjögren’s syndrome and its functional role in innate immunity. Eur J Immunol. 2011;41(7):2029–2039. doi: 10.1002/eji.201040757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sonkoly E, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2(7):e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang F, et al. MicroRNA-146a induced by hypoxia promotes chondrocyte autophagy through Bcl-2. Cell Physiol Biochem. 2015;37(4):1442–1453. doi: 10.1159/000438513. [DOI] [PubMed] [Google Scholar]
- 98.de Pontual L, et al. Germline deletion of the miR-17~92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43(10):1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H, Miao D, Zhu Q, Huang J, Lu G, Xu W. MicroRNA-17-5p contributes to osteoarthritis progression by binding p62/SQSTM1. Exp Ther Med. 2018;15(2):1789–1794. doi: 10.3892/etm.2017.5622. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.He W, Cheng Y. Inhibition of miR-20 promotes proliferation and autophagy in articular chondrocytes by PI3K/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;97:607–615. doi: 10.1016/j.biopha.2017.10.152. [DOI] [PubMed] [Google Scholar]
- 101.Singh RP, et al. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12(12):1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 102.Stanczyk J, et al. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 103.Long L, et al. Upregulated microRNA-155 expression in peripheral blood mononuclear cells and fibroblast-like synoviocytes in rheumatoid arthritis. Clin Dev Immunol. 2013;2013:296139. doi: 10.1155/2013/296139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Trenkmann M, Brock M, Gay RE, Michel BA, Gay S, Huber LC. Tumor necrosis factor α-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-κB signaling. Arthritis Rheum. 2013;65(4):916–927. doi: 10.1002/art.37834. [DOI] [PubMed] [Google Scholar]
- 105.Gantier MP, et al. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res. 2012;40(16):8048–8058. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stanczyk J, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63(2):373–381. doi: 10.1002/art.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Le LT, et al. The microRNA-29 family in cartilage homeostasis and osteoarthritis. J Mol Med. 2016;94(5):583–596. doi: 10.1007/s00109-015-1374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ko JY, et al. MicroRNA-29a counteracts synovitis in knee osteoarthritis pathogenesis by targeting VEGF. Sci Rep. 2017;7(1):3584. doi: 10.1038/s41598-017-03616-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maurer B, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62(6):1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 110.Ge FX, Li H, Yin X. Upregulation of microRNA-125b-5p is involved in the pathogenesis of osteoarthritis by downregulating SYVN1. Oncol Rep. 2017;37(4):2490–2496. doi: 10.3892/or.2017.5475. [DOI] [PubMed] [Google Scholar]
- 111.Tokar T, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brown KR, et al. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinformatics. 2009;25(24):3327–3329. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu H, McKenna LA, Dean MF. An N-terminal peptide from link protein can stimulate biosynthesis of collagen by human articular cartilage. Arch Biochem Biophys. 2000;378(1):116–122. doi: 10.1006/abbi.2000.1758. [DOI] [PubMed] [Google Scholar]
- 114.Nguyen Q, Liu J, Roughley PJ, Mort JS. Link protein as a monitor in situ of endogenous proteolysis in adult human articular cartilage. Biochem J. 1991;278(Pt 1):143–147. doi: 10.1042/bj2780143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.He R, et al. Link protein N-terminal peptide as a potential stimulating factor for stem cell-based cartilage regeneration. Stem Cells Int. 2018;2018:3217895. doi: 10.1155/2018/3217895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Holmes MW, Bayliss MT, Muir H. Hyaluronic acid in human articular cartilage. Age-related changes in content and size. Biochem J. 1988;250(2):435–441. doi: 10.1042/bj2500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hiscock DR, Caterson B, Flannery CR. Expression of hyaluronan synthases in articular cartilage. Osteoarthr Cartil. 2000;8(2):120–126. doi: 10.1053/joca.1999.0280. [DOI] [PubMed] [Google Scholar]
- 118.Konishi T, Sasaki S, Watanabe T, Kitayama J, Nagawa H. Overexpression of hRFI (human ring finger homologous to inhibitor of apoptosis protein type) inhibits death receptor-mediated apoptosis in colorectal cancer cells. Mol Cancer Ther. 2005;4(5):743–750. doi: 10.1158/1535-7163.MCT-05-0020. [DOI] [PubMed] [Google Scholar]
- 119.Sasaki S, Watanabe T, Konishi T, Kitayama J, Nagawa H. Effects of expression of hRFI on adenoma formation and tumor progression in colorectal adenoma-carcinoma sequence. J Exp Clin Cancer Res. 2004;23(3):507–512. [PubMed] [Google Scholar]
- 120.Rahmati S, Abovsky M, Pastrello C, Jurisica I. pathDIP: an annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucleic Acids Res. 2017;45(D1):D419–D426. doi: 10.1093/nar/gkw1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kotlyar M, Pastrello C, Sheahan N, Jurisica I. Integrated interactions database: tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2016;44(D1):D536–D541. doi: 10.1093/nar/gkv1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jia H, et al. EGFR signaling is critical for maintaining the superficial layer of articular cartilage and preventing osteoarthritis initiation. Proc Natl Acad Sci USA. 2016;113(50):14360–14365. doi: 10.1073/pnas.1608938113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang X, et al. Reduced EGFR signaling enhances cartilage destruction in a mouse osteoarthritis model. Bone Res. 2014;2:14015. doi: 10.1038/boneres.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Keshet Y, Seger R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 2010;661:3–38. doi: 10.1007/978-1-60761-795-2_1. [DOI] [PubMed] [Google Scholar]
- 125.Oh CD, et al. Opposing role of mitogen-activated protein kinase subtypes, erk-1/2 and p38, in the regulation of chondrogenesis of mesenchymes. J Biol Chem. 2000;275(8):5613–5619. doi: 10.1074/jbc.275.8.5613. [DOI] [PubMed] [Google Scholar]
- 126.Stanton LA, Underhill TM, Beier F. MAP kinases in chondrocyte differentiation. Dev Biol. 2003;263(2):165–175. doi: 10.1016/S0012-1606(03)00321-X. [DOI] [PubMed] [Google Scholar]
- 127.Sun HY, Hu KZ, Yin ZS. Inhibition of the p38-MAPK signaling pathway suppresses the apoptosis and expression of proinflammatory cytokines in human osteoarthritis chondrocytes. Cytokine. 2017;90:135–143. doi: 10.1016/j.cyto.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 128.Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153(1):35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van der Kraan PM. The changing role of TGFβ in healthy, ageing and osteoarthritic joints. Nat Rev Rheumatol. 2017;13(3):155–163. doi: 10.1038/nrrheum.2016.219. [DOI] [PubMed] [Google Scholar]
- 130.Finnson KW, Chi Y, Bou-Gharios G, Leask A, Philip A. TGF-b signaling in cartilage homeostasis and osteoarthritis. Front Biosci (Schol Ed) 2012;4:251–268. doi: 10.2741/S266. [DOI] [PubMed] [Google Scholar]
- 131.Bush JR, Beier F. TGF-β and osteoarthritis--the good and the bad. Nat Med. 2013;19(6):667–669. doi: 10.1038/nm.3228. [DOI] [PubMed] [Google Scholar]
- 132.Zhen G, et al. Inhibition of TGF-β signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med. 2013;19(6):704–712. doi: 10.1038/nm.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lories RJ, Corr M, Lane NE. To Wnt or not to Wnt: the bone and joint health dilemma. Nat Rev Rheumatol. 2013;9(6):328–339. doi: 10.1038/nrrheum.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Murata K, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12(3):R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Redova M, et al. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med. 2012;10:55. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu H, et al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316(2):196–203. doi: 10.1016/j.canlet.2011.10.034. [DOI] [PubMed] [Google Scholar]
- 137.Ntoumou E, et al. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenetics. 2017;9:127. doi: 10.1186/s13148-017-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yin CM, Suen WC, Lin S, Wu XM, Li G, Pan XH. Dysregulation of both miR-140-3p and miR-140-5p in synovial fluid correlate with osteoarthritis severity. Bone Joint Res. 2017;6(11):612–618. doi: 10.1302/2046-3758.611.BJR-2017-0090.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ali SA, et al. Regulation of cholesterol homeostasis by hedgehog signaling in osteoarthritic cartilage. Arthritis Rheumatol. 2016;68(1):127–137. doi: 10.1002/art.39337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yamasaki K, et al. Expression of microRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60(4):1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kung LHW, et al. Cartilage microRNA dysregulation during the onset and progression of mouse osteoarthritis is independent of aggrecanolysis and overlaps with candidates from end-stage human disease. Arthritis Rheumatol. 2018;70(3):383–395. doi: 10.1002/art.40378. [DOI] [PubMed] [Google Scholar]
- 142.Kung LHW, Ravi V, Rowley L, Bell KM, Little CB, Bateman JF. Comprehensive expression analysis of microRNAs and mRNAs in synovial tissue from a mouse model of early post-traumatic osteoarthritis. Sci Rep. 2017;7(1):17701. doi: 10.1038/s41598-017-17545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bader AG, Lammers P. The therapeutic potential of microRNAs. Innovations in Pharmaceutical Technology. 2011:52–55.
- 144.Wiggins JF, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31(7):577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 147.Nejadnik H, Hui JH, Feng Choong EP, Tai BC, Lee EH. Autologous bone marrow-derived mesenchymal stem cells versus autologous chondrocyte implantation: an observational cohort study. Am J Sports Med. 2010;38(6):1110–1116. doi: 10.1177/0363546509359067. [DOI] [PubMed] [Google Scholar]
- 148.Sekiya I, Muneta T, Horie M, Koga H. Arthroscopic transplantation of synovial stem cells improves clinical outcomes in knees with cartilage defects. Clin Orthop Relat Res. 2015;473(7):2316–2326. doi: 10.1007/s11999-015-4324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim YS, Choi YJ, Koh YG. Mesenchymal stem cell implantation in knee osteoarthritis: an assessment of the factors influencing clinical outcomes. Am J Sports Med. 2015;43(9):2293–2301. doi: 10.1177/0363546515588317. [DOI] [PubMed] [Google Scholar]
- 150.Lamo-Espinosa JM, et al. Intra-articular injection of two different doses of autologous bone marrow mesenchymal stem cells versus hyaluronic acid in the treatment of knee osteoarthritis: multicenter randomized controlled clinical trial (phase I/II) J Transl Med. 2016;14(1):246. doi: 10.1186/s12967-016-0998-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Soler R, et al. Final results of a phase I-II trial using ex vivo expanded autologous mesenchymal stromal cells for the treatment of osteoarthritis of the knee confirming safety and suggesting cartilage regeneration. Knee. 2016;23(4):647–654. doi: 10.1016/j.knee.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 152.Vega A, et al. Treatment of knee osteoarthritis with allogeneic bone marrow mesenchymal stem cells: a randomized controlled trial. Transplantation. 2015;99(8):1681–1690. doi: 10.1097/TP.0000000000000678. [DOI] [PubMed] [Google Scholar]
- 153.Orozco L, et al. Treatment of knee osteoarthritis with autologous mesenchymal stem cells: a pilot study. Transplantation. 2013;95(12):1535–1541. doi: 10.1097/TP.0b013e318291a2da. [DOI] [PubMed] [Google Scholar]
- 154.Maumus M, et al. Adipose mesenchymal stem cells protect chondrocytes from degeneration associated with osteoarthritis. Stem Cell Res. 2013;11(2):834–844. doi: 10.1016/j.scr.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 155.Yang X, et al. Intraarticular injection of allogenic mesenchymal stem cells has a protective role for the osteoarthritis. Chin Med J. 2015;128(18):2516–2523. doi: 10.4103/0366-6999.164981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chiang ER, Ma HL, Wang JP, Liu CL, Chen TH, Hung SC. Allogeneic mesenchymal stem cells in combination with hyaluronic acid for the treatment of osteoarthritis in rabbits. PLoS ONE. 2016;11(2):e0149835. doi: 10.1371/journal.pone.0149835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mei L, et al. Culture-expanded allogenic adipose tissue-derived stem cells attenuate cartilage degeneration in an experimental rat osteoarthritis model. PLoS One. 2017;12(4):e0176107. doi: 10.1371/journal.pone.0176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hermeto LC, et al. Effects of intra-articular injection of mesenchymal stem cells associated with platelet-rich plasma in a rabbit model of osteoarthritis. Genet Mol Res. 2016;15(3):gmr.15038569. doi: 10.4238/gmr.15038569. [DOI] [PubMed] [Google Scholar]
- 159.Yun S, Ku SK, Kwon YS. Adipose-derived mesenchymal stem cells and platelet-rich plasma synergistically ameliorate the surgical-induced osteoarthritis in beagle dogs. J Orthop Surg Res. 2016;11:9. doi: 10.1186/s13018-016-0342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ozeki N, et al. Not single but periodic injections of synovial mesenchymal stem cells maintain viable cells in knees and inhibit osteoarthritis progression in rats. Osteoarthr Cartil. 2016;24(6):1061–1070. doi: 10.1016/j.joca.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 161.Saulnier N, et al. Intra-articular administration of xenogeneic neonatal mesenchymal stromal cells early after meniscal injury down-regulates metalloproteinase gene expression in synovium and prevents cartilage degradation in a rabbit model of osteoarthritis. Osteoarthr Cartil. 2015;23(1):122–133. doi: 10.1016/j.joca.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 162.Lai RC, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4(3):214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 163.Tamura R, Uemoto S, Tabata Y. Immunosuppressive effect of mesenchymal stem cell-derived exosomes on a concanavalin A-induced liver injury model. Inflamm Regen. 2016;36:26. doi: 10.1186/s41232-016-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xin H, Li Y, Cui Y, Yang JJ, Zhang ZG, Chopp M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J Cereb Blood Flow Metab. 2013;33(11):1711–1715. doi: 10.1038/jcbfm.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang Y, et al. Effect of exosomes derived from multipluripotent mesenchymal stromal cells on functional recovery and neurovascular plasticity in rats after traumatic brain injury. J Neurosurg. 2015;122(4):856–867. doi: 10.3171/2014.11.JNS14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang S, Chu WC, Lai RC, Lim SK, Hui JH, Toh WS. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthr Cartil. 2016;24(12):2135–2140. doi: 10.1016/j.joca.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 167.Wang Y, et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res Ther. 2017;8(1):189. doi: 10.1186/s13287-017-0632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38(1):215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mead B, Tomarev S. Bone marrow-derived mesenchymal stem cells-derived exosomes promote survival of retinal ganglion cells through miRNA-dependent mechanisms. Stem Cells Transl Med. 2017;6(4):1273–1285. doi: 10.1002/sctm.16-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tao SC, Yuan T, Zhang YL, Yin WJ, Guo SC, Zhang CQ. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics. 2017;7(1):180–195. doi: 10.7150/thno.17133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.