Abstract
Long noncoding RNAs (lncRNAs) play an important role in the proliferation and metastasis of osteosarcoma. Identification of the pathogenesis of osteosarcoma and development of new therapeutic strategies against osteosarcoma are urgently needed. In this study, we evaluated the expression of TUG1 (Taurine Upregulated Gene 1) in osteosarcoma tissues and selected it as our target for further analyses. In vitro, we found that TUG1 was upregulated by FOXM1 (Forkhead Box M1) in osteosarcoma cells. TUG1 accelerated osteosarcoma proliferation, migration, and invasion by competitively sponging miR‐219a‐5p, leading to upregulation of Phosphatidylinositol‐4, 5‐Bisphosphate 3‐Kinase Catalytic Subunit Alpha and activation of the protein kinase B (AKT) signaling pathway. In addition, the AKT pathway activation promoted TUG1 expression by upregulating the expression of FOXM1, forming a positive feedback loop in osteosarcoma. Furthermore, we designed and synthesized therapeutic locked nucleic acids targeting TUG1. The proliferation of osteosarcoma was significantly repressed. Hence, TUG1 may be a potential biomarker and therapeutic target for osteosarcoma.
Keywords: FOXM1, miR‐219a‐5p, osteosarcoma, PIK3CA, TUG1
1. INTRODUCTION
Osteosarcoma is the most common malignant bone tumor that affects children, young adults and adolescents, and the median age is 20 years in all cases.1 The primary sites of osteosarcoma are the distal femur (43%), proximal tibia (23%) and humerus (10%), and more than 85% of metastatic lesions migrate to the lungs.2 After the application of chemotherapy in the early 1980s, the long‐term survival rate with this disease improved from less than 20% to 65%.3 Since then, no significant progress has been made in improving osteosarcoma diagnosis and treatment.4 In studies exploring treatments for osteosarcoma, signaling pathways related to tumor formation, proliferation and metastasis have received increased attention,5, 6, 7, 8 but the outlook remains unclear. Therefore, it is imperative to clarify the intrinsic mechanism of osteosarcoma and identify potential diagnostic and prognostic biomarkers of osteosarcoma.
An extensive array of functional mutations with profound effects on the expression of long noncoding RNAs (lncRNAs) was identified by genome mutation analysis of cancer within the noncoding genome.9, 10 As a multifarious class of transcripts, lncRNAs have a length >200 nucleotides and a limited protein‐coding ability11, 12 and display a unique expression pattern in different tissues, organs or specific tumor types.13 lncRNAs participate in multiple cell phenomena, such as regulation of gene expression, including recruiting transcription‐modifying complexes to influence specific gene expression14, 15, 16 and interacting with microRNAs (miRNAs), mRNAs or proteins for post‐transcriptional regulation.17, 18, 19 Increasing research has shown that lncRNAs serve as signals of cell behavior, identify the intrinsic pathogenesis of cancer, and have prognostic or therapeutic value.20 However, the basic function of lncRNAs in osteosarcoma is still unclear. In this study, we explored the roles of lncRNAs in osteosarcoma and investigated the diagnostic and treatment implications for osteosarcoma.
Taurine upregulated gene 1 (TUG1), a 7542 bp highly conserved lncRNA, was originally identified in taurine‐induced retinal cells21 and are expressed in many human tumors, such as glioblastoma, prostate cancer, and hepatocellular carcinoma.22, 23, 24 TUG1 is distributed in the cytoplasm and nucleus, and its function begins with its cellular localization. In the nucleus, TUG1 can interact with Polycomb Repressive Complex 2 (PRC2) and suppress cell‐cycle‐related gene expression.25 However, TUG1's cytoplasmic function and its influence on osteosarcoma are not clear. In this study, we found that TUG1, which is upregulated by the protein kinase B / Forkhead Box M1 (AKT/FOXM1) signaling pathway, has high expression in osteosarcoma tissues and cell lines and maintains tumor proliferation and invasion as a competing endogenous RNA (ceRNA) to upregulate Phosphatidylinositol‐4, 5‐Bisphosphate 3‐Kinase Catalytic Subunit Alpha (PIK3CA) through competition with miR‐219a‐5p. Furthermore, we designed the locked nucleic acids (LNAs) targeting TUG1 and demonstrated that LNAs can effectively inhibit the proliferation of osteosarcoma in vivo.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
Frozen osteosarcoma tissues and paratumor samples were obtained with informed consent from patients who underwent thorough resections at the First Affiliated Hospital of Harbin Medical University, Heilongjiang, China. All samples were confirmed by histopathological evaluation, and stored at −80°C before total RNA was extracted. This study was approved by the Committees for Ethical Review of Research Involving Human Subjects of Harbin Medical University.
2.2. Cell lines and culture conditions
The human osteosarcoma cell lines U2OS, SaOS2, MNNG/HOS (abbreviated HOS), and MG63 and the human colon carcinoma cell line HCT116 were obtained from the Cell Resource Center, Peking Union Medical College. The human normal osteoblast cell line hFOB1.19 was obtained from the Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human colon carcinoma cell line HCT116 dicerex5 was a kind gift of Dr. Yu from Harbin Medical University. The HCT116 dicerex5 cell line has a mutant DICER with an insertion disruption in the N‐terminal helicase domain (exon 5). This hypomorphic mutation in DICER impairs its function in the maturation of the vast majority of miRNAs.23 All cell lines were grown in penicillin/streptomycin at 37°C in a humidified atmosphere with 5% CO2.
2.3. RNA extraction and quantitative real‐time PCR
The cytoplasmic and nuclear fractions were separated using the PARIS protocol (Life Technologies, Carlsbad, CA, USA). RNA was extracted from tissues, cell lines and subcellular fractions using RNAiso Plus reagent (TaKaRa, Shiga, Japan), and the concentration was then tested by NanoDrop 2000c (Thermo Scientific, Waltham, MA, USA). For reverse transcription, 1 μg of total RNA was used to synthesize complementary DNA (cDNA) with an RT reagent Kit with gDNA Eraser (#RR047A, TaKaRa). For measurement of the levels of target genes, quantitative real‐time PCR was performed in a reaction mix of SYBR Green (Roche, Basel, Switzerland) with at least three replicates with the ABI 7500 Sequence Detection System (Life Technologies), and GAPDH was used for normalization. For miRNA expression, quantitative real‐time PCR was performed using the TaqMan MiRNA Reverse Transcription Kit and Universal Master Mix (Applied Biosystems, Life Technologies). All the results were calculated by the 2−ΔΔCT method. The sequences of the primers and the TaqMan Assays IDs of the miRNAs are listed in Table S1.
2.4. Plasmid construction, cell transfection, and RNA interference
The cDNAs encoding FOXM1 and PIK3CA were PCR‐amplified by a PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Japan) and subcloned into the EcoRI/XbaI and KpnI/XhoI sites of the pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA), yielding pcDNA3.1‐FOXM1 and pcDNA3.1‐PIK3CA. The DNA binding domain‐truncated mutant of the FOXM1 sequence was synthesized by Sangon (Shanghai, China) and subcloned into a pcDNA3.1 vector to obtain pcDNA3.1‐FOXM1‐mut. The wild‐type TUG1 promoter region was PCR‐amplified and subcloned into the KpnI/XhoI sites of the pGL3‐basic vector (Promega, Madison, WI, USA), yielding pGL3‐TUG1. The pGL3‐TUG1 constructs with point mutations in FOXM1 binding sites were synthesized by Sangon (Shanghai, China) and named pGL3‐TUG1‐mutP1, pGL3‐TUG1‐mutP2, and pGL3‐TUG1‐mutR1. The 3′‐UTR of the PIK3CA fragment containing the miR‐219a‐5p binding site was amplified and subcloned into the XhoI/NotI sites of the psiCHEK2‐vector (Promega) and named psiCHECK2‐PIK3CA. The binding sites of miR‐219a‐5p in the 3′‐UTR of the PIK3CA fragment were mutated and synthesized by Sangon (Shanghai, China) to obtain psiCHECK2‐PIK3CA‐mut. The same methods were used to obtain psiCHECK2‐TUG1 and psiCHECK2‐TUG1‐mut reporter plasmids. All constructs were confirmed by DNA sequencing. Transfection of the plasmids was achieved using Lipofectamine 2000 Transfection Reagent (Life Technologies).
The lentiviral vector expressing shRNA targeting TUG1 was designed and constructed by Genpharma (Shanghai, China). siRNAs, miRNA mimics, and miRNA inhibitors were designed and synthesized by Sangon (Shanghai, China). The sequences of siRNAs, LNAs and shRNAs are listed in Table S1. Transfection of siRNAs, miRNA mimics or inhibitors was performed using Lipofectamine RNAiMAX (Invitrogen) at a final concentration of 100 nmol/L.
2.5. Dual‐luciferase reporter assay
In summary, 1 × 105 cells were seeded per well in 24‐well plates (Corning, Corning, NY, USA) 24 hours before transfection. Cells were incubated for 48 hours before lysis. Then, the luciferase activity was evaluated by the Dual‐luciferase Reporter Assay System (Promega). The firefly luciferase activity of the pGL3‐vector was normalized against pRL‐TK Renilla luciferase activity, and the Renilla luciferase activity of psiCHECK2‐vector was standardized against firefly luciferase activity. All experiments were performed in triplicate and repeated at least three times.
2.6. Cell proliferation assay and colony‐forming assay
For Cell Counting Kit‐8 (CCK8, Beyotime, Shanghai, China) assay, the cells were seeded in 96‐well plates at 5 × 103 cells/well for 24 hours. Then, the medium of each well was replaced with 100 μL fresh culture medium with 10% CCK8 at different times. The cells were incubated for 2 hours, and then, absorbance was measured at 450 nm by a microplate reader. For colony‐forming assay, 6 replicate wells were used for each sample, and the experiments were repeated independently three times. Cells were resuspended in medium with 10% FBS and seeded into 6‐well plates at a density of 1 × 103 cells per well. Colonies were counted by ImageJ software (National Institutes of Health, Bethesda, MD, USA) after 3 weeks.
2.7. Wound healing assay
After transfection for 24 hours, cells were seeded into 6‐well plates (Corning). An artificial wound was made using a 200 μL pipette tip across the cell monolayer. For measurement of wound healing, images were taken at 0, 24, and 48 hours, and the percentage of wound healing was calculated using ImageJ software. Each experiment was carried out in triplicate independent experiments.
2.8. Transwell migration assay
The cells were resuspended in serum‐free culture medium at a density of 2.5 × 105 cells/mL. Then, 200 μL cells were seeded onto the upper Matrigel Invasion Chambers (size 8 μm; BD Biosciences, Franklin Lakes, NJ, USA), and the chambers were moved to 24‐well plates (Corning) containing 600 μL of medium with 10% FBS. After 12 hours of incubation, the cells that had invaded the lower chambers were fixed with 4% paraformaldehyde and stained with crystal violet (Sangon, China). Stained cells were counted using an inverted microscope (Olympus, Tokyo, Japan), and the cell numbers were averaged with five randomly chosen fields.
2.9. Western blot assay
For the western blot assay, cells were harvested and lysed in radio‐immunoprecipitation Assay (RIPA, Beyotime) with protease and phosphatase inhibitors. Each sample was separated in 12% SDS‐PAGE and then transferred to PVDF membranes (Sigma, St Louis, MO, USA). The membranes were blocked in Tris‐buffered saline with 0.05% Tween‐20 milk for 2 hours and immunoblotted with specific primary and secondary antibodies. Sample blots were visualized with a chemiluminescence detection reagent (Amersham, Piscataway, NJ, USA) and analyzed using ImageJ software; The primary antibodies used in this study are listed in Table S2.
2.10. Nude mice xenograft and treatment experiments
For subcutaneous xenograft research, 4‐week‐old female nu/nu nude mice (Vital River Laboratory Animal Technology Co., Beijing, China) were fed in standard pathogen‐free conditions. Sodium pentobarbital anesthesia was performed in all surgeries to minimize suffering. HOS cells (1 × 106) were injected subcutaneously into the right back area of mice in the LNA treatment groups (n = 12), or both sides of the back in the shTUG1 group (n = 6). Tumor sizes were measured by Vernier calipers and calculated with the following formula: Volume = (a × b 2)/2 (a represents the length and b represents the width). When the size of the tumor reached 0.01 cm3, the 6 mice in the LNA treatment groups were randomly injected with TUG1 LNA or control LNA (50 nmol, 3 times a week) for 6 weeks. The LNAs targeting TUG1 or scrambled LNAs were designed and synthesized by Sangon (China). The procedures for care and use of animals were granted by the Ethics Committee of Harbin Medical University and all applicable institutional and governmental regulations concerning the ethical use of animals were followed.
2.11. Statistical analysis
SPSS 18.0 software (SPSS inc., Chicago, IL, USA) was used for all statistical analyses in our study. Data are presented as the mean ± SD and the experiments were repeated at least three times or more. Student's t test (two‐tailed) or one‐way ANOVA was used for statistical comparisons between experimental groups. A P < .05 was considered statistically significant.
3. RESULTS
3.1. TUG1 is highly expressed in osteosarcoma tissues and cell lines
We extracted RNAs from 19 osteosarcoma samples with adjacent tissues from patients with no therapy history and analyzed them by quantitative real‐time PCR. As shown in Figure 1A, TUG1 levels were higher in osteosarcoma tissues than adjacent paratumor tissues. Consistent with the upregulation of TUG1 in osteosarcoma tissues, TUG1 levels were substantially higher in osteosarcoma cell lines than osteoblast cell lines (Figure 1B). According to the standards set by the National Comprehensive Cancer Network (NCCN) guidelines, high levels of TUG1 were observed in high‐grade and metastatic patients (Figure 1C,D). Moreover, according to the overall survival data obtained from the GEPIA database (http://gepia.cancer-pku.cn/), high TUG1 levels in sarcoma patients were correlated with reduced survival percentages (Figure S1).
Figure 1.

Expression of TUG1 (Taurine Upregulated Gene 1) in human osteosarcoma tissues and cell lines. (A) TUG1 levels in human osteosarcoma tissues and paired adjacent paratumor tissues (n = 19). (B) TUG1 expression levels in human osteosarcoma cell lines (MG63, HOS, SaOS2, U2OS) compared with the human osteoblast cell line (hFOB1.19). (C) TUG1 expression levels in different grades of osteosarcoma tissues (n = 19). (D) Relative expression of TUG1 was examined in metastatic (n = 11) and non‐metastatic (n = 8) osteosarcoma patients. For A, B, C and D, error bars indicate ± SD. *P < .05, **P < .001, ***P < .0001
3.2. TUG1 is upregulated by the AKT/FOXM1 axis in osteosarcoma
We examined the intrinsic mechanism for high TUG1 expression in the osteosarcoma cell line. DNA methyltransferase inhibition had little effect on TUG1 expression in osteosarcoma cells (Figure 2A). Analysis of the promoter region (−2000 to 200 bp) of TUG1 using the bioinformatics web tool GTRD (http://gtrd16-07.biouml.org/) predicted 2 DNA binding elements (DBEs), named P1 and P2, for FOXM126 (Figure 2B). Transfection of pcDNA3.1‐FOXM1 significantly upregulated TUG1 levels in osteosarcoma cell lines, while pcDNA3.1‐FOXM1‐mut had no influence on TUG1 expression. Furthermore, FOXM1 siRNA downregulated TUG1 levels (Figure 2C, D). To verify the relationship between FOXM1 and TUG1, we performed dual luciferase reporter assays using co‐transfection of pGL3‐TUG1, pRL‐TK and an increasing number of pcDNA3.1‐FOXM1 plasmids in U2OS cells. As shown in Figure 2E, FOXM1 over‐expression increased the activity of the TUG1 promoter in a dose‐dependent manner. To confirm which putative site influenced the transactivation ability of FOXM1, we individually mutated the two putative sites and one random site of the TUG1 promoter in pGL3‐TUG1 (Figure 2F). The mutation of P2 significantly decreased the transactivation of the TUG1 promoter by FOXM1. AKT was reported to promote FOXM1 activation by inducing the phosphorylation of FOXO3 for protein degradation.27, 28 Knockdown of AKT in osteosarcoma cells restrained the expression of TUG1 (Figure 2G). According to previous reports, FOXM1 is highly expressed in osteosarcoma,29, 30 and these data showed that the enhancement of FOXM1 by AKT in osteosarcoma cells, at least, activates TUG1 transcription by direct binding to the TUG1 promoter.
Figure 2.
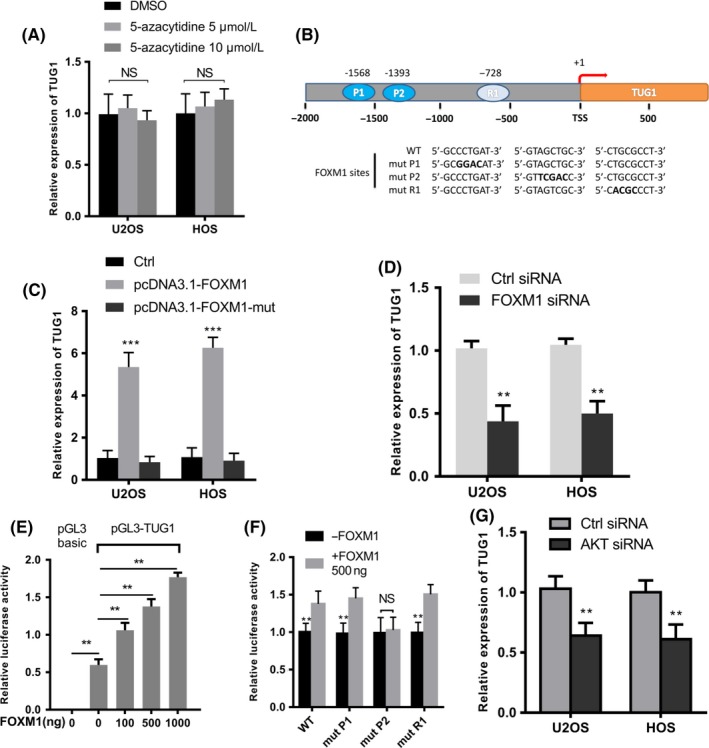
Identification of TUG1 (Taurine Upregulated Gene 1) in an protein kinase B / Forkhead Box M1 (AKT/FOXM1) axis regulated in osteosarcoma cells. (A) Quantitative real‐time PCR analysis of TUG1 in U2OS and HOS treated with DMSO or 5‐azacytidine (5 μmol/L or 10 μmol/L) for 48 h (n = 3). (B) A schematic illustration of the TUG1 promoter region. The wild‐type and mutant sequences of two predicted binding sites, P1 (‐1568) and P2 (‐1393), and one random site, R1 (‐728), are underlined. (C) Quantitative real‐time PCR analysis of TUG1 in U2OS and HOS cells transfected with 500 ng indicated plasmids after 48 h (n = 3). (D) Quantitative real‐time PCR analysis of TUG1 in U2OS and HOS cells after transfection with control or FOXM1 siRNA (n = 3). (E) A combination of 500 ng pGL3‐TUG1 (or pGL3‐Basic as a negative control), 50 ng pRL‐TK and an increasing number of pcDNA3.1‐FOXM1 plasmids were co‐transfected into U2OS cells. Luciferase activity was tested after 48 h (n = 3). pGL3‐basic was used as a negative control. (F) A combination of 500 ng pGL3‐TUG1 promoter carrying either wild‐type sequence or mutations in two putative FOXM1 binding sites and one random site, 50 ng pRL‐TK and 500 ng pcDNA3.1‐FOXM1 were co‐transfected in U2OS cells. Luciferase activity was tested after 48 h (n = 3). (G) The expression levels of TUG1 in U2OS and HOS cells transfected with control or si‐AKT for 48 h (n = 3). Error bars indicate the mean ± SD. **P < .001, ***P < .0001
3.3. TUG1 is necessary for the proliferation and metastasis of osteosarcoma cells
To evaluate the function of TUG1, we designed three different lentiviral shRNA sequences (shRNA #1, #2, #3) to stably knock down TUG1 in osteosarcoma cells, and chose the sh‐TUG1 #2 for further experiments (Figure 3A). Because of the large size of TUG1 (7542 bp), we could not overexpress it. The CCK8 assay showed that the proliferation of osteosarcoma cells was impaired when TUG1 was knocked down (Figure 3B). Colony formation assays similarly concluded that the number of colonies was decreased in TUG1 knockdown cells (Figure 3C). Then, we evaluated the influence of TUG1 knockdown on osteosarcoma cells by wound healing and transwell assays. As shown in Figure 3D and E, knocking down TUG1 significantly repressed the migration of osteosarcoma cells. The number of invading cells was also decreased in TUG1 knockdown cells (Figure 3E). These results indicated that TUG1 was indispensable for the proliferation and metastasis of osteosarcoma cells.
Figure 3.
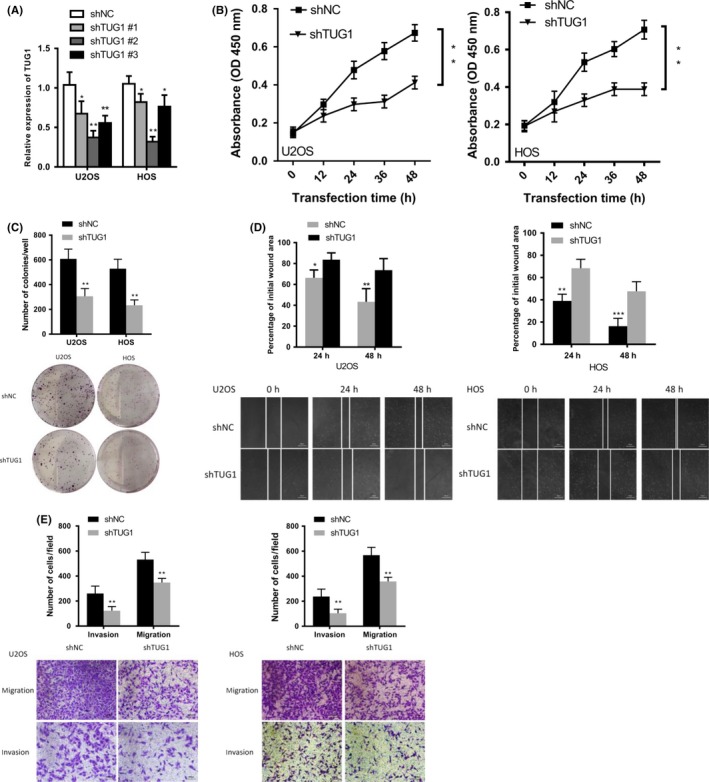
TUG1 (Taurine Upregulated Gene 1) silencing inhibited cell proliferation, colony formation, migration, and invasion. (A) Expression levels of TUG1 in U2OS and HOS cells treated with either control or shTUG1 (shTUG1 #1, #2, and #3) (n = 3). (B) The proliferation of U2OS and HOS cells was determined by Cell Counting Kit‐8 (CCK‐8) assays after transfection with either control or shTUG1 after 48 h. (C) Colony‐forming assays evaluated the proliferation of TUG1‐knockdown and control U2OS/HOS cells in 6‐well dishes (1 × 103 cells per well) for 3 wk (n = 3). (D) Wound healing assays assessed the migration of U2OS and HOS cells transfected with control or shTUG1. (E) Transwell assays assessed the migration and invasion of U2OS and HOS cells transfected with control or shTUG1. Error bars indicate the mean ± SD. *P < .05, **P < .001, ***P < .0001
3.4. PIK3CA contributes to TUG1‐mediated osteosarcoma cell tumorigenesis and metastasis
Increasing evidence has shown that activation of the PI3K/AKT pathway, MAPK/ERK pathway, and JAK/STAT (signal transducers and activators of transcription) pathway has key roles in osteosarcoma tumorigenesis and metastasis.31, 32, 33 To elucidate the possible mediators through which TUG1 drives osteosarcoma cell tumorigenesis and metastasis, we performed a western blot assay to analyze the core proteins of each pathway in TUG1 knockdown osteosarcoma cells. As shown in Figure 4A, TUG1 knockdown only decreased AKT at the phosphorylation level but not at the total protein level, consistent with reports that AKT was overexpressed in osteosarcoma cells and tissues.31 We analyzed the proteins that influence AKT phosphorylation levels in osteosarcoma cells, and only PIK3CA protein levels, as well as mRNA levels, were dramatically decreased in TUG1 knockdown osteosarcoma cells (Figure 4B, C). We further investigated whether PIK3CA contributed to TUG1‐mediated osteosarcoma cell tumorigenesis and metastasis. Overexpression of PIK3CA restored proliferation and metastasis in TUG1 knockdown osteosarcoma cells (Figure 4D‐F). According to the correlation data obtained from the GEPIA database (http://gepia.cancer-pku.cn/), there was a positive correlation between TUG1 and PIK3CA expression (Figure S2). Taken together, these data indicate that PIK3CA contributes to TUG1‐mediated osteosarcoma cell tumorigenesis and metastasis.
Figure 4.

Phosphatidylinositol‐4, 5‐Bisphosphate 3‐Kinase Catalytic Subunit Alpha (PIK3CA) contributes to TUG1 (Taurine Upregulated Gene 1)‐mediated osteosarcoma cell tumorigenesis and metastasis. (A and B) Western blot analysis of the indicated proteins in TUG1‐knockdown and control U2OS cells for 72 h. (C) Quantitative real‐time PCR analysis of the indicated RNA expression in TUG1‐knockdown and control U2OS cells (n = 3). (D) Cell Counting Kit‐8 (CCK‐8) assays evaluated the proliferation of TUG1‐knockdown U2OS cells transfected with pcDNA3.1‐PIK3CA after 48 h (n = 3). (E) Colony‐forming assays evaluated the proliferation of TUG1‐knockdown U2OS cells transfected with pcDNA3.1‐PIK3CA in 6‐well dishes (1 × 103 cells per well) for 3 wk (n = 3). (F) Transwell assays assessed the migration and invasion of U2OS co‐transfected with shTUG1 and pcDNA3.1‐PIK3CA. The results are presented as the mean ± SD. **P < .01
3.5. TUG1 functions as a sponge for miR‐219a‐5p to upregulate PIK3CA levels in osteosarcoma cells
The function of lncRNAs is closely related to their subcellular location.34 It has been reported that TUG1 is distributed within the nucleus and cytoplasm, but the ratio was higher in the cytoplasm.34 We applied nuclear and cytoplasmic experiments followed by quantitative real‐time PCR to test the subcellular localization of TUG1 in osteosarcoma cells. Certainly, TUG1 was mostly localized within the cytoplasm in U2OS and HOS cell lines (Figure 5A). As for the location of TUG1, it might function as a competitive endogenous RNA to sponge miRNA, quenching miRNA ability and liberating the miRNA target. To test this premise, we transfected HCT116 and HCT116 dicerex5 cells with shTUG1. HCT116 cell line of colon cancer has a wild‐type DICER, and HCT116 dicerex5 has a mutant DICER that impairs its function in the maturation of miRNAs. As shown in Figure 5B and C, downregulation of PIK3CA mediated by TUG1 knockdown was abrogated in HCT116 dicer‐ex5. These results suggest that TUG1‐mediated PIK3CA downregulation is mostly dependent on miRNA activity.
Figure 5.

TUG1 (Taurine Upregulated Gene 1) functions as a sponge for miR‐219a‐5p to up‐regulate Phosphatidylinositol‐4, 5‐Bisphosphate 3‐Kinase Catalytic Subunit Alpha (PIK3CA) levels. (A) Quantitative real‐time PCR analysis of the expression levels of TUG1 in the nuclear and cytoplasmic fractions of HOS and U2OS cells. (B) Quantitative real‐time PCR analysis of PIK3CA RNA levels in HCT116 and HCT116 dicerex5 cells transfected with control or shTUG1 (n = 3). (C) Western blot for PIK3CA protein levels in HCT116 and HCT116 dicerex5 cells transfected with control or shTUG1. (D) Relative miRNA levels in U2OS cells treated with shTUG1 or control (n = 3). (E) Putative miR‐219a‐5p miRNA response elements (MREs) in TUG1 and the 3′UTR of PIK3CA. (F) Luciferase activity of psiCHECK2‐TUG1 and psiCHECK2‐PIK3CA reporters containing wild‐type or mutant miR‐219a‐5p MREs transfected with miR‐219a‐5p mimics or control in U2OS cells (n = 3). (G) Luciferase activity of psiCHECK2‐PIK3CA and psiCHECK2‐PIK3CA‐mut with indicated treatment in U2OS cells (n = 3). (H) Quantitative real‐time PCR analysis of PIK3CA in U2OS cells transfected with control or TUG1 siRNA with miR‐219a‐5p inhibitor. (I) Western blot analysis of PIK3CA expression in U2OS cells co‐transfected with TUG1 siRNA and miR‐219a‐5p inhibitor (n = 3). (J) Cell Counting Kit‐8 (CCK‐8) assay of U2OS cells co‐transfected with TUG1 siRNA and miR‐219a‐5p inhibitor for 48 h (n = 3). (K) Colony‐forming assay evaluated the proliferation of TUG1‐knockdown and control U2OS cells transfected with miR‐219a‐5p inhibitor in 6‐well dishes (1 × 103 cells per well) for 3 wk (n = 3). (L) Transwell assays of U2OS cells co‐transfected with shTUG1 and miR‐219a‐5p inhibitor. Error bars indicate the mean ± SD. *P < .05, **P < .001, ***P < .0001, NS, not significant
A bioinformatics analysis of Starbase (http://starbase.sysu.edu.cn/) and miRanda (http://www.microrna.org/microrna/home.do) revealed 9 putative miRNAs shared by TUG1 and PIK3CA. We evaluated these miRNA levels in TUG1 knockdown osteosarcoma cells using quantitative real‐time PCR. Among the upregulated miRNAs, miR‐219a‐5p was the most significantly upregulated following inhibition of TUG1 (Figure 5D). We mutated the binding sites on TUG1 and the 3′‐UTR of PIK3CA (Figure 5E) and constructed dual luciferase reporters. Co‐transfection of miR‐219a‐5p mimics downregulated the luciferase activity of psiCHECK2‐TUG1/PIK3CA. Conversely, psiCHECK2‐TUG1mt/PIK3CAmt reporter activity was not obviously decreased (Figure 5F). These results demonstrated that TUG1 and the 3′‐UTR of PIK3CA contained binding sites for miR‐219a‐5p.
Then we explored whether TUG1‐mediated downregulation of miR‐219a‐5p was the reason for the upregulation of PIK3CA. The luciferase activity of PIK3CA was decreased with TUG1 knockdown and was rescued by miR‐219a‐5p inhibitors, but the luciferase activity of the mutant reporter was not obviously changed (Figure 5G). These data were not only confirmed by the RNA and protein levels of PIK3CA (Figure 5H, I) but were also confirmed by CCK8, colony formation and transwell assays (Figure 5J‐L). All these data showed that TUG1 functions as a sponge for miR‐219a‐5p to upregulate PIK3CA levels in osteosarcoma cells.
3.6. Targeting TUG1 promotes tumorigenesis of osteosarcoma in vivo
To assess the potential of TUG1 in the treatment of osteosarcoma in vivo, we synthesized and selected an optimized LNA targeting TUG1 for intravenous injection. We generated a subcutaneous xenograft model of HOS cells in nu/nu mice and injected LNA‐TUG1 for 6 weeks. As shown in Figure 6A and B, there were significant changes in the size and weight of tumors between the LNA treatment group and the normal group. Knockdown of TUG1 by LNA was verified by quantitative real‐time PCR assays, along with inhibition of PIK3CA at the mRNA and protein levels (Figure 6C, D). In addition, we established a model of TUG1‐knockdown HOS cells in nu/nu mice and reached a similar conclusion (Figure 6A‐D). Collectively, the data showed that TUG1 could promote tumorigenesis of osteosarcoma in vivo and that LNA targeting TUG1 had a therapeutic potential for osteosarcoma.
Figure 6.
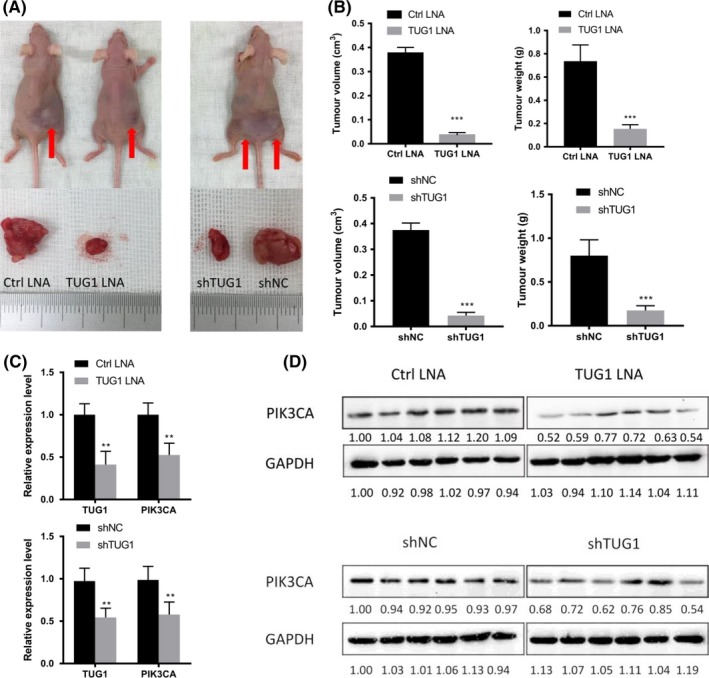
Targeting TUG1 (Taurine Upregulated Gene 1) promotes tumorigenesis of osteosarcoma in vivo. Silencing of TUG1 by locked nucleic acid (LNA) or shRNA inhibited HOS xenograft proliferation in nu/nu mice (n = 6 per group). (A) The two left groups are the LNA group, and the one right is the shRNA group. Arrows indicate tumors in situ. (B) Average volume and weight of the subcutaneous xenografts. (C) Quantitative real‐time PCR analysis of the indicated transcripts in HOS xenografts after the indicated treatment. (D) Western blot analysis of Phosphatidylinositol‐4, 5‐Bisphosphate 3‐Kinase Catalytic Subunit Alpha (PIK3CA) in the different treatment groups. All results are presented as the mean ± SD. **P < .001, ***P < .0001, NS, not significant
4. DISCUSSION
The disease‐free survival (DFS) rate is obviously poorer in patients with metastatic disease than those with nonmetastatic disease.35 In this study, we observed high levels of TUG1 in osteosarcoma tissues and cell lines. TUG1 accelerated osteosarcoma proliferation, migration, and invasion by competitively sponging miR‐219a‐5p, leading to the upregulation of PIK3CA and the activation of the PI3K/AKT signaling pathway. In addition, the activation of the PI3K/AKT pathway promoted TUG1 expression by upregulating the expression of FOXM1, forming a positive feedback loop in osteosarcoma (Figure 7). Hence, TUG1 was shown to play an important role in osteosarcoma. Clarifying the molecular basis of osteosarcoma could contribute to the development of rationally designed combination therapies to control the related signaling pathways. Our results showed that knockdown of TUG1 by LNA inhibited the proliferation of osteosarcoma, indicating that lncRNA‐targeted therapy could be implemented as a potential therapeutic strategy for osteosarcoma.
Figure 7.

A schematic diagram of a TUG1 (Taurine Upregulated Gene 1)‐based signaling circuit in osteosarcoma. TUG1 accelerated osteosarcoma proliferation, migration, and invasion by competitively sponging miR‐219a‐5p, leading to the up‐regulation of Phosphatidylinositol‐4, 5‐Bisphosphate 3‐Kinase Catalytic Subunit Alpha (PIK3CA) and the activation of the protein kinase B (AKT) signaling pathway. In addition, the activation of the AKT pathway promoted TUG1 expression by up‐regulating the expression of Forkhead Box M1 (FOXM1), forming a positive feedback loop in osteosarcoma
TUG1 was shown to be overexpressed in osteosarcoma tissues and cell lines.36 We verified this conclusion according to the stage of the osteosarcoma and the presence or absence of metastasis and reached a similar conclusion. Through the in vitro study, we evaluated the influence of TUG1‐knockdown on osteosarcoma cells and confirmed the conclusion of clinical sample statistics. According to previous reports, TUG1 expression is induced by p53 in non‐small cell lung cancer cells,36 Sp1 in hepatocellular carcinoma,24 and RBPJ (recombination signal binding protein for immunoglobulin kappa J region) and MYC in glioma.22 We overexpressed these transcription factors in osteosarcoma cell lines, and none of these could significantly promote the expression of TUG1. Through bioinformatics prediction of the TUG1 promoter region, we confirmed that FOXM1 induced high expression of TUG1 in osteosarcoma cell lines. FOXM1 is overexpressed in many different tumors, including osteosarcoma, and contributes to cell proliferation and metastasis.37, 38 It has been reported that the PI3K/AKT pathway can stimulate the phosphorylation of FOXO3 at Ser294, Ser253, and Thr32 and then eliminate the negative regulation of FOXM1.28 We assumed that FOX proteins may influence the expression of TUG1 and showed that FOXM1 could bind to the promoter region of TUG1 by luciferase reporter assays. These data indicated that the expression and activity of TUG1 are controlled by different transcription factors in various tissues.
We evaluated changes in proliferation and metastasis‐related signaling pathways in TUG1 knockdown osteosarcoma cell lines and found that only the PI3K/AKT pathway was obviously suppressed. We found that PIK3CA was downregulated not only at the mRNA level but also at the protein level. The PI3K/AKT signaling pathway can influence the proliferation and metastasis of tumors. This pathway was found to be overactivated in many cancers.39, 40 PI3K receives many cell signals from numerous receptor tyrosine kinases (RTKs) and interactions with the AKT pathway. PIK3CA, a component of PI3K, has been found to be overexpressed in osteosarcoma tissues31, 41 and described as an oncogene. In our study, the overexpression of PIK3CA in TUG1‐knockdown osteosarcoma cells reversed the cell proliferation and metastasis phenotype. Therefore, PIK3CA may be responsible for the TUG1‐mediated cell proliferation and metastasis.
Cellular distribution has a crucial influence on the basic function of lncRNAs. It has been reported that some special lncRNAs, such as Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) and Nuclear Enriched Abundant Transcript 1 (NEAT1), participate in the formation of the nucleus and regulate gene transcription.42, 43 In this study, we showed that high levels of TUG1 were distributed in the cytoplasm, indicating that it might act as a ceRNA to regulate PIK3CA in osteosarcoma. A recent study showed that by competing with SOX2 in glioma and phosphatase and tensin homolog (PTEN) in prostate cancer, TUG1 acts as either a tumor suppressor or oncogenic factor. As shown by our data, we assessed this hypothesis and found that miR‐219a‐5p was the link between TUG1 and PIK3CA. There may be a regulatory network of osteosarcoma‐related genes, which is mediated by TUG1 sponging miRNAs. Through this network, TUG1 can widely influence the expression of pivotal components of osteosarcoma driver genes and serve as an oncogene or a tumor suppressor.
Understanding the basic mechanism of osteosarcoma could be beneficial for the improvement and development of therapies for osteosarcoma patients. Our in vivo data showed that inhibition of TUG1 by therapeutic LNAs repressed the proliferation of osteosarcoma cells, indicating that TUG1 targeting may be useful as a therapeutic method for osteosarcoma patients. In summary, our studies showed that TUG1 could function as a clinical biomarker for the diagnosis of osteosarcoma but also as a promising target for therapy.
CONFLICT OF INTEREST
The authors have no conflict of interests.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the Heilongjiang Provincial Ordinary Undergraduate College Young Innovative Talents Training Plan (No. UNPYSCT‐2016111), the Heilongjiang Provincial Academy of Medical Sciences (No. 201715, 201814), and the Fundamental Research Funds for the Provincial Universities (No. 2017LCZX35).
Li Y, Zhang T, Zhang Y, Zhao X, Wang W. Targeting the FOXM1‐regulated long noncoding RNA TUG1 in osteosarcoma. Cancer Sci. 2018;109:3093‐3104. 10.1111/cas.13765
REFERENCES
- 1. Ottaviani G, Jaffe N. The etiology of osteosarcoma. Cancer Treat Res. 2009;152:15‐32. [DOI] [PubMed] [Google Scholar]
- 2. Bielack SS, Kempf‐Bielack B, Delling G, et al. Prognostic factors in high‐grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20:776‐790. [DOI] [PubMed] [Google Scholar]
- 3. Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment ‐ where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523‐532. [DOI] [PubMed] [Google Scholar]
- 4. Smith MA, Seibel NL, Altekruse SF, et al. Outcomes for children and adolescents with cancer: challenges for the twenty‐first century. J Clin Oncol. 2010;28:2625‐2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perry JA, Kiezun A, Tonzi P, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci USA. 2014;111:E5564‐E5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chawla SP, Staddon AP, Baker LH, et al. Phase II study of the mammalian target of rapamycin inhibitor ridaforolimus in patients with advanced bone and soft tissue sarcomas. J Clin Oncol. 2012;30:78‐84. [DOI] [PubMed] [Google Scholar]
- 7. Tao J, Chen S, Lee B. Alteration of Notch signaling in skeletal development and disease. Ann N Y Acad Sci. 2010;1192:257‐268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Warzecha J, Gottig S, Chow KU, et al. Inhibition of osteosarcoma cell proliferation by the Hedgehog‐inhibitor cyclopamine. J Chemother. 2007;19:554‐561. [DOI] [PubMed] [Google Scholar]
- 9. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775‐1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559‐1563. [DOI] [PubMed] [Google Scholar]
- 11. Cech TR, Steitz JA. The noncoding RNA revolution‐trashing old rules to forge new ones. Cell. 2014;157:77‐94. [DOI] [PubMed] [Google Scholar]
- 12. Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689‐693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang L, Lin C, Jin C, et al. lncRNA‐dependent mechanisms of androgen‐receptor‐regulated gene activation programs. Nature. 2013;500:598‐602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med. 2008;14:723‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang P, Xue Y, Han Y, et al. The STAT3‐binding long noncoding RNA lnc‐DC controls human dendritic cell differentiation. Science. 2014;344:310‐313. [DOI] [PubMed] [Google Scholar]
- 20. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452‐463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501‐512. [DOI] [PubMed] [Google Scholar]
- 22. Katsushima K, Natsume A, Ohka F, et al. Targeting the Notch‐regulated non‐coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7:13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA‐mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang MD, Chen WM, Qi FZ, et al. Long non‐coding RNA TUG1 is up‐regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667‐11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482‐495. [DOI] [PubMed] [Google Scholar]
- 27. Ho C, Wang C, Mattu S, et al. AKT (v‐akt murine thymoma viral oncogene homolog 1) and N‐Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c‐Myc pathways. Hepatology. 2012;55:833‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao S, Fan LY, Lam EW. The FOXO3‐FOXM1 axis: a key cancer drug target and a modulator of cancer drug resistance. Semin Cancer Biol. 2018;50:77‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalinichenko VV, Major ML, Wang X, et al. FOXM1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830‐850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang IC, Chen YJ, Hughes DE, et al. FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J Biol Chem. 2008;283:20770‐20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joseph CG, Hwang H, Jiao Y, et al. Exomic analysis of myxoid liposarcomas, synovial sarcomas, and osteosarcomas. Genes Chromosome Cancer. 2014;53:15‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang R, Piperdi S, Gorlick R. Activation of the RAF/mitogen‐activated protein/extracellular signal‐regulated kinase kinase/extracellular signal‐regulated kinase pathway mediates apoptosis induced by chelerythrine in osteosarcoma. Clin Cancer Res. 2008;14:6396‐6404. [DOI] [PubMed] [Google Scholar]
- 33. Tu B, Du L, Fan QM, Tang Z, Tang TT. STAT3 activation by IL‐6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012;325:80‐88. [DOI] [PubMed] [Google Scholar]
- 34. van Heesch S, van Iterson M, Jacobi J, et al. Extensive localization of long noncoding RNAs to the cytosol and mono‐ and polyribosomal complexes. Genome Biol. 2014;15:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bacci G, Briccoli A, Rocca M, et al. Neoadjuvant chemotherapy for osteosarcoma of the extremities with metastases at presentation: recent experience at the Rizzoli Institute in 57 patients treated with cisplatin, doxorubicin, and a high dose of methotrexate and ifosfamide. Ann Oncol. 2003;14:1126‐1134. [DOI] [PubMed] [Google Scholar]
- 36. Zhang EB, Yin DD, Sun M, et al. P53‐regulated long non‐coding RNA TUG1 affects cell proliferation in human non‐small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang W, Duan N, Zhang Q, et al. DNA methylation mediated down‐regulation of miR‐370 regulates cell growth through activation of the Wnt/beta‐catenin signaling pathway in human osteosarcoma cells. Int J biol Sci. 2017;13:561‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koo CY, Muir KW, Lam EW. FOXM1: from cancer initiation to progression and treatment. Biochem Biophys Acta. 2012;1819:28‐37. [DOI] [PubMed] [Google Scholar]
- 39. Cheng JQ, Lindsley CW, Cheng GZ, Yang H, Nicosia SV. The Akt/PKB pathway: molecular target for cancer drug discovery. Oncogene. 2005;24:7482‐7492. [DOI] [PubMed] [Google Scholar]
- 40. Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497‐5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Teicher BA. Searching for molecular targets in sarcoma. Biochem Pharmacol. 2012;84:1‐10. [DOI] [PubMed] [Google Scholar]
- 42. Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B‐MYB. PLoS Genet. 2013;9:e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


