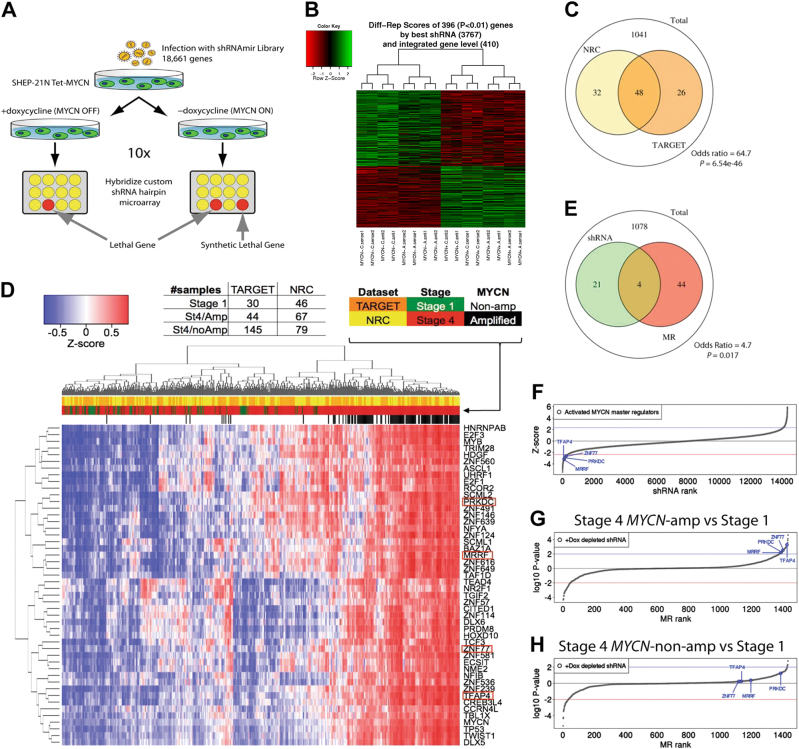
Fig. 1.
TFAP4 is a synthetic lethal candidate and master regulator of MYCN-amplified neuroblastoma. a Schema of whole-genome shRNA screening in SHEP21N neuroblastoma cells. Abundance of an individual shRNA was determined by hybridization to a customized microarray. Synthetic lethal candidates are the shRNAs depleted in MYCN ON population only; b Heatmap of differentially expressed genes in MYCN ON vs. MYCN OFF conditions. Abundance of shRNAs against 396 genes significantly changed (P < 0.01), with 218 genes depleted in the MYCN ON population. c Venn diagram of overlapping MRs activated in MYCN-amplified tumors (P < 0.01), between NRC (yellow circle) and TARGET (orange circle) cohorts. There are 1041 genes tested in both data sets. d VIPER single sample activity heatmap for 48 MRs of MYCN-amplified neuroblastoma from TARGET and NRC intersected results. Blue–red colors indicate z-score scaled enrichment scores from single sample analysis. The four MRs that are synthetic lethal with MYCN are highlighted in red rectangles. e Venn diagram of overlapping transcriptional regulators in the synthetic lethal screen and MRs in MYCN-amplified neuroblastoma. Twenty-five transcriptional regulators that were significantly depleted in the shRNA screen (green circle) and 48 activated MRs identified by MARINA algorithm (red circle). There are 1078 genes tested in both data sets. f Four candidate activated MYCN MRs (TFAP4, MRRF, PRKDC, ZNF77), were depleted in the shRNA screen. shRNA differential change was ranked by z-score with significantly depleted shRNAs having a z-score of < −2.326 (one-tiled P < 0,01) (red line). g 1344 transcriptional regulators were ranked by the significance of differential expression of its regulon in MYCN-amplified compared with Stage 1. The curve represents the distribution with the most repressed MRs at left tail end and the most activated MRs at right tail end. The candidate MYCN MRs (TFAP4, MRRF, PRKDC, ZNF77), were significantly activated (log10 P value, > 2, blue line). h 1344 transcriptional regulators were ranked by the significance of differential expression of its regulon in non-MYCN-amplified stage 4 compared with Stage 1. None of the candidate MYCN MRs (TFAP4, MRRF, PRKDC, ZNF77) were significantly activated (log10 P value, > 2, blue line)