Abstract
Aberrant expression of TRIM‐containing protein 44 (TRIM44) acts as a promoter in multiple cancers. Here, we investigated the biological functions and clinical significance of TRIM44 in human esophageal cancer (HEC). TRIM44 expression was significantly higher in HEC tissues than corresponding normal tissues at both the mRNA (2.42 ± 0.52 vs 0.99 ± 0.25) and protein (1.01 ± 0.27 vs 0.30 ± 0.13) levels. Patients with high TRIM44 expression showed poor differentiation (P = 1.39 × 10−5), advanced TNM stage (P = 3.87 × 10−4) and, most importantly, significantly poorer prognosis (P = 2.80 × 10−5). TRIM44 played a crucial role in epithelial mesenchymal transition (EMT). A significant correlation was observed between TRIM44 and Ki67 expression. We demonstrated that TRIM44 markedly enhanced HEC cell proliferation, migration, and invasion. Additionally, TRIM44 was involved in the AKT/mTOR signaling pathway and its downstream targets, such as STAT3 phosphorylation. Thus, elevated TRIM44 expression promotes HEC development by EMT via the AKT/mTOR pathway, and TRIM44 may be a novel prognostic indicator for HEC patients after curative resection.
Keywords: EMT, HEC, prognosis, survival, TRIM44
1. INTRODUCTION
Esophageal cancer, which exhibits male‐dominant mortality, continues to be a leading cause of cancer death worldwide. Approximately 480 000 cases are diagnosed annually worldwide.1, 2 Despite technological advances in surgical treatment and systemic therapy during the past few decades, over 400 000 individuals have died from human esophageal cancer (HEC) within the last 5 years.3 The predicted 5‐year survival rate of HEC, which ranges from 15% to 20%, has barely improved in recent decades due to high recurrence rates and an early metastatic tendency.4 Thus, exploring the mechanism of HEC invasion and metastasis would help to identify new biomarkers that can accurately predict prognosis and improve targeted treatments.
Ubiquitination is among the most abundant post‐translational protein modifications and is involved in almost all fundamental cellular processes, including cell cycle regulation, DNA repair, signal transduction, antigen processing and apoptosis.5, 6 Recent studies have indicated that alterations in the ubiquitin and ubiquitin‐like protein (Ubl) systems underlie the genesis of different tumor types.7 TRIM‐containing protein 44 (TRIM44), a member of the tripartite motif (TRIM) protein family, contains B‐box, coiled‐coil domains and a zinc‐finger domain that is also found in ubiquitin hydrolases.8 TRIM44, a protein that may function as a “USP‐like TRIM”, was originally isolated from a mouse brain cDNA library.8, 9 Accumulating data suggest that TRIM44 plays stimulatory and cancerogenic roles in multiple cancers. TRIM44 overexpression facilitates cell proliferation, migration, and invasion in gastric cancer,10 hepatocellular carcinoma,11 testicular germ cell tumors,12 and non‐small cell lung cancer (NSCLC).13, 14 In addition, knocking down endogenous TRIM44 expression inhibits proliferation and motility in cancer cells.10, 11, 12, 13 Moreover, TRIM44 has been identified as an independent prognostic marker in patients with different types of cancer.10, 11, 14 Mechanistically, TRIM44 functions as a vital regulator of carcinogenesis through different downstream signaling pathways. For example, TRIM44 promotes non‐small cell lung cancer via the mammalian target of rapamycin (mTOR) and nuclear factor (NF)‐κB signaling pathways.13, 14 In 2010, FITZGERALD, on behalf of the Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Study Group15 revealed the prognostic value of TRIM44 in adenocarcinoma of the esophageal junction and gastric cardia. In 2014, these authors also demonstrated that TRIM44 can serve as a potential therapeutic marker for relevant cancers.16 However, the function and mechanism of TRIM44 in other types of esophageal cancer remain largely unknown. Considering the similarities in cancer pathogenesis between esophageal and gastric cancer,17 we investigated the clinical and molecular mechanisms of TRIM44 in HEC.
In the present study, we explored the altered expression of TRIM44 in HEC and examined its relationship with clinicopathological characteristics and biological function. Finally, we determined the molecular mechanisms of TRIM44 in HEC progression.
2. MATERIALS AND METHODS
2.1. HEC tissue samples
A total of 100 pairs of esophageal tissue samples were randomly collected from HEC patients who underwent curative resection from January 2005 to January 2006 at the Second Affiliated Hospital of Nanchang University (Nanchang, China) and the Affiliated Hospital of Hainan Medical University (Haikou, China). The distances between the corresponding adjacent normal tissues and HEC lesions were at least 3‐4 cm. Detailed clinical data and information on the storage of the specimens were previously described.18
Informed consent was obtained from all patients. Ethics approval was given by the Research Ethics Committees of the Second Affiliated Hospital of Nanchang University and the Affiliated Hospital of Hainan Medical University.
2.2. Cell lines
The human normal esophageal epithelial cell line HEEC and esophageal squamous cell carcinoma cell lines, including KYSE150, EC109 and KYSE510, were obtained from the American Type Culture Collection and maintained in our laboratory. The human esophageal squamous cell carcinoma cell lines KYSE140 and EC9706 were purchased from the Chinese Academy of Sciences. Cell lines including ECA109, HEEC and KYSE510 were maintained in Dulbecco's modified Eagle's medium (DMEM) (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA), streptomycin sulfate (100 IU/mL) and penicillin (100 μg/mL). KYSE150, KYSE140 and EC9706 cells were cultured in RPMI 1640 (HyClone) with the same supplements. All cell lines were incubated at 37°C in a humidified incubator with 5% CO2.
2.3. Tissue microarray construction, immunohistochemistry and evaluation
The construction and detailed information on the tissue microarray (TMA) utilized in the present study have been previously described.18 Immunohistochemistry (IHC) was performed as described.19, 20 Briefly, a total of 100 cases of HEC were included, and all samples were reviewed and confirmed via hematoxylin and eosin (H&E) staining. The slides were deparaffinized in xylene and rehydrated with alcohol. Diluted hydrogen peroxide (3%) was used to inactivate endogenous peroxidase activity, followed by washing with phosphate‐buffered saline (PBS). After being blocked with 5% bovine serum albumin (BSA, YESEN, Shanghai, China), the sections were treated with primary rabbit antibodies against TRIM44 (11511‐1‐AP; 1:150 dilution; Proteintech, Chicago, IL, USA) and mouse antibodies against Ki67 (#9449; 1:400 dilution; Cell Signaling Technology, Beverly, MA, USA) and incubated overnight at 4°C. The next day, the sections were incubated for 1 hour with horseradish peroxidase (HRP)‐labeled secondary antibody (Gene Tech; Shanghai, China) and then stained using diaminobenzidine (DAB, Gene Tech). Finally, the slides were imaged on a microscope (Olympus, Tokyo, Japan) with the same settings.
TRIM44 staining was evaluated based on intensity scores (0, no staining; 1, weak staining; 2, moderate staining; and 3±, strong staining) and percentage scores (0, 0%; 1 ≤25%; 2, >25%‐≤50%; 3, 50%‐≤75%; 4, >75%). Final scores were evaluated using a combination of intensity and percentage scores: scores ≤3 were defined as the low expression group and those ≥4 were considered the high expression group.
2.4. RNA isolation and quantitative real‐time PCR
Total RNA was extracted from both tissue samples and cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. cDNA was synthesized using the PrimeScript RT Reagent Kit (RRO47A; TaKaRa, Dalian, China). An ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) and SYBR Green Real‐Time PCR Master Mix (TaKaRa) were used in the quantitative real‐time PCR according to the manufacturer's instructions. Glyceraldehyde‐3‐phosphate dehydrogenase (GADPH) was used as an internal standard. mRNA levels were calculated using GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA) based on the Ct values. The following primers were designed: TRIM44, forward: 5′‐AGGCAGCTCATCTGTGTCCT‐3′ and reverse: 5′‐GCCTTCAGTCCAC CTGAGTC‐3′; and GADPH, forward: 5′‐GGTATGACAACGAATTTGGC‐3′ and reverse 5′‐ GAGCACAGGGTACTTTATTG‐3′.
2.5. Western blotting
Tissue or cell lysates were prepared in RIPA buffer (Beyotime, Shanghai, China) and measured using a BCA kit (Beyotime). Protein samples were separated via sodium dodecyl sulfate – polyacrylamide gel electrophoresis (SDS‐PAGE) and electrotransferred onto poly(vinylidene difluoride) (PVDF) membranes. After being blocked with 5% nonfat milk in Tris‐buffered saline with Tween‐20 (TBST) (Sangon Biotech, Shanghai, China) for 1 hour, the membranes were incubated in antibody diluent at 4°C overnight with the following primary antibodies: rabbit anti‐human TRIM44 (11511‐1‐AP; 1:1000 dilution; Proteintech), rabbit anti‐human E‐cadherin (#3195, 1:1000 dilution; Cell Signaling Technology), rabbit anti‐human Vimentin (ab92547, 1:1000 dilution, Abcam, Cambridge, UK), rabbit anti‐human N‐cadherin (ab18203,1:100 dilution, Abcam), rabbit anti‐Phospho‐mTOR (Ser2448) (#5536, 1:1000 dilution; Cell Signaling Technology), rabbit anti‐human AKT (#4685, 1:1000 dilution; Cell Signaling Technology), rabbit anti‐Phospho‐AKT (Ser473) (#4060, 1:1000 dilution; Cell Signaling Technology), rabbit anti‐human STAT3 (#9132, 1:1000 dilution; Cell Signaling Technology), rabbit anti‐Phospho‐Stat3 (Tyr705) (#9145, 1:1000 dilution; Cell Signaling Technology) and mouse anti‐GADPH (AG019, 1:1000 dilution, Beyotime) as internal controls. The membranes were then washed three times with TBST and incubated with HRP‐conjugated secondary antibody (YESEN) for 1 hour at room temperature. Detection was performed using a chemiluminescent HRP substrate (Millipore, Billerica, MA, USA) and an electrogenerated chemiluminescence (ECL) imaging system (Tanon, Shanghai, China). Image‐Pro Plus software version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA) was used for analysis.
2.6. Proliferation, colony formation and wound‐healing assays
Cell proliferation was detected using the Cell Counting Kit‐8 (CCK8, YESEN) according to the manufacturer's instructions. Colony formation assays were performed to assess the ability of a single cell to grow into a colony. Cells were plated at a low density (500‐1000 cells/well) onto 6‐well plates and observed for 2 weeks. Colonies were fixed in 4% paraformaldehyde, stained with 0.5% crystal violet staining solution and washed with PBS. For the wound‐healing assay, transfected cells were seeded onto 6‐well plates and incubated at 37°C with 5% CO2. Upon reaching 90% confluence the following day, cells in each well were scratch‐wounded using a sterile 200‐μL pipette tip. The detached cells were rinsed with PBS, after which the remaining cells were incubated with DMEM (HyClone) or 1640 (HyClone) medium containing 10% FBS. Micrographs were acquired at 0 and 48 hours. Three independent experiments were performed.
2.7. Cell migration and Matrigel invasion assays
Migration and invasion assays were performed using Transwell chambers with an 8‐μm pore size (24‐well format, Corning, Corning, NY, USA). For invasion assays, cells (1 × 105) suspended in serum‐free medium were added to the upper chamber, which had been pre‐coated with 50 μL of diluted Matrigel (1:6; BD Biosciences, San Jose, CA, USA). DMEM or 1640 medium supplemented with 30% FBS was added to the lower chambers, and the cells were cultured in a thermostatic incubator with 5% CO2 at 37°C for 48 hours. For the migration assay, the cells were directly added to the upper chamber without diluted Matrigel and cultured under the same conditions. Finally, the cells that migrated to or invaded the underside of the Transwell chambers were stained with 0.5% crystal violet and counted on an inverted microscope.
2.8. Lentivirus construction and transfection
pGMLV‐SC5‐shRNA‐TRIM44 (shRNA: short hairpin RNA)‐ and PGMLV‐PE3‐TRIM44‐expressing lentivirus carrying green fluorescent protein (GFP) and red fluorescent protein (RFP), respectively, were designed and provided by Genomeditech (Shanghai, China). The target shRNA sequences are shown in Table S1. KYSE510 and KYSE140 cells were transfected with the lentiviral vectors according to the manufacturer's instructions. Control cells were transfected with empty vectors carrying GFP or RFP. For establishment of stable cell lines, 2 μg/mL puromycin was used for 1 week after transfection with the lentiviral vectors. Knockdown and overexpression efficiency was validated via fluorescence microscopy, Western blotting and quantitative real‐time PCR. The stable TRIM44‐knockdown cell line was named KYSE510‐sh, and KYSE510‐ncl was used as a control. The TRIM44‐overexpression cell line was named KYSE140‐oe, and KYSE140‐nc was used as a control.
2.9. Immunofluorescence
For immunofluorescence assays, stably transfected cells were fixed with 4.0% paraformaldehyde, permeabilized with 0.3% diluted Triton X‐100 (Beyotime) and washed three times with PBS. After being blocked with 10% BSA, the slides were treated overnight with the following primary antibodies at 4°C: rabbit anti‐human Vimentin (ab92547, 1:100 dilution, Abcam), mouse anti‐human E‐cadherin (ab1416, 1:100 dilution, Abcam), rabbit anti‐human N‐cadherin (ab18203,1:100 dilution, Abcam) or Ki67 (#9449; 1:400 dilution; Cell Signaling Technology). Then, the cells were incubated with a fluorescent secondary antibody (1:200 dilution, YESEN) for 1 hour at room temperature, followed by three washes with PBS (5‐10 minutes each). After the slides were mounted in 4ʹ,6‐diamidino‐2‐phenylindole, fluorescence was observed under a microscope (Olympus).
2.10. Statistical analysis
SPSS 21.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism software version 5.0 (La Jolla, CA, USA) were used for statistical analysis. The Chi‐square or Fisher exact tests were used to assess the association between TRIM44 expression and clinicopathological features. Survival curves were constructed according to the Kaplan‐Meier method (log‐rank test used to test for differences between different groups). A Cox proportional hazards model, which was used for univariate and multivariate analyses, was applied to identify important prognostic factors for overall survival (OS). All P values were two‐sided, and P < .05 was considered statistically significant.
3. RESULTS
3.1. TRIM44 is overexpressed in HEC
To elucidate the potential role of TRIM44 in cancer, we used Oncomine (a publicly available data source) to investigate TRIM44 mRNA expression in cancer samples. Representative results suggested that TRIM44 mRNA levels were frequently elevated in colorectal carcinoma21 and gastric cancer22 (Figure 1A,B). Elevated TRIM44 expression is therefore related to multiple human cancers.
Figure 1.
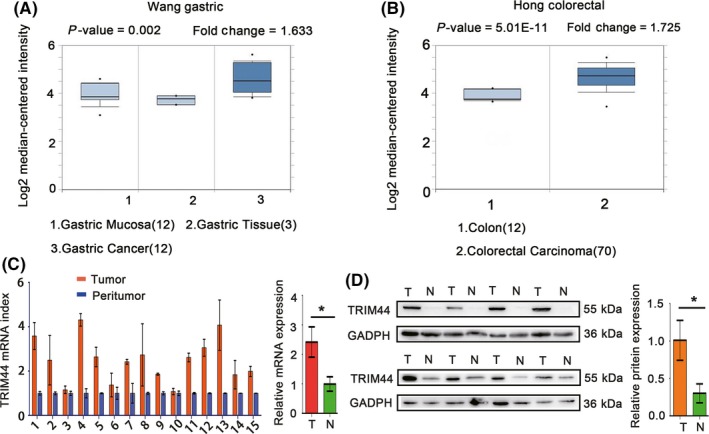
Increased TRIM44 expression levels in different types of cancer and prediction of an unfavorable outcome (data retrieved from Oncomine and Kaplan‐Meier Plotter). A, TRIM44 transcript levels in gastric mucosa and gastric cancer vs gastric tissues (P = .002, fold change = 1.633); B, TRIM44 transcript levels in colorectal carcinoma vs colon tissues (P = 5.01E‐11, fold change = 1.725); C, relative TRIM44 mRNA expression in 15 paired human esophageal cancer (HEC) tissues (T) and adjacent non‐tumor tissues (N) (P < .01); D, relative TRIM44 protein levels in 8 paired HEC tissues (T) and adjacent non‐tumor tissues (N) (P < .01). (T: tumor, N: peritumor; *P < .05, **P < .01, and ***P < .001)
We next examined TRIM44 expression in HEC using quantitative real‐time PCR, western blotting and IHC. Consistent with the Oncomine data, TRIM44 mRNA levels were significantly higher in the 15 HEC samples than in the corresponding peritumoral tissues (Figure 1C; 2.42 ± 0.52 vs 0.99 ± 0.25). Eight pairs of fresh HEC specimens and matched adjacent non‐tumor tissues were randomly selected for western blot analysis.
The results also demonstrated that TRIM44 protein levels were indeed increased in HEC tissues (Figure 1D; 1.01 ± 0.27 vs 0.30 ± 0.13). These results revealed that TRIM44 mRNA and protein levels are frequently elevated in HEC.
3.2. TRIM44 overexpression is associated with aggressive phenotypes and poor prognosis in HEC patients
We then focused on the clinical characteristics of TRIM44 expression in HEC patients. The expression of TRIM44 varies greatly in HEC tissues (Figure 2A). Based on the TRIM44 immunostaining intensity and percentage scores mentioned above, the cohort of 100 HEC patients was divided into TRIM44high and TRIM44low groups. We found that elevated TRIM44 expression was more frequently detected in HEC tissues than in corresponding non‐tumorous tissues in this cohort of patients (Figure 2B). As shown in Table 1, high TRIM44 expression correlated significantly with aggressive tumor characteristics, including poor tumor differentiation (P = 1.39 × 10−5) and advanced tumor stage (P = 3.87 × 10−4). High TRIM44 levels were more common in HEC with poor differentiation (88%, 22/25) than in HEC with good (25%, 8/32) or moderate (53%, 23/20) differentiation (Figure 2C). Elevated TRIM44 expression in TNM stages III‐IV (76.3%, 29/38) was more frequent than that in TNM stages I‐II (38.7%, 24/62, Figure 2E). In addition, patients with a higher TNM stage and poorer differentiation had a more unfavorable OS (Figure 2D,F).
Figure 2.
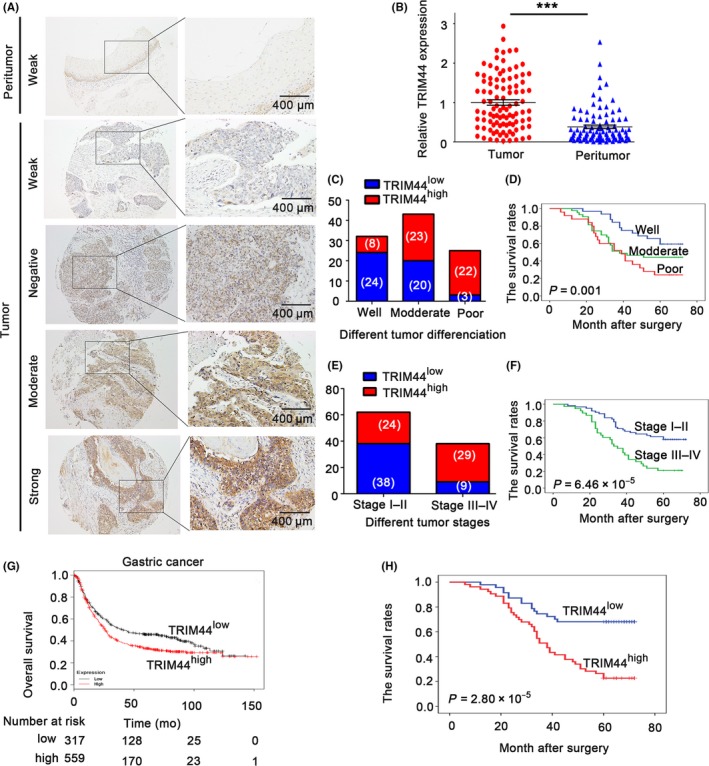
Increased TRIM44 expression in human esophageal cancer (HEC) patients is associated with poor prognosis. A, Representative photos of TRIM44 staining in 100 paired HEC tissues and matched adjacent normal tissue. The top figures: adjacent non‐tumor tissues; and the bottom figures: different staining intensities in HEC tumors; B, analysis of the integrated optical density (IOD) value of TRIM44 immunostaining in 100 paired HEC tissues adjacent normal tissue; C,D, high TRIM44 correlated with poor tumor differentiation (P = .001) and overall survival (OS) was also found; E,F, high TRIM44 correlated with high TNM stage (P = 6.46E‐05) and OS was also found; G, Kaplan‐Meier plots showing the OS associated with TRIM44 expression in gastric cancer. In red: patients with high TRIM44 expression and in black: patients with low TRIM44 expression (data retrieved from Kaplan‐Meier Plotter); H, overexpression of TRIM44 correlated with poor tumor differentiation and higher TNM stage (TRIM44high: high expression of TRIM44, TRIM44low: low expression of TRIM44; *P < .05, **P < .01, and ***P < .001)
Table 1.
Correlation between TRIM44 and clinicopathological characteristics for 100 human esophageal cancer (HEC) patients
| Variables | No. of patients | TRIM44 expression level | ||
|---|---|---|---|---|
| Low | High | P | ||
| Age (years) | ||||
| ≤65 | 74 | 35 | 39 | 1.00 |
| >65 | 26 | 12 | 14 | |
| Gender | ||||
| Male | 73 | 31 | 42 | .177 |
| Female | 27 | 16 | 11 | |
| Histological type | ||||
| Squamous cell carcinoma | 89 | 42 | 47 | .984 |
| Adenocarcinomas | 9 | 4 | 5 | |
| Othera | 2 | 1 | 1 | |
| Localization | ||||
| Upper | 22 | 9 | 13 | .627 |
| Middle | 44 | 23 | 21 | |
| Lower | 34 | 15 | 19 | |
| Lymphatic metastatis | ||||
| Yes | 48 | 13 | 35 | 1.46 × 10−4 a |
| No | 52 | 34 | 18 | |
| T stage | ||||
| I‐II | 56 | 34 | 22 | .002a |
| III‐IV | 44 | 13 | 31 | |
| Tumor stage | ||||
| I‐II | 62 | 38 | 24 | 3.87 × 10−4 a |
| III‐IV | 38 | 9 | 29 | |
| Differentiation | ||||
| Well | 32 | 24 | 8 | 1.39 × 10−5 a |
| Moderate | 43 | 20 | 23 | |
| Poor | 25 | 3 | 22 | |
P values < .05 were considered statistically significant. The Pearson Chi‐square test was used.
aSmall‐cell undifferentiated carcinoma.
We next observed the prognostic value of TRIM44 using an online tool (http://kmplot.com/analysis/). We found that gastric cancer patients with high TRIM44 expression exhibit unfavorable OS (Figure 2G). Consistent with the kmplot analysis data in gastric cancer, patients in the high TRIM44 expression group had obviously worse OS than those in the low TRIM44 expression group (26.4% vs 68%, P = 2.80 × 10−5, Figure 2H). Univariate analysis revealed that poor differentiation, lymph node metastasis, high T stage, advanced TNM stage and TRIM44 overexpression were significant indicators of poor OS in HEC. In the multivariate analysis, TRIM44 expression (HR, 2.240; 95% CI, 1.182‐4.246; P = .013) remained significantly correlated with 5‐year OS; however, tumor differentiation (HR, 1.292; 95% CI, 0.884‐1.890; P = .186) lost its significance as an independent prognostic factor for OS. Taken together, these data predicted that TRIM44 overexpression indicated significantly worse prognosis in HEC patients (Table 2).
Table 2.
Univariate and multivariate analysis of factors associated with OS
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Gender | ||||||
| Male vs female | 1.076 | 0.595‐1.944 | 0.809 | |||
| Age (years) | ||||||
| ≤65 vs >65 | 0.995 | 0.972‐1.018 | 0.659 | |||
| Location | ||||||
| Upper/middle vs lower | 1.033 | 0.716‐1.492 | 0.862 | |||
| Lymphatic metastatis | ||||||
| Yes vs no | 2.635 | 1.527‐4.547 | 0.001 | 1.720 | 0.920‐3.212 | 0.009 |
| T stage | ||||||
| III‐IV vs I‐II | 2.094 | 1.234‐3.555 | 0.006 | 0.217 | 0.029‐1.648 | 0.140 |
| Tumor stage | ||||||
| III‐IV vs I‐II | 2.790 | 1.642‐4.742 | 1.49 × 10−4 | 6.747 | 0.914‐49.817 | 0.001 |
| Differentiation | ||||||
| Well/moderate vs poor | 1.875 | 1.327‐2.650 | 3.70 × 10−4 | 1.302 | 0.895‐1.895 | 0.167 |
| TRIM44 level | ||||||
| Low vs high | 3.286 | 1.812‐5.960 | 8.98 × 10−5 | 2.240 | 1.182‐4.246 | 0.013 |
OS, overall survival; 95% CI, 95% confidence interval; multivariate analysis, Cox proportional hazards regression model. Variables were adopted for their prognostic significance by univariate analysis with forward stepwise selection (forward, likelihood ratio). Variables were adopted for their prognostic significance by univariate analysis (P < .05).
3.3. TRIM44 induces cell migratory and invasive abilities in HEC cell lines by regulating epithelial‐mesenchymal transition (EMT)
The clinical characteristics and prognostic value of TRIM44 in HEC prompted us to further examine its potential biological functions. Constitutive expression of TRIM44 in 5 HEC cell lines, as well as the normal esophageal epithelial cell HEEC, was confirmed (Figure 3A). We stably down‐regulated TRIM44 in KYSE510 cells with high TRIM44 expression (Figure 3B). We also successfully overexpressed the TRIM44 gene in KYSE140 cells with low TRIM44 expression (Figure 3C). The different levels of TRIM44 were confirmed via western blotting.
Figure 3.
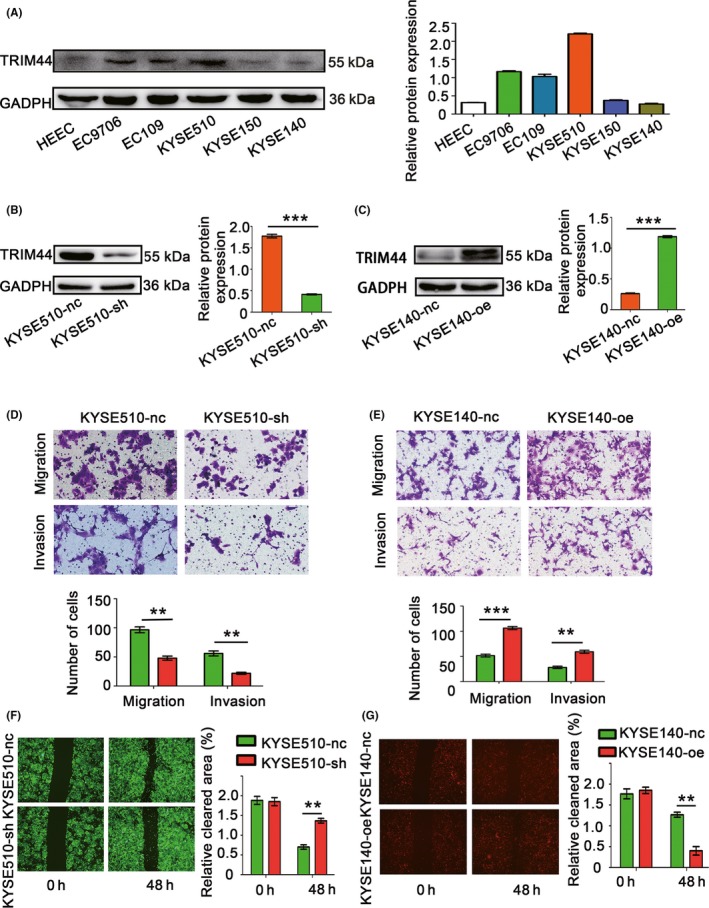
Overexpression of TRIM44 accelerates human esophageal cancer (HEC) cell migration and invasion. A, Relative TRIIM44 protein levels in HEC cell lines. Glyceraldehyde‐3‐phosphate dehydrogenase (GADPH) was used as a loading control; B,C, efficiency of TRIM44 overexpression and down‐regulation in HEC cells; D,E, Transwell assays were used to measure the effects of TRIM44 overexpression and down‐regulation on migration and invasion; F,G, wound‐healing assays were used to evaluate the migration of KYSE510 and KYSE140 cells. (*P < .05, **P < .01, and ***P < .001)
Published studies have also reported that increased TRIM44 expression promoted cell migration and invasion in NSCLC and hepatocellular carcinoma by regulating key components of EMT.11, 13 We therefore investigated whether TRIM44 similarly mediated EMT to promote cell migration and invasion in HEC cell lines. Transwell migration and invasion assays, as well as wound‐healing assays, were used to determine the effects of TRIM44 on migration and invasion in both TRIM44 gain‐of‐function (KYSE140‐oe) and loss‐of‐function (KYSE510‐sh) HEC cell lines. The results showed a 50.4% reduction in migration rates (P = 1.40 × 10−3) and a 60.7% reduction in invasion rates (P = 1.90 × 10−3) of KYSE510‐sh cell lines compared with the rates in the control cells (KYSE510‐nc) (Figure 3D). Conversely, TRIM44‐overexpressing cells (KYSE140‐oe) showed a 51.4% increase in migration rates (P = 2.00 × 10−4) and a 52.3% increase in invasion rates (P = 1.20 × 10−3) compared with the rates in control cells (KYSE140‐nc) (Figure 3E). Similar results were also observed in the wound‐healing assays. The down‐regulation of TRIM44 by shRNA in KYSE510 cells resulted in slower closure of scratch wounds in these cells than control cells (Figure 3F), and KYSE140 cells in which TRIM44 was overexpressed showed faster closure than KYSE140‐nc cells (Figure 3G). The acquisition of an EMT phenotype is essential for tumor cells to acquire increased migratory and invasive abilities.23 We next evaluated alterations in key components of EMT via western blotting and immunofluorescence in HEC cells with both TRIM44 down‐regulation and overexpression. E‐cadherin expression was increased in TRIM44‐silenced HEC cells, and N‐cadherin and Vimentin were both decreased compared with the levels in control cells (Figure 4A‐C). Increased E‐cadherin and down‐regulated N‐cadherin and Vimentin expression were observed in TRIM44‐overexpressing HEC cells (Figure 4B,D). We also investigated the function of TRIM44 in other cells (EC9706 and KYSE150) by overexpressing TRIM44 or knocking down TRIM44 (Figure S1). Together, these observations suggest that TRIM44 promotes HEC cell migration and invasion by regulating EMT. High TRIM44 expression was associated with impaired E‐cadherin expression and high levels of Vimentin. However, low TRIM44 expression was associated with increased E‐cadherin expression and low levels of Vimentin; representative images are shown in Figure 4E. A significant negative association was observed between TRIM44 and E‐cadherin expression in this cohort of patients (R 2 = 0.5072 and P < .001; Figure 4F), and the TRIM44 expression was positively correlated with Vimentin (R 2 = 0.4904 and P < .001; Figure 4F).
Figure 4.
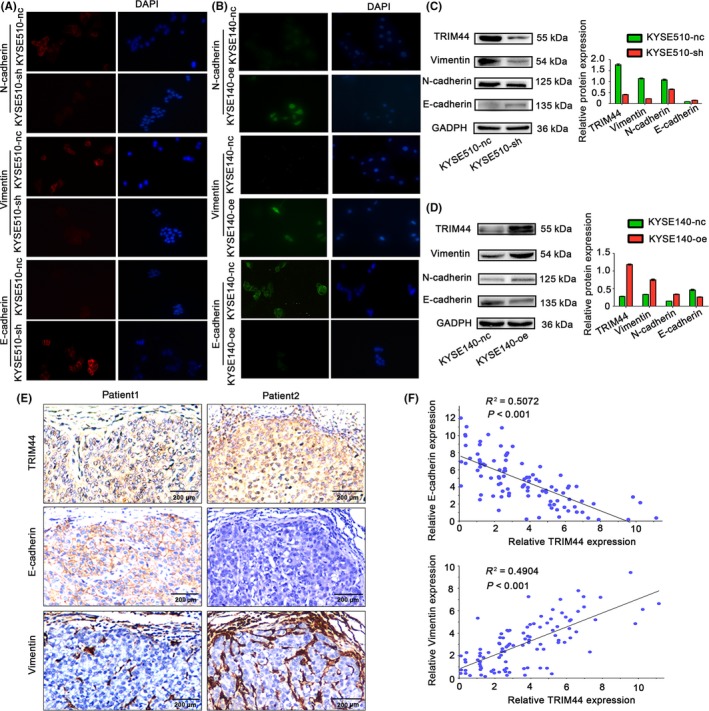
TRIM44 promotes human esophageal cancer (HEC) cell migration and invasion by promoting epithelial mesenchymal transition (EMT) and histopathological correlation of TRIM44 expression and EMT phenotypes in the tissue. A,B, Effects of TRIM44 overexpression and down‐regulation on epithelial and mesenchymal makers (including E‐cadherin, Vimentin and N‐cadherin) using fluorescence staining; C,D, changes in key molecules of EMT determined by western blot analyses after overexpression and down‐regulation of TRIM44 in KYSE140 and KYSE510 cells, respectively. E, High TRIM44 expression was associated with impaired E‐cadherin expression and high levels of Vimentin. Low TRIM44 expression was associated with increased E‐cadherin expression and low levels of Vimentin. F, The correction between TRIM44 and E‐cadherin expression according to the integrated optical density of the immunostaining. B, The correction between TRIM44 and Vimentin expression according to the integrated optical density of the immunostaining
3.4. TRIM44 modulates Ki67 expression and promotes HEC cell proliferation
CCK8 and colony formation assays were performed to determine the role of TRIM44 in HEC cell proliferation. The CCK8 assays indicated that compared with the control cells, TRIM44‐overexpressing HEC cells exhibited significantly increased proliferation, whereas significantly decreased proliferation was observed in HEC cells with TRIM44 down‐regulation (Figure 5A). Statistical significance was first observed after the cells were incubated for 36 hours (P = .0398) and 48 hours (P = 1.79 × 10−5). Similarly, colony formation was significantly elevated in KYSE140‐oe cells compared with KYSE140‐nc cells and was significantly reduced in KYSE510‐sh cells compared with KYSE510‐nc cells (Figure 5B). A recent study in hepatocellular carcinoma (HCC) reported that TRIM44 expression was tightly correlated with Ki67.11 Considering the strict association between cell proliferation and Ki67,24 we next examined the correlation between Ki67 and TRIM44 in HEC. We randomly selected 8 pairs of fresh HEC specimens for IHC analysis to assess the connection between TRIM44 and Ki67. Figure 5C is a representative picture of the results. Immunohistochemical staining of TRIM44 was similar to Ki67 staining (Figure 5C). In addition, TRIM44‐positive expression was more frequently detected in HEC tissues than in non‐tumorous tissues (Figure 5C). Immunofluorescence assays showed that TRIM44‐overexpressing KYSE140 cells exhibited significant up‐regulation of Ki67, whereas TRIM44‐silenced KYSE150 cells showed a loss of Ki67 (Figure 5D).
Figure 5.
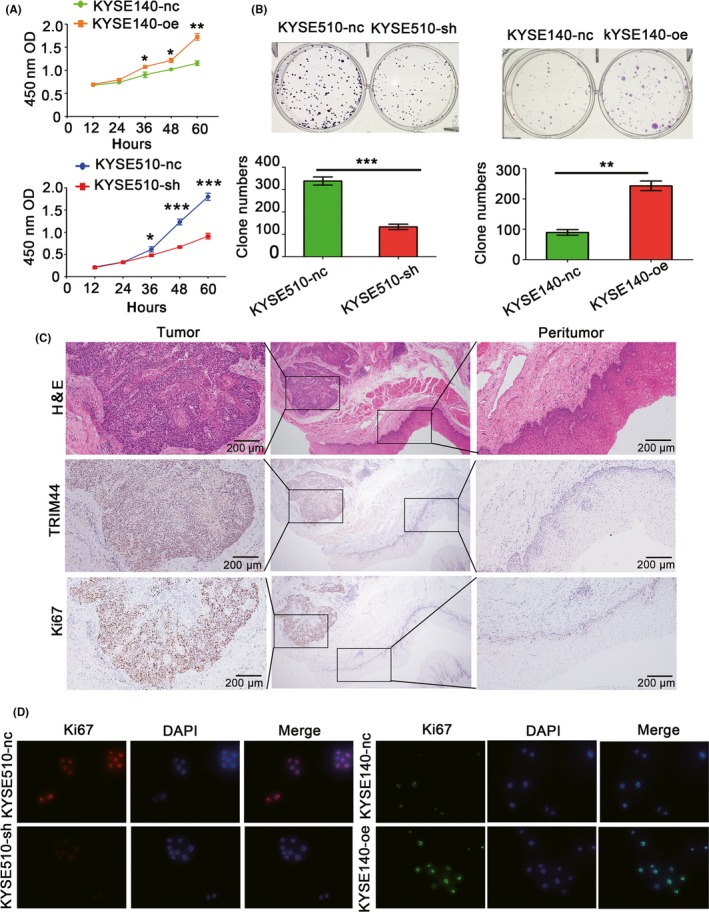
TRIM44 modulates Ki67 expression and promotes human esophageal cancer (HEC) cell proliferation. A,B, Cell Counting Kit‐8 (CCK8) and colony formation assays were used to detect cell proliferation in TRIM44 overexpression and down‐regulation of HEC cells; C, representative images of TRIM44 and Ki67 protein expression in the same tissues; D, representative fluorescence staining pictures of Ki67 expression changes induced by TRIM44 overexpression and down‐regulation in HEC cells. (*P < .05, **P < .01, and ***P < .001)
3.5. High levels of TRIM44 induce HEC cell EMT by the PI3K pathway
w?>Previous studies have revealed that TRIM44 is highly associated with the PI3K pathway in esophagogastric cancer (EGC)25 and NSCLC.13 Here, we also showed that shRNA knockdown of TRIM44 in KYSE510 cells caused a decrease in p‐AKT (Ser473) levels, as well as weak p‐mTOR activity. In contrast, TRIM44 overexpression in KYSE140 cells led to increased p‐AKT (Ser473) and p‐mTOR activity (Figure 6A). Interestingly, the phosphorylation levels of STAT3, which represents a point of convergence for numerous oncogenic pathways,25, 26, 27 were markedly enhanced in KYSE140‐oe cells and down‐regulated in KYSE510‐sh cells compared with the levels in their respective controls (Figure 6B).
Figure 6.
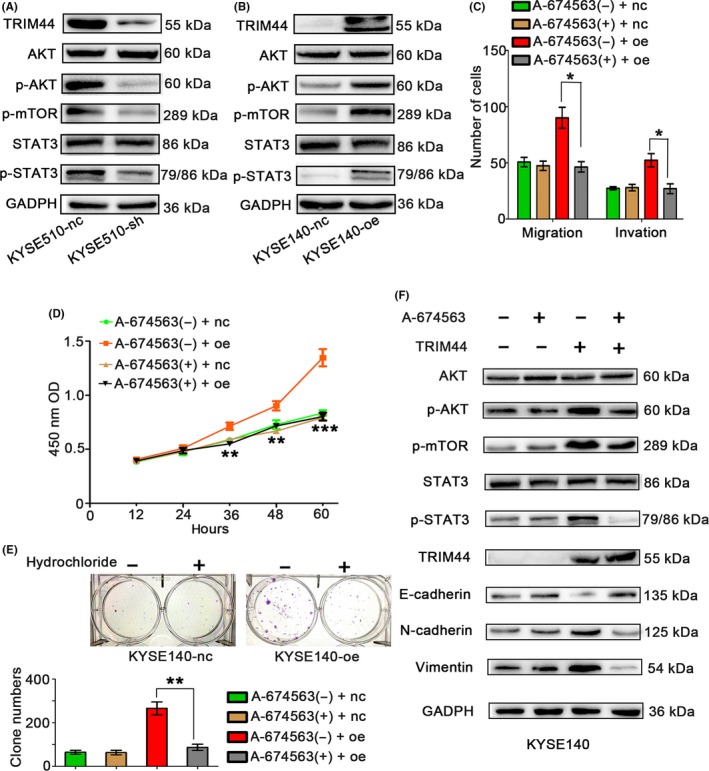
Association between TRIM44 overexpression and PI3K signaling activity. A, Western blot detection of p‐AKT, AKT, p‐mTOR, p‐STAT3 and STAT3 in KYSE510‐nc and KYSE510‐sh cell lines. B, Western blot detection of p‐AKT, AKT, p‐mTOR, p‐STAT3 and STAT3 in KYSE140‐nc and KYSE140‐oe cell lines. C, Transwell assays were performed to investigate changes in KYSE140‐oe cells after treatment with hydrochloride (A‐674563, an AKT inhibitor). D,F, Proliferation of KYSE140‐nc and KYSE140‐oe cells was measured by CCK8 and colony formation assays after treatment with hydrochloride. E, Western blots analyzed molecules of the PI3K pathway and epithelial mesenchymal transition markers in KYSE140‐nc and ‐oe cells treated by hydrochloride or phosphate‐buffered saline. (*P < .05, **P < .01, and ***P < .001)
Next, to further determine whether the PI3K signaling pathway plays an essential role in TRIM44‐induced cell proliferation, migration and invasion in HEC, we performed Transwell, CCK8 and colony formation assays of KYSE140‐nc and KYSE140‐oe cells treated by hydrochloride (A‐674563, an AKT inhibitor). As shown in Figure 6C‐E, KYSE140‐oe cell migration, invasion and proliferation were significantly reduced under A‐674563 treatment compared with the PBS treatment. Additionally, hydrochloride inhibited the phosphorylation of AKT and its downstream molecules, such as STAT3 (Figure 6F). Furthermore, hydrochloride was found to down‐regulate N‐cadherin and Vimentin expression but up‐regulate E‐cadherin expression in KYSE140‐oe cells (Figure 6F). These findings further indicated that high levels of TRIM44 induced HEC cell EMT by PI3K signaling.
4. DISCUSSION
In the present study, we reported that TRIM44 was commonly amplified in HEC specimens compared with its expression in corresponding normal tissues, consistent with the Oncomine analysis of prostate cancer, colorectal carcinoma21 and gastric cancer.22 Patients with TRIM44 overexpression showed poor differentiation, advanced TNM stage and, most importantly, significantly poorer prognosis. TRIM44 down‐regulation suppressed HEC cell proliferation, migration and invasion. In contrast, overexpressing TRIM44 in HEC cells promoted these activities. We also observed that TRIM44 played a crucial role in EMT. TRIM44 staining was similar to that of Ki67 by IHC. Additionally, overexpressing or knocking down TRIM44 successfully reduced or enhanced Ki67 expression, respectively, in immunofluorescence assays. These results revealed a significant association between TRIM44 and the AKT/mTOR signaling pathway. Collectively, our preliminary study strongly suggests that TRIM44 overexpression may represent an important factor in HEC progression and a be candidate prognostic marker.
Epithelial mesenchymal transition, which is characterized by the loss of cell‐cell adhesion and increased cell motility and invasion, is a major process underlying tumor metastasis.23, 28 Previous studies have reported that TRIM44 promotes NSCLC cell invasion and metastasis by inducing EMT.14 To this end, we evaluated changes in key components of EMT in both TRIM44‐overexpressing and TRIM44‐silenced HEC cells. TRIM44 expression was positively associated with N‐cadherin and Vimentin, whereas TRIM44 and E‐cadherin expression levels were inversely correlated. Thus, TRIM44 may regulate HEC cell migration and invasion by activating EMT. However, the precise mechanism underlying TRIM44‐mediated EMT in HEC remains unknown and requires further exploration.
Accumulating evidence indicates that the activation of the AKT/mTOR pathway plays a key role in controlling fundamental cellular processes, including cell growth, proliferation, apoptosis and metabolism in different cancer types.29, 30 A previous study showed a statistically significant correlation between TRIM44 and the mTOR signaling pathway in EGC.31 The TRIM44‐overexpression gene signature of EGC was reversed by AKT/mTOR pathway inhibitors.31 Similarly, TRIM44 overexpression resulted in high mTOR activity in NSCLC.13 In light of these previous reports, we examined the relationship between TRIM44 and the AKT/mTOR signaling pathway in HEC. We observed significant alterations in the phosphorylation levels of AKT/mTOR signaling pathway molecules after TRIM44 up‐regulation or knockdown. Interestingly, STAT3 phosphorylation levels were also affected. In addition, the phosphorylation levels of AKT/mTOR signaling pathway members and its downstream targets were re‐inhibited by AKT inhibitors. STAT3 is constitutively activated in multiple cancers, such as breast, lung, colorectal and prostate cancers.26, 32, 33 Moreover, STAT3 is a point of convergence for many oncogenic pathways, including AKT/mTOR, that are involved in cell proliferation, apoptosis, cell proliferation, migration, survival and tumorigenesis.34 However, the mechanism by which TRIM44 regulates the AKT/mTOR signaling pathway in HEC remains to be determined.
Our results demonstrate that TRIM44 promotes HEC cell proliferation, migration and invasion. TRIM44 might function by modulating the AKT/mTOR signaling pathway and contributing to EMT. Moreover, TRIM44 expression may serve as a promising prognostic indicator and a potential therapeutic target for HEC patients.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Research Ethics Committees of the Second Affiliated Hospital of Nanchang University and the Affiliated Hospital of Hainan Medical University.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation (No. 81560401).
Xiong D, Jin C, Ye X, et al. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci. 2018;109:3080–3092. 10.1111/cas.13762
Xiong and Jin are equally contributed to this work; none of the material that appears in the article has been previously presented or published in any form.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Wu CC, Chen CJ. Esophageal carcinoma. N Engl J Med. 2015;372:1472. [DOI] [PubMed] [Google Scholar]
- 3. Sohda M, Kuwano H. Current status and future prospects for Esophageal cancer treatment. Ann Thorac Cardiovasc Surg. 2017;23:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271‐289. [DOI] [PubMed] [Google Scholar]
- 5. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425‐479. [DOI] [PubMed] [Google Scholar]
- 6. Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol. 2003;4:491‐497. [DOI] [PubMed] [Google Scholar]
- 7. Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin‐like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776‐788. [DOI] [PubMed] [Google Scholar]
- 8. Urano T, Usui T, Takeda S, et al. TRIM44 interacts with and stabilizes terf, a TRIM ubiquitin E3 ligase. Biochem Biophys Res Commun. 2009;383:263‐268. [DOI] [PubMed] [Google Scholar]
- 9. Boutou E, Matsas R, Mamalaki A. Isolation of a mouse brain cDNA expressed in developing neuroblasts and mature neurons. Brain Res Mol Brain Res. 2001;86:153‐167. [DOI] [PubMed] [Google Scholar]
- 10. Kashimoto K, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci. 2012;103:2021‐2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu X, Wu Y, Miao X, et al. High expression of TRIM44 is associated with enhanced cell proliferation, migration, invasion, and resistance to doxorubicin in hepatocellular carcinoma. Tumour Biol. 2016;37:14615‐14628. [DOI] [PubMed] [Google Scholar]
- 12. Yamada Y, Takayama KI, Fujimura T, et al. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci. 2017;108:32‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xing Y, Meng Q, Chen X, et al. TRIM44 promotes proliferation and metastasis in nonsmall cell lung cancer via mTOR signaling pathway. Oncotarget. 2016;7:30479‐30491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luo Q, Lin H, Ye X, Huang J, Lu S, Xu L. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF‐kappaB signaling pathway. Int J Clin Oncol. 2015;20:508‐517. [DOI] [PubMed] [Google Scholar]
- 15. Peters CJ, Rees JR, Hardwick RH, et al. A 4‐gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology. 2010;139:1995‐2004. e15 [DOI] [PubMed] [Google Scholar]
- 16. Ong CA, Shapiro J, Nason KS, et al. Three‐gene immunohistochemical panel adds to clinical staging algorithms to predict prognosis for patients with esophageal adenocarcinoma. J Clin Oncol. 2013;31:1576‐1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quante M, Bhagat G, Abrams JA, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett‐like metaplasia. Cancer Cell. 2012;21:36‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu YB, Huang YS, Xu YP, et al. A high level of TM4SF5 is associated with human esophageal cancer progression and poor patient survival. Dig Dis Sci. 2013;58:2623‐2633. [DOI] [PubMed] [Google Scholar]
- 19. Liao YF, Wu YB, Long X, et al. High level of BRD4 promotes non‐small cell lung cancer progression. Oncotarget. 2016;7:9491‐9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao GY, Ding JY, Lu CL, Lin ZW, Guo J. The overexpression of 14‐3‐3zeta and Hsp27 promotes non‐small cell lung cancer progression. Cancer. 2014;120:652‐663. [DOI] [PubMed] [Google Scholar]
- 21. Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis‐prone’ signature for early‐stage mismatch‐repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83‐90. [DOI] [PubMed] [Google Scholar]
- 22. Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77‐83. [DOI] [PubMed] [Google Scholar]
- 23. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial‐mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scholzen T, Gerdes J. The Ki‐67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311‐322. [DOI] [PubMed] [Google Scholar]
- 25. Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41‐51. [DOI] [PubMed] [Google Scholar]
- 26. Yu CL, Meyer DJ, Campbell GS, et al. Enhanced DNA‐binding activity of a Stat3‐related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81‐83. [DOI] [PubMed] [Google Scholar]
- 27. Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4:97‐105. [DOI] [PubMed] [Google Scholar]
- 28. Acloque H, Adams MS, Fishwick K, Bronner‐Fraser M, Nieto MA. Epithelial‐mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438‐1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dey N, De P, Leyland‐Jones B. PI3K‐AKT‐mTOR inhibitors in breast cancers: from tumor cell signaling to clinical trials. Pharmacol Ther. 2017;175:91‐106. [DOI] [PubMed] [Google Scholar]
- 30. Zhou H, Huang S. Role of mTOR signaling in tumor cell motility, invasion and metastasis. Curr Protein Pept Sci. 2011;12:30‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ong CA, Shannon NB, Ross‐Innes CS, et al. Amplification of TRIM44: pairing a prognostic target with potential therapeutic strategy. J Natl Cancer Inst. 2014;106:2504‐2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Banerjee K, Resat H. Constitutive activation of STAT3 in breast cancer cells: a review. Int J Cancer. 2016;138:2570‐2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siveen KS, Sikka S, Surana R, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845:136‐154. [DOI] [PubMed] [Google Scholar]
- 34. Kortylewski M, Yu H. Stat3 as a potential target for cancer immunotherapy. J Immunother. 2007;30:131‐139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


