Abstract
Gastric cancer patients positive for peritoneal cytology are at increased risk of tumor recurrence, but although a certain proportion of cytology‐positive patients relapse rapidly with aggressive progression, others survive longer with conventional chemotherapies. This heterogeneity makes it difficult to stratify patients for more intensive therapy and poses a substantial challenge for the implementation of precision medicine. We developed a new approach to identify biologically malignant subpopulations in cytology‐positive gastric cancer patients, using a green fluorescent protein (GFP)‐expressing attenuated adenovirus in which the telomerase promoter regulates viral replication (TelomeScan, OBP‐401). The fluorescence emitted from TelomeScan‐positive cells was successfully quantified using a multi‐mode microplate reader. We then analyzed clinical peritoneal washes obtained from 68 gastric cancer patients and found that patients positive for TelomeScan had a significantly worse prognosis. In 21 cytology‐positive patients, the median survival time of those who were TelomeScan positive (235 days) was significantly shorter than that for those who were TelomeScan negative (671 days; P = 0.0062). This fluorescent virus‐guided cytology detects biologically malignant cancer cells from the peritoneal washes of gastric cancer patients and may thus be useful for both therapy stratification and precision medicine approaches based on genetic profiling of disseminated cells.
Keywords: fluorescent virus, gastric cancer, peritoneal dissemination, peritoneal lavage cytology, precision medicine
Abbreviations
- EMT
epithelial‐mesenchymal transition
- hTERT
human telomerase reverse transcriptase
- RBC
red blood cells
1. INTRODUCTION
As gastric cancer advances, tumor cells invade the serosa, and eventually exfoliate from the primary lesion. A few of these cells evade anoikis and the immune system to survive in the peritoneal cavity, where they develop metastatic lesions in the peritoneum.1 Peritoneal dissemination is the most common form of metastasis in gastric cancer and is associated with an extremely poor prognosis.2 Even in the absence of macroscopic peritoneal lesions, the simple detection of free cancer cells in the peritoneal cavity has proven to be one of the worst prognostic factors for gastric cancer. Thus, peritoneal lavage cytology is routinely performed during surgery for patients with advanced gastric cancer,3, 4, 5, 6 with positive cases classified as stage IV disease in the Japanese staging system.7 Such patients are treated with systemic palliative chemotherapy.8
Peritoneal cytology is thus an indispensable examination for stratifying the appropriate treatment of gastric cancer,9 but the detection of low numbers of cancer cells in the peritoneal wash is laborious. In addition, because diagnosis is made based on cytomorphological criteria, it inevitably relies on professional competence and, in part, on subjective judgement. The accuracy, sensitivity, and specificity of conventional cytology have been reported to range between 73% and 92%, 11% and 80%, and 86% and 100%, respectively, and some improvement is needed.10 Moreover, even in cytology‐positive cases, there are significant differences in outcome, with some patients succumbing rapidly to the disease, but others surviving longer.11 One of the reasons for this may due to the technical limits of conventional cytology in detecting small populations of cancer cells among the larger number of normal peritoneal cells.12 Furthermore, it is difficult to distinguish viable from dying (or non‐competent) cancer cells with this technique. Thus, novel methods to detect biologically active cancer cells in the peritoneal cavity would be conducive to cytological assessment and staging.
In this study we applied our unique nano‐biologic, a telomerase‐specific virus, to the problem of detecting cancer cells in peritoneal cytology. TelomeScan (OBP‐401) is a telomerase‐specific replication‐selective adenovirus,13, 14, 15, 16 which uses the human telomerase reverse transcriptase (hTERT) promoter to drive the expression of the E1A and E1B genes for viral replication and contains the green fluorescent protein (GFP) gene under the control of the cytomegalovirus (CMV) promoter. Given that telomerase is known to be active in most cancers, including in gastric cancer,17 and its activity correlates with the promotor activity of the hTERT gene,18 TelomeScan can theoretically visualize viable cancer cells with green fluorescence even among numerous normal cells.
Here, we investigated whether TelomeScan technology is capable of detecting cancer cells in peritoneal wash samples from patients with gastric cancer and analyzed the correlation between the presence of TelomeScan‐positive cells in the peritoneal wash and patient prognosis. We also developed a next‐generation sequencing (NGS) strategy involving conventional multi‐laser fluorescence‐activated cell sorting (FACS) to capture TelomeScan‐labelled GFP‐positive disseminated cells in the peritoneal wash.
2. MATERIALS AND METHODS
2.1. Cell line and recombinant adenovirus
The human non‐small cell lung cancer cell line H1299 was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured according to the manufacturer's specifications. The cell line authentication was performed and reported by the Japanese Collection of Research Bioresources (JCRB) Cell Bank. TelomeScan is a telomerase‐specific replication‐competent adenovirus variant, in which the hTERT gene promoter drives the expression of the E1A and E1B genes linked by an internal ribosome entry site, and in which the GFP gene is inserted into the deleted E3 region of the CMV promoter (Figure 1A).13, 16, 19 TelomeScan was purified by ultracentrifugation using CsCl step gradients. Viral titers were determined by a plaque‐forming assay using 293 cells, and the virus was stored at −80°C. This study was approved by the Recombinant DNA Experiment Safety Committee and carried out in accordance with the approved protocol (approval ID: 12015).
Figure 1.
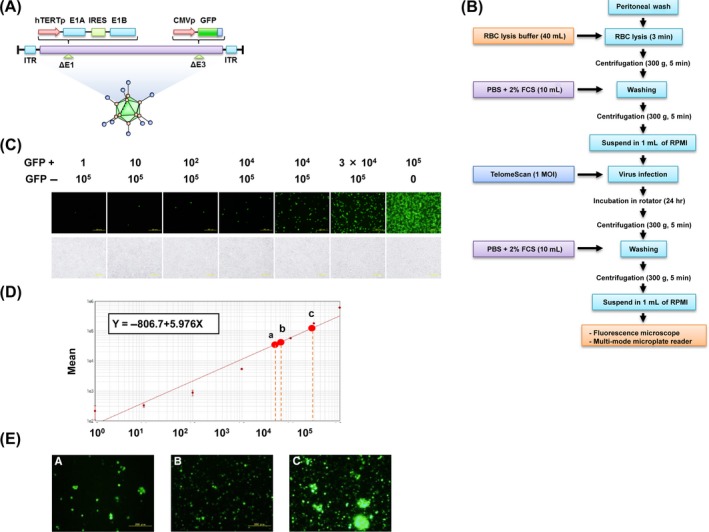
A simple quantification of TelomeScan‐positive cells using a multi‐mode microplate reader. A, Schematic DNA structure of TelomeScan (OBP‐401). TelomeScan is a telomerase‐specific replication‐competent adenovirus variant, in which the human telomerase reverse transcriptase promoter (hTERTp) element drives expression of the E1A and E1B genes linked by an internal ribosome entry site, and the green fluorescent protein (GFP) gene is inserted under the cytomegalovirus promoter (CMVp) into the deleted E3 region. B, Steps in the sample preparation for GFP‐fluorescence detection. Samples were collected and initially incubated with red blood cell (RBC) lysis buffer for 3 min when bloody. After centrifugation and washing, cell pellets were resuspended in 1 mL RPMI‐1640 medium, mixed with various concentrations of TelomeScan (finally fixed at 1 multiplicity of infection [MOI]), and incubated at 37°C with gentle rolling for another 24 h. Cells were subsequently resuspended in 1 mL RPMI‐1640 medium following centrifugation and counted under a fluorescence microscope or enumerated with a multi‐mode microplate reader. C, GFP‐fluorescence intensity was measured using a fluorescence microplate reader with excitation/emission at 473 nm/505 nm, optimized using TelomeScan‐infected H1299 cells. For optimization GFP‐positive cells and GFP‐negative cells were mixed at various ratios. D, GFP‐positive cells (TelomeScan‐infected H1299 cells) were mixed with GFP‐negative cells (H1299 cells) at various ratios. The fluorescence intensity of GFP was then quantified using a microplate reader (SpectraMax i3). The relationship between the number of GFP‐positive cells and GFP fluorescence intensity was expressed using the equation of the line derived from these calibration experiments. E, The GFP intensity of clinical samples was substituted into the formula, and the total numbers of GFP‐expressing cells in the indicated samples a (6995 cells), b (7291 cells), and c (29 013 cells) were estimated
2.2. Patients and clinical samples
A total of 68 patients with histologically proven gastric cancer were enrolled in this clinical study from March 2011 to October 2015. Overall, 491 gastric cancer patients underwent the operation in Okayama University Hospital at the same time points. Of these, peritoneal washes were obtained at the time of operation from 68 patients according to the advanced clinical stages. Briefly, on either laparotomy or laparoscopy, approximately 100‐200 mL of saline was introduced into the Douglas pouch (and occasionally into the left subphrenic space) and gently stirred. A wash sample was then aspirated, with half subjected to conventional pathological cytology and the other half examined on TelomeScan‐guided cytology. The institutional review board at Okayama University Graduate School approved the study protocol, and written informed consent was obtained from all patients.
2.3. Quantification of GFP‐positive cells
The protocol of TelomeScan infection for clinical samples is shown in Figure 1B. When samples were bloody, red blood cells (RBC) were removed in advance using RBC lysis buffer (BioLegend, San Diego, CA, USA). Samples were centrifuged, and cell pellets resuspended in 1 mL RPMI‐1640 medium. The total number of viable cells was counted, and then they were infected with TelomeScan at a multiplicity of infection (MOI) of 1 and incubated for 24 hours at 37°C with gentle rolling. GFP‐expressing cells were subsequently counted under a fluorescence microscope in a black 96‐well plate with a clear flat bottom. When there were >1000 GFP‐expressing cells, the total number of GFP‐expressing cells was arbitrarily recorded as 1000. The fluorescence intensity of GFP was quantified using the SpectraMax i3 multi‐mode microplate reader (Molecular Devices, Sunnyvale, CA, USA). The total number of GFP‐expressing cells was calculated relative to the GFP intensity of TelomeScan‐infected H1299 cells.
2.4. Immunofluorescence staining
The cells were stained with anti‐CD45 mouse IgG1 (HI30; BioLegend) and anti‐calretinin rabbit polyclonal IgG (PAD: DC8; Invitrogen, Carlsbad, CA, USA). For epithelial marker staining, cells were stained with anti‐cytokeratin (anti‐CK; pan‐reactive) mouse IgG1 (C‐11; BioLegend) and anti‐CK 19 mouse IgG2a (A53‐B/A2; BioLegend). For macrophage marker staining, cells were stained with phycoerythrin (PE)‐conjugated anti‐CD14 mouse IgG2a (M5E2; BioLegend). As secondary antibodies, Alexafluor647‐goat anti‐mouse IgG (Invitrogen; Life Technologies Corporation; Carlsbad, CA, USA) and Alexafluor568‐goat anti‐mouse IgG (Invitrogen) were used. The anti‐CK antibody was labelled using the Zenon labelling kit (Invitrogen). Immunofluorescence staining was evaluated under an inverted fluorescence microscope (IX71; Olympus, Tokyo Japan).
2.5. DNA extraction from GFP‐positive, CD45‐negative cells
The cells collected from peritoneal wash samples were infected with TelomeScan at an MOI of 1 and incubated for 24 hours. Thereafter, cell pellets were labeled with anti‐CD45 antibody conjugated with allophycocyanin (APC; BioLegend), and the GFP‐positive CD45‐negative cells were sorted using the FACS Aria (Becton Dickinson, San Jose, CA, USA). DNA was extracted from captured cells using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instruction.
2.6. Gene mutation NGS analysis
The DNA extracted from captured cells was quantified and assessed on quantitative PCR (qPCR) using a GeneRead DNA QuantiMIZE kit (Qiagen), and input DNA volume for subsequent NGS adjusted according to the manufacturer's recommendations. Target exon enrichment was performed with the GeneRead DNAseq Targeted Panel V2 for gastric cancer designed to analyze 29 human gastric cancer‐relevant genes (Qiagen). Following the qPCR‐based quantification of NGS libraries using a GeneRead DNAseq Library Quant Array (Qiagen), deep sequencing was performed on the MiSeq platform according to the manufacturer's instructions (Illumina, San Diego, CA, USA) at Okayama University Hospital Biobank.
2.7. Statistical analysis
The number of GFP‐expressing cells was compared with cytology results and clinicopathological data. Survival curves were estimated using the Kaplan‐Meier method and compared using Wilcoxon test. P < 0.05 was considered statistically significant. Statistical analysis was performed with JMP® version 11 (SAS Institute, Cary, NC, USA). Patient clinicopathological data were obtained from medical records and analyzed using Student's t test and Pearson's chi‐squared test.
3. RESULTS
3.1. Optimal conditions for TelomeScan‐mediated cancer cell visualization
TelomeScan was constructed by inserting the GFP gene under the control of the CMV promoter at the deleted E3 region of the CMV promoter of the telomerase‐specific replication‐selective type 5 adenovirus (Figure 1A). We first examined whether TelomeScan could visualize human cancer cells among the numerous co‐existing cells in the peritoneal wash. As illustrated in Figure 1B, following the lysis of RBC in peritoneal wash samples obtained from gastric cancer patients, cell pellets were incubated with various doses of TelomeScan for 24 hours and sequentially counted for the GFP signals under the fluorescence microscope. A pilot study of clinical samples using various MOI of TelomeScan ranging from 2.8 × 10−4 to 10 (Table S1) and a preliminary cell line‐based study (Figure S1) confirmed that an MOI of 1 would be optimal for TelomeScan infection.
3.2. Fluorescent intensity‐based quantification of TelomeScan‐positive cells
The biggest advantage of the fluorescence‐based detection of cancer cells by TelomeScan is the elimination of the need for subjective cytological assessments. To quantify cancer cells, samples were placed into 96‐well plates and GFP‐fluorescence intensity measured using a microplate reader with excitation/emission at 473 nm/505 nm (optimized using the human cancer cell line, H1299). TelomeScan‐infected GFP‐positive cells and uninfected GFP‐negative cells were mixed at various ratios (Figure 1C). The GFP intensity was measured over 21 points per well, and a calibration curve prepared (Figure 1D). According to the calibration curve, the number of TelomeScan‐positive cells could be estimated from GFP intensity when they numbered >100. This calibration curve was then used to compute the total number of TelomeScan‐positive cells in clinical samples based on their GFP fluorescence (Figure 1E). This confirmed the feasibility of automated enumeration of TelomeScan‐positive cells with a multi‐mode fluorescence microplate reader.
3.3. Comprehensive multicolor profiling of TelomeScan‐positive peritoneal cells
In order to positively confirm that the cells detected by TelomeScan were indeed cancer cells, we performed multicolor immunocytochemistry of cells obtained from a subset of cytology‐positive patients. The GFP‐positive cells in peritoneal wash samples were positive for CK but negative for CD45 (Figure 2A), meaning that these cells were epithelial in origin rather than hematopoietic, that is, that they were cancer cells. We further confirmed that these GFP (+)/CD45 (−) cells did not express the mesothelial marker, calretinin, to rule out the possibility that they might be mesothelial cells (Figure S2). GFP (−)/CK (−)/CD45 (+) cells were found to co‐exist with these viable cancer cells, meaning that leukocytes, such as macrophages, were abundantly present in the peritoneal cavity (Figure 2B). TelomeScan‐positive cancer cells were sometimes found in a cluster with smaller TelomeScan‐negative/CD45 (+) cells, suggesting that cancer cells in the peritoneal cavity occasionally form clusters with hematopoietic cells (Figure 2C). Although most of the TelomeScan‐positive (ie, viable) cancer cells expressed CK, some occasionally lost the expression of this marker (Figure 2A), suggesting that these cells might have undergone epithelial‐mesenchymal transition (EMT).19 We also observed other staining patterns, for example, GFP (−)/CK (+)/CD45 (−) cells (Figure 2B), which were presumed to be cancer cells that were dead or dying. TelomeScan‐guided cytology was therefore successful in detecting cancer cells from peritoneal wash samples, including those undergoing EMT, and discriminated biologically active, viable, cancer cells from those that were dead or dying.
Figure 2.
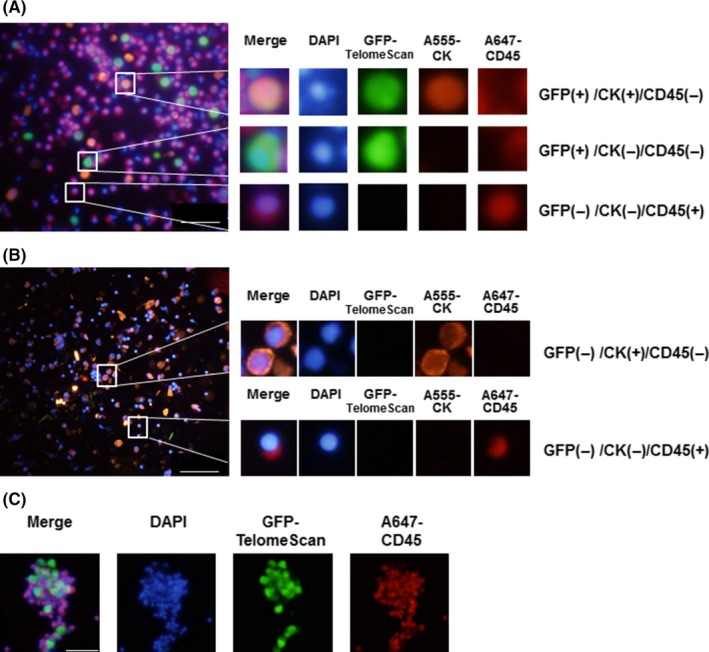
Multicolor immunofluorescence assay of TelomeScan‐positive peritoneal cells. A, Peritoneal wash samples obtained from cytology‐positive patients were analyzed for green fluorescent protein (GFP) expression following TelomeScan infection at a multiplicity of infection (MOI) of 1. TelomeScan‐positive samples were then stained with A647‐labeled anti‐CD45 and A555‐labeled anti‐cytokeratin (anti‐CK) antibodies. The merged images are shown. The GFP‐positive, CK‐negative, and CD45‐negative cells could be viable cancer cells with an epithelial‐mesenchymal transition phenotype; CD45, leukocytes marker; CK, epithelial marker (scale bar, 100 μm). B, TelomeScan‐negative peritoneal cells obtained from cytology‐positive cases were also stained for CD45 and CK. Dead cancer cells were GFP negative, CK positive, and CD45 negative (scale bar, 100 μm). C, Formation of cell clusters with TelomeScan‐positive and ‐negative cells. TelomeScan‐negative cells were relatively small and expressed CD45, suggesting that these cells were leukocytes (scale bar, 60 μm)
3.4. TelomeScan‐guided cytology and conventional cytology: Comparative analysis
From March 2011 to October 2015, peritoneal washings were obtained from patients undergoing gastric cancer surgery and analyzed on TelomeScan‐guided cytology. We examined a total of 68 clinical samples. Receiver operating characteristic (ROC) curve analysis was performed in regard to the number of GFP‐positive cells in the sample (Figure S3), and sensitivity/specificity were calculated based on positive cytology results. Based on this analysis, the threshold for declaring a positive sample with high sensitivity and specificity in this study was set at 100 GFP‐positive cells. The patient clinicopathological features are listed in Table 1. Most patients received a second or additional subsequent treatment such as S‐1 monotherapy, S‐1 plus cisplatin, paclitaxel, S‐1 and docetaxel in combination with trastuzumab, and so on. We found that T, N, P factors defined using gastric cancer classification rules correlated significantly with the positivity of the TelomeScan test, indicating that patients with advanced cases were more prone to having viable cancer cells in the peritoneal cavity. Of the 68 cases, 21 were conventional cytology positive, and the remaining 47 were cytology negative. Of the 21 cytology‐positive cases, 14 (67%) were TelomeScan positive. Of the 47 cytology‐negative cases, 36 (77%) were TelomeScan negative (Figure 3). Thus, we found a significant correlation (P = 0.0006). Based on the cytology results, the specificity, sensitivity, and accuracy of TelomeScan‐guided cytology were 76.6%, 66.7%, and 73.5%, respectively.
Table 1.
Gastric cancer patients: Clinicopathological features and correlation with TelomeScan‐guided cytology
| Factors | Total (n) | GFP | GFP | P‐value |
|---|---|---|---|---|
| Positive (n) | Negative (n) | |||
| T‐stagea | ||||
| T1 | 3 | 2 | 1 | .02 |
| T2 | 3 | 0 | 3 | |
| T3 | 16 | 2 | 14 | |
| T4 | 42 | 18 | 24 | |
| TX | 4 | 3 | 1 | |
| N‐stagea | ||||
| 0 | 17 | 5 | 12 | .064 |
| 1 | 12 | 7 | 5 | |
| 2 | 17 | 2 | 15 | |
| 3 | 15 | 7 | 8 | |
| X | 7 | 4 | 3 | |
| Hepatic metastasis | ||||
| 0 | 64 | 22 | 42 | .108 |
| 1 | 4 | 3 | 1 | |
| Peritoneal metastasis | ||||
| 0 | 54 | 14 | 40 | 0 |
| 1 | 14 | 11 | 3 | |
| Cytology | ||||
| 0 | 47 | 11 | 36 | .001 |
| 1 | 21 | 14 | 7 | |
| Any distant metastasis | ||||
| 0 | 38 | 8 | 30 | .002 |
| 1 | 30 | 17 | 13 | |
| Histology | ||||
| Differentiated | 18 | 6 | 12 | .724 |
| Undifferentiated | 50 | 19 | 31 | |
| Stage | ||||
| 0‐2 | 16 | 3 | 13 | .038 |
| 3 | 22 | 5 | 17 | |
| 4 | 30 | 17 | 13 | |
According to the Japanese classification of gastric carcinoma.
Figure 3.
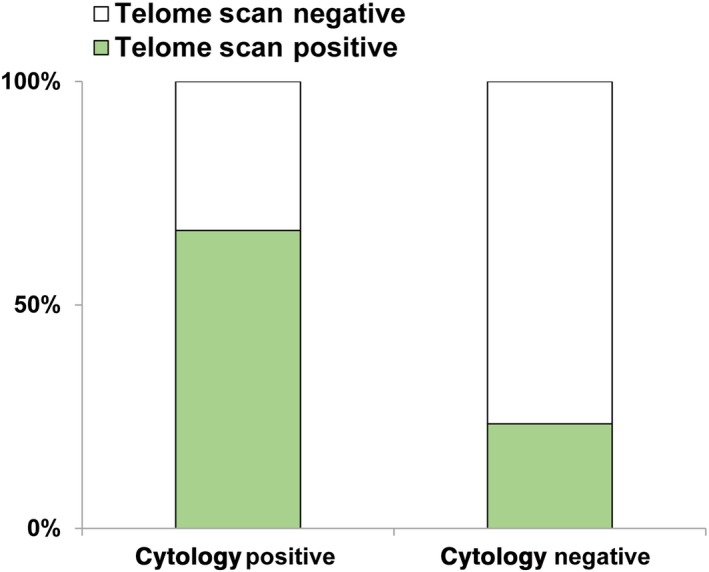
Correlation of TelomeScan‐guided cytology with conventional cytology, in clinical samples of peritoneal wash obtained from 68 gastric cancer patients. Cytology positive, n = 21; cytology negative, n = 47
3.5. TelomeScan‐guided peritoneal cytology stratifies gastric cancer prognosis
As expected, there were significant differences in overall survival stratified by conventional peritoneal wash cytology‐positive or ‐negative status (P = 0.0011; Figure S4A), and there were also significant differences in survival between TelomeScan‐positive and ‐negative patients (P = 0.0126; Figure S4B). Moreover, of the 21 patients who were positive on conventional cytology, the 14 patients who were also TelomeScan positive had significantly worse prognosis than the seven who were TelomeScan negative (P = 0.0062; Figure 4A). The median survival time of TelomeScan‐positive patients was 235 days, whereas that of TelomeScan‐negative patients was 671 days.
Figure 4.
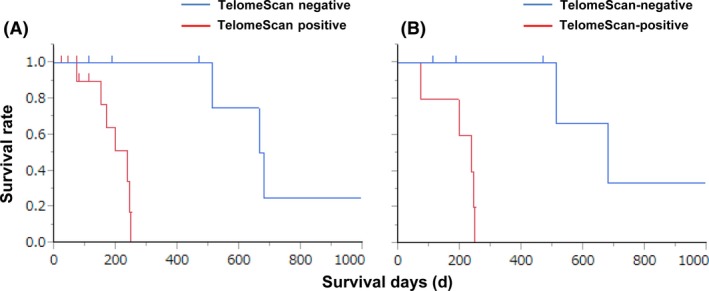
Kaplan‐Meier survival curves of overall survival in A, 21 conventional cytology‐positive gastric cancer patients and B, after exclusion of patients with macroscopic peritoneal metastasis, according to TelomeScan‐guided cytology stratification. There was a significantly poorer survival in TelomeScan‐positive than TelomeScan‐negative cases (A, P = 0.0062; B, P = 0.0086).
No significant differences in age, sex, histological type, or liver metastasis were observed between the TelomeScan‐positive and ‐negative groups, but the TelomeScan‐positive group included nine patients who already had peritoneal metastasis, while in the TelomeScan‐negative group there was only one patient with peritoneal metastasis (Table S2). Interestingly, when patients with peritoneal metastasis were excluded from the analysis, there was still a significant difference in prognosis between TelomeScan‐positive and ‐negative patients (Figure 4B), suggesting that TelomeScan‐based cytology was able to identify a subpopulation with particularly poor prognosis in conventional cytology‐positive patients, even in the absence of macroscopic peritoneal dissemination. Based on these findings, and the functional profiling of TelomeScan‐positive cells that indicated that GFP‐expressing cancer cells are viable and carry malignant markers (Figure 2A), we propose a model for the combined application of conventional and TelomeScan‐guided cytology to identify gastric cancer patients at risk of poor prognosis (Figure S5).
3.6. Fluorescence‐guided capture of potentially malignant cancer cells for genetic profiling
Finally, we used TelomeScan‐guided cytology to capture viable tumor cells in the peritoneal wash for genetic profiling. The cell pellets obtained from four gastric cancer patients were incubated with TelomeScan at an MOI of 1, labeled with anti‐CD45 antibody conjugated with PE, and sequentially sorted on multi‐laser FACS (Figure 5A). The extracted DNA was sufficient, although its concentration varied, for NGS of 29 human gastric cancer‐relevant genes using a targeted platform. The assay successfully detected 774 genetic variants, including single‐nucleotide polymorphisms (SNP), deletions, insertions, and point mutations, although most were variants of uncertain significance (Figure 5B). Although larger‐scale trials are required to confirm pathogenic variants related to poor prognosis, TelomeScan‐guided cytology and its capture system may also be a useful technology for the genetic analysis of disseminated tumor cells in the peritoneal cavity.
Figure 5.
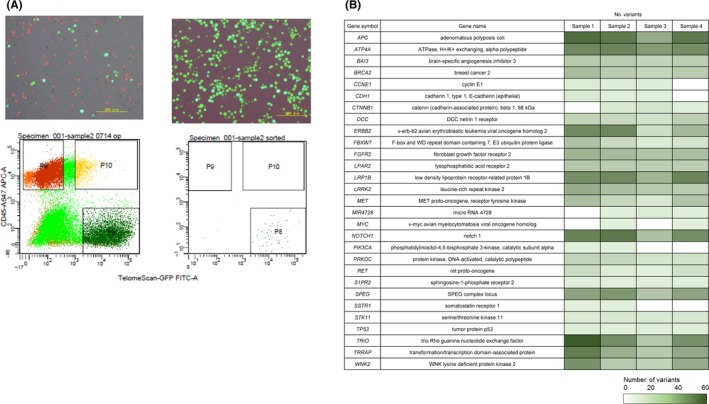
Simple TelomeScan‐guided capture system for potentially malignant disseminated tumor cells in the peritoneal wash. A, Following TelomeScan infection at 1 multiplicity of infection (MOI), cells were incubated with anti‐CD45 antibody, and the cell pellets sorted on fluorescence‐activated cell sorting (FACS). The gates were set to isolate green fluorescent protein(GFP)‐positive cells, but to exclude background fluorescence‐positive cells and CD45‐positive normal blood cells. Scale bars, 200 μm. B, Next‐generation sequencing analysis of the number of genetic variants identified in each of the 29 human gastric cancer‐relevant genes, in clinical samples from four gastric cancer patients positive for TelomeScan‐guided cytology
4. DISCUSSION
We have demonstrated that TelomeScan is successful in detecting and quantifying viable cancer cells in the peritoneal wash and can significantly improve the prediction of outcome for patients with gastric cancer. Two applications of this technology are suggested by the data. First, we propose a new stratification model to more accurately identify gastric cancer patients with poor prognosis, by combining TelomeScan‐guided fluorescent cytology with conventional cytology (Figure S5). Second, we propose that this TelomeScan‐guided, GFP‐based capture technology can used for genetic profiling of disseminated tumor cells in the peritoneal cavity.
To make this technology broadly available to health‐care providers, we wished to develop a simple and objective detection method, allowing the fluorescence intensity of TelomeScan‐positive cells to be quantified using a multi‐mode microplate reader. This means that quantification of TelomeScan‐positive cancer cells can be automated, and the assessment of positivity made using an appropriate cut‐off. Given that we have confirmed the utility of this test in only a limited number of samples, it will be necessary to confirm this in a larger number of clinical samples. Nevertheless, the method is potentially very valuable because it does not require laborious techniques, such as the extraction of nucleic acids or staining with expensive antibodies, and does not rely on professional competence for diagnosis.
Although conventional cytology of peritoneal wash samples is the most reliable procedure at present, investigators have assessed a number of methods to improve the diagnostic accuracy of cytology using various detection assays.10 Of these, PCR‐based detection systems have been the most extensively investigated.20, 21 The potential efficiency, high sensitivity, and objectiveness of this approach have been demonstrated by multiple investigators, but the lack of appropriate marker genes universally and specifically expressed in gastric cancer presents an important challenge for this approach.22, 23 In contrast, the present virus‐based system targets telomerase, which is known to be specifically and universally active in cancer cells, including gastric cancer cells.13, 14, 15, 16 In addition, the TelomeScan test requires viral infection and translation of the GFP protein, which can occur only in viable cells.16 Therefore, this virus can not only discriminate viable cancer cells from normal cells, but also from dead cancer cells. In cytology positive but TelomeScan‐negative cases, we found GFP (−) and CK (+) cells that we presume to be dead cancer cells. Thus, TelomeScan‐guided cytology provides additional biological information over conventional cytology. We hypothesize that these unique features of the TelomeScan approach would assist in identifying a subpopulation of patients with particularly poor prognosis among those who are cytology positive on conventional methods. Therefore, combining TelomeScan‐guided cytology with conventional cytology may have the potential to improve prognostic discrimination and patient stratification, to determine, for example, the appropriate application of intraperitoneal chemotherapy or more intensive combination chemotherapy, or avoiding non‐beneficial invasive surgery.24, 25, 26, 27
Immunocytochemistry involving the use of epithelial markers for the identification of cancer cells has been reported to improve the accuracy of cytology. Cancer cells, however, do not always express epithelial marker proteins such as EpCAM and E‐cadherin, particularly during metastasis, when cancer cells may undergo the process of EMT.28 In the present samples, immunofluorescence indicated that TelomeScan‐positive cells sometimes expressed CK, but others did not, suggesting that some TelomeScan‐positive cells had lost the expression of their epithelial marker and had undergone EMT. The TelomeScan detection system thus has the potential to identify tumor cells with both EMT and non‐EMT status, as previously reported.19
The current clinical classification for malignant potential in cancer patients is mostly based on the genetic analysis of tissues obtained from primary tumors. Given, however, that tumors are heterogeneous in regard to migration and/or invasion abilities,29 the genetic features of the primary tumors may not reflect the malignant potential of distant metastatic tumor sites. Therefore, to predict outcome precisely, it is important to obtain samples of disseminated cells or metastatic tissues.30 The increasing application of NGS makes it an important tool for both cancer diagnosis and therapeutic decision‐making31 the NGS platform, however, requires certain amounts of target DNA. By excluding the hematopoietic CD45‐positive cells, TelomeScan amplified the fluorescent signal and increased the tumor‐specific signal‐to‐background ratio.19 We detected a number of variants on 29 human gastric cancer‐relevant genes, demonstrating that sufficient amounts of DNA can be recovered from disseminated tumor cells using the TelomeScan‐guided capturing technology, although the correlation between these particular genetic changes and clinical outcome has yet to be assessed.
It is important to recognize some limitations of this technology. First, the TelomeScan procedure currently takes 24 hours for viral infection and protein expression, and it is therefore unsuitable for the rapid cytological diagnosis required, for example, in making decisions of surgical strategy in the operating room, where conventional cytology predominates over other diagnostics. This system would instead be useful in combination with conventional cytology when treating patients after surgery. After staging laparoscopy in particular, it is important to decide which type of anti‐cancer drug is suitable for each patient from the point of conversion chemotherapy.32 Second, the present sample size was relatively small, and patient background was not uniform. Although significant differences in prognosis were noted between TelomeScan‐positive and ‐negative cases in the positive conventional cytology cohort, the most important clinical benefit would be the sub‐stratification of those with negative conventional cytology, or to identify patients who might benefit from personalized therapy based on genetic profiling. Indeed, two cytology‐negative patients with T1 tumors had TelomeScan‐positive cells, but the origin of these cells has not been identified yet, even on multicolor immunohistochemistry. Moreover, in the cytology‐negative cohort, the survival curves were not significantly different between the TelomeScan‐positive and ‐negative population, presumably due to the small sample sizes (M. Watanabe, T. Fujiwara, et al., unpublished data). Therefore, a large‐scale multicenter prospective clinical study will soon be conducted to definitively establish the clinical significance of TelomeScan‐guided fluorescent cytology for gastric cancer patients.
In conclusion, our unique nano‐biologic, TelomeScan, successfully detected biologically active cancer cells in the peritoneal washes from gastric cancer patients, findings that were correlated with patient prognosis, demonstrating the potential utility of automated cytology diagnostics based on fluorescence intensity. We hope that this novel technology can add value to the information obtained from clinical peritoneal washes and can assist in improving the dismal prognosis of advanced gastric cancer in the near future.
CONFLICT OF INTEREST
Yasuo Urata is the president and CEO of Oncolys BioPharma, Inc., the manufacturer of TelomeScan (OBP‐401). Hiroshi Tazawa and Toshiyoshi Fujiwara are consultants for Oncolys BioPharma, Inc. The other authors declare no conflicts of interest.
Supporting information
ACKNOWLEDGMENTS
This study was supported by grants‐in‐aid from the Ministry of Education Culture, Sports, Science and Technology, Japan (to T. Fujiwara; No. 16673992), grants from the Ministry of Health, Labor and Welfare, Japan (to T. Fujiwara; No. 11949927, No. 13801458, No. 14525167), and grants from the Japan Agency for Medical Research and Development (to T. Fujiwara; No. 15652856). We thank Tomoko Sueishi and Yuko Hoshijima for their excellent technical support.
Watanabe M, Kagawa S, Kuwada K, et al. Integrated fluorescent cytology with nano‐biologics in peritoneally disseminated gastric cancer. Cancer Sci. 2018;109:3263–3271. 10.1111/cas.13760
REFERENCES
- 1. Sodek KL, Murphy KJ, Brown TJ, Ringuette MJ. Cell‐cell and cell‐matrix dynamics in intraperitoneal cancer metastasis. Cancer Metastasis Rev. 2012;31:397‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roviello F, Marrelli D, de Manzoni G, et al. Prospective study of peritoneal recurrence after curative surgery for gastric cancer. Br J Surg. 2003;90:1113‐1119. [DOI] [PubMed] [Google Scholar]
- 3. Leake PA, Cardoso R, Seevaratnam R, et al. A systematic review of the accuracy and utility of peritoneal cytology in patients with gastric cancer. Gastric Cancer. 2012;15(Suppl 1):S27‐S37. [DOI] [PubMed] [Google Scholar]
- 4. Bentrem D, Wilton A, Mazumdar M, Brennan M, Coit D. The value of peritoneal cytology as a preoperative predictor in patients with gastric carcinoma undergoing a curative resection. Ann Surg Oncol. 2005;12:347‐353. [DOI] [PubMed] [Google Scholar]
- 5. Bando E, Yonemura Y, Takeshita Y, et al. Intraoperative lavage for cytological examination in 1,297 patients with gastric carcinoma. Am J Surg. 1999;178:256‐262. [DOI] [PubMed] [Google Scholar]
- 6. Kodera Y, Yamamura Y, Shimizu Y, et al. Peritoneal washing cytology: prognostic value of positive findings in patients with gastric carcinoma undergoing a potentially curative resection. J Surg Oncol. 1999;72:60‐65. [DOI] [PubMed] [Google Scholar]
- 7. Sano T, Aiko T. New Japanese classifications and treatment guidelines for gastric cancer: revision concepts and major revised points. Gastric Cancer. 2011;14:97‐100. [DOI] [PubMed] [Google Scholar]
- 8. Isobe Y, Nashimoto A, Akazawa K, et al. Gastric cancer treatment in Japan: 2008 annual report of the JGCA nationwide registry. Gastric Cancer. 2011;14:301‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jamel S, Markar SR, Malietzis G, Acharya A, Athanasiou T, Hanna GB. Prognostic significance of peritoneal lavage cytology in staging gastric cancer: systematic review and meta‐analysis. Gastric Cancer. 2018;21:10‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kagawa S, Shigeyasu K, Ishida M, et al. Molecular diagnosis and therapy for occult peritoneal metastasis in gastric cancer patients. World J Gastroenterol. 2014;20:17796‐17803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki T, Ochiai T, Hayashi H, et al. Importance of positive peritoneal lavage cytology findings in the stage grouping of gastric cancer. Surg Today. 1999;29:111‐155. [DOI] [PubMed] [Google Scholar]
- 12. Wong J, Coit D. Detection of gastric cancer peritoneal metastases by peritoneal lavage: current limitations and future perspectives. Surgery. 2012;152:1‐4. [DOI] [PubMed] [Google Scholar]
- 13. Kishimoto H, Kojima T, Watanabe Y, et al. In vivo imaging of lymph node metastasis with telomerase‐specific replication‐selective adenovirus. Nat Med. 2006;12:1213‐1219. [DOI] [PubMed] [Google Scholar]
- 14. Kishimoto H, Zhao M, Hayashi K, et al. In vivo internal tumor illumination by telomerase‐dependent adenoviral GFP for precise surgical navigation. Proc Natl Acad Sci USA. 2009;106:14514‐14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kishimoto H, Urata Y, Tanaka N, Fujiwara T, Hoffman RM. Selective metastatic tumor labeling with green fluorescent protein and killing by systemic administration of telomerase‐dependent adenoviruses. Mol Cancer Ther. 2009;8:3001‐3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kojima T, Hashimoto Y, Watanabe Y, et al. A simple biological imaging system for detecting viable human circulating tumor cells. J Clin Invest. 2009;119:3172‐3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011‐2015. [DOI] [PubMed] [Google Scholar]
- 18. Nakayama J‐I, Tahara H, Tahara E, et al. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet. 1998;18:65‐68. [DOI] [PubMed] [Google Scholar]
- 19. Shigeyasu K, Tazawa H, Hashimoto Y, et al. Fluorescence virus‐guided capturing system of human colorectal circulating tumour cells for non‐invasive companion diagnostics. Gut. 2015;64:627‐635. [DOI] [PubMed] [Google Scholar]
- 20. Kodera Y, Nakanishi H, Ito S, et al. Quantitative detection of disseminated free cancer cells in peritoneal washes with real‐time reverse transcriptase‐polymerase chain reaction: a sensitive predictor of outcome for patients with gastric carcinoma. Ann Surg. 2002;235:499‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dalal KM, Woo Y, Kelly K, et al. Detection of micrometastases in peritoneal washings of gastric cancer patients by the reverse transcriptase polymerase chain reaction. Gastric Cancer. 2008;11:206‐213. [DOI] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong SS, Kim KM, Ting JC, et al. Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole‐genome sequencing. Nat Commun. 2014;5:5477. [DOI] [PubMed] [Google Scholar]
- 24. Ishigami H, Yamaguchi H, Yamashita H, Asakage M, Kitayama J. Surgery after intraperitoneal and systemic chemotherapy for gastric cancer with peritoneal metastasis or positive peritoneal cytology findings. Gastric Cancer. 2017;20:128‐134. [DOI] [PubMed] [Google Scholar]
- 25. Iizumi S, Takashima A, Narita Y, et al. Efficacy and safety of taxane monotherapy in advanced gastric cancer refractory to triplet chemotherapy with docetaxel, cisplatin, and S‐1: a multicenter retrospective study. Cancer Chemother Pharmacol. 2017;80(3):575‐582. [DOI] [PubMed] [Google Scholar]
- 26. Kitayama J, Ishigami H, Yamaguchi H, et al. Treatment of patients with peritoneal metastases from gastric cancer. Ann Gastroenterol Surg. 2018;2:116‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimada H. Intra‐abdominal chemotherapy for peritoneal metastases comes of age? Ann Gastroenterol Surg. 2018;2:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larue L, Bellacosa A. Epithelial‐mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24:7443‐7454. [DOI] [PubMed] [Google Scholar]
- 29. Padmanabhan N, Ushijima T, Tan P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat Rev Gastroenterol Hepatol. 2017;14:467‐478. [DOI] [PubMed] [Google Scholar]
- 30. Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy‐number profiles in patients with chemosensitive and chemorefractory small‐cell lung cancer. Nat Med. 2017;23:114‐119. [DOI] [PubMed] [Google Scholar]
- 31. Ulahannan D, Kovac MB, Mulholland PJ, Cazier JB, Tomlinson I. Technical and implementation issues in using next‐generation sequencing of cancers in clinical practice. Br J Cancer. 2013;109:827‐835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Andrade JP, Mezhir JJ. The critical role of peritoneal cytology in the staging of gastric cancer: an evidence‐based review. J Surg Oncol. 2014;110:291‐297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


