Abstract
Telomere maintenance by telomerase activity supports the infinite growth of cancer cells. MST-312, a synthetic telomerase inhibitor, gradually shortens telomeres at non-acute lethal doses and eventually induces senescence and apoptosis of telomerase-positive cancer cells. Here we report that MST-312 at higher doses works as a dual inhibitor of telomerase and DNA topoisomerase II and exhibits acute anti-proliferative effects on cancer cells and xenografted tumours in vivo. Our cell-based chemical fingerprinting approach revealed that cancer cells with shorter telomeres and lower expression of lamin A, a nuclear architectural protein, exhibited higher sensitivity to the acute deleterious effects of MST-312, accompanied by formation of telomere dysfunction-induced foci and DNA double-strand breaks. Telomere elongation and lamin A overexpression attenuated telomeric and non-telomeric DNA damage, respectively, and both conferred resistance to apoptosis induced by MST-312 and other DNA damaging anticancer agents. These observations suggest that sufficient pools of telomeres and a nuclear lamina component contribute to the cellular robustness against DNA damage induced by therapeutic treatment in human cancer cells.
Introduction
Telomeres are protective structures at the ends of eukaryotic linear chromosomes that consist of telomeric DNA, (TTAGGG)n, and binding proteins, such as shelterin1. Telomeric DNA has a single-stranded 3′-overhang that forms a protective t-loop structure2. Because the DNA polymerase-dependent replication machinery cannot replicate the ends of linear DNAs (the end replication problem), telomeres shorten after each round of DNA replication. Critically shortened telomeres lose the ability to form the t-loop structure and are then recognised as deleterious DNA double-strand breaks3. This condition leads to a block in DNA replication and cell division, resulting in a permanent growth arrest, called replicative senescence. Thus, in normal cells, telomere shortening and the accompanying inhibition of cell division blocks the growth of cells and functions as a tumour suppressor mechanism4. In contrast, dysfunction of telomeres can drive genomic instability, which can facilitate tumour progression5.
In vast majority of human cancer cells, the end replication problem is solved by the maintenance of telomeres by the telomerase enzyme6, which consists of the hTERT catalytic subunit and the RNA template (called hTR or hTERC, in humans). While hTR is ubiquitously expressed in broad ranges of tissues, hTERT is the limiting factor for telomerase activity7. Recent cancer genome analyses have identified hTERT promoter mutations that lead to elevated transcriptional activation of the gene in many cancers8,9. These cancer cells harbouring mutations in hTERT acquire unlimited proliferative potential10. Therefore, telomerase has been postulated as a therapeutic target for cancer. Indeed, various telomerase inhibitors, including epigallocatechin gallate (EGCG), a major tea catechin, and its synthetic derivative, MST-312, have been found or developed11–15. These inhibitors gradually shorten telomeres in cancer cells to eventually induce cell crisis11–16. Among them, imetelstat is currently under clinical investigation17–19.
In general, most anticancer drugs exhibit rapid anti-proliferative activities in cancer cells. However, telomerase inhibitors are an exception, for the following reasons. First, because the end replication problem only occurs during DNA synthesis, telomere shortening depends on cell growth. Therefore, the process of inhibiting telomerase to allow for telomere shortening requires the treatment of cancer cells with telomerase inhibitors at low concentrations that do not induce inhibition of cell growth. In the presence of telomerase inhibitors at such low concentrations, cancer cells would continue to grow but their telomeres would gradually shorten, suggesting that the anti-proliferative effect of telomerase inhibitors on cancer cells would only be seen after the shortened telomeres lose their end-capping functions. This suggests that the initial telomere length and the rate of telomere shortening would determine the duration of the inhibitor treatment required for the induction of telomere crisis in target cells. We previously showed that elevated expression of a telomeric poly(ADP-ribose) polymerase (PARP) tankyrase 1, which enhances telomerase access to telomeres20,21, blocks efficient telomere shortening by the telomerase inhibitor MST-312 at non-toxic low doses and confers resistance to the compound16. Conversely, tankyrase 1 blockade by PARP inhibitors enhances telomere shortening by MST-312 and induces earlier crisis of cells16.
Interestingly, RNA interference-mediated knockdown of the hTR template RNA component of telomerase results in a rapid deleterious effect on cancer cell growth22,23. This effect is observed independently of telomere length and is not accompanied by telomere shortening, suggesting a telomere-independent mechanism in blocking cell growth. Furthermore, accumulating evidence has shown that telomerase supports tumourigenesis and normal stem cell growth in a telomere-independent manner24–26. These observations suggest that human cancer cells are “addicted” to telomerase activity even if they have sufficiently long telomeres. By contrast, telomerase knockout mice can be born and grow fertile until the telomeres shorten over generations to reach the minimal length that is required for chromosome end capping27. Together this suggests that a telomerase inhibitor at higher doses may promptly abrogate cancer cell growth. In this situation, however, it will likely be difficult to discriminate any off-target effects of treatment with the inhibitor from its specific effect on telomerase inhibition.
In this study, we examined the effects of the synthetic telomerase inhibitor MST-312 at higher doses than those in our previous reports14,16 and focused on the molecular mechanism for the acute cancer cell growth inhibition induced by high MST-312 concentrations. Exploiting the in silico data analysis with our newly developed Telomere Fingerprint Database, we found that MST-312 at high doses works as a dual inhibitor of telomerase and DNA topoisomerase II, an enzyme that regulates the topological state of DNA. Furthermore, using MST-312 as a probe, we demonstrated that both telomere length and lamin A, an inner nuclear membrane protein, are the functional determinants for cancer cell sensitivity to MST-312 and other DNA damaging anticancer agents.
Results
MST-312 inhibits human cancer cell growth in mouse xenograft models
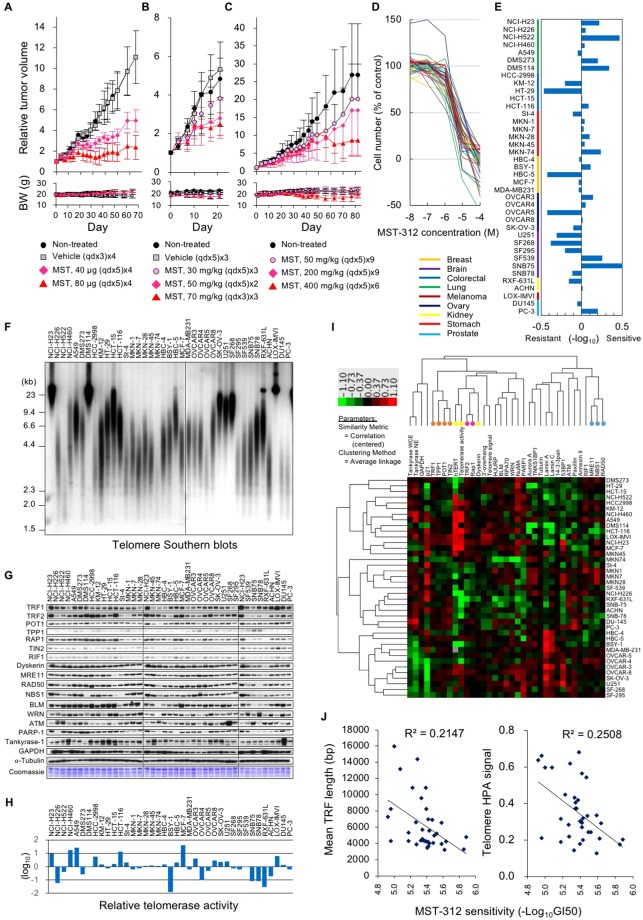
Non-acute cytotoxic doses of the telomerase inhibitor MST-312 shorten telomeres and induce senescence and apoptosis of telomerase-positive human leukaemia and solid tumour cells14,16. Meanwhile, MST-312 at higher doses promptly inhibits the proliferation of leukaemia cells14. To examine the in vivo antitumour efficacy of MST-312, we subcutaneously injected human breast cancer HBC-4 cells into nude mice and treated mice with various doses of MST-312 through various routes. In non-treated and vehicle-treated mice, the tumours grew extensively (Fig. 1A–C). In contrast, intratumoural, intravenous or oral administration of MST-312 at the maximum tolerated or lower doses retarded tumour growth. In mice receiving oral administration of 400 mg/kg MST-312, while a maximum 16.7% body weight loss was observed at day 56, the body weight recovered and returned to levels higher than the original body weight at day 82 (Fig. 1C). Reduction in body-weight was less than 10% during the course of treatments in case of the intratumoural (Fig. 1A) and intravenous (Fig. 1B) MST-312 administration.
Figure 1.
Acute anticancer effect of MST-312 inversely correlates with telomere length of cancer cells. (A–C) In vivo anti-tumour effect of MST-312 in mouse xenograft models. Human breast cancer HBC-4 cells were subcutaneously injected into nude mice. Mice were treated with intratumoural (A), intravenous (B) or oral administration (C) of vehicle or MST-312. Error bar indicates standard deviation. Upper and lower graphs indicate the relative tumour volume and body weight (BW) of the mice, respectively. (D) In vitro anti-proliferative effect of MST-312 on the JFCR39 panel of 39 human cancer cell lines. Cells were treated with indicated concentrations (molar) of MST-312 for 48 h and then cell numbers were quantitated. (E) Fingerprint of MST-312 sensitivity. GI50 values of MST-312 quantitated by (D) and the average of all cell lines was defined as zero. (F) Telomere blot analysis of JFCR39. Genomic DNA was prepared and subjected to Southern blot analysis with the [32P]-labelled telomeric probe to detect telomeric restriction fragments (TRFs). Two different blots were derived from the same experiment and were processed in parallel. Their border was indicated by a dotted line. (G) Expression of telomere-related proteins in JFCR39. Cell lysates were prepared and subjected to western blot analyses with indicated primary antibodies. For each antibody blot and Coomassie stain, three different blots or gels were derived from the same experiment and were processed in parallel. Their borders were indicated by dotted lines. Each blot/gel contains NCI-H23 cells as a calibration standard. Full-length blots were presented in Supplementary Fig. S4. (H) Telomerase activity in JFCR39 cells. Cell lysates were prepared and subjected to TRAP assay. Average telomerase activity of all cell lines was defined as zero. (I) Two-dimensional hierarchical cluster analysis of the telomere-related bioparameters. The clustering result was generated by Cluster (ver. 3.0) and Java TreeView (Ver. 1.1.6r4). Orange dots: four of six shelterin components (TRF1, POT1, TIN2 and TPP1); yellow dots: telomerase components (hTERT and Dyskerin) and telomerase activity; pink dots: two shelterin components (TRF2, RAP1); light blue dots: MRN complex (MRE11, NBS1 and RAD50). (J) Correlation between the cell sensitivity to MST-312 and the length of the mean TRF length (left) or telomere signals quantitated by HPA assay (right). In these graphs, the super long telomere cells (NCI-H23, DMS114 and LOX-IMVI) were excluded from the calculation of correlation coefficient.
Telomere Fingerprint Database reveals a correlation between shorter telomeres and higher susceptibility to MST-312-induced acute cell growth inhibition
To explore the acute anti-proliferative effects of MST-312, we used a human cancer cell line panel that consists of 39 cell lines derived from various tissues (JFCR39)28. Each cell line was treated with various concentrations of MST-312 for 48 h and the anti-proliferative effects were determined (Fig. 1D). The average GI50 value of the 39 cell lines was 4.7 μM. The GI50 deviation from the average was calculated for each cell line and aligned in a fingerprint (Fig. 1E).
To analyse the relationship between MST-312 sensitivity and telomere status, we quantitated various telomere-related parameters, including the single-stranded (i.e., G-tail/3′-overhang) and the double-stranded telomere length (Fig. 1F); the protein expression for shelterin components (TRF1, TRF2, POT1, TPP1, RAP1, TIN2), DNA damage response factors (RIF1, MRE11, RAD50, NBS1, 53BP1, PARP-1), tankyrase and its binding proteins (TNKS1BP1, NuMA), RecQ helicases (WRN, BLM) and other related proteins (Fig. 1G); telomerase activity (Fig. 1H); and hTERT gene expression (Fig. 1I, column 9 from the left). Two-dimensional hierarchical clustering grouped several factors according to their functional relevance (Fig. 1I). For example, components for the MRN complex, MRE11, NBS1 and RAD50 (light blue dots), were classified into the same cluster. In addition, four of six shelterin components, TRF1, POT1, TIN2 and TPP1 (orange dots), were within the same cluster, whereas the other two direct binding components, TRF2 and RAP1 (pink dots), were closely bound. Another larger cluster contained hTERT, telomerase activity, and its binding protein dyskerin (yellow dots). These observations suggest that our newly constructed telomere fingerprint database might be able to predict the relative intensity of the physical or functional interaction between various combinations of the fingerprints. There was no significant similarity between the fingerprints of MST-312 and telomere-related parameters. For example, while MST-312 is a telomerase inhibitor, the cell sensitivity to this compound did not correlate with telomerase activity (r = −0.094, P = 0.582) or hTERT mRNA expression (r = 0.131, P = 0.439) (Supplementary Fig. S1A,B).
When three cell lines, DMS114, NCI-H23 and LOX-IMVI, which possess very long telomeres [more than 20 kb, designated as super long telomere (SLT) type cells] (Fig. 1F), were excluded, we found that acute sensitivity to MST-312 correlated with the length of the double-stranded telomere DNA (Fig. 1J). The cells with shorter telomeres tended to be more susceptible to the acute deleterious effect of MST-312. Furthermore, among the cell lines with very short telomeres (i.e., the mean TRF length is less than 4 kb), there was an inverse correlation between telomerase activity and MST-312 sensitivity (r = −0.826, P = 0.0415, Supplementary Fig. S1C).
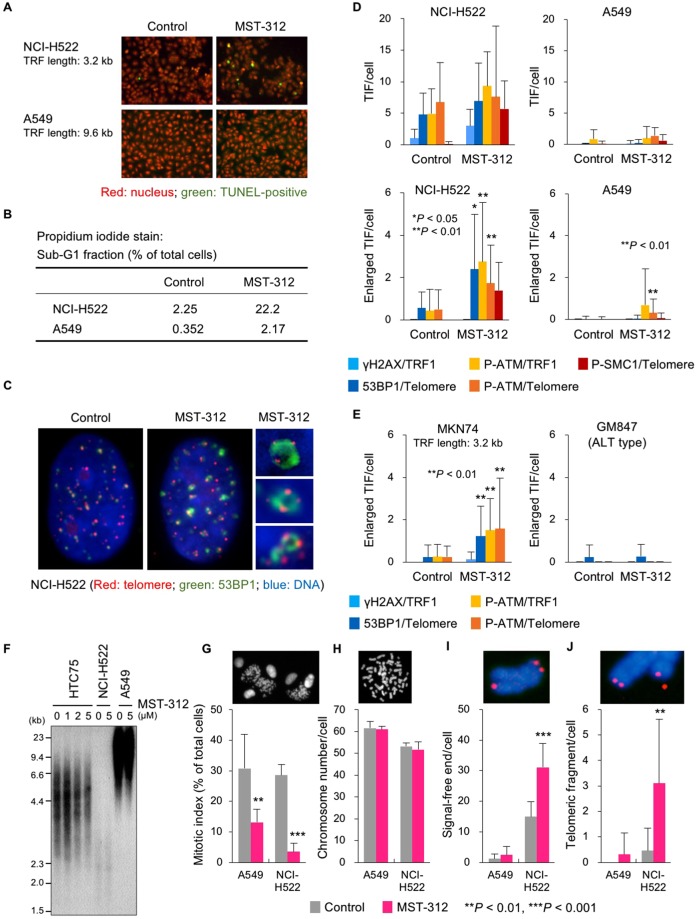
We further examined the effect of MST-312 on lung cancer NCI-H522 and A549 cells, which retain short and long telomeres with a mean telomeric restriction fragment (TRF) length of 3.2 kb and 9.6 kb, respectively. The GI50 values for MST-312 in NCI-H522 and A549 cells were 1.4 μM and 4.6 μM, respectively. Treatment with 5 μM MST-312 for 48 h induced apoptosis of NCI-H522 more frequently than A549 cells (Fig. 2A,B). Consistent with these observations, NCI-H522 cells showed enlarged telomere dysfunction-induced foci (TIF) of DNA damage response factors, such as γH2AX, 53BP1, phosphorylated ATM kinase and phosphorylated SMC1, upon treatment with 5 μM MST-312, whereas A549 did not (Fig. 2C,D). MST-312-induced enlarged TIF formation was also observed in another cell line with short telomeres, stomach MKN74 cells (mean TRF length: 3.2 kb), but not in telomerase-independent immortalised, alternative lengthening of telomere (ALT) type GM847 cells, which possess a homologous recombination-based telomere maintenance mechanism29 (Fig. 2E). Under these short term treatment conditions, telomere shortening was not detectable by Southern blot analysis (Fig. 2F). Furthermore, MST-312 reduced the mitotic index in A549 and NCI-H522 cells (Fig. 2G) without affecting chromosome numbers (Fig. 2H) and caused telomeric signal-free ends (Fig. 2I) and telomere fragmentation (Fig. 2G) only in NCI-H522 cells. These observations indicate that MST-312 treatment at high doses for 48 h causes telomere dysfunction in cancer cells with short telomeres.
Figure 2.
Telomere dysfunction induced by MST-312. (A) TUNEL assay of MST-312-treated cells. Human lung cancer NCI-H522 [mean telomeric restriction fragment (TRF) length = 3.2 kb] and A549 (mean TRF length = 9.6 kb) cells were treated with 5 μM MST-312 for 48 h and then subjected to TUNEL assay for detection of apoptotic cells (green). (B) Quantification of apoptotic cells. Cells were treated as in (A) and then stained with propidium iodide; the apoptotic sub-G1 fraction was quantitated by flow cytometry. (C) Immuno-FISH analysis of NCI-H522 cells treated with 5 μM MST-312 for 48 h. Red: telomere DNA; green: 53BP1; blue: DAPI. Right panels are magnified views of enlarged telomere dysfunction-induced foci (TIF). (D,E) Quantitation of TIF. Cells were treated as in (A), and telomeres and the indicated proteins were detected by FISH and immunofluorescence staining, respectively. Because the cells with short telomeres gave high background levels of TIF even without MST-312 treatment (D, upper left), enlarged TIF were quantitated as a hallmark of the MST-312-induced telomeric DNA damage response. Error bar indicates standard deviation. Asterisk indicates statistical significance in the difference between control and MST-312-treated cells (unpaired two-tailed t test). ALT: alternative lengthening of telomeres. (F) Telomere southern blot analysis. Cells were treated with indicated doses of MST-312 for 48 h. HTC75 fibrosarcoma cells were analysed as a control because the telomere length fluctuation of this cell line can be detected by southern blot analysis. (G) Cells were treated as in (A) and mitotic index was quantitated. (H–J) Cells in (G) were further incubated with colcemid. Metaphase spreads of chromosomes were subjected to telomere FISH, and chromosome number (H), telomeric signal-free ends (I) and telomeric fragments (J) were quantitated. Red: telomere DNA, blue: DAPI stain of chromosome DNA. Asterisk indicates statistical significance (vs. control, unpaired two-tailed t test).
MST-312 induces DNA double-strand breaks in the drug-sensitive cancer cells
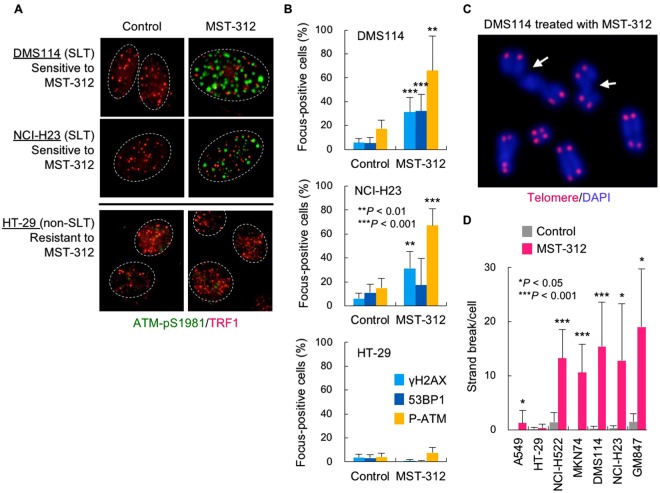
Having confirmed telomere dysfunction upon MST-312 treatment of cells that have short telomeres, we next examined the effect of MST-312 on two SLT type cell lines, DMS114 and NCI-H23. We found that both cell lines were sensitive to MST-312 (GI50 values: 2.5 μM and 1.9 μM, respectively) even though these cells possess very long telomeres (mean TRF length: > 20 kb). As shown in Fig. 3A,B, MST-312 treatment of these cells caused DNA damage foci that were not colocalised with telomeres. MST-312-resistant colorectal cancer HT-29 cells (GI50: 12 μM; mean TRF length: 7.3 kb) did not exhibit foci upon treatment. Consistent with these observations, metaphase spreads of chromosomes showed that MST-312 caused DNA double-strand breaks in MST-312-sensitive cells (NCI-H522, MKN74, DMS114 and NCI-H23) but not in MST-312-resistant cells (A549 and HT-29) (Fig. 3C,D). Furthermore, the telomerase negative (ALT type) GM847 cells also exhibited DNA double-strand breaks in response to MST-312 (Fig. 3D). These results suggest that MST-312 at a higher dose (e.g., 5 μM) than that for telomerase inhibition (~1 μM) can induce DNA double-strand breaks in a telomerase-independent manner.
Figure 3.
MST-312 induces DNA double-strand breaks in a telomerase-independent manner. (A) Non-telomeric DNA damage response induced by MST-312. MST-312-sensitive NCI-H23 [super long telomere (SLT) type: mean TRF length = 22 kb] and DMS114 cells (SLT type: mean TRF length = 23 kb) and MST-312-resistant HT-29 cells (mean TRF length = 7.3 kb) were treated with 5 μM MST-312 for 48 h. Cells were subjected to indirect immunofluorescence staining with anti-phospho-ATM (p1981) (green) and anti-TRF1 (red) antibodies. (B) Quantitative graphs of non-telomeric DNA damage foci in NCI-H23, DMS114 and HT-29 cells. Cells were treated as in (A) and immunofluorescence staining was performed with the indicated primary antibodies. Cells with more than four foci were counted as positive. (C) DNA strand breaks induced by MST-312. DMS114 cells were treated as in (A) and further incubated with colcemid for 5 h. Metaphase spreads of chromosomes were subjected to telomere FISH analysis. Red: telomere DNA; blue: DAPI stain of chromosome DNA; arrow: chromatid break. (D) Quantitation of DNA strand breaks in MST-312-treated cells. Cells were treated as in (A) and subjected to telomere FISH analysis. GM847 is a telomerase-independent ALT type cell line. Error bar indicates standard deviation. Asterisk indicates statistical significance (vs. control, unpaired two-tailed t test).
Then, we next knocked down hTERT in A549 and NCI-H522 cells and evaluated their MST-312 sensitivities. In the absence of MST-312, siRNA-mediated knockdown of hTERT reduced the growth of A549 and NCI-H522 cells to 77% and 58%, respectively, of the non-silencing control siRNA-treated cells. It has been reported that knockdown, instead of enzymatic inhibition, of telomerase exerts such an acute anticancer effect in an enzyme activity-independent manner30. We confirmed that this siRNA had no effect on the growth of telomerase-independent GM847 cells, excluding the possibility of an off-target effect (Kyotaro Hirashima and HS, unpublished observation). Under these hTERT-depleted conditions, MST-312 further inhibited the cell growth in a dose-dependent manner (Supplementary Fig. S1D,E).
COMPARE analysis of chemical fingerprints identifies MST-312 as a DNA topoisomerase II inhibitor
The ability of MST-312 to induce DNA damage in GM847 cells suggests that MST-312 has another molecular target in addition to telomerase. To identify potential targets, we performed COMPARE analysis12,31 using chemical fingerprints of MST-312 and various anticancer compounds. We found that MST-312 has a similar fingerprint to fingerprints of DNA topoisomerase II inhibitors, such as ICRF-193, ICRF-154, TAS-103 and NC190 (Fig. 4A,B). This result prompted us to perform DNA decatenation assays, which monitor DNA topoisomerase II activity to decatenate the kinetoplast DNA. As shown in Fig. 4C, MST-312 inhibited DNA topoisomerase II with an IC50 value of 2 μM, which was three-fold higher than the IC50 value for telomerase inhibition14. Downregulation of DNA topoisomerase II is a well-established mechanism for resistance to DNA topoisomerase II inhibitors32. MST-312 at 150 μM inhibited the growth of Saccharomyces cerevisiae YFK30, a drug-hypersensitive strain33 (Fig. 4D). In contrast, the DNA topoisomerase II-mutant in the YFK30 background, top2, K414N, was resistant to the growth inhibitory effect of MST-312. In the control treatment, this mutant strain was also resistant to doxorubicin, another DNA topoisomerase II inhibitor. iFISH analysis revealed that DNA topoisomerase II inhibitors, such as ICRF-193, TAS-103 and etoposide, induced non-telomeric DNA damage foci that were distinct from the TIF induced by MST-312 in NCI-H522 cells (Fig. 4E, left panel). While the percentages of the ATM-pS1981 DNA damage focus-positive cells were comparable between the drug-treated cells, the percentages of the TIF-positive cells were significantly higher in MST-312-treated cells than in other drug-treated cells (Fig. 4E, right panel). MST-295, a telomerase inhibitor with telomerase inhibiting activity comparable to MST-31214, only marginally inhibited DNA topoisomerase II (Fig. 4F). MST-295 induced TIF and enlarged TIF in NCI-H522 cells, indicating telomerase inhibition (Fig. 4G). However, MST-295 did not induce DNA double-stranded breaks (Fig. 4H). These observations indicate that MST-312 used at 5 μM and higher doses works as a dual inhibitor for telomerase and DNA topoisomerase II and induces telomere dysfunction in cancer cells with short telomeres and non-telomeric DNA damage in cancer cells that do not necessarily have short telomeres.
Figure 4.
MST-312 inhibits DNA topoisomerase II. (A) COMPARE analysis of MST-312 with anticancer compounds. Similarities between chemical fingerprints of MST-312 and anticancer compounds in the JFCR39 cancer cell line panel were evaluated. (B) Correlation between sensitivities to MST-312 and DNA topoisomerase II (Topo II) inhibitors ICRF-193 (blue, R² = 0.2101), ICRF-154 (dark blue, R² = 0.24561), NC-190 (orange, R² = 0.22231) and TAS-103 (purple, R² = 0.17742). (C) In vitro Topo II enzyme assay. Kinetoplast DNA was incubated with Topo II in the presence of the indicated concentrations of the compounds and subjected to agarose gel electrophoresis. Decatenated DNA is a marker of Topo II reaction. Border of two different gels, which were derived from the same experiment and processed in parallel, was indicated by a dotted line. (D) Spotting assay of 10-fold serial dilution of the yeast parental strain YFK30 and top2 K414N mutant. The 10-fold dilutions of log-phase cells were spotted onto YPD plates containing the indicated concentrations of MST-312 or the positive control doxorubicin and then cells were cultivated at 30 °C. In the right panel, border of two different parts of the same plate was indicated by a dotted line. (E) Telomere immuno-FISH analysis. NCI-H522 cells were treated with 5 μM MST-312 or Topo II inhibitors (150 μM ICRF-193, 0.2 μM TAS-103 or 8.5 μM etoposide) for 48 h and then subjected to telomere immuno-FISH analysis. Left: representative photos. Red: telomere DNA; green: phospho-ATM (p1981); blue: DAPI stain of DNA. Right: quantitative graphs of phospho-ATM (p1981) and TIF-positive cells. Cells with more than four foci were counted as positive. (F) Effect of MST-295 on Topo II enzyme activity. (G) TIF induced by MST-295 in NCI-H522 cells. Cells were treated with 5 μM MST-295 or MST-312 for 48 h and then subjected to telomere immuno-FISH. Numbers of TIF and enlarged TIF were quantitated. (H) Effect of MST-295 on DNA double strand breaks in DMS114 cells. Cells were treated as in (G) and metaphase spreads of chromosomes were analysed for DNA double strand breaks. Error bar indicates standard deviation. Asterisk indicates statistical significance (Tukey-Kramer test).
Lamin A is a functional determinant for MST-312-induced DNA double-strand breaks
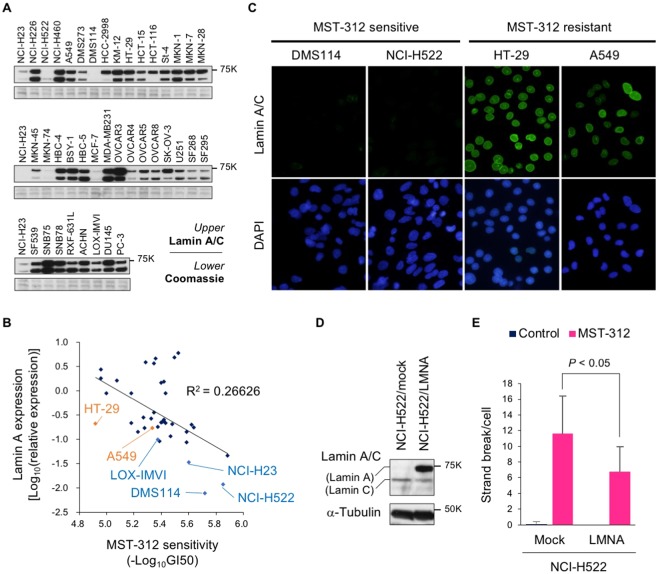
As described above, MST-312 did not induce the DNA damage response in some cancer cell lines, such as A549 and HT-29 cells. To identify the functional determinant for the susceptibility to MST-312-induced non-telomeric DNA damage, we performed BIO-COMPARE analysis34 and examined the similarity between the chemical fingerprint of MST-312 sensitivity and biological fingerprints of protein expression. We found that the expression levels for lamin A, an inner nuclear membrane protein, inversely correlated with MST-312 sensitivity with the highest correlation (r = −0.518, P = 0.0006) (Fig. 5A,B). Meanwhile, there was a weak correlation between expression levels of lamin C, a splicing valiant of LMNA gene, and MST-312 sensitivity (r = −0.389, P = 0.0137) (Supplementary Fig. S2A). Importantly, SLT-type DMS114, NCI-H23 and LOX-IMVI cells, which retain very long telomeres and are sensitive to MST-312, expressed the lowest levels of lamin A protein. NCI-H522 cells, which retain very short telomeres and are sensitive to MST-312, also expressed very low levels of lamin A. MST-312-resistant cells, such as A549 and HT-29 cells, expressed high levels of lamin A. These expression patterns were confirmed by indirect immunofluorescent staining for lamin A (Fig. 5C).
Figure 5.
Lamin A prevents MST-312-induced DNA double strand breaks. (A) Western blot analysis of lamin A/C in the JFCR39 cancer cell line panel. Coomassie brilliant blue gel staining is shown to confirm equal loading. Three different blots or gels were derived from the same experiment and were processed in parallel. Each blot/gel contains NCI-H23 cells as a calibration standard. Full-length blots/gels were presented in Supplementary Fig. S5. (B) Correlation between MST-312 sensitivity and lamin A protein expression level in JFCR39. Representative cell lines are shown in orange and blue. GI50 values were determined by the cell growth inhibition curve of 48-h treated cells in Fig. 1D. (C) Indirect immunofluorescence staining with anti-lamin A/C antibody (green). MST-312-sensitive [NCI-H522 (short telomeres) and DMS114 cells (super long telomeres)] and MST-312-resistant cells (HT-29, A549) are shown. DNA was counterstained with DAPI (blue). (D) Ectopic expression of lamin A in NCI-H522 cells. Cells were infected with retrovirus for expression of lamin A (LMNA) exogene and selected by hygromycin. The resulting cells, NCI-H522/mock and NCI-H522/LMNA, were subjected to western blot analysis. α-tubulin served as loading control. Full-length blots were presented in Supplementary Fig. S5. (E) Cells in (D) were treated with 5 μM MST-312 for 48 h and further incubated with colcemid. Metaphase spreads of chromosomes were analysed for DNA double strand breaks. Error bar indicates standard deviation. Statistical significance was evaluated by Tukey-Kramer test.
To examine whether lamin A is functionally involved in the susceptibility to MST-312-induced DNA damage, NCI-H522 cells were infected with retrovirus containing lamin A (LMNA) cDNA and selected with hygromycin (Fig. 5D). The resulting NCI-H522/LMNA cells were treated with 5 μM MST-312 for 48 h and DNA double-stranded breaks were quantitated. As shown in Fig. 5E, lamin A overexpression in NCI-H522 cells reduced the number of MST-312-induced double-stranded breaks. By contrast, lamin C overexpression did not affect the formation of DNA damage foci upon treatment with MST-312 (Supplementary Fig. S2B). These results suggest that lamin A expression is a functional determinant of the sensitivity to the DNA damage acutely induced by higher doses of MST-312.
Telomere elongation and lamin A expression alleviate MST-312-induced apoptosis
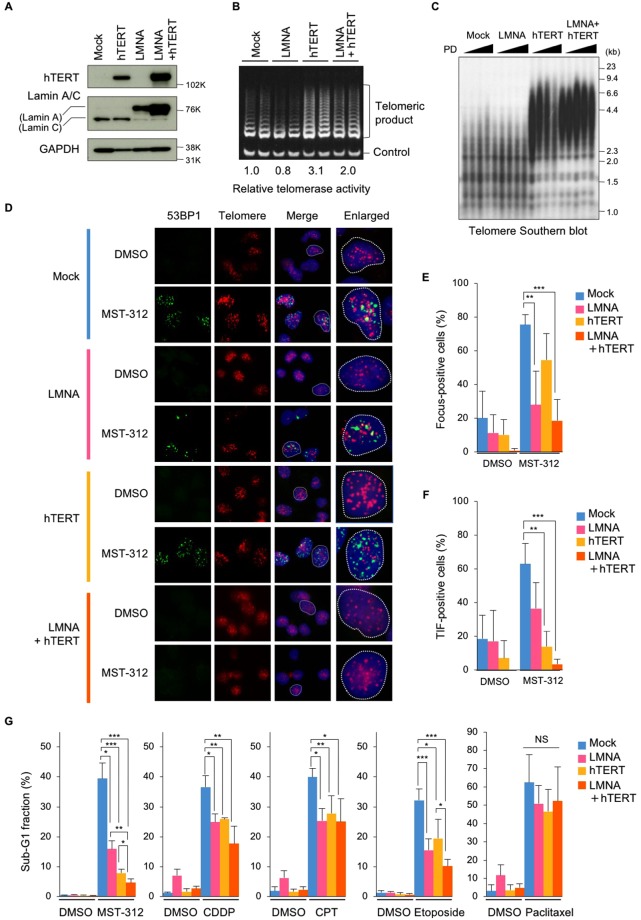
Because cancer cells with shorter telomeres tended to be more sensitive to MST-312 than cells with longer telomeres (Fig. 1J), we next transfected NCI-H522 or NCI-H522/LMNA cells with an expression vector encoding the telomerase catalytic subunit, hTERT, and upregulated the telomerase activity in the cells (Fig. 6A,B). The resulting NCI-H522/hTERT and NCI-H522/LMNA + hTERT cells exhibited elongated telomeres (Fig. 6C). Immuno-FISH analysis revealed that the telomeric and non-telomeric DNA damage induced by MST-312 were reduced by lamin A and hTERT expression, respectively, and simultaneous overexpression of the two proteins eliminated the DNA damage foci in the nuclei (Fig. 6D–F). Consistent with these observations, expression of either lamin A or hTERT protein reduced the levels of apoptosis induced by MST-312 in NCI-H522 cells, and simultaneous overexpression of both proteins further inhibited apoptosis (Fig. 6G). Lamin A and hTERT also reduced the rates of apoptosis induced by DNA damaging agents, such as cisplatin, camptothecin and etoposide, but not by the anti-microtubule agent paclitaxel (Fig. 6G). These observations indicate that telomere length and lamin A expression are the critical determinants for telomeric and non-telomeric DNA damaging agents.
Figure 6.
Telomere elongation and lamin A expression confers resistance to MST-312. (A,B) Telomere elongation and lamin A expression in NCI-H522 cells. Cells were infected with hTERT and/or LMNA expressing retrovirus. Cells were subjected to western blot analysis for detection of hTERT and lamin A expression (A) and TRAP assay for detection of telomerase activity (B). For (A) full-length blots were presented in Supplementary Fig. S5. (C) Telomere southern blot analysis. Genomic DNA was prepared and subjected to southern blot analysis with [32P]-labelled telomeric probe. (D) Telomere immuno-FISH analysis. Cells were treated with 5 μM MST-312 for 48 h and subjected to immuno-FISH analysis. Green: 53BP1; red: telomere DNA; blue: DAPI stain of DNA. (E) Quantitation of DNA damage foci in (D). Cells with more than four 53BP1 foci were counted as positive. (F) Quantitation of telomeric DNA damage (TIF and enlarged TIF) in (D). Cells with more than four TIF were counted as positive. (G) Apoptosis induced by MST-312 and anticancer drugs. Cells were treated with 5 μM MST-312, 3.3 μM cisplatin (CDDP), 10 nM camptothecin (CPT), 1.6 μM etoposide or 5 nM paclitaxel for 96 h. Cells were stained with propidium iodide and the apoptotic sub-G1 fraction of the cell cycle was quantitated by flow cytometry. Error bar indicates standard deviation of four (E,F) or at least three (G) experiments. Statistical significance was evaluated by Tukey-Kramer test. *P < 0.05, **P < 0.01, ***P < 0.001. NS: not significant.
Lamin A overexpression not only reduced lamin C expression but also slightly reduced telomerase activity (Fig. 6A,B), suggesting the possibility that lamin C expression plays a supportive role for telomerase activity. Then, we reconstituted lamin C expression in NCI-H522/mock and NCI-H522/LMNA cells and quantified the telomerase activity (Supplementary Fig. S3). The results showed that lamin C expression does not enhance telomerase activity in NCI-H522/mock cells (lanes 1/2 and 5/6). Meanwhile, a slight downregulation of telomerase activity in NCI-H522/LMNA cells was marginally rescued by lamin C expression (lanes 3/4 and 7/8). These observations suggest that lamin C supports telomerase activity to some extent, but it is not sufficient to rescue the MST-312-induced double-stranded breaks (Supplementary Fig. S2).
Discussion
In cultured cancer cells, telomerase inhibitors can allow for telomerase shortening, restoring the end replication problem, and inducing eventual telomere crisis14. This pharmacological effect is achieved by using intentionally low doses of telomerase inhibitors that would not induce acute cytotoxicity. Here we abandoned the classical idea that telomerase inhibitors should be evaluated at such low doses. As shown in Fig. 1D, treatment with the telomerase inhibitor MST-312 at higher doses exhibited acute anti-proliferative effects. Upon investigating the molecular mechanism for this rapid anticancer effect of MST-312, we found that the compound, when used at high doses, is a dual inhibitor of telomerase and DNA topoisomerase II and can induce telomeric and non-telomeric DNA damage.
Exploiting the telomere fingerprints of 39 human cancer cell lines, we found that cancer cell lines with shorter telomeres are more sensitive to the acute deleterious effects of MST-312. This result makes sense because shorter telomeres would allow fewer numbers of cell division to occur and promptly cause telomere crisis in the absence of telomerase activity. In fact, telomere elongation by overexpression of telomerase (this study) or tankyrase16, a positive regulator of telomerase, confers resistance to MST-312. Consistent with these observations, a preclinical study suggested that telomere length could be a predictive biomarker of the telomerase inhibitor imetelstat35. Furthermore, a randomised phase II study of imetelstat has demonstrated a trend toward longer median progression-free survival and overall survival in imetelstat-treated patients with non-small cell lung cancer with short telomeres19. Intriguingly, cancer cells often maintain shorter telomeres than those in cells in surrounding normal tissues36,37, and short telomeres are associated with a less-differentiated status of tumours in vivo38,39. In this sense, telomerase inhibitors may be suitable for targeting such malignant tumours.
We cannot precisely quantitate the level of telomerase activity in MST-312-treated cells. Because MST-312 is a reversible telomerase inhibitor, it is washed out during the course of lysate preparation from the drug-treated cells. Accordingly, telomerase activity is restored in the cell lysate14. For these reasons, it is difficult to evaluate the relationship between telomerase activity in MST-312-treated cells and their drug sensitivities. Telomerase can be also inhibited by siRNA-mediated knockdown. However, effects of hTERT knockdown and telomerase inhibition are not necessarily equivalent each other and, therefore, it was difficult by the knockdown approach to check if MST-312 sensitivity is partially mediated by the inhibition of telomerase activity. Still, however, MST-312 resistance by hTERT overexpression (Fig. 6) would support that this drug sensitivity is partially mediated by telomerase inhibition.
We have also demonstrated that the expression levels of lamin A protein are inversely correlated with the susceptibility to apoptosis by treatment with MST-312 and DNA damaging anticancer drugs but not by paclitaxel. Lamin A is one of the nuclear matrix proteins and interacts with several proteins, such as LAPs 1A, 1B40, emerin41, retinoblastoma transcription factor pRB42 and myne-143. Among these proteins, both emerin and pRB associate with DNA, indicating that lamin A interacts with several DNA-binding proteins and plays important roles in the nucleus. In fact, loss of lamin A function increases the chromatin motion44–47. Mutations in the LMNA gene, which encodes the two nuclear lamins, lamin A and C, produced by alternative splicing, cause a wide range of diseases called laminopathies48,49. With regard to the association between senescence and lamin A function, much can be learned about the premature aging syndrome Hutchinson-Gilford progeria syndrome (HGPS)50,51. In cases with a splicing mutation of LMNA, the mutant lamin A protein, progerin, accumulates in HGPS patient cells. Progerin has a 50-amino-acid internal deletion and lacks the proteolytic cleavage site necessary to remove the carboxy-terminal 18 amino acids to generate mature lamin A50,51. Progerin acts as a dominant negative mutant of lamin A and causes dysregulation of the DNA damage response48,49,52–55. HGPS patient-derived cells are highly sensitive to ionising radiation-induced DNA damage, and ectopic expression of progerin mimics this phenotype54. At the molecular level, progerin traps DNA-dependent protein kinase, a critical protein in double-strand break repair56. Loss of wild-type lamin A can also result in perturbed non-homologous end joining and homologous recombination57. Our BIO-COMPARE analysis of the JFCR39 panel showed that the expression level of lamin A protein inversely correlates with the sensitivity to various DNA damaging drugs (YM, TY and HS, unpublished observations). These observations support the functional importance of lamin A for proper DNA damage response and repair.
hTERT-mediated immortalisation of HGPS cells improves nuclear morphology, decreases progerin protein levels and diminishes the number of persisting 53BP1 foci, alleviating the dysregulation in the DNA damage response and repair machinery54. Furthermore, loss of lamin A alters the nuclear distribution and heterochromatin status of telomeres58. These observations suggest a functional linkage between telomeres and lamin A. Consistent with these data, we found that lamin A overexpression in NCI-H522 cells repressed the telomeric DNA damage induced by MST-312 (Fig. 6F).
Our chemical fingerprinting analysis demonstrates that the MST-312 sensitivity profile at the acute phase is similar to the profiles of DNA topoisomerase II inhibitors, especially ICRF-193. One study showed that ICRF-193 preferentially attacks telomeres in a telomeric protein TRF2-dependent manner59. However, we found that ICRF-193 or other DNA topoisomerase II inhibitors, which have no telomerase inhibitory activities, induced less telomeric DNA damage than MST-312 (Fig. 4E). Thus, the mixed phenotype of the telomeric and non-telomeric DNA damage would be caused by the dual inhibition of telomerase and DNA topoisomerase II by MST-312 treatment in NCI-H522 cells. Inhibitors with multiple nuclear target, as demonstrated in this study, may suggest new treatment strategies against cancer.
Materials and Methods
Chemicals
MST-312 and MST-295 were synthesised as described previously14. ICRF-193 and ICRF-154 were provided by Zenyaku Kogyo Co., Ltd. (Tokyo, Japan). TAS-103 was provided by Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan). Etoposide and cisplatin were purchased from Cosmo Bio Co., Ltd. (Tokyo, Japan). Camptothecin, doxorubicin and paclitaxel were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture, cell growth and apoptosis assays
The JFCR39 human cancer cell line panel was described previously60. Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated calf serum and 100 μg/ml of kanamycin at 37 °C in a humidified atmosphere of 5% CO2. Cell growth inhibition was assessed by sulforhodamine B assay61, the CellTiter 96 AQueous One Solution cell proliferation assay kit (Promega, Fitchburg, WI, USA) or MTT assay62. To predict molecular target of MST-312, COMPARE analysis using JFCR39 panel was performed as described previously12,31. Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling (TUNEL) assay for detection of apoptotic cells was performed using the ApoAlert DNA Fragmentation Assay Kit (Takara Bio, Shiga, Japan) according to the manufacturer’s instructions. Quantitation of the sub-G1 fractions was performed by propidium iodide staining followed by flow cytometry, as described previously16.
In vivo xenografts
Human breast cancer HBC-4 cells were implanted subcutaneously in the right flank region of 9-week-old BALB/cAJcl-nu nude mice (Charles River Laboratories Japan, Kanagawa, Japan). Experiments were started when tumours reached 50–150 mm3 as measured with callipers (day 0). MST-312 was administered by either intratumoural injection at 40 or 80 μg/day; intravenous injection at 30, 50 or 70 mg/kg/day; or orally at 50, 200 or 400 mg/kg/day. In the intratumoural injection experiment, vehicle group was administrated three times of once daily (qd) per week for four weeks and 40 and 80 μg/day groups were administrated five times of qd per week for four weeks. In the intravenous injection experiment, vehicle and 30 mg/kg groups were administrated five times of qd per week for three weeks, 50 mg/kg group was administrated five times of qd per week for two weeks and 70 mg/kg group was three times of qd per week for three weeks. In the orally administrated group, 50 and 200 mg/kg groups were administrated five times of qd per week for nine weeks and 400 mg/kg group was administrated five times of qd per week for 6 weeks. Control mice (n = 5–6) received the same volume of saline as experimental mice (n = 5–6 per group). Tumour growth was monitored. The length (L) and width (W) of the tumours were measured, and the tumour volume (TV) was calculated as TV = LW2/2. All animal procedures were performed in the animal experiment room of the Japanese Foundation for Cancer Research (JFCR) using protocols approved by the JFCR Animal Care and Use Committee.
Western blot analysis
Whole cell and nuclear extracts were prepared as described previously63. Western blot analysis was performed as described64 with the following primary antibodies: anti-TRF1 (#5745, 0.5 μg/ml)63, anti-TRF2 (4A794, 2.5 μg/ml, Novus Biologicals, Littleton, CO, USA), anti-POT1 (#978, 1:1,000, provided by Dr. Titia de Lange), anti-TIN2 (1:1,500, provided by Dr. Zhou Songyang), anti-TPP1 (#467, 1:1,000, provided by Zhou Songyang), anti-RAP1 (1:1,000, provided by Zhou Songyang), anti-RIF1 (#1060, 1:1,000, provided by Titia de Lange), anti-dyskerin (H-300, 2 μg/ml, Santa Cruz Biotechnology, Dallas, TX, USA), anti-MRE11 (12D7, 1:500, GeneTex, Irvine, CA, USA), anti-RAD50 (13B3, 1:1,000, GeneTex), anti-NBS1 (#3002, 1:1,000, Cell Signaling Technology, Danvers, MA, USA), anti-BLM (ab476, 1:2,000, Abcam, Cambridge, UK), anti-WRN (ab200, 1:2,000, Abcam), anti-ATM (2C1, 2 μg/ml, Santa Cruz Biotechnology), anti-PARP-1 (C2-10, 1:2,000, Pharmingen, Franklin Lakes, NJ, USA), anti-tankyrase-1 (H-350, 2 μg/ml, Santa Cruz Biotechnology), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; RDI-TRK5G4-6C5, 0.5 μg/ml, Research Diagnostics, Flanders, NJ, USA), anti-α-tubulin (B-5-1-2, 1:1,000, Sigma), anti-lamin A/C (636, 2 μg/ml, Santa Cruz Biotechnology) and anti-hTERT (1531-1/Y182, 1:1,000, Abcam). Images were processed with Photoshop CS5 (Adobe).
Telomere and telomerase assays
Genomic DNA was isolated and telomere restriction fragments (TRFs) were detected by Southern blot analysis as previously described38. The mean length of TRFs was determined with an Atto densitoscan analyser (Tokyo, Japan). All procedures were performed in accordance with the institutional guideline and regulation under approval by the JFCR Radiation Safety Committee. Telomeric DNA signals and the telomeric 3′-overhang were quantitated by HPA assay as described previously65. TRAP assay for estimation of telomerase activity was performed as previously described38.
RT-PCR
Total RNAs were prepared using the RNeasy Mini kit (Qiagen, Venlo, Netherlands), and hTERT expression was monitored by RT-PCR using Ready-To-Go beads (Amersham Pharmacia, Piscataway, NJ, USA) and a pair of primers, 5′-TTGGTGCACACCGTCTGGAGG-3′ and 5′-CTGGAGGTGCAGAGCGACTAC-3′. The PCR conditions were 29 temperature cycles of 94 °C for 45 sec, 60 °C for 45 sec, and 72 °C for 90 sec. Signals were detected by agarose gel electrophoresis and normalised by β-microglobulin expression, which was detected by primers 5′-ACCCCCACTGAAAAAGATGA-3′ and 5′-ATATTCAAACCTCCATGATG-3′, under conditions of 24 temperature cycles of 94 °C for 30 sec, 55 °C for 1 min and 72 °C for 2 min. In pilot studies, we determined the quantitative range of the reaction by modifying the number of temperature cycle.
Immunofluorescence and immuno-FISH (iFISH) assays
Immunofluorescence staining and iFISH assays were performed as described64. The following primary antibodies were used: anti-human TRF1 (5747, 1 μg/ml)63, anti-ATM pS1981 (200-301-400 or 600-401-400, 7.5 μg/ml, Rockland Immunochemicals, Limerick, PA, USA), anti-53BP1 (#4937, 1:100, Cell Signaling Technology), anti-SMC1 pSer966 (BL311, 2 μg/ml, Bethyl Laboratories, Montgomery, TX, USA) and anti-γH2AX (JBW301, 2 μg/ml, BD Biosciences, San Jose, CA, USA). For telomere PNA FISH, cells were treated with 0.25 μg/ml colcemid for 3 h, trypsinised and swollen in 0.6% sodium citrate for 30 min at 37 °C. Metaphase spreads were prepared on slide glass and telomere PNA FISH was performed as described16. Images were processed with Photoshop CS5 (Adobe).
DNA topoisomerase II assay
In vitro DNA topoisomerase II assay was performed using the Topo II Assay Kit (TopoGEN, Buena Vista, CO, USA) according to the manufacturer’s instructions. In brief, kinetoplast DNA was incubated with 2 units of DNA topoisomerase II for 1 h at 37 °C in the presence of test compounds. DNA decatenation was assessed by agarose gel electrophoresis.
Budding yeast viability assay
For spot assays, the S. cerevisiae YFK30 strain (MATa TRP1 erg3::HIS3 pdr1::hisG pdr3::hisG pdr5::LEU2) and top2 K414N strain (MATa TRP1 erg3::HIS3 pdr1::hisG pdr3::hisG pdr5::LEU2 top2-K414N) were grown to a concentration of 2.5 × 107 cells/ml in YPD medium (glucose 2%, yeast extract 1%, polypeptone 2%, adenine 0.04%, uracil 0.02%). Serial dilutions (1:10) were prepared, and aliquots (4 μl) were spotted onto plates containing the indicated concentration of drugs (MST-312 or doxorubicin); plates were then cultivated at 30 °C. All procedures were performed in accordance with the institutional guideline and regulation under approval by Hiroshima University Safety Committee for Recombinant DNA Experiments.
siRNA-mediated knockdown
hTERT siRNA (5′-CATTCCTGCTCAAGCTGACTCGACA-3′) was designed by BLOCK-iT RNAi Designer (Thermo Fisher Scientific, Waltham, MA, USA). Negative control siRNA was Stealth RNAi Negative Control Medium GC Duplex #2. All siRNAs were purchased from Thermo Fisher Scientific and introduced into cells with the reverse transfection method using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions.
Plasmid transfection and Retroviral infection
pLNCX2/lamin C was generated by amplifying lamin C cDNA from total cDNA of A549 cells as a template and subsequently inserting it into the pLNCX2 vector (BD Biosciences). NCI-H522 cells were transiently transfected pLNCX2 or pLNCX2/lamin C with Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. pLNCX2/hTERT was generated by cloning the hTERT fragment66 into the pLNCX2 vector. pLHCX/lamin A was generated by cloning the lamin A fragment (Open Biosystems, Cambridge, UK) into the pLHCX vector (BD Biosciences). Amphotropic retroviruses were produced by transfecting the plasmids into Phoenix amphotropic cells using standard calcium phosphate precipitation. NCI-H522 cells were infected with the retrovirus essentially as described67. Infected cells were selected with 400 μg/ml of G418 and/or 200 μg/ml of hygromycin and cultured as described68. All procedures were performed in accordance with the institutional guideline and regulation under approval by the JFCR Safety Committee for Recombinant DNA Experiments.
Statistical analysis
Unpaired t tests were performed for comparison of the groups with and without MST-312 treatment. Tukey-Kramer tests were performed to examine every combination of multiple experimental groups.
Electronic supplementary material
Acknowledgements
We would like to thank Dr. Kyotaro Hirashima for evaluating siRNAs, Tomoko Oh-hara and Michiko Sakamoto for performing TRAP assays and Hiroko Sawada for performing RT-PCR. We are also grateful to Dr. Zou Songyang for providing anti-TIN2, anti-TPP1 and anti-RAP1 antibodies; Dr. Titia de Lange for providing anti-POT1 and anti-RIF1 antibodies; Dr. Tokichi Miyakawa for providing yeast strains. We thank the Screening Committee of New Anticancer Agents supported by Grant-in-Aid for Scientific Research from The Ministry of Education, Culture, Sports, Science and Technology, Japan for evaluation of JFCR39 cell sensitivities to MST-312, and the Molecular Profiling Committee, Grant-in-Aid for Scientific Research on Innovative Areas “Platform of Advanced Animal Model Support” from The Ministry of Education, Culture, Sports, Science and Technology, Japan (KAKENHI 16H06276) for evaluation of the effect of MST-312 on cell phenotype. We also thank members of the Seimiya lab for invaluable discussions. This work was supported in part by Grants-in-Aid for Scientific Research, Japan Society for the Promotion of Science (JSPS) [Scientific Research (B) 16H04716 to HS, Challenging Exploratory Research 24650639 to HS and Challenging Research (Exploratory) 18K19487 to HS] and Grants-in-Aid for Scientific Research, The Ministry of Education, Culture, Sports, Science and Technology (MEXT) (Innovative Areas 18H04633 to HS), a research grant from Kobayashi Foundation for Cancer Research (to HS) and Japan Agency for Medical Research and Development (AMED, 16cm0106102h0001 to HS). Funding for open access charge: [Japan Society for the Promotion of Science/16H04716]. A draft of this manuscript was commercially edited by Edanz Group (www.edanzediting.com/ac).
Author Contributions
C.F. and H.S. conceived and designed the study; C.F., Y.M., S.S., M.N., H.T., M.U., T.Y. and H.S. performed the experiments; K.T. established and provided the yeast strains to M.U., C.F., H.T., M.U., T.Y., Y.S. and H.S. analysed the data; and C.F. and H.S. wrote the manuscript. All authors reviewed and approved the manuscript.
Data Availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
KT is an employee of Nippon Kayaku Co., Ltd. Other authors have no competing interests to disclose.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33139-x.
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/S0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 3.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg RA, et al. Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/S0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 5.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 7.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 8.Bell RJ, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell RJ, et al. Understanding TERT Promoter Mutations: A Common Path to Immortality. Mol Cancer Res. 2016;14:315–323. doi: 10.1158/1541-7786.MCR-16-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shay JW. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 2016;6:584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naasani I, Seimiya H, Tsuruo T. Telomerase inhibition, telomere shortening, and senescence of cancer cells by tea catechins. Biochem Biophys Res Commun. 1998;249:391–396. doi: 10.1006/bbrc.1998.9075. [DOI] [PubMed] [Google Scholar]
- 12.Naasani I, Seimiya H, Yamori T, Tsuruo T. FJ5002: a potent telomerase inhibitor identified by exploiting the disease-oriented screening program with COMPARE analysis. Cancer Res. 1999;59:4004–4011. [PubMed] [Google Scholar]
- 13.Damm K, et al. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seimiya H, et al. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-199. Mol Cancer Ther. 2002;1:657–665. [PubMed] [Google Scholar]
- 15.Asai A, et al. A novel telomerase template antagonist (GRN163) as a potential anticancer agent. Cancer Res. 2003;63:3931–3939. [PubMed] [Google Scholar]
- 16.Seimiya H, Muramatsu Y, Ohishi T, Tsuruo T. Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell. 2005;7:25–37. doi: 10.1016/j.ccr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 18.Thompson PA, et al. A phase I trial of imetelstat in children with refractory or recurrent solid tumors: a Children’s Oncology Group Phase I Consortium Study (ADVL1112) Clin Cancer Res. 2013;19:6578–6584. doi: 10.1158/1078-0432.CCR-13-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiappori AA, et al. A randomized phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann Oncol. 2015;26:354–362. doi: 10.1093/annonc/mdu550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 21.Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000;10:1299–1302. doi: 10.1016/S0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 22.Li S, et al. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Crothers J, Haqq CM, Blackburn EH. Cellular and gene expression responses involved in the rapid growth inhibition of human cancer cells by RNA interference-mediated depletion of telomerase RNA. J Biol Chem. 2005;280:23709–23717. doi: 10.1074/jbc.M502782200. [DOI] [PubMed] [Google Scholar]
- 24.Stewart SA, et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarin KY, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 27.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/S0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 28.Kong D, Yamori T. JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs. Bioorg Med Chem. 2012;20:1947–1951. doi: 10.1016/j.bmc.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 30.Lombardo GE, et al. Anti-hTERT siRNA-loaded nanoparticles block the growth of anaplastic thyroid cancer xenograft. Mol Cancer Ther. 2018;17:1187–1195. doi: 10.1158/1535-7163.MCT-17-0559. [DOI] [PubMed] [Google Scholar]
- 31.Yaguchi S, et al. Antitumor activity of ZSTK474, a new phosphatidylinositol 3-kinase inhibitor. J Natl Cancer Inst. 2006;98:545–556. doi: 10.1093/jnci/djj133. [DOI] [PubMed] [Google Scholar]
- 32.Long BH, et al. Mechanisms of resistance to etoposide and teniposide in acquired resistant human colon and lung carcinoma cell lines. Cancer Res. 1991;51:5275–5283. [PubMed] [Google Scholar]
- 33.Kobayashi Y, Mizunuma M, Osada H, Miyakawa T. Identification of Saccharomyces cerevisiae ribosomal protein L3 as a target of curvularol, a G1-specific inhibitor of mammalian cells. Biosci Biotechnol Biochem. 2006;70:2451–2459. doi: 10.1271/bbb.60186. [DOI] [PubMed] [Google Scholar]
- 34.Mashima T, et al. p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target. J Natl Cancer Inst. 2005;97:765–777. doi: 10.1093/jnci/dji133. [DOI] [PubMed] [Google Scholar]
- 35.Frink RE, et al. Telomerase inhibitor imetelstat has preclinical activity across the spectrum of non-small cell lung cancer oncogenotypes in a telomere length dependent manner. Oncotarget. 2016;7:31639–31651. doi: 10.18632/oncotarget.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommerfeld HJ, et al. Telomerase activity: a prevalent marker of malignant human prostate tissue. Cancer Res. 1996;56:218–222. [PubMed] [Google Scholar]
- 37.Barthel FP, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirashima K, et al. Telomere length influences cancer cell differentiation in vivo. Mol Cell Biol. 2013;33:2988–2995. doi: 10.1128/MCB.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirashima K, Seimiya H. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Res. 2015;43:2022–2032. doi: 10.1093/nar/gkv063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-T. [DOI] [PubMed] [Google Scholar]
- 41.Lee KK, et al. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- 42.Ozaki T, et al. Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene. 1994;9:2649–2653. [PubMed] [Google Scholar]
- 43.Mislow JM, Kim MS, Davis DB, McNally EM. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J Cell Sci. 2002;115:61–70. doi: 10.1242/jcs.115.1.61. [DOI] [PubMed] [Google Scholar]
- 44.Solovei I, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 46.Bronshtein I, et al. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat Commun. 2015;6:8044. doi: 10.1038/ncomms9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lammerding J, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B, et al. Genomic instability in laminopathy-based premature aging. Nat Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 49.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 51.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Rusinol A, Sinensky M, Wang Y, Zou Y. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J Cell Sci. 2006;119:4644–4649. doi: 10.1242/jcs.03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benson EK, Lee SW, Aaronson SA. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J Cell Sci. 2010;123:2605–2612. doi: 10.1242/jcs.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noda A, et al. Progerin, the protein responsible for the Hutchinson-Gilford progeria syndrome, increases the unrepaired DNA damages following exposure to ionizing radiation. Genes Environ. 2015;37:13. doi: 10.1186/s41021-015-0018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinoshita D, et al. Progerin impairs vascular smooth muscle cell growth via the DNA damage response pathway. Oncotarget. 2017;8:34045–34056. doi: 10.18632/oncotarget.15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu GH, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redwood AB, et al. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011;10:2549–2560. doi: 10.4161/cc.10.15.16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalez-Suarez I, et al. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009;28:2414–2427. doi: 10.1038/emboj.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L, et al. The topoisomerase II catalytic inhibitor ICRF-193 preferentially targets telomeres that are capped by TRF2. Am J Physiol Cell Physiol. 2015;308:C372–377. doi: 10.1152/ajpcell.00321.2014. [DOI] [PubMed] [Google Scholar]
- 60.Yamori T, et al. Potent antitumor activity of MS-247, a novel DNA minor groove binder, evaluated by an in vitro and in vivo human cancer cell line panel. Cancer Res. 1999;59:4042–4049. [PubMed] [Google Scholar]
- 61.Skehan P, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka N, et al. APC Mutations as a potential biomarker for sensitivity to tankyrase inhibitors in colorectal cancer. Mol Cancer Ther. 2017;16:752–762. doi: 10.1158/1535-7163.MCT-16-0578. [DOI] [PubMed] [Google Scholar]
- 63.Seimiya H, Muramatsu Y, Smith S, Tsuruo T. Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol Cell Biol. 2004;24:1944–1955. doi: 10.1128/MCB.24.5.1944-1955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamura T, et al. Targeting glioma stem cells in vivo by a G-quadruplex-stabilizing synthetic macrocyclic hexaoxazole. Sci Rep. 2017;7:3605. doi: 10.1038/s41598-017-03785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahara H, Kusunoki M, Yamanaka Y, Matsumura S, Ide T. G-tail telomere HPA: simple measurement of human single-stranded telomeric overhangs. Nat Methods. 2005;2:829–831. doi: 10.1038/nmeth797. [DOI] [PubMed] [Google Scholar]
- 66.Seimiya H, et al. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.