Abstract
Studies demonstrate that Tropheryma whipplei (T. whipplei) is present in the lungs of healthy individuals without acute respiratory symptoms or acute respiratory infection and is more common in the lungs of HIV-infected individuals and in smokers. The impact of T. whipplei colonization in the lung on local inflammation and pulmonary dysfunction in HIV-infected individuals is currently unknown. In this study, we performed specific polymerase chain reaction (PCR) and sequencing for T. whipplei in bronchoalveolar lavage (BAL) and induced sputum (IS) samples in 76 HIV-infected participants from three clinical sites. Pulmonary function and proinflammatory cytokine and chemokine levels in BAL were measured. Frequency of T. whipplei in either BAL or IS was 43.4%. The sensitivity and specificity of IS compared to BAL for detection of T. whipplei was 92.3% and 84.2%, respectively, and isolates of T. whipplei in the BAL and IS in the same subject shared genetic identity. Pulmonary function measures were not associated with T. whipplei colonization, and proinflammatory cytokine and chemokine levels in BAL and plasma as well as percentages of inflammatory cells in BAL and IS were not higher in colonized individuals. Overall, these results indicate that T. whipplei colonization in the lung is common, but may not be associated with decreased pulmonary function or inflammation in HIV-infected individuals.
Introduction
The organism Tropheryma whipplei (T. whipplei) causes Whipple’s disease, an infectious disease that primarily involves the gastrointestinal tract [1]. The prevalence of pulmonary involvement in Whipple’s disease is about 13 percent, and its clinical respiratory features include shortness of breath, dry cough and chest pain[2]. Recent microbiome studies demonstrate that T. whipplei is present in the lungs of healthy individuals without acute respiratory symptoms or acute respiratory infection and more common in the lungs of HIV-infected individuals and in smokers with antiretroviral therapy (ART) significantly reducing its relative abundance [3–5].
Chronic obstructive pulmonary disease (COPD) is a common complication in HIV-infected individuals, as 7–21% of HIV-infected individuals have obstructive ventilatory defects and 50–64% have reduced diffusing capacity for carbon monoxide (DLco) [6–10]. Despite its frequency in HIV, understanding of the pathogenesis of COPD in HIV is incomplete. Immune dysfunction caused by HIV infection could lead to microbial changes in the lung, and alterations of the lung microbiome could be involved in the development and progression of COPD [11,12]. It has been reported that low levels of organisms such as Pneumocystis jirovecii (P. jirovecii) are associated with COPD in HIV-infected individuals [13,14], but associations between T. whipplei colonization in the lung, local and systemic inflammation, and pulmonary dysfunction in HIV-infected individuals are unknown. If T. whipplei contributes to inflammation and lung damage in HIV, it might be a reversible contributor to COPD as it can be treated with antibiotics. We therefore investigated the detection of T. whipplei by bronchoscopy and induced sputum and the relationship to lung and systemic inflammation and pulmonary function in HIV-infected individuals.
Methods
Participants and sample collection
We included 76 HIV-infected participants who had undergone bronchoscopy with bronchoalveolar lavage (BAL) as part of the Pittsburgh, San Francisco and Los Angeles sites of the Lung HIV Microbiome Project (LHMP) cohort (https://lhmp.bsc.gwu.edu/) or the Pittsburgh Lung HIV Study cohort and had sufficient BAL samples available for analysis [15–17]. Inclusion criteria in the LHMP were (1) age 18 to 75 years old; (2) HIV-infected. Exclusion criteria were (1) an upper or lower respiratory tract infection within the past month, acute onset of shortness of breath, cough, fever; or use of antibiotics within the past six months; (2) Heart conditions such as tachycardia, angina, or arrhythmias; (3) Significant or uncontrolled systemic diseases. The study protocol was approved by the Institutional Review Boards at the University of Pittsburgh, the University of California San Francisco, and University of California Los Angeles. Informed consent was obtained from each participant.
Demographic and clinical data were collected by standardized participant interview including age, gender, smoking history, current antiretroviral therapy (ART), and history of prior pneumonia. CD4 cell count, and HIV RNA level (viral load) were confirmed by chart review or direct testing. Bronchoscopy was performed according to a standardized protocol developed to minimize oral contamination [3,18]. Bronchoalveolar lavage was performed in a subsegment of the right middle lobe or lingula with up to a maximum of 200 ml of 0.9% sterile saline instilled. Pre- and post-bronchodilator spirometry and DLco were performed per American Thoracic Society/European Respiratory Society guidelines [19,20]. Hankinson and Neas equations were used to determine percent predicted values of spirometry [21,22] and DLco, respectively. DLco was corrected for hemoglobin and carboxyhemoglobin [22]. Induced sputum (IS) samples were obtained from 64 of 76 (84.2%) participants as previously described [23].
T. whipplei PCR and sequence analysis
DNA was isolated from 5ml of whole BAL and 1ml of IS using the PowerSoil DNA isolation kit (MoBio, Carlsbad, CA). T. whipplei was detected by using T. whipplei hsp65 specific nested-PCR [24]. Negative and positive T. whipplei controls were included in all reactions. Amplified products were purified using Agencourt AMPure XP PCR Purification kit (Beckman Coulter, Brea, CA), and then sequenced using specific primers [24] by the Genomics and Proteomics Core laboratories at the University of Pittsburgh. To determine the genotype of T. whipplei, we performed Sanger sequencing of amplified products of the partial T. whipplei hsp65 gene. CLC Main Workbench 6.5 and the Molecular Evolutionary Genetics Analysis version 6 (MEGA6) software packages [25] were used for analyses of T. whipplei hsp65 partial gene sequences.
Measurement of lung and peripheral circulating proinflammatory cytokines and chemokines
BAL supernatant samples were obtained by centrifuging 50ml of BAL fluid at 300g for 7 minutes at 4°C. Levels of proinflammatory cytokines and chemokines in BAL supernatant and plasma were measured using a Bio-Plex human cytokine, chemokine 9-plex assay kit (Bio-Rad, Hercules, California, USA) per the manufacturer’s protocol utilizing recommended standard curve concentrations. The 9-plex assay kit quantified interleukin-1 receptor antagonist (IL-1RA), interleukin (IL)-4, IL-6, IL-13, CCL3, CCL4, CCL5, CCL11 and tumor necrosis factor alpha (TNF-α). Luminex data were analyzed using Bio-Plex Manager software (Bio-Rad). Cytokine and chemokine concentrations in the BAL supernatant were determined after correction of dilution factor as ratio of serum urea/BAL urea [26].
Cytology of BAL fluid and IS samples
Cytospin slides (25,000 cells per slide) of BAL and IS samples were generated. The slides were stained with Diff Quick and 300–500 cell differential counts were performed to determine the numbers and percentages of inflammatory cells.
Statistical analysis
Sensitivity of IS compared to BAL for detection of T. whipplei was determined. Demographic and clinical characteristics including pulmonary function variables, and levels of proinflammatory cytokines and chemokines in BAL supernatant and plasma as well as cytology of BAL fluid and IS were compared between participants with and without detectable T. whipplei in either BAL or IS using Welch's t-test or Kruskal-Wallis test for continuous variables and Fisher exact test for categorical variables. We used ANOVA to test the gender and smoking adjusted effect of T. whipplei on pulmonary function tests (as continuous variables). We also used Firth’s penalized logistic regression to test this effect on FEV1/FVC less than 70 percent and DLco less than 80 percent predicted. We had 80% power to detect a difference of 11% or more between T. whipplei groups for Post FVC%. All analyses were performed in Stata 14.2 (StataCorp LLC, College Station, TX).
Results
Frequency and genotype of T. whipplei in the lung
Thirty-three of 76 HIV-infected participants (43.4%) had T. whipplei detected in either BAL or IS (Table 1). T. whipplei was detected in 27 of 76 BAL samples (35.5%) and 30 of 64 IS samples (46.9%) by both specific nested PCR and sequencing techniques. Sixty-four participants had both BAL and IS samples collected; 24 of 26 (92.3%) T. whipplei-positive BAL samples also had matching T. whipplei-positive IS samples (Table 2) and one subject with BAL T. whipplei positive did not have an IS sample. There were 6 T. whipplei-negative BAL samples that had matching positive IS samples (Table 2). Sensitivity and specificity of IS compared to BAL for detection of T. whipplei was 92.3% and 84.2%, respectively. We analyzed and compared our previous 16s rRNA and current PCR data on frequency of T. whipplei in 39 BAL samples from this study and found that T. whipplei was detected in 33.3% (13/39) and 30.8% (12/39) BAL samples by T. whipplei PCR and 16s rRNA gene sequencing, respectively, indicating that T. whipplei hsp65 specific-PCR assay has greater sensitivity than 16S rRNA gene sequencing.
Table 1. Demographic and clinical characteristics of participants.
| Characteristic |
T. whipplei- (n = 43) |
T. whipplei+ (n = 33) |
P-value* |
|---|---|---|---|
| Age, median year (IQR) | 53 (46–59) | 52 (46–58) | 0.87 |
| Female, sex, n (%) | 8 (19) | 16 (48) | 0.007 |
| CD4, median (IQR) | 616 (410–892) | 630 (379–876) | 0.89 |
| Viral load, median (IQR) copies per ml, | 109 (27–2478) | 102 (31–4965) | 0.85 |
| ART, n (%) | 41 (95) | 30 (91) | 0.65 |
| Prior pneumonia, n (%) | 9 (21%) | 5 (15%) | 0.57 |
| Smoker, n (%) | 24 (56) | 24 (73) | 0.16 |
| Cigarette pack year, median (IQR) | 3.2 (0–13.5) | 11.9 (0–21.8) | 0.23 |
| Post-FEV1/FVC<0.7, n (%) | 6 (15) | 6 (19) | 0.75 |
| DLco<0.8, n (%) | 26 (67) | 18 (58) | 0.62 |
IQR, interquartile range; ART, antiretroviral therapy.
* p values were calculated using t-test test for continuous variables and Fisher exact test for categorical.
Table 2. Frequency of T. whipplei in bronchoalveolar lavage and induced sputum from 64 participants with both bronchoalveolar lavage and induced sputum available.
|
T. whipplei (Bronchoalveolar lavage) |
T. whipplei (Induced sputum) Positive |
T. whipplei (Induced sputum) Negative |
Total |
|---|---|---|---|
| Positive | 24 (92.3%) | 2 | 26 |
| Negative | 6 | 32 (84.2%) | 38 |
| Total | 30 | 34 | 64 |
BAL, Bronchoalveolar lavage, IS, Induced sputum
The majority of T. whipplei detected in BAL (63.0%, 17/27) or IS (66.7%, 20/30) were identical to the reference sequence (wild-type). T. whipplei containing one or more mutations in the hsp65 gene was found in 37.0% of BAL samples (10/27) and in 33.3% of IS (10/30), respectively. In addition, we compared the genotype of T. whipplei in 24 participants who had T. whipplei in both BAL and IS and found 100% concordance (Table 3). Fifteen participants had wild-type T. whipplei, and nine had the same mutations in the hsp65 gene in both BAL and IS (Table 3 and Fig 1).
Table 3. Comparison of partial hsp65 gene sequence of T. whipplei in bronchoalveolar lavage and induced sputum samples.
| Subject | Bronchoalveolar lavage | Induced sputum |
|---|---|---|
| 1 | Wild-type | Wild-type |
| 2 | Wild-type | Wild-type |
| 3 | Mutation (1nt) | Mutation (1nt) |
| 4 | Wild-type | Wild-type |
| 5 | Mutation (7nt) | Mutation (7nt) |
| 6 | Wild-type | Wild-type |
| 7 | Wild-type | Wild-type |
| 8 | Wild-type | Wild-type |
| 9 | Wild-type | Wild-type |
| 10 | Mutation (6nt) | Mutation (6nt) |
| 11 | Mutation (7nt) | Mutation (7nt) |
| 12 | Wild-type | Wild-type |
| 13 | Mutation (4nt) | Mutation (4nt) |
| 14 | Mutation (9nt) | Mutation (9nt) |
| 15 | Wild-type | Wild-type |
| 16 | Mutation (4nt) | Mutation (4nt) |
| 17 | Wild-type | Wild-type |
| 18 | Wild-type | Wild-type |
| 19 | Wild-type | Wild-type |
| 20 | Mutation (6nt) | Mutation (6nt) |
| 21 | Wild-type | Wild-type |
| 22 | Wild-type | Wild-type |
| 23 | Mutation (6nt) | Mutation (6nt) |
| 24 | Wild-type | Wild-type |
BAL, Bronchoalveolar lavage; 1nt and 7nt, 1 and 7 nucleotides different compared to the reference sequence; hsp65, heat-shock protein 65 gene
Fig 1. Partial hsp65 gene sequence of T. whipplei in bronchoalveolar lavage and induced sputum samples from the same participant.
The gene sequences of the reference T. whipplei hsp65 and the isolates of T. whipplei from number 5 participant’s BAL and IS were analyzed using Molecular Evolutionary Genetics Analysis version 6 (MEGA6) software packages. Both isolates of T. whipplei in the BAL and IS in the same participant had mutations in the hsp65 gene and shared genetic identity. BAL, Bronchoalveolar lavage; IS, induced sputum; hsp65, heat-shock protein 65 gene.
Association between T. whipplei colonization, pulmonary dysfunction, and inflammation
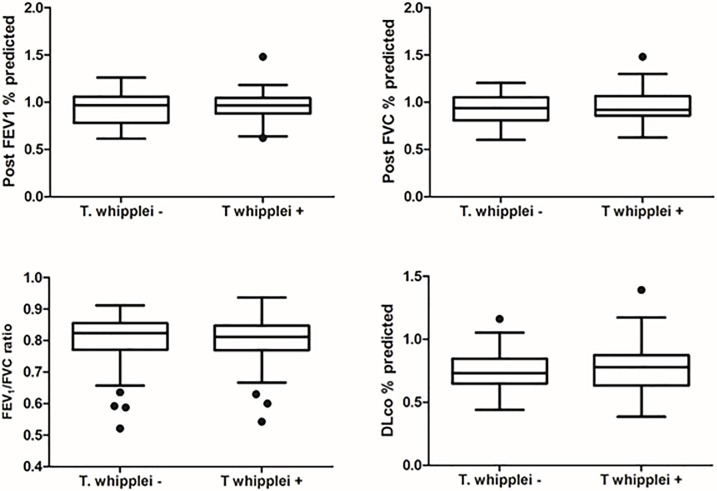
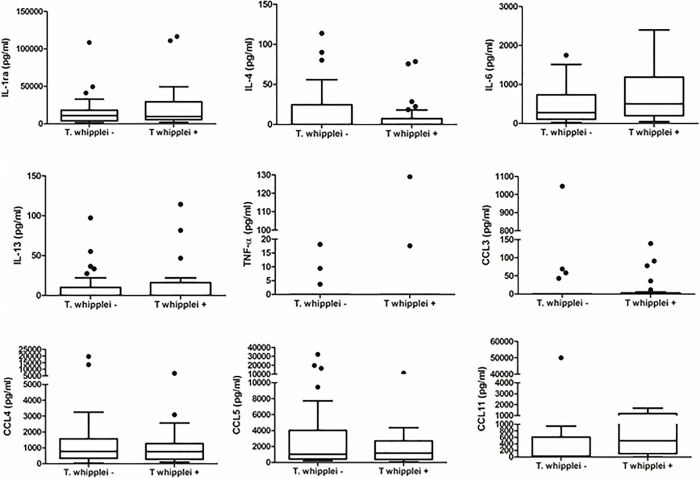
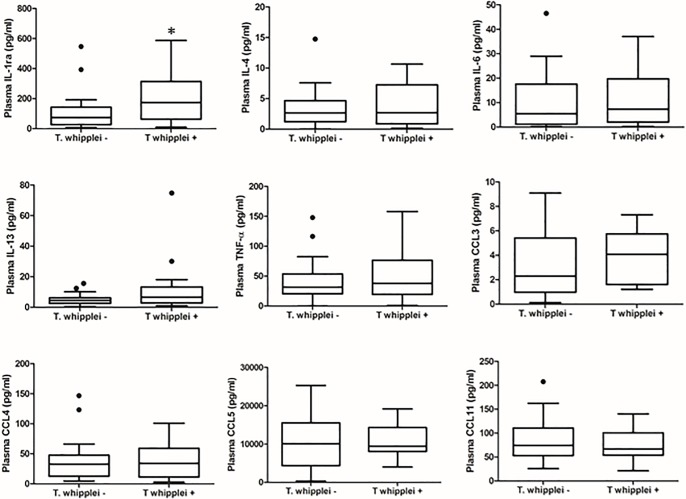
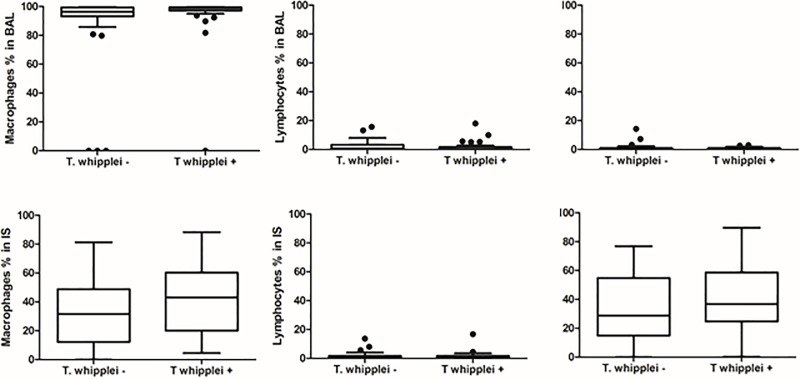
Demographic data, peripheral CD4+ cell count, plasma HIV RNA viral level, ART use and history of prior pneumonia did not differ significantly by T. whipplei status (Table 1). There was a trend for T. whipplei-colonized participants to be more likely to be female and smokers. Overall, 49 of 76 participants (64.5%) had at least one abnormality in pulmonary function tests (PFTs) with 12 of 76 (15.8%) having a post-bronchodilator forced expiratory volume in one second (FEV1)/forced vital capacity (FVC) <0.70 and 44 of 76 (57.9%) having a DLco percent predicted <80%. Analyses of pulmonary function parameters, specifically demonstrated that there were no significant differences in FEV1, FVC, FEV1/FVC and DLco between the T. whipplei-positive and negative groups based on either BAL or IS (Fig 2). Twenty-one of 49 (42.9%) participants with abnormal lung function had T. whipplei detected in the lungs, similar to the overall prevalence of T. whipplei (43.4%) in the cohort. After adjusting for effects of gender and smoking, there was no statistically significant difference between T whipplei-positive and negative participants forFEV1% (P = 0.61), FVC% (P = 0.37), FEV1/FVC (P = 0.63) and DLco% (P = 0.44) as well as FEV1/FVC below 70 percent (P = 0.40) and DLco below 80 percent predicted (P = 0.59). In a penalized logistic regression and after adjusting for smoking status and gender, the effect of T. whipplei on FEV1/FVC<0.7 (Odds ratio = 1.73, 95%CI = 0.49–6.13) and DLco<0.8 (Odds ratio = 0.76, 95%CI = 0.27–2.08) was not significant. We did not detect significant differences between T. whipplei-positive and negative participants in the levels of 9 proinflammatory biomarkers in BAL and plasma except plasma IL-1RA (Figs 3 and 4). In addition, we did not find significant differences in percentages of inflammatory cells in BAL fluid and IS samples by T. whipplei status (Fig 5).
Fig 2. Measures of pulmonary function in HIV-infected individuals by T. whipplei status.
Post-bronchodilator spirometry and diffusing capacity for DLco were performed for each participant per American Thoracic Society guidelines. Measures of pulmonary function were analyzed by T. whipplei status using Kruskal-Wallis test for continuous variables and Fisher exact test for categorical. There were no significant differences in pulmonary function test parameters by T. whipplei status. FEV1, forced expiratory volume in one second; FVC, forced vital capacity; DLco, diffusing capacity for carbon monoxide.
Fig 3. Levels of proinflammatory biomarkers in bronchoalveolar lavage by T. whipplei status.
Levels of proinflammatory cytokines and chemokines in BAL supernatant were measured using a Luminex assay kit from Bio-Rad per the manufacturer’s protocol. Data were analyzed by T. whipplei status using Kruskal-Wallis test for continuous variables and Fisher exact test for categorical. There were no significant differences in levels of cytokine and chemokine by T. whipplei status.IL-1RA, interleukin-1 receptor antagonist.
Fig 4. Levels of proinflammatory biomarkers in plasma by T. whipplei status.
Levels of proinflammatory cytokines and chemokines in plasma were measured using a Luminex assay kit from Bio-Rad per the manufacturer’s protocol. Data were analyzed by T. whipplei status using Kruskal-Wallis test for continuous variables and Fisher exact test for categorical. There were no significant differences in levels of cytokine and chemokine except IL-1RA by T. whipplei status. IL-1RA, interleukin-1 receptor antagonist. *P = 0.025.
Fig 5. Cytology of bronchoalveolar lavage and induced sputum samples by T. whipplei status.
Differential cell counts in BAL fluid and IS were performed to determine the numbers and percentages of inflammatory cells. Data were analyzed by T. whipplei status using Kruskal-Wallis test for continuous variables and Fisher exact test for categorical. There were no significant differences in percentages of different inflammatory cell types by T. whipplei status.
Discussion
This study investigated the concordance of BAL and IS detection of T. whipplei and the relationship between T. whipplei colonization in the lung, pulmonary dysfunction, and lung and systemic inflammation in HIV-infected individuals. We used T. whipplei hsp65 specific nested-PCR and sequencing approaches to identify and confirm T. whipplei in BAL and IS samples and found that 43.4% of HIV-infected individuals had T. whipplei colonization in the lungs. We found concordance between BAL and IS samples in the detection of T. whipplei. We did not find independent associations between pulmonary T. whipplei colonization and lung and systemic inflammation or pulmonary dysfunction.
The T. whipplei hsp65 specific-PCR assay is based on the T. whipplei hsp65 gene sequence and has very high sensitivity and specificity [24]. Using this assay, Morgebegg and colleagues detected T. whipplei in gastric aspirates, intestinal tissue and saliva samples from ten individuals without active Whipple’s disease [24]. In this study, we detected a higher frequency (43.4%) of T. whipplei colonization in the lungs of HIV-infected individuals compared to the 18.5% frequency in healthy individuals reported previously using PCR [27]. Using 16S rRNA gene sequencing, frequency of T. whipplei in BAL was previously determined to be about 13.0% in healthy individuals and 31.7% in HIV-infected individuals [4]. In bronchial brush samples, the frequency was 23.4% in HIV-infected individuals [28]. In 39 BAL samples from this study, 33.3% (13/39) and 30.8% (12/39) were detected T. whipplei by T. whipplei PCR and 16s rRNA gene sequencing, respectively, indicating that T. whipplei hsp65 specific-PCR assay has greater sensitivity than 16S rRNA gene sequencing. Simpson reported that the T. whipplei PCR assay was more sensitive than 16S rRNA sequencing for T. whipplei detection in IS of individuals with asthma [29]. These results indicate that T. whipplei hsp65 specific-PCR assay can be used for detection of T. whipplei colonization in lung.
Relative characteristics of BAL versus IS for detection of T. whipplei are unknown, but sensitivity and specificity were high for IS when compared to BAL. In addition, there was strong genetic identity of T. whipplei detected in the BAL and IS samples from the same participants, and individuals with a positive BAL were more likely to have positive IS. These results indicate that IS may also be useful to detect T. whipplei colonization in the lung and may detect some cases not sequenced in BAL as it samples a wider area of alveoli.
HIV-infected individuals have a high prevalence of pulmonary function abnormalities. In our cohort, 15.8% of participants had a post-bronchodilator FEV1/FVC less than 70 percent, 57.9% had a DLco percent predicted less than 80 percent predicted, and 64.5% had at least one pulmonary abnormality. Detection of certain organisms, such as P. jirovecii, at low levels in the lungs of HIV-infected individuals has been associated with pulmonary inflammation and COPD [13,14]. Whether T. whipplei in the lungs of HIV-infected individuals increases local and systemic inflammation and worsens pulmonary function has not previously been investigated. In this study, we did not find significant associations between T. whipplei colonization, lung and systemic inflammation and pulmonary dysfunction in HIV-infected individuals. These results add to prior work conducted in HIV-uninfected individuals and animal studies[12,27]. We previously showed that T. whipplei was detectable in the lung in HIV-uninfected healthy individuals, but there was not a significant association between presence of T. whipplei and decreased pulmonary function [27]. Sze and colleagues have detected T. whipplei in bronchial brush samples in HIV-infected individuals, but did not find any difference in microbial diversity and richness between HIV-infected individuals with and without COPD [28]. In a SHIV-infected macaque model, we also did not find a relationship between T. whipplei colonization and the development of COPD [12].
There are several limitations to our study. First, we may have lacked power to see some associations. The current study has 80% power to detect a difference of 11% or more between T. whipplei groups for post FVC%. A total sample size of 540 (including 310 T. whipplei—and 230 T. whipplei +) is required to gain 80% power for detecting the current difference of 4% significantly. Our previous work in determining the impact of Pneumocystis colonization in HIV-infected individuals showed a statistically significant effect in 32 individuals[11]. The current study is more than twice as large as this previous cohort and still fails to find an association with T. whipplei. In addition, we do not see any trend towards differences in pulmonary function with colonization. There is not a strict definition of a large bronchoscopy study although given the difficulties and expense associated with research bronchoscopy, 76 participants are bigger than many prior studies [11,27]. In addition, selection of different inflammatory markers might have shown a relationship to T. whipplei. However, we chose these inflammatory markers because studies have shown that plasma levels of these cytokines are negatively associated with lung function in HIV-infected individuals [16,17]. In this cohort, 93.4% participants received ART, median CD4+ cell count was more than 600 cells/μl, and median plasma HIV viral load was about 100 copies per ml. Thus, we do not know if these results are consistent with those from the individuals with severe immune suppression. Finally, we only investigated presence or absence of T. whipplei. Gene activity of the organism, abundance of T. whipplei, or interaction of T. whipplei with other members of the microbial community may have more impact on lung inflammation and function.
Although we detected T. whipplei in the lungs of 43.4% participants of an HIV-infected cohort, there was no relationship of T. whipplei to either pulmonary and systemic inflammation or pulmonary function. Thus, there does not seem to be a basis to suggest that prevention or treatment of T. whipplei would impact lung function in HIV-infected individuals. However, future larger longitudinal studies with serial BAL, serial measurement of biomarkers and pulmonary function tests are needed.
Acknowledgments
A subset of subjects in this cohort was recruited from sites of the Multicenter AIDS Cohort Study (MACS) with centers University of California, Los Angeles (Roger Detels) and University of Pittsburgh (Charles R. Rinaldo, Lawrence Kingsley). The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute. UO1-AI-35042, UL1-RR025005 (GCRC), UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041. Website located at http://www.statepi.jhsph.edu/macs/macs.html. A subset of subjects in this cohort was recruited from the Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt, Phyllis Tien and Bradley Aouizerat) of the Women’s Interagency HIV Study (WIHS) Collaborative Study Group. The WIHS is funded by the National Institute of Allergy and Infectious Diseases (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (UO1-HD-32632). The WIHS is cofunded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders. Funding is also provided by the National Center for Research Resources (UCSF-CTSI Grant Number UL1 RR024131).
Data Availability
16S microbiota sequence data are available in NCBI sequence read archive (SRA) under accession SRP065274 and The European Bioinformatics Institute (EMBL-EBI) under study accession PRJNA253761.
Funding Statement
This work was supported by grants from NIH U01 HL098962, R01 HL120398, R01 HL125049, K24 HL023342 and University of Pittsburgh CTRC UL1 TR001857 (AM). Dr. Huang was partially supported by NIH K24 HL087713 (LH). San Francisco WIHS data used for this study were supported by U01-AI-103390 (RG). MACS (UO1-AI-35042, UL1-RR025005, UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041); WIHS (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, UO1-AI-42590). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Desnues B, Al Moussawi K, Fenollar F (2010) New insights into Whipple's disease and Tropheryma whipplei infections. Microbes Infect 12: 1102–1110. 10.1016/j.micinf.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 2.Durand DV, Lecomte C, Cathebras P, Rousset H, Godeau P (1997) Whipple disease. Clinical review of 52 cases. The SNFMI Research Group on Whipple Disease. Societe Nationale Francaise de Medecine Interne. Medicine (Baltimore) 76: 170–184. [DOI] [PubMed] [Google Scholar]
- 3.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. (2013) Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 187: 1067–1075. 10.1164/rccm.201210-1913OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, et al. (2013) Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am J Respir Crit Care Med 187: 1110–1117. 10.1164/rccm.201211-2145OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck JM, Schloss PD, Venkataraman A, Twigg H 3rd, Jablonski KA, Bushman FD, et al. (2015) Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. Am J Respir Crit Care Med 192: 1335–1344. 10.1164/rccm.201501-0128OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingo MR, He J, Wittman C, Fuhrman C, Leader JK, Kessinger C, et al. (2014) Contributors to diffusion impairment in HIV-infected persons. Eur Respir J 43: 195–203. 10.1183/09031936.00157712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gingo MR, George MP, Kessinger CJ, Lucht L, Rissler B, Weinman R, et al. (2010) Pulmonary function abnormalities in HIV-infected patients during the current antiretroviral therapy era. Am J Respir Crit Care Med 182: 790–796. 10.1164/rccm.200912-1858OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George MP, Kannass M, Huang L, Sciurba FC, Morris A (2009) Respiratory symptoms and airway obstruction in HIV-infected subjects in the HAART era. PLoS One 4: e6328 10.1371/journal.pone.0006328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick ME, Gingo MR, Kessinger C, Lucht L, Kleerup E, Greenblatt RM, et al. (2013) HIV infection is associated with diffusing capacity impairment in women. J Acquir Immune Defic Syndr 64: 284–288. 10.1097/QAI.0b013e3182a9213a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond MB, Kirk GD (2014) HIV-associated obstructive lung diseases: insights and implications for the clinician. Lancet Respir Med 2: 583–592. 10.1016/S2213-2600(14)70017-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui L, Lucht L, Tipton L, Rogers MB, Fitch A, Kessinger C, et al. (2015) Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med 191: 932–942. 10.1164/rccm.201409-1583OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris A, Paulson JN, Talukder H, Tipton L, Kling H, Cui L, et al. (2016) Longitudinal analysis of the lung microbiota of cynomolgous macaques during long-term SHIV infection. Microbiome 4: 38 10.1186/s40168-016-0183-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris A, Wei K, Afshar K, Huang L (2008) Epidemiology and clinical significance of pneumocystis colonization. J Infect Dis 197: 10–17. 10.1086/523814 [DOI] [PubMed] [Google Scholar]
- 14.Morris A, Norris KA (2012) Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev 25: 297–317. 10.1128/CMR.00013-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crothers K, McGinnis K, Kleerup E, Wongtrakool C, Hoo GS, Kim J, et al. (2013) HIV infection is associated with reduced pulmonary diffusing capacity. J Acquir Immune Defic Syndr 64: 271–278. 10.1097/QAI.0b013e3182a9215a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzpatrick ME, Nouraie M, Gingo MR, Camp D, Kessinger CJ, Sincebaugh JB, et al. (2016) Novel relationships of markers of monocyte activation and endothelial dysfunction with pulmonary dysfunction in HIV-infected persons. AIDS 30: 1327–1339. 10.1097/QAD.0000000000001092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzpatrick ME, Singh V, Bertolet M, Lucht L, Kessinger C, Michel J, et al. (2014) Relationships of pulmonary function, inflammation, and T-cell activation and senescence in an HIV-infected cohort. AIDS 28: 2505–2515. 10.1097/QAD.0000000000000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwai S, Fei M, Huang D, Fong S, Subramanian A, Grieco K, et al. (2012) Oral and airway microbiota in HIV-infected pneumonia patients. J Clin Microbiol 50: 2995–3002. 10.1128/JCM.00278-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macintyre N, Crapo RO, Viegi G, Johnson DC, van der Grinten CP, Brusasco V, et al. (2005) Standardisation of the single-breath determination of carbon monoxide uptake in the lung. Eur Respir J 26: 720–735. 10.1183/09031936.05.00034905 [DOI] [PubMed] [Google Scholar]
- 20.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, et al. (2005) Standardisation of spirometry. Eur Respir J 26: 319–338. 10.1183/09031936.05.00034805 [DOI] [PubMed] [Google Scholar]
- 21.Hankinson JL, Odencrantz JR, Fedan KB (1999) Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med 159: 179–187. 10.1164/ajrccm.159.1.9712108 [DOI] [PubMed] [Google Scholar]
- 22.Neas LM, Schwartz J (1996) The determinants of pulmonary diffusing capacity in a national sample of U.S. adults. Am J Respir Crit Care Med 153: 656–664. 10.1164/ajrccm.153.2.8564114 [DOI] [PubMed] [Google Scholar]
- 23.Gershman NH, Wong HH, Liu JT, Mahlmeister MJ, Fahy JV (1996) Comparison of two methods of collecting induced sputum in asthmatic subjects. Eur Respir J 9: 2448–2453. [DOI] [PubMed] [Google Scholar]
- 24.Morgenegg S, Dutly F, Altwegg M (2000) Cloning and sequencing of a part of the heat shock protein 65 gene (hsp65) of "Troaheryma whippelii" and its use for detection of "T-whippelii" in clinical specimens by PCR. Journal of Clinical Microbiology 38: 2248–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsao TC, Hong J, Li LF, Hsieh MJ, Liao SK, Chang KS (2000) Imbalances between tumor necrosis factor-alpha and its soluble receptor forms, and interleukin-1beta and interleukin-1 receptor antagonist in BAL fluid of cavitary pulmonary tuberculosis. Chest 117: 103–109. [DOI] [PubMed] [Google Scholar]
- 27.Qin S, Clausen E, Lucht L, Michael H, Beck JM, Curtis JL, et al. (2016) Presence of Tropheryma whipplei in Different Body Sites in a Cohort of Healthy Subjects. Am J Respir Crit Care Med 194: 243–245. 10.1164/rccm.201601-0162LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sze MA, Xu S, Leung JM, Vucic EA, Shaipanich T, Moghadam A, et al. (2016) The bronchial epithelial cell bacterial microbiome and host response in patients infected with human immunodeficiency virus. BMC Pulm Med 16: 142 10.1186/s12890-016-0303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, et al. (2016) Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J 47: 792–800. 10.1183/13993003.00405-2015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
16S microbiota sequence data are available in NCBI sequence read archive (SRA) under accession SRP065274 and The European Bioinformatics Institute (EMBL-EBI) under study accession PRJNA253761.