Abstract
Belimumab has therapeutic benefit in active systemic lupus erythematosus (SLE), especially in patients with high-titer anti-dsDNA antibodies. We asked whether the profound B cell loss in belimumab-treated SLE patients is accompanied by shifts in the immunoglobulin repertoire. We enrolled 15 patients who had been continuously treated with belimumab for more than 7 years, 17 matched controls, and 5 patients who were studied before and after drug initiation. VH genes of sort-purified mature B cells and plasmablasts were subjected to next-generation sequencing. We found that B cell–activating factor (BAFF) regulates the transitional B cell checkpoint, with conservation of transitional 1 (T1) cells and approximately 90% loss of T3 and naive B cells after chronic belimumab treatment. Class-switched memory B cells, B1 B cells, and plasmablasts were also substantially depleted. Next-generation sequencing revealed no redistribution of VH, DH, or JH family usage and no effect of belimumab on representation of the autoreactive VH4-34 gene or CDR3 composition in unmutated IgM sequences, suggesting a minimal effect on selection of the naive B cell repertoire. Interestingly, a significantly greater loss of VH4-34 was observed among mutated IgM and plasmablast sequences in chronic belimumab–treated subjects than in controls, suggesting that belimumab promotes negative selection of activated autoreactive B cells.
Keywords: Autoimmunity, Immunology
Keywords: B cells, Tolerance
Belimumab minimally influences selection of the naïve B cell repertoire but may promote negative selection of autoreactive activated B cells.
Introduction
The TNF-like cytokines B cell–activating factor (BAFF) and a proliferating ligand (APRIL) are important regulators of B cell survival and function. The physiologic effects of BAFF and APRIL are influenced by the availability of the individual cytokines in each microenvironment, the relative expression of the 3 receptors, BAFF-R, transmembrane activator and CAML interactor (TACI), and B cell maturation antigen (BCMA) on each B cell subset, and differences in receptor binding specificity and downstream signaling pathways of the 2 cytokines (1, 2).
BAFF excess causes lupus-like autoreactivity by several proposed mechanisms. First, the binding of BAFF to BAFF-R cooperates with BCR signaling to set the threshold for selection of transitional B cells (3). Second, the binding of BAFF to TACI promotes the selection and T cell–independent differentiation of a subpopulation of autoreactive B cells that may include transitional B cells and/or B1b B cells (4–6). Third, BAFF-mediated signals augment the B cell expression of endosomal Toll-like receptors (TLRs) and rescue B cells from death mediated by TLR9 signals, thereby amplifying immune responses to nucleic acids and regulating B cell tolerance to DNA (7, 8). In the germinal center milieu, BAFF produced by T follicular helper cells enhances the selection of high-affinity germinal center B cells via its binding to TACI (9), whereas in the extrafollicular milieu and bone marrow both BAFF and APRIL support plasma cell survival via binding to TACI and BCMA (9–11). Finally, BAFF regulates inflammation through its effects on cytokine release by antigen-presenting cells (12). Conversely, BAFF deficiency delays the onset of lupus through mechanisms that may include altered B cell selection, B cell depletion, and decreased target organ inflammation (13–18).
The human anti-BAFF antibody belimumab is therapeutic for active systemic lupus erythematosus (SLE), especially in patients with high titers of anti-dsDNA antibodies and low serum levels of complement (19–23). Belimumab has modest and variable effects on anti-DNA antibody synthesis; however, it has not yet been shown in humans whether the basis of its therapeutic effect is an alteration in selection of either the naive or antigen-induced immunoglobulin repertoire. The current study was designed to address whether the profound loss of B cells in belimumab-treated patients is accompanied by a shift in the immunoglobulin repertoire of either mature B cells or plasma cells.
Results
Human subjects.
The demographic characteristics of the SLE patients and controls are shown in Table 1. There were no differences between the belimumab-treated patients and the SLE controls except in disease duration, which was slightly shorter in the lupus controls (P < 0.05). Patients receiving belimumab chronically and lupus controls had quiescent disease with limited use of immunosuppressive medications. Patients with active disease newly starting on belimumab were on significantly higher doses of prednisone than either the patients on chronic belimumab or the lupus controls (P < 0.001 and P < 0.0001, respectively).
Table 1. Demographic characteristics of lupus patients and healthy donors.
B cell phenotype.
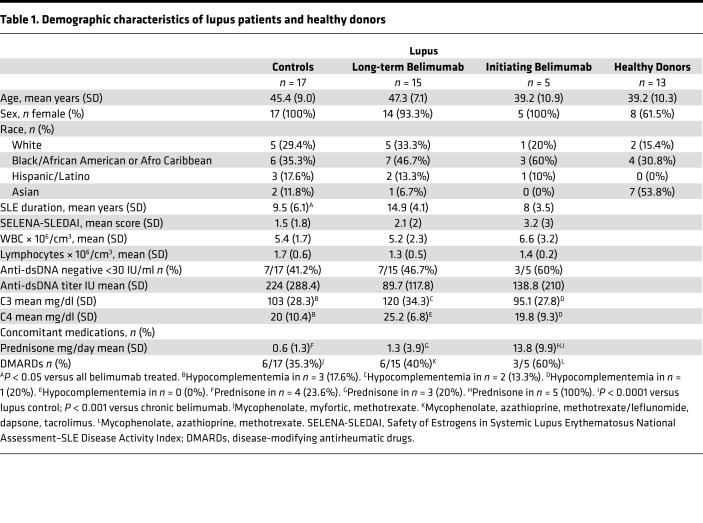
The gating strategy for B cell phenotyping is shown in Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122525DS1). Patients receiving chronic belimumab had an average depletion of 88% of all B cells compared with SLE controls (Figure 1, A and B). In agreement with our previous study (24), not all B cell subsets were depleted to the same degree, resulting in a redistribution of B cell subsets. Mature CD27–IgD+ B cells constituted a lower percentage and class-switched memory B cells a higher percentage of the remaining B cells. Class-switched memory B cells and B1 cells are BAFF independent and take longer to deplete after belimumab treatment than naive B cells (10, 24, 25) (Supplemental Figure 2). Nevertheless, memory subsets were significantly depleted in the peripheral blood after long-term belimumab treatment (Figure 1, C and D) as were plasmablasts and B1 cells (Figure 1, E and F), although to a lesser degree than memory cells.
Figure 1. Most B cell subsets are depleted after chronic belimumab therapy.
PBMCs from healthy donors (n = 13), lupus controls (n = 17), and chronic belimumab–treated subjects (n = 15) were stained with a cocktail of antibodies (Supplemental Table 1 – Panel 1) and analyzed by flow cytometry. Cells were gated as shown in Supplemental Figure 1. (A and B) Plots display frequency (A) and absolute cell count/ml (B) of CD19+ B cells in gated live singlet lymphocytes. (C–F) Plots display frequency (C and E) and absolute cell count/ml (D and F) of major B cell subsets in gated CD19+ B cells. Average percentage depletion of each cell subset compared with lupus controls is shown above the plots. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant. Comparisons were performed using Kruskal-Wallis test (A, C, and E) and Mann-Whitney analysis (B, D, and F).
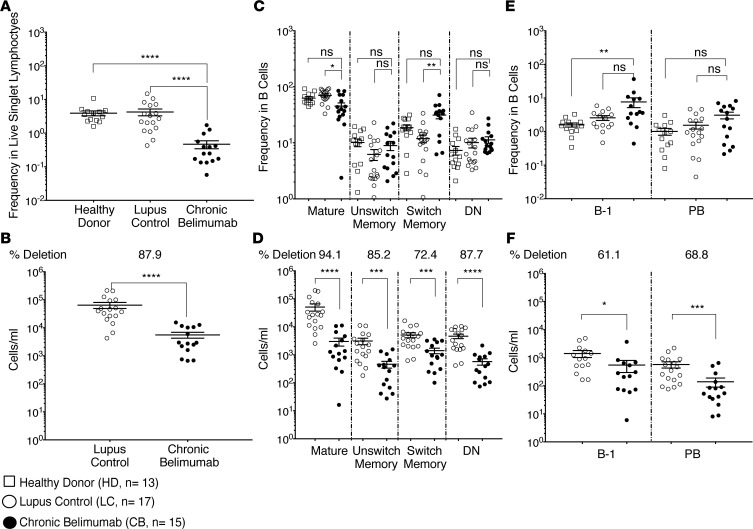
To investigate how BAFF regulates the early development of human B cells, we utilized the ABCB1 transporter and other B cell developmental markers (26–29) to rigorously separate CD27–IgD+ B cells into their different subsets (Supplemental Figure 1). We found no difference in the number of transitional 1 (T1) B cells between chronic belimumab–treated patients and lupus controls. By contrast, there was 79% deletion of the T2 subset and 93% deletion of the T3 subset (Figure 2, A and B). Similarly, patients newly treated with belimumab had lost most of their T3 cells by the 6-month visit (7 treatments) while retaining their T1 cells (Supplemental Figure 2). Notably, a large population of circulating T1 cells was detected in 5 chronic belimumab–treated patients, constituting from 11% to 60% of surviving B cells. A large population of T1 cells was similarly observed at the 6-month visit in the 1 patient that had a large number of T1 cells (0.75% of all B cells) at the initial visit (Supplemental Figure 2). These data suggest that the high T1 cell number observed in a subset of chronically treated patients reflects a high starting number of T1 cells that are unaffected by belimumab treatment.
Figure 2. Belimumab regulates survival of B cells as they transition from the T1 stage to the naive phenotype.
Mature B cells were stained with a cocktail of antibodies (Supplemental Table 1 – Panel 2), and gated as shown in Supplemental Figure 1. Cells were subsetted on the basis of CD24 and CD38 expression and the ability to extrude MitoTracker Green (MTG). (A and B) Plots display frequency (A) and absolute cell count/ml (B) of transitional type 1 (T1), T2, T3, naive, activated naive (29), and MTG+CD24+CD38– subsets in gated CD19+ B cells. The number of subjects in each group was healthy donor (n = 13), lupus control (n = 14), chronic belimumab (n = 14). (C and D) Pseudocolor plots showing changes in B cell subsets of a representative lupus donor before (C) and after (D) belimumab therapy. Samples with fewer than 200 total B cells collected were excluded from some of the subset analyses due to insufficient cells in the subgates. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant. Comparisons were performed using Kruskal-Wallis test (A) and Mann-Whitney analysis (B).
CD24+ activated naive B cells that fail to extrude MitoTracker Green (MTG+) are clonally related to antibody-secreting cells, contain mutated immunoglobulin genes, and contribute to the autoantibody repertoire in active SLE patients (29). We found that these cells were depleted to the same degree as late transitional and naive B cells (Figure 2, A and B, and Supplemental Figure 2). In addition, the MTG+CD24+CD38+ subpopulation whose function is not documented (28) was present in both healthy donors and lupus controls and was depleted by 93% in belimumab-treated patients, similar to the naive population (Figure 2, A and B, and Supplemental Figure 2).
Signaling through the Fas receptor is an important mechanism for apoptosis of autoreactive B cells (30, 31). Fas expression was lower in naive B cells than in the other subsets, with no differences between belimumab-treated subjects and lupus controls or healthy donors in the naive or memory compartments. Unexpectedly, significantly more B-1 B cells from chronic belimumab–treated patients expressed Fas than from lupus controls or healthy donors, with a trend towards a difference for double-negative (DN) B cells (LC vs. CB, P < 0.08; Supplemental Figure 3A). No differences were observed, however, when comparing the paired pre- and posttreatment samples.
CD21lo B cells are frequent in autoimmune patients and can be found in multiple B cell compartments including the DN compartment and the overlapping subset of age-associated B cells that are induced in inflammatory conditions (32, 33). No differences were observed in the frequencies of CD21lo B cells in any B cell subset between chronic belimumab–treated subjects and the other groups (Supplemental Figure 3B), as the reductions of CD21lo B cells were similar across observed populations (Supplemental Figure 3C). Similarly, although CD21lo B cells decreased in subjects newly treated with belimumab (Supplemental Figure 2), no changes in the frequencies of CD21lo cells per subset were observed.
VH repertoire of unmutated IgM sequences from mature B cells.
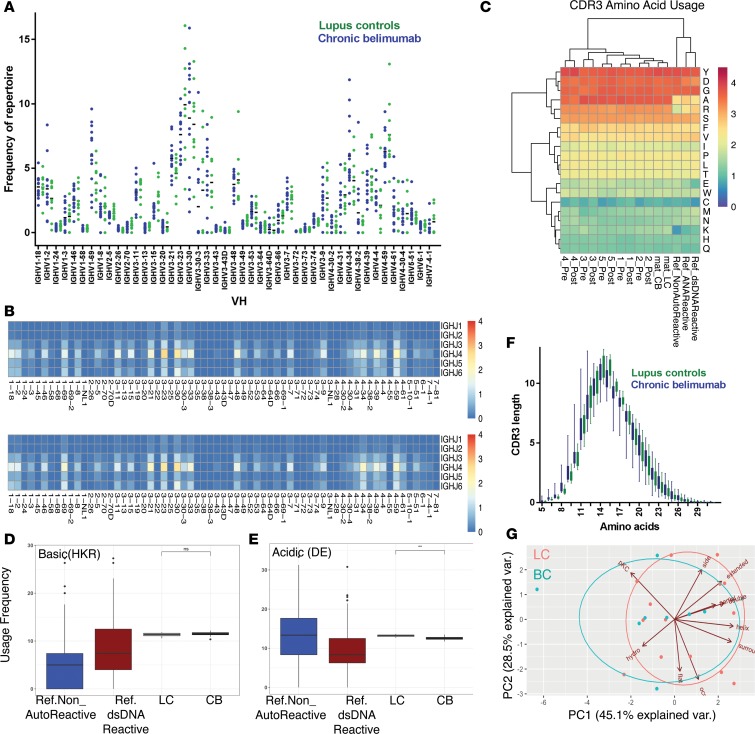
Naive B cells from healthy subjects are reported to have an increased ratio of VH1/VH3, increased expression of IgHD3, and an increase in the ratio of JH4/JH6 compared with transitional B cells (34), reflecting selection at this checkpoint. These changes were not exaggerated in the unmutated IgM sequences from chronic belimumab–treated patients compared with lupus controls (Supplemental Figure 4, A, C, and E) or between pre- and post-belimumab samples from 5 subjects (Supplemental Figure 4, B, D, and F). At the individual VH gene level, naive B cells from healthy subjects express more VH1-69 and 5-51 and less VH3-30 than transitional cells (34). We observed an increase in VH1-69, VH2-26, and VH3-15 expression in chronic belimumab–treated patients compared with lupus controls (raw P values < 0.05), although not in the posttreatment versus pretreatment samples (Figure 3, A and B). However, these changes did not reach statistical significance after correcting for multiple comparisons. Furthermore, using Simpson’s diversity index, we found no difference in clonal diversity between the 2 groups (Supplemental Table 2 and Supplemental Figure 5A).
Figure 3. VH repertoire of unmutated IgM sequences from mature B cells.
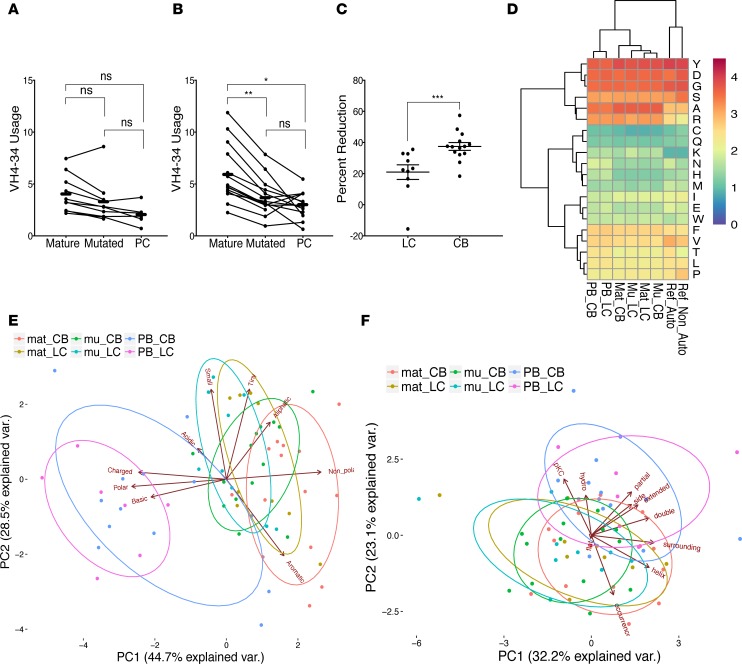
(A) Frequency of VH usage of the functional unmutated IgM repertoire from SLE patients treated with chronic belimumab (CB, blue) and lupus controls (LC, green) is shown. Each data point represents an individual subject. Heatmaps show the average frequency of VH-JH paired usage of LC and CB groups (B, left and right, respectively). (C) Clustered heatmap shows the amino acid usage of the CDR3 from LC, CB, reference non-autoreactive, and ANA-reactive groups, and of 5 individuals before (Pre) and 6 months after (Post) belimumab treatment. Distribution of CDR3 physicochemical properties (acidic [D], basic [E]) for LC and CB groups compared with the reference non-autoreactive and ANA-reactive groups. **P < 0.01; ns, not significant (linear mixed model). (F) Distribution of CDR3 amino acid length usage of LC (green) and CB (blue) groups. (G) Principle component analysis (PCA) of Kidera factors. For A, D, and F, the number of subjects in each group was LC (n = 12) and CB (n = 15).
Certain VH region genes are known to encode for autoreactivity in their germline configuration. Of these, VH4-34, which is negatively selected from the memory and plasmablast compartments of healthy individuals, is overrepresented among activated naive B cells as well as among memory B cells and plasmablasts of lupus patients (29, 35–37). VH4-34 was expressed in 4% of the naive B cell repertoire from lupus controls and was not preferentially deleted in the unmutated sequences of chronic belimumab–treated patients (Figure 3, A and B). Similarly, there was no significant decrease in its representation among unmutated IgM sequences 6 months after belimumab initiation in 5 newly treated patients (Supplemental Figure 4, G–K).
These data in sum show that the approximately 90% depletion of naive B cells found after belimumab treatment involves B cells encoded by virtually all VH genes without preferential deletion or enrichment of a particular VH.
The heavy chain D region makes an important contribution to antigen specificity, and particular charged amino acids, especially arginine (R), have been shown to confer specificity for dsDNA (38, 39). No particular pattern of CDR3 amino acid distribution distinguished chronic belimumab–treated patients from lupus controls. Similarly, when we examined pre- and post-belimumab samples, sequential samples from individual subjects clustered together regardless of their treatment status (Figure 3C). Among a reference set of autoreactive single cells isolated from naive and immature B cells of healthy donors and lupus patients (40–42), those binding dsDNA had a significant increase in basic residues H, K, and R and a decrease in acidic residues D and E compared with non-autoreactive cells. We found a statistically significant decrease in acidic residues in the CDR3 of belimumab-treated subjects compared with lupus controls, contrary to what would be expected, but no difference in basic residues (Figure 3, D and E). When this analysis was restricted to VH4-34–encoding sequences, no differences were found either in acidic or basic residues. It has been further suggested that longer heavy chain CDR3 length contributes to polyreactivity (40, 43). We found no change in the CDR3 length of sequences in patients treated with chronic belimumab compared with lupus controls (Figure 3F). We next examined the biophysical features of the CDR3 regions using Kidera factors that reduce over 100 different biophysical characteristics of peptides into 10 independent factors (44). We found no difference in any of the tested characteristics of the CDR3 regions in belimumab-treated subjects compared with lupus controls when we examined all factors individually or together (Figure 3G). Similarly, no changes in acidic or basic residues or in Kidera factors were observed in unmutated IgM sequences of 5 subjects examined before and after belimumab treatment. One characteristic of B cell selection is the loss of hydrophobicity in CDR3 between transitional and mature recirculating B cells (45). Using either Boman’s index or Kidera factors, we found no differences in hydrophobicity between chronic belimumab–treated subjects and lupus controls or between pre- and posttreatment samples (Figure 3G).
VH repertoire of mutated IgM sequences from mature B cells.
Mutated IgM sequences derived from CD27– B cells provide a window into the antigen-selected B cell repertoire that includes activated naive B cells. The repertoire of these cells was clearly different from that of their low-mutated counterparts both in lupus controls and in chronic belimumab–treated subjects, with strikingly similar alterations in the landscape of VH genes in both groups (Figure 4, A and B, and Supplemental Table 3). Compared with unmutated mature B cells, there was loss of VH4-34 among mutated IgM sequences in both lupus controls and chronic belimumab–treated subjects; after correcting for multiple comparisons, this loss was only significant in the belimumab-treated subjects (37% ± 9% loss vs. 21% ± 15%, P < 0.001; Figure 5C). No consistent changes were found, however, between pre- and post-belimumab samples from the 5 newly treated subjects (Supplemental Figure 4, G–K). There were no significant differences in length, amino acid composition, hydrophobicity, or biophysical characteristics of the CDR3 regions between mutated IgM sequences of belimumab-treated patients compared with lupus controls, or between pre- and posttreatment samples (Figure 5, D–F).
Figure 4. VH repertoire of mutated mature IgM sequences and plasmablasts.
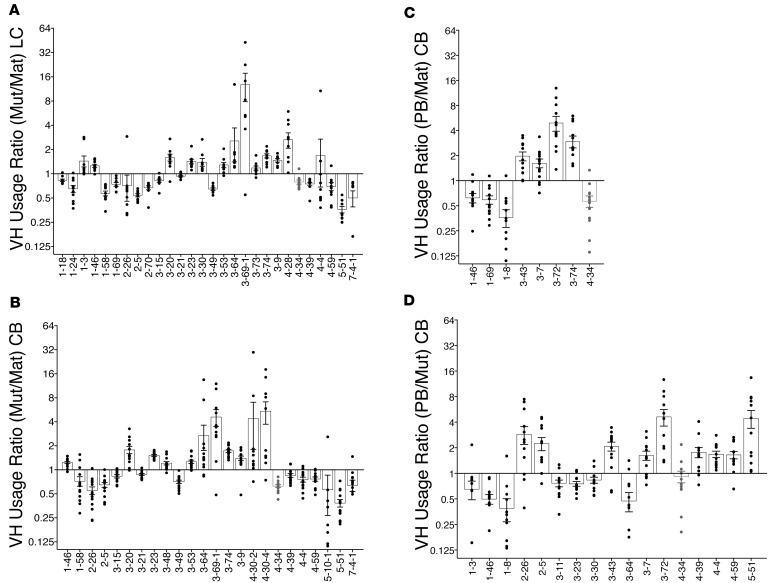
Changes in the VH usage of mutated mature (Mut) versus unmutated mature (Mat) IgM sequences of individual lupus control (LC) (n = 12) and chronic belimumab (CB) patients (n = 14) are shown as ratios in A and B, respectively. Each dot represents one patient. Changes in the VH usage of plasmablast (PB) versus unmutated mature (Mat) of and mutated mature (Mut) of individual CB patients are shown as ratios in C and D, respectively. Only those VH with statistically significant differences performed using a linear mixed model with crossing between cell populations and treatment groups to look for interactions are shown (see Supplemental Table 3).
Figure 5. VH4-34 and CDR3 usage across groups.
Frequency of VH4-34 in each subset for (A) lupus control (LC) (n = 6–8) and (B) chronic belimumab (CB) (n = 13) patients are shown in connected dot plots, with each line representing 1 patient. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Comparisons were performed using Wilcoxon’s rank-sum test. (C) Plot displays the percentage reduction of VH4-34 usage in mutated mature B cells compared with unmutated mature B cells. Comparisons were performed using Mann-Whitney analysis. (D) Clustered heatmap shows the amino acid usage of the CDR3 from unmutated, mutated, and plasmablast (PB) sequences of LC and CB compared with reference non-autoreactive and autoreactive sequences. (E and F) Principle component analysis (PCA) of CDR3 amino acid composition (E) and physicochemical properties (Kidera factors, F) are shown for unmutated, mutated, and PB sequences.
Two of the 14 belimumab-treated subjects had clonal expansions within the mutated IgM sequences, only some of which were shared with the plasmablast sequences (Supplemental Table 2 and Supplemental Figure 5B). In one of these patients, a VH4-34 clone was present but was not shared with the plasmablast compartment from the same patient. There were no differences in clonal diversity of mutated IgM sequences between the pre- and posttreatment samples (Supplemental Table 2 and Supplemental Figure 5C).
VH repertoire of plasmablasts.
Sufficient cells were recovered from 7 lupus controls and 13 belimumab-treated subjects to examine the repertoire and clonal diversity of plasmablasts. The repertoire of these cells differed from that of both unmutated and mutated IgM B cells in the chronic belimumab group (Figure 4, C and D, and Supplemental Table 3); because of the small number of subjects with matched samples, we may not have had enough power to detect differences in the lupus control group. Representation of VH4-34 was lower among plasmablasts than among matched unmutated IgM sequences in the chronic belimumab–treated group, but did not differ from mutated IgM B cells (Figure 4, C and D, and Figure 5, A and B).
Plasmablasts manifested clonal expansion similar to that previously reported for SLE patients (29); the degree of clonality of the plasmablast repertoire was variable in both groups, with no differences between the lupus control and chronic belimumab–treated groups (Supplemental Table 2 and Supplemental Figure 5D). We also asked whether the repertoire of the most overrepresented clones differed between belimumab and control subjects, but there were no differences between groups. In contrast to previous observations in highly active SLE patients (29), VH4-34 contributed only 2%–3% of the plasmablast repertoire in either group, perhaps reflecting the inactive status of the patients in our cohort (Supplemental Figure 6 and Supplemental Figure 7, A and B). Nevertheless, anti-DNA antibodies were still present in the serum of the belimumab-treated patients, with variable changes in the levels of these antibodies compared with the pre-belimumab values (Supplemental Figure 7C). The biophysical properties of the CDR3 regions of the plasmablast sequences from both belimumab-treated subjects and lupus controls were clearly different from those of unmutated and mutated IgM, but not different from each other. This difference was accounted for mainly by an increase in polar and charged residues, both acidic and basic, in the CDR3 regions of the plasmablasts (Figure 5, D–F).
Discussion
This study extends our knowledge of the effects of BAFF inhibition on the survival and selection of human B cells. We show here that BAFF regulates the checkpoint between transitional B cell stage 1 and transitional stage 3 in humans, with conservation of transitional type 1 cells and approximately 90% loss of transitional type 3 and naive B cells after chronic belimumab treatment. Despite the known expression of BAFF-R on T1 cells (28, 46), BCR- and BAFF-mediated signals are not well integrated in these cells and they manifest low BAFF responsiveness, making them less sensitive to changes in BAFF availability. This is similar to what has been reported in mouse T1 cells (47, 48). We examined naive activated B cells and CD21lo B cells that have a phenotype that overlaps with the recently described age-associated B cell subset, and that can give rise to autoreactive plasmablasts in response to innate B cell signals such as endosomal TLR ligands (32, 33). Neither subset was preferentially depleted by belimumab compared with naive B cells, suggesting that these cells require BAFF but do not have a generally increased BAFF requirement compared with the naive population. We found that the increase in CD27+ class-switched cells observed early after belimumab treatment is due to an increase in class-switched memory B cells rather than B1 cells. After more than 7 years of treatment, there was a substantial loss of BAFF-independent subsets, such as class-switched memory B cells, B1 B cells, and plasmablasts; this may reflect the failure to replenish these cells over time as naive B cells are lost.
In T cell–dependent responses, both CD40 ligation and FDC exposure in the germinal center enhance expression of Fas, thus priming B cells for death; however, these cells can be rescued by strong BCR or costimulatory signals (49, 50). The interaction of BAFF with BAFF-R can similarly protect B cells from Fas-mediated apoptosis and may therefore rescue low-affinity B cells from death (51). On the other hand, the interaction of BAFF with TACI in innate B cells has been reported to facilitate upregulation of both Fas and FasL on B cells and render them more susceptible to Fas-mediated apoptosis by downregulating inhibitors of the apoptotic pathway (52). While there was no change in Fas expression in naive and class-switched memory B cells, we observed an increase in the percentage of Fas-expressing B1 B cells and DN B cells in chronic belimumab–treated patients compared with controls; these subsets are thought to represent T cell–independent B cells. This finding is puzzling and could reflect either a difference in how BAFF regulates FasL expression in these cells or the failure to downregulate apoptotic inhibitors in a low-BAFF environment, thus protecting the cells from Fas-mediated death when other B cells are diminished. Whether these cells are a source of the plasmablasts that continue to be generated in belimumab-treated subjects remains to be determined.
One explanation for the therapeutic efficacy of BAFF inhibition is counterselection of autoreactive B cells. While extreme BAFF overexpression in mouse models is associated with altered selection of the naive B cell repertoire and/or induction of autoimmunity driven by T cell–independent B cells (4, 53, 54), studies of lupus mice bearing autoreactive V region transgenes have revealed variability in the response to BAFF antagonists, especially when there is physiologic competition from non-autoreactive B cells (13). We have shown in lupus-prone mice that negative selection of the D42 autoreactive V region gene occurs between the T1 and T2 stages and is 2–3 times more stringent in mice treated with a BAFF antagonist. Nevertheless, the light chain repertoire and clonal diversity of those D42 cells that escape deletion following BAFF inhibition is not substantially altered, and high-affinity autoreactive D42 B cells are still positively selected into the germinal center where they mature to autoreactive plasma cells (55). By contrast, B cells expressing the 3H9 autoreactive V region gene are deleted at the pre–B cell stage in the bone marrow with no additive effect of BAFF inhibition on the stringency of selection of naive 3H9 B cells that escape central deletion (16). There is a similar lack of effect of BAFF over- or underexpression on regulation of the autoreactive HKIR transgene, which is also regulated by central tolerance (56). BAFF inhibition has a modest effect on germinal center entry and proliferation of 3H9-positive autoreactive B cells and on selection of the plasma cell repertoire; these effects vary between lupus strains (16). The effect of BAFF inhibition on selection of the human B cell repertoire has not been previously reported.
Next-generation sequencing (NGS) is a quantitative method to perform both cross-sectional and longitudinal analyses of immunoglobulin repertoires and to identify expanded B cell clones. Here we used several analyses of NGS data from belimumab-treated subjects to determine whether there might be loss of autoreactive V region genes in the naive repertoire of patients treated with belimumab. We first asked whether the normal pattern of changes in VH gene usage observed at the transitional to naive checkpoint is exaggerated following belimumab therapy, suggesting more stringent negative selection. Although approximately 90% of the naive B cell population is depleted following chronic belimumab therapy, we did not observe a redistribution of V, D, or J family usage among unmutated IgM sequences from treated patients. Among VH genes, VH4-34 is associated with the anti-dsDNA response and its expression is increased during disease activity. Nevertheless, the VH4-34 gene is frequently expressed in the naive repertoire both in healthy individuals and lupus patients (37), indicating that B cells expressing this gene are not regulated by central deletion or deletion at the transitional checkpoints. Consistent with this concept, we found no effect of belimumab on the frequency of VH4-34 among unmutated IgM sequences.
The heavy chain CDR3 region makes a large contribution to the antigen binding site, and its composition is thought to be heavily influenced by selection events. Counterselection of B cells containing VH genes with long hydrophobic CDR3s occurs during naive B cell selection and autoreactive B cells have been reported to contain longer, more hydrophobic heavy chain CDR3 regions than non-autoreactive B cells (40, 43). No differences in length or hydrophobicity of CDR3s from the unmutated sequences were found between lupus controls and chronic belimumab–treated subjects, suggesting that BAFF availability does not further influence this aspect of negative selection. Anti-dsDNA antibodies use heavy chain CDR3 regions with more basic and fewer acidic amino acids than non-autoreactive antibodies; our analyses did not detect selection against these characteristics in belimumab-treated subjects. These findings, in sum, confirm that although BAFF highly regulates survival of naive B cells past the T1 stage in humans, we were unable to identify an effect of belimumab on selection of the naive B cell repertoire. These data are consistent with a recent report showing no difference in the percentage of naive anti–nuclear antigen–reactive (ANA-reactive) B cells identified by flow cytometry between belimumab-treated subjects and lupus controls (57).
Naive activated B cells from patients with active SLE are enriched in VH4-34 genes and can directly give rise to plasmablasts (29). In subjects treated with chronic belimumab, we found a significantly greater loss of VH4-34 among mutated IgM sequences compared with unmutated sequences from the same subjects, suggesting that BAFF inhibition promotes negative selection of autoreactive activated naive B cells. VH4-34 is counterselected in the germinal center and plasma cell compartments of healthy donors, but this counterselection fails in lupus (37). We found a significant loss of VH4-34 in the plasmablast compartment of chronic belimumab–treated subjects compared with the naive compartment of the same patients, further confirming a potential role for BAFF inhibition in regulating activated B cells. Nevertheless, clonal expansions of mutated IgM sequences were observed in 2 chronic belimumab–treated patients, with matched expansions of some of these clones in the plasmablast compartment showing that belimumab does not prevent general clonal activation of B cells.
Overall, our study indicates that BAFF regulates B cell survival at the T1 to T3 checkpoint in humans as it does in mice, resulting in greater than 90% depletion of naive B cells and 60%–95% deletion of all other B cell subsets after chronic treatment. We were unable to identify changes in the naive resting B cell VH or heavy chain CDR3 repertoire to suggest effects on the stringency of negative selection of this subset. By contrast, belimumab may promote negative selection of autoreactive activated B cells and plasmablasts.
The strengths of our study are the inclusion of patients treated with belimumab for more than 7 years compared with closely matched lupus controls as well as patients studied longitudinally. We were able to perform careful phenotyping and to sample a large number of naive B cells, class-switched and mutated CD27– cells and plasmablasts for repertoire analysis. There are, however, a number of caveats to our study. First, we chose to analyze only the immunoglobulin heavy chain since this is responsible for most of the differences in repertoire selection in normal individuals (34); however, we are missing information about the light chains, especially in the plasmablasts in which clonal expansions occur. Second, we are unable to distinguish whether a small number of anergic cells might contribute to the repertoire of surviving naive B cells in the belimumab-treated patients, as has been previously suggested (57). This would be of interest to explore further since naive VH4-34–encoded B cells (9G4+ B cells) appear to be anergic both in healthy individuals and in lupus patients. Rescue of these anergic cells by excess BAFF may be one mechanism by which abnormal activation and expansion of VH4-34–encoded B cells is permitted in SLE patients (37). Restoration of anergy could help explain the observed decrease in representation of VH4-34 among activated B cell subsets in the belimumab-treated subjects reported here. Another potential mechanism would be a decrease in endosomal TLRs that drive activation and expansion of autoreactive B cells and whose expression is enhanced by BAFF (7). It would be ideal to study the frequency of autoreactive B cells in belimumab-treated patients before and after treatment by re-expressing the genes derived from single cells and testing for autospecificities; these studies are currently ongoing. Other affected functions of the B cells that survive under conditions of chronic BAFF depletion remain to be determined in future studies.
Methods
Human subjects.
The following subjects were recruited to this investigator-initiated study: 15 SLE patients who had been continuously treated with 10 mg/kg belimumab monthly for more than 7 years as part of GSK-sponsored belimumab clinical studies; 17 SLE controls matched as closely as possible for disease activity, medications, gender, and ethnicity; and 13 healthy donors whose cells were used to set the gates for the B cell phenotyping. Five SLE patients newly starting on belimumab were studied before and 6 months after drug initiation. One additional patient had samples available only at 0 and 2 months. Demographic characteristics of SLE patients and controls are shown in Table 1.
Flow cytometry panels for B cell phenotype.
Peripheral blood mononuclear cells (PBMCs) were stained with fluorophore-conjugated anti-human antibody cocktails in PBS/0.2% BSA/2 mM EDTA at 4°C for 30 minutes (Supplemental Figure 1 and Supplemental Table 1). To distinguish naive from transitional cells, PBMCs were first loaded with 5 nM MitoTracker Green FM (Molecular Probes, Thermo Fisher Scientific) at 37°C for 30 minutes, washed, and then stained with the relevant antibody cocktail. Absolute B cell counts were calculated using lymphocyte counts and the B cell gate.
B cell VH library generation.
Mature B cells were sorted as live CD3–CD11b–CD56–CD19+CD27–CD10–CD43– cells. This sorting strategy included naive B cells, activated naive B cells, and DN cells that are thought to represent extrafollicularly activated B cells, but excluded transitional type 1 B cells that are not influenced by BAFF inhibition. Plasmablasts/plasma cells were sorted as live CD19loCD3–CD11b–CD56–CD19loCD27hiCD38hi cells (Supplemental Table 1) and are referred to throughout the manuscript as plasmablasts. Pellets were washed, and RNA was made using RNeasy (Qiagen) followed by PCR using a One-Step RT-PCR Kit (Qiagen). First-round VH primers HBHI-M 01-10 (iRepertoire) conferred a unique barcode from 1 to 10. A second round of PCR was performed using a Qiagen Multiplex PCR Kit and communal primers (iRepertoire). Gel bands corresponding to 490–570 bp were purified using a QIAquick Gel extraction Kit (Qiagen). Purified samples (120 ng of each) were used to generate pooled libraries, each containing 10 individual libraries marked with a different barcode. Pooled libraries were then sequenced using miSeq. Data were deposited into the NCBI’s Sequence Read Archive (SRA BioProject SAMN099549-601; https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP159206&go=go).
Analysis of sequencing data.
After initial filtering using iRepertoire algorithms, sequence data from mature B cells was filtered to separate IgM from non-IgM sequences. Unproductive sequences and/or sequences without identifiable CDR3 and/or D genes were filtered out using customized Perl and R scripts. Sequences were genotyped using IMGT High V-QUEST. IgM sequences with more than 3 mutations (29) were analyzed separately. Sequences with identical VH, DH, JH, and CDR3 were considered as a clone/single cell. For mutated sequences, identical CDR3 length and CDR3 similarity of greater than 85% were considered to define clonotype (29, 58).
Since antibody-secreting cells are highly mutated and contain high mRNA content, sequence data from the plasmablasts was filtered to exclude single-copy reads so as to minimize mutations introduced by PCR or sequencing. Customized Perl and R scripts were applied to filter out unproductive sequences and/or sequences without identifiable CDR3 and/or D genes. Sequences with identical VH, DH, JH, and CDR3 length with greater than 85% CDR3 similarity were identified using alakazam and shazam packages from immcantation toolbox (https://immcantation.readthedocs.io) and considered as a clone.
Numbers of sorted cells, sequence counts, and unique sequences at each step of the filtering analysis are shown in Supplemental Table 2. After filtering, the number of unique sequences recovered from each individual patient correlated closely with the starting number of B cells.
CDR3 count, length, and amino acid frequencies were calculated using customized Perl and R scripts. The biophysical properties of CDR3 were analyzed using R package (Peptides: kidera) to calculate Kidera factors (44) and a customized Perl script was used to calculate amino acid composition of the CDR3 sequences from each donor (Tiny = ACGST, Small = ACDGNPSTV, Aliphatic = AILV, Aromatic = FHWY, Non-polar = ACFGILMPVWY, Polar = DEHKNQRST, Charged = DEHKR, Basic = HKR, and Acidic = DE). Mean values of the Kidera factors from each VH gene subfamily on CDR3 were calculated, and pricipal component analysis (PCA) plots were graphed using the prcomp R package. Figures were generated using either PRISM or R scripts using ggplot2 packages. Heatmaps were generated with log2-transformed data (percentage of usage) using the pheatmap package and unsupervised hierarchical clustering of the 20 amino acids according to Euclidian distance.
Reference sequence data of non-autoreactive and autoantigen-specific reactive antibodies.
The heavy sequences of 445 non-autoreactive and 287 autoreactive B cells of different specificities (ANA [n = 274], dsDNA [n = 124]) from early immature, immature, new immigrant, and mature naive B cells derived from 3 healthy subjects and 6 lupus patients were used as a reference repertoire (40, 41, 59).
Statistics.
Comparisons in Figures 1 and 2, Table 1, and Supplemental Figures 3 and 6 were performed using 2-tailed Mann-Whitney analysis for 2-group comparisons or Kruskal-Wallis test for 3-group comparisons. For 3-group problems, multiple comparisons were carried out using the the Dwass-Steel-Critchlow-Fligner method. Paired comparisons in Figure 5 and Supplemental Figure 7 were performed using a 2-tailed Wilcoxon’s rank-sum test. Comparisons for VH repertoire analyses in Figures 3 and 4 and Supplemental Figures 4 and 6 were performed using a linear mixed model with main effects of cell populations and treatment groups as well as cell population × treatment groups interaction. Cell populations within subjects are the repeated factors. A P value less than 0.05 was considered statistically significant.
Study approval.
The study was approved by the Feinstein Institute IRB and written informed consent was received from participants prior to inclusion in the study.
Author contributions
WH, TDQ, ML, JH, TLR, CA, and AD designed the research studies. WH, TDQ, CD, and ZL conducted experiments. RF, MM, CA, and AD recruited and consented patients. WH, TDQ, CD, ZL, MBS, WP, and QY acquired data. TDQ, CD, TL, MBS, WP, QY, JH, ML, and AD analyzed data. WH, TDQ, CD, and AD wrote the manuscript.
Supplementary Material
Acknowledgments
Patients taking chronic belimumab were enrolled in GSK-sponsored clinical studies. We acknowledge the assistance of Ferva Abidi and Sanita Kandasami with recruiting patients and collecting and coordinating transfer of samples. Samples from lupus controls and healthy donors were obtained from the Feinstein Rheumatology Specimen and Clinical Data Bank. We thank the patients and healthy donors who agreed to donate samples for these studies. This work was funded by NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases R01 AR064811-01.
Version 1. 09/06/2018
Electronic publication
Footnotes
Conflict of interest: RF and CA have received consulting fees from GSK.
Reference information: JCI Insight. 2018;3(17):e122525. https://doi.org/10.1172/jci.insight.122525.
Contributor Information
Weiqing Huang, Email: whuang@northwell.edu.
Cosmin Dascalu, Email: cosmin.dascalu2007@gmail.com.
Zheng Liu, Email: steve.zheng.liu@gmail.com.
Miranda Byrne-Steele, Email: msteele@irepertoire.com.
Wenjing Pan, Email: wpan@irepertoire.com.
Qunying Yang, Email: qyang@irepertoire.com.
Jian Han, Email: jhan@hudsonalpha.org.
Richard Furie, Email: RFurie@northwell.edu.
Meggan Mackay, Email: mmackay@nshs.edu.
Cynthia Aranow, Email: caranow@nshs.edu.
Anne Davidson, Email: adavidson1@nshs.edu.
References
- 1.Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9(7):491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- 2.Naradikian MS, Perate AR, Cancro MP. BAFF receptors and ligands create independent homeostatic niches for B cell subsets. Curr Opin Immunol. 2015;34:126–129. doi: 10.1016/j.coi.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Cancro MP. Tipping the scales of selection with BAFF. Immunity. 2004;20(6):655–656. doi: 10.1016/j.immuni.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Groom JR, et al. BAFF and MyD88 signals promote a lupuslike disease independent of T cells. J Exp Med. 2007;204(8):1959–1971. doi: 10.1084/jem.20062567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs HM, et al. Cutting Edge: BAFF promotes autoantibody production via TACI-dependent activation of transitional B cells. J Immunol. 2016;196(9):3525–3531. doi: 10.4049/jimmunol.1600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairfax KA, et al. BAFF-driven autoimmunity requires CD19 expression. J Autoimmun. 2015;62:1–10. doi: 10.1016/j.jaut.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Treml LS, et al. TLR stimulation modifies BLyS receptor expression in follicular and marginal zone B cells. J Immunol. 2007;178(12):7531–7539. doi: 10.4049/jimmunol.178.12.7531. [DOI] [PubMed] [Google Scholar]
- 8.Sindhava VJ, et al. A TLR9-dependent checkpoint governs B cell responses to DNA-containing antigens. J Clin Invest. 2017;127(5):1651–1663. doi: 10.1172/JCI89931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goenka R, et al. Local BLyS production by T follicular cells mediates retention of high affinity B cells during affinity maturation. J Exp Med. 2014;211(1):45–56. doi: 10.1084/jem.20130505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson MJ, et al. Cutting edge: the dependence of plasma cells and independence of memory B cells on BAFF and APRIL. J Immunol. 2008;180(6):3655–3659. doi: 10.4049/jimmunol.180.6.3655. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Plasma cell output from germinal centers is regulated by signals from Tfh and stromal cells. J Exp Med. 2018;215(4):1227–1243. doi: 10.1084/jem.20160832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rochas C, et al. Transmembrane BAFF from rheumatoid synoviocytes requires interleukin-6 to induce the expression of recombination-activating gene in B lymphocytes. Arthritis Rheum. 2009;60(5):1261–1271. doi: 10.1002/art.24498. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Davidson A. BAFF and selection of autoreactive B cells. Trends Immunol. 2011;32(8):388–394. doi: 10.1016/j.it.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramanujam M, et al. Similarities and differences between selective and nonselective BAFF blockade in murine SLE. J Clin Invest. 2006;116(3):724–734. doi: 10.1172/JCI26385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob CO, et al. Development of systemic lupus erythematosus in NZM 2328 mice in the absence of any single BAFF receptor. Arthritis Rheum. 2013;65(4):1043–1054. doi: 10.1002/art.37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boneparth A, Woods M, Huang W, Akerman M, Lesser M, Davidson A. The effect of BAFF inhibition on autoreactive B cell selection in murine SLE. Mol Med. 2016;22:173–182. doi: 10.2119/molmed.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson A. Targeting BAFF in autoimmunity. Curr Opin Immunol. 2010;22(6):732–739. doi: 10.1016/j.coi.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob CO, et al. Paucity of clinical disease despite serological autoimmunity and kidney pathology in lupus-prone New Zealand mixed 2328 mice deficient in BAFF. J Immunol. 2006;177(4):2671–2680. doi: 10.4049/jimmunol.177.4.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furie R, et al. A phase III, randomized, placebo-controlled study of belimumab, a monoclonal antibody that inhibits B lymphocyte stimulator, in patients with systemic lupus erythematosus. Arthritis Rheum. 2011;63(12):3918–3930. doi: 10.1002/art.30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarra SV, et al. Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, phase 3 trial. Lancet. 2011;377(9767):721–731. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- 21.van Vollenhoven RF, et al. Belimumab in the treatment of systemic lupus erythematosus: high disease activity predictors of response. Ann Rheum Dis. 2012;71(8):1343–1349. doi: 10.1136/annrheumdis-2011-200937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria A, et al. Efficacy safety of subcutaneous belimumab in anti-dsDNA-positive, hypocomplementemic patients with systemic lupus erythematosus. Arthritis Rheumatol. 2018;70(8):1256–1264. doi: 10.1002/art.40511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furie RA, et al. Long-term safety and efficacy of belimumab in patients with systemic lupus erythematosus: A continuation of a seventy-six-week phase III parent study in the United States. Arthritis Rheumatol. 2018;70(6):868–877. doi: 10.1002/art.40439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobi AM, et al. Effect of long-term belimumab treatment on B cells in systemic lupus erythematosus: extension of a phase II, double-blind, placebo-controlled, dose-ranging study. Arthritis Rheum. 2010;62(1):201–210. doi: 10.1002/art.27189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naradikian MS, Hao Y, Cancro MP. Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunol Rev. 2016;269(1):118–129. doi: 10.1111/imr.12380. [DOI] [PubMed] [Google Scholar]
- 26.Wirths S, Lanzavecchia A. ABCB1 transporter discriminates human resting naive B cells from cycling transitional and memory B cells. Eur J Immunol. 2005;35(12):3433–3441. doi: 10.1002/eji.200535364. [DOI] [PubMed] [Google Scholar]
- 27.Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005;105(11):4390–4398. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palanichamy A, et al. Novel human transitional B cell populations revealed by B cell depletion therapy. J Immunol. 2009;182(10):5982–5993. doi: 10.4049/jimmunol.0801859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tipton CM, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16(7):755–765. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akagi T, Yoshino T, Kondo E. The Fas antigen and Fas-mediated apoptosis in B-cell differentiation. Leuk Lymphoma. 1998;28(5-6):483–489. doi: 10.3109/10428199809058355. [DOI] [PubMed] [Google Scholar]
- 31.Alabyev B, Vuyyuru R, Manser T. Influence of Fas on the regulation of the response of an anti-nuclear antigen B cell clonotype to foreign antigen. Int Immunol. 2008;20(10):1279–1287. doi: 10.1093/intimm/dxn087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karnell JL, Kumar V, Wang J, Wang S, Voynova E, Ettinger R. Role of CD11c+ T-bet+ B cells in human health and disease. Cell Immunol. 2017;321:40–45. doi: 10.1016/j.cellimm.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Isnardi I, et al. Complement receptor 2/CD21- human naive B cells contain mostly autoreactive unresponsive clones. Blood. 2010;115(24):5026–5036. doi: 10.1182/blood-2009-09-243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin VG, et al. Transitional B cells in early human B cell development - time to revisit the paradigm? Front Immunol. 2016;7:546. doi: 10.3389/fimmu.2016.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isenberg DA, et al. Correlation of 9G4 idiotope with disease activity in patients with systemic lupus erythematosus. Ann Rheum Dis. 1998;57(9):566–570. doi: 10.1136/ard.57.9.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson FK, et al. Utilization of the VH4-21 gene segment by anti-DNA antibodies from patients with systemic lupus erythematosus. J Autoimmun. 1993;6(6):809–825. doi: 10.1006/jaut.1993.1066. [DOI] [PubMed] [Google Scholar]
- 37.Cappione A, et al. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J Clin Invest. 2005;115(11):3205–3216. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radic MZ, Mackle J, Erikson J, Mol C, Anderson WF, Weigert M. Residues that mediate DNA binding of autoimmune antibodies. J Immunol. 1993;150(11):4966–4977. [PubMed] [Google Scholar]
- 39.Radic MZ, Weigert M. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu Rev Immunol. 1994;12:487–520. doi: 10.1146/annurev.iy.12.040194.002415. [DOI] [PubMed] [Google Scholar]
- 40.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 41.Yurasov S, et al. Persistent expression of autoantibodies in SLE patients in remission. J Exp Med. 2006;203(10):2255–2261. doi: 10.1084/jem.20061446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yurasov S, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005;201(5):703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larimore K, McCormick MW, Robins HS, Greenberg PD. Shaping of human germline IgH repertoires revealed by deep sequencing. J Immunol. 2012;189(6):3221–3230. doi: 10.4049/jimmunol.1201303. [DOI] [PubMed] [Google Scholar]
- 44.Laffy JMJ, et al. Promiscuous antibodies characterised by their physico-chemical properties: From sequence to structure and back. Prog Biophys Mol Biol. 2017;128:47–56. doi: 10.1016/j.pbiomolbio.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schelonka RL, Tanner J, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW. Categorical selection of the antibody repertoire in splenic B cells. Eur J Immunol. 2007;37(4):1010–1021. doi: 10.1002/eji.200636569. [DOI] [PubMed] [Google Scholar]
- 46.Darce JR, Arendt BK, Chang SK, Jelinek DF. Divergent effects of BAFF on human memory B cell differentiation into Ig-secreting cells. J Immunol. 2007;178(9):5612–5622. doi: 10.4049/jimmunol.178.9.5612. [DOI] [PubMed] [Google Scholar]
- 47.Scholz JL, Oropallo MA, Sindhava V, Goenka R, Cancro MP. The role of B lymphocyte stimulator in B cell biology: implications for the treatment of lupus. Lupus. 2013;22(4):350–360. doi: 10.1177/0961203312469453. [DOI] [PubMed] [Google Scholar]
- 48.Hoek KL, Carlesso G, Clark ES, Khan WN. Absence of mature peripheral B cell populations in mice with concomitant defects in B cell receptor and BAFF-R signaling. J Immunol. 2009;183(9):5630–5643. doi: 10.4049/jimmunol.0901100. [DOI] [PubMed] [Google Scholar]
- 49.Rathmell JC, et al. CD95 (Fas)-dependent elimination of self-reactive B cells upon interaction with CD4+ T cells. Nature. 1995;376(6536):181–184. doi: 10.1038/376181a0. [DOI] [PubMed] [Google Scholar]
- 50.Rathmell JC, Goodnow CC. The in vivo balance between B cell clonal expansion and elimination is regulated by CD95 both on B cells and in their micro-environment. Immunol Cell Biol. 1998;76(5):387–394. doi: 10.1046/j.1440-1711.1998.00774.x. [DOI] [PubMed] [Google Scholar]
- 51.Hancz A, Hérincs Z, Neer Z, Sármay G, Koncz G. Integration of signals mediated by B-cell receptor, B-cell activating factor of the tumor necrosis factor family (BAFF) and Fas (CD95) Immunol Lett. 2008;116(2):211–217. doi: 10.1016/j.imlet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Figgett WA, et al. The TACI receptor regulates T-cell-independent marginal zone B cell responses through innate activation-induced cell death. Immunity. 2013;39(3):573–583. doi: 10.1016/j.immuni.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 53.Lesley R, et al. Reduced competitiveness of autoantigen-engaged B cells due to increased dependence on BAFF. Immunity. 2004;20(4):441–453. doi: 10.1016/S1074-7613(04)00079-2. [DOI] [PubMed] [Google Scholar]
- 54.Thien M, et al. Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity. 2004;20(6):785–798. doi: 10.1016/j.immuni.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Huang W, et al. BAFF/APRIL inhibition decreases selection of naive but not antigen-induced autoreactive B cells in murine systemic lupus erythematosus. J Immunol. 2011;187(12):6571–6580. doi: 10.4049/jimmunol.1101784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikbakht N, Migone TS, Ward CP, Manser T. Cellular competition independent of BAFF/B lymphocyte stimulator results in low frequency of an autoreactive clonotype in mature polyclonal B cell compartments. J Immunol. 2011;187(1):37–46. doi: 10.4049/jimmunol.1003924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malkiel S, et al. Checkpoints for autoreactive B cells in the peripheral blood of lupus patients assessed by flow cytometry. Arthritis Rheumatol. 2016;68(9):2210–2220. doi: 10.1002/art.39710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hershberg U, Luning Prak ET. The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos Trans R Soc Lond, B, Biol Sci. 2015;370(1676):20140239. doi: 10.1098/rstb.2014.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yurasov S, Hammersen J, Tiller T, Tsuiji M, Wardemann H. B-cell tolerance checkpoints in healthy humans and patients with systemic lupus erythematosus. Ann N Y Acad Sci. 2005;1062:165–174. doi: 10.1196/annals.1358.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.