Abstract
Human induced pluripotent stem cells (hiPSCs) represent promising raw materials of human cell-based therapeutic products (hCTPs). As undifferentiated hiPSCs exhibit intrinsic tumorigenicity properties that enable them to form teratomas, hCTPs containing residual undifferentiated hiPSCs may cause tumor formation following transplantation. We first established quantitative and sensitive tumorigenicity testing of hiPSCs dissociated into single cells using NOD/Shi-scid IL2Rγnull (NOG) mice by inhibiting apoptosis of hiPSCs with a Rho kinase inhibitor. To examine different features in tumorigenicity of various hiPSCs, 10 commonly available hiPSC lines were subjected to in vivo tumorigenicity testing. Transplanted hiPSC lines showed remarkable variation in tumor incidence, formation latency, and volumes. Most of the tumors formed were classified as immature teratomas. However, no signs of malignancies, such as carcinoma and sarcoma, were recognized in the tumors. Characteristics associated tumorigenicity of hiPSCs were investigated with microarray analysis, karyotype analysis, and whole exome sequencing. Gene expression profiling and pathway analysis supported different features of hiPSC lines in tumorigenicity. hiPSC lines showed chromosomal abnormalities in some lines and 61–77 variants of cancer-related genes carrying effective nonsynonymous mutations, which were confirmed in the COSMIC databases. In this study, the chromosomal abnormalities and cancer-related gene mutations observed in hiPSC lines did not lead to the malignancy of tumors derived from hiPSCs. Our results suggest that the potential tumorigenicity risk of hCTPs containing residual undifferentiated hiPSCs is dependent on not only amounts of undifferentiated hiPSCs but also features of the cell lines used as raw materials, a finding that should be considered from the perspective of quality of hCTPs used.
Introduction
Human pluripotent stem cells (hPSCs), such as human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs), are used as the raw materials of human cell-based therapeutic products (hCTPs) due to their infinite self-renewal capacity and ability to differentiate into various cell types in vitro. Practically, several clinical trials and studies using hPSC-derived hCTPs have already started to treat spinal cord injuries, age-related macular degeneration, heart failure, and type I diabetes around the world [1]. However, undifferentiated hPSCs are known to possess intrinsic tumorigenicity properties and to form teratomas in immunodeficient animals. In addition, safety concerns associated with teratoma derived from hPSCs have been raised for development of hCTPs [2]. The possibility cannot be excluded that a trace amount of residual undifferentiated hPSCs in hCTPs leads to tumor development following transplantation. Thus, contamination of hCTPs with residual undifferentiated hPSCs is a potential concern in terms of certain hazardous factors as well as the quality attributes of hPSC-derived hCTPs.
The World Health Organization (WHO) Technical Report Series (TRS) No. 978 Annex 3 is the only international guideline that addresses tumorigenicity testing for medicinal products [3]. However, the in vivo tumorigenicity testing proposed in WHO TRS 978 covers only viable animal cells used as cell substrates for manufacturing biological products but not cell products used directly for therapy by transplantation into patients. No international guideline has been issued for tumorigenicity testing of hCTPs even though its establishment is urgently needed for the development of hCTPs.
We have previously reported the performance of in vivo tumorigenicity testing using severe immunodeficient NOD/Shi-scid IL2Rγnull (NOG) mice for the detection of HeLa cells used to spike human mesenchymal stem cells as a model of tumorigenic cellular impurity in hCTPs [4]. This in vivo tumorigenicity testing using NOG mice and Matrigel serves as a highly sensitive and quantitative method to detect tumorigenic cells contained in hCTPs. On the other hand, in order to form teratomas, hPSCs generally need to be injected into immunodeficient mice in the form of clumps, as hPSCs dissociated into single cells readily undergo apoptosis [5]. Thus, tumorigenicity testing has a number of challenges with regard to transplanting defined numbers of hPSCs into mice. To overcome this issue, single dissociated hESCs were spiked into mitomycin C-treated fibroblasts for transplantation into SCID mice [6] and NOD SCID mice [7] with Matrigel, which revealed that efficient engraftment of single dissociated hPSCs in immunodeficient mice requires both feeders and a basement membrane matrix in the transplanted cell mixture. Additionally, to understand the risk of tumorigenicity of hPSC-derived hCTPs, quality attributes related to tumorigenicity of hPSCs should also be taken into consideration as well as quantitative evaluation for residual undifferentiated hPSCs in hCTPs. Although the amounts of residual hPSCs in hCTPs is appropriately controlled in the manufacturing process, any different features in the tumorigenicity of hPSCs per se may influence tumor formation and their potential risks resulting from residual hPSCs in hCTPs. However, different features in tumorigenicity, including benign and malignancy, and its associated quality attributes of hPSCs has not been well studied.
Here, to address the issues regarding tumorigenicity of hPSCs used as raw materials for hCTPs, we established a quantitative method for detecting hiPSCs by in vivo tumorigenicity testing using NOG mice. Using this method, we performed tumorigenicity testing of 10 hiPSC lines and also pathologically evaluated the tumor formed in the mice to examine difference in tumorigenicity features of the hiPSC lines. A comprehensive gene expression analysis of the transplanted hiPSC lines identified genes statistically correlated with tumorigenicity of hiPSCs for the pathway analysis. In addition, we examined the mutations observed in the hiPSC lines against the cancer-related genes shown in public databases using exome sequence analysis.
Materials and methods
Cell culture
hiPSC lines 201B7 [8], 253G1 [9], 409B2 [10], 454E2 [10], HiPS-RIKEN-1A, HiPS-RIKEN-2A, and HiPS-RIKEN-12A were provided by the RIKEN BRC through the Project for Realization of Regenerative Medicine and the National Bio-Resource Project of the MEXT, Japan. hiPSC lines ATCC-DYR0100 and ATCC-HYR0103 were obtained from the ATCC. The human cell line mc-iPS was purchased from System Biosciences [11]. Information for the hiPSCs used in this study is described in S1 Table. hiPSCs were first cultured on mitomycin C-treated SNL cells, a mouse fibroblast STO cell line expressing the neomycin-resistance gene cassette and LIF, in primate ES cell medium (ReproCell) supplemented with 4 ng/ml human basic fibroblast growth factor (R&D Systems). hiPSCs were then passaged onto Matrigel-coated dishes with feeder-free cell-culture medium mTeSR1 (Stem Cell Technologies) for at least three passages to remove feeder cells before transplantation into mice. Undifferentiated colonies were passaged as small clumps once in every 5–6 days using CTK dissociation solution (ReproCell) and a StemPro EZPassage device (Invitrogen). Normal human neonatal dermal fibroblasts (NHDF) were obtained from Lonza and maintained in Eagle’s minimum essential medium (Sigma), supplemented with 10% fetal bovine serum (FBS; Sigma), 2 mM GlutaMax™ (Gibco), 50 U/ml penicillin, and 50 μg/ml streptomycin (Nacalai Tesque). The NHDF were inactivated by treatment with 10 μg/ml mitomycin C (Sigma-Aldrich) one day before transplantation into mice. Normal human bone marrow-derived mesenchymal stem cells (hMSCs) were purchased from Lonza and cultured in MSCGM™ medium (Lonza). All cells were cultured in a humidified atmosphere of 5% CO2 and 95% air at 37 °C.
Preparation of hiPSC suspension for transplantation
Cells to be transplanted into immunodeficient mice were prepared according to the method reported by Gropp et al. with modification [7]. Cultured hiPSCs were treated with CTK dissociation solution and a StemPro EZPassage device or with StemPro Accutase Cell Dissociation Reagent (Gibco) to dissociate them into smaller cell clumps or single cells, respectively. To count the cell number of clumps, we used aliquots of clump suspension and enzymatically dissociated cell clamps into single cells. Mitomycin C-treated NHDFs and hMSCs were detached and dissociated with 0.25% trypsin-EDTA solution (Gibco). The hiPSCs, and mitomycin C-treated NHDFs or hMSCs were suspended together in a 1:1 mixture of mTeSR1 medium and Matrigel (product #354234, Corning) in the presence or absence of 10 μM Y-27632 (Wako) just prior to transplantation. Various numbers of hiPSCs combined with 1 × 106 mitomycin C-treated NHDFs or 1 × 106 hMSCs were transplanted into one mouse.
Tumorigenicity tests
Male NOD.Cg-Prkdcscid Il2rgtm1Sug/Jic (NOD/Shi-scid IL2Rγnull; NOG) mice maintained in the Central Institute for Experimental Animals (CIEA, Kanagawa, Japan) were used for in vivo tumorigenicity studies. All mice were housed in polycarbonate plastic cages (W155 x D245 x H149 mm; 2–3 mice/cage) with soft bedding (GLP Aspen Bedding, CLEA Japan) in an air-conditioned room at a temperature of 23 ± 2°C and a humidity of 50 ± 10% with a 12-hr light and 12-hr dark cycle. The animals were given irradiation sterilized commercial diet (CA-1, CLEA Japan) and sterilized water filled in plastic bottles ad libitum. Prepared cell suspensions (100 μl) were subcutaneously injected using 1 ml syringes with a 25 G needle (Terumo) into 8-week-old mice. The mice were palpated weekly for 16 weeks to observe nodule formation at the injection site. Tumor size was assessed by external measurement of the length and width of the tumors in two dimensions using a caliper as soon as tumors reached measurable size. The tumor volume was calculated using the formula: volume (mm3) = 1/2 length (mm) × (width [mm])2. The engraftment was determined according to progressive nodule growth at the injection site. Mice were euthanized and necropsied when tumors grew greater than 10% of their body weight or when a sign of deconditioning was noted according to the categories of Scientists Center for Animal Welfare (SCAW) [12] and National Institutes of Health (NIH) Animal Research Advisory Committee Guideline (https://oacu.oir.nih.gov/animal-research-advisory-committee-guidelines). Tumor-forming capacity is defined as 50% tumor-producing dose (TPD50), which represents the threshold dose of cells forming tumors in 50% of the animals. TPD50 values and their 95% confidence intervals were calculated using the Spearman-Kärber method, as implemented in the R statistical application with the package ‘tsk’ version 1.2 (https://r-forge.r-project.org/R/?group_id=1169), at each time point. For the Spearman-Kärber method to be applicable, it is necessary to use a range of doses wide enough to include both the dose at and below which 100% of the animals will be positive and the dose at and above which 100% of the animals will be negative [13]. If each of these conditions was not fulfilled, it was assumed that the next higher (10 times) or lower (1/10) dose step (a dummy set of data) to the last one tested would have produced the predicted incidence.
The protocol of the present study was reviewed beforehand and approved by the Animal Ethics Committees of CIEA and the National Institute of Health Sciences (NIHS, Kanagawa, Japan). All animal experiments were performed according to the Ethical Guidelines for Animal Experimentation from the CIEA and the NIHS. All animals were euthanized under isoflurane inhalation anesthesia.
Histology
The resulting tumors were dissected and fixed with 10% neutral buffered formalin. The paraffin-embedded tissues were sliced into 5 μm sections and stained with hematoxylin and eosin. The specimens at different depths through the tumor mass were subjected to histological analysis by certified pathologists. The formation of differentiated tumors comprising three germ layers was evaluated, and immature teratomas were graded from 0 to 3 on the basis of the amount of immature neuroepithelium found in the tumor specimen according to the system developed by O’Connor and Norris [14].
GeneChip and statistical analysis
Total RNA was isolated from hiPSCs using an RNeasy Mini Kit with DNase I treatment (QIAGEN) and converted to biotinylated cRNA using a GeneChip 3´ IVT Express kit (Affymetrix). Labeled RNA was processed for microarray hybridization to a GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix), which contains 54,675 probe sets (~47,400 transcripts). An Affymetrix GeneChip Fluidics Station was used to perform streptavidin/phycoerythrin staining. The hybridization signals on the microarray were scanned using a GeneChip Scanner 3000 (Affymetrix) and analyzed using Expression Console Software ver.1.1 (Affymetrix). Normalization was performed using MAS5 algorithm, and the MAS5 data were scaled to a Target Intensity of 500. The hybridization experiments were performed on 3 samples of each hiPSC line. The NCBI GEO accession number for the microarray data is GSE108566.
To extract the informationally significant probe sets from the data set, we filtered probe sets using the following two steps. First, probe sets were regarded as “absent” not when indicated as “present” by “absolute analysis” using GCOS software in all 30 samples. Probe sets regarded as “absent” were eliminated from the data set. Secondly, when no significant difference was observed among cell lines using ANOVA (p ≥ 0.05), probe sets were also eliminated from the data set. Tumorigenicity of each hiPSC line was evaluated by the latency of tumor formation and by tumor incidence. To identify probe sets related to the tumorigenicity of the hiPSC lines, the correlation between the intensity values of the filtered probe sets and the two variables (latency of tumor formation and tumor incidence) was determined by calculating Spearman’s rank correlation coefficients and their p-values [15]. Probe sets exhibiting statistically significant correlations with both of two variables (p < 0.01) were selected. The resulting probe sets were analyzed with Ingenuity Pathway Analysis software (Ingenuity Systems).
Karyotype analysis
Chromosomal G-band analyses of hiPSCs were performed at the Nihon Gene Research Laboratories (Sendai, Japan). At least 20 metaphase spreads were examined for each cell line.
Exome sequencing analysis
Whole exome sequencing was performed by Takara Bio Inc. Genomic DNA was isolated from hiPSCs using a NucleoSpin Tissue Kit (Macherey-Nagel). DNA (3 μg) was sheared into peak fragment size of 300 bp using a Covaris S2 sonicator (Covaris) and used to conduct end-repair, A-addition, and adaptor ligation with SureSelect XT reagents (Agilent Technologies). Target sequences were enriched with SureSelect XT Human All Exon Kit V5 (Agilent Technologies). Sequencing was performed on a HiSeq 2500 System (Illumina) using a 100 bp paired-end protocol. More than 15 Gbp of read data were generated for each sample of hiPSC line. After trimming the adaptor sequences and quality filtering using Trimmomatic [16], the reads were mapped to the reference genome (hg 19) using BWA-MEM [17], followed by duplicate read removal. Genomic variants, including SNV and short indels, were annotated using SnpEff [18], Human genetic variation database (HGVD) [19] and the Exome Aggregation Consortium (ExAC) database [20], and related to a gene list including the Cancer Gene Census provided in the COSMIC database (S2 Table). The NCBI Sequence Read Archive (SRA) accession number for the whole exome sequencing data is SRP134676.
Results
The in vivo tumorigenicity tests reflecting quantification of hiPSCs
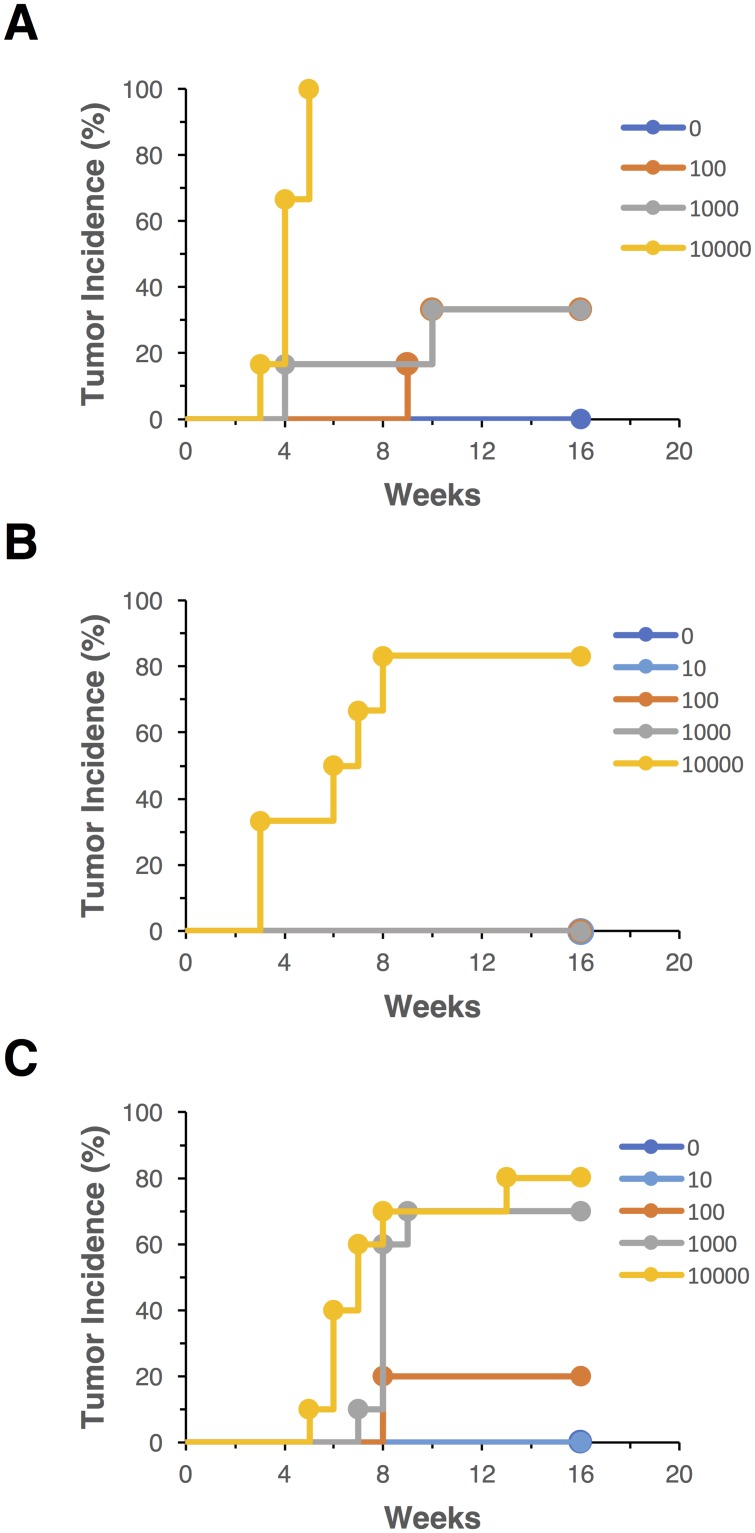
The tumorigenicity of hiPSC line 201B7 was first evaluated when these cells were transplanted into NOG mice as clumps with Matrigel. The hiPSC suspension was subcutaneously injected into NOG mice at a dose of 0, 1 × 102, 1 × 103, and 1 × 104, and tumor formation was monitored at the transplanted sites for 16 weeks. Transplantation of 1 × 102, 1 × 103, and 1 × 104 hiPSCs caused tumor formation within 10 weeks at an incidence of 33% (2/6 animals), 33% (2/6 animals), and 100% (6/6 animals), respectively, whereas no mice showed any tumor formation at a dose of 0 hiPSCs (Fig 1A). The TPD50 value was calculated as 681 when hiPSCs were transplanted into NOG mice as clumps in Matrigel (Table 1). On the other hand, to precisely define the number of hiPSCs transplanted into mice, single cell dissociation of hiPSCs would be required for cell counting. Gropp et al. reported a defined number of dissociated hESCs should be subcutaneously transplanted with mitotically inactivated feeder cells and Matrigel into NOD/SCID mice for a quantitative and sensitive teratoma assay [7]. Thus, we attempted to transplant single dissociated hiPSCs into NOG mice to ensure quantitativity of the tumorigenicity test. The 201B7 hiPSCs dissociated into single cells (0, 1 × 10, 1 × 102, 1 × 103, and 1 × 104) were mixed with mitomycin C-treated NHDFs, 1 × 106) and subcutaneously transplanted into NOG mice with Matrigel. Tumors formed in 83% mice (5/6 animals) at a dose of 1 × 104 hiPSCs. On the other hand, no mice showed tumor formation at doses of 0, 1 × 10, 1 × 102, or 1 × 103 hiPSCs (Fig 1B). Its TPD50 value was calculated as 4642 and approximately 6.8-fold higher than that when transplanted with clumps of hiPSCs (Table 1). These results indicated that single cell dissociation of hiPSCs impairs their engraftment at the subcutaneous sites of NOG mice even in the presence of fibroblasts. Y-27632, a Rho kinase inhibitor, is well known to effectively inhibit dissociation-induced apoptosis of human pluripotent stem cells [5]. Next, using Y-27632, we performed tumorigenicity tests of hiPSCs dissociated into single cells. Single dissociated hiPSCs (0, 1 × 10, 1 × 102, 1 × 103, and 1 × 104) were mixed with 1 × 106 of mitotically inactivated NHDFs and Matrigel in the presence of 10 μM Y-27632. The prepared cell suspension was subcutaneously transplanted into NOG mice, and their tumor formation was monitored for 16 weeks. The tumor incidence of NOG mice transplanted with 1 × 10, 1 × 102, 1 × 103, and 1 × 104 hiPSCs was 0% (0/10 animals), 20% (2/10 animals), 60% (6/10 animals), and 70% (7/10 animals), respectively (Fig 1C). The TPD50 value was calculated as 631 and effectively improved by mixing with Y-27632 (Table 1). We next examined subcutaneous tumor formation of hiPSCs (0, 1 × 10, 1 × 102, 1 × 103, and 1 × 104 cells) spiked into 1 × 106 hMSCs instead of NHDFs with Matrigel and Y-27632 in NOG mice. Even when injected with 1 × 104 hiPSCs, no mice showed formation of tumors derived from hiPSCs in the presence of hMSCs (S3 Table). These results suggested that hMSCs suppress tumor formation of hiPSCs under some defined condition in vivo. Thus, we have established in vivo tumorigenicity tests reflecting the quantification of hiPSCs to transplant mixtures of fibroblasts, Matrigel, and a Rho kinase inhibitor into NOG mice.
Fig 1. Tumor incidence of the 201B7 hiPSC line.
201B7 cells were subcutaneously injected into NOG mice at the indicated doses (0, 1 × 102, 1 × 103, or 1 × 104 cells) with Matrigel. The 201B7 cells were transplanted as clumps (A), single cells with 1 × 106 mitomycin C-treated normal human neonatal dermal fibroblasts (NHDF) (B), or single cells with 1 × 106 mitomycin C-treated NHDF and 10 μM Y-27632 (C). Tumor formation was examined for 16 weeks. Six (A, B) or ten (C) mice were used in each group.
Table 1. Tumor formation capacity of 201B7 hiPSCs in NOG mice.
| Group | Tumor incidence at indicated hiPSC dose at 16 wk | TPD50 | 95% confidence interval for the TPD50 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 × 10 | 1 × 102 | 1 × 103 | 1 × 104 | 1 × 105 | Lower | Upper | ||
| hiPSC clumps | 0/6a | — (0/6)b |
2/6 | 2/6 | 6/6 | — | 681 | 199 | 2327 |
| Single hiPSCs/NHDF | 0/6 | 0/6 | 0/6 | 0/6 | 5/6 | — (6/6)c |
4642 | 2336 | 9223 |
| Single hiPSCs/NHDF + Y27632 | 0/10 | 0/10 | 2/10 | 7/10 | 8/10 | — (10/10)c |
631 | 223 | 1783 |
—: not tested
aNumber of mice in which tumor formed/total number of mice inoculated.
bSince not all mice inoculated with the lowest dose (1x102) showed the negative (non-tumor bearing) results, it was assumed that the tumor incidence of mice at an even lower dose step (a dummy set of data) would have been 0% for the Spearman-Kärber method to be applicable.
cSince not all mice inoculated with the highest dose (1x104) formed tumors, it was assumed that the tumor incidence of mice at an even higher dose step (a dummy set of data) would have been 100% for the Spearman-Kärber method to be applicable.
Difference in tumorigenic potential of 10 hiPSC lines
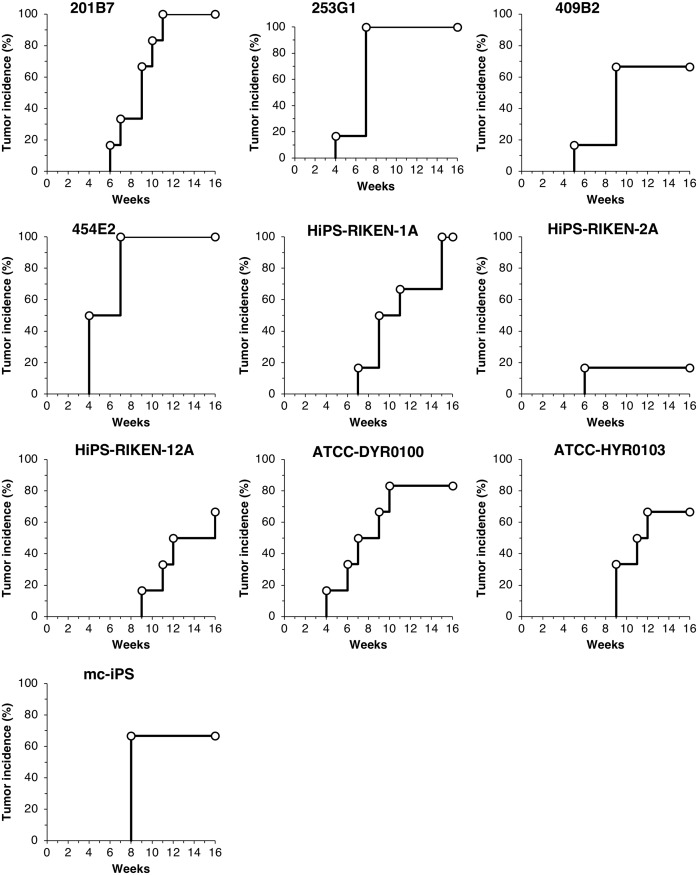
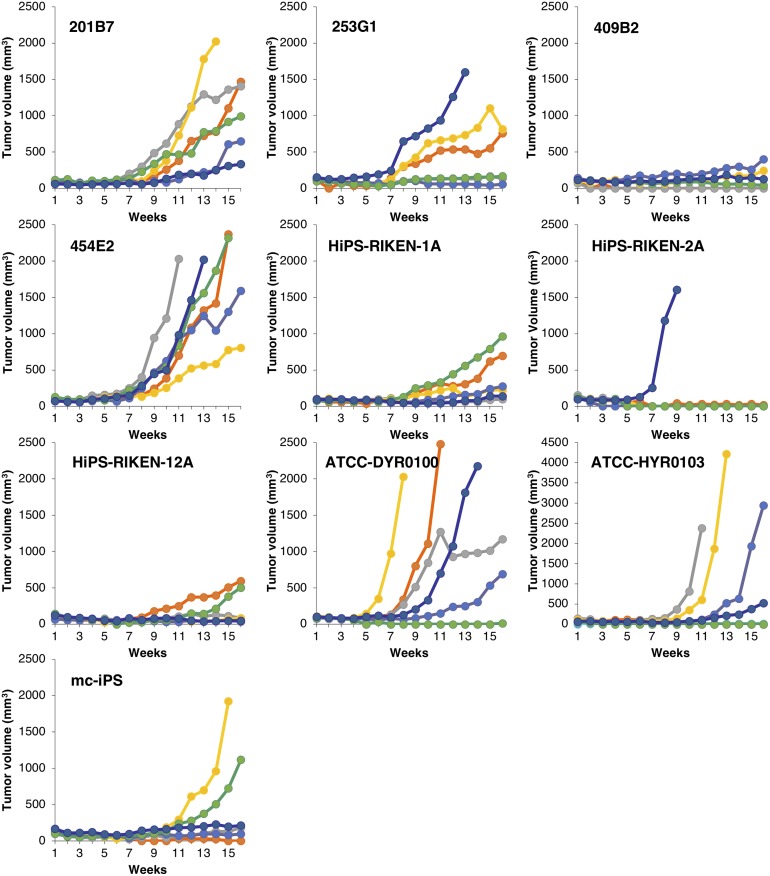
To examine the difference in tumorigenic potential of 10 hiPSC lines (201B7, 253G1, 409B2, 454E2, HiPS-RIKEN-1A, HiPS-RIKEN-2A, HiPS-RIKEN-12A, ATCC-DYR0100, ATCC-HYR0103, mc-iPS), 3 × 104 of single dissociated hiPSCs were inoculated at subcutaneous sites of NOG mice with 1 × 106 of mitotically inactivated NHDFs and Matrigel in the presence of Y-27632. We continuously monitored their tumor formation for 16 weeks to obtain the data of tumor incidence, latency, and growth. The ratio of the number of mice showing tumor formation to that of their group determined tumor incidence, following 16 weeks of observation. Tumor incidence of each hiPSC line was 100% (201B7), 100% (253G1), 67% (409B2), 100% (454E2), 100% (HiPS-RIKEN-1A), 17% (HiPS-RIKEN-2A), 67% (HiPS-RIKEN-12A), 83% (ATCC-DYR0100), 67% (ATCC-HYR0103), and 67% (mc-iPS), as shown in Fig 2. With regard to tumor latency, we found symptoms of tumor formation within 9 weeks in at least one mouse in each hiPSC line group. The tumor latency of each hiPSC line was 6 weeks (201B7), 4 weeks (253G1), 5 weeks (409B2), 4 weeks (454E2), 7 weeks (HiPS-RIKEN-1A), 6 weeks (HiPS-RIKEN-2A), 9 weeks (HiPS-RIKEN-12A), 4 weeks (ATCC-DYR0100), 9 weeks (ATCC-HYR0103), and 8 weeks (mc-iPS). These results indicated that tumor incidence and latency distinctly varied in the transplanted hiPSC lines. Tumor sizes increased almost in a time-dependent manner, and growth rates of tumor mass were quite different among hiPSC lines as well as individual mice (Fig 3). Engrafted tumors of 454E2, ATCC-DYR0100, and ATCC-HYR0103 hiPSCs grew progressively to the size of more than 800, 600, and 500 mm3, respectively. In contrast, none of the tumors derived from 409B2, HiPS-RIKEN1A, or HiPS-RIKEN12A hiPSCs exceeded 500, 1000, or 600 mm3, respectively, even after 16 weeks of observation. The growth rates of tumors derived from 201B7, 253G1, and mc-iPS hiPSCs appeared to be moderate compared with the other hiPSC lines indicated above. Exclusively, rapid growth of only one tumor was seen, and no other tumor was formed in the HiPS-RIKEN-2A hiPSC group. Indeed, tumor sizes of 454E2, ATCC-DYR0100, and ATCC-HYR0103 hiPSCs began to show statistically significant differences from those of the three hiPSC groups, 409B2, HiPS-RIKEN1A, and HiPS-RIKEN12A, at 11, 11, and 13 weeks of transplantation, respectively, using two-way repeated measures ANOVA followed by Bonferroni t-test. Taken together, these results suggest that tumorigenic potential is largely dependent on the property of each hiPSC line.
Fig 2. Tumor incidence of mice subcutaneously injected with 10 hiPSC lines.
Dissociated single cells of hiPSC lines (201B7, 253G1, 409B2, 454E2, HiPS-RIKEN-1A, HiPS-RIKEN-2A, HiPS-RIKEN-12A, DYR0100, HYR0103, and mc-iPS) were subcutaneously transplanted into NOG mice at 3 × 104 cells with Matrigel and 1 × 106 mitomycin C-treated NHDF in the presence of 10 μM Y-27632. Tumor formation was examined for 16 weeks. Six mice were used in each group.
Fig 3. Time course of tumor volume in mice subcutaneously injected with 10 hiPSC lines.
Dissociated single cells of hiPSC lines (201B7, 253G1, 409B2, 454E2, HiPS-RIKEN-1A, HiPS-RIKEN-2A, HiPS-RIKEN-12A, DYR0100, HYR0103, and mc-iPS) were subcutaneously transplanted into NOG mice at 3 × 104 cells with Matrigel and 1 × 106 mitomycin C-treated NHDF in the presence of 10 μM Y-27632. Tumor size was observed for 16 weeks. Six mice were used in each group.
Histological analysis of formed tumor
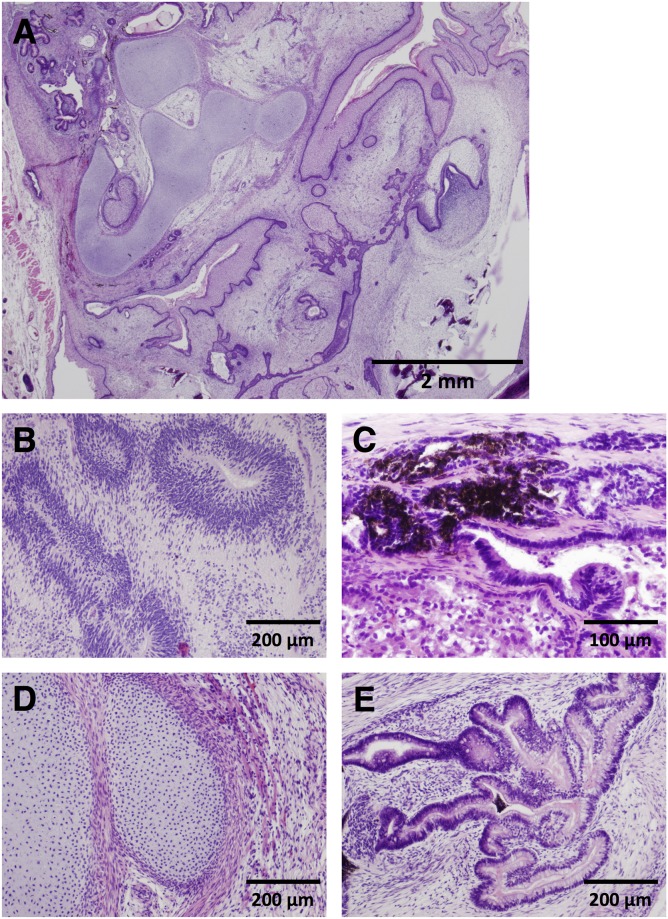
To observe tumors formed in NOG mice transplanted with the 10 hiPSC lines, we prepared tumor samples isolated from all mice showing tumor formation for histological analysis. Evaluating these H&E stained specimens, we focused on three points: a) differentiation to more than one of three germ layers (endoderm, mesoderm, and ectoderm) to define the tumor sample as a teratoma, b) grading based on the amount of neuroepithelial cell component immaturity [14], and c) formation of carcinoma and sarcoma involved in malignant tumor. Fig 4 shows typical images of teratoma with ectodermal neuroepithelia and melanocytes, mesodermal cartilage, and endodermal intestinal tract-like ducts. The observed tumor samples clearly showed differentiation into either two germ layers (mesoderm and ectoderm) or three germ layers, indicating that all of the tumor samples are first defined as teratoma (Table 2, S1 Fig). Next, we examined whether the tumor samples were categorized into immature or mature teratoma. No immature neuroepithelial cell components were found in only two samples derived from the mc-iPS line out of 47 tumor samples. Therefore, almost all of the tumor samples (45 samples) were recognized as immature teratomas, and each sample was graded from 1 to 3 (Table 2). However, we did not find any significant difference in the grades among hiPSC line groups using the Kruskal–Wallis test. In addition, neither carcinoma nor sarcoma was histopathologically observed in tumors derived from any of the hiPSC lines.
Fig 4. Representative images of teratomas subcutaneously formed in NOG mice injected with hiPSCs in a mixture of Matrigel, mitomycin C-treated NHDF, and 10 μM Y-27632.
Excised tumor samples were stained with hematoxylin and eosin. Low power view of teratoma (A), ectodermal neuroepithelia (B), melanocytes (C), mesodermal cartilage (D), and endodermal intestinal tract-like ducts (E).
Table 2. Sizes and grades of tumor derived from hiPSCs transplanted into NOG mice.
| hiPSC lines (passages) | No. | Tumor size (mm3) | Time after transplantation (wk) | Differentiation into germ layers | Grade of teratoma |
|---|---|---|---|---|---|
| 201B7 (p35) | 1 | 1468 | 16 | Three germ layers | 3 |
| 2 | 1406 | 16 | Three germ layers | 1 | |
| 3 | 2026 | 14 | Three germ layers | 3 | |
| 4 | 648 | 16 | Three germ layers | 1 | |
| 5 | 991 | 16 | Ectoderm/mesoderm | 3 | |
| 6 | 332 | 16 | Three germ layers | 1 | |
| 253G1 (p46) | 1 | 760 | 16 | Three germ layers | 1 |
| 2 | 139 | 16 | Ectoderm/mesoderm | 1 | |
| 3 | 814 | 16 | Ectoderm/mesoderm | 1 | |
| 4 | 58 | 16 | Three germ layers | 1 | |
| 5 | 168 | 16 | Ectoderm/mesoderm | 1 | |
| 6 | 1599 | 13 | Ectoderm/mesoderm | 3 | |
| 409B2 (p38) | 3 | 244 | 16 | Ectoderm/mesoderm | 2 |
| 4 | 400 | 16 | Ectoderm/mesoderm | 3 | |
| 5 | 43 | 16 | Three germ layers | 1 | |
| 6 | 128 | 16 | Ectoderm/mesoderm | 1 | |
| 454E2 (p44) | 1 | 2369 | 15 | Three germ layers | 3 |
| 2 | 2030 | 11 | Three germ layers | 1 | |
| 3 | 804 | 16 | Ectoderm/mesoderm | 1 | |
| 4 | 1592 | 16 | Ectoderm/mesoderm | 1 | |
| 5 | 2319 | 15 | Three germ layers | 1 | |
| 6 | 2021 | 13 | Three germ layers | 1 | |
| HiPS-RIKEN-1A (p24) | 1 | 697 | 16 | Three germ layers | 3 |
| 2 | 99 | 16 | Three germ layers | 1 | |
| 3 | 226 | 16 | Ectoderm/mesoderm | 3 | |
| 4 | 273 | 16 | Three germ layers | 2 | |
| 5 | 961 | 16 | Three germ layers | 1 | |
| 6 | 141 | 16 | Three germ layers | 1 | |
| HiPS-RIKEN-2A (p27) | 6 | 1603 | 9 | Three germ layers | 2 |
| HiPS-RIKEN-12A (p32) | 1 | 595 | 16 | Ectoderm/mesoderm | 1 |
| 2 | 85 | 16 | Three germ layers | 1 | |
| 5 | 506 | 16 | Three germ layers | 3 | |
| 6 | 49 | 16 | Three germ layers | 1 | |
| ATCC-DYR0100 (unknown; >p11) | 1 | 2481 | 11 | Three germ layers | 3 |
| 2 | 1172 | 16 | Three germ layers | 1 | |
| 3 | 2030 | 8 | Three germ layers | 1 | |
| 4 | 690 | 16 | Three germ layers | 1 | |
| 6 | 2179 | 14 | Three germ layers | 1 | |
| ATCC-HYR0103 (unknown; >p10) | 2 | 2380 | 11 | Three germ layers | 1 |
| 3 | 4218 | 13 | Three germ layers | 2 | |
| 4 | 2950 | 16 | Three germ layers | 2 | |
| 6 | 524 | 16 | Three germ layers | 1 | |
| mc-iPS (unknown; >p27) | 2 | 181 | 16 | Three germ layers | 0 |
| 3 | 1921 | 15 | Three germ layers | 0 | |
| 5 | 1115 | 16 | Three germ layers | 3 | |
| 6 | 213 | 16 | Three germ layers | 1 |
Gene expression correlated with tumorigenicity of hiPSC lines
Next, we attempted to find marker genes, the transcript expression of which in hiPSCs is correlated with their tumorigenicity. We examined their transcriptional profiles using microarray analysis, and the defined filtering criteria (see Material and methods) identified a set of 16,454 probes with significantly different expression levels among 10 hiPSC lines (S2 Fig). To isolate genes associated with tumorigenicity of the hiPSC lines, we compared GeneChip signal intensities of the hiPSC lines with the latency of tumor formation and tumor incidence in NOG mice transplanted with these cells as described in the Materials and Methods section. The correlations of signal intensities of the filtered probe sets with two variables, i.e., (i) latency of tumor formation and (ii) tumor incidence, were determined by calculating Spearman’s rank correlation coefficients: The rank order of tumor formation latency, 253G1 = 454E2 = ATCC-DYR0100 < 409B2 < 201B7 = HiPS-RIKEN-2A < HiPS-RIKEN-1A < mc-iPS < HiPS-RIKEN-12A = ATCC-HYR0103; the rank order of tumor incidence, HiPS-RIKEN-2A < 409B2 = HiPS-RIKEN-12A = ATCC-HYR0103 = mc-iPS < ATCC-DYR0100 < 201B7 = 253G1 = 454E2 = HiPS-RIKEN-1A. Finally, 289 resulting probe sets derived from 258 genes exhibited statistically significant positive or negative correlation (p < 0.01) with both two variables (S4 Table). The expression of pluripotency markers, OCT3/4, NANOG, SOX2, and LIN28A, did not exhibit significant correlation with tumor formation latency and incidence, and the integration of reprogramming factor genes into genome of hiPSCs did not also affect the transcript expression (S3 Fig). Pathway analysis using these genes showed top 5 networks (S5 Table), and the top diseases and functions included cellular assembly and organization, developmental disorder, and hematological diseases. In addition, 5-fluorouracil (5-FU), LIN28A, let-7, RASSF5, and ALKBH5 were shown as top 5 upstream regulators, all of which have been reported to be related to tumor formation (Table 3). These results suggest that the networks of our identified genes would play central roles in the in vivo tumorigenicity of hiPSCs.
Table 3. Top 5 of upstream regulators.
| Upstream Regulator | Genes | p-value of overlap |
|---|---|---|
| 5-fluorouracil | ATP5C1, ATP5O, CCT4, GTF2I, HNRNPAB, IARS, RPS16, RPS4X, SMS, UNG, YAP1 | 4.12 × 10−4 |
| LIN28A | CDC20, CDC25A, TIA1 | 1.53 × 10−3 |
| let-7 | CDC16, CDC20, CDC25A, E2F5, GTF2I, MCM7, YAP1 | 5.89 × 10−3 |
| RASSF5 | LSM7, NAP1L3, PSMD1 | 6.36 × 10−3 |
| ALKBH5 | ADGRG2, CELSR2, CLDN11, PAFAH1B1, PRDX4 | 6.49 × 10−3 |
Karyotype of transplanted hiPSC lines
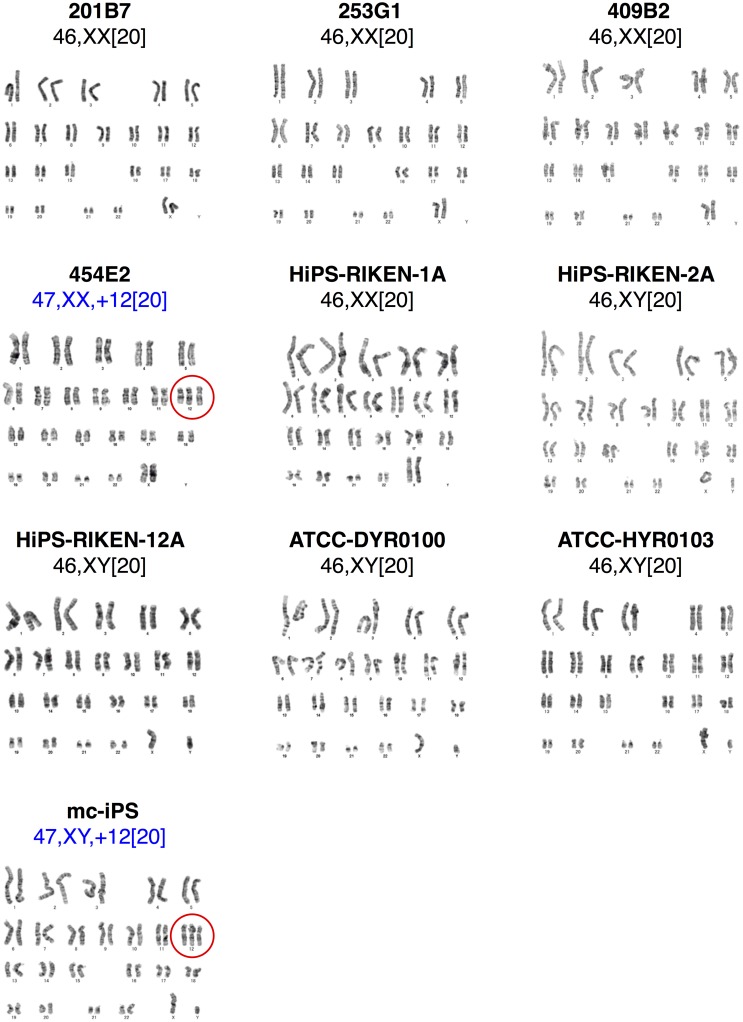
Karyotype analysis is frequently used for quality assessment of PSCs to examine chromosomal abnormalities in structure and number [21]. According to the karyotype analysis of the 10 hiPSC lines cultured under our conditions for transplantation into mice, a normal karyotype was observed in 8 hiPSC lines but not in 2 hiPSC lines (mc-iPS and 454E2). Both of these hiPSC lines exhibited the same type of chromosomal abnormality, trisomy 12, in all tested cells (Fig 5). It is likely that hiPSCs having trisomy 12 exclusively grew in vitro because trisomy 12 has been reported to increase proliferation rates of hPSCs [22]. However, we did not observe distinct effects of trisomy 12 on the tumorigenicity potential of hiPSCs and histological properties of hiPSC-derived tumors.
Fig 5. Karyotype analysis of the 10 hiPSC lines.
In vitro cultured hiPSC lines (201B7, 253G1, 409B2, 454E2, HiPS-RIKEN-1A, HiPS-RIKEN-2A, HiPS-RIKEN-12A, DYR0100, HYR0103, and mc-iPS) were subjected to karyotype analysis. 454E2 and mc-iPS lines showed trisomy 12. Chromosomal aberrations are encircled.
Exome sequence analysis of cancer-related genes in hiPSCs
The analysis of mutations in cancer-related genes has recently been applied in the quality evaluation of hiPSC lines using next-generation sequencing (NGS). We, therefore, performed exome sequence analysis and examined SNV and indel mutations of 613 cancer-related genes publically shown in databases including the COSMIC Cancer Gene Census. These 613 cancer-related genes are listed in S2 Table. A mutation rate of greater than 9% is considered to be reliable with more than 15 Gb read data per sample under the condition used in this exome sequencing analysis (Miura et al., unpublished observation). Therefore, we first searched for mutations with a mutation rate of more than 9% in cancer-related genes. Second, read depth was set at 40 or more than 40, which covered more than 97.3% of the target sequences in all analyzed samples. Third, we focused on mutations that would highly or moderately impact the amino acid sequence of the encoding protein in cancer-related genes as indicated by the possible functional change of the translated proteins [18]. Fourth, we further extracted gene mutations as classified into variants of COSMIC database. We identified 61–77 variants of cancer-related genes that satisfy the criteria per hiPSC line (Table 4, S6 Table). Mutation profiles of 201B7, 253G1, and 409B2 hiPSC lines were quite similar, likely due to the common fibroblasts used for establishing hiPSCs. The allele frequency reported in public exome sequencing database, HGVD and ExAC, was exhibited on the mutations observed in hiPSCs in the COSMIC database (S7 Table). Our findings showed that the identified mutations in the indicated cancer-related genes did not directly lead to malignancy of the teratomas derived from the hiPSCs under our experimental conditions.
Table 4. Genes having mutations, which were assessed as a high or moderate impact and confirmed in the COSMIC cancer database.
| hiPSC lines | Genes having mutations reported in the COSMIC database |
|---|---|
| 201B7 | TNFRSF14, SPEN, NOTCH2, PIK3CA, TET2, FAT1, TERT, DROSHA, IL7R, APC, HLA-A*, NFKBIE, KMT2C, PCM1, NCOA2, OMD, NOTCH1, KAT6B, CREB3L1, MEN1, MAML2, ATM, KCNJ5, PTPRB, SH2B3, HNF1A, NCOR2, FLT3, ERCC5, FOXA1, HIF1A, ZFHX3, TP53, BRCA1, AXIN2, SETBP1, MAP2K2, JAK3, EP300, ZRSR2, FANCB |
| 253G1 | TNFRSF14, SPEN, NOTCH2, PIK3CA, TET2, FAT1, TERT, DROSHA, IL7R, APC, HLA-A, NFKBIE, CREB3L2, KMT2C, PCM1, NCOA2, OMD, NOTCH1, KAT6B, CREB3L1, MEN1, MAML2, ATM, KCNJ5, PTPRB, SH2B3, HNF1A, NCOR2, FLT3, ERCC5, FOXA1, HIF1A, ZFHX3, TP53, BRCA1, AXIN2, SETBP1, JAK3, EP300, AR, FANCB |
| 409B2 | TNFRSF14, SPEN, NOTCH2, PIK3CA, TET2, FAT1, TERT, DROSHA, IL7R, APC, HLA-A, NFKBIE, CREB3L2, KMT2C, PCM1, NCOA2, OMD, NOTCH1, KAT6B, CREB3L1, MEN1, MAML2, ATM, KCNJ5, PTPRB, SH2B3, HNF1A, NCOR2, FLT3, ERCC5, FOXA1, HIF1A, ZFHX3, TP53, BRCA1, AXIN2, SETBP1, JAK3, EP300, AR, FANCB |
| 454E2 | NOTCH2, ALK, AFF3, FANCD2, ATR, WWTR1, TET2, FAT4, FAT1, SDHA, IL6ST, APC, RANBP17, NSD1, NFKBIE, EGFR CREB3L2, EZH2, KMT2C, FANCG, NCOA4, KAT6B, FGFR2, CREB3L1, MEN1, MAML2, ATM, KCNJ5, ZNF384, SH2B3, HNF1A, NCOR2, ERCC5, FOXA1, ZFHX3, TP53, ERBB2, RNF43, RNF43, AXIN2, SETBP1, KDM6A, ATRX |
| HiPS-RIKEN-1A | SPEN, NOTCH2, ALK, FANCD2, FOXL2, KIT, KDR, PTPN13, TET2, FAT4, FAT1, SDHA, IL7R, APC, NSD1, HLA-A, NFKBIE, TRRAP, CREB3L2, KMT2C, NCOA2, CDKN2A, FANCG, PTCH1, KAT6B, FGFR2, CREB3L1, MEN1, MAML2, ATM, CBL, KCNJ5, ZNF384, SH2B3, HNF1A, NCOR2, FLT3, BRCA2, ERCC5, FOXA1, BLM, ZFHX3, ERBB2, RNF43, AXIN2, SETBP1, AR, ATRX |
| HiPS-RIKEN-2A | TNFRSF14, SPEN, NOTCH2, ALK, FANCD2, FOXL2, ATR, WWTR1, KDR, TET2, FAT4, FAT1, SDHA, APC, RANBP17, HLA-A, EGFR, CREB3L2, KMT2C, PTCH1, KAT6B, FGFR2, CREB3L1, MEN1, MAML2, ATM, KCNJ5, ZNF384, COL2A1, SH2B3, HNF1A, NCOR2, FLT3, BRCA2, ERCC5, FOXA1, ZFHX3, TP53, NF1, BRCA1, RNF43, AXIN2, SETBP1, TCF3, KDM6A, ATRX |
| HiPS-RIKEN-12A | TNFRSF14, NOTCH2, ALK, FANCD2, GATA2, ATR, WWTR1, KDR, PTPN13, TET2, FAT4, FAT1, APC, RANBP17, HLA-A, NFKBIE, EGFR, CREB3L2, KMT2C, PTCH1, RET, KAT6B, TCF7L2, CREB3L1, MEN1, MAML2, ATM, KCNJ5, ZNF384, COL2A1, SH2B3, HNF1A, NCOR2, POLE, FLT3, BRCA2, ERCC5, FOXA1, HIF1A, ZFHX3, TP53, RNF43, AXIN2, STK11, TCF3, ATRX |
| ATCC-DYR0100 | TNFRSF14, NOTCH2, ALK, FANCD2, ATR, WWTR1, KDR, PTPN13, TET2, FAT1, DROSHA, IL7R, APC, NSD1, HLA-A, ELN, CREB3L2, KMT2C, KAT6B, CREB3L1, MEN1, MAML2, ATM, KCNJ5, CHD4, ZNF384, SH2B3, HNF1A, NCOR2, POLE, FLT3, ERCC5, FOXA1, ZFHX3, TP53, BRCA1, RNF43, SETBP1, ASXL1, MN1, EP300 |
| ATCC-HYR0103 | TNFRSF14, SPEN, NOTCH2, ALK, MSH2, KDR, FAT4, FAT1, IL7R, APC, KMT2C, NRG1, PDCD1LG2, OMD, TSC1, CREB3L1, MEN1, MAML2, ATM, KCNJ5, ZNF384, COL2A1, KMT2D, SH2B3, HNF1A, NCOR2, FLT3, FOXA1, ZFHX3, TP53, BRCA1, RNF43, AXIN2, MN1, EP300, AR, ATRX |
| mc-iPS | TNFRSF14, SPEN, NOTCH2, FANCD2, FOXL2, ATR, KIT, KDR, PTPN13, TET2, FAT4, FAT1, IL7R, APC, HLA-A, CREB3L2, KMT2C, PTCH1, NOTCH1, CREB3L1, MEN1, MAML2, ATM, KCNJ5, CHD4, ZNF384, SH2B3, HNF1A, NCOR2, FLT3, BRCA2, FOXA1, ZFHX3, TP53, RNF43, AXIN2, SETBP1, TCF3, JAK3, KDM6A, ATRX |
*Variants of gene symbols indicated in bold letters were assessed as a “high” impact by SnpEff software.
Discussion
We have established in vivo testing to quantify undifferentiated hPSCs using severe immunodeficient NOG mice. When defined numbers of dissociated single cells of the hiPSC line 201B7 were mixed with feeder cells, Matrigel and a Rho kinase inhibitor and subcutaneously transplanted into NOG mice, the TPD50 was calculated as 631. In the tumorigenicity testing, the 10 hiPSC lines differed in tumor incidence, formation latency, and volumes, indicating variety of tumorigenicity in these hiPSC lines. Almost of the formed tumors were categorized as immature teratomas and did not exhibit any signs of carcinoma or sarcoma, although the transplanted hiPSC lines possessed mutations putatively impacting the translated structure of cancer-related genes and in part showed chromosomal aberrations. We identified genes that were expressed in the hiPSCs and that significantly correlated with tumorigenicity and found their upregulators using pathway analysis.
Regarding the methods for quantifying undifferentiated hPSCs by in vivo tumorigenicity testing, Gropp et al. previously reported the sensitive teratoma assay of single cell-dissociated hESCs with mitotically inactivated feeder cells and Matrigel using NOD/SCID mice [7]. This paper described that addition of a Rho kinase inhibitor had no effect on the timing of tumor detection or its size, which was inconsistent with our results using NOG mice. Although the reason for this discrepancy is still unknown, differences in the immunosuppression of the mouse strains used for assaying would possibly affect the efficiency of hPSC engraftment. On the other hand, in this study, no tumor formation was detected in NOG mice transplanted with the mixture of hiPSCs when hMSCs were used instead of fibroblasts. This observation suggests that hMSCs suppress the growth of hiPSCs in vivo, although hMSCs are well known feeder cells used for in vitro co-culture with hPSCs [23]. Recently, Chang et al. reported that human umbilical cord mesenchymal stem cells abolish in vivo tumorigenicity of co-cultured hESCs by suppressing their WNT/β-catenin signaling [24]. hMSCs may possibly inhibit tumor formation of hiPSCs in mice by the same mechanism as in in vitro culture. In this context, Kanemura et al. reported that retinal pigment epithelium induced apoptosis in co-cultured hiPSCs in vitro and also suppressed tumor formation potential of co-transplanted iPSCs in vivo [25], suggesting that some micro-environment constituted of cells influences the survival of hiPSCs.
The hiPSC lines used in this study are commonly available and were established with several types of primary cells and reprogramming methods. The transplanted hiPSC lines markedly varied in tumorigenicity features, including in tumor incidence, formation latency, and volumes. Thus, the capacity to form tumors is largely dependent on each hiPSC line. It seems that the tumorigenicity of residual hiPSCs contained in hCTPs is evaluated differently according to the hiPSC lines used as raw materials even if the number of residual hiPSCs is estimated to be the same as those levels used in in vitro methods. Histopathological analysis showed that 96% of the tumors derived from the transplanted hiPSCs were evaluated as immature teratomas composed of mature and immature tissues. Indeed, immature neuroepithelium is commonly observed in teratomas produced from hPSCs xenografted in immunodeficient mice [26]. Interestingly, Akutsu et al. reported that teratomas derived from hESCs histologically matured in vivo over time [27]. Therefore, the formation of immature teratomas in vivo is regarded as a quality attribute of hPSCs, and immaturity of teratomas largely depends on the duration of hPSC transplantation.
Here we extracted 258 genes, the expression of which positively and negatively correlated with the tumorigenicity properties of the hiPSCs, and pathway analysis with these genes revealed upstream regulators including transcription factors, microRNAs, and drugs. 5-fluorouracil is an analog of uracil used for the treatment of cancer. LIN28A is known to block the expression of let-7 microRNA which has tumor suppressor function. RASSF5 is a member of the Ras association domain and serves as tumor suppressor. ALKBH5 is N6-methyladenosine demethylase and recently reported to be involved in the tumorigenicity of glioblastoma stem-like cells [28–31]. These upstream regulators would reflect gene clusters constructing several central networks in the tumorigenicity of hiPSCs. Our identified genes are expected to be useful in evaluating the tumorigenicity properties of hiPSCs as the raw materials of hCTPs.
Exome and whole genome sequence analysis with NGS in the search for genetic variants is now attempted to be applied in the quality assessment of clinical-grade hPSCs as raw materials of hCTPs [32]. Using exome sequence analysis with hiPSCs, we detected approximately 150 cancer-related genes per hiPSC line having nonsynonymous mutations that were predicted to affect the function of the translated proteins, and 61–77 mutations in the cancer-related genes were confirmed in the COSMIC databases. Moreover, 2 of the 10 hiPSC lines showed chromosomal abnormality. However, in spite of the variable mutations and chromosomes in hiPSCs, no malignant signs, i.e., sarcoma and carcinoma, were observed in all of hiPSC-derived tumors. These results suggest that the assessment of mutations in cancer-related genes and chromosomal abnormalities is not so useful in predicting malignancy of hiPSCs transplanted into immunodeficient mice at present. Studies to reveal the relationship of tumor malignancy with specific cancer-related gene mutations and chromosomal abnormalities are required in future to provide cancer-related gene mutations and karyotypes for the assessment of hiPSCs as raw materials of hCTPs.
The qualification of hPSCs as raw materials is one of the issues in the development and manufacturing of hCTPs. In our experiments, mutations in cancer-related genes and chromosomal abnormalities found in hiPSC lines did not predict malignancy in these cell lines, which was confirmed by in vivo tumorigenicity testing. Qualification of hPSCs should be mainly discussed in terms of suitability for manufacturing the final products. Our in vivo tumorigenicity testing method followed by pathological evaluation will contribute to the assessment of hCTPs as well as raw materials, in terms of tumorigenic cell impurities and malignancy. Furthermore, to internationally standardize tumorigenicity testing methods for hCTPs, it would be necessary to establish standard protocols and to validate the testing methods according to reports. Hopefully, our method will support international standardization as a reference of the testing methods to detect residual undifferentiated cells of hCTPs in the future.
Supporting information
(DOCX)
(XLSX)
(DOCX)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
NOG mice were subcutaneously injected with 3 × 104 hiPSCs in a mixture of Matrigel, 1 × 106 mitomycin C-treated NHDF, and 10 μM Y-27632. Typical images of teratomas derived from 10 hiPSC lines are shown stained with hematoxylin and eosin [201B7 #3 (A, B), 253G1 #6 (C, D), 409B2 #3 (E, F), 454E2 #6 (G, H), HiPS-RIKEN-1A #6 (I, J), HiPS-RIKEN-2A #6 (K, L), HiPS-RIKEN-12A #5 (M, N), ATCC-DYR0100 #6 (O, P), ATCC-HYR0103 #4 (Q, R), and mc-iPS #5 (S, T)]. Low power view (× 1.25) of teratoma represents two or three germ layer components (A, C, E, G, I, K, M O, Q and S). Higher power view (× 10) shows mesodermal cartilage and endodermal intestinal tract-like duct (B), ectodermal glial tissues, melanocytes and choroid-like tissues (D), ectodermal glial tissues (F), mesodermal smooth muscle and endodermal intestinal tract-like duct (H), ectodermal choroid-like tissues and immature neuroepithelia (J), ectodermal choroid-like tissues and melanocytes (L), ectodermal stratified squamous epithelia and endodermal duct structures (N), ectodermal glial and neural cells and mesodermal blood vessels (P), endodermal duct structures accompanied with intestinal and respiratory epithelium-like cells (R), and mesodermal smooth muscle and endodermal intestinal tract structures (T).
(PDF)
A set of 16,454 probes on GeneChip Human Genome U133 Plus 2.0 Array was statistically identified with significantly different expression levels among 10 hiPSC lines (one-way ANOVA, p < 0.05). Hierarchical clustering analysis was performed using R version 3.5.1 software.
(TIFF)
Transcript expression of OCT3/4 (A), NANOG (B), SOX2 (C), and LIN28A(D) in 10 hiPSC lines is shown with microarray data. Data are represented as mean ± SD (n = 3).
(TIFF)
Acknowledgments
We thank Dr. Emiko Ito (Osaka University Graduate School of Medicine) for helpful advice on tumorigenicity testing methods.
Data Availability
The NCBI GEO accession number for the microarray data is GSE108566. The NCBI Sequence Read Archive (SRA) accession number for the whole exome sequencing data is SRP134676.
Funding Statement
The present work was supported by a Research Grant from the Japanese Ministry of Health, Labour and Welfare (http://www.mhlw.go.jp/english/) (H25-JITSUYOKA(SAISEI)-IPPAN-008) and the Japan Agency for Medical Research and Development (https://www.amed.go.jp/en/) (15bk0104014h0103 to YS, 15bk0104014h0203 to SY, 16bk0104006h0104 to YS, 16bk0104006h0204 to SY, 17bk0104006j0105 to YS, and 17bk0104006j0205 to SY). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Trounson A, McDonald C. Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell stem cell. 2015;17(1):11–22. Epub 2015/07/04. 10.1016/j.stem.2015.06.007 . [DOI] [PubMed] [Google Scholar]
- 2.Cunningham JJ, Ulbright TM, Pera MF, Looijenga LH. Lessons from human teratomas to guide development of safe stem cell therapies. Nature biotechnology. 2012;30(9):849–57. Epub 2012/09/12. 10.1038/nbt.2329 . [DOI] [PubMed] [Google Scholar]
- 3.WHO Expert Committee on Biological Standardization: Sixty-first Report. WHO Technical Report Series, No. 978 Annex 3. Recommendations for the evaluation of animal cell cultures as substrates for the manufacture of biological medicinal products and for the characterization of cell banks: World Health Organization; 2013. 79–187 p.
- 4.Kusakawa S, Machida K, Yasuda S, Takada N, Kuroda T, Sawada R, et al. Characterization of in vivo tumorigenicity tests using severe immunodeficient NOD/Shi-scid IL2Rgnull mice for detection of tumorigenic cellular impurities in human cell-processed therapeutic products. Regen Ther. 2015;1:30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature biotechnology. 2007;25(6):681–6. Epub 2007/05/29. 10.1038/nbt1310 . [DOI] [PubMed] [Google Scholar]
- 6.Hentze H, Soong PL, Wang ST, Phillips BW, Putti TC, Dunn NR. Teratoma formation by human embryonic stem cells: evaluation of essential parameters for future safety studies. Stem Cell Res. 2009;2(3):198–210. 10.1016/j.scr.2009.02.002 . [DOI] [PubMed] [Google Scholar]
- 7.Gropp M, Shilo V, Vainer G, Gov M, Gil Y, Khaner H, et al. Standardization of the teratoma assay for analysis of pluripotency of human ES cells and biosafety of their differentiated progeny. PLoS One. 2012;7(9):e45532 10.1371/journal.pone.0045532 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. Epub 2007/11/24. 10.1016/j.cell.2007.11.019 . [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26(1):101–6. Epub 2007/12/07. 10.1038/nbt1374 . [DOI] [PubMed] [Google Scholar]
- 10.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nature methods. 2011;8(5):409–12. Epub 2011/04/05. 10.1038/nmeth.1591 . [DOI] [PubMed] [Google Scholar]
- 11.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, et al. A nonviral minicircle vector for deriving human iPS cells. Nature methods. 2010;7(3):197–9. Epub 2010/02/09. 10.1038/nmeth.1426 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consensus recommendations on effective institutional Animal Care and Use Committees. Laboratory animal science. 1987;37 Spec No:11–3. Epub 1987/01/01. . [PubMed] [Google Scholar]
- 13.Lorenz R, Bogel K. Methods of calculation In: Kaplan M, Koprowski H, editors. Laboratory techniques in rabies. 3rd ed: World Health Organization; 1973. p. 321–35. [PubMed] [Google Scholar]
- 14.O'Connor DM, Norris HJ. The influence of grade on the outcome of stage I ovarian immature (malignant) teratomas and the reproducibility of grading. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists. 1994;13(4):283–9. Epub 1994/10/01. . [DOI] [PubMed] [Google Scholar]
- 15.Glantz SA. Primer of biostatistics. 5th ed: McGraw-Hill; 2002. [Google Scholar]
- 16.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114–20. Epub 2014/04/04. 10.1093/bioinformatics/btu170 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997. 2013.
- 18.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. Epub 2012/06/26. 10.4161/fly.19695 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higasa K, Miyake N, Yoshimura J, Okamura K, Niihori T, Saitsu H, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. Journal of human genetics. 2016;61(6):547–53. Epub 2016/02/26. 10.1038/jhg.2016.12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. Epub 2016/08/19. 10.1038/nature19057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nature biotechnology. 2011;29(12):1132–44. Epub 2011/11/29. 10.1038/nbt.2051 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-David U, Arad G, Weissbein U, Mandefro B, Maimon A, Golan-Lev T, et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat Commun. 2014;5:4825 10.1038/ncomms5825 . [DOI] [PubMed] [Google Scholar]
- 23.Cheng L, Hammond H Fau—Ye Z, Ye Z Fau—Zhan X, Zhan X Fau—Dravid G, Dravid G. Human adult marrow cells support prolonged expansion of human embryonic stem cells in culture. Stem cells (Dayton, Ohio). 2003;21(2):131–42. [DOI] [PubMed] [Google Scholar]
- 24.Chang YH, Chu TY, Ding DC. WNT/beta-Catenin signaling pathway regulates non-tumorigenesis of human embryonic stem cells co-cultured with human umbilical cord mesenchymal stem cells. Sci Rep. 2017;7:41913 10.1038/srep41913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanemura H, Go MJ, Nishishita N, Sakai N, Kamao H, Sato Y, et al. Pigment epithelium-derived factor secreted from retinal pigment epithelium facilitates apoptotic cell death of iPSC. Sci Rep. 2013;3:2334 Epub 2013/08/02. 10.1038/srep02334 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damjanov I, Andrews PW. Teratomas produced from human pluripotent stem cells xenografted into immunodeficient mice—a histopathology atlas. The International journal of developmental biology. 2016;60(10-11-12):337–419. Epub 2016/12/22. 10.1387/ijdb.160274id . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akutsu H, Nasu M, Morinaga S, Motoyama T, Homma N, Machida M, et al. In vivo maturation of human embryonic stem cell-derived teratoma over time. Regen Ther. 2016;5:31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocrine-related cancer. 2010;17(1):F19–36. Epub 2009/09/26. 10.1677/ERC-09-0184 . [DOI] [PubMed] [Google Scholar]
- 29.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nature reviews Cancer. 2003;3(5):330–8. Epub 2003/05/02. 10.1038/nrc1074 . [DOI] [PubMed] [Google Scholar]
- 30.Volodko N, Gordon M, Salla M, Ghazaleh HA, Baksh S. RASSF tumor suppressor gene family: biological functions and regulation. FEBS letters. 2014;588(16):2671–84. Epub 2014/03/13. 10.1016/j.febslet.2014.02.041 . [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer cell. 2017;31(4):591–606.e6. Epub 2017/03/28. 10.1016/j.ccell.2017.02.013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews PW, Ben-David U, Benvenisty N, Coffey P, Eggan K, Knowles BB, et al. Assessing the Safety of Human Pluripotent Stem Cells and Their Derivatives for Clinical Applications. Stem Cell Reports. 2017;9(1):1–4. Epub 2017/07/13. 10.1016/j.stemcr.2017.05.029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
(DOCX)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
NOG mice were subcutaneously injected with 3 × 104 hiPSCs in a mixture of Matrigel, 1 × 106 mitomycin C-treated NHDF, and 10 μM Y-27632. Typical images of teratomas derived from 10 hiPSC lines are shown stained with hematoxylin and eosin [201B7 #3 (A, B), 253G1 #6 (C, D), 409B2 #3 (E, F), 454E2 #6 (G, H), HiPS-RIKEN-1A #6 (I, J), HiPS-RIKEN-2A #6 (K, L), HiPS-RIKEN-12A #5 (M, N), ATCC-DYR0100 #6 (O, P), ATCC-HYR0103 #4 (Q, R), and mc-iPS #5 (S, T)]. Low power view (× 1.25) of teratoma represents two or three germ layer components (A, C, E, G, I, K, M O, Q and S). Higher power view (× 10) shows mesodermal cartilage and endodermal intestinal tract-like duct (B), ectodermal glial tissues, melanocytes and choroid-like tissues (D), ectodermal glial tissues (F), mesodermal smooth muscle and endodermal intestinal tract-like duct (H), ectodermal choroid-like tissues and immature neuroepithelia (J), ectodermal choroid-like tissues and melanocytes (L), ectodermal stratified squamous epithelia and endodermal duct structures (N), ectodermal glial and neural cells and mesodermal blood vessels (P), endodermal duct structures accompanied with intestinal and respiratory epithelium-like cells (R), and mesodermal smooth muscle and endodermal intestinal tract structures (T).
(PDF)
A set of 16,454 probes on GeneChip Human Genome U133 Plus 2.0 Array was statistically identified with significantly different expression levels among 10 hiPSC lines (one-way ANOVA, p < 0.05). Hierarchical clustering analysis was performed using R version 3.5.1 software.
(TIFF)
Transcript expression of OCT3/4 (A), NANOG (B), SOX2 (C), and LIN28A(D) in 10 hiPSC lines is shown with microarray data. Data are represented as mean ± SD (n = 3).
(TIFF)
Data Availability Statement
The NCBI GEO accession number for the microarray data is GSE108566. The NCBI Sequence Read Archive (SRA) accession number for the whole exome sequencing data is SRP134676.