Abstract
Organ transplant recipients (OTRs) on cyclosporine A (CSA) are prone to catastrophic cutaneous squamous cell carcinoma (SCC). Allograft-sparing, cancer-targeting systemic treatments are unavailable. We have shown increased risk for catastrophic SCC in OTRs via CSA-mediated induction of IL-22. Herein, we found that CSA drives SCC proliferation and tumor growth through IL-22 and JAK/STAT pathway induction. We in turn inhibited SCC growth with an FDA-approved JAK1/2 inhibitor, ruxolitinib. In human SCC cells, the greatest proliferative response to IL-22 and CSA treatment occurred in nonmetastasizing lines. IL-22 treatment upregulated JAK1 and STAT1/3 in A431 SCC cells. JAK/STAT pathway genes were highly expressed in tumors from a cohort of CSA-exposed OTRs and in SCC with high risk for metastasis. Compared with immunocompetent SCC, genes associated with innate immunity, response to DNA damage, and p53 regulation were differentially expressed in SCC from OTRs. In nude mice engrafted with human A431 cells, IL-22 and CSA treatment increased tumor growth and upregulated IL-22 receptor, JAK1, and STAT1/3 expression. Ruxolitinib treatment significantly reduced tumor volume and reversed the accelerated tumor growth. CSA and IL-22 exacerbate aggressive behavior in SCC. Targeting the IL-22 axis via selective JAK/STAT inhibition may reduce the progression of aggressive SCC in OTRs, without compromising immunosuppression.
Keywords: Dermatology, Transplantation
Keywords: Drug therapy, Organ transplantation, Skin cancer
JAK1/2 blockade with ruxolitinib inhibits cyclosporine A–induced proliferation in primary cutaneous squamous cell carcinoma and is amenable for use in immunosuppressed organ transplant patients.
Introduction
Nonmelanoma skin cancers (NMSCs) are the most commonly diagnosed cancers in the US, and their incidence is increasing annually (1, 2). Primary cutaneous squamous cell carcinomas (SCCs) account for only 20% of these diagnoses, yet cause the vast majority of NMSC-related metastases and deaths (1, 3). In areas with high UV exposure, SCC is estimated to account for 4,000 to 9,000 deaths annually — a rate that is comparable to that for renal and oropharyngeal carcinomas and melanoma (1). While we recently observed promising results in treating advanced SCC with a new immunotherapy agent (4), systemic treatments for inoperable, extensive, or metastatic SCC are relatively limited; display efficacy in only a subset of tumors; and are not suitable or may not be tolerated in a number of patients (4–6).
Advanced-stage SCC and overall tumor burden in immunosuppressed organ transplant recipients (OTRs) remain a significant cause of morbidity and mortality, contributing to the steeply rising costs of skin cancer management in the US (2). The number of solid-organ transplantations performed in the US increased 3-fold between 1988 and 2015, with over 30,000 transplants performed in 2015 (7). OTRs are at a greater risk of malignancy, owing at least in part to the long-term immunosuppression required to prevent rejection of the transplanted organ, with skin cancers accounting for approximately 40%–50% of these malignancies (8–10). Not only are skin cancers the most common malignancy in this population, those that occur in OTRs can also be more extensive and aggressive and are more likely to recur following resection (1, 3). OTRs are 65–250 times more likely than the general population to develop SCC (1, 9, 11, 12). They are also much more likely to develop SCCs compared with basal cell carcinomas; in the general population, this ratio is reversed (13). The mechanism for increased and aggressive SCC in OTRs has not been completely characterized. However, there is emerging evidence that calcineurin inhibitors (CNIs), such as cyclosporin A (CSA) and tacrolimus, which continue to be a mainstay of transplant immunosuppression regimes, may contribute directly to SCC development and progression via multiple mechanisms, including interference with cell morphology, DNA repair mechanisms, apoptosis, and p53 function (14–17).
We previously found that current use, or history of use of CSA in transplant immunosuppression, was a significant risk for catastrophic SCC in our cohort of 118 OTRs (18). We examined freshly isolated tumor tissue from OTRs and found a T cell polarization profile that favors decreased tumor surveillance via an elevated Treg/CD8 ratio, increased IL-22 production, and increased expression of the IL-22 receptor complex in transplant-related SCC (TSCC) compared with immunocompetent SCC (19). Expression of IL-22 and the IL-22 receptor complex were found to be induced by CSA treatment of A431 SCC cells in vitro, and treatment of cells with CSA and IL-22, both alone and in combination, drove accelerated cell proliferation, migration, and invasion (18, 19).
IL-22 is produced by a number of cells of both the adaptive and innate immune systems, primarily activated T22 cells, and group 3 innate lymphoid cells (ILCs) (18–23). While conventional transplant immunosuppression regimes have become quite effective in suppressing adaptive immunity, the role of the innate immune system is less clear and is thought to be a factor in persisting rates of chronic rejection, despite adequate immunosuppression (24–27). Thus, even in the context of appropriate immune suppression, IL-22–producing cells may still remain active. While IL-22 is produced exclusively by immune cells, its receptor, IL-22R1, is expressed primarily on epithelial cells in the skin, gut, liver, and pancreas (21, 22). Upon binding to its receptor complex of IL-22R1 and IL-10R2, IL-22 signals via the JAK/STAT pathway, activating JAK1 and tyrosine kinase 2 (TYK2) to predominantly induce phosphorylation of STAT3, with emerging evidence supporting IL-22–induced phosphorylation of STAT1 and, to a lesser extent, STAT5 (23, 28, 29). In keratinocytes, IL-22 has also been shown to activate the PI3K/AKT/mTOR pathway (30). Physiologically, IL-22 signaling aids in epithelial tissue regeneration and immunity via increasing cell proliferation and migration, decreasing differentiation, and aiding angiogenesis (22, 23). These actions have been implicated in many inflammatory and autoimmune conditions of various IL-22R1–expressing target organs (21, 22). Further, in the context of cancer, the IL-22 axis can be “hijacked” by malignant cells to hasten growth and progression (21).
Systemic treatment options for SCC in OTRs are limited by the need to maintain sufficient immunosuppression so as to permit tolerance of the allograft. Ruxolitinib, an oral JAK1/JAK2 inhibitor, is FDA approved for treatment of myelofibrosis and polycythemia vera. Ruxolitinib is in phase II trials for various malignancies and steroid refractory graft-versus-host disease (31–33). Ruxolitinib also has immunosuppressive properties (34), which makes it an attractive candidate for JAK/STAT inhibition in OTRs. Given our previous findings demonstrating upregulation of the IL-22 axis in OTRs and its induction following CSA treatment in vitro, along with catastrophic outcomes in our CSA-exposed OTR population, we hypothesized that JAK/STAT pathways may also be upregulated in CSA-exposed OTRs. We sought to clarify the downstream mechanisms involved in CSA-mediated aggressive SCC behavior and to inhibit those pathways in order to assess new targeted treatment options for tumor reduction and prevention in this challenging population.
Results
Early-stage human SCC cell lines show the greatest proliferative response to IL-22 treatment.
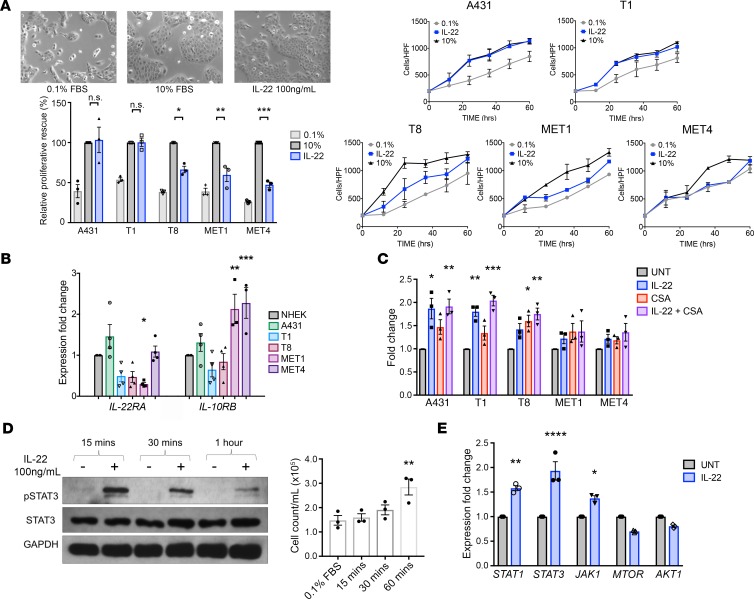
Five SCC cell lines derived from patient tumors with increasing metastatic potential (A431, primary nonmetastasizing SCC from an immunocompetent patient; T1, well differentiated, nonmetastasizing primary SCC from an immunosuppressed OTR; T8 and MET1, aggressive, poorly differentiated primary SCCs from OTRs that subsequently metastasized; and MET4, from a metastatic deposit of the MET1 SCC, Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120750DS1) were serum starved for 24 hours to synchronize proliferation at G0 phase and then rescued with either 10% FBS or IL-22 (100 ng/ml). Cells were photographed and counted 24 hours after treatment (Figure 1A), and comparisons made between treated and serum-starved cells of each cell line. Treatment of the nonmetastasizing A431 and T1 cells with IL-22 caused a proliferative response of a similar magnitude as serum replacement (Figure 1A). The degree of proliferative response due to IL-22 in comparison to full growth media sequentially decreased with increasing metastatic potential of the cell line, indicating that the greatest proproliferative effects of IL-22 may occur on cells at an early stage of tumor development.
Figure 1. IL-22 treatment causes the greatest proliferative response in the least aggressive cell lines and signals via JAK1 and STAT1/3.
(A) Five human squamous cell carcinoma (SCC) cell lines with increasingly aggressive properties (A431 and T1, nonmetastatic; T8 and MET1, aggressive primary SCC that metastasized; MET4, metastatic deposit of MET1 primary) were studied. A proliferation assay of serum-starved A431 cells was performed to compare IL-22 treatment (100 ng/ml) with serum replacement (10% FBS) in each cell line, and growth curves were constructed. Images show the A431 cell response to IL-22 and 10% FBS (original magnification, ×10). (B) PCR for IL-22 receptor complex expression on mRNA extracted from each cell line. (C) Proliferation assay of all SCC cell lines in full growth media (10% FBS) comparing treatment with 50 ng/ml cyclosporin A (CSA), 100 ng/ml IL-22, and a combination of both. (D) A431 cells were treated with IL-22 (100 ng/ml) for 15, 30, and 60 minutes. Cell counts and Western blot for STAT3 phosphorylation at each time point are shown. (E) PCR of IL-22–treated A431 cells examining expression of genes downstream of IL-22. Data represent the mean of 3 experiments ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, determined by 1-way ANOVA with Dunnett’s multiple comparisons test, as compared with each sample’s respective control.
RNA was extracted from each cell line, and qPCR for IL-22 receptor complex performed. Normal human epidermal keratinocytes were used as a baseline comparison for relative expression. IL22RA1 mRNA expression was increased compared with normal keratinocytes in A431 SCC cells (1.4-fold, NS) and decreased in T1, T8, and MET1 cell lines. IL10RB expression was significantly increased in MET1 and MET4 cell lines (2.1-fold, P < 0.01 and 2.3-fold, P < 0.001, respectively, Figure 1B). Relative to normal keratinocytes, the ratio of IL22RA1 to IL10RB expression was increased in A431s (1.1) and reduced in the other cell lines (T1; 0.8, T8; 0.6, MET1; 0.1, and MET4; 0.5).
Cell lines were seeded in full growth media (10% FBS) and treated for 24 hours with vehicle, 100 ng/ml IL-22, 50 ng/ml CSA, or a combination of IL-22 and CSA. The greatest proliferative response to IL-22 alone was seen in the early-stage lines (P < 0.001, both A431 and T1), to CSA alone in T8 cells (P < 0.05), and to the combination in A431, T1, and T8 cells (P < 0.0001, P < 0.0001 and P < 0.01 respectively, Figure 1C). The MET1 and MET4 lines did not demonstrate a significant proliferative increase. Thus, IL-22 treatment acts on the least aggressive lines most effectively.
IL-22 induces rapid STAT3 phosphorylation, early proliferation, and downstream JAK1 and STAT1/3 activation.
In order to assess response time and downstream mechanisms of IL-22–related SCC proliferation, A431 SCC cells were treated with IL-22 (100 ng/ml) and then harvested and counted at 15, 30, and 60 minutes after treatment. IL-22 treatment triggered rapid STAT3 phosphorylation by 15 minutes, which decreased over the course of 1 hour (Figure 1D; see complete unedited blots in the supplemental material). By 60 minutes, cell counts had significantly increased (1.9-fold, P < 0.01, Figure 1D). qPCR performed on RNA harvested from IL-22–treated A431 cells at 24 hours demonstrated upregulation of STAT1, STAT3, and JAK1 compared with untreated cells (1.6-fold, P < 0.01; 1.9-fold, P < 0.0001; and 1.4-fold, P < 0.05, respectively, Figure 1E). These data demonstrate the IL-22–related increased proliferative behavior occurs early and is accompanied by JAK/STAT activation.
JAK/STAT-related genes are highly expressed in tissue from CSA-treated OTRs and in high-risk SCCs.
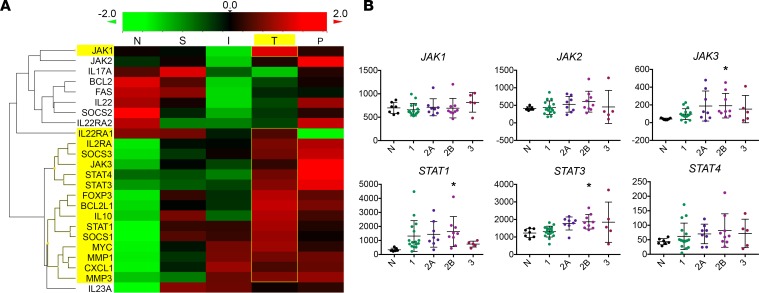
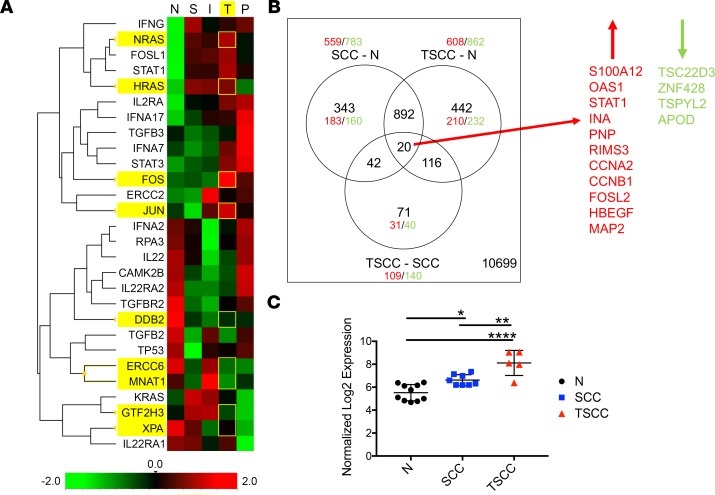
To examine potential downstream mechanisms and to determine if the findings with cell lines were replicated in patient tumors, NanoString gene expression analysis was performed on RNA extracted from 45 formalin-fixed, paraffin-embedded (FFPE) samples from patient biopsies with varying histology and clinical features: normal tissue (n = 7), superficial SCC (n = 6), locally invasive SCC (n = 12), SCC with perineural invasion (PNI) (n = 10), and SCC from immunosuppressed OTRs (n = 10). Z-score transformation of grouped expression data was represented via heatmap (Figure 2A) and analyzed in order to compare expression of IL-22– and JAK/STAT-related genes between diagnostic groups. Genes downstream of IL-22 pathway activation were highly expressed in CSA-exposed OTRs, with greatest expression of JAK1 and STAT1 in this group, along with antiapoptotic BCL2L1 (BCL-XL), mitogenic MYC (c-Myc), and Treg marker FOXP3. The greatest expression of JAK2, JAK3, STAT3, and STAT4 was in the PNI group, followed by OTRs. Other IL-22–regulated genes that were also differentially expressed in the OTR, invasive, and PNI tumors included cell motility genes, MMP1 and MMP3, and chemokine CXCL1, with downregulation of inhibitory IL22RA2 (IL-22BP) and FAS cell death receptor seen in the invasive SCCs and the OTRs (Figure 2A).
Figure 2. JAK/STAT-related genes are highly expressed in cyclosporine A–exposed organ transplant recipients.
(A) NanoString gene expression analysis was performed on mRNA extracted from 45 samples with varying histology and clinical features: normal tissue (N, n = 7), superficial squamous cell carcinoma (SCC) (S, n = 6), locally invasive SCC (I, n = 12), SCC with PNI (P, n = 10), and SCC from immunosuppressed transplant patients (T, n = 10). Heatmap displaying IL-22 and downstream related gene expression by diagnosis group. (B) Normalized expression values for JAK/STAT genes from NanoString classified by Brigham and Woman’s Hospital’s (BWH’s) staging system (as described previously, ref. 70) (mean with SD). *P < 0.05, determined by 1-way ANOVA with Dunnett’s multiple comparisons test.
The Brigham and Woman’s Hospital (BWH) SCC tumor staging system classifies tumors from 1 to 3, with increasing risk of poor outcome represented by a higher stage number (35). We examined the correlation between IL-22 and JAK/STAT gene expression and BWH staging. STAT3 expression increased with each level of increasing risk of poor outcome according to BWH stage, with a similar pattern seen in JAK1 expression (Figure 2B). Of note, BWH stage 2B tumors, a small but distinct subgroup that has been identified to be at high risk of metastasis (36), had significantly greater expression of JAK3 (P < 0.05), STAT1 (P < 0.05), and STAT3 (P < 0.05) compared with normal skin (Figure 2B). Therefore, increased JAK/STAT expression may also be associated with SCCs at high risk of relapse and metastasis, in addition to SCCs from immunosuppressed OTRs.
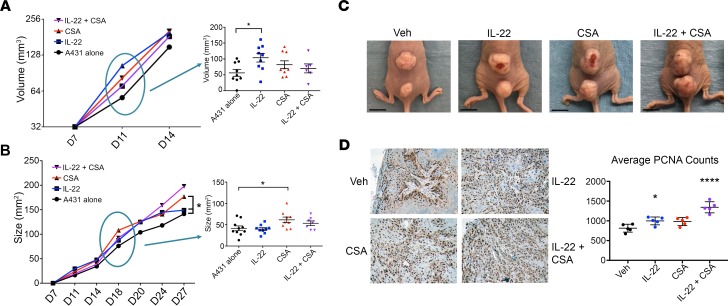
Treatment with IL-22 and CSA increases human SCC tumor growth in immunosuppressed mice, both alone and in combination.
A431 xenograft studies were performed to examine the effect of exogenous IL-22 and CSA on tumor behavior. 2 × 104 A431 SCC cells were engrafted intradermally in the midline back skin of athymic nude mice. Mice with palpable tumors were randomized at day 7 to 4 treatment groups: IL-22 (4 μg daily; 5 days per week; n = 9), CSA (20 mg/kg daily; n = 8), IL-22 plus CSA (n = 6) or vehicle alone (n = 9) (Figure 3A). Tumor size and volume were measured via a digital caliper, and any tumor ulceration documented. By the fifth day of treatment, day 11, IL-22–treated mice had significantly increased tumor growth from baseline compared with control mice (1.8-fold increase, P < 0.05, Figure 3A). This growth difference decreased over the course of the experiment, suggesting the greatest effect at an early tumor stage (Figure 3, A and B). CSA-treated mice had significantly greater tumor growth compared with control mice by day 18, following 12 days of treatment (1.7-fold, P < 0.05, Figure 3B). By the endpoint at day 27, CSA-treated tumors were significantly larger than those of A431 controls and those treated with IL-22 alone (P < 0.05, Figure 3, B and C) and had a significantly higher growth fraction, as measured by proliferating cell nuclear antigen (PCNA) immunohistochemistry with quantification of positive staining (P < 0.0001, Figure 3D).
Figure 3. The combination of IL-22 and cyclosporine A produced larger, more proliferative tumors in a murine xenograft model.
(A) Following engraftment with A431 squamous cell carcinoma (SCC) cells, nude mice were randomized to 4 treatment groups at day 7: vehicle (n = 9), IL-22 (4 μg/d i.p. M–F, n = 9), cyclosporine A (CSA) (20 mg/kg i.p., n = 8), or both IL-22 and CSA (n = 6). The difference in tumor volume at day 11 is shown. (B) Difference in tumor growth at day 18. (C) Representative tumors at endpoint, day 27. Scale bar: 1 cm. (D) Immunohistochemistry (shown at original magnification, ×10) and quantification of positively stained cells for proliferative cell nuclear antigen (PCNA) performed on paraffin-embedded tumors harvested at endpoint. Data represent mean of 5 experiments ± SEM. *P < 0.05, ****P < 0.0001, determined by 1-way ANOVA with Dunnett’s multiple comparisons test.
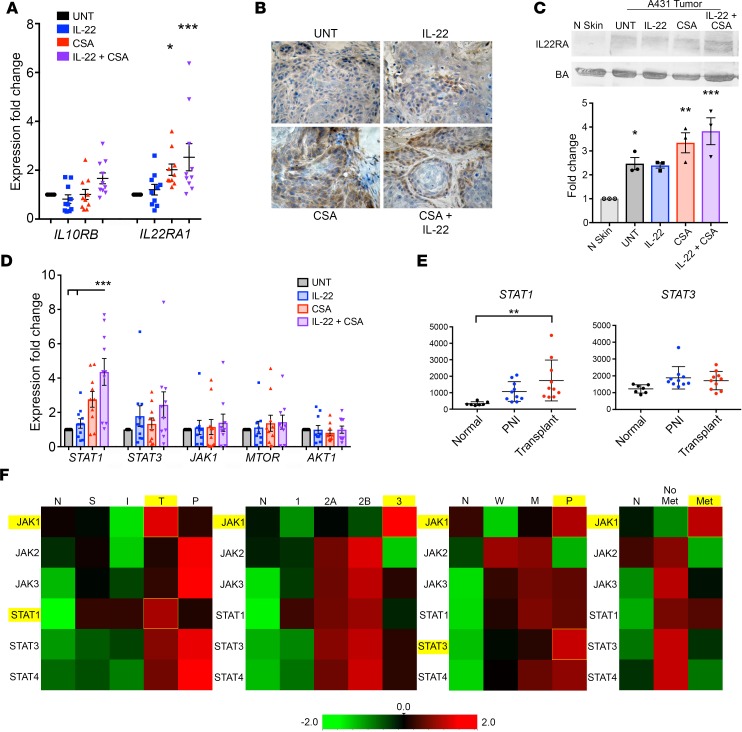
CSA exposure upregulates IL-22R1 expression and increases STAT1 expression in xenograft and patient tumors.
Tumors from mice treated with either CSA alone or with CSA in combination with IL-22 showed significant upregulation of IL22RA1 via qPCR (2- and 2.5-fold, P < 0.05 and P < 0.001, respectively, Figure 4A). Immunohistochemistry (Figure 4B) and Western blot with quantification (Figure 4C) confirmed increased IL-22RA protein expression in CSA-treated tumors. qPCR performed on mRNA extracted from treated tumors demonstrated a trend toward upregulation of STAT1, STAT3, JAK1, and MTOR, with the greatest effect for CSA-treated tumors seen in STAT1 (Figure 4D). Tumors exposed to combined treatment with IL-22 and CSA showed significantly increased expression of STAT1 compared with both those exposed to vehicle and IL-22 treatment alone (P < 0.05, Figure 4D). This correlated with our observations in human tissue from NanoString assessment, where we found significantly increased STAT1 expression in CSA-exposed transplant SCC compared with normal tissue (P < 0.01, Figure 4E). To further explore differences in expression of JAK/STAT genes, we compared NanoString expression data by diagnosis group, BWH stage, histological differentiation, and presence or absence of metastasis (Figure 4F). STAT1 expression was greater than STAT3 in OTRs and moderately differentiated tumors. Whereas, STAT3 expression was greater than STAT1 expression in tumors with PNI and in poorly differentiated tumors, JAK1 expression was greatest in transplant patients, BWH stage 3 tumors, poorly differentiated tumors, and in tumors that metastasized. These data suggest that CSA exposure may prompt additional signaling pathways in TSCC that favor STAT1 activation.
Figure 4. Cyclosporine A exposure upregulates IL22RA1 expression and increases STAT1 expression in xenograft and patient tumors.
(A) PCR of mRNA extracted from cyclosporine A– (CSA–) and IL-22–treated tumor samples. (B) Immunohistochemistry demonstrated IL-22 receptor α (IL-22RA) expression in all tumors (shown at original magnification, ×10), with stronger staining in the CSA-treated tumors, (C) confirmed by immunoblot with quantification. (D) PCR of tumor mRNA was assessed for expression of markers downstream to IL-22. (E) NanoString digital gene expression analysis showed greater expression of STAT1 in tumors from CSA-exposed organ transplant recipients (OTRs) compared with normal skin (P < 0.01) and with high-risk tumors with perineural invasion (PNI). (F) Comparison of JAK/STAT gene expression via NanoString according to diagnosis group (normal, N; superficial, S; invasive, I; transplant, T; PNI, P), Brigham and Women’s Hospital (BWH) stage, histological differentiation (normal, N; well differentiated, W; moderately differentiated, M; poorly differentiated, P), and presence or absence of metastasis. Data represent the mean of 3+ samples ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, determined by 1-way ANOVA with Dunnett’s multiple comparisons test.
CSA-exposed OTRs show increased STAT1, IFN, RAS, and AP-1 expression, with loss of multiple tumor-suppressor genes.
To further investigate additional pathways that may be activated in the CSA-exposed OTRs, a heatmap was constructed of Z-score–transformed NanoString expression data from FFPE samples and probed for genes relating to CNI-related interference with DNA damage repair mechanisms, STAT1, IL-22, and IFN pathways (Figure 5A). Several proto-oncogenes were found to be highly expressed in the CSA-exposed OTR group, including subunits of the AP-1 transcription factor JUN, FOS, and FOSL1 (P < 0.05 for each), along with NRAS and HRAS (P < 0.01 for both, Figure 5A). Nucleotide excision repair–related genes DDB2, ERCC6, MNAT1, GTF2H3, and XPA, along with genes related to TGF-β signaling (TGFB2, TGFBR2) were also reduced in the OTRs compared with normal tissue (Figure 5A).
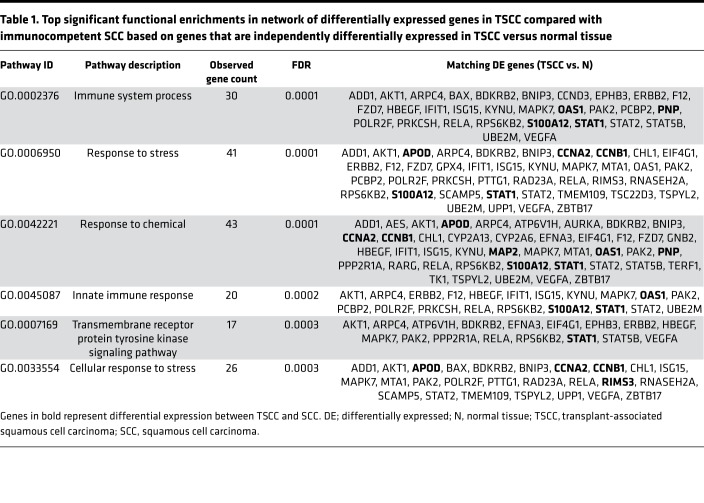
Figure 5. Cyclosporine A–exposed organ transplant recipients show increased STAT1, IFN, RAS oncogene, and AP-1 transcription factor subunit expression and TGF-β dysregulation, with loss of multiple tumor-suppressor genes.
(A) Heatmap of genes relevant to DNA damage, nucleotide excision repair (NER), and IFN pathways based on NanoString data, demonstrating high expression of HRAS, KRAS, JUN, and FOS oncogenes in organ transplant recipients and downregulation of NER and TGF-β–related gene expression. (B) Venn diagram representation of microarray expression data for differentially expressed genes in squamous cell carcinoma (SCC, n = 8) and transplant-associated SCC (TSCC, n = 5) versus normal tissue (N, n = 10) and in TSCC versus SCC. (C) STAT1 microarray expression reveals significant upregulation in TSCC compared with both immunocompetent SCC and normal tissue (mean with SD). *P < 0.05, **P < 0.01, ****P < 0.0001, determined by 1-way ANOVA with Dunnett’s multiple comparisons test.
We also analyzed microarray gene expression data from prospectively obtained fresh tissue of 8 immunocompetent SCCs, 5 TSCCs from OTRs, and 10 normal skin samples. A total of 1,470 probes were differentially expressed in TSCC compared with normal skin (608 upregulated and 862 downregulated, Figure 5B), and 149 probes were differentially expressed in TSCC compared with SCC (109 upregulated and 140 downregulated, Figure 5B). Differentially expressed genes in TSCC compared with SCC, based on those that were also differentially expressed in TSCC versus normal skin, were analyzed in the STRING database of known and predicted protein-protein interactions (37) to produce a visual network of associations in our data set (Supplemental Figure 1). Top significant functional enrichments in this network included genes associated with response to chemicals (43 genes, FDR = 0.0001), response to stress (41 genes, FDR = 0.0001), and innate immune system response (20 genes, FDR = 0.0002, Table 1).
Table 1. Top significant functional enrichments in network of differentially expressed genes in TSCC compared with immunocompetent SCC based on genes that are independently differentially expressed in TSCC versus normal tissue.
Of the genes that were differentially expressed in both SCC and TSCC compared with normal skin, there were 20 genes that were further differentially expressed in TSCC compared with SCC (Figure 5B and Table 2). Differentially upregulated genes included STAT1 (2.82-fold higher in TSCC than SCC, P < 0.01), AP-1 subunit FOSL2 (2.1-fold, P < 0.01), and several genes induced by IL-22, STAT3, and IFN, with the majority of upregulated genes known to be involved in regulating cell proliferation, migration, and/or invasion (Table 2). STAT1 microarray expression revealed significant upregulation in TSCC compared with both immunocompetent SCC and normal tissue (P < 0.01 and P < 0.0001, respectively, Figure 5C). Of note, 3 of 4 of the significantly downregulated genes in TSCC compared with SCC have established tumor-suppressive functions (Table 2). Further, TSPYL2, a regulator of TGF-β signaling, was 3.4-fold downregulated in TSCC compared with SCC (Table 2).
Table 2. Top differentially expressed genes between TSCC and immunocompetent SCC that are also differentially expressed in both TSCC and SCC compared with normal tissue.
Ruxolitinib suppresses SCC cell proliferation and counteracts the proproliferative effects of CSA in vitro via STAT1/3 inhibition.
Given our findings suggesting that CSA causes increased SCC proliferation by JAK1/STAT1/3 signaling, we sought to examine the effects of interrupting this axis on SCC behavior. A431 cells were seeded in full growth media for 24 hours and then treated with increasing concentrations of the JAK1/2 inhibitor, ruxolitinib. Cells were harvested and counted at 48 hours, demonstrating a dose-dependent decrease in proliferation, with ruxolitinib (20 μM) reducing cell counts by 44% compared with vehicle (P < 0.01, Figure 6, A and B). A431 cells were then treated in full growth media with vehicle, CSA (50 ng/ml), ruxolitinib (20 μM), or both ruxolitinib and CSA for 36 hours. Ruxolitinib-treated cells failed to proliferate compared with DMSO vehicle, an effect that was sustained even in combination with CSA (P < 0.01, Figure 6C), which demonstrated a significant increase in proliferation compared with vehicle when used alone (P < 0.01, Figure 6C). Protein harvested from ruxolitinib-treated cells demonstrated reduced STAT1, STAT3 and IL-22R1 expression (Figure 6D). In order to further examine the effect of STAT1 and STAT3 inhibition in SCC, A431 cells were treated with STAT1α-specific inhibitor, S14-95, and STAT3-specific inhibitor, HO-3867, both alone and in combination, and in combination with 25 ng/ml CSA (Figure 6E). Cells were seeded in full growth media, exposed to treatments for 48 hours, and counted following staining with trypan blue to determine cell viability. Compared with CSA alone, ruxolitinib (10 μM) and ruxolitinib with CSA significantly reduced cell counts in these conditions by 50% and 59% (P < 0.001 and P < 0.0001, respectively) (Figure 6E). The STAT1 inhibitor S14-95 also significantly reduced cell counts at 1 μg/ml and 2.5 μg/ml (by 41 and 44%, P < 0.01 for both), as did STAT3 inhibitor HO-3867 at 2.5 μM (33% reduction, P < 0.05, Figure 6E). Compared with CSA alone, STAT1 inhibitor in combination with CSA reduced proliferation by 30% and STAT3 inhibitor in combination with CSA reduced proliferation by 18%; however, these reductions were not statistically significant. STAT1 and STAT3 inhibitors used in combination reduced proliferation by 42% (P < 0.01), and this effect was maintained with the addition of CSA (46% reduction compared with CSA alone, P < 0.01, Figure 6E). There were no significant differences in cell viability observed in any treatment condition compared with vehicle (Supplemental Figure 2). Therefore, STAT1 and STAT3 inhibition each reduce proliferation of SCC cells, and when used in combination, they maintain this reduction, even in the presence of proproliferative CSA.
Figure 6. Ruxolitinib reduces squamous cell carcinoma cell proliferation and counteracts the proproliferative effects of cyclosporine A via STAT1/3 inhibition.
(A) A431 squamous cell carcinoma (SCC) cells were treated with increasing concentrations of ruxolitinib and assessed for proliferation over 24 hours. (B) The increased proliferation of A431 cells treated with cyclosporine A (CSA) is inhibited when cells are treated with ruxolitinib (20 μM) and CSA (50 ng/ml) in combination. Representative images are shown (original magnification, ×10) (B) and quantified by cell count (C). (D) Western blot of protein from treated cells demonstrates reduction in STAT1 and STAT3 expression and phosphorylation with reduced IL-22RA expression. (E) Proliferation assay of A431 cells exposed to ruxolitinib (10 μM); STAT1α-specific inhibitor, S14-95; and STAT3-specific inhibitor, HO-3867; alone and in combination, with and without 25 ng/ml CSA. Significance relative to CSA-treated cells is shown. Data for all panels represent mean of 3 experiments ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, determined by 1-way ANOVA with Dunnett’s multiple comparisons test.
Ruxolitinib reduces SCC tumor growth and counteracts the IL-22R1 upregulation and proproliferative effects caused by CSA in vivo.
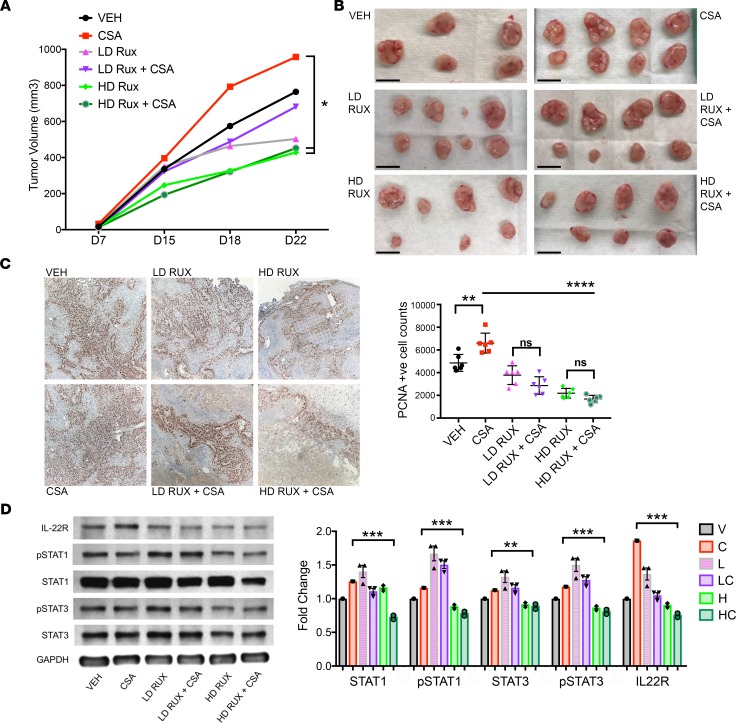
Mice with A431 xenografts were randomized at day 7 (as described above) to 6 treatment groups: CSA (20 mg/kg daily; n = 8), high-dose ruxolitinib (HD Rux; 100 mg/kg/d via oral gavage; n = 8), low-dose ruxolitinib (LD Rux; 10 mg/kg/d via oral gavage; n = 8), HD Rux with CSA (same doses as above; n = 8), LD Rux with CSA (same doses as above; n = 8), or vehicle alone (n = 5). Determination of the high dose was based on published murine experiments in the literature (38), and the low dose was calculated using murine body surface area and current FDA-approved human doses (39). Ruxolitinib had a striking effect on growth of A431 SCC tumors in nude mice. The higher-dose ruxolitinib treatment reduced tumor volume on average by 44% compared with vehicle (P < 0.05, Figure 7, A and B), with 2 tumors demonstrating regression following initiation of treatment. One tumor could not be palpated at endpoint. Addition of HD Rux attenuated the accelerated tumor growth induced by CSA (average 53% volume reduction from CSA alone, P < 0.05, Figure 6A). One tumor also regressed completely in the combined HD Rux with CSA group, and, in 2 others, there was a volume reduction of 71% and 85% compared with vehicle (Figure 7B). The LD Rux group showed a trend of decreased growth by endpoint; however, this was not significant. LD Rux in the presence of CSA treatment did not significantly reduce tumor growth compared with CSA alone (Figure 7, A and B). No adverse effects, defined by weight loss or signs of distress, were observed in ruxolitinib-treated mice at any dose, and all mice survived to endpoint.
Figure 7. The JAK1/2 inhibitor ruxolitinib reduces squamous cell carcinoma tumor growth and counteracts the proproliferative effects and IL-22R upregulation caused by cyclosporine A in vivo.
(A) Nude mice were engrafted with A431 squamous cell carcinoma (SCC) cells and randomized to treatment on day 7 with cyclosporine A (CSA) (20 mg/kg d, i.p., n = 8), low-dose ruxolitinib (LD Rux, 10 mg/kg/d via oral gavage, n = 8), high-dose ruxolitinib (HD Rux, 100 mg/kg/d via oral gavage, n = 8), LD Rux with CSA (n = 8), HD Rux with CSA (n = 8), or vehicle (n = 5). Tumor growth was significantly reduced in mice treated with high-dose ruxolitinib, and the proproliferative effect of CSA treatment on tumor growth was negated by concurrent treatment at this dose. (B) Excised tumors at endpoint, day 22. Scale bars: 1 cm. (C) Immunohistochemistry (IHC) (original magnification, ×10) and quantification of positively staining cells for proliferating cell nuclear antigen (PCNA), performed on tumors harvested from mice at sacrifice following 16 days of treatment. (D) Western blot of protein extracted from tumors and ImageJ quantification shows decreased STAT1, STAT3, and IL-22RA protein expression in combination CSA/HD Rux–treated tumors compared with CSA alone. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, determined by 1-way ANOVA with Dunnett’s multiple comparisons test.
Tumors harvested at sacrifice following 16 days of treatment were fixed, and immunohistochemistry was performed for PCNA as a marker of cell proliferation (Figure 7C). Quantification of PCNA staining showed significantly increased proliferation in the CSA group (P < 0.01, Figure 7C), with a significant reduction in positively stained proliferating cells seen in tumors from HD Rux–treated mice when given alone and in combination with CSA (P < 0.0001, Figure 7C). Protein extracted from treated tumors demonstrated a significant reduction of STAT1 and STAT3 signaling in tumors treated with CSA and HD Rux in combination with CSA compared with CSA treatment alone (P < 0.001, Figure 7D). HD Rux exposure significantly suppressed IL-22R1 protein expression in comparison to the increase seen with CSA treatment alone (P < 0.001, Figure 7D). These data suggest that the CSA-mediated increase of IL-22R1 expression and downstream STAT1 and STAT3 activation is reversed when treatment is combined with ruxolitinib at therapeutic doses. Ruxolitinib is effective in reducing SCC tumor growth via reduced cell proliferation, potentially by its action of JAK1/2 inhibition and subsequent reduction in STAT1 and STAT3 signaling.
Discussion
OTRs have an increased risk of unresectable locally advanced and metastatic SCC; they are also subject to an increased mortality rate once this occurs (40). There are no effective systemic treatments for advanced or metastatic SCC that spare the allograft (41, 42). The present results suggest that ruxolitinib may be a viable therapy in select patients. With these findings, we have identified a strategy for targeted treatment of CSA-induced SCC, along with an application for an existing FDA-approved drug that may be additionally beneficial to use in OTRs, owing to its immunosuppressive properties.
There is a distinct difference in biological behavior between TSCC and immunocompetent SCC. While it is generally accepted that immunosuppressive medications contribute to this, the direct and indirect pathways that lead to an elevated risk for SCC in OTRs have not been fully characterized. We previously found IL-22 axis–related changes between these two populations (19), and CSA exposure was associated with increased risk for catastrophic SCC in our OTRs (18). In the present study, we found that the proproliferative effects of IL-22 on SCC occur via STAT3 phosphorylation and JAK1/STAT3 upregulation. JAK/STAT-related genes were upregulated in OTRs and in SCC with higher risk staging. CSA exposure hastened tumor growth, upregulated IL-22R1 in vivo, and caused increased JAK1, STAT1, and STAT3 expression. STAT1- and STAT3-targeted inhibition reduced SCC cell proliferation in vitro and, when used in combination, negated the proproliferative effects of CSA. Using ruxolitinib, an FDA-approved JAK1/2 inhibitor, we demonstrated inhibition of JAK1, STAT1, and STAT3 in human SCC cells and xenografts, with marked reduction in proliferation both in vitro and in vivo. We were also able to reverse the accelerated tumor growth profile associated with CSA exposure, along with the associated increase in IL-22R expression.
Several other findings stand out from the present studies. We found evidence of TGF-β signaling disruption, a known driver of cutaneous SCC (43, 44), in TSCC compared with SCC in both our NanoString and microarray data sets. We also found a previously unreported increase in STAT1 expression associated with CSA exposure in both xenograft tumors and in patient samples. When we looked for causes for this in both prospectively obtained tissue and FFPE samples from our OTR population, we discovered an IFN-related gene expression profile in TSCC patients and signs of DNA damage with reduced repair mechanisms. STAT1 has been found to have both protumorigenic and antitumorigenic actions, depending on the tumor type and context (45). STAT1 activation and expression can be induced by various stimuli, including IFNs, actions of oncogenes or loss of tumor suppressors, interaction with tumor stroma, and by genotoxic agents, acting to contribute to an immunosuppressive tumor microenvironment via induction of PD-L1, inhibition of apoptosis, and promotion of invasion and metastasis (45). Calcineurin inhibition, the mechanism of action of CSA, has been shown to induce AP-1 transcription factor subunit ATF3, which counteracts p53-dependent cancer cell senescence, therefore increasing tumorigenic potential (46). ATF3 is also a potent inducer of STAT1 (47). Our data demonstrate upregulation of CNI-related oncogenes, including other subunits of AP-1, which may also directly effect STAT1 signaling. Additionally, IL-22 has been found to mediate STAT1 activation and the induction of IFN-stimulated genes to produce an antiviral state (23), and IL-22R1 can also be induced to signal via STAT1 in the presence of IFN-α in primary keratinocytes (48). A study comparing invasive SCC in immunocompetent patients versus OTRs found an increase in STAT1-positive cells in the perineoplastic inflammatory infiltrate from OTRs, thought to be induced by IFN-γ from activated T cells and NK cells in response to infection or graft rejection (49). Our data demonstrate upregulation of both type 1 IFNs and IFN-γ, along with several IFN-inducible genes, in CNI-exposed OTRs providing support to this theory. Since OTRs are immunosuppressed and in a virus-susceptible state, the antiviral activation of IFNs and STAT1 intended to protect the host may also play a role in tumorigenesis.
We found the most prominent proliferative effect from IL-22 stimulation occurred in cell lines that are yet to develop metastatic potential. We also found that the proproliferative effects on SCC occur rapidly following treatment with IL-22 via STAT3 phosphorylation and JAK1/STAT3 upregulation, and demonstrated an early proliferative response to IL-22 exposure in A431 xenograft tumors. Owing to the small effect sizes seen in this study, the data, despite being statistically significant, should be interpreted with some degree of caution. However, our findings highlight a potential role for the CSA/IL-22 proliferation axis in early stages of SCC and may relate to the varying sources of IL-22 production in different cell types. Group 3 ILCs have been found to be the main producers of IL-22 in early stages of inflammation in murine models of infection (21, 50). Conversely, conventional αβT cells take over this function in later stages (51). Therefore, in OTRs where T cell function is predominantly depleted, innate sources of IL-22 production may prove important in the context of carcinogenesis. Inhibiting JAK/STAT activation downstream of innate immune cell IL-22 production may reduce the increased proliferative stimulus present in immunosuppressed OTRs.
The choice of immunosuppressive agents in OTRs may effect the composition of residual immunity and even promote some immune functions — for example, memory T cells are not inhibited by the immunosuppressants sirolimus or mycophenolic acid, and it has been suggested that this may contribute to a lower risk of skin carcinogenesis with those agents (52). Sirolimus and CNIs are also known to have differing effects on Treg numbers, function, and phenotype (53, 54). Given our previous findings of an altered T cell phenotype in fresh tissue from CSA-treated OTRs, this is of interest (18, 19). In SCC from OTRs, T cell production was polarized toward IL-22–producing T22 cells, and the ratio of Treg cells to cytotoxic CD8 cells was increased (18, 19). There is also evidence that high Treg numbers predict recurrent SCC in kidney transplant patients (55) and that Tregs directly regulate IL-22 expression via regulation of IL-22–producing innate and adaptive immune cells (56). Ruxolitinib has been shown to inhibit dendritic cells, NK cells, and T cells (57); induce long-lasting Treg reduction in myelofibrosis patients (58); and prevent cytokine release syndrome associated with CAR-T therapy, without impairing its antitumor efficacy (59). Promising results are also emerging with JAK inhibition in other IL-22–related hyperproliferative cutaneous conditions, such as psoriasis (60, 61). Therefore, ruxolitinib may help balance downstream effects of proproliferative cytokine release, while also assisting long-term allograft survival by promoting immune tolerance of the transplanted organ.
Ruxolitinib is mostly well tolerated; however, there have been reports of increased skin cancer among those with hematological malignancies exposed to the drug (62, 63). This has been partially attributed to high recurrence rates of NMSC in the hematological malignancy population, especially in the context of previous exposure to hydroxyurea, an independent risk factor for development of NMSC (63, 64). A recent case series reported 5 patients being treated with ruxolitinib who developed skin cancers with aggressive biological behavior (62). Each of these patients, however, had multiple identifiable preexisting significant risk factors for aggressive cutaneous carcinogenesis (62). Further, we previously found hematological malignancy to be an independent risk factor associated with metastatic SCC (65), and patients with CLL have been shown to have distinct T cell changes that are different from those seen in immunosuppressed OTRs (66). Therefore, the effects of ruxolitinib in the context of hematological malignancy may not be applicable to OTRs.
In summary, CSA may promote SCC development in OTRs via a direct drug effect on DNA repair mechanisms to exacerbate UV-induced DNA damage, along with creating a proproliferative cytokine signature caused by an imbalance of innate and adaptive immunity. JAK/STAT signaling was observed to be a common downstream pathway in both of these mechanisms of increased proliferative stimulus. Ruxolitinib, a safe and tolerable oral JAK1/2 inhibitor with preexisting FDA approval and immunosuppressant properties, may present a feasible option for systemic SCC treatment or prevention in OTRs requiring long-term immunosuppression. We have demonstrated significant reductions in SCC proliferation and xenograft tumor growth following treatment with this drug. Further, owing to its mechanism of action, the addition of ruxolitinib negates the stimulatory effects of CSA on the JAK/STAT pathway mediated by IL-22R1 upregulation. In theory, there may also be additional beneficial effects of adding ruxolitinib to the OTR immunosuppressant regimen in terms of prevention of chronic rejection, supported by its actions on Tregs and efficacy in steroid refractory graft-versus-host disease. Our data suggest that application of this existing agent may be of particular benefit in the OTR population at high risk of aggressive SCC and provide evidence to support human trials in this area.
Methods
Cell lines and culture.
The human epithelial epidermoid carcinoma cell line A431 was purchased from ATCC, cultured in DMEM supplemented with 10% FBS, and incubated in 5% CO2 at 37°C. Normal human epidermal keratinocytes were purchased from Lifeline Cell Technology and cultured in DermaLife K cell culture medium. T1, T8, MET1, and MET4 cell lines, originally isolated from fresh human tumor, were obtained from the Proby lab in Dundee and cultured as previously described (67–69). T1 was derived from a well-differentiated SCC on the forearm of a renal transplant patient that did not metastasize, and T8 was derived from a poorly differentiated SCC on the ear of a renal transplant patient that did metastasize (Supplemental Table 1) (69). MET1 and MET4 cell lines were derived from a single immunosuppressed patient, from a primary invasive SCC on the back of the hand that subsequently recurred locally twice and from a distant axillary lymph node metastasis that was deemed to have occurred from the MET1 primary tumor (Supplemental Table 1) (67).
Proliferation assay.
Cells were cultured in 0.1% FBS for serum starvation studies for 24–48 hours to synchronize cellular proliferation at G0 phase. In full serum proliferation experiments, cells were seeded in 10% FBS for 12–24 hours prior to treatment. Subsequently, cells were divided into treatment groups: 0.1% or 10% vehicle control, 50 ng/ml CSA (MilliporeSigma), and 100 ng/ml IL-22 (R&D Systems). Both CSA and IL-22 were reconstituted in PBS prior to administration according to the manufacturer’s protocol. Ruxolitinib and HO-3867 were purchased from MedChemExpress, S14-95 was purchased from Santa Cruz Biotechnology, and all three were reconstituted as per the manufacturer’s instructions. At the stated time point, cells were photographed under light microscopy, harvested by trypsinization, and stained with trypan blue for viability exclusion testing; live cells were counted using an automated cell counter (Countess Automated Cell Counter, Invitrogen). Fold change calculations were made in comparison with untreated cells of the same cell line, unless otherwise stated.
qPCR.
Cells were placed in RLT lysis buffer with 1% β-mercaptoethanol (Qiagen) and then mRNA was extracted using the RNeasy micro kit (Qiagen). Tumors were harvested, snap frozen in liquid nitrogen at sacrifice, and stored in –80 degrees. Tumor samples were homogenized in TRIzol (Invitrogen), and total RNA was extracted according to the manufacturer’s protocol. The Qiagen RNeasy mini kit was used to extract mRNA according to the manufacturer’s instructions. Total mRNA concentration and purity were evaluated with the Thermo Fisher Scientific NanoDrop 2000C (cells) and the Pharmacia GeneQuant Pro RNA/DNA calculator (tumors). The SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen) was used according to the manufacturer’s instructions. The following primers were used: STAT3 forward, CAGCAGCTTGACACACGGTA, and reverse, AAACACCAAAGTGGCATGTGA; STAT1 forward, CGGCTGAATTTCGGCACGT, and reverse, CAGTAACGATGAGAGGACCCT; JAK1 forward, CTTTGCCCTGTATGACGAGAAC, and reverse, ACCTCATCCGGTAGTGGAGC; AKT1 forward, AGCGACGTGGCTATTGTGAAG, and reverse, GCCATCATTCTTGAGGAGGAAGT; MTOR forward, CACCAGAATTGGCAGATTTGC, and reverse, CTTGGACGCCATTTCCATGAC; IL-22R1 forward, CCGGCTAACCCTGGACAC, and reverse, TCCAAGGTGCATTTGGTAGGT; IL-10R2 forward, ATGAGCATTCAGACTGGGTAAAC, and reverse, TTTTAGGGGCTAAGAAACGCAT; GAPDH forward, GGAGCGAGATCCCTCCAAAAT, and reverse, GGCTGTTGTCATACTTCTCATGG; and BA forward, AGAGCTACGAGCTGCCTG, and reverse, AGCACTGTGTTGGCGTACAG. All data were normalized to GAPDH and β-actin.
Western blots.
Proteins were extracted from cells (lysed in RIPA buffer with protease inhibitor) and tumors (homogenized and then lysed using the M-PER Mammalian Protein Extraction Reagent, Thermo Fisher Scientific) and then quantified using a commercial protein assay (Bio-Rad DC protein assay). The protein samples (10–20 μg) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. Immunoblot analysis was conducted using antibodies against total STAT3 (9139S, CST), p-STAT3 (Ser727) (9134S, CST), p-STAT3 (Tyr705) (9145S, CST), STAT1 (14994S, CST), p-STAT1 (44-382G, Thermo Fisher Scientific), IL-22RA (bs-2624R, Bioss Inc.), GAPDH (2118S, CST), and β-actin (ab119716, Abcam). The results were visualized using either the chemiluminescent Pierce ECL Substrate Western blotting detection system (Thermo Fisher Scientific) and exposure to autoradiography film (HyBlot ES, Denville Scientific Inc.) or the amplified Opti-C4N substrate kit (Bio-Rad). Bands were quantified using ImageJ (NIH) and normalized to GAPDH or β-actin.
Patients and samples.
Cutaneous SCC samples were obtained from debulking during Mohs micrographic surgery. All tumors were obtained from sun-exposed skin. Normal specimens were obtained from non-sun-exposed areas of patients without skin cancer.
FFPE samples.
Archived FFPE blocks were obtained from 7 normal skin specimens and 38 SCCs. These 38 tumors came from 24 patients, with 10 patients in this cohort having more than one tumor included in the analysis (Supplemental Table 2). This cohort has been previous analyzed in separate studies assessing potential biomarkers in SCC (70). Four consecutive 15-μm sections were obtained from each FFPE block using a standard microtome (Leica Instruments), with microtome blade replacement and equipment sterilization using 100% isopropanol and Terminator RNase remover (Denville) between blocks.
RNA extraction from FFPE.
Total RNA was extracted from FFPE tissue samples using the Agencourt FormaPure kit (Beckman Coulter) as per the manufacturer’s protocol (protocol 000385v005). The final samples were suspended in 80 μl RNase/DNase-free water, and purity level was measured using a NanoDrop Reader (NanoDrop 2000C, Thermo Fisher Scientific).
NanoString analysis.
Total RNA quality was assessed using the Agilent 2200 TapeStation Bioanalyzer. Total RNA samples were processed using a custom probe set via the nCounter Analysis System (NanoString Technologies) as per company’s protocol. The raw quantification that resulted from the nCounter System’s barcode analysis was normalized using the nSolver software (NanoString Technologies) with relation to housekeeping genes. Data obtained via the NanoString nCounter system were analyzed using NanoString nSolver software.
Murine studies.
2 × 104 A431 cells in a 50% Matrigel (Corning) suspension were engrafted via intradermal injection in 2 midline back skin regions of 6-week-old female athymic nude mice (Hsd:Athymic Nude-FOXn1nu, Envigo). Once tumors were palpable, mice were randomized to treatment groups (day 7). The first set of experimental treatment groups are as follows: 4 μg IL-22 daily, 5 days per week (GenScript, catalog Z03243-1, n = 9); 20 mg/kg CSA daily (Perrigo, n = 9); IL-22 plus CSA (n = 7); and vehicle (n = 9). The second set of experimental treatment groups are as follows: 20 mg/kg CSA daily (n = 8), HD Rux (100 mg/kg/d via oral gavage; n = 8), LD Rux (10 mg/kg/d via oral gavage; n = 8), HD Rux with CSA (n = 8), LD Rux with CSA (n = 8), and vehicle alone (n = 5). Determination of the high dose was based on published murine experiments in the literature (38), and the low dose was calculated using murine body surface area and current FDA-approved human doses (39). Tumors were detected by palpation and measured using a digital caliper, and tumor volume was calculated as follows: 1/2 × l × w2, where l represents length in mm and w represents width in mm.
Immunohistochemistry.
Paraffin-embedded 5-μm tumor sections were deparaffinized and rehydrated. Antigen retrieval was performed in Tris base (pH 9) at 100°C for 30 minutes. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide solution. Slides were blocked with 5% NGS. Primary antibodies used were PCNA (PC10, Abcam) and IL-22RA (bs-2624R, Bioss Inc.) at 1:100 dilution, incubated at room temperature for 1 hour. Sections were then incubated with biotinylated rabbit anti-mouse (PK6102, Vector Laboratories) or anti-rabbit (PK6101, Vector Laboratories) secondary antibody and then avidin-biotin peroxidase complex (Vector Laboratories) and developed with diaminobenzidine. Counterstaining was performed with 10% hematoxylin.
Microarray analysis.
Total RNA was extracted from tissue samples of normal skin (n = 10) and immunocompetent SCC (n = 8) and SCC from OTRs (n = 5) and converted to biotinylated cRNA as previously described (71). After fragmentation and quality confirmation with the Affymetrix Test-3 Array, 15 μg of the biotinylated cRNA was hybridized to Affymetrix Human Genome U95A GeneChip (~12,000 probe sets) (Affymetrix Inc.). The chips were washed, stained with streptavidin–phycoerythrin, and scanned with a probe array scanner (HP GeneArray Scanner, Hewlett-Packard Company). Microarray data were analyzed using R/Bioconductor packages (https://www.r-project.org/). Normalized expression values were obtained using the GCRMA algorithm. The data have been deposited to the Gene Expression Omnibus repository (GSE117247). Expression values were linearly modeled in the limma package framework (72). For the comparison of interest, the moderated t test was used to assess differential expression. P values for each comparison were adjusted for multiple hypotheses using the Benjamini-Hochberg approach. Genes with FDR < 0.05 and fold change > 2.0 were considered as differentially expressed. Differentially expressed gene lists were also analyzed using the STRING database v10.5 (http://string-db.org) (37).
Provision of materials.
T1, T8, MET1 and MET4 cell lines were provided by CP’s lab.
Statistics.
Unless otherwise specified, statistical analyses were performed using GraphPad Prism 7 software. One-way ANOVA tests, supplemented with Bonferroni post hoc analysis, were used for the statistical comparison of grouped data. Tukey’s multiple comparison test was used as appropriate to account for multiple comparisons, with P < 0.05 considered significant. Averaged results of multiple experiments are presented as the arithmetic mean ± SEM. For the microarray data, a 2-tailed moderated t test was used to calculate differential expression, and adjusted P values were calculated using the Benjamini-Hochberg approach.
Study approval.
All human studies were reviewed and approved by the institutional review board at NYU Langone Medical Center. Written informed consent was obtained for all patients before their participation, and the study was performed with strict adherence to the Declaration of Helsinki Principles. Animal studies described were reviewed and approved by the Institutional Animal Care and Use Committee at Weill Cornell Medicine and were conducted according to the requirements established by the American Association for Accreditation of Laboratory Animal Care. All procedures were approved by the Institutional Animal Care and Use Committee before the initiation of any studies.
Author contributions
MAB, NR, DF, and JAC designed research studies. MAB, NR, and JC conducted experiments. MAB, NR, ALS, ND, DF, and JAC analyzed data. CP provided study materials. MAB, DF, and JAC wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Cancer Research Institute Clinic and Laboratory Integration Program grant. The authors wish to thank Jeffrey S. Weber for review of manuscript and insightful comments.
Version 1. 09/06/2018
Electronic publication
Footnotes
Conflict of interest: JAC has received income from Dermatology TA, GlaxoSmithKline, and Regeneron for other studies in SCC.
Reference information: JCI Insight. 2018;3(17):e120750. https://doi.org/10.1172/jci.insight.120750.
Contributor Information
Melody Abikhair Burgo, Email: melodyburgo@gmail.com.
Nazanin Roudiani, Email: naz.roudiani@gmail.com.
Jie Chen, Email: jic2002@med.cornell.edu.
Alexis L. Santana, Email: alexis.santana@nyumc.org.
Nicole Doudican, Email: nicole.doudican@nyumc.org.
Diane Felsen, Email: dfelsen@med.cornell.edu.
References
- 1. Karia PS. Epidemiology and outcomes of cutaneous squamous cell carcinoma. In: Schmults CD, ed. High-Risk Cutaneous Squamous Cell Carcinoma: A Practical Guide for Patient Management. Berlin, Heidelberg: Springer Berlin Heidelberg; 2016:3–28. [Google Scholar]
- 2.Guy GP, Machlin SR, Ekwueme DU, Yabroff KR. Prevalence and costs of skin cancer treatment in the U.S., 2002-2006 and 2007-2011. Am J Prev Med. 2015;48(2):183–187. doi: 10.1016/j.amepre.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santana AL, Felsen D, Carucci JA. Interleukin-22 and cyclosporine in aggressive cutaneous squamous cell carcinoma. Dermatol Clin. 2017;35(1):73–84. doi: 10.1016/j.det.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevenson ML, et al. Expression of programmed cell death ligand in cutaneous squamous cell carcinoma and treatment of locally advanced disease with pembrolizumab. JAMA Dermatol. 2017;153(4):299–303. doi: 10.1001/jamadermatol.2016.5118. [DOI] [PubMed] [Google Scholar]
- 5.Barroso-Sousa R, et al. Incidence of endocrine dysfunction following the use of different immune checkpoint inhibitor regimens: a systematic review and meta-analysis. JAMA Oncol. 2018;4(2):173–182. doi: 10.1001/jamaoncol.2017.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kao JC, et al. Neurological complications associated with anti-programmed death 1 (PD-1) antibodies. JAMA Neurol. 2017;74(10):1216–1222. doi: 10.1001/jamaneurol.2017.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrett GL, et al. Incidence of and risk factors for skin cancer in organ transplant recipients in the United States. JAMA Dermatol. 2017;153(3):296–303. doi: 10.1001/jamadermatol.2016.4920. [DOI] [PubMed] [Google Scholar]
- 8.Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N Engl J Med. 2003;348(17):1681–1691. doi: 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg JN, Zwald FO. Management of skin cancer in solid-organ transplant recipients: a multidisciplinary approach. Dermatol Clin. 2011;29(2):231–241. doi: 10.1016/j.det.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Mittal A, Colegio OR. Skin cancers in organ transplant recipients. Am J Transplant. 2017;17(10):2509–2530. doi: 10.1111/ajt.14382. [DOI] [PubMed] [Google Scholar]
- 11.Krynitz B, Olsson H, Lundh Rozell B, Lindelöf B, Edgren G, Smedby KE. Risk of basal cell carcinoma in Swedish organ transplant recipients: a population-based study. Br J Dermatol. 2016;174(1):95–103. doi: 10.1111/bjd.14153. [DOI] [PubMed] [Google Scholar]
- 12.Moloney FJ, Comber H, O’Lorcain P, O’Kelly P, Conlon PJ, Murphy GM. A population-based study of skin cancer incidence and prevalence in renal transplant recipients. Br J Dermatol. 2006;154(3):498–504. doi: 10.1111/j.1365-2133.2005.07021.x. [DOI] [PubMed] [Google Scholar]
- 13. doi: 10.1007/978-0-387-77574-6_19. Xie J. Molecular biology of basal and squamous cell carcinomas. Sunlight, Vitamin D and Skin Cancer. New York, NY; Springer; 2008:241–251. [DOI] [PubMed] [Google Scholar]
- 14.Athar M, Walsh SB, Kopelovich L, Elmets CA. Pathogenesis of nonmelanoma skin cancers in organ transplant recipients. Arch Biochem Biophys. 2011;508(2):159–163. doi: 10.1016/j.abb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norman KG, Canter JA, Shi M, Milne GL, Morrow JD, Sligh JE. Cyclosporine A suppresses keratinocyte cell death through MPTP inhibition in a model for skin cancer in organ transplant recipients. Mitochondrion. 2010;10(2):94–101. doi: 10.1016/j.mito.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarosh DB, Pena AV, Nay SL, Canning MT, Brown DA. Calcineurin inhibitors decrease DNA repair and apoptosis in human keratinocytes following ultraviolet B irradiation. J Invest Dermatol. 2005;125(5):1020–1025. doi: 10.1111/j.0022-202X.2005.23858.x. [DOI] [PubMed] [Google Scholar]
- 17.Wheless L, Jacks S, Mooneyham Potter KA, Leach BC, Cook J. Skin cancer in organ transplant recipients: more than the immune system. J Am Acad Dermatol. 2014;71(2):359–365. doi: 10.1016/j.jaad.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abikhair M, et al. Cyclosporine A immunosuppression drives catastrophic squamous cell carcinoma through IL-22. JCI Insight. 2016;1(8):e86434. doi: 10.1172/jci.insight.86434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, et al. Increased Tc22 and Treg/CD8 ratio contribute to aggressive growth of transplant associated squamous cell carcinoma. PLoS ONE. 2013;8(5):e62154. doi: 10.1371/journal.pone.0062154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita H, Nograles KE, Kikuchi T, Gonzalez J, Carucci JA, Krueger JG. Human Langerhans cells induce distinct IL-22-producing CD4+ T cells lacking IL-17 production. Proc Natl Acad Sci USA. 2009;106(51):21795–21800. doi: 10.1073/pnas.0911472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim C, Savan R. The role of the IL-22/IL-22R1 axis in cancer. Cytokine Growth Factor Rev. 2014;25(3):257–271. doi: 10.1016/j.cytogfr.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13(1):21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez P, Gronke K, Diefenbach A. A catch-22: Interleukin-22 and cancer. Eur J Immunol. 2018;48(1):15–31. doi: 10.1002/eji.201747183. [DOI] [PubMed] [Google Scholar]
- 24.Julliard W, Owens LA, O’Driscoll CA, Fechner JH, Mezrich JD. Environmental exposures-the missing link in immune responses after transplantation. Am J Transplant. 2016;16(5):1358–1364. doi: 10.1111/ajt.13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodhi SA, Lamb KE, Meier-Kriesche HU. Solid organ allograft survival improvement in the United States: the long-term does not mirror the dramatic short-term success. Am J Transplant. 2011;11(6):1226–1235. doi: 10.1111/j.1600-6143.2011.03539.x. [DOI] [PubMed] [Google Scholar]
- 26.Solhjou Z, Athar H, Xu Q, Abdi R. Emerging therapies targeting intra-organ inflammation in transplantation. Am J Transplant. 2015;15(2):305–311. doi: 10.1111/ajt.13073. [DOI] [PubMed] [Google Scholar]
- 27. Zhao Y, Lan P, Li XC. Modulation of innate immune cells to create transplant tolerance. In: Corradetti B, ed. The Immune Response to Implanted Materials and Devices: The Impact of the Immune System on the Success of an Implant. Cham, Switzerland: Springer International Publishing; 2017:125–150. [Google Scholar]
- 28.Buchert M, Burns CJ, Ernst M. Targeting JAK kinase in solid tumors: emerging opportunities and challenges. Oncogene. 2016;35(8):939–951. doi: 10.1038/onc.2015.150. [DOI] [PubMed] [Google Scholar]
- 29.Huynh J, Etemadi N, Hollande F, Ernst M, Buchert M. The JAK/STAT3 axis: A comprehensive drug target for solid malignancies. Semin Cancer Biol. 2017;45:13–22. doi: 10.1016/j.semcancer.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Mitra A, Raychaudhuri SK, Raychaudhuri SP. IL-22 induced cell proliferation is regulated by PI3K/Akt/mTOR signaling cascade. Cytokine. 2012;60(1):38–42. doi: 10.1016/j.cyto.2012.06.316. [DOI] [PubMed] [Google Scholar]
- 31.Jagasia M, Zeiser R, Arbushites M, Delaite P, Gadbaw B, Bubnoff NV. Ruxolitinib for the treatment of patients with steroid-refractory GVHD: an introduction to the REACH trials. Immunotherapy. 2018;10(5):391–402. doi: 10.2217/imt-2017-0156. [DOI] [PubMed] [Google Scholar]
- 32.Hurwitz HI, et al. Randomized, double-blind, phase II study of Ruxolitinib or placebo in combination with capecitabine in patients with metastatic pancreatic cancer for whom therapy with gemcitabine has failed. J Clin Oncol. 2015;33(34):4039–4047. doi: 10.1200/JCO.2015.61.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu HA, et al. A phase 1/2 trial of ruxolitinib and erlotinib in patients with egfr-mutant lung adenocarcinomas with acquired resistance to erlotinib. J Thorac Oncol. 2017;12(1):102–109. doi: 10.1016/j.jtho.2016.08.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heine A, et al. The JAK-inhibitor ruxolitinib impairs dendritic cell function in vitro and in vivo. Blood. 2013;122(7):1192–1202. doi: 10.1182/blood-2013-03-484642. [DOI] [PubMed] [Google Scholar]
- 35.Karia PS, Jambusaria-Pahlajani A, Harrington DP, Murphy GF, Qureshi AA, Schmults CD. Evaluation of American Joint Committee on Cancer, International Union Against Cancer, and Brigham and Women’s Hospital tumor staging for cutaneous squamous cell carcinoma. J Clin Oncol. 2014;32(4):327–334. doi: 10.1200/JCO.2012.48.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwald F, et al. Recommendations for solid organ transplantation for transplant candidates with a pretransplant diagnosis of cutaneous squamous cell carcinoma, merkel cell carcinoma and melanoma: a consensus opinion from the International Transplant Skin Cancer Collaborative (ITSCC) Am J Transplant. 2016;16(2):407–413. doi: 10.1111/ajt.13593. [DOI] [PubMed] [Google Scholar]
- 37.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallath S, et al. CADM1 inhibits squamous cell carcinoma progression by reducing STAT3 activity. Sci Rep. 2016;6:24006. doi: 10.1038/srep24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm. 2016;7(2):27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez JC, et al. Defining the clinical course of metastatic skin cancer in organ transplant recipients: a multicenter collaborative study. Arch Dermatol. 2003;139(3):301–306. doi: 10.1001/archderm.139.3.301. [DOI] [PubMed] [Google Scholar]
- 41.Lipson EJ, et al. Tumor regression and allograft rejection after administration of anti-PD-1. N Engl J Med. 2016;374(9):896–898. doi: 10.1056/NEJMc1509268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller DM, Faulkner-Jones BE, Stone JR, Drews RE. Complete pathologic response of metastatic cutaneous squamous cell carcinoma and allograft rejection after treatment with combination immune checkpoint blockade. JAAD Case Rep. 2017;3(5):412–415. doi: 10.1016/j.jdcr.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cammareri P, et al. Inactivation of TGFβ receptors in stem cells drives cutaneous squamous cell carcinoma. Nat Commun. 2016;7:12493. doi: 10.1038/ncomms12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Epping MT, et al. TSPYL2 is an essential component of the REST/NRSF transcriptional complex for TGFβ signaling activation. Cell Death Differ. 2015;22(8):1353–1362. doi: 10.1038/cdd.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meissl K, Macho-Maschler S, Müller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. 2017;89:12–20. doi: 10.1016/j.cyto.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Wu X, et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature. 2010;465(7296):368–372. doi: 10.1038/nature08996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JY, et al. The induction of STAT1 gene by activating transcription factor 3 contributes to pancreatic beta-cell apoptosis and its dysfunction in streptozotocin-treated mice. Cell Signal. 2010;22(11):1669–1680. doi: 10.1016/j.cellsig.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Bachmann M, Ulziibat S, Härdle L, Pfeilschifter J, Mühl H. IFNα converts IL-22 into a cytokine efficiently activating STAT1 and its downstream targets. Biochem Pharmacol. 2013;85(3):396–403. doi: 10.1016/j.bcp.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Mühleisen B, et al. Progression of cutaneous squamous cell carcinoma in immunosuppressed patients is associated with reduced CD123+ and FOXP3+ cells in the perineoplastic inflammatory infiltrate. Histopathology. 2009;55(1):67–76. doi: 10.1111/j.1365-2559.2009.03324.x. [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 51.Basu R, et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37(6):1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung JW, et al. Does the nature of residual immune function explain the differential risk of non-melanoma skin cancer development in immunosuppressed organ transplant recipients? Int J Cancer. 2016;138(2):281–292. doi: 10.1002/ijc.29450. [DOI] [PubMed] [Google Scholar]
- 53.Noris M, et al. Regulatory T cells and T cell depletion: role of immunosuppressive drugs. J Am Soc Nephrol. 2007;18(3):1007–1018. doi: 10.1681/ASN.2006101143. [DOI] [PubMed] [Google Scholar]
- 54.Akimova T, et al. Differing effects of rapamycin or calcineurin inhibitor on T-regulatory cells in pediatric liver and kidney transplant recipients. Am J Transplant. 2012;12(12):3449–3461. doi: 10.1111/j.1600-6143.2012.04269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hope CM, Grace BS, Pilkington KR, Coates PT, Bergmann IP, Carroll RP. The immune phenotype may relate to cancer development in kidney transplant recipients. Kidney Int. 2014;86(1):175–183. doi: 10.1038/ki.2013.538. [DOI] [PubMed] [Google Scholar]
- 56.Lin S, Yang X, Liang D, Zheng SG. Treg cells: a potential regulator for IL-22 expression? Int J Clin Exp Pathol. 2014;7(2):474–480. [PMC free article] [PubMed] [Google Scholar]
- 57.Cornez I, Yajnanarayana SP, Wolf AM, Wolf D. JAK/STAT disruption induces immuno-deficiency: Rationale for the development of JAK inhibitors as immunosuppressive drugs. Mol Cell Endocrinol. 2017;451:88–96. doi: 10.1016/j.mce.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 58.Massa M, Rosti V, Campanelli R, Fois G, Barosi G. Rapid and long-lasting decrease of T-regulatory cells in patients with myelofibrosis treated with ruxolitinib. Leukemia. 2014;28(2):449–451. doi: 10.1038/leu.2013.296. [DOI] [PubMed] [Google Scholar]
- 59.Kenderian SS, et al. Ruxolitinib prevents cytokine release syndrome after car T-cell therapy without impairing the anti-tumor effect in a xenograft model. Blood. 2017;23(3):S19–S20. [Google Scholar]
- 60.Damsky W, King BA. JAK inhibitors in dermatology: The promise of a new drug class. J Am Acad Dermatol. 2017;76(4):736–744. doi: 10.1016/j.jaad.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu L, Armstrong AW. JAK inhibitors: treatment efficacy and safety profile in patients with psoriasis. J Immunol Res. 2014;2014:283617. doi: 10.1155/2014/283617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blechman AB, et al. Aggressive skin cancers occurring in patients treated with the janus kinase inhibitor ruxolitinib. J Drugs Dermatol. 2017;16(5):508–511. [PubMed] [Google Scholar]
- 63.Harrison CN, et al. Long-term findings from COMFORT-II, a phase 3 study of ruxolitinib vs best available therapy for myelofibrosis. Leukemia. 2016;30(8):1701–1707. doi: 10.1038/leu.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gómez M, et al. Risk factors for non-melanoma skin cancer in patients with essential thrombocythemia and polycythemia vera. Eur J Haematol. 2016;96(3):285–290. doi: 10.1111/ejh.12588. [DOI] [PubMed] [Google Scholar]
- 65.Stevenson ML, Kim R, Meehan SA, Pavlick AC, Carucci JA. Metastatic cutaneous squamous cell carcinoma: the importance of t2 stratification and hematologic malignancy in prognostication. Dermatol Surg. 2016;42(8):932–935. doi: 10.1097/DSS.0000000000000798. [DOI] [PubMed] [Google Scholar]
- 66.Riches JC, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612–1621. doi: 10.1182/blood-2012-09-457531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Proby CM, et al. Spontaneous keratinocyte cell lines representing early and advanced stages of malignant transformation of the epidermis. Exp Dermatol. 2000;9(2):104–117. doi: 10.1034/j.1600-0625.2000.009002104.x. [DOI] [PubMed] [Google Scholar]
- 68.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 1981;41(5):1657–1663. [PubMed] [Google Scholar]
- 69.Watt SA, et al. Integrative mRNA profiling comparing cultured primary cells with clinical samples reveals PLK1 and C20orf20 as therapeutic targets in cutaneous squamous cell carcinoma. Oncogene. 2011;30(46):4666–4677. doi: 10.1038/onc.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abikhair M, et al. MAGEA3 expression in cutaneous squamous cell carcinoma is associated with advanced tumor stage and poor prognosis. J Invest Dermatol. 2017;137(3):775–778. doi: 10.1016/j.jid.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 71.Haider AS, et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J Invest Dermatol. 2006;126(4):869–881. doi: 10.1038/sj.jid.5700157. [DOI] [PubMed] [Google Scholar]
- 72.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.