Abstract
In mammals, the central circadian clock is located in the suprachiasmatic nucleus (SCN) of the hypothalamus and it orchestrates peripheral clocks in the whole body to organize physiological and behavioral rhythms. Light-induced phase-shift of the SCN clock enables synchronization of the circadian clock system with 24-h environmental light/dark cycle. We previously found that adenosine deaminase acting on RNA 2 (Adar2), an A-to-I RNA editing enzyme catalyzing rhythmic A-to-I RNA editing, governs a wide range of mRNA rhythms in the mouse liver and regulates the circadian behavior. In brain, ADAR2-mediated A-to-I RNA editing was reported to occur in various transcripts encoding ion channels and neurotransmitter receptors, which could influence neuronal function of the SCN. Here we show that ADAR2 plays a crucial role for light-induced phase-shift of the circadian clock. Intriguingly, exposure of Adar2-knockout mice to a light pulse at late night caused an aberrant phase-advance of the locomotor rhythms. By monitoring the bioluminescence rhythms of the mutant SCN slices, we found that a phase-advance induced by treatment with pituitary adenylyl cyclase-activating polypeptide (PACAP) was markedly attenuated. The present study suggests that A-to-I RNA editing in the SCN regulates a proper phase response to light in the mouse circadian system.
Introduction
The circadian clock is a self-sustained time-measuring system that generates daily rhythms of behavior, physiology and gene expression with a period of approximately 24 h1. In mammals, a wide range of physiological and behavioral processes are orchestrated by the central clock in the SCN2,3. To maintain synchrony with the environmental conditions, the SCN receives photic information from the retina through retinohypothalamic tract (RHT) projection4, to which glutamate and PACAP predominantly contribute as neurotransmitters5–7. Photic stimulation received by the retina at early night induces phase-delay of the SCN clock, while the stimulation at late night induces phase-advance. These light-induced bidirectional phase-shifts of the circadian clock are thought to be evoked by acute induction of a subset of clock gene expression3, but the signaling pathway directing either phase-advance or phase-delay is still elusive.
The cell-autonomous circadian oscillation is driven by a mechanism based on transcriptional-translational feedback loops (TTFLs)8–11, where CLOCK and BMAL1 heterodimer activates transcription of Per and Cry genes12–15. Translated PER and CRY proteins bind to the CLOCK-BMAL1 complex and inhibit their transactivation9. In addition to the TTFLs, the importance of post-transcriptional regulation in the circadian clockwork has come under the spotlight in the last decade16–20. We recently demonstrated that adenosine deaminase acting on RNA 2 (Adar2), an A-to-I RNA editing enzyme, generates circadian rhythmicities of a wide range of mRNAs and ADAR2 deficiency shortened the free-running period of the cellular and behavioral rhythms21. ADAR2 binds to double-stranded RNAs (dsRNA) and catalyzes hydrolytic conversion of adenosine (A) to inosine (I), a process termed A-to-I RNA editing22,23. When A-to-I RNA editing occurs in coding region of mRNA, it can cause an amino acid change of the coded protein because the translational machinery recognizes inosine as guanosine (G). Our previous study on the physiological role of ADAR2-mediated A-to-I RNA editing was mainly performed in the mouse liver21. On the other hand, it is known that A-to-I editing is most prevalent in the central nervous system and plays a significant role in neuronal function by modifying activities of receptors involved in neurotransmission24,25. In fact, dysregulation in A-to-I RNA editing causes neurological disorders26 such as amyotrophic lateral sclerosis (ALS)27 and glioblastoma multiforme28. These studies prompted us to investigate a role of ADAR2 for the circadian clock in the SCN.
Here we found that ADAR2 deficiency caused a larger phase-advance in response to light in the wheel-running activity rhythms, while leaving the phase-delay unaffected. The larger light-induced phase-advance in the Adar2-knockout mice was verified by jet lag experiments. Interestingly, PACAP signaling pathway was implicated in the mechanism of the ADAR2-regulated phase-advance in the SCN clock. Our data demonstrate an important role of ADAR2 in regulating the phase-advance of the circadian rhythms and suggest that RNA modification regulates the light input pathway in the SCN.
Results
ADAR2-mediated A-to-I RNA editing in the SCN
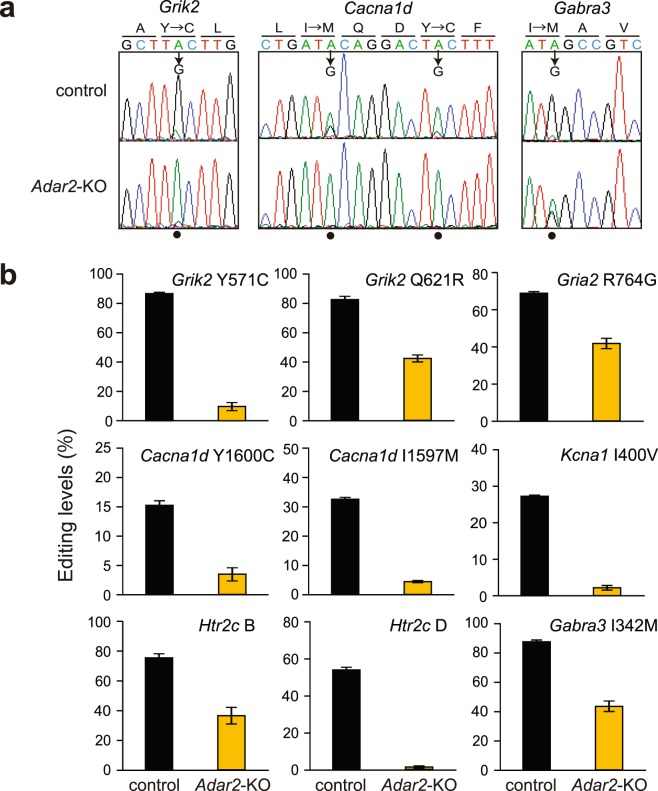
ADAR-mediated A-to-I RNA editing is known to regulate activities of a range of ion channels and neurotransmitter receptors by recoding amino acid sequences (reviewed in refs24–26). Among three members of mammalian ADAR family, ADAR1, ADAR2 and ADAR3, the editing activity of ADAR3 has not been demonstrated yet. In the present study, contribution of ADAR2-mediated A-to-I RNA editing to the SCN neuronal function was investigated by using Adar2-deficient (Adar2−/−) mice. Adar2−/− mice exhibit a postnatal lethal phenotype because Q607R substitution in GRIA2 does not occur due to a loss of the A-to-I editing activity29. The lethality is rescued by introducing a mutation replacing the codon for Q607 with arginine in Gria2 (termed Gria2R/R)29. In this study, Adar2−/−Gria2R/R and Adar2+/+Gria2R/R mice were referred to as Adar2-knockout and control mice, respectively. We quantified A-to-I RNA editing levels [G/(A + G) (%)] in the SCN punch-out as the change of A peaks to G peaks in direct sequencing chromatograms of the RT-PCR products30,31. In control mice SCN, direct sequencing analysis detected substantial levels of A-to-I RNA editing in the transcripts coding ion channels and receptors, such as glutamate receptor ionotropic AMPA2 (Gria2), glutamate receptor ionotropic kainate 2 (Grik2), calcium channel voltage-dependent L type alpha 1D subunit (Cacna1d), potassium voltage-gated channel shaker-related subfamily member 1 (Kcna1), 5-hydroxytryptamine receptor 2 C (Htr2c) and gamma-aminobutyric acid A receptor subunit alpha 3 (Gabra3) (Fig. 1a, Supplementary Fig. 1a). Among these transcripts, ADAR2 deficiency significantly reduced the editing levels at particular sites (Fig. 1a,b, Supplementary Fig. 1a). Our results indicated that ADAR2 regulates considerable levels of the A-to-I editing of the mRNAs encoding these ion channels and receptors expressed in the SCN.
Figure 1.
ADAR2-mediated A-to-I RNA editing in mouse SCN. (a) Direct sequencing chromatograms from RT-PCR products in control (top) and Adar2-KO (bottom) mouse SCN at CT22. The solid circles indicate the editing sites in the transcripts. (b) A-to-I RNA editing level at each editing sites in direct sequencing analysis (mean ± s.e.m.; n = 3).
The SCN receives and transduces photic signals released from the terminal of RHT via several receptors and channels3,32. A major neurotransmitter, glutamate, released from the RHT terminals stimulates NMDA, AMPA (Gria2) and kainate (Grik2) receptors for membrane depolarization, which indirectly activates voltage-gated calcium channels such as Cav 1.3 (Cacna1d) expressed in the SCN neurons3. It should be noted that some of the A-to-I RNA editing events in these transcripts regulate their characteristics and neuronal activities31,33–35. These facts, together with our findings (Fig. 1), raise the possibility that ADAR2-mediated A-to-I RNA editing regulates the photic signal transduction in the SCN.
Light-induced phase-shifts in Adar2-KO mouse
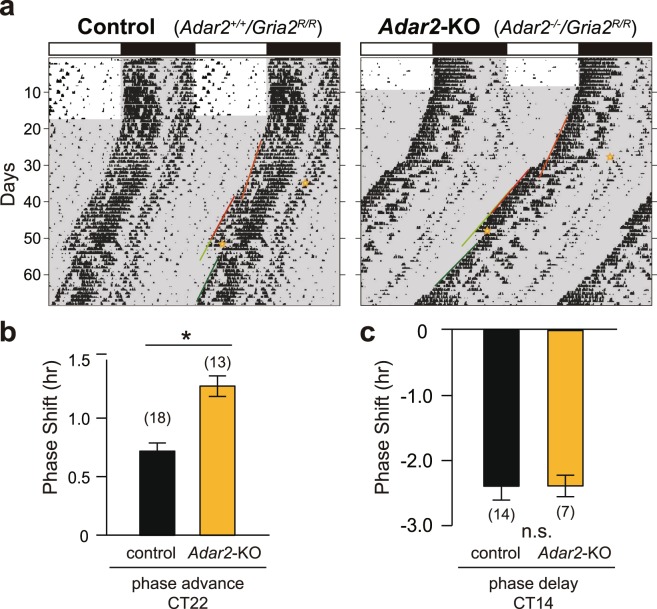
In vivo role of ADAR2 in photic regulation of the SCN was examined by monitoring light-induced phase-shifts of the locomotor activity rhythms of Adar2-knockout mice. A 30-min light pulse given at early or late subjective night is respectively known to induce phase-delay or phase-advance of the circadian behavioral rhythms in mice36. Wheel-running activities of Adar2-knockout mice were recorded under the constant dark (DD) condition after entrainment under 12-h/12-h light/dark (LD) cycle. After two weeks in DD, mice were exposed to the 30-min light pulse at subjective night. When compared to control mice, Adar2-knockout mice showed significantly larger phase-advance by the light pulse given at CT22, where the activity onset was designated as CT12 (Fig. 2a,b). In contrast, no significant difference was observed in the phase-delaying effects induced by the light pulse at CT14 between control and Adar2-knockout mice (Fig. 2a,c). These observations revealed an important role of ADAR2 specifically in the light-induced phase-advance of the SCN clock. The normal phase-delay of the mutant mice by the light pulse given at CT14 suggests no significant difference in retinal light sensitivity between Adar2-knockout and control mice, though we cannot rule out the possibility that ADAR2 deficiency caused aberrance of photic signaling specific to the phase-advance.
Figure 2.
Effects of light pulses on the circadian phase at two different circadian times in Adar2-KO mice. (a) Representative double-plotted actograms of wheel-running activity. The area with gray shading indicates the dark period. The light pulses (30 min, 300 lux) indicated by yellow stars were given first at CT22 and subsequently given at CT14. The red and green lines indicate the onset of activity used to calculate phase-advance and phase-delay of locomotor activity rhythms, respectively. (b,c) Phase-shift induced by a light pulse given at CT22 (b) or CT14 (c) (mean ± s.e.m.; n.s. P ≥ 0.05, *P < 0.05 by Student’s t-test). The numbers of animals are indicated in parentheses.
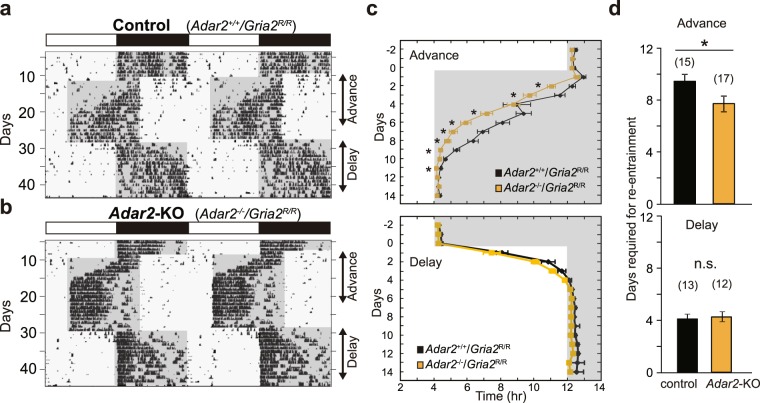
The role of ADAR2 in the phase shifting mechanisms of the SCN was validated by jet lag experiments. Control and Adar2-knockout mice were subjected to an abrupt shift of the 12-h/12-h LD cycles with 8-h advance and subsequently to the reverse shift of 8-h delay (Fig. 3a,b). The re-entrainment to the new LD cycle was visualized by plotting the activity onsets of the locomotor rhythms (Fig. 3c). In control mice, the 8-h advance of the LD cycle evoked a gradual shift of the behavioral rhythms and it took 9.5 days in average for complete re-entrainment to the new LD cycle (Fig. 3a,c,d). Noticeably, it took 7.7 days in average in Adar2-knockout mice, thus showing faster re-entrainment to the advance of LD cycle (Fig. 3b–d). In contrast, ADAR2 deficiency caused no marked difference in the re-entrainment when the mice were subjected to an 8-h delay (Fig. 3a–d). These results are consistent with the light-pulse experiments showing that ADAR2 deficiency caused an increase of the phase-advance while having no measurable effect on the phase-delay (Fig. 2b,c).
Figure 3.
Re-entrainment of Adar2-KO mouse activities to a new LD cycle. (a,b) Representative double-plotted actograms of wheel-running activities in the jet lag paradigm. The area with gray shading indicates the dark period. (c) Activity onset in the jet lag paradigm with an 8-h advance and an 8-h delay (mean ± s.e.m.; *P < 0.05 by Student’s t-test). (d) Days required for re-entrainment to the new LD cycles after an abrupt shift of the 12-h/12-h LD cycles with an 8-h advance and an 8-h delay (mean ± s.e.m.; n.s. P ≥ 0.05, *P < 0.05 by Student’s t-test). The numbers of animals are indicated in parentheses.
Then we compared the A-to-I editing levels between CT14 and CT22 in transcripts encoding several receptors in the SCN punch-out, but no significant difference in the editing levels was found between the two time points (Supplementary Fig. 1b). In addition, the light pulse at CT22 caused no measurable change in the editing levels of these transcripts in both control and Adar2-knockout mice (Supplementary Fig. 1c). These results suggest that ADAR2 regulates photic signal transduction in a light-independent manner throughout the day. However, the possibility is not eliminated that we were unable to detect rhythmic (or light-dependent) change in A-to-I RNA editing levels because the SCN punch-out contains a mixed population of SCN neurons and many surrounding cells, and hence any changes of the RNA editing could be masked by the heterogeneity of their circadian phases.
Of note, we found no significant difference in not only the phase-delay but also the phase-advance between wild-type (Gria2Q/Q) and control (Gria2R/R) mice on the Adar2+/+ background (Supplementary Fig. 2a–c). These results indicate that the knock-in allele of Gria2R/R has a marginal effect, if any, on the light-induced phase-shift.
Effect of PACAP on the SCN slice rhythms of Adar2-KO mouse
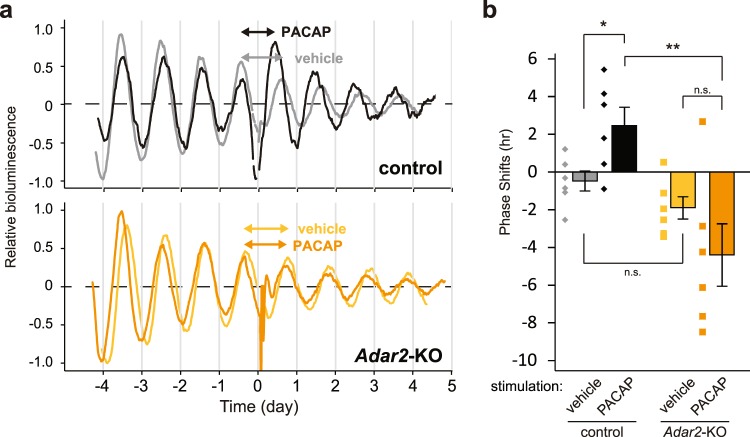
Light exposure of mice during the subjective night is known to stimulate retinal ganglion cells to release their neurotransmitters, glutamate and PACAP, from the RHT terminals5–7. These neurotransmitters induce the phase-shifts of the SCN rhythms for synchronization with external light/dark cycle3. To investigate the signaling pathway responsible for the ADAR2-regulated phase-advance, Adar2-knockout mice were crossed with PER2::LUC knock-in mice37, and the circadian oscillation of the bioluminescence signals were recorded from the SCN slices of PER2::Luc/Adar2−/− mice. Compounds to be tested were dissolved in the recording medium and applied to the SCN slices for 60 min (60-min pulse treatment) at the late declining phase of the bioluminescence rhythms in order to mimic the photic stimulation at late night. When PACAP was applied to the SCN slice prepared from control mice, the next peak of the bioluminescence signal was significantly phase-advanced as compared to that after vehicle treatment with the recording medium (Fig. 4a, top, 4b), and this result is consistent with previous studies7,38. In contrast, ADAR2 deficiency abolished the phase-advancing effect of PACAP stimulation (Fig. 4a, bottom, 4b), indicating significant reduction of the PACAP-triggered signaling in the SCN of Adar2-knockout mice. Importance of the PACAP signaling in the phase shifting mechanism was demonstrated by previous studies describing that mutant mice deficient for PACAP39,40 or its specific receptor PAC141 showed abnormal light-induced phase-shifts in the locomotor activities. Collectively, our data demonstrate that ADAR2 regulates the phase-shift mechanisms of the SCN clock through modifying PACAP signaling pathway. In addition, ADAR2 plays an important role in the appropriate light-induced phase-advance in mouse behavioral rhythms, though the detailed mechanism is yet to be determined.
Figure 4.
PACAP-induced phase-shift of bioluminescence rhythm recorded from the SCN slice in culture. (a) A representative set of bioluminescence rhythms of control (top) and Adar2-knockout (bottom) mouse SCN. The time of the PACAP application (1 μM, 1 h) was set to 0. The double-headed arrows indicate the peak intervals of the bioluminescence rhythms. (b) The phase-shifts induced by the PACAP application (mean ± SEM; n = 6; n.s. P ≥ 0.05, *P < 0.05, **P < 0.01 by Student’s t-test).
Discussion
Recent studies on the circadian oscillation ant its output mechanisms have shed light on the important roles of the post-transcriptional regulation, such as RNA methylation18, alternative splicing19, polyadenylation42 and miRNA-mediated gene silencing20. Our previous study demonstrated that A-to-I RNA editing enzyme ADAR2 is a key player for not only determining the circadian period of mouse behavioral rhythm but also generating a wide range of mRNA rhythms in the mouse liver21. Since A-to-I RNA editing events have been identified in key components of synaptic transmission and is known to have a strong impact on neuronal signaling24, we speculate that A-to-I RNA editing should play an important role in the SCN when receiving light signals from retina. The present study undertook to investigate the role of ADAR2 in light-dependent phase shifting mechanisms. We found augmentation of light-induced phase-advance in the wheel-running activity rhythms of Adar2-knockout mice (Fig. 2b). Our molecular level analysis indicates that PACAP signaling in the SCN should be regulated by ADAR2 (Fig. 4b).
It is reported that, in concert with glutamate, PACAP mediates transmission of light information to the SCN clock as a neurotransmitter released from the RHT terminal43. PACAP stimulates its specific receptor PAC1 to activate adenylyl cyclase signaling. Preceding studies on PACAP-deficient39,40 or PAC1-deficient41 mice demonstrated significant attenuation of phase-advance and phase-delay induced by light exposure. However, the A-to-I RNA editing databases44,45 in mouse transcriptome revealed no editing sites in PAC1, VPAC1 and VPAC2 receptors which are sensitive to PACAP. Here, it should be noted that binding of ADAR2 to dsRNAs by itself can modulate RNA processing independently of its RNA editing activity46,47. Such an editing-independent action of ADAR2 might contribute to expression levels of PACAP receptor(s) and/or the downstream signaling components, and consequently affect the light-induced phase-advance. In this study, we found that ADAR2 deficiency affected phase-advance but not phase-delay, suggesting that ADAR2 suppresses light signaling pathways specific to the phase-advancing process. It is also possible that the light signaling may be rhythmically regulated by ADAR2 in an editing activity-dependent or independent manner, because we found rhythmic expressions of Adar2 and the related members in the SCN (Supplementary Fig. 1d).
As compared to control mice, Adar2-knockout mice exhibited a significantly larger phase-advance induced by a light pulse (Fig. 2b). In contrast, PACAP application did not significantly evoke the phase-shift in Adar2-knockout SCN slices, while PACAP caused the phase-advance in the SCN slices of control mice (Fig. 4). These apparently contradictory results could be associated with the cooperation of various receptors in ADAR2-regulated photic signaling pathways. Indeed, we found that several ion channels and receptors are subjected to A-to-I RNA editing in their mRNAs, which cause alteration in amino acid sequences (Fig. 1a,b, Supplementary Fig. 1a), and these changes might modulate neuronal activities in the SCN. For example, a recent study indicated that ADAR2-mediated RNA editing in voltage-dependent calcium channel (Cacna1d) modulates potential firing rates of the SCN slice31. In addition, A-to-I RNA editing at the R764G site in AMPA-type glutamate receptor (Gria2) regulates recovery rates from desensitization34, and editing at the Y571C and Q621R sites in kainate-type glutamate receptor (Grik2) changes calcium permeability of the receptors35. Interestingly, it is known that PACAP enhances the glutamatergic signaling by increasing glutamate-evoked currents48. Not only PACAP pathway but also these various ADAR2-regulated receptors and channels including glutamatergic pathways might orchestrate the phase-shifting mechanism in the SCN49. Further study is needed to explore how ADAR2 suppresses the light-induced phase-advance in mouse behavioral rhythm.
ADAR family members play key roles in adaptation to altered environmental conditions such as temperature, hypoxia and UV-irradiation in various species50,51. Collectively, our findings not only provide a new insight into the physiological role of ADAR2 in the SCN but also demonstrate that ADAR2 is important for synchronization between the circadian rhythms and environmental light/dark cycle through regulating the light-dependent phase shifting mechanism.
Methods
Animals
C57BL/6 J mice, PER2::LUC knock-in mice (C57BL/6 J background, obtained from The Jackson Laboratory)37 and Adar2−/−/Gria2R/R mice (C57BL/6 J background, obtained from Mutant Mouse Regional Resource Centers (MMRRC))29, were housed in cages with commercial chow (CLEA Japan) and tap water available ad libitum under a controlled environment (temperature 23 ± 1 °C). All experiments were approved by the animal ethics committee of the University of Tokyo, and all experimental procedures were conducted in accordance with the University of Tokyo guidelines for use of experimental animals.
Behavioral experiments
Young adult male mice were housed individually in cages equipped with running wheels and were entrained to the 12 h/12 h LD cycles for at least 2 weeks. The spontaneous locomotor activities were recorded by wheel-running revolutions in 5-min bins and the data were analyzed by using ClockLab software (Actimetrics). For light pulse experiments, the LD-entrained mice were released into the DD conditions for 3 weeks, and single light pulses (30 min, 300 lux) were given at circadian time (CT) 22, where the activity onset was designated as CT12. After the first light pulses, mice were maintained under the DD conditions for at least 15 days, and subsequently, second light pulses (30 min, 300 lux) were given at CT14. Phase-shifts were calculated from the phase-difference between the two regression lines, one fitted to 12 consecutive activity onsets immediately before the light pulse and the other to those after the light pulse excluding transient periods of first 3 days. For jet lag experiments, the entrained mice were subjected to an abrupt shift of the 12 h/12 h LD cycles with an 8-h advance and subsequently to the second shift of that with an 8-h delay.
RNA preparation
Mouse SCN was isolated at each time point per day after 38 hr from the beginning of DD conditions (projected CT). Bilateral punches of the SCN were prepared from each frozen section (0.6 mm-thick) by using a stainless needle with an inside diameter of 0.6 mm. For quantitative RT-PCR and direct sequencing, total RNA was prepared from the SCN punch-out by using TRIzol (Thermo Fisher Scientic) according to the manufacturer’s protocol.
Direct Sequencing
To quantify the A-to-I RNA editing levels, we performed direct sequencing analysis as described previously21. Briefly, total RNA was reverse transcribed by Go Script Reverse Transcriptase (Promega) with both anchored (dT)15 primers and random oligo primers. The cDNAs were amplified by KOD-Plus-Neo (TOYOBO) with gene specific primers (Supplementary Table S1, Fw1 and Rv1). Second PCR reactions were performed with the second sets of gene specific primers (Supplementary Table S1, Fw2 and Rv2) and with the first RT-PCR products (diluted 80-fold) as templates. Subsequently, the amplified cDNA fragments from RT-PCR were treated with ExoSAP-IT (USB) and directly sequenced with the gene specific primer, Fw2 or Rv2. The editing levels were quantified by measuring heights of A peaks (unedited) and G peaks (edited) and calculating percentage of the population edited at each site (100% × [G height/(A height + G height)]).
Quantitative RT-PCR (qRT-PCR)
For quantification of gene expression, total RNA was reverse transcribed by Go Script Reverse Transcriptase (Promega) with both anchored (dT)15 primers and random oligo primers. The cDNA was subjected to real-time PCR with the gene specific primers (Supplementary Table S1).
Real-time monitoring of luciferase expression in the SCN slices
Bioluminescence signals from the SCN cultures were recorded as described previously with minor modifications52. The coronal SCN slices (300 μm thick) were cultured on a membrane (Millicell-CM, Millipore) in a sealed 35-mm petri dish with the recording medium [DMEM (Sigma Aldrich) with B-27 Supplement, 0.1 mM luciferin (Promega), and 4.2 mM NaHCO3]. Bioluminescence signals were recorded at 37 °C in air with LumiCycle (Actimetrics). After recording the bioluminescence for 4 days, the SCN slices on a membrane were transferred to the medium with or without PACAP (final 1 μM) for an hour, and then washed twice with the recording medium for 5 min at 37 °C. After the wash, the SCN slices were transferred to the original culture medium. The PACAP stimulation was performed at a late declining phase of the luminescence rhythms. The raw data of bioluminescence rhythms were detrended by subtracting the 24-h moving average. To determine the degree of phase-shift induced by PACAP pulse treatment, the time points of three consecutive peaks of the bioluminescence rhythms before and after the PACAP application were fitted to regression lines. The two regression lines were extrapolated to a time when the pulse treatment was given, and the difference in time between the extrapolated time points were calculated as the phase difference.
Electronic supplementary material
Acknowledgements
Adar2−/−/Gria2R/R mice were obtained from MMRRC 8U42OD010924-13. This work was partially supported by Grants-in-Aid for Scientific Research (S) to Y.F. (24227001) and for Specially Promoted Research to Y.F. (17H06096) from MEXT of Japan, by the PRIME from Japan Agency for Medical Research and Development to H.Y. (17937210), by the Japan Prize foundation to H.Y. and by the INAMORI foundation to H.Y.
Author Contributions
H.T. performed almost all the experiments. T.Y. performed the phase-shifts experiments by PACAP stimulation to the SCN slices. Y.S. supervised the experiments of the SCN slices culture. H.Y. and Y.F. supervised the whole project. H.T., H.Y. and Y.F. co-wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33114-6.
References
- 1.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore RY, Lenn NJ. A retinohypothalamic projection in the rat. J. Comp. Neurol. 1972;146:1–14. doi: 10.1002/cne.901460102. [DOI] [PubMed] [Google Scholar]
- 5.Hannibal J. Neurotransmitters of the retino-hypothalamic tract. Cell Tissue Res. 2002;309:73–88. doi: 10.1007/s00441-002-0574-3. [DOI] [PubMed] [Google Scholar]
- 6.Ebling FJ. The role of glutamate in the photic regulation of the suprachiasmatic nucleus. Prog. Neurobiol. 1996;50:109–132. doi: 10.1016/S0301-0082(96)00032-9. [DOI] [PubMed] [Google Scholar]
- 7.Hannibal J, et al. Pituitary adenylate cyclase-activating peptide (PACAP) in the retinohypothalamic tract: a potential daytime regulator of the biological clock. J. Neurosci. 1997;17:2637–2644. doi: 10.1523/JNEUROSCI.17-07-02637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 10.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 11.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshitane H, et al. CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol. Cell. Biol. 2014;34:1776–1787. doi: 10.1128/MCB.01465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike N, et al. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vollmers C, et al. Circadian Oscillations of Protein-Coding and Regulatory RNAs in a Highly Dynamic Mammalian Liver Epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J. Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 2013;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 18.Fustin J-M, et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 19.McGlincy NJ, et al. Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 2012;13:R54. doi: 10.1186/gb-2012-13-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du N-H, Arpat AB, De Matos M, Gatfield D. MicroRNAs shape circadian hepatic gene expression on a transcriptome-wide scale. eLife. 2014;3:e02510. doi: 10.7554/eLife.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terajima H, et al. ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat. Genet. 2017;49:146–151. doi: 10.1038/ng.3731. [DOI] [PubMed] [Google Scholar]
- 22.Hogg M, Paro S, Keegan LP, O’Connell MA. RNA editing by mammalian ADARs. Adv. Genet. 2011;73:87–120. doi: 10.1016/B978-0-12-380860-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 23.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behm, M. & Öhman, M. RNA Editing: A Contributor to Neuronal Dynamics in the Mammalian Brain. Trends Genet. TIG (2016). [DOI] [PubMed]
- 25.Tariq A, Jantsch MF. Transcript Diversification in the Nervous System: A to I RNA Editing in CNS Function and Disease Development. Front. Neurosci. 2012;6:99. doi: 10.3389/fnins.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 28.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 30.Barbon A, Vallini I, La Via L, Marchina E, Barlati S. Glutamate receptor RNA editing: a molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Res. Mol. Brain Res. 2003;117:168–178. doi: 10.1016/S0169-328X(03)00317-6. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, et al. RNA editing of the IQ domain in Ca(v)1.3 channels modulates their Ca2+-dependent inactivation. Neuron. 2012;73:304–316. doi: 10.1016/j.neuron.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-J. [DOI] [PubMed] [Google Scholar]
- 34.Lomeli H, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 35.Köhler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-P. [DOI] [PubMed] [Google Scholar]
- 36.Daan S, Pittendrigh CS. A Functional analysis of circadian pacemakers in nocturnal rodents. J. Comp. Physiol. 1976;106:253–266. doi: 10.1007/BF01417857. [DOI] [Google Scholar]
- 37.Yoo S-H, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrington ME, Hoque S, Hall A, Golombek D, Biello S. Pituitary Adenylate Cyclase Activating Peptide Phase Shifts Circadian Rhythms in a Manner Similar to Light. J. Neurosci. 1999;19:6637–6642. doi: 10.1523/JNEUROSCI.19-15-06637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colwell CS, et al. Selective deficits in the circadian light response in mice lacking PACAP. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;287:R1194–1201. doi: 10.1152/ajpregu.00268.2004. [DOI] [PubMed] [Google Scholar]
- 40.Kawaguchi C, et al. Changes in light-induced phase shift of circadian rhythm in mice lacking PACAP. Biochem. Biophys. Res. Commun. 2003;310:169–175. doi: 10.1016/j.bbrc.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Hannibal J, et al. Dissociation between light-induced phase shift of the circadian rhythm and clock gene expression in mice lacking the pituitary adenylate cyclase activating polypeptide type 1 receptor. J. Neurosci. 2001;21:4883–4890. doi: 10.1523/JNEUROSCI.21-13-04883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26:2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannibal J. Roles of PACAP-containing retinal ganglion cells in circadian timing. Int. Rev. Cytol. 2006;251:1–39. doi: 10.1016/S0074-7696(06)51001-0. [DOI] [PubMed] [Google Scholar]
- 44.Kiran AM, O’Mahony JJ, Sanjeev K, Baranov PV. Darned in 2013: inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 2012;41:D258–261. doi: 10.1093/nar/gks961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heale BSE, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ota H, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michel S, Itri J, Han JH, Gniotczynski K, Colwell CS. Regulation of glutamatergic signalling by PACAP in the mammalian suprachiasmatic nucleus. BMC Neurosci. 2006;7:15. doi: 10.1186/1471-2202-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol. Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 50.Ma E, Gu XQ, Wu X, Xu T, Haddad GG. Mutation in pre-mRNA adenosine deaminase markedly attenuates neuronal tolerance to O2 deprivation in Drosophila melanogaster. J. Clin. Invest. 2001;107:685–693. doi: 10.1172/JCI11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakurai, M. et al. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. (2017). [DOI] [PMC free article] [PubMed]
- 52.Kon N, et al. CaMKII is essential for the cellular clock and coupling between morning and evening behavioral rhythms. Genes Dev. 2014;28:1101–1110. doi: 10.1101/gad.237511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.