Abstract
BACKGROUND. The extent of weight loss among patients undergoing bariatric surgery is highly variable. Herein, we tested the contribution of genetic background to such interindividual variability after biliopancreatic diversion with duodenal switch.
METHODS. Percentage of excess body weight loss (%EBWL) was monitored in 865 patients over a period of 48 months after bariatric surgery, and two polygenic risk scores were constructed with 186 and 11 (PRS186 and PRS11) single nucleotide polymorphisms previously associated with body mass index (BMI).
RESULTS. The accuracy of the %EBWL logistic prediction model — including initial BMI, age, sex, and surgery modality, and assessed as the area under the receiver operating characteristics (ROC) curve adjusted for optimism (AUCadj = 0.867) — significantly increased after the inclusion of PRS186 (ΔAUCadj = 0.021; 95% CI of the difference [95% CIdiff] = 0.005–0.038) but not PRS11 (ΔAUCadj= 0.008; 95% CIdiff= –0.003–0.019). The overall fit of the longitudinal linear mixed model for %EBWL showed a significant increase after addition of PRS186 (–2 log-likelihood = 12.3; P = 0.002) and PRS11 (–2 log-likelihood = 9.9; P = 0.007). A significant interaction with postsurgery time was found for PRS186 (β = –0.003; P = 0.008) and PRS11 (β = –0.008; P = 0.03). The inclusion of PRS186 and PRS11 in the model improved the cost-effectiveness of bariatric surgery by reducing the percentage of false negatives from 20.4% to 10.9% and 10.2%, respectively.
CONCLUSION. These results revealed that genetic background has a significant impact on weight loss after biliopancreatic diversion with duodenal switch. Likewise, the improvement in weight loss prediction after addition of polygenic risk scores is cost-effective, suggesting that genetic testing could potentially be used in the presurgical assessment of patients with severe obesity.
FUNDING. Heart and Stroke Foundation of Canada (G-17-0016627) and Canada Research Chair in Genomics Applied to Nutrition and Metabolic Health (no. 950-231-580).
Keywords: Genetics
Keywords: Obesity, Surgery
Genetic background has a significant impact on weight loss prediction after bariatric surgery, suggesting a potential use in the pre-surgical assessment of patients with obesity.
Introduction
Bariatric surgery in any of its modalities represents nowadays one of the most effective weight loss treatments for individuals with severe obesity (1). However, the degree of weight loss in response to the surgery is subject to a high variability (2). A number of factors have been previously identified as having a significant role in such interindividual variability. Among them, presurgical body mass index (BMI), age, or sex consistently stand as the most relevant determinants of the heterogeneous response to the weight loss surgery (3, 4).
Other underlying factors such as genetic background have emerged as potentially relevant for bariatric surgery outcomes (5–15). Studies reporting on the impact of genetic makeup on postsurgery weight loss have generated variable results, depending on the type of surgery (14, 15) or the statistical methods used to evaluate weight loss (5, 6, 8, 9) and the genetic variants analyzed (10–13).
In the present study, we aimed to test the contribution of genetic factors to the interindividual variability observed in weight loss after bariatric surgery, specifically biliopancreatic diversion with duodenal switch. For that, two different statistical methods previously validated for the study of weight loss dynamics were applied to analyze weight loss after biliopancreatic diversion with duodenal switch (5, 6). In order to focus on a relatively large and straightforward sample of genetic variants, we selected SNPs previously associated to BMI in prior GWAS and meta-analyses. Regarding the type of surgery, Roux-en-Y gastric bypass and laparoscopic adjustable gastric banding stand among the most extensively studied (16, 17). In the present study, we focused on biliopancreatic diversion with duodenal switch, a surgery recognized for its particular effectiveness in inducing durable weight loss and metabolic improvements, including type 2 diabetes remission and lipid profile normalization (18, 19). Yet not all patients undergoing biliopancreatic diversion with duodenal switch show equal benefits after surgery, with a number of patients exhibiting reduced weight loss (20). We tested the hypothesis that the contribution of genetic factors in the form of polygenic risk scores to postsurgery weight loss is significant, even in response to one of the most effective bariatric operations.
Results
Bariatric patients.
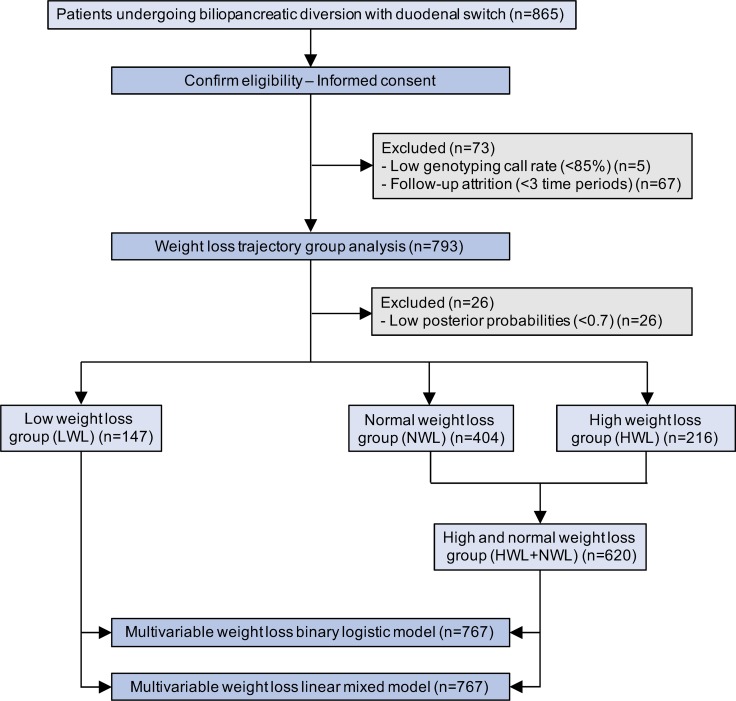
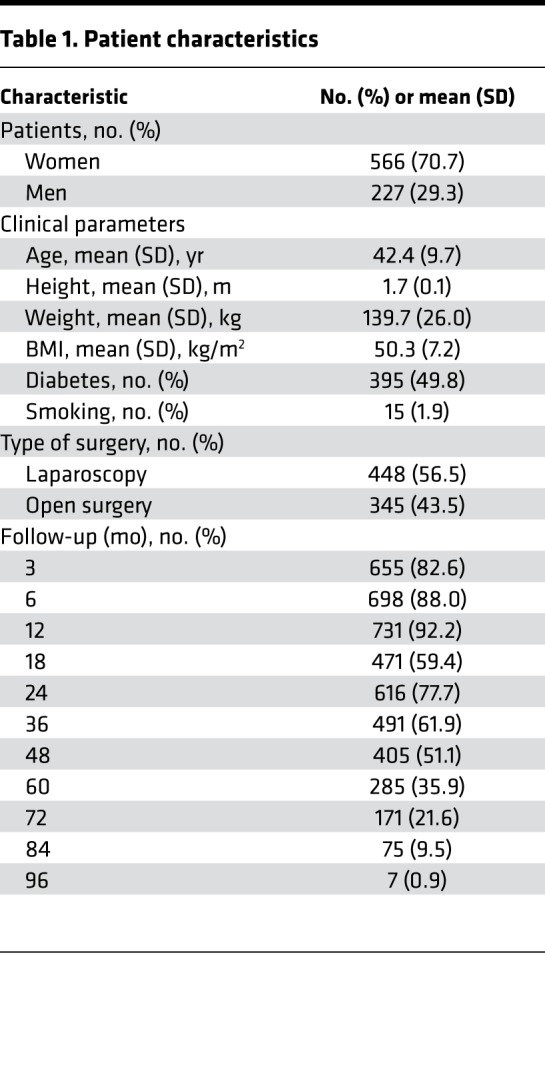
Of the initial 865 patients undergoing biliopancreatic diversion with duodenal switch, 5 were excluded from follow-up analyses due to a low overall call rate (<85%) after analysis of genotyping data (Figure 1). During the follow-up, 11 deaths occurred (1.4%), including two within the first 3 months after surgery (duodenal fistula after open surgery and suicide after laparoscopy). Since patient recruitment extended from 2008 to 2015, several time points were missing for certain subjects, with initial postsurgery weight data completeness varying from a maximum of 89.4% at 12 months to a minimum of 0.9% at 96 months after surgery. Thus, a total of 67 patients were excluded from trajectory analysis due to follow-up attrition (Figure 1). After patient exclusion, the first 7 follow-up time points (3, 6, 12, 18, 24, 36, and 48 months) fulfilled the 50% completeness requirement and were further analyzed (Table 1). The final cohort of 793 bariatric patients was composed of 566 women and 227 men with a mean initial BMI of 50.3 kg/m2 (SD 7.2), and a mean age of 42.4 years (SD 9.7) (Table 1). At the time of surgery, a total of 395 patients (49.8%) were diagnosed with type 2 (n = 392) or type 1 diabetes (3), and 15 were smokers (1.9%). Of the 793 patients, 448 (56.5%) underwent biliopancreatic diversion with duodenal switch by laparoscopy and 345 (43.5%) by open surgery. Open surgery was gradually replaced by laparoscopy throughout the recruitment period, from 97% in 2008, 68% in 2009, 62% in 2010, 44% in 2011 and 30% in 2012, to complete replacement from 2013 onward.
Figure 1. Flow diagram of study participants.
Table 1. Patient characteristics.
Weight loss trajectory group analysis.
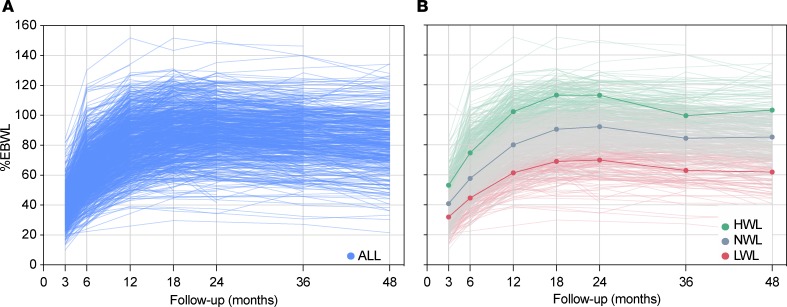
The fitting procedure resulted in a final model where Bayesian information criterion (BIC) was minimized by estimating the percentage of excess body weight loss (%EBWL) trajectories of all the 793 patients for 3 different groups and by fitting the trajectories with cubic functions of time. Significant variables influencing the probability of %EBWL trajectory group assignment (initial BMI, sex, age, and surgery modality) were included in the final model. A total of 26 patients showed posterior probabilities of group membership less than 0.7 and were excluded from further analyses (Figure 1). The 767 remaining patients (Figure 2A) were assigned to 3 trajectory groups depending on %EBWL as follows: high weight loss (HWL), normal weight loss (NWL), and low weight loss (LWL), representing 28.2%, 52.7%, and 19.2% of patients, respectively (Figure 2B). The mean posterior probabilities for belonging to each group were 0.94 HWL, 0.95 NWL, and 0.96 LWL. A Wald test showed that the 3 trajectory groups were significantly and mutually different (χ2 = 369.0; P = 2.3 × 10–16). In order to test the ability of the prediction model to identify patients with a reduced weight loss response to the surgery, patients from the LWL group were compared with patients from the HWL and NWL groups, who were reassigned into a unique group (HWL+NWL) (Figure 1).
Figure 2. Postsurgery weight loss trajectory analysis.
(A) Spaghetti plot showing the percentage of excess body weight loss (%EBWL) of the 767 bariatric patients studied (ALL) over the first 4 years postsurgery (6, 12, 18, 24, 36, and 48 months). (B) Trajectory groups of %EBWL estimated by latent class growth analysis. Weight loss data from the follow-up period of 48 months were used to create groups depending on %EBWL. The 3 resulting trajectory groups were high weight loss (HWL; green lines), representing 28.2% of subjects; normal weight-loss (NWL, gray lines), composed of 52.7% of subjects; and low weight-loss (LWL, red lines), which encompassed 19.2% of subjects.
Multivariable weight loss binary logistic model.
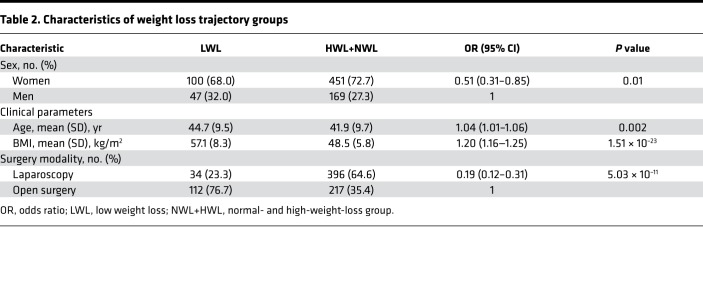
A significant association of surgery modality, sex, initial BMI, and age with %EBWL trajectory group assignment was found in the multivariable binary logistic regression. The probability of belonging to the LWL group was significantly lower in women as compared with men (odds ratio [OR] = 0.51; 95% CI = 0.31–0.85; P = 0.01), as well as in laparoscopy patients as compared with open surgery patients (OR = 0.19; 95% CI = 0.12–0.31; P = 5.03 × 10–11) (Table 2). By contrast, the linear trend test used to analyze the effect of quantitative variables showed that the probability of belonging to the LWL group rose with increasing initial BMI (ORtrend = 1.20; 95% CI = 1.16–1.25; P = 1.51 × 10–23) and age (ORtrend = 1.04; 95% CI = 1.01–1.06; P = 0.002) (Table 2). In other words, patients showing a reduced weight loss response to bariatric surgery were older and had higher initial BMI than patients exhibiting a normal or high-weight-loss response. Moreover, patients with a reduced weight loss response were overrepresented by men undergoing open surgery (Table 2).
Table 2. Characteristics of weight loss trajectory groups.
Impact of polygenic risk scores on weight loss trajectory prediction.
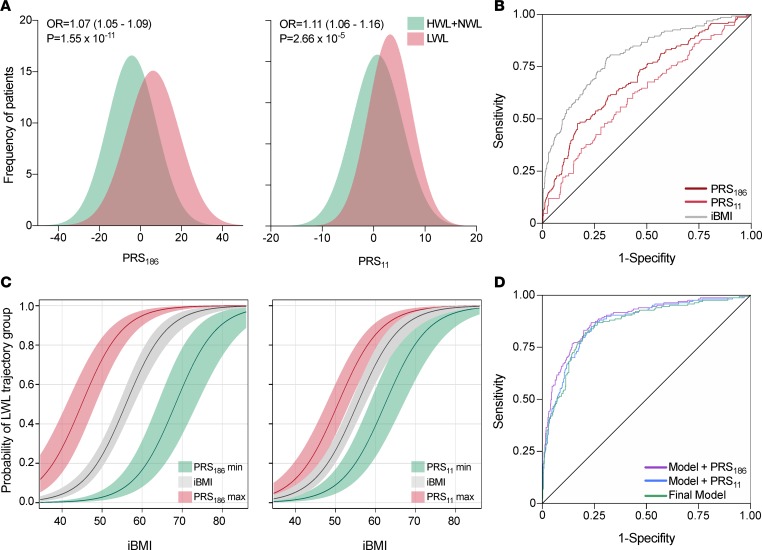
Independent association tests carried out by binary logistic regression revealed that of the 186 SNPs analyzed, only 11 showed a significant association (P < 0.05) with %EBWL trajectory group assignment (Supplemental Table 3; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122011DS1). Among the 5 SNPs significantly associated to the LWL group, 3 were mapped to the MC4R locus. On the other hand, the most highly represented locus within the 6 SNPs significantly associated to the HWL+NWL group was BDNF, with 4 SNPs mapped to this gene. Two weighed polygenic risk scores were then constructed, one including all the SNPs tested (PRS186) and the other including only the 11 significantly associated SNPs (PRS11). Both polygenic risk scores were independently tested into the final %EBWL logistic model, including all variables significantly associated with %EBWL trajectory group assignment (initial BMI, sex, age, and surgery modality) as confounding factors. The linear trend test demonstrated a significant association of PRS186 (ORtrend = 1.06; 95% CI = 1.04–1.08; P = 1.02 × 10–9) and PRS11 (ORtrend = 1.10; 95% CI = 1.04–1.16; P = 4.53 × 10–4) with an increased probability of being in the LWL trajectory group (Figure 3A). Figure 3B shows the predictive value of the %EBWL trajectory group, determined as the area under the ROC curve adjusted for optimism by bootstrapping (AUCadj), displayed by PRS186 alone (AUCadj = 0.711) and PRS11 alone (AUCadj = 0.632). The predictive value of PRS186 was significantly higher than that of PRS11 (ΔAUCadj = 0.121; 95% CI of the difference [95% CIdiff] = 0.03–0.14). On the other hand, initial BMI showed the most elevated predictive value among clinical predictors (AUCadj = 0.807), and, as shown in Figure 3C, the risk of belonging to the LWL group rose with increasing initial BMI. In parallel, increasing values of both PRS186 and PRS11 enhanced the risk of belonging to the LWL group at lower initial BMI, while decreasing values led to the opposite effect. The final model including all the significant predictors (surgery modality, sex, age, and initial BMI) displayed a strong predictive power (AUCadj = 0.867) (Figure 3D). The addition of the PRS186 significantly increased the predictive value of the final model (ΔAUCadj = 0.021; 95% CIdiff = 0.005–0.038), but the inclusion of the simplified PRS11 did not (ΔAUCadj = 0.008; 95% CIdiff = –0.003–0.019) (Figure 3D). Significant improvements in net reclassification index (NRI = 0.638; 95% CI = 0.466-0.809; P = 2.9 × 10–13) and integrated discrimination index (IDI = 0.066; 95% CI = 0.041–0.091; P = 1.7 × 10–7) were also observed after the addition of PRS186 to the final model. Similar but weaker improvements in reclassification indexes were observed after the addition of PRS11 in both NRI (0.279; 95% CI = 0.103–0.455; P = 0.002) and IDI (0.016; 95% CI = 0.003–0.029; P = 0.01). Bootstrapped calibration curves relating predicted and actual probabilities (Supplemental Figure 1) showed that the mean absolute error (MAE) exhibited by the model without genetic information (MAE = 0.014) were minimized after the inclusion of PRS186 (MAE = 0.004) but not PRS11 (MAE = 0.017).
Figure 3. Effect of polygenic risk scores on weight loss trajectory groups.
(A) Histograms displaying the different distribution of polygenic risk scores PRS186 and PRS11 in patients assigned to LWL or HWL+NWL trajectory groups. As shown, increasing values of PRS186 and PRS11 are significantly associated with a greater probability of being in the LWL group. Odds ratios (OR) and P values are for the linear trend test in the multivariable binary logistic model. HWL, NWL, and HWL: high-, normal-, and low-weight-loss trajectory groups. (B) Graphical representation of the area under the ROC curve (AUC) for PRS186 and PRS11. The AUC for initial BMI (iBMI) is also shown. (C) Impact of maximum (red lines) and minimum (green lines) values of PRS186 and PRS11 on the probability of being in the LWL group as a function of iBMI (gray lines). Gray, red, and green shaded areas represent 95% confidence intervals. (D) The AUC of the final logistic prediction models, including all the demographic and clinical predictors before (Final Model: sex, age, type of surgery, and iBMI) and after the inclusion of polygenic risk scores (Model + PRS186 and Model + PRS11) are shown.
Multivariable weight loss linear mixed model.
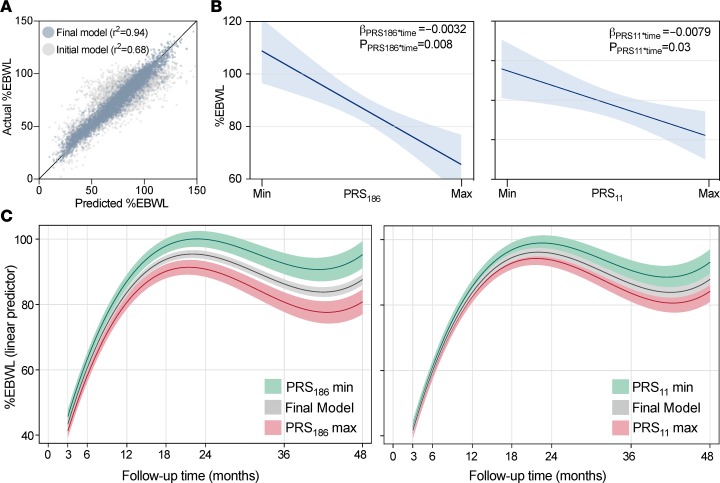
The accuracy of different multivariable linear mixed models of %EBWL over time was estimated by maximum likelihood. Different weight loss models with %EBWL as the dependent outcome were fitted by including predictor variables showing statistical significance when tested in a univariate model, as well as postsurgery time interactions. The initial model only considering significant fixed effects showed a relatively inaccurate fit (r2 = 0.68) (Figure 4A). A large and significant improvement in model accuracy was shown when random effects were added to the model (–2 log-likelihood = 1,050.1; P < 0.0001) (Figure 4A). The final model showing the best goodness of fit (r2 = 0.94) included postsurgery cubic time, age, sex, initial BMI, and surgery modality, as well as time interactions for initial BMI and sex as fixed effects. Intercept and postsurgery cubic time terms were included as random effects with an unstructured covariance matrix. A significant increase in the overall model fit was observed after the inclusion of both PRS186 (–2 log-likelihood = 12.26; P = 0.002) and PRS11 (–2 log-likelihood = 9.87; P = 0.007). Moreover, a significant interaction with postsurgery time was shown for both PRS186 (β = –0.0032; P = 0.008) and PRS11 (β = –0.0079; P = 0.03), revealing a significant difference in %EBWL change over time across increasing polygenic risk score values. Figure 4B illustrates the impact of both PRS186 and PRS11 on %EBWL at the end of the follow-up period. At 48 months and for the standard patient, defined as a 42 years old female laparoscopy patient with an initial BMI of 50, PRS186 represents a 14.2%EBWL (3.6 BMI units) difference between its extreme values, while PRS11 was responsible for 8.5%EBWL (2.4 BMI units). The significant postsurgery time interaction shown by PRS186 and PRS11 is also shown in Figure 4C, where the dynamic effect of PRS186 and PRS11 over time on the %EBWL of the standard patient is shown.
Figure 4. Effect of polygenic risk scores on weight loss evolution over time.
(A) Predicted versus actual longitudinal percentage of excess body weight loss (%EBWL) according to an initial model considering only fixed effects (postsurgery cubic time, age, sex, initial BMI, and surgery modality) and the final multivariable linear mixed model considering fixed and random effects (intercept and postsurgery cubic time terms). (B) Graphical representation of the quantitative impact of the extreme values (Min and Max) of polygenic risk scores PRS186 and PRS11 on %EBWL at the end of the follow-up period (48 months). Estimates are calculated for the standard patient, defined as a 42-year-old woman with a mean initial BMI of 50 undergoing laparoscopic surgery. β Estimates and P values of time interaction are shown. Blue shaded areas are 95% confidence intervals. (C) Gray lines show the linear predictor of %EBWL inferred from the final model (fixed effects: postsurgery cubic time, age, sex, initial BMI, and surgery modality; random effects: intercept and postsurgery cubic time terms). Green and red lines show the linear predictor of %EBWL estimated for the maximum and minimum PRS186 and PRS11 values. Gray, red, and green shaded areas are 95% CIs.
Cost-effectiveness of polygenic risk score inclusion into the prediction model.
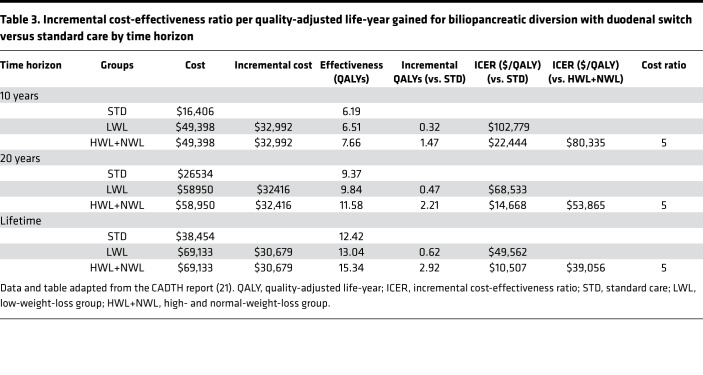
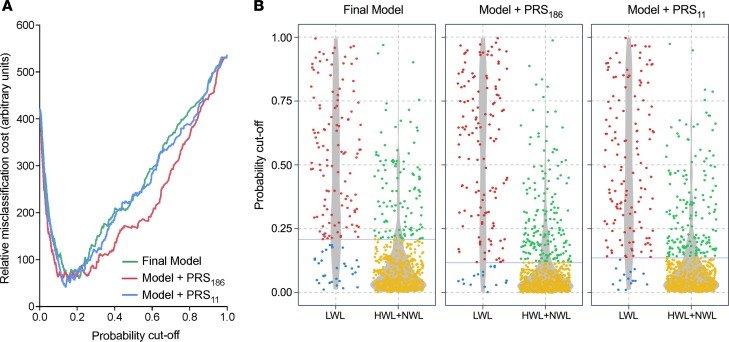
Finally, we further estimated the clinical significance of the inclusion of PRS186 and PRS11 in the predictive model. Using the mean 23%EBWL (5.8 BMI units) difference observed between LWL and HWL+NWL patients at the end of the follow-up period, we first adjusted bariatric surgery effectiveness data, determined as quality-adjusted life-years (QALY), from the Canadian Agency for Drugs and Technologies in Health (CADTH) report (21) (Table 3). In health and economic terms, and assuming a sustained 23%EBWL difference, HWL+NWL patients would represent incremental cost-effectiveness ratio (ICER) reductions of $80,335, $53,865, and $39,056 per QALY, at time horizons of 10 years, 20 years, and lifetime, respectively, as compared with LWL patients (Table 3). In other words, the incorrect assignment of LWL patients to the HWL+NWL group, or false negatives (FNs), was estimated as 5 times as costly as false positives (FPs), i.e., HWL+NWL patients incorrectly assigned to the LWL group (Table 3). Cost curves were then created including this misclassification cost ratio. As shown in Figure 5A, optimal cutoff values minimizing misclassification cost were 0.21 for the final model before adding genetic information, 0.12 after the inclusion of PRS186, and 0.14 for the model including PRS11. At these probability thresholds, the sensitivity of the prediction model including a polygenic risk score was higher than that of the model without it, with the number of LWL patients incorrectly assigned to the HWL+NWL group decreasing around 50% after addition of both polygenic risk scores. In concrete terms, the number of FNs decreased from 20.4% to 10.9% (from 30 to 16 patients) after the inclusion of PRS186, and from 20.4% to 10.2% (from 30 to 15 patients) after the inclusion of PRS11 (Figure 5B).
Table 3. Incremental cost-effectiveness ratio per quality-adjusted life-year gained for biliopancreatic diversion with duodenal switch versus standard care by time horizon.
Figure 5. Cost-effectiveness analysis of polygenic risk scores.
(A) Cost curves of the final logistic prediction model including all the demographic and clinical predictors (Final Model: sex, age, type of surgery, and initial BMI), Model + PRS186, and Model + PRS11 are shown. Minimal misclassification cost was obtained by estimating a false negative (FN) decision (actual LWL patients assigned to HWL+NWL group) to be 5 times as costly as a false positive (FP) decision (actual HWL+NWL assigned to LWL group). (B) Confusion matrix of each model showing the different distribution of correctly (true positives, TP, and true negatives, TN) and incorrectly (FN and FP) assigned observations. Blue lines indicate the probability cutoff with minimal misclassification cost for each model (final model = 0.21, model + PRS186 = 0.12, and model + PRS11 = 0.14). Red and blue points represent TN and FN; green and yellow points, TP and FP, respectively. Gray shaded areas are violin plots representing the density of observations across probability cutoffs. HWL, NWL, and LWL: high-, normal-, and low-weight-loss groups.
Discussion
In the present study, we took a straightforward approach to construct two BMI-associated polygenic risk scores and test their predictive ability toward weight loss trajectory after biliopancreatic diversion with duodenal switch. Our results show that genetic background has a significant impact on weight loss after bariatric surgery and improves weight loss prediction before the surgery.
A dual methodology was used to analyze the impact of BMI-associated polygenic risk scores on postsurgery weight loss. First, a semiparametric trajectory–based analysis was used to stratify patients into high and low responders to the surgery in terms of %EBWL. This methodology has been broadly used to analyze longitudinal weight loss data (2, 5, 6) and has been demonstrated to be an accurate method to estimate weight loss over time. In this study, 3 different %EBWL groups were generated by trajectory analysis. The group of low responders (LWL) was composed of about 20% of patients. Similar results have been obtained in previous studies analyzing clinical outcomes of bariatric surgery (20, 22), which makes biliopancreatic diversion with duodenal switch a very successful weight loss therapeutic option with regard to the extent of weight loss and long-term weight loss maintenance (23). The relatively moderate impact of polygenic risk scores on weight loss in the present study could be due to the fact that biliopancreatic diversion with duodenal switch is a very effective surgery, which probably explains why the benefit of adding a polygenic risk score to prediction models, though significant, is not as strong as expected. Indeed, the individuals labeled as low responders in the present study had an estimated %EBWL of 67.6%. We propose that the genetic scores generated in the present study are likely to be even more significant with surgeries or other weight loss approaches generating more variable responses. In any case, initial BMI was the strongest variable affecting trajectory group assignment, as previously reported (24), leaving only a small portion of the variance to be explained by other predictors.
The second methodology used to test the impact of polygenic risk scores on postsurgery weight loss was a multivariable linear mixed model that returned a more comprehensive value of the actual genetic effect on %EBWL. Moreover, testing the genetic effect over several postoperative time points allowed us to reveal a significant interaction between polygenic risk scores and time after surgery. This is particularly interesting, since it shows that the observed predictive value of polygenic risk scores on %EBWL group trajectory assignment is mediated not by their potential association with initial BMI, but through a continuous effect on weight loss over time. Thus, one interpretation for the significant interaction between postsurgery time and polygenic scores is that weight loss variability could be predicted presurgically by polygenic risk score levels, independently of initial BMI, as illustrated in the present study. Similar results but of lower magnitude have been observed in previous studies (25–28). In this regard, the main distinctive feature of the present study resides in its use of genome-wide BMI-associated SNPs, which a priori should warrant an eventual impact of polygenic risk scores on weight loss dynamics. Interestingly, a large percentage of BMI-associated SNPs analyzed in this study showed a significant association with greater weight loss over time. In this regard, it is worth highlighting that the MC4R and BDNF genes carried the highest number SNPs independently associated to %EBWL, but with opposite directionality of effect. Interestingly, whereas the SNPs mapped to MC4R were associated to a lower predisposition to weight loss, those mapped to BDNF showed a significant association with greater weight loss in response to surgery. Such within-gene directional consistency of SNP effects supports the notion that biliopancreatic diversion with duodenal switch induces pathway alterations. Accordingly, previous studies have shown that metabolic improvements obtained through bariatric surgery are mediated by the induction/suppression of specific metabolic pathways, which would lead to uncovering/masking SNP effects after surgery (29–31).
Presurgical evaluation of patients with severe obesity markedly depends on cost-effectiveness analysis. In this regard, the clinical interpretation of the significant improvements in the predictive power of the model observed after the inclusion of PRS186 and PRS11 must be taken with some caution. In order to assess the clinical utility of polygenic risk scores, an estimation of ICER-per-QALY gain was derived from CADTH report data (21). Assuming a linear association between QALY gain and BMI decrease, a sustained BMI difference between LWL and HWL+NWL groups, and no further changes in BMI occurring in any group beyond 10 years (21), the incorrect assignment of a LWL patient to the HWL+NWL group was estimated to be 5 times as costly as to assign an HWL+NWL patient to the LWL group at each time horizon analyzed. One interpretation of these results is that health benefits from bariatric surgery in LWL patients are less evident than in HWL+NWL patients, possibly leading to increasing health costs. Although the actual cost of genotyping has not been included as a factor in this analysis, the growing demand for diagnostic genotyping should tend to decrease it, improving the cost-effectiveness of genetic testing in the presurgical evaluation of bariatric patients. Taken together, these promising results emphasize the need to deepen the clinical utility of polygenic risk scores, by validating results in larger and heterogeneous study samples, as well as by using a large number of genetic variants, which will help in the decision-making about whether it is worth including genetic background in presurgical evaluation procedures.
Since the exclusion of patients with any of the follow-up data missing could have introduced selection bias and given the ability of the Proc Traj modeling to handle random missing data, we resolved to include those patients with at least 3 follow-up time points. Moreover, in order to minimize misclassification error, a posterior probability of class assignment equal or greater than 0.7 was established (32). As previously stated, patient recruitment for the present study was extended from 2008 to 2015, leading to a high level of attrition for patients enlisted at the end of the recruitment period. In order to keep most follow-up time points without losing too much data, we set an additional 50% completeness as inclusion threshold per time period. We acknowledge this as a limitation of the dataset, since long-term applicability of the prediction model cannot be determined based on available data. Nevertheless, although more genetics-based long-term weight-loss analyses are expected in the field, results presented herein are relevant enough to point out genetic background as a factor worth considering when assessing bariatric surgery eligibility.
In conclusion, genetic background in the form of BMI-associated polygenic risk scores has shown to be a significant predictor of weight loss trajectory after biliopancreatic diversion with duodenal switch. On the one hand, the significant association found between polygenic risk scores and %EBWL trajectory group assignment supports a role for genetic background in the interindividual variability observed in weight loss after biliopancreatic diversion with duodenal switch. On the other hand, the significant interaction found between polygenic risk scores and weight loss evolution over time suggests that patients with different genetic backgrounds are losing weight over time at different rates, supporting a significant impact of genetics on postsurgery weight loss. Taken together, these results open the door to more in-depth studies about the impact of genetic background on postsurgery weight loss dynamics. Finally, the results suggest that genetic testing could potentially be included as a complementary tool in the presurgical assessment of patients with severe obesity.
Methods
Study participants.
A total of 865 patients with severe obesity (BMI ≥35 kg/m2) undergoing biliopancreatic diversion with duodenal switch between February 2008 and March 2015 at the Bariatric Surgery Clinic of the Quebec Heart and Lung Institute were included in the present study. Fasting blood samples for measurement of metabolic parameters and DNA isolation were taken preoperatively. On the eve of the surgery, height and body weight were measured and used to calculate the initial BMI as the weight in kilograms divided by the square of the height in meters. Body weight was further measured during postoperative visits or phone calls thorough a follow-up period of 8 years. %EBWL was then calculated at each follow-up time point (3, 6, 12, 24, 36, 48, 60, 72, 84, and 96 months) as the difference between actual body weight loss (calculated initial BMI minus actual BMI) and ideal body weight loss (calculated as the initial BMI minus ideal BMI fixed at 25 kg/m2). Patients underwent one of the two surgical modalities of biliopancreatic diversion with duodenal switch: open surgery or laparoscopic surgery. The surgical protocol, blood sample collection, and standardized procedures to measure anthropometric and metabolic parameters are described elsewhere (33). All study participants provided written informed consent. Blood samples for DNA extraction were obtained from the Biobank of the Quebec Heart and Lung Institute according to institutionally approved management modalities.
SNP selection and genotyping.
SNPs previously associated with BMI were selected from the NHGRI-EBI GWAS catalog (34) by using “body mass index” as both search term and disease/trait filter. A total of 167 significantly BMI-associated SNPs (P < 5 × 10−8) and 24 SNPs suggestively BMI-associated SNPs (P < 1 × 10−6) were selected from 15 previous GWAS and 5 GWAS meta-analyses. The complete list of SNPs and GWAS is shown in Supplemental Table 1. Selected SNPs were then genotyped in the entire cohort of 865 patients by using TaqMan probes (Applied Biosystems). Genomic DNA was extracted from the blood buffy coat using the GenElute Blood Genomic DNA kit (MilliporeSigma). Genotypes were determined using 7500 Fast Real-Time PCR System (Applied Biosystems) and analyzed using a high-throughput array technology QuantStudio 12K Flex System, coupled with TaqMan OpenArray Technology (Life Technologies). Inclusion criteria for BMI-associated SNPs were a minor allele frequency (MAF) >1%, call rate >95%, and Hardy-Weinberg equilibrium (HWE) P > 2.6 × 10–4. Sample quality, call rate, allele frequencies, and HWE tests were assessed using PLINK 1.9 (35). A total of 5 SNPs were excluded based on HWE and MAF criteria, leaving 186 SNPs for statistical analyses and polygenic risk score construction (Supplemental Table 2).
Statistics.
In order to identify patients with a reduced postsurgery weight loss response, bariatric patients following similar %EBWL trajectories after surgery were first grouped by semiparametric trajectory analysis. Second, based on %EBWL trajectory groups, a predictive model of weight loss response to the bariatric surgery, including clinical, demographic, and genetic factors (polygenic risk scores), was assessed by binary logistic regression. Finally, a multivariate linear mixed model was further fitted in order to assess %EBWL evolution over time and to test the impact of polygenic risk scores on %EBWL interindividual variability. Clinical and demographic variables (surgery modality — open surgery vs. laparoscopy — sex, age, initial BMI, presence of diabetes, and smoking status) were tested independently and added to the models when showing statistical significance (P < 0.05).
Weight loss trajectory analysis.
Postsurgery weight loss (%EBWL) trajectories were estimated for each patient by latent class growth analysis implemented using Proc Traj (36, 37) in SAS 9.4 (SAS Institute Inc.). Briefly, the Traj procedure is a semiparametric, group-based modelling technique able to identify groups of individuals following distinct longitudinal trajectories for a given repeatedly measured variable (37). Weight loss trajectory groups identified by latent class growth analysis are then estimations of multiple patterns of change within the population, and not reified groups. Thus, in order to mitigate misclassification, we set the posterior probability threshold to 0.7 to determine a correct class membership assignment, as previously reported by Nagin (32). Proc Traj is able to handle data that are missing completely at random (32), yet 3 time points per subject was set as inclusion criteria. Moreover, in order to minimize classification error, follow-up time points with less than 50% completeness were excluded from weight loss trajectory analysis, leaving 7 postsurgery time points (3, 6, 12, 18, 24, 36, and 48 months) to be used for class estimation. Trajectory model selection was iterative, and class assignment was estimated based on minimizing BIC, which is the model log-likelihood adjusted for sample size and number of parameters (37). Briefly, model fitting started with one group on a linear pattern, and complexity (number of groups and equation degrees) was added until BIC was minimized. The number of groups was limited to a minimum class membership of 5% of patients. The model with the smallest BIC, satisfying class membership criteria and including all the significant predictor variables, was used to estimate the posterior probabilities of %EBWL group assignment. Finally, the resulting groups allowed categorization of patients as a function of %EBWL.
Weight loss binary logistic model.
Results from trajectory analysis were further used to develop a binary logistic regression model for predicting the probability of %EBWL class assignment. First, %EBWL trajectory groups were reassigned into a binary category, in order to build a prediction model able to accurately identify patients having a reduced weight loss response to surgery. Then, clinical and demographic factors were tested and added to the model when significant (P < 0.05). Finally, the impact of genetic background, in the form of polygenic risk scores, on the model’s predictive value was tested. Results were adjusted for optimism by bootstrapping (n = 1,000), obtaining bias-corrected 95% CIdiff for further AUCadj comparison, ultimately used to determine the overall performance of the different predictive models. The added predictive value of different models was tested by calculating the differences between bootstrapped AUCadj, which was considered significant when the 95% CIdiff did not contain zero. The NRI and IDI were also used as complementary indexes to evaluate the net effect of adding a polygenic risk score to the prediction model (38). Calibration was evaluated through calibration plots obtained after 1,000 bootstrap replications, in order to determine the MAE of model predictions (39). Model comparison was performed in R by using the “Hmisc” and “rms” packages.
Longitudinal weight loss linear mixed model.
Longitudinal %EBWL data were further fitted by using a linear mixed model with an unstructured covariance matrix where patients were nested by %EBWL trajectory group. Linear mixed models including significant predictor variables, and time interactions were fitted by using maximum likelihood estimation. A likelihood ratio test (LRT), computed as twice the difference in the log-likelihood between models (–2 log-likelihood), was used to determine whether the inclusion of polygenic risk scores significantly increased the goodness of fit of the model.
Polygenic risk scores.
The most accurate binary logistic regression model was used to test the predictive value of polygenic risk scores in determining %EBWL group assignment. Univariate associations between genotyped SNPs and %EBWL trajectory groups were first tested by binary logistic regression. Results from association tests led to the construction of two weighted polygenic risk scores by summing the number of alleles of all the 186 BMI-associated SNPs (PRS186) multiplied by their OR. A simplified polygenic risk score, including only the 11 SNPs (PRS11) significantly associated with %EBWL group, was also constructed. The sum of weighed alleles resulted in two continuous polygenic risk scores, whose predictive value of %EBWL was subsequently and independently tested in both the logistic and the linear mixed models. First, the linear trend of association of polygenic risk scores with %EBWL group was evaluated, as well as the increase in the predictive power of the logistic model after the inclusion of either PRS186 or PRS11. Further, the effect of polygenic risk scores on the overall fit of the linear mixed model and their interaction with time postsurgery were also tested to evaluate their impact on the evolution of %EBWL over time. The logistic model and the linear mixed model were implemented with Proc Logistic and Proc Mixed, respectively, in SAS 9.4.
Cost-effectiveness analysis of polygenic risk scores.
In order to test whether the potential improvements of the predictive power observed after the inclusion of polygenic risk scores were clinically relevant, a cost-effectiveness analysis was carried out. Before performing the analysis, two relevant aspects were taken into account given the nature of the data. First, biliopancreatic diversion with duodenal switch is a highly successful bariatric surgery, thus leading to an unbalanced study population, with the vast majority of patients being assigned to the HWL+NWL trajectory group. Then, instead of comparing the performance of models by means of Youden’s index, which is not appropriate for unbalanced datasets (40), prediction models were compared at optimal probability cutoff values minimizing misclassification costs. Second, the different health and economic cost of incorrectly assigning a patient to the LWL or the HWL+NWL trajectory group (FN) and vice versa (FP) was estimated by using data from the CADTH report on the clinical and cost-effectiveness of bariatric surgery procedures (21). In concrete terms, ICER per QALY gained for biliopancreatic diversion compared with standard care by time horizon was used herein for assigning different costs to FN and FP. The %EBWL difference between the LWL and HWL+NWL groups at the end of the follow-up period was used to perform ICER (cost/QALY) estimates from the CADTH report. The misclassification cost ratio was obtained as the ICER ratio difference between LWL and HWL+NWL groups, and further included in a cost function, leading to the construction of cost curves for each model. Probability cutoff values were selected from cost curves at the lowest misclassification cost point for model comparison purposes. All cost-effectiveness analyses were performed using R (version 3.4.3).
Study approval.
This study was conducted in accordance with the Declaration of Helsinki and approved by the Université Laval and Quebec Heart and Lung Institute ethics committee. Written informed consent was received from participants prior to inclusion in the study.
Author contributions
JTM performed statistical analysis, interpreted the data, and wrote the manuscript; MCV and FG conceived and designed the research; AT and LP participated in the elaboration of the study design and critically reviewed the manuscript. SM participated in clinical care and patient recruitment, sampled blood from the study subjects, and critically reviewed the manuscript.
Supplementary Material
Acknowledgments
This study was supported by a grant-in-aid from the Heart and Stroke Foundation of Canada (G-17-0016627) and by the Canada Research Chair in Genomics Applied to Nutrition and Metabolic Health, held by MCV. JTM received a postdoctoral fellowship from the Fonds de Recherche de Québec-Santé. We thank all participants for their collaboration, and the surgeons of the Bariatric Surgery Center of the Quebec Heart and Lung Institute for their invaluable collaboration in clinical care and patient recruitment (Laurent Biertho, Simon Biron, Léonie Bouvet-Bouchard, Frédéric-Simon Hould, François Julien, Stéfane Lebel, Odette Lescelleur, Simon Marceau). We are grateful to Paule Marceau for data management, Catherine Raymond for genotyping and technical assistance, and the staff of the Quebec Heart and Lung Institute Biobank for patient consent and sample processing. We also sincerely thank our long-term collaborator Picard Marceau, with whom this project was initiated.
Version 1. 09/06/2018
Electronic publication
Footnotes
Conflict of interest: André Tchernof receives research funding from Johnson & Johnson Medical companies, Pfizer, and Medtronic for studies on bariatric surgery.
Reference information: JCI Insight. 2018;3(17):e122011. https://doi.org/10.1172/jci.insight.122011.
Contributor Information
Juan de Toro-Martín, Email: juan.de-toro-martin.1@ulaval.ca.
Frédéric Guénard, Email: Frederic.Guenard@fsaa.ulaval.ca.
André Tchernof, Email: andre.tchernof@criucpq.ulaval.ca.
Louis Pérusse, Email: Louis.Perusse@kin.ulaval.ca.
Simon Marceau, Email: mirsim@videotron.ca.
Marie-Claude Vohl, Email: Marie-Claude.Vohl@fsaa.ulaval.ca.
References
- 1.Terranova L, Busetto L, Vestri A, Zappa MA. Bariatric surgery: cost-effectiveness and budget impact. Obes Surg. 2012;22(4):646–653. doi: 10.1007/s11695-012-0608-1. [DOI] [PubMed] [Google Scholar]
- 2.Courcoulas AP, et al. Seven-year weight trajectories and health outcomes in the Longitudinal Assessment of Bariatric Surgery (LABS) study. JAMA Surg. 2018;153(5):427–434. doi: 10.1001/jamasurg.2017.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood GC, et al. Evaluation of the association between preoperative clinical factors and long-term weight loss after Roux-en-Y gastric bypass. JAMA Surg. 2016;151(11):1056–1062. doi: 10.1001/jamasurg.2016.2334. [DOI] [PubMed] [Google Scholar]
- 4.Rojo-Tirado MA, Benito PJ, Atienza D, Rincón E, Calderón FJ. Effects of age, sex, and treatment on weight-loss dynamics in overweight people. Appl Physiol Nutr Metab. 2013;38(9):967–976. doi: 10.1139/apnm-2012-0441. [DOI] [PubMed] [Google Scholar]
- 5.Dallal RM, Quebbemann BB, Hunt LH, Braitman LE. Analysis of weight loss after bariatric surgery using mixed-effects linear modeling. Obes Surg. 2009;19(6):732–737. doi: 10.1007/s11695-009-9816-8. [DOI] [PubMed] [Google Scholar]
- 6.Baldridge AS, et al. Factors associated with long-term weight loss following bariatric surgery using 2 methods for repeated measures analysis. Am J Epidemiol. 2015;182(3):235–243. doi: 10.1093/aje/kwv039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatoum IJ, Greenawalt DM, Cotsapas C, Reitman ML, Daly MJ, Kaplan LM. Heritability of the weight loss response to gastric bypass surgery. J Clin Endocrinol Metab. 2011;96(10):E1630–E1633. doi: 10.1210/jc.2011-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobourn C, Chapman MA, Ali A, Amrhein J. Five-year weight loss experience of outpatients receiving laparoscopic adjustable gastric band surgery. Obes Surg. 2013;23(7):903–910. doi: 10.1007/s11695-013-0881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corcelles R, et al. Total weight loss as the outcome measure of choice after Roux-en-Y gastric bypass. Obes Surg. 2016;26(8):1794–1798. doi: 10.1007/s11695-015-2022-y. [DOI] [PubMed] [Google Scholar]
- 10.Käkelä P, et al. Genetic risk score does not predict the outcome of obesity surgery. Obes Surg. 2014;24(1):128–133. doi: 10.1007/s11695-013-1080-2. [DOI] [PubMed] [Google Scholar]
- 11.Rinella ES, et al. Genome-wide association of single-nucleotide polymorphisms with weight loss outcomes after Roux-en-Y gastric bypass surgery. J Clin Endocrinol Metab. 2013;98(6):E1131–E1136. doi: 10.1210/jc.2012-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matzko ME, et al. Association of ghrelin receptor promoter polymorphisms with weight loss following Roux-en-Y gastric bypass surgery. Obes Surg. 2012;22(5):783–790. doi: 10.1007/s11695-012-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Still CD, et al. High allelic burden of four obesity SNPs is associated with poorer weight loss outcomes following gastric bypass surgery. Obesity (Silver Spring) 2011;19(8):1676–1683. doi: 10.1038/oby.2011.3. [DOI] [PubMed] [Google Scholar]
- 14.Still CD, et al. Clinical factors associated with weight loss outcomes after Roux-en-Y gastric bypass surgery. Obesity (Silver Spring) 2014;22(3):888–894. doi: 10.1002/oby.20529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courcoulas AP, et al. Weight change and health outcomes at 3 years after bariatric surgery among individuals with severe obesity. JAMA. 2013;310(22):2416–2425. doi: 10.1001/jama.2013.280928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seip RL, et al. Comparative physiogenomic analyses of weight loss in response to 2 modes of bariatric surgery: demonstration with candidate neuropsychiatric and cardiometabolic genes. Surg Obes Relat Dis. 2016;12(2):369–377. doi: 10.1016/j.soard.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Salminen P, et al. Effect of laparoscopic sleeve gastrectomy vs laparoscopic Roux-en-Y gastric bypass on weight loss at 5 years among patients with morbid obesity: The SLEEVEPASS randomized clinical trial. JAMA. 2018;319(3):241–254. doi: 10.1001/jama.2017.20313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebel S, Dion G, Marceau S, Biron S, Robert M, Biertho L. Clinical outcomes of duodenal switch with a 200-cm common channel: a matched, controlled trial. Surg Obes Relat Dis. 2016;12(5):1014–1020. doi: 10.1016/j.soard.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Piché MÈ, et al. Changes in predicted cardiovascular disease risk after biliopancreatic diversion surgery in severely obese patients. Metab Clin Exp. 2014;63(1):79–86. doi: 10.1016/j.metabol.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Marceau P, et al. Long-term metabolic outcomes 5 to 20 years after biliopancreatic diversion. Obes Surg. 2015;25(9):1584–1593. doi: 10.1007/s11695-015-1599-5. [DOI] [PubMed] [Google Scholar]
- 21. Klarenbach S, et al. Bariatric Surgery for Severe Obesity: Systematic Review and Economic Evaluation. Ottawa, Ontario, Canada: Canadian Agency for Drugs and Technologies in Health. Ottawa, Canada: Canadian Agency for Drugs and Technologies in Health; 2010. [Google Scholar]
- 22.Eldar S, Heneghan HM, Brethauer SA, Schauer PR. Bariatric surgery for treatment of obesity. Int J Obes (Lond) 2011;35(Suppl 3):S16–S21. doi: 10.1038/ijo.2011.142. [DOI] [PubMed] [Google Scholar]
- 23.Strain GW, et al. The impact of biliopancreatic diversion with duodenal switch (BPD/DS) over 9 years. Obes Surg. 2017;27(3):787–794. doi: 10.1007/s11695-016-2371-1. [DOI] [PubMed] [Google Scholar]
- 24.Hatoum IJ, Stein HK, Merrifield BF, Kaplan LM. Capacity for physical activity predicts weight loss after Roux-en-Y gastric bypass. Obesity (Silver Spring) 2009;17(1):92–99. doi: 10.1038/oby.2008.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sevilla S, Hubal MJ. Genetic modifiers of obesity and bariatric surgery outcomes. Semin Pediatr Surg. 2014;23(1):43–48. doi: 10.1053/j.sempedsurg.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Resende CMM, et al. The polymorphism rs17782313 near MC4R gene is related with anthropometric changes in women submitted to bariatric surgery over 60 months. Clin Nutr. 2018;37(4):1286–1292. doi: 10.1016/j.clnu.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen-Torvik LJ, et al. rs4771122 predicts multiple measures of long-term weight loss after bariatric surgery. Obes Surg. 2015;25(11):2225–2229. doi: 10.1007/s11695-015-1872-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandstein M, et al. A genetic risk score is associated with weight loss following Roux-en Y gastric bypass surgery. Obes Surg. 2016;26(9):2183–2189. doi: 10.1007/s11695-016-2072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poitou C, et al. Bariatric surgery induces disruption in inflammatory signaling pathways mediated by immune cells in adipose tissue: a RNA-Seq study. PLoS ONE. 2015;10(5):e0125718. doi: 10.1371/journal.pone.0125718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barres R, et al. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3(4):1020–1027. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Vohl MC, et al. Differential methylation of inflammatory and insulinotropic genes after metabolic surgery in women. J Clin Epigenetics. 2015;1(1):1–9. [Google Scholar]
- 32. Nagin DS. Group-Based Modeling of Development. Cambridge, Massachusetts, USA: Harvard University Press; 2005: [Google Scholar]
- 33.Biertho L, Lebel S, Marceau S, Hould FS, Julien F, Biron S. Biliopancreatic diversion with duodenal switch: surgical technique and perioperative Care. Surg Clin North Am. 2016;96(4):815–826. doi: 10.1016/j.suc.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 34.MacArthur J, et al. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones BL, Nagin DS. Advances in group-based trajectory modeling and an SAS procedure for estimating them. Sociol Methods Res. 2007;35(4):542–571. doi: 10.1177/0049124106292364. [DOI] [Google Scholar]
- 37.Jones BL, Nagin DS, Roeder K. A SAS procedure based on mixture models for estimating developmental trajectories. Sociol Methods Res. 2001;29(3):374–393. doi: 10.1177/0049124101029003005. [DOI] [Google Scholar]
- 38.Pencina MJ, D’Agostino RB, D’Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 39. Harrell F. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. New York, New York, USA: Springer; 2001. [Google Scholar]
- 40.Smits N. A note on Youden’s J and its cost ratio. BMC Med Res Methodol. 2010;10:89. doi: 10.1186/1471-2288-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.