Abstract
Over the past several years, long non‐coding RNAs (lncRNAs) have attracted more and more attention due to their special functions. They are vital biomarkers in multiple diseases. LncRNA HOMEOBOX A11 (HOXA11) has been found to be aberrantly expressed in some kinds of malignant tumors. In this study, we mainly discuss the oncogenic role of it in promoting malignant progression and chemoresistance in lung adenocarcinoma (LUAD) cells. The expression of HOXA11‐AS was much stronger in cisplatin‐resistant LUAD cells. Based on The Cancer Genome Atlas database, patients with high expression of HOXA11‐AS had shorter survival time. Additionally, knockdown of HOXA11‐AS caused positive changes in cell activities of LUAD. For example, cell proliferation and migration were weakened, the epithelial mesenchymal transition process was reversed, and apoptosis was induced. These changes were more obvious in cells treated with cisplatin. Next, the HOXA11‐AS/miR‐454‐3p/Stat3 (signal transducer and activator of transcription 3) pathway was found to influence the cisplatin resistance of LUAD cells. HOXA11‐AS specifically acted as a competing endogenous RNA (ceRNA) in LUAD cells. The combinations among these three genes were demonstrated. Finally, rescue assays were applied to demonstrate the ceRNA pattern consisting of HOXA11‐AS, miR‐454‐3p and Stat3. In conclusion, lncRNA HOXA11‐AS acted as a ceRNA to promote cisplatin resistance of human LUAD cells via the miR‐454‐3p/Stat3 axis.
Keywords: Cisplatin resistance, HOXA11‐AS, lung adenocarcinoma, miR‐454‐3p, Stat3
1. INTRODUCTION
Lung cancer is considered to be one of the commonest malignant tumors all over the world.1, 2 It has almost become the first major factor which causes death for people in urban areas of China. After several years’ research, people have divided lung cancer into several subtypes. Non‐small cell lung cancer (NSCLC), taking charge of around 85% of all lung cancers, is the most frequent one among these subtypes. Furthermore, NSCLC can be classified to three types (adenocarcinoma, squamous cell carcinoma, and large cell carcinoma),3 among which, lung adenocarcinoma (LUAD) is the most common.4 Despite great efforts having been made in developing novel treatments, the prognosis for patients is still unsatisfactory in malignant LUAD, and the 5‐year survival rate is still under 10%.5 LUAD keeps a relatively high sensitivity to primary chemotherapy, but in fact, extensive metastasis and rapid progression of chemoresistance to chemotherapy result in mortality for most patients.6, 7 Chemoresistance is a huge problem in chemotherapy. So in this study, we focused on how to sensitize LUAD to cisplatin.
Long non‐coding RNA (lncRNA) is known as a type of noncoding RNA with over 200 nucleotides in length and non‐protein coding ability.8 It has been certified that abnormal expression of lncRNAs plays a vital role in the progression of various cancers such as ovarian cancer,9 colon cancer10 and gastric cancer.11 Therefore, studying lncRNAs may be of great value in exploring the occurrence and development of tumors. Although so many lncRNAs have been reported in human cancers, countless lncRNAs still await investigation. LncRNA HOMEOBOX A11 (HOXA11) antisense RNA (HOXA11‐AS) has been recorded in some cancer reports. According to all previous reports, we knew that HOXA11‐AS could positively modulate progression and development of several types of human cancers, such as colorectal cancer,12 hepatocellular carcinoma,13 glioma14 and breast cancer.15 The specific role of HOXA11‐AS in lung adenocarcinoma still lacks investigation. In this study, we tried to reveal the specific function and mechanism of HOXA11‐AS in lung adenocarcinoma. Functional experiments were utilized to analyze the oncogenic role of HOXA11‐AS1. The investigation was designed to develop a molecular mechanism centered around HOXA11‐AS1. Finally, rescue assays were used to demonstrate the effects of the HOXA11‐AS1‐miR‐454‐3p‐Stat3 pathway on the progression and cisplatin resistance of LUAD.
2. MATERIALS AND METHODS
2.1. TCGA database
The expression profile of HOXA11‐AS1 in LUAD tissues and corresponding normal tissues from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) were analyzed. Tissues samples in TCGA database were divided into two groups (HOXA11‐AS high expression group: n = 267; HOXA11‐AS low expression group: n = 268). A survival curve was generated to analyze the correlation between HOXA11‐AS expression and the overall survival of LUAD patients.
2.2. Cell lines
LUAD cells (A549 and H157) and HEK‐293T cells used in this study were obtained from the Institute of Cell Research, Chinese Academy of Sciences, Shanghai, China. Cells were cultured in RPMI‐1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% of fetal bovine serum (FBS) added. Then, cells were maintained in RPMI‐1640 medium (Invitrogen) which was also supplemented with 10% FBS. Plates used to seed cells were incubated at 37°C in a humidified atmosphere with 5% CO2.
2.3. Development of cisplatin‐resistant cell lines
According to the methods introduced by Barr et al.,16 we continuously treated parental LUAD cells (A549 and H157) with a gradually increasing dose of cisplatin for half a year. In short, cells were treated with 1 μmol/L cisplatin for 3 days. After this step was repeated one more time, cells were then treated with 2 μmol/L cisplatin. During the progress of cisplatin resistance induction, the 50% inhibitory concentration (IC50) value of every 5 passages was examined and continuously compared with those of parental cells until the IC50 value stopped increasing. The cisplatin‐resistant cells were established and harvested when the IC50 value stopped increasing at 10 μmol/L.
2.4. Transfection
We bought miR‐454‐3p mimic, miR‐454‐3p inhibitor and negative control from Applied Biological Materials (ABM, Vancouver, British Columbia, Canada). According to the instructions of the manufacturer, the Lipofectamine 2000 kit (Invitrogen) was utilized for all necessary transfections.
2.5. Plasmid construction
The cDNA encoding HOXA11‐AS was PCR amplified and subcloned into the pLent‐GIII‐CMV‐Puro vector (ABM, Canada), which was named pLent‐HOXA11‐AS. The short hairpin RNA (shRNA) was constructed with sequence specifically targeted to HOXA11‐AS, which was obtained from ABM (Canada). The specific shRNAs for HOXA11‐AS and signal transducer and activator of transcription 3 (Stat3) were synthesized by ABM (Canada). The plasmid pLent‐HOXA11‐AS was stably transfected into cells and screened with Puromycin (2 μg/mL) for 4 weeks. The sequences of sh‐HOXA11‐AS are shown in Table S1.
2.6. Isolation of RNA and quantitative RT‐PCR
According to the user's manual, Trizol reagent (Invitrogen) was adopted to isolate total RNA from cells. Reverse transcription (RT) was finished by means of PrimeScript RT reagent Kit (Takara, Dalian, China). Based on the instructions for use, we conducted quantitative RT‐PCR with SYBR Prime Script RT‐PCR Kits (Takara, Dalian, China). Relative quantification (2−ΔΔCt method) was employed to calculate fold changes, which was normalized to GAPDH. All assays were carried out in triplicate. All primers are displayed in Table S1.
2.7. MTT assay
In order to detect cell viability, 96‐well plates (3 × 103 cells per well) were utilized to immediately seed cells or 1 day after transfection. After the indicated drug combinations were treated for 2 days, cell viability was assessed by means of MTT assay. Chemosensitivity was measured by MTT (Sigma, St Louis, MO, USA). Cells were cultured in 96‐well plates which were treated with cisplatin. After 2 days, the MTT solution (5 mg/mL, 20 μL) was added into each well. After incubation for 4 hours, the media were taken away and 100 μL DMSO was added into each well. We obtained the relative number of surviving cells at 560 nm by means of calculating the optical density (OD) of cell lysates.
2.8. Colony‐formation assay
Colony‐formation assay was utilized for analyzing cell proliferation. At 48 hours after transfection, 500 cells were seeded in a 6‐well plate. The cells were incubated for about 2 weeks at 37°C, and then the cells were stained with 0.5% crystal violet solution in 20% methanol. Then, the number of colonies was manually calculated.
2.9. Cell migration assays
Cell migration was measured by means of a transwell chamber (8 μm pore size, Corning, Corning, NY, USA). At 48 hours after transfection, we placed 2 × 104 cells that were previously cultured in serum‐free media into the upper chamber. Against this, in the lower chamber 10% FBS was added to the media. After incubation for 2 days, we wiped off cells remaining in the upper membrane, and migratory cells were fixed in methanol. After the above steps were finished, we used 0.1% crystal violet to stain those cells and counted them under a microscope.
2.10. Immunofluorescence
Glass coverslips of 6‐well plates were applied to seed cells, for the fixation and permeation of cells; 4% formaldehyde solution and 0.5% Triton X‐100/PBS were separately put into use. In order to block cells, we added 5% BSA PBS for 1 hour at room temperature. Next, the primary antibody was incubated at 4°C overnight, followed with 1‐hour incubation of fluorescent‐dye conjugated secondary antibody (Invitrogen), and then, we stained cells with DAPI. Finally, images were obtained with a reverse‐fluorescence microscope.
2.11. Flow cytometry analysis assay
Flow cytometry was applied to measure cell apoptosis. Briefly, cells were cultured in a normal medium and transfected with corresponding plasmids. Cells were collected after transfection for 48 hours. According to the user's guide, PE Annexin V apoptosis detection kits (BD Pharmingen, San Diego, CA, USA) were used for determining cell apoptosis. Finally, cell apoptosis was assessed by using flow cytometry (EPICS, XL‐4, Beckman, Brea, CA, USA).
2.12. Subcellular fractionation assay
We utilized NE‐PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA) for extracting cytoplasmic and nuclear elements from LUAD cells. Quantitative RT–PCR was applied to analyze RNAs extracted from each of the fractions, and to examine the levels of nuclear and cytoplasmic control transcript (U6 and GAPDH), lncRNA HOXA11‐AS and miR‐454‐3p.
2.13. RNA‐binding protein immunoprecipitation
In accordance with the manufacturer's protocol, both the Magna RNA Immunoprecipitation (RIP) RNA‐binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) and the Ago2 antibody (Abcam, Cambridge, MA, USA) were utilized for RIP experiment. We analyzed co‐precipitated RNAs by means of quantitative RT–PCR analysis.
2.14. Dual‐luciferase reporter assay
The sequence of wild type HOXA11‐AS (HOXA11‐AS‐WT) and the sequence of mutant type HOXA11‐AS (HOXA11‐AS‐MUT) were inserted into pmirGLO reporter vector (GenePharma, Shanghai, China). Next, pmirGLO‐HOXA11‐AS‐WT and pmirGLO‐HOXA11‐AS‐MUT as well as miR‐454‐3p mimics and miR‐NC were transfected into HEK‐293T and A549‐CR cell lines with Lipofectamine 2000 (Invitrogen). According to the instructions, the luciferase activity was detected by the Dual‐Luciferase Reporter Assay System (Promega, Madison, WI, USA).
2.15. Western blotting analysis
Total cell lysates were maintained in a 1× sodium dodecyl sulfate buffer for preparation. Total protein was dissolved by sodium dodecyl sulfate‐polyacrylamide gelelectrophoresis and then was transferred onto nitrocellulose membranes. Next, the membrane was sealed with 5% non‐fat milk and incubated with primary antibodies at 4°C overnight. After incubation with antibodies (E‐cadherin, β‐catenin, N‐cadherin, Vimentin, Snail, Twist, Cleaved caspase‐3, Cleaved caspase‐9, Total‐caspase‐3, Total caspase‐9, Stat3, GAPDH) (Abcam, Hong Kong, China), the blots were incubated with goat anti‐rabbit secondary antibody (Abcam). Chemiluminescence brought them out.
2.16. Statistical analysis
Data are displayed as the means ± SD error. All data were obtained from more than two independent experiments. Statistical analysis was made by SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Student's t test was performed to make comparisons between two groups. On the other hand, one‐way ANOVA was used to analyze the comparisons among multiple groups. All tests were bidirectional. All these data with P values less than .05 were recognized as statistically significant.
3. RESULTS
3.1. Dysregulation of HOXA11‐AS is associated with cisplatin resistance of LUAD cells
The expression level of LUAD in the LUAD samples of TCGA database was analyzed. Obviously, HOXA11‐AS was expressed much higher in LUAD tissues (Figure 1A). Subsequently, LUAD samples in TCGA database were divided into two groups in accordance with the mean value of HOXA11‐AS expression. A survival curve was generated to reveal the correlation between HOXA11‐AS expression and the overall survival of LUAD patients. It could be observed that the overall survival rate in the high expression group (n = 267) was lower than that in the low expression group (n = 268) (Figure 1B). Since we aimed to study the effect of HOXA11‐AS on the cisplatin resistance of LUAD cells, quantitative RT‐PCR was used for detection of the expression level of HOXA11‐AS in both LUAD cell lines (A549 and H157) and their matched cisplatin‐resistant cells (A549‐CR and H157‐CR). Unsurprisingly, HOXA11‐AS was highly expressed in the cisplatin‐resistant cells (Figure 1C). To make further confirmation, we applied the MTT kit to examine the IC50 value of parental LUAD cells and corresponding cisplatin‐resistant cells. As expected, the IC50 values of A549‐CR and H157‐CR cells were significantly higher than that of A549 and H157 cells (Figure 1D). Subsequently, HOXA11‐AS was overexpressed in A549 and H157 cells through transfecting with pLent‐HOXA11‐AS (Figure 1E), whereas, A549‐CR and H157‐CR cells were transfected with shRNAs especially targeted to HOXA11‐AS (sh‐HOXA11‐AS#1, sh‐HOXA11‐AS#2, sh‐HOXA11‐AS#3, sh‐HOXA11‐AS#4). The highest transfection efficiency was observed when cisplatin‐resistant cells were transfected with sh‐HOXA11‐AS#2 (sh‐HOXA11‐AS) (Figure 1F). After transfection, the IC50 values of parental cells and cisplatin‐resistant cells were tested with MTT assay. Not surprisingly, the IC50 values of A549 and H157 cells were increased by pLent‐HOXA11‐AS (Figure 1G) and the IC50 values of A549‐CR and H157‐CR cells were decreased by sh‐HOXA11‐AS (Figure 1H). All these findings indicated that HOXA11‐AS is a poor prognostic factor for LUAD patients and a potential biomarker for cisplatin resistance.
Figure 1.
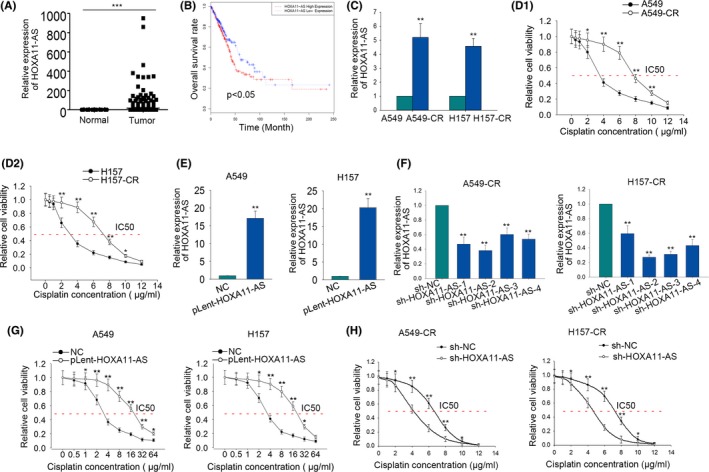
Dysregulation of HOMEOBOX A11 antisense RNA (HOXA11‐AS) is associated with cisplatin resistance of lung adenocarcinoma (LUAD) cells. (A) The expression level of HOXA11‐AS in LUAD tissues and non‐tumorous tissues of The Cancer Genome Atlas (TCGA) database was analyzed and shown. (B) Based on TCGA dataset, a survival curve was generated to analyze the correlation between HOXA11‐AS expression and the overall survival of LUAD patients. (C) Quantitative RT‐PCR was used for detection of the expression level of HOXA11‐AS in parental cells (A549 and H157) and cisplatin‐resistant cells (A549‐CR and H157‐CR). (D) The 50% inhibitory concentration (IC50) value of parental cells and corresponding cisplatin‐resistant cells was tested with MTT assay. (E) HOXA11‐AS was overexpressed in A549 and H157 cells by transfecting with pLent‐HOXA11‐AS. (F) HOXA11‐AS was downregulated in cisplatin‐resistant cells by transfecting with short hairpin RNA (sh)‐HOXA11‐AS. (G) The IC50 value of parental cells was examined after HOXA11‐AS was overexpressed. (H) The IC50 value of cisplatin‐resistant LUAD cells was detected after HOXA11‐AS was knocked down. Error bars represent the mean ± SD of at least three independent experiments. *P < .05, **P < .01, ***P < .001 vs control group
3.2. Effects of HOXA11‐AS on chemoresistance of LUAD cells
Considering the potential oncogenic role of HOXA11‐AS, we designed and conducted functional assays in both parental cells and cisplatin‐resistant cells which were treated with or without cisplatin. The parental cells were treated with cisplatin at a density of 1 μg/mL, while the cisplatin‐resistant cells were treated with cisplatin at a concentration of 3 μg/mL. At first, colony formation assays revealed that the proliferative ability was enhanced after A549 and H157 cells were transfected with pLent‐HOXA11‐AS. The cell proliferation was affected in a dose‐dependent manner (Figures 2A and 3A). As for A549‐CR and H157‐CR cells, the proliferative ability was weakened after cells were transfected with sh‐HOXA11‐AS. The inhibitory effect was more obvious in the cisplatin group (Figures 2B and 3B). Flow cytometry analysis disclosed that the apoptotic rate of A547 and H157 cells transfected with pLent‐HOXA11‐AS was decreased in a dose‐dependent manner (Figures 2C and 3C). As for cisplatin‐resistant cells (A549‐CR and H157‐CR), the apoptotic rate in the cisplatin group was higher than that in the non‐cisplatin group when both of the groups were simultaneously treated with sh‐HOXA11‐AS (Figures 2D and 3D). For further confirmation, we also examined the levels of apoptosis‐related proteins with western blotting assay. The results showed that the levels of cleaved‐caspase‐3 and cleaved‐caspase‐9 in the cisplatin group and non‐cisplatin group were decreased when transfected with pLent‐HOXA11‐AS (Figures 2E and 3E). Western blot analysis also showed the protein levels of cleaved‐caspase‐3 and cleaved‐caspase‐9 were much higher in the cisplatin group than that the in non‐cisplatin group (Figures 2F and 3F). Therefore, we concluded that HOXA11‐AS can affect the chemoresistance of LUAD cells.
Figure 2.
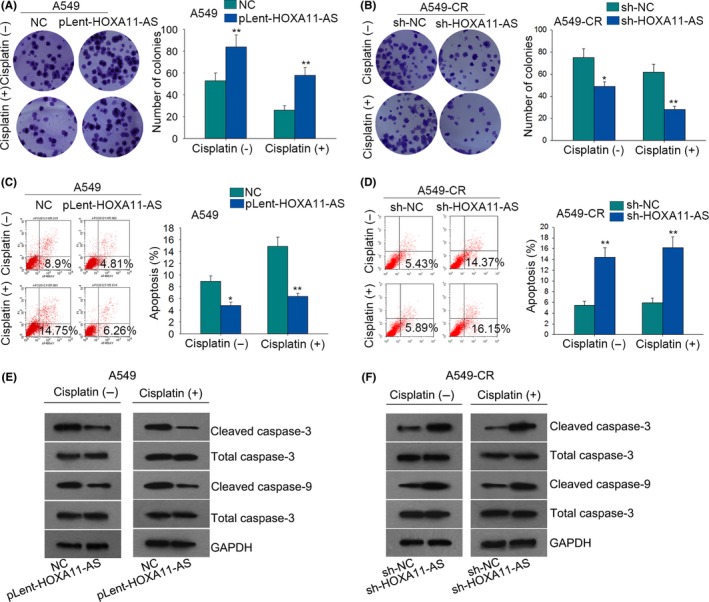
The effects of HOMEOBOX A11 antisense RNA (HOXA11‐AS) on the chemoresistance of lung adenocarcinoma (LUAD) cells. (A) Colony formation assays measured the proliferation ability of A549 cells in the cisplatin group (1 μg/mL) and non‐cisplatin group after HOXA11‐AS was overexpressed. (B) The proliferation of A549‐CR cells was evaluated in the cisplatin group (3 μg/mL) and non‐cisplatin group after HOXA11‐AS was downregulated. (C) Flow cytometry analysis examined the apoptotic rate in both cisplatin and non‐cisplatin groups after A549 cells were transfected with pLent‐HOXA11‐AS. (D) Flow cytometry analysis examined the apoptotic rate in both cisplatin and non‐cisplatin groups after A549‐CR cells were transfected with short hairpin RNA (sh)‐HOXA11‐AS. (E) Western blot analysis tested the protein levels of apoptosis‐related proteins in the cisplatin group or in the non‐cisplatin group after A549 cells were treated with pLent‐HOXA11‐AS. (F) Western blot analysis tested the protein levels of apoptosis‐related proteins in the cisplatin group or in the non‐cisplatin group after A549‐CR cells were treated with sh‐HOXA11‐AS. Error bars represent the mean ± SD of at least three independent experiments. *P < .05, **P < .01 vs control group
Figure 3.
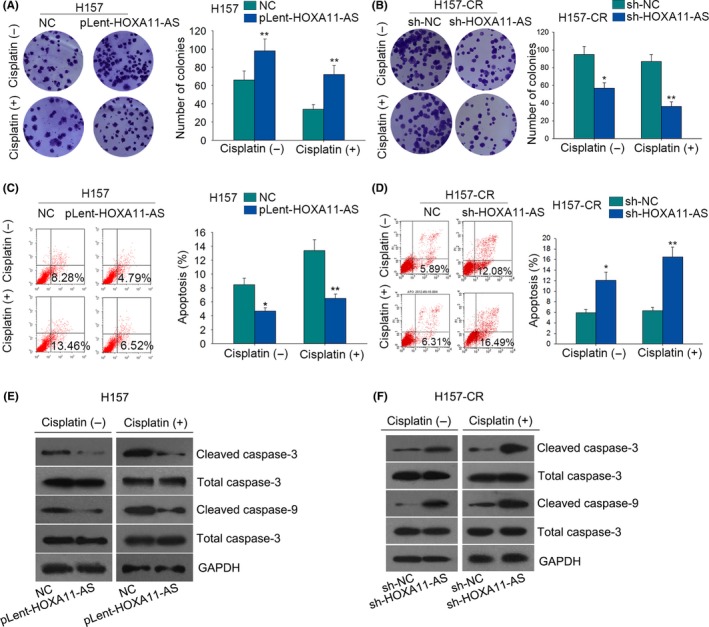
The effects of HOMEOBOX A11 antisense RNA (HOXA11‐AS) on the chemoresistance of lung adenocarcinoma (LUAD) cells. (A) Colony formation assays measured the proliferation ability of H157 cells in the cisplatin group (1 μg/mL) and non‐cisplatin group after HOXA11‐AS was overexpressed. (B) The proliferation of H157‐CR cells was evaluated in the cisplatin group (3 μg/mL) and non‐cisplatin group after HOXA11‐AS was downregulated. (C) Flow cytometry analysis examined the apoptotic rate in both cisplatin and non‐cisplatin groups after H157 cells were transfected with pLent‐HOXA11‐AS. (D) Flow cytometry analysis examined the apoptotic rate in both cisplatin and non‐cisplatin groups after H157‐CR cells were transfected with short hairpin RNA (sh)‐HOXA11‐AS. (E) Western blot analysis tested the protein levels of apoptosis‐related proteins in the cisplatin group or in the non‐cisplatin group after H157 cells were treated with pLent‐HOXA11‐AS. (F) Western blot analysis tested the protein levels of apoptosis‐related proteins in the cisplatin group or in the non‐cisplatin group after H157‐CR cells were treated with sh‐HOXA11‐AS. Error bars represent the mean ± SD of at least three independent experiments. *P < .05, **P < .01 vs control group
3.3. HOXA11‐AS affects the metastasis and EMT process in parental cells and cisplatin‐resistant cells
Here, we continued to explore the influence of HOXA11‐AS on cell metastasis and epithelial mesenchymal transition (EMT). According to the results of the transwell assay, metastatic ability of A459 and H157 cells was significantly enhanced by HOXA11‐AS overexpression (Figure 4A). However, the metastatic ability of A459‐CR and H157‐CR cells was weakened by silenced HOXA11‐AS (Figure 4B). To investigate the effects of HOXA11‐AS on EMT process, quantitative RT‐PCR and western blot assay were used to test the level of EMT markers (E‐cadherin, β‐catenin, N‐cadherin, Vimentin) and EMT‐related transcription factors (Snail, Twist). As a result, overexpression of HOXA11‐AS led to increased levels of Snail, Twist, N‐cadherin, Vimentin but decreased the levels of E‐cadherin and β‐catenin (Figure S1A‐B). In parental cells, we observed that HOXA11‐AS overexpression promoted the formation of EMT phenotype. As for cisplatin‐resistant cells, silenced HOXA11‐AS induced EMT to MET (Figure 4C‐D). Finally, the result of immunofluorescence is consistent with that of quantitative RT‐PCR and western blot assay (Figure 4E‐F).
Figure 4.
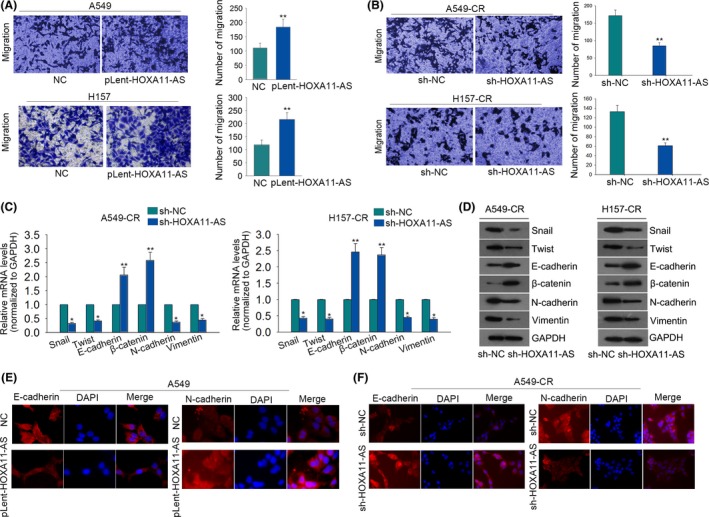
HOMEOBOX A11 antisense RNA (HOXA11‐AS) affects the metastasis and epithelial mesenchymal transition (EMT) process in parental cells and cisplatin‐resistant cells. (A) Metastatic ability of A549 and H157 cells was assessed in response to HOXA11‐AS overexpression. (B) The effect of short hairpin RNA (sh)‐HOXA11‐AS on migratory ability of A459‐CR and H157‐CR cells was measured with transwell assay. (C‐D) EMT‐related transcription factors and EMT markers were detected in HOXA11‐AS‐downregulated A459‐CR and H157‐CR cells. (E‐F) Immunofluorescence was used to validate the effects of HOXA11‐AS overexpression or knockdown on EMT progress. Error bars represent the mean ± SD of at least three independent experiments. *P < .05, **P < .01 vs control group
3.4. HOXA11‐AS is a sponge of miR‐454‐3p
It is widely acknowledged that lncRNAs can act as ceRNA to increase the expressions of the underlying target mRNAs through sponging with miRNAs in various cancers. To determine the possible mechanism of HOXA11‐AS, we first figured out the location of HOXA11‐AS. By applying subcellular fractionation assays (Figure 5A), we revealed that HOXA11‐AS was mainly located in the cytoplasm, indicating that HOXA11‐AS exerted its function at the post‐transcriptional level. To obtain the miRNAs possibly binding to HOXA11‐AS, we utilized online bioinformatics analysis (http://starbase.sysu.edu.cn/index.php) and found that about 30 miRNAs could sponge with HOXA11‐AS (Table S2). To make further confirmation, we applied quantitative RT‐PCR analysis to measure the expression changes of these miRNAs when HOXA11‐AS was downregulated. It was found that only miR‐454‐3p could be observably affected by silenced HOXA11‐AS (Table S2), indicating that miR‐454‐3p might be involved in the function mediated by HOXA11‐AS. Later, RIP assay was performed in A549‐CR and H157‐CR cells by using the antibody against Ago2. As elucidated in Figure 5B, HOXA11‐AS and miR‐454‐3p were obviously enriched in Ago2‐immunoprecipitation. According to the bioinformatics analysis result, we uncovered that miR‐454‐3p could bind to the wild type HOXA11‐AS, but not to mutated HOXA11‐AS (Figure 5C). Furthermore, the sequence of wild type HOXA11‐AS (HOXA11‐AS‐WT) and the sequence of mutant type HOXA11‐AS (HOXA11‐AS‐MUT) were inserted into pmirGLO vector. Next, pmirGLO‐HOXA11‐AS‐WT and pmirGLO‐HOXA11‐AS‐MUT were transfected into HEK‐293T and A549‐CR cells along with miR‐454‐3p mimics and miR‐NC: 48 hours later, the luciferase activity of wild type HOXA11‐AS (HOXA11‐AS‐WT) was largely decreased by miR‐454‐3p mimics, while the luciferase activities of the mutant HOXA11‐AS (HOXA11‐AS‐MUT) and empty vector were almost unchanged (Figure 5D). Therefore, we confirmed the combination between HOXA11‐AS and miR‐454‐3p. Subsequently, quantitative RT‐PCR revealed that miR‐454‐3p was expressed lower in cisplatin‐resistant LUAD cells than that in the parental LUAD cells (Figure 5E). Then, we continued to detect whether miR‐454‐3p can affect the expression of HOXA11‐AS. The expression of HOXA11‐AS was decreased in A549‐CR and H157‐CR cells in response to miR‐454‐3p mimics (Figure 5F), whereas, the expression of HOXA11‐AS in A549 and H157 cells was enhanced by miR‐454‐3p inhibitor (Figure 5G). Taken together, HOXA11‐AS is a sponge of miR‐454‐3p.
Figure 5.
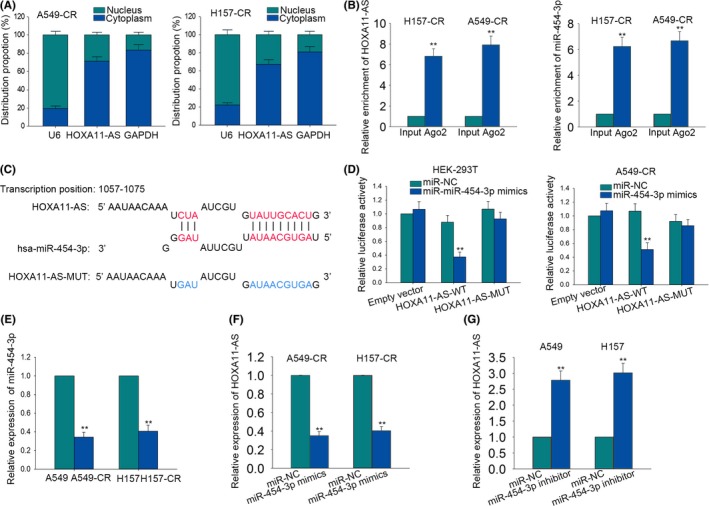
HOMEOBOX A11 antisense RNA (HOXA11‐AS) is a sponge of miR‐454‐3p. (A) Subcellular fractionation assays revealed that HOXA11‐AS was mainly located in cytoplasm. (B) RNA immunoprecipitation (RIP) assays were performed in A549‐CR and H157‐CR cells by using anti‐Ago2. (C) The binding sites between wild type HOXA11‐AS (HOXA11‐AS‐WT) or mutant type HOXA11‐AS (HOXA11‐AS‐MUT) and miR‐454‐3p were predicted with bioinformatics analysis. (D) Dual luciferase reporter assay was applied to demonstrate the combination between HOXA11‐AS and miR‐454‐3p in A549‐CR and H157‐CR cells. (E) The higher expression level of miR‐454‐3p was tested in parental cells by using quantitative RT‐PCR. (F) The expression level of HOXA11‐AS was examined in A549‐CR and H157‐CR cells in response to miR‐454‐3p mimics. (G) The expression level of HOXA11‐AS was examined in parental cells in response to miR‐454‐3p inhibitor. Error bars represent the mean ± SD of at least three independent experiments. **P < .01 vs control group
3.5. HOXA11‐AS positively modulates Stat3 through sequestering miR‐454‐3p
It has been reported that miR‐454‐3p can inhibit chondrosarcoma cell growth by targeting Stat3.17 Here, we hypothesized that the miR‐454‐3p‐Stat3 axis can interact with HOXA11‐AS under a ceRNA pattern. We obtained the binding sites between miR‐454‐3p and Stat3 by searching on the internet (http://34.236.212.39/microrna/microrna/getGeneForm.do) (Figure 6A). Next, the sequence of wild type Stat3 (Stat3‐WT) and the sequence of mutant type Stat3 (Stat3‐MUT) were inserted into pmirGLO vector. Next, pmirGLO‐ Stat3‐WT and pmirGLO‐Stat3‐MUT were transfected into HEK‐293T and A549‐CR cells along with miR‐NC, miR‐454‐3p mimics and pLent‐HOXA11‐AS1 or pLent‐HOXA11‐AS‐MUT. MiR‐454‐3p mimics decreased the luciferase activity of Stat3‐WT. However, the luciferase activity of Stat3‐MUT and empty vector was not obviously changed by miR‐454‐3p mimics. (Figure 6B). Moreover, the effects of miR‐454‐3p mimics on the luciferase activity of Stat3‐WT were partially abolished by pLent‐HOXA11‐AS but not pLent‐HOXA11‐AS‐MUT (Figure 6B). Next, quantitative RT‐PCR and western blot analysis were conducted in parental cells and cisplatin‐resistant cells. The level of Stat3 in parental cells was increased by pLent‐HOXA11‐AS and was decreased again by miR‐454‐3p mimics (Figure 6C‐D). The level of Stat3 in cisplatin‐resistant cells was decreased by sh‐HOXA11‐AS and was increased again by miR‐454‐3p inhibitor (Figure 6E‐F). These results showed that HOXA11‐AS regulated Stat3 by sequestering miR‐454‐3p.
Figure 6.
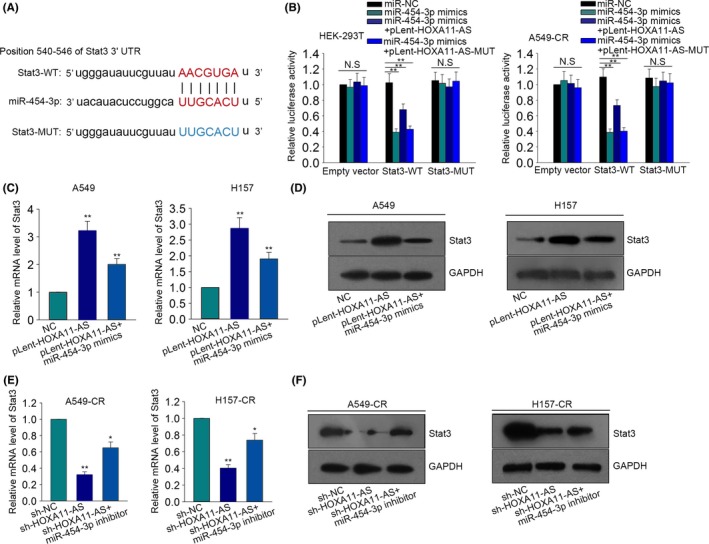
HOMEOBOX A11 antisense RNA (HOXA11‐AS) positively modulates signal transducer and activator of transcription 3 (Stat3) through sequestering miR‐454‐3p. (A) We obtained the binding sites between Stat3 and miR‐454‐3p by searching the internet. (B), The dual luciferase reporter assay was made in HEK293T cells and A549‐CR cells to further prove the combination among HOXA11‐AS, miR‐454‐3p and Stat3. (C‐D) Quantitative RT‐PCR and western blot assay were designed and performed to detect the expression of Stat3 in A549 and H157 cells in which pLent‐HOXA11‐AS and miR‐454‐3p mimics were transfected. (E‐F) The expression of Stat3 was detected in A549‐CR and H157‐CR cells which were transfected with short hairpin RNA (sh)‐HOXA11‐AS and miR‐454‐3p inhibitor. Error bars represent the mean ± SD of at least three independent experiments. *P < .05, **P < .01 vs control group
3.6. Effect of HOXA11‐AS‐miR‐454‐3p‐Stat3 axis on cisplatin resistance of LUAD cells
To prove the function of this ceRNA pathway in our study, we designed a series of rescue assays in parental cells (A459 and H157) transfected with pLent‐HOXA11‐AS and cisplatin‐resistant cells (A459‐CR and H157‐CR) transfected with sh‐HOXA11‐AS. As illustrated in Figure 7A, increased IC50 value of HOXA11‐AS‐upregulated parental cells was decreased by miR‐454‐3p mimics, which was increased again by Stat3, whereas, the decreased IC50 value of HOXA‐11‐AS‐downregulated cisplatin‐resistant cells was increased by miR‐454‐3p inhibitor, which was reversed again by silenced Stat3 (Figure 7B). Subsequently, we performed a western blot assay to analyze the protein level of apoptosis‐related proteins (cleaved caspase‐3 and cleaved caspase‐9) in parental cells and cisplatin‐resistant cells. The decreased levels of cleaved caspase‐3 and cleaved caspase‐9 in HOXA11‐AS‐upregulated parental cells were increased again by miR‐454‐3p mimics, which was attenuated by Stat3 (Figure 7C). The increased protein level of cleaved caspase‐3 and cleaved caspase‐9 in HOXA11‐AS‐downregulated cisplatin‐resistant cells was cut down by the transfection with miR‐454‐3p inhibitor, while this tendency was reversed by sh‐Stat3 (Figure 7D). Western blot was also utilized to detect the changes in EMT process. The EMT process in transfected parental cells was reversed by miR‐454‐3p mimics, while this tendency was attenuated by Stat3 (Figure 7E). In cisplatin‐resistant cells, the inhibitory effects of sh‐HOXA11‐AS on EMT progress were partially attenuated by miR‐454‐3p inhibitor, while sh‐Stat3 reversed the effects of miR‐454‐3p inhibitor (Figure 7F).
Figure 7.
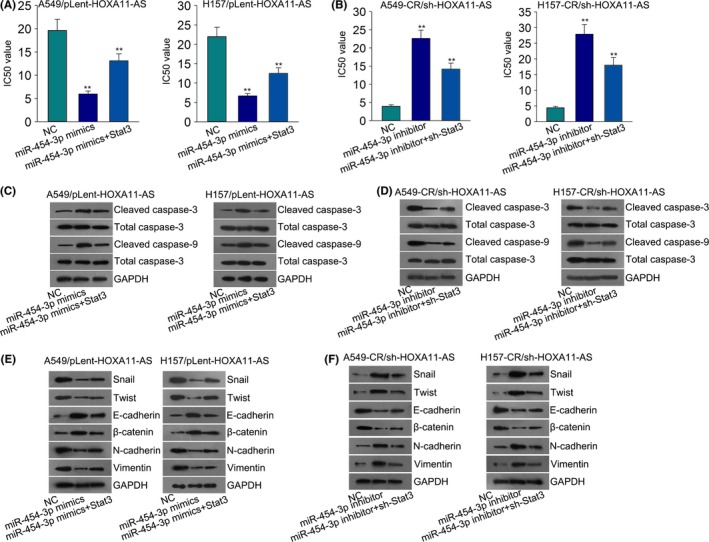
The effect of HOMEOBOX A11 antisense RNA (HOXA11‐AS)‐miR‐454‐3p‐Stat3 (signal transducer and activator of transcription 3) axis on cisplatin resistance of lung adenocarcinoma (LUAD) cells. (A) The 50% inhibitory concentration (IC50) value of A549/pLent‐HOXA11‐AS and H157/pLent‐HOXA11‐AS cells was measured after co‐transfection with miR‐454 mimics and Stat3 expression vector. (B) The IC50 value of indicated cisplatin‐resistant cells was measured. (C‐D) Apoptosis‐related proteins were detected in indicated parental cells and cisplatin‐resistant cells after transfection. (E‐F) Epithelial mesenchymal transition (EMT)‐related transcription factors and EMT‐markers were detected in indicated cells after the same transfection. Error bars represent the mean ± SD of at least three independent experiments. **P < .01 vs control group
4. DISCUSSION
As one of the most frequent malignant tumors for human beings, NSCLC is well‐known for its high mortality and poor survival rate worldwide.1 Although various chemotherapy drugs have been developed to treat lung cancers, the resistance to these drugs is a vital factor in the poor prognosis of lung cancer patients.18, 19 Cisplatin is a common chemical compound, which is widely used in chemotherapy for its high efficiency in the early therapeutic stage. Nevertheless, with the development of malignance, the sensitivity of cancerous cells to cisplatin is weakened, thus resistance to this drug emerges. This situation can be observed in many therapeutic processes of cancers. Therefore, it is urgent to study the specific molecular mechanism by which cisplatin resistance is promoted. The inhibition of the apoptosis and the promotion of proliferation have all been identified as possible mechanisms of drug resistance for cancers. Much evidence has suggested that lncRNAs partly affect drug resistance in cancer cells through these two mechanisms.20, 21, 22, 23, 24, 25 According to TCGA database, lncRNA HOXA11‐AS was expressed much higher in LUAD tissues than that in corresponding non‐tumor tissues. Upregulation of HOXA11‐AS predicts poor overall survival for LUAD patients. Therefore, we chose HOXA11‐AS for further study. Functional assays demonstrated that HOXA11‐AS enhanced resistance of parental LUAD cells and cisplatin‐resistant LUAD cells to cisplatin.
LncRNAs have been widely reported to target to miRNAs, thus promoting cisplatin resistance in various cancers. For instance, lncRNA UCA1 intensifies cisplatin resistance in oral squamous cell carcinoma by repressing the expression of miR‐18426 lncRNA CCAT1 increases cisplatin resistance in cell lines of gastric cancer via targeting miR‐130a‐3p.27 CASC2/miR‐21 pathway has been found to be able to affect the sensitivity to cisplatin in cervical cancer.28 All the above reports provide proof to demonstrate the interaction between lncRNAs and miRNAs in the cisplatin resistance of cancers. Here, miR‐454‐3p was certified to be a target miRNA of HOXA11‐AS by applying bioinformatics analysis and mechanism experiments. Since lncRNAs can act as a ceRNA by competitively binding miRNAs, we detected the localization of HOXA11‐AS. As a result, quantitative RT‐PCR revealed that the expression of HOXA11‐AS was enriched in the cytoplasm of cisplatin‐resistant LUAD cells. Subsequently, Stat3 was demonstrated to be a target of miR‐454‐3p and positively regulated by HOXA11‐AS. All these results demonstrated that HOXA11‐AS enhanced cisplatin resistance of LUAD cells via regulating the miR‐454‐3p‐Stat3 axis. Finally, rescue assays were designed and carried out in parental cells and cisplatin‐resistant cells. In conclusion, HOXA11‐AS exerts an oncogenic role to affect cisplatin resistance in LUAD via acting as a ceRNA of the miR‐454‐3p‐Stat3 axis. All findings in this study may help to reverse chemoresistance of LUAD, thereby providing novel therapeutic targets for LUAD.
CONFLICT OF INTEREST
There are no conflicts of interest to disclose.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81372396 and 81570186).
Zhao X, Li X, Zhou L, et al. LncRNA HOXA11‐AS drives cisplatin resistance of human LUAD cells via modulating miR‐454‐3p/Stat3. Cancer Sci. 2018;109:3068–3079. 10.1111/cas.13764
Xia Zhao, Xiaoyou Li and Leilei Zhou contributed equally to this work.
Contributor Information
Jifeng Feng, Email: feng_jifeng@126.com.
Ping Chen, Email: doc_pchen@163.com.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9‐29. [DOI] [PubMed] [Google Scholar]
- 3. Cheng L, Alexander RE, Maclennan GT, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol 2012;25:347‐369. [DOI] [PubMed] [Google Scholar]
- 4. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277‐300. [DOI] [PubMed] [Google Scholar]
- 5. Riaz SP, Luchtenborg M, Coupland VH, Spicer J, Peake MD, Moller H. Trends in incidence of small cell lung cancer and all lung cancer. Lung Cancer. 2012;75:280‐284. [DOI] [PubMed] [Google Scholar]
- 6. Murray N, Turrisi AT III. A review of first‐line treatment for small‐cell lung cancer. J Thorac Oncol. 2006;1:270‐278. [DOI] [PubMed] [Google Scholar]
- 7. Sarvi S, Mackinnon AC, Avlonitis N, et al. CD133 + cancer stem‐like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res. 2014;74:1554‐1565. [DOI] [PubMed] [Google Scholar]
- 8. Lu Q, Yu T, Ou X, Cao D, Xie T, Chen X. Potential lncRNA diagnostic biomarkers for early gastric cancer. Mol Med Rep. 2017;16:9545‐9552. [DOI] [PubMed] [Google Scholar]
- 9. Vafaee F, Colvin EK, Mok SC, Howell VM. Functional prediction of long non‐coding RNAs in ovarian cancer‐associated fibroblasts indicate a potential role in metastasis. Sci Rep. 2017;7:10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J, Xue W, Lv J, Han P, Liu Y, Cui B. Identification of potential long non‐coding RNA biomarkers associated with the progression of colon cancer. Oncotarget. 2017;8:75834‐75843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Wen X, Wang L, et al. LncRNA ZEB1‐AS1 predicts unfavorable prognosis in gastric cancer. Surg Oncol. 2017;26:527‐534. [DOI] [PubMed] [Google Scholar]
- 12. Chen D, Sun Q, Zhang L, et al. The lncRNA HOXA11‐AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR‐125a‐5p in liver metastasis of colorectal cancer. Oncotarget. 2017;8:70642‐70652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu B, Li J, Liu X, et al. Long non‐coding RNA HOXA11‐AS promotes the proliferation HCC cells by epigenetically silencing DUSP5. Oncotarget. 2017;8:109509‐109521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11‐AS/miR‐214‐3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504‐1513. [DOI] [PubMed] [Google Scholar]
- 15. Li W, Jia G, Qu Y, Du Q, Liu B, Liu B. Long Non‐Coding RNA (LncRNA) HOXA11‐AS Promotes Breast Cancer Invasion and Metastasis by Regulating Epithelial‐Mesenchymal Transition. Med Sci Monitor. 2017;23:3393‐3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barr MP, Gray SG, Hoffmann AC, et al. Generation and characterisation of cisplatin‐resistant non‐small cell lung cancer cell lines displaying a stem‐like signature. PLoS One. 2013;8:e54193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang J, Li L, Huang W, et al. MiR‐429 increases the metastatic capability of HCC via regulating classic Wnt pathway rather than epithelial‐mesenchymal transition. Cancer Lett. 2015;364:33‐43. [DOI] [PubMed] [Google Scholar]
- 18. Orfanelli U, Wenke AK, Doglioni C, Russo V, Bosserhoff AK, Lavorgna G. Identification of novel sense and antisense transcription at the TRPM2 locus in cancer. Cell Res. 2008;18:1128‐1140. [DOI] [PubMed] [Google Scholar]
- 19. Voulgari A, Pintzas A. Epithelial‐mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochem Biophys Acta. 2009;1796:75‐90. [DOI] [PubMed] [Google Scholar]
- 20. Ma YF, Liang T, Li CR, Li YJ, Jin S, Liu Y. Long non‐coding RNA HNF1A‐AS1 up‐regulation in non‐small cell lung cancer correlates to poor survival. Eur Rev Med Pharmacol Sci. 2016;20:4858‐4863. [PubMed] [Google Scholar]
- 21. Xiong J, Li J, Yang Q, Wang J, Su T, Zhou S. Gossypol has anti‐cancer effects by dual‐targeting MDM2 and VEGF in human breast cancer. Breast Cancer Res. 2017;19:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Idogawa M, Ohashi T, Sasaki Y, Nakase H, Tokino T. Long non‐coding RNA NEAT1 is a transcriptional target of p53 and modulates p53‐induced transactivation and tumor‐suppressor function. Int J Cancer 2017;140:2785‐2791. [DOI] [PubMed] [Google Scholar]
- 23. Zhang M, Dong BB, Lu M, et al. miR‐429 functions as a tumor suppressor by targeting FSCN1 in gastric cancer cells. Onco Targets and Therapy. 2016;9:1123‐1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang J, Liu Y, He A, et al. Hsa‐miR‐429 promotes bladder cancer cell proliferation via inhibiting CDKN2B. Oncotarget. 2017;8:68721‐68729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao P, Liu W, Zhou H. miR‐429 promotes the proliferation of non‐small cell lung cancer cells via targeting DLC‐1. Oncol Lett. 2016;12:2163‐2168. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Wang P, Cao J, Liu S, et al. Upregulated microRNA‐429 inhibits the migration of HCC cells by targeting TRAF6 through the NF‐kappaB pathway. Oncol Rep. 2017;37:2883‐2890. [DOI] [PubMed] [Google Scholar]
- 27. Liu D, Xia P, Diao D, et al. MiRNA‐429 suppresses the growth of gastric cancer cells in vitro. J Biomed Res. 2012;26:389‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu W, An J, Li K, Hou H. MiR‐429 regulates gastric cancer cell invasiveness through ZEB proteins. Tumour Biol. 2015;37:15575‐15581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


